UNC-89 is a giant protein, homologous to vertebrate obscurin, located at the sarcomeric M-lines of Caenorhabditis elegans muscle. A yeast two-hybrid screen shows that the SH3 domain of UNC-89 interacts with paramyosin, a coiled-coil thick filament protein. The interaction is verified using biochemical and genetic approaches.
Abstract
UNC-89 is a giant polypeptide located at the sarcomeric M-line of Caenorhabditis elegans muscle. The human homologue is obscurin. To understand how UNC-89 is localized and functions, we have been identifying its binding partners. Screening a yeast two-hybrid library revealed that UNC-89 interacts with paramyosin. Paramyosin is an invertebrate-specific coiled-coil dimer protein that is homologous to the rod portion of myosin heavy chains and resides in thick filament cores. Minimally, this interaction requires UNC-89’s SH3 domain and residues 294–376 of paramyosin and has a KD of ∼1.1 μM. In unc-89 loss-of-function mutants that lack the SH3 domain, paramyosin is found in accumulations. When the SH3 domain is overexpressed, paramyosin is mislocalized. SH3 domains usually interact with a proline-rich consensus sequence, but the region of paramyosin that interacts with UNC-89’s SH3 is α-helical and lacks prolines. Homology modeling of UNC-89’s SH3 suggests structural features that might be responsible for this interaction. The SH3-binding region of paramyosin contains a “skip residue,” which is likely to locally unwind the coiled-coil and perhaps contributes to the binding specificity.
INTRODUCTION
To carry out their functions, most proteins interact with multiple other proteins in a time- and space-dependent manner. The quest to identify and understand the functions of interactors is most challenging when considering extraordinarily large polypeptides such as those found in striated muscle. The sarcomere, the unit of muscle contraction, consists of several hundred different proteins, among them being a number of giant polypeptides (>700,000 Da), which combine structural with signaling functions, and in some cases, provide muscle compliance through highly elastic regions (Kontrogianni-Konstantopoulos et al., 2009). Much is known about the function of the largest of these polypeptides, vertebrate titin: its role in sarcomere assembly, muscle elasticity, protein turnover and quality control, and muscle-specific signaling pathways. Identification of titin’s binding partners has been one of the keys to understanding titin’s function. A nearly complete analysis identified >25 interacting proteins (Linke and Hamdani, 2014). Much less is known about the function and binding partners of the newest member of this family of proteins, UNC-89 of Caenorhabditis elegans and Drosophila, and its human homologue, obscurin (for a comparative overview, see Benian and Mayans, 2015). This type of protein was first discovered in the model genetic system C. elegans (Waterston et al., 1980; Benian et al., 1996). Loss-of-function unc-89 mutants have reduced locomotion, disorganized sarcomeres usually lacking M-lines, and disorganized myosin thick filaments (Waterston et al., 1980; Benian et al., 1999; Qadota et al., 2008a). unc-89 is a complex gene; through the use of three promoters and alternative splicing, at least eight major polypeptides are generated, ranging in size from 156,000 to 900,000 Da (Benian et al., 1996; Small et al., 2004; Ferrara et al., 2005). The largest of these isoforms, UNC-89-B consists of 53 immunoglobulin (Ig) domains, two Fn3 domains, a triplet of SH3, DH, and PH domains near its N-terminus, and two protein kinase domains (PK1 and PK2) near its C-terminus. Antibodies generated to three different regions of UNC-89 localize the proteins to M-lines, structures in the middle of the sarcomere where thick filaments are cross-linked.
