Abstract
Current theories of muscle contraction propose that the power stroke of a myosin motor is the sole source of mechanical energy driving the sliding filaments of a contracting muscle. These models exclude titin, the largest protein in the human body, which determines the passive elasticity of muscles. Here, we show that stepwise unfolding/folding of titin Ig domains occurs in the elastic I band region of intact myofibrils at physiological sarcomere lengths and forces of 6-8 pN. We use single molecule techniques to demonstrate that unfolded titin Ig domains undergo a spontaneous stepwise folding contraction at forces below 10 pN, delivering up to 105 zJ of additional contractile energy, which is larger than the mechanical energy delivered by the power stroke of a myosin motor. Thus, it appears inescapable that folding of titin Ig domains is an important, but so far unrecognized contributor to the force generated by a contracting muscle.
INTRODUCTION
The sliding filament hypothesis explains muscle contraction as resulting solely from the force generated by ATP driven myosin motors that form reversible cross-bridges with actin filaments and generate force through a rotation of the myosin head: the power stroke (Huxley and Simmons, 1971). In these models, ATP hydrolysis stores elastic energy in the compliance of the cross-bridges themselves, delivering >38 zJ of contractile energy with each shortening step (Finer et al., 1994; Piazzesi et al., 2007). However, recent work has shown that only small amounts of elastic energy can be stored in the cross-bridges themselves (~5 zJ (Piazzesi et al., 2014)). Thus, where does the returning elastic energy of a contracting muscle comes from? Remarkably, none of these studies considers the largest human protein found thus far: titin. Titin, the giant tandem modular protein anchored to myosin and the Z-disc in muscle sarcomeres (Figure 1A) is responsible for the passive elasticity of striated muscle tissue (Granzier and Labeit, 2002; Linke and Fernandez, 2002). Although titin has now been recognized as the third filament of muscle, its role is largely viewed as that of a passive spring that provides elastic recoil in relaxed sarcomeres, without playing a role during active muscle contraction.
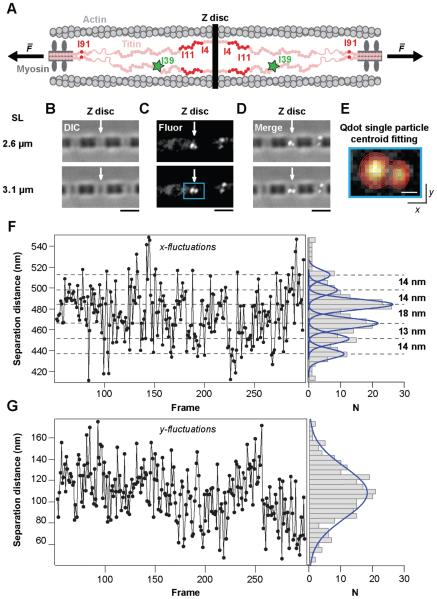
Figure 1. Single-molecule tracking in a stretched myofibril reveals discrete extension states.
(A) Schematic of the sarcomeric I-band. (B) Single rabbit psoas myofibril stretched between two glass microneedles and shown at two sarcomere lengths (2.6 µm; upper panel and 3.1 µm; lower panel). (C) Fluorescence imaging of I39 specific Qdot labels see (A); same view as in (B). (D) Merged view including (B) and (C). Pairs of Qdots are visible, spaced symmetrically around a Z-disc. (E) Magnified view of the area highlighted in (C) showing an individual fit used to track the Qdot positions. (F-G) Separation distance for individual Qdot pairs along the axis of applied force (F) and in the perpendicular direction (G). Right panels: histograms of separation distances. Qdots were tracked at 10 fps. Scale bars, 2 µm (B,C,D) and 300 nm (E).
Force spectroscopy studies have uncovered the basic elements of how titin proteins respond to a stretching force; domains unfold and refold in a time and force dependent manner, while any unfolded polypeptide simply follows the laws of polymer elasticity (Kellermayer et al., 1997; Rief et al., 1997; Tskhovrebova et al., 1997). A significant discovery was the finding that unfolded proteins could actively collapse and refold while under force, thereby generating positive mechanical work (Fernandez and Li, 2004). However, domain unfolding/refolding of titin has been thought not to take place in intact muscle tissue, and only operates as a safety mechanism to limit high forces in the sarcomere (Li et al., 2002; Tskhovrebova and Trinick, 2003). Thus, the folding of titin domains has not been considered as a plausible mechanism for delivering elastic energy in conjunction with the cycling myosin motors during muscle contraction.
Here, we directly measured the displacement of single titin molecules in intact myofibrils at physiological sarcomere lengths. We used fluorescence microscopy combined with titin specific quantum dot labels to track the length of the proximal tandem Ig region of titin in stretched myofibrils. Our results demonstrate that proximal Ig domains undergo stepwise unfolding and refolding in situ, within the working range of the sarcomere. We further show that folding of a single proximal Ig domain of titin can deliver up to 105 zJ of elastic energy at 7.9 pN of force, which is more than the contractile energy supplied by the power stroke of a myosin motor (Piazzesi et al., 2007). We conclude that passive stretching of muscle tissue stores elastic energy by unfolding Ig domains, and that refolding of these domains supplies an important, hitherto unrecognized, component of the force delivered by a contracting muscle.