To learn how UNC-89 is localized and performs its functions, we are systematically identifying its binding partners. Our approach has been to screen a yeast two-hybrid library using as bait several-hundred-residue segments of UNC-89. We have obtained some significant biological insights. For example, near the N-terminus of UNC-89, Ig1-3 interacts with CPNA-1, a copine-domain-containing protein that is required not for the initial assembly of UNC-89 or myosin into sarcomeres but instead for their stable association once muscle activity begins in the embryo (Warner et al., 2013). Ig2-3 and Ig53-Fn2 interact with MEL-26, a substrate recognition protein for cullin 3 that acts as a scaffold for assembly of the ubiquitination machinery. Our studies (which include use of temperature-sensitive mutants) suggest that UNC-89 inhibits the MEL-26/CUL-3 complex from promoting the degradation of MEI-1 (katanin, which severs microtubules), and this is important for proper thick filament assembly or maintenance (Wilson et al., 2012b). A similar finding was made independently for mouse obscurin (UNC-89): degradation of small ankyrin 1.5 is dependent upon obscurin and is promoted by a cullin 3 substrate recognition protein, KCTD6 (Lange et al., 2012). Another intriguing result is that the two protein kinase domains of UNC-89 lying at its C-terminus interact with SCPL-1, a CTD-type protein phosphatase (Qadota et al., 2008b). This was the first time that this class of phosphatase, well known to have roles in transcriptional regulation, was found to have a role in the sarcomere. Of interest, the phenotype of loss of function of unc-89 includes less ability to bend, and loss of function of scpl-1 has greater ability to bend (Nahabedian et al., 2012). The idea that the protein kinase domains of UNC-89 interact with other enzymes and may act as signaling hubs was bolstered by finding that the Drosophila UNC-89 Kin1 kinase domain interacts with another kinase called Ball, and Ball is required for proper sarcomere assembly (Katzemich et al., 2015). We reported that both the PK1 region and the “interkinase region” interact with LIM-9 (Xiong et al., 2009), the closest nematode homologue of human protein FHL (Lecroisey et al., 2013). lim-9(RNAi) shows disorganization of myosin thick filaments (Meissner et al., 2009). Similarly, obscurin has been reported to interact with FHL2 (Hu and Kontrogianni-Konstantopoulos, 2013). We also reported that the DH-PH region of UNC-89 activates RHO-1 (RhoA) specifically, and attenuated RNA interference for rho-1 results in disorganization of muscle thick filaments (Qadota et al., 2008a). A similar interaction between RhoA and obscurin was also demonstrated (Ford-Speelman et al., 2009). Early studies showed that the C-terminal region of some isoforms of vertebrate obscurin interact with the sarcoplasmic reticulum membrane proteins small ankyrin-1 isoform 5 (sAnk1.5) and ankyrin-2 (Ank2; Bagnato et al., 2003; Kontrogianni-Konstantopoulos et al., 2003). The functional importance of this interaction is demonstrated by the obscurin-knockout mouse, in which longitudinal SR architecture is disrupted (Lange et al., 2009). The role of obscurin in linking the sarcomere to the SR may be conserved for UNC-89; UNC-89 is required for the proper organization of the ryanodine receptor and sarco/endoplasmic reticulum Ca2+-ATPase, as well as for optimal calcium signaling (Spooner et al., 2012).
Here we report results of using the SH3-DH-PH–domain region of UNC-89 to screen a yeast two-hybrid library. We discovered that the SH3 domain interacts with UNC-15 (paramyosin), an invertebrate-specific thick filament protein with significant homology with myosin rods. We demonstrate that UNC-89’s SH3 domain interacts with an 82-residue segment of the mostly coiled-coil 873-residue-long paramyosin. unc-89 mutants lacking expression of giant isoforms that contain the SH3 domain show large aggregates of paramyosin. Overexpression of the SH3 domain results in mislocalization of paramyosin. Finally, we use homology modeling and bioinformatics analyses to explore the molecular features of the UNC-89 SH3 domain and its interaction with this α-helical segment of paramyosin. We speculate that although the SH3 domain of vertebrate obscurin is unlikely to interact with myosin, it may interact with another coiled-coil protein.
RESULTS
Identification of paramyosin as a binding partner of UNC-89
A segment of UNC-89 containing the SH3-DH-PH domains (Figure 1A) was used to screen a yeast two-hybrid library of C. elegans cDNAs. After screening of 442,000 colonies, 22 prey clones were found to be positive upon retransformation (Supplemental Table S2). Twenty of these clones represented portions of paramyosin. Paramyosin is an invertebrate-specific coiled-coil dimer that bears close resemblance to the rod domain of myosin (∼40% identical in amino acid sequence) and is located in the cores of thick filaments (Cohen et al., 1971; Levine et al., 1976). Paramyosin is encoded by the unc-15 gene in C. elegans and is 873 residues long (Waterston et al., 1977; Kagawa et al., 1989). In C. elegans adult body-wall striated muscle, thick filaments are ∼10 μm long and composed primarily of myosin heavy chain A (MHC A), myosin heavy chain B (MHC B), and paramyosin. Within the thick filament, these components are differentially localized. MHC A is located in the middle, whereas MHC B is in the outer or polar regions of the thick filament (Miller et al., 1983). MHC A and MHC B myosins and a portion of paramyosin are organized around a tubular core consisting of paramyosin and nematode-specific core proteins called filagenins in a specific geometry (Deitiker and Epstein, 1993; Epstein et al., 1995; Liu et al., 1998, 2000; Muller et al., 2001). As shown in Figure 1B, comparison of the spans of the prey clones indicates that the minimal portion of paramyosin required to interact with UNC-89 is 579 residues long (294–873).