RESULTS
Proximal Ig region of titin undergoes in stepwise changes in length
In order to measure the dynamics of single titin molecules in intact sarcomeres, we isolated single myofibrils from rabbit psoas muscles and attached them between two microneedles in an inverted light microscope. The sarcomere length of the myofibril was adjusted by changing the position of one of these needles using a piezoelectric stage. Sarcomeres were identified by differential interference contrast microscopy (DIC, Figure 1B). Individual titin molecules were labeled in situ using antibody-conjugated quantum dots (Qdots) targeted towards Ig-domain I39 in the proximal Ig region (Figure 1A). At high labeling density, two closely spaced fluorescent stripes could be seen symmetrically around each Z-disc, verifying the specificity of the I39 antibodies (Figure S1). At low labeling density, individual Qdots could be detected in the myofibrillar I-bands. Qdot pairs (hereafter referred to as doublets) were occasionally seen under these low labeling density conditions, spaced equidistantly around a Z-disc and thus likely marking two epitopes in adjoining sarcomeres (Figure 1C and D). Along the myofibrillar axis, two such epitopes are separated only by the Z-disc and proximal Ig-domains, without any major flexible linkers in between (Figure 1A). Since the Z-disc behaves as a rigid object at low forces (Luther, 2009), we speculated that it would be possible to directly detect folding/unfolding of Ig domains by measuring the separation distance between the doublets. We tracked the Qdot separation distance along the axis of applied force using a centroid-fitting algorithm (Figure 1E, see experimental methods).
When stretching the sarcomere to physiological extensions such as 3.1 µm (Lieber et al., 1994; Llewellyn et al., 2008; Wang et al., 1991), we observed that the Qdot doublets jumped along a discrete distribution of distances as shown by the accumulated length histograms (Figure 1F, right panel). Similar results were found for several Qdot doublets in separate myofibrils, showing that the result is reproducible (Figure S2). A multipeak fixed-width Gaussian fit to the data revealed two independent populations of separation distances centered at 13 ± 3 nm and 22 ± 3 nm (Figure S2 and Figure S3C). The y-fluctuations measured for the same Qdot doublets did not show discrete populations in the accumulated histograms (Figure 1G and Figure S2). Here, the fluctuations are larger in the x-direction than perpendicular to it, suggesting force-driven transitions within the titin molecules along pulling coordinate (Figure S4). These measurements therefore indicate that the end-to-end distance of the proximal Ig region of titin undergoes discrete changes in length at a sarcomere length of 3.1 µm. These steps and the lack of a flexible unstructured segment between the Z-disk and domain I39 suggest that Ig-domains from the proximal Ig region undergo stepwise unfolding-refolding transitions. Earlier studies have inferred the force on single titin molecules from measurements of the passive sarcomere length-tension relationship for single myofibrils and from estimates of the number of parallel titin molecules per myofibrillar cross-sectional area. Accordingly, from 2.1 to 3.1 µm sarcomere length, the force per titin grows from <1 pN to 6-8 pN assuming 6 titin molecules per half thick filament (Linke et al., 1998). Therefore, the in situ measurements of ~13 and ~22 nm steps are predicted to arise from folding/unfolding reactions of the proximal Ig domains at 6-8 pN.
Titin Ig domains unfold and refold in a stepwise manner at physiological forces
In order to test these predictions we studied the behavior of isolated titin molecules using single molecule force spectroscopy techniques. We combined magnetic tweezers force-spectroscopy (Chen et al., 2015; Liu et al., 2009; Yao et al., 2014) together with HaloTag anchoring (Popa et al., 2013), allowing us to record the extension of Ig domains for several hours at nanometer resolution in passive force-clamp mode (Figure 2A). We selected two well-characterized titin Ig domains from the proximal and distal regions of human titin, I10 and I91 (Anderson et al., 2013; Li et al., 2002) to measure their dynamics and folding/unfolding steps over a wide force range. Both proteins were engineered as polyproteins containing eight repeats of the Ig domain, flanked at the N-terminus by a HaloTag and at the C-terminus by AviTag (Figure 2A). The polyprotein constructs were covalently linked to a glass coverslip functionalized with HaloTag ligand (Popa et al., 2013) and to a magnetic bead coated with streptavidin. The polyproteins were exposed to forces varying between 3 and 100 pN by manipulation of a pair of permanent magnets positioned above the fluid cell.
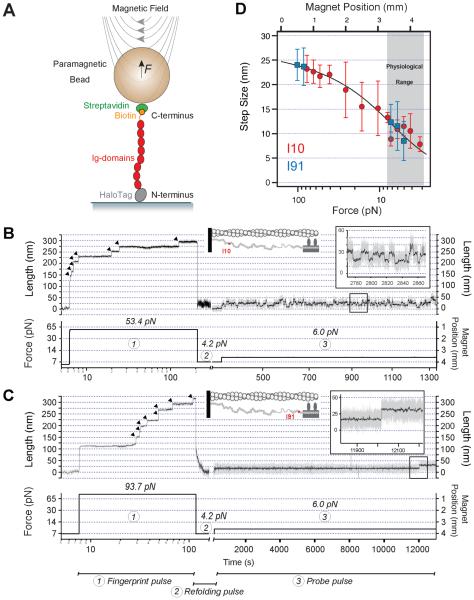
Figure 2. Dynamics of Ig domains I10 and I91 at physiological forces.