FIGURE 1:
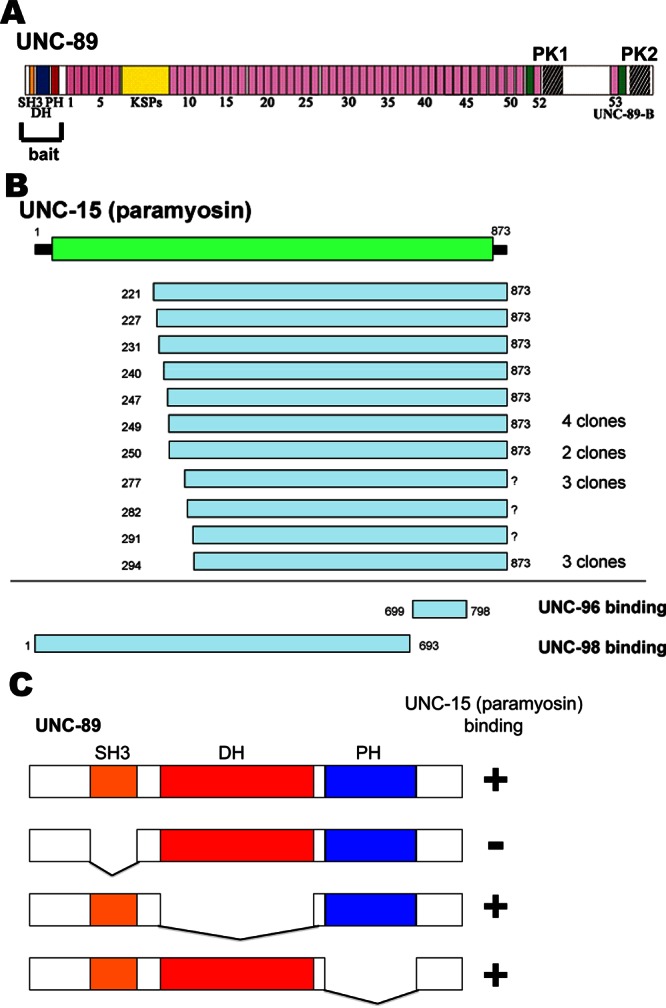
A yeast two-hybrid library screen revealed that UNC-89 interacts with paramyosin. (A) Schematic representation of domains within the largest isoform of UNC-89 (UNC-89B) and the portion, SH3-DH-PH, used as bait to screen the library. (B) Schematic representation of the paramyosin polypeptide and boundaries of paramyosin prey clones that were isolated. Paramyosin is almost entirely composed of α-helical coiled-coil structure (green). In C. elegans, paramyosin is encoded by the unc-15 gene. In contrast to the region of paramyosin that interacts with UNC-89 (minimally residues 294–873), the binding regions of paramyosin for UNC-96 and UNC-98 are different, as indicated (Miller et al., 2008). (C) Domain mapping using yeast two-hybrid assays suggests that the minimal region of UNC-89 that interacts with paramyosin is the SH3 domain. The indicated portions of SH3-DH-PH were used as bait to test for interaction with paramyosin (221–873) prey. +, growth, –, no growth of yeast on His– media.
As indicated in Figure 1C, various portions of the SH3-DH-PH region were used to further map the region of UNC-89 responsible for interaction with paramyosin by yeast two-hybrid assays. Results suggest that the SH3 domain is required for interaction with paramyosin. When a paramyosin clone containing residues 221–873 was used in two-hybrid assays against clones representing the entire largest UNC-89 isoform (isoform B), the only region with which UNC-89 was found to interact was the SH3-DH-PH region (unpublished data). This result further suggests the specificity of the interaction.
Minimally, the SH3 domain of UNC-89 interacts with UNC-15 (294–376)
To obtain additional and independent evidence for an UNC-89/paramyosin interaction, we used a biochemical assay with purified recombinant proteins. Paramyosin (221–873) was expressed as a maltose-binding protein (MBP) fusion, and portions of UNC-89—residues 1–500 containing the SH3-DH-PH region—were expressed as glutathione S-transferase (GST) fusions (Figure 2B). A far Western assay was conducted with MBP or MBP-paramyosin (221–873) on the blot and GST, GST-UNC-89 fusions in solution. As shown in Figure 2A, MBP-paramyosin, but not MBP, reacted with GST-a, GST-c, and GST-d but not GST-b. GST-b lacks the SH3 domain. We concluded that UNC-89 can interact directly with paramyosin, and that the minimal region of UNC-89 that reacts with paramyosin is the SH3 domain (contained in GST-d, spanning UNC-89 (58–131)). We also noted that the apparent interaction was increased (shorter enhanced chemiluminescence [ECL] exposure time during the final step of the far Western assay) when the portion of UNC-89 included an additional 57 residues N-terminal and 20 residues C-terminal of the SH3 domain (contained in GST-c). These additional sequences may contribute to binding or alternatively help to stabilize the SH3 structure.
FIGURE 2:
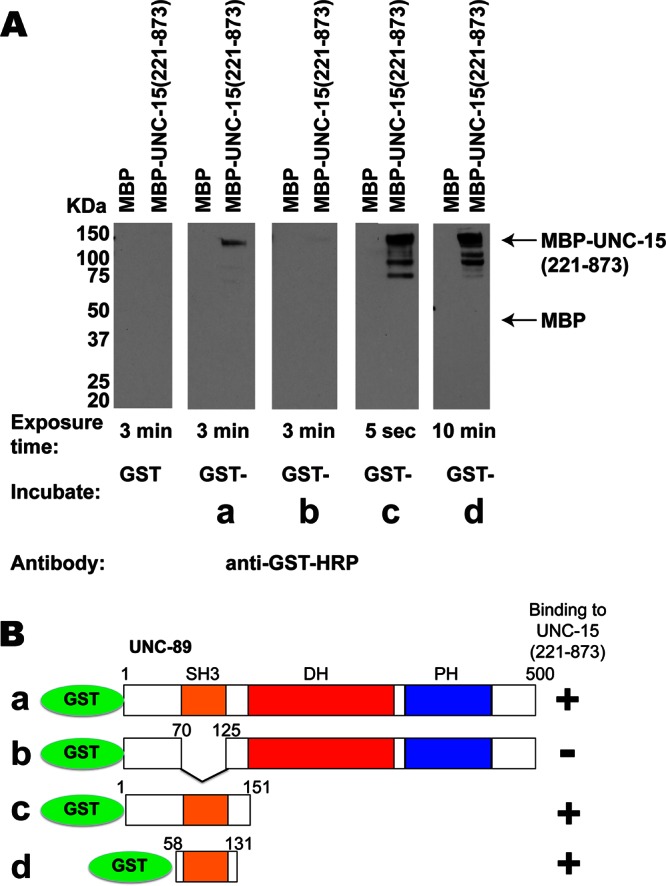
Far Western assays verify that UNC-89 interacts with paramyosin and show that only the SH3 domain of UNC-89 is required. (A) Pairs of MBP and MBP-UNC-15 (221–873) were separated by SDS–PAGE, transferred to membrane, reacted with GST or various GST fusions (B), washed, incubated with anti–GST-horseradish peroxidase, washed, and detected by ECL. As shown, MBP-UNC-15 (221–873), but not MBP, reacted with fusion proteins a, c, and d but not with fusion protein b. Arrows indicate positions of MBP and full-length MBP-UNC-15 (221–873) on the blot. The minimal portion of UNC-89 that interacts with paramyosin is UNC-89 (58–131), which is essentially the SH3 domain, as summarized in B.
To further map the segment of paramyosin responsible for interaction, we made a series of deletion derivatives of the paramyosin 221–873 region (Figure 3B). As shown in Figure 3A, these fragments were tested by far Western assay for interaction with GST-UNC-89 (1–151) and, as negative control, with GST itself (Supplemental Figure S1). Results summarized in Figure 3B indicate that the minimal region of paramyosin responsible spans residues 294–376, totaling 82 residues. This region of paramyosin that interacts with UNC-89 is different from the regions of paramyosin that have been reported to interact with two other thick filament/M-line proteins, UNC-96 and UNC-98 (699–798 and 1–693, respectively; Miller et al., 2008; Figure 1B).
FIGURE 3:
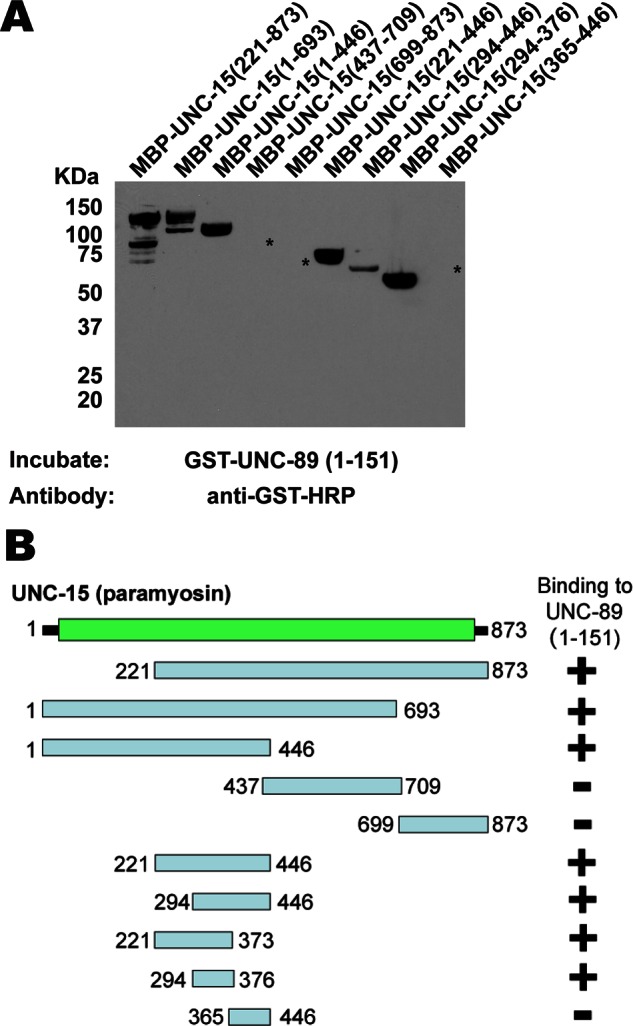
Far Western assays with purified proteins show that the minimal portion of paramyosin that interacts with UNC-89 spans paramyosin residues 294–376. (A) Portions of paramyosin (B) were separated by SDS–PAGE, transferred to a membrane, reacted with GST-UNC-89 (1–151), washed, incubated with anti–GST-horseradish peroxidase, washed, and detected by ECL. Asterisks indicate the positions on the blot of the three paramyosin fusion proteins (revealed by Ponceau S staining) that did not react with GST-UNC-89 (1–151). An identical blot showed no reaction with GST (Supplemental Figure S1). (B) Schematic summary of results in A. Note that the data for MBP-UNC-15 (221–373) are not shown in A.