(A) Schematics of the magnetic tweezers experiment showing a polyprotein tethered between a glass coverslip and a magnetic bead. Force is applied by varying the separation between a pair of permanent magnets and the magnetic bead. (B) and (C) Dynamics of I10 and I91 (which also known as I27) polyproteins under force; first fully unfolded by a fingerprint pulse (1), followed by a refolding pulse at low force (2), and then examined by a probe pulse (3). (Insets: positions of I10 and I91 in the half sarcomere, and zoom-in of folding/unfolding events). Arrowheads mark Ig domain unfolding events. (D) Measured step sizes for I10 (red symbols) and I91 (blue symbols) are well fit by equation S3 (solid black line). Data includes both unfolding steps and refolding steps pooled together. The error bars represent S.D. The experiments were recorded at ~260 fps then smoothed.
Exposing these protein constructs to a high force caused the rapid unfolding of individual domains, which manifested as stepwise increases in the measured length (Figure 2B and C). In agreement with previous reports (Li et al., 2002), the I91 domain from the distal region was more mechanically stable than the proximal Ig domain I10, requiring higher pulling forces to unfold at a rate comparable to I10 (92.7 pN vs 53.7 pN; Figure 2B and C). More to the point, after allowing for complete refolding at 4.2 pN, pulling up to 6.0 pN showed these differences more starkly. In the example shown in Figure 2, at 6.0 pN, I10 underwent 202 stepwise unfolding/collapse events over a period of 23 minutes (Figure 2B). By contrast, at the same force only one unfolding event was recorded for I91 over a four hour-long recording (Figure 2C). Notwithstanding their differing mechanical stability, both proteins showed step sizes that varied steeply with the pulling force (Figure 2D: I10 red circles; I91 blue squares). At the same force, unfolding and collapse step sizes were identical (Figure 2D and S5). Using similar pulse protocols we collected a large number of unfolding/collapse steps for both proteins over the 3 pN to 100 pN range (Figure 2D). By combining our magnetic tweezer’s magnet-law (equation S1) together with the Worm Like-Chain (WLC) model of polymer elasticity (equation S2) we derived an expression (equation S3) that was readily fit to the measured step sizes vs magnet position (MP, Figure 2D). A fit of equation S3 to the data gave values of 27.8 nm for the contour length (LC) and 0.55 nm for the persistence length (p) (solid line in Figure 2D and Figure S6B).
Ig domains collapse to a highly compliant common state
After unfolding all eight Ig domains at a high force, quenching the force always resulted in a fast recoil, as the extended polypeptide adjusted to the new pulling force. At forces below 10 pN, this recoil was followed by a series of downward steps that we interpret as folding events. In contrast to the large differences observed in the rates of unfolding, the rate of descent of the folding steps was similar for both the I10 and I91 domains (Figure 3A and B). If these steps are associated with the folding of the Ig domains, one would expect the same number of unfolding events during a subsequent high-force probe pulse (Figure 3A and B). For the majority of recordings, in which 0-8 downward steps were observed during the refolding pulse, there was a 1:1 relationship between the number of downward steps and the number of unfolding events (Figure 3C). However, in many cases there were fewer unfolding events than downward steps, indicated by the points below the 1:1 curve (Figure 3C). This observation indicates that a downward step does not necessarily lead to a natively folded domain. Most likely, there is an intermediate state that precedes the mechanically stable native fold. This intermediate state corresponds to the molten globule state that was observed before for Ig domain folding under force (Garcia-Manyes et al., 2009). The molten globule state shows low mechanical stability, readily converting back and forth from a collapsed state to the shorter molten globule state. The molten globule state is particularly apparent in the case of I91, which extended and collapsed rapidly to the molten globule state (Figure 3D), but required several hours to show even one unfolding event from a vastly more mechanically stable native state (Figure 2C). Although we expected I91 to rarely unfold and extend in vivo, we found that it repeatedly interconverted between the extended and molten globule states at low forces (<10 pN) as long as it did not acquire the native state (Figure 3D).
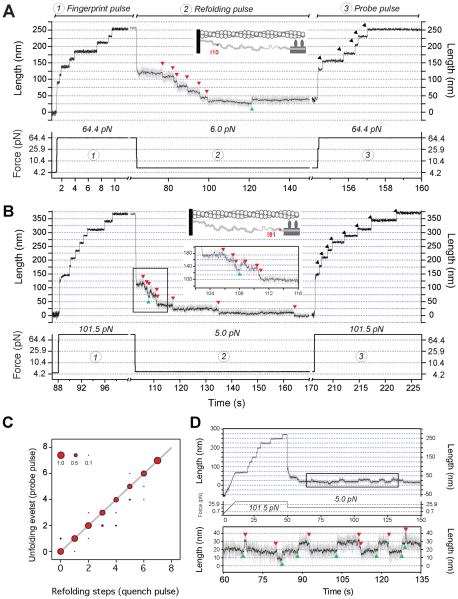
Figure 3. Titin Ig domains from the distal and proximal region refold in a stepwise manner.
(A) Seven I10 domains are unfolded at high forces. Reducing the force to 6 pN results in the sequential folding of each of six domains indicated by 11 nm steps downwards (red arrows). At this force, a single upwards step (green arrow) indicates one of the folded domains again unfolding. A subsequent high force probe pulse shows that five domains have acquired their native fold. (B) I91 domains exhibit equivalent folding dynamics at 5 pN: three domains fold first, followed by unfolding and refolding of a single domain, and then folding of the remaining five domains. The subsequent high force probe pulse shows that all 8 domains have acquired their native fold. (C) Downwards steps at low force are highly correlated with unfolding steps in the subsequent probe pulse, indicating that the native fold is acquired after the Ig domain collapses. (D) I91 domains proceed through a molten globule intermediate before acquiring their native folded structure. Contrasted with the slow unfolding dynamics of I91 in the folded state (Figure 2C), the I91 molten globule has low stability and is highly extensible. The I91 molten globule is easily unfolded (green arrows) at 5 pN and therefore contributes to the elasticity of titin.
Intact myofibril and single molecule measurements match at 6 pN
Our in situ measurements showed that at 6-8 pN of force, the proximal Ig region of titin underwent step changes in length of 13 ± 3 nm and 22 ± 3 nm (Figure S3). Our single molecule studies with isolated I10 and I91 domains showed that 13 ± 3 nm steps occurred in the 5-12 pN range in good agreement with our in situ measurements. However, we also observed much larger steps (22 ± 3 nm) in the single myofibril QDot experiments (Figures 1F and S2), which could not be explained by the step sizes observed in the single molecule experiments with I10 and I91. We reasoned that perhaps this mechanical behavior was unique to the native sequence of titin. To test this hypothesis, we cloned the segment of titin spanning Ig domains I4 through I11 (Figure 1A), which is contained within the proximal Ig region and was therefore tracked with our QDot experiments. Given the heterogeneous mechanical stability of the Ig domains present in this native segment (Li et al., 2002), we used a force-ramp protocol to unfold the individual Ig domains (Figure S3A). After unfolding the I4-I11, the force was quenched for about 100 s to allow the domains to refold. The force was then increased to 6 pN and held constant for several minutes (Figure S3A and S7). At 6 pN we readily observed mixed folding and unfolding steps that were similar in size to those observed in situ. Higher forces rapidly shifted the balance toward unfolding, making the observation of folding steps difficult. At 6.0 pN, I4-I11 showed numerous unfolding/refolding steps on a time scale of seconds (Figures S3B and S7). Hundreds of such stepwise transitions were recorded from several molecules (n = 5 molecules). Figure S3C compares histograms of the folding/unfolding step sizes collected in situ at 3.1 µm SL, and from single I4-I11 molecules at 6.0 pN. Remarkably, the single molecule force spectroscopy data showed two principal peaks centered at 10 ± 4 and 22 ± 5 nm, which are very similar to those measured in situ. Although the 22 nm steps observed in the single myofibril data were originally thought to be due to the unfolding of two individual domains during the integration time of the CCD camera, the force spectroscopy data suggested that these are distinct transitions (e.g. Figure S3B). Alternatively, it is unlikely that these steps are due to a calibration error because several quantum dot tracking experiments and several magnetic tweezers experiments showed a mixture of both step sizes during the same recording. The high occurrence of these step sizes in I4-I11 is not surprising given that this segment probably contains many endogenous interactions that are lost in the engineered homopolyproteins.
Stepwise Ig domain folding provides a large amount of elastic energy to a contracting muscle
The remarkable agreement between the in situ and single molecule data provides strong support for the view that folding/unfolding of titin Ig domains occurs in intact muscle tissue at physiological forces of 6-8 pN. Thus, we examined in more detail the folding behavior of Ig domains in this force range. Figure 4A summarizes our single molecule findings. Quenching the force down to 5.0 pN in a previously unfolded I10 polyprotein triggered first elastic recoil, followed by a stepwise folding contraction that eventually reached a steady state (Figure 4A). The stepwise folding contraction was found to be very sensitive to force. After unfolding all eight I10 domains at high force, the force was quenched to a value ranging from 4 to 10 pN (Figure 4B). Above 10 pN, no folding contraction was observed. Below 10 pN, the number of folding steps and their rate of appearance increased as the force was reduced. Reducing the force to 4 pN or less always resulted in complete refolding of all eight domains. From these data we calculated the folding probability as a function of force for an unfolded I10 domain (Figure 4C, red circles). We did similar experiments with I91, which demonstrated nearly identical refolding probability over this range of forces (Figure 4C, red squares). For both domains, the transition from fully unfolded to fully folded was narrow. We fit a sigmoid to the folding probability data (Figure 4C, red curve) where the half point is at 6.6 pN and the 5% to 95% range occurs over 4.1 pN. Therefore, reducing the load on titin by only a few pN triggered the refolding of Ig domains, which is likely the case during active muscle contraction.
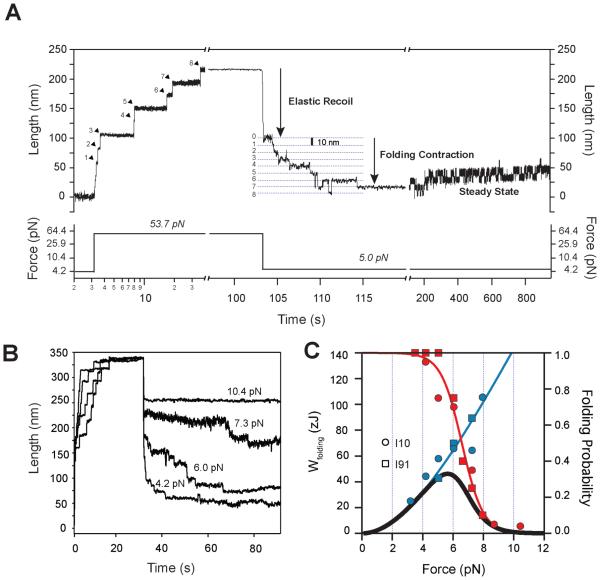
Figure 4. Titin Ig domains perform mechanical work during folding.