Measurement of kinetics and affinity of binding
To further explore the interaction between the SH3 domain of UNC-89 (residues 58–131) and UNC-15 (294–376), we used biolayer interferometry (BLI; Abdiche et al., 2008; Concepcion et al., 2009). Much like surface plasmon resonance, BLI permits measurement of protein–protein interactions in real time. One protein, the ligand, is tethered to a fiber-optic sensor and then exposed to various concentrations of a second protein, the analyte, in solution. Binding is measured as a shift of the interference pattern of white light reflected from the end of the sensor. After a binding phase, the sensor is moved to buffer only, and dissociation is measured. By observing several different concentrations of the analyte and fitting the data to association and disassociation models, rate and affinity constants can be determined. We first examined the specificity of the interaction using GST, GST-UNC-89(58–131), MBP, and MBP-UNC-15 (294–376). As shown in Figure 4A, binding was observed only between tethered GST-UNC-89 (58–131) and analyte MBP-UNC-15 (294–376). Fusion partner–only controls generated shifts that were indistinguishable from GST-UNC-89 versus buffer only, that is, nonspecific binding was negligible. To quantitatively analyze the interaction between GST-UNC-89 (58–131) and MBP-UNC-15 (294–376), we performed kinetic characterization using MBP-UNC-15 (294–376) concentrations ranging from 0.45 to 7.2 μM. The sensorgram (Figure 4B) was fit to a global 1:1 simple binding model. KD was 1.1 μM, as determined by the ratio of koff (1.5 × 10−4 s−1) to kon (143 M−1 s−1). Parameters had low standard errors and tight 95% confidence intervals, indicating that fits are good; for example, the 95% confidence interval for KD is 0.96–1.14 μM.
FIGURE 4:
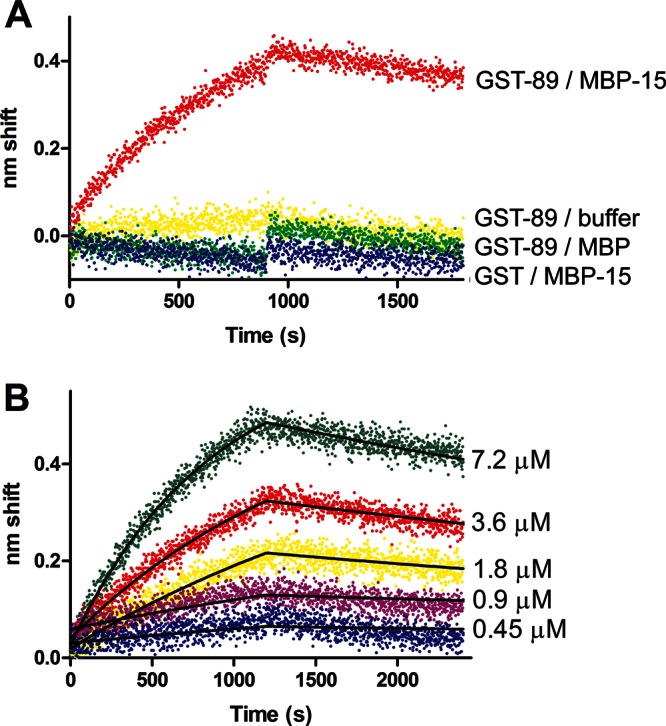
Biolayer interferometry analysis of binding of UNC-89 (58–131, SH3 domain) to UNC-15 (294–376). (A) Qualitative binding. Ligand GST-UNC-89 (SH3) was tethered to anti-GST sensors and exposed to 7.2 μM MBP-UNC-15 (red), MBP (green), or binding buffer only (yellow). Tethered GST exhibited no response to analyte MBP-UNC-15 (blue). Transition from association to dissociation phase occurred at 900 s. (B) Kinetic characterization of GST-UNC-89/MBP-UNC-15 binding. Concentrations of the analyte MBP-UNC-15 are noted to the right of each trace. Fits to a global 1:1 model are shown as black lines. Both association and dissociation phases were 1200 s in length.