(A) When the stretching force is quenched, unfolded I10 domains shorten first with an elastic recoil, followed by a stepwise folding contraction. (B) The folding contraction occurs over a narrow range of forces (4-10 pN). (C) Folding work done by titin Ig domains (blue circles: I10, and squares: I91). The blue line is the work predicted by the worm-like chain model (Lc= 27.8 nm; p=0.55 nm). The folding probability measured at 60s (red circles: I10, and squares: I91) is well described by a sigmoid (red line). The expected value of the energy delivered by folding (blue line × red line) peaks at 5.7 pN delivering 46 zJ of contractile energy.
Owing to its force dependency (Figure 2D), a folding step can deliver varying amounts of elastic energy. We measured the amount of contractile work done by the folding of a single I10 or I91 domain, calculated as F·ΔL(Figure 4C, blue squares for I10; blue circles for I91). We showed that the force dependency of the folding step size closely follows the WLC model of polymer elasticity (Figure 2D). We use this model to fit the data of Figure 4C, calculated as FWLC·ΔL (Figure 4C, blue line). Folding steps that occur at higher forces are also larger (Figure 2D) and hence do more work and recover more energy (e.g. 105 zJ at 7.9 pN; Figure 4C). However, at higher forces, there is a lower probability of folding (Figure 4C, red curve). A more useful measure for evaluating the delivery of elastic folding energy in a contracting muscle is to compute the expected value of the work performed by a single titin Ig domain, calculated as the product of the work performed and the probability of folding (Figure 4C, black curve). We conclude that for muscle to recover the maximum amount of elastic potential energy from Ig domain folding (46 zJ peak expected value), the force on titin during an extension/contraction cycle should vary between 8.6 pN (5% folding) and 5.7 pN (peak of the expected value function).
DISCUSSION
Muscle tissue derives its properties directly from the dynamics of its single molecules. In a muscle fiber, there are ~2.4·109 titin molecules per mm2 operating in parallel and acting independently of each other to provide elastic recoil to the sarcomere (Higuchi et al., 1993). Intimately connected to titin is the thick filament from which myosin heads protrude to interact with actin filaments in a Ca++ and ATP dependent manner. These two molecular systems have been thought to operate independently from one another, with titin playing a role only during the passive extension of muscle, and the myosin motors as the sole source of mechanical force during a contraction. In this paper we provide evidence to support the view that the titin and myosin molecular systems both work together over a narrow range of forces, to store and recover elastic mechanical energy from folding Ig domains during muscle contraction.
Our data demonstrates that unfolded proximal Ig domains can do a surprisingly large amount of contractile mechanical work. Indeed, we show that an unfolded I10 or I91 domain can deliver up to W = 105 zJ of contractile work at 7.9 pN (Figure 4C); which is nearly three times as much contractile work as that of a single myosin motor (~38 zJ; Piazzesi et al, 2007, Finer et al, 1994). However, folding steps are relatively rare at 7.9 pN of force, so a better measure of the recovery of elastic energy is its expected value, E(W), which we calculate as the positive work done by the folding steps multiplied by their probability of occurrence (see supplementary notes). Thus by dropping the force per titin down to 5.7 pN, at the peak of the E(W) distribution, a large number of folding Ig domains would be expected to deliver ~46 zJ /folding domain of contractile mechanical work. These calculations were done based on the folding steps of isolated I10 and I91 modules which were observed to be as large as 13 nm at 8 pN (Figures 2 and 3). However, the folding steps measured in situ as well as those measured from the native I4-I11 tandem Ig region both showed steps found to be almost twice as big (~22 nm) producing ~132 zJ of contractile mechanical work at an operating force of 6 pN (Figure 4 and S3). Although the origin of the larger steps remains speculative, their high frequency of occurrence suggests that in native titin there are additional mechanisms in place to increase the delivery of elastic energy during Ig domain folding. It also appears that the measured Ig domain folding probability curve is a generic property of all unfolded domains. We chose to compare I10 and I91, from the proximal and distal regions of titin, as two well studied extremes in mechanical stability. While the native state of I91 is significantly more stable (Figure 2), both domains demonstrate the same folding probability as a function of force. This result implies that recruitment of Ig domain unfolding will be complex and time dependent, whereas the contractile delivery of the folding energy will occur in synchrony and exactly over the same range of forces for all unfolded domains. If titin has unfolded a few of its Ig domains at a stretched length prior to the contraction, then the extra "kick" from titin folding should have a significant function in supporting muscle contraction.