unc-89 mutants that fail to express UNC-89 isoforms with the SH3 domain show aggregates of paramyosin
We reasoned that if UNC-89 interacts with paramyosin in vivo, then in nematode mutants lacking the paramyosin-binding region of UNC-89, paramyosin should show an abnormal localization pattern. To examine this question, we determined the localization of paramyosin in two mutant alleles of unc-89: 1) unc-89(su75), an unc-89 mutant in which all the giant isoforms are missing (Small et al., 2004) and thus the paramyosin-binding SH3 domain is missing; and 2) unc-89(tm752), an unc-89 mutant in which all of the kinase-containing isoforms are missing but the giant isoforms containing the SH3 domain (UNC-89-A and -E) are present (Ferrara et al., 2005). Indirect immunofluorescence was used to assess the localization of paramyosin and the two body-wall muscle cell myosin heavy chains MHC A and MHC B in wild type and unc-89(su75) and unc-89(tm752). As shown in Figure 5A, unc-89(su75) has a disorganization of both MHC A and MHC B, whereas unc-89(tm752) has a disorganization of MHC A but normal organization of MHC B. Most importantly, however, large accumulations of paramyosin were found in unc-89(su75) but not in unc-89(tm752). These accumulations of paramyosin were found in unc-89(su75) even in the muscle of L1-stage animals (Figure 5B). We conclude that when the paramyosin-binding SH3 domain of UNC-89 is missing, paramyosin forms large accumulations. This result is further indication that paramyosin interacts with UNC-89 in muscle cells.
FIGURE 5:
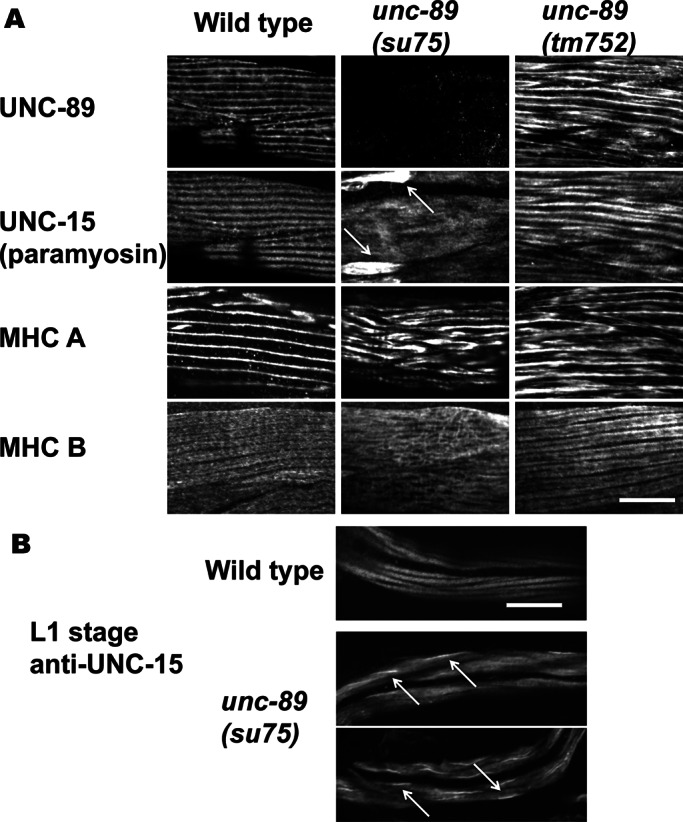
Localization of thick filament proteins in the body wall muscle of wild type and two categories of unc-89 mutants. (A) Adult muscle. Each box contains an immunofluorescence image of portions of several body wall muscle cells reacted with antibodies to UNC-89, paramyosin, or one of the two myosin heavy chains (MHC A or MHC B). unc-89(su75) lacks all giant UNC-89 isoforms (Small et al., 2004), and thus the paramyosin-binding SH3 domain is missing. unc-89(tm752) is an unc-89 mutant in which all of the kinase-containing isoforms are missing but giant isoforms (UNC-89-A and -E) are present (Ferrara et al., 2005). The antibody used to detect UNC-89, EU30, was developed to Ig domains 3–6 and thus detects the giant isoforms (Small et al., 2004). Note the large accumulations of paramyosin in unc-89(su75), indicated by arrows. Although unc-89(tm752) shows disorganization of UNC-89, paramyosin, and MHC A, no accumulations of paramyosin are detected. Therefore, when the paramyosin-binding SH3 domain is missing, paramyosin forms large accumulations outside the normal thick filaments. (B) L1 larval-stage muscle reacted with anti-paramyosin. In wild-type animals, paramyosin is localized to A-bands, each body wall muscle cell having two A-bands. In unc-89(su75), abnormal accumulations of paramyosin are seen, indicted by arrows (examples of two animals). Scale bars, 10 μm.