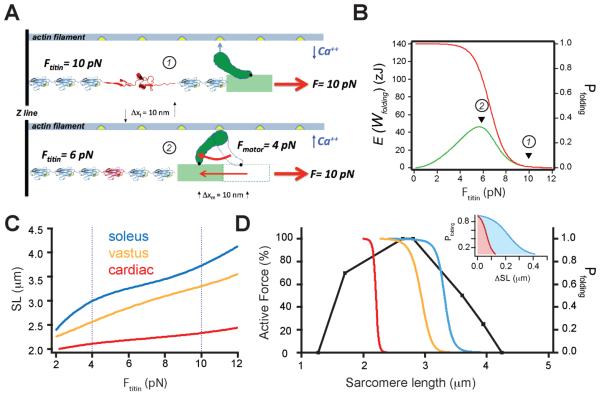
For simplicity, we consider the case where there may be one myosin motor cycling while attached to a single titin molecule composed solely of tandem Ig domains (Figure 5A). In the extension phase of a relaxed sarcomere (low Ca++), the myosin head is disengaged from actin and the load of 10 pN is transmitted directly to the titin molecule, triggering the unfolding and extension of an Ig domain (Figure 5A and B). Upon elevation of intracellular Ca++, the myosin head engages the actin filament, generating a force of 4 pN during the power stroke. This force is immediately subtracted from the force on the titin molecule, such that titin now experiences a stretching force of 6 pN, where the probability of folding is high. Folding of a single domain at 6 pN takes place in steps of ~10 nm; thus, the folding energy delivered would be 60 zJ which is larger than the energy delivered by the motor itself. The coincident force ranges over which both the motors and titin folding operate, and the distance over which folding Ig domains and myosin travel, are striking. Earlier studies had shown that the power stroke of a single myosin motor can generate forces ranging from 3 to 7 pN (Finer et al., 1994). Whereas more recent data shows that in vivo, the force of an active myosin motor remains closer to 6 pN (Piazzesi et al., 2007). These forces are sufficient to fully straddle the folding probability curve of titin Ig domains where a 4 pN reduction in the stretching force increases the folding probability from 5% up to 95% (Figure 4C). To place the single molecule folding data in perspective, we calculate that the mechanism shown in Figure 5A would readily lift a ~6 kg load (e.g. human vastus lateralis muscle, ~2500 mm2, 2.4·109 titin/mm2) (Beneke et al., 1991; Higuchi et al., 1993), whereas the force generated by the motor alone could lift less than half that weight (see note in supplementary information).
Figure 5. Mechanical energy delivered by a folding contraction occurs at an optimal sarcomere length that is muscle-type specific.
(A) Oversimplified sarcomere consisting of a single myosin motor and a single titin molecule. A relaxed sarcomere is stretched by 10 pN, unfolding an Ig domain, which extends by 10 nm (top; 1). Upon activation, the myosin head engages actin and the resulting power stroke generates 4 pN of force, reducing the load on titin from 10 pN down to 6 pN, at which force the previously unfolded Ig domain will fold in a single 10 nm step generating 60 zJ in contractile elastic energy (bottom; 2). Note that both folding and the power stroke shorten with matching step sizes (10 nm). (B) Folding probability (red) and contractile elastic energy (green) shown in relation to the conditions in A (1 and 2). (C) Sarcomere length (SL) versus force relationships for titin measured from three types of human muscles; soleus, vastus lateralis, and cardiac, and using a value of 2.4 × 109 titin molecules/mm2 cross-sectional area (solid lines are fits to the experimental data from (Kotter et al., 2014; Olsson et al., 2006; Trombitas et al., 2003). (D) Plot of Ig domain folding probability as a function of sarcomere length calculated using the measured Ig domain folding probability and the three relationships between sarcomere length and force shown in (C). The calculated range of sarcomere lengths that are optimal for delivering a maximum amount of folding energy closely matches the peak of the (human) active force-length relationship (black line). The inset shows that cardiac muscle only needs to extend about 100 nm/sarcomere to fully straddle the Ig domain folding probability (5%-95%). By contrast, the much softer soleus muscle needs to extend by about 500 nm/sarcomere to cover a similar range of forces.
A directly measurable parameter in striated muscle is sarcomere length. Many studies on isolated muscles have confirmed the close relationship between sarcomere length and the maximal amount of force generated during tetanus. These active (isometric) force-length relationships have been found to remain constant amongst many different types of muscles in support of the sliding filament hypothesis. Human muscles develop a maximal force around 2.8 µm sarcomere length. Cardiac muscles operate on the ascending limb of the force-length relationship whereas soleus operates on the descending limb. These two muscles represent the extremes of muscle stiffness, and express both the shortest and longest human titin isoforms, respectively. For each of these muscles, the relationship between passive tension and sarcomere length has been measured (Kotter et al., 2014; Trombitas et al., 2003). The relationship between force per titin molecule and sarcomere length for both cardiac muscle and soleus are shown in Figure 5C. In addition, we also show data for vastus lateralis which falls somewhere in between (Olsson et al., 2006). The stiff cardiac muscle needs only to extend by remarkably little, from 2.1 µm up to 2.2 µm in sarcomere length, to double the force per titin from 4 pN to 8 pN. By contrast, soleus needs a much larger extension from 2.9 µm to 3.4 µm to have the same effect. Thus it is clear that these two muscles will activate Ig domain folding in different regions of the active force versus sarcomere length curve. We use the relationships shown in Figure 5C to plot the titin domain folding probability versus sarcomere length for all three muscle types. Figure 5D shows these results in relation to the active-force versus sarcomere length relationship for human muscles. It is remarkable that the titin folding probability curves are near the optimal sarcomere lengths for all three types of muscles. The figure shows that the contribution that titin folding makes to muscle contraction is largest near the descending limb of the active length-tension relationship of skeletal muscles. In cardiac muscles, which have a much stiffer titin isoform, this contribution would support active contraction already on the ascending limb. Because skeletal muscle operates in vivo on the early descending limb of the active length tension curve, whereas cardiac muscle operates on the ascending limb, the titin "kick" is largest within the physiological working range of either muscle type. It is also interesting to consider that the titin isoform present in the muscle tissue determines the relationship between force and sarcomere length (Figure 5C). Thus, isoform switching will cause a shift in the position of the folding probability curve, and most likely in the operating range of sarcomere lengths. Finally, the observation that only small sarcomere length changes are needed to fully activate the delivery of a titin folding contraction, agrees well with direct measurements of sarcomere length changes during the full range of motion of a human limb (Llewellyn et al., 2008). From the evidence presented here, it appears inescapable that the folding of titin Ig domains generates a substantial component of the force in a contracting muscle. From this perspective, both the myosin motors and titin operate as an inextricable molecular system for the storage and delivery of mechanical energy.