Next we wondered whether we could find a second unc-89 mutant allele that lacks all the giant UNC-89 isoforms and, if so, whether this mutant would also show large accumulations of paramyosin. Previous Western blot analysis of 20 different unc-89 alleles using antibodies that detect the giant isoforms (unpublished data) revealed one additional allele, r452, which also lacks all detectable giant isoforms (Figure 6B). Muscle staining with anti-paramyosin shows that r452, like su75, has large accumulations of paramyosin, whereas an additional mutant allele, e1460, and tm752 do not (Figure 6A). Thus there are two independently generated mutant alleles of unc-89 that lack the paramyosin-binding SH3 domain and show accumulations of paramyosin. Therefore the interaction of UNC-89 with paramyosin is crucial for the organization (assembly or maintenance) of paramyosin into sarcomeres in body wall muscle.
FIGURE 6:
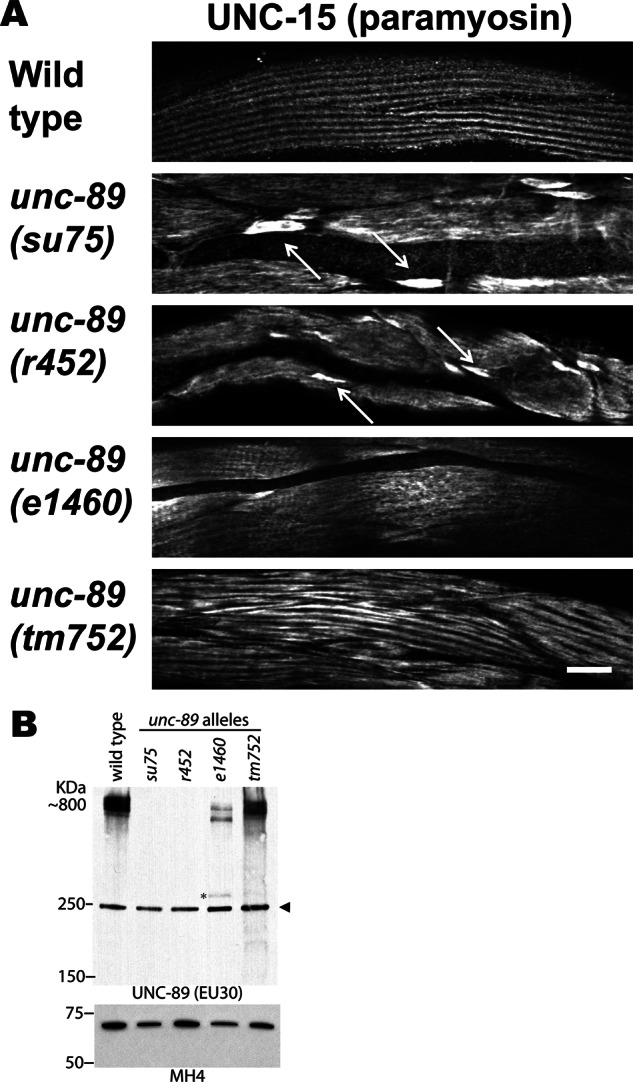
A second unc-89 mutant that lacks expression of all giant UNC-89 isoforms also shows accumulations of paramyosin in body wall muscle. (A) Each image is of several body wall muscle cells stained with antibodies to paramyosin from wild type and the indicated unc-89 mutant alleles. Wider fields of view are shown for unc-89(su75) and unc-89(tm752) than shown in Figure 3. Paramyosin accumulations are prominent in unc-89(su75) and unc-89(r452) but not in unc-89(e1460) or unc-89(tm752). Arrows point to some of the paramyosin accumulations. Scale bar, 10 μm. (B) Immunoblots of total nematode SDS-soluble proteins separated on a 4.5% polyacrylamide–SDS gel reacted with anti–UNC-89 EU30, which detects the giant ∼800-kDa isoforms of UNC-89. As shown, these giant isoforms are not detectable in su75 or r452 but are detectable at nearly wild-type levels from tm752 and at reduced levels in e1460. The asterisk indicates an ∼300-kDa protein that corresponds in size to a truncated protein terminating at the premature stop mutation in e1460. The arrowhead indicates a protein of nonspecific reactivity for this antibody. Bottom, portion of a blot in which equal amounts of protein extracts were separated by a 10% polyacrylamide gel, blotted, and reacted with monoclonal MH4 as a loading control.