EXPERIMENTAL PROCEDURES
For more details regarding the procedures used in this work, see the Supplemental Experimental Procedures.
Single myofibril mechanics
Single myofibrils were isolated from freshly excised rabbit psoas muscle as described previously (Linke et al., 1996). Single myofibrils were held for several seconds at or near slack SL, then rapidly stretched (typically within 1 s) and held at the stretched SL for 10-30 s during data acquisition. When the myofibrils were released back to slack SL after the stretch, the slack SL was unchanged compared to before the stretch.
Single-particle tracking of Qdots
Single Qdot tracking was accomplished by simultaneously fitting 2D Gaussians to pairs of Qdots for each frame. The separation distance between paired Qdots was calculated as the distance between the centroids of the Gaussians along the X-axis. Multi-Gaussian fits to the histograms shown in Figure 1F and Figure S2 were carried out using the built-in multi-peak procedure in Igor Pro.
Magnetic-tweezers experiments
The single molecule experiments were carried out in fluid cells, where the bottom glass was functionalized with HaloTag (O4) amine ligand (Promega). Recombinant halotagged-proteins immobilized on glass linked to paramagnetic streptavidin beads (Invitrogen), were pulled by changing the position of a pair of permanent magnets. The relationship between magnet position (MP) and force (F) is given by the equation F(MP)= 65·e0.91(0.91(0.99–MP)) (see Equation S1 in supplemental information).
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by NIH grants GM116122 and HL061228 (J.M.F.), NSF grant DBI-1252857 (J.M.F.), and Deutsche Forschungsgemeinschaft grant SFB1002/TPB03 (W.A.L.).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes supplemental experimental procedures, seven figures and supplementary notes, which can be found with this article online at http://cell.com.
AUTHOR CONTRIBUTION
J.M.F and W.L designed the project. P.K., W.L. and J.M.F. did the single-particle tracking experiments. J.A.R-P., E.C.E., I.P. and J.M.F did the single-molecule experiments. All authors wrote the paper.
REFERENCES
- Anderson BR, Bogomolovas J, Labeit S, Granzier H. Single molecule force spectroscopy on titin implicates immunoglobulin domain stability as a cardiac disease mechanism. The Journal of biological chemistry. 2013;288:5303–5315. doi: 10.1074/jbc.M112.401372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beneke R, Neuerburg J, Bohndorf K. Muscle cross-section measurement by magnetic resonance imaging. European journal of applied physiology and occupational physiology. 1991;63:424–429. doi: 10.1007/BF00868073. [DOI] [PubMed] [Google Scholar]
- Chen H, Yuan G, Winardhi RS, Yao M, Popa I, Fernandez JM, Yan J. Dynamics of equilibrium folding and unfolding transitions of titin immunoglobulin domain under constant forces. Journal of the American Chemical Society. 2015;137:3540–3546. doi: 10.1021/ja5119368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez JM, Li H. Force-clamp spectroscopy monitors the folding trajectory of a single protein. Science. 2004;303:1674–1678. doi: 10.1126/science.1092497. [DOI] [PubMed] [Google Scholar]
- Finer JT, Simmons RM, Spudich JA. Single myosin molecule mechanics: piconewton forces and nanometre steps. Nature. 1994;368:113–119. doi: 10.1038/368113a0. [DOI] [PubMed] [Google Scholar]
- Garcia-Manyes S, Dougan L, Badilla CL, Brujic J, Fernandez JM. Direct observation of an ensemble of stable collapsed states in the mechanical folding of ubiquitin. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:10534–10539. doi: 10.1073/pnas.0901213106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granzier H, Labeit S. Cardiac titin: an adjustable multi-functional spring. The Journal of physiology. 2002;541:335–342. doi: 10.1113/jphysiol.2001.014381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi H, Nakauchi Y, Maruyama K, Fujime S. Characterization of beta-connectin (titin 2) from striated muscle by dynamic light scattering. Biophysical journal. 1993;65:1906–1915. doi: 10.1016/S0006-3495(93)81261-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huxley AF, Simmons RM. Proposed mechanism of force generation in striated muscle. Nature. 1971;233:533–538. doi: 10.1038/233533a0. [DOI] [PubMed] [Google Scholar]
- Kellermayer MS, Smith SB, Granzier HL, Bustamante C. Folding-unfolding transitions in single titin molecules characterized with laser tweezers. Science. 1997;276:1112–1116. doi: 10.1126/science.276.5315.1112. [DOI] [PubMed] [Google Scholar]
- Kotter S, Unger A, Hamdani N, Lang P, Vorgerd M, Nagel-Steger L, Linke WA. Human myocytes are protected from titin aggregation-induced stiffening by small heat shock proteins. The Journal of cell biology. 2014;204:187–202. doi: 10.1083/jcb.201306077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Linke WA, Oberhauser AF, Carrion-Vazquez M, Kerkvliet JG, Lu H, Marszalek PE, Fernandez JM. Reverse engineering of the giant muscle protein titin. Nature. 2002;418:998–1002. doi: 10.1038/nature00938. [DOI] [PubMed] [Google Scholar]
- Lieber RL, Loren GJ, Friden J. In vivo measurement of human wrist extensor muscle sarcomere length changes. Journal of neurophysiology. 1994;71:874–881. doi: 10.1152/jn.1994.71.3.874. [DOI] [PubMed] [Google Scholar]
- Linke WA, Fernandez JM. Cardiac titin: molecular basis of elasticity and cellular contribution to elastic and viscous stiffness components in myocardium. Journal of muscle research and cell motility. 2002;23:483–497. doi: 10.1023/a:1023462507254. [DOI] [PubMed] [Google Scholar]
- Linke WA, Ivemeyer M, Olivieri N, Kolmerer B, Ruegg JC, Labeit S. Towards a molecular understanding of the elasticity of titin. Journal of molecular biology. 1996;261:62–71. doi: 10.1006/jmbi.1996.0441. [DOI] [PubMed] [Google Scholar]
- Linke WA, Stockmeier MR, Ivemeyer M, Hosser H, Mundel P. Characterizing titin's I-band Ig domain region as an entropic spring. J Cell Sci. 1998;111:1567–1574. doi: 10.1242/jcs.111.11.1567. Pt 11. [DOI] [PubMed] [Google Scholar]
- Liu R, Garcia-Manyes S, Sarkar A, Badilla CL, Fernandez JM. Mechanical characterization of protein L in the low-force regime by electromagnetic tweezers/evanescent nanometry. Biophysical journal. 2009;96:3810–3821. doi: 10.1016/j.bpj.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llewellyn ME, Barretto RP, Delp SL, Schnitzer MJ. Minimally invasive high-speed imaging of sarcomere contractile dynamics in mice and humans. Nature. 2008;454:784–788. doi: 10.1038/nature07104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luther PK. The vertebrate muscle Z-disc: sarcomere anchor for structure and signalling. Journal of muscle research and cell motility. 2009;30:171–185. doi: 10.1007/s10974-009-9189-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson MC, Kruger M, Meyer LH, Ahnlund L, Gransberg L, Linke WA, Larsson L. Fibre type-specific increase in passive muscle tension in spinal cord-injured subjects with spasticity. The Journal of physiology. 2006;577:339–352. doi: 10.1113/jphysiol.2006.116749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piazzesi G, Dolfi M, Brunello E, Fusi L, Reconditi M, Bianco P, Linari M, Lombardi V. The myofilament elasticity and its effect on kinetics of force generation by the myosin motor. Archives of biochemistry and biophysics. 2014;552-553:108–116. doi: 10.1016/j.abb.2014.02.017. [DOI] [PubMed] [Google Scholar]
- Piazzesi G, Reconditi M, Linari M, Lucii L, Bianco P, Brunello E, Decostre V, Stewart A, Gore DB, Irving TC, et al. Skeletal muscle performance determined by modulation of number of myosin motors rather than motor force or stroke size. Cell. 2007;131:784–795. doi: 10.1016/j.cell.2007.09.045. [DOI] [PubMed] [Google Scholar]
- Popa I, Berkovich R, Alegre-Cebollada J, Badilla CL, Rivas-Pardo JA, Taniguchi Y, Kawakami M, Fernandez JM. Nanomechanics of HaloTag tethers. Journal of the American Chemical Society. 2013;135:12762–12771. doi: 10.1021/ja4056382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rief M, Gautel M, Oesterhelt F, Fernandez JM, Gaub HE. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science. 1997;276:1109–1112. doi: 10.1126/science.276.5315.1109. [DOI] [PubMed] [Google Scholar]
- Trombitas K, Wu Y, McNabb M, Greaser M, Kellermayer MS, Labeit S, Granzier H. Molecular basis of passive stress relaxation in human soleus fibers: assessment of the role of immunoglobulin-like domain unfolding. Biophysical journal. 2003;85:3142–3153. doi: 10.1016/S0006-3495(03)74732-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tskhovrebova L, Trinick J. Titin: properties and family relationships. Nature reviews Molecular cell biology. 2003;4:679–689. doi: 10.1038/nrm1198. [DOI] [PubMed] [Google Scholar]
- Tskhovrebova L, Trinick J, Sleep JA, Simmons RM. Elasticity and unfolding of single molecules of the giant muscle protein titin. Nature. 1997;387:308–312. doi: 10.1038/387308a0. [DOI] [PubMed] [Google Scholar]
- Wang K, McCarter R, Wright J, Beverly J, Ramirez-Mitchell R. Regulation of skeletal muscle stiffness and elasticity by titin isoforms: a test of the segmental extension model of resting tension. Proceedings of the National Academy of Sciences of the United States of America. 1991;88:7101–7105. doi: 10.1073/pnas.88.16.7101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao M, Goult BT, Chen H, Cong P, Sheetz MP, Yan J. Mechanical activation of vinculin binding to talin locks talin in an unfolded conformation. Scientific reports. 2014;4:4610. doi: 10.1038/srep04610. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.