To verify that mutations in unc-89(su75) and unc-89(r452) result in lack of expression of isoforms containing the SH3 domain and are not simply missing the epitopes (Ig3-6) for anti–UNC-89 antibodies (EU30) used for the Western blot, we determined the sequence alterations in both mutant alleles. Because the unc-89 gene is large (∼60 kb) and its coding sequence is also large (∼25 kb), we determined the whole genomic sequences of each allele by next-generation sequencing and limited our analysis to the unc-89 gene. unc-89(su75) contains an insertion of a C in the coding sequence for Ig38, resulting in a frameshift encoding 38 novel residues followed by a stop codon. unc-89(r452) has a deletion of a T in the coding sequence for Ig39, resulting in a frameshift encoding 92 novel residues followed by a stop codon. The fact that we could not detect UNC-89 large isoforms from these mutant alleles using an antibody (EU30) generated to Ig3-6 suggests that the mutant mRNAs containing these premature stop codons are degraded by non–sense-mediated decay. Consistent with this interpretation, using reverse transcription PCR with primers situated 5′ and 3′ of the 5.8-kb exon that encodes Ig27–Ig47, we could not detect any small cDNAs that would be evidence of alternative splicing across this region, which contains the su75 and r452 mutation sites. We previously reported that unc-89 (e1460) has a stop codon in the coding sequence for Ig21 (Spooner et al., 2012), and we have now determined by reverse transcription PCR that the 2.1-kb exon that encodes Ig20–Ig26 can be alternatively spliced. This explains why on Western blot (Figure 6B) for e1460, 1) we detect large isoforms that are somewhat smaller than wild type and 2) a low level of a protein of ∼300 kDa (indicated with an asterisk in Figure 6B), corresponding to the size of an expected truncated polypeptide ending at the premature stop codon. tm752 is a 531–base pair deletion (WormBase, www.wormbase.org) that removes most of intron 40 (including its 3′ acceptor site) and 33 base pairs of exon 41, which encodes part of the interkinase region.
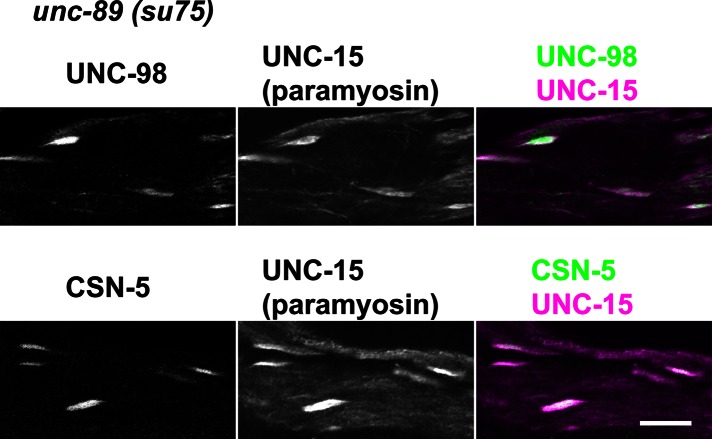
Loss-of-function mutations in genes encoding two additional M-line components, UNC-98 and UNC-96, also result in abnormal accumulations of paramyosin (Mercer et al., 2006; Miller et al., 2008). UNC-98 is a 310-residue polypeptide without obvious human homologue and contains four C2H2 Zn-finger domains. These Zn-finger domains interact with UNC-97 (PINCH), which is part of a conserved four-protein complex that interacts with the cytoplasmic tail of integrin (Mercer et al., 2003); the N-terminal 110 residues of UNC-98 interact with MHC A, which resides in the middle of the nematode thick filament (Miller et al., 2006). UNC-96 is a 418-residue polypeptide without human homologue and contains no recognizable domains (Mercer et al., 2006). UNC-96 also interacts with MHC A (Qadota et al., 2007). UNC-98 and UNC-96 interact with each other, and each also interacts with paramyosin (Mercer et al., 2006; Miller et al., 2008). Paramyosin accumulations in unc-98 mutants contain UNC-96, and paramyosin accumulations in unc-96 contain UNC-98 (Mercer et al., 2006). Moreover, the paramyosin accumulations of unc-96 and unc-98 mutants contain CSN-5, a highly conserved component of the COP9 signalosome, which is implicated in regulating ubiquitin-mediated proteolysis and is located at A-bands (Miller et al., 2009). CSN-5 interacts directly with UNC-98 and UNC-96. Paramyosin interacts directly with UNC-96 and UNC-98; UNC-96 interacts with UNC-98. We wondered whether the paramyosin aggregates in unc-89(su75) might also contain these additional proteins. As shown in Figure 7, both UNC-98 and CSN-5 colocalize with paramyosin accumulations in unc-89(su75). Our interpretation is that when the paramyosin- binding portion of UNC-89 is missing, some paramyosin is not incorporated into thick filaments and instead accumulates, dragging with it its direct and indirect partners, UNC-96, UNC-98, and CSN-5.
FIGURE 7:
Paramyosin accumulations in unc-89(su75) also contain UNC-98 and CSN-5. Each row shows images of several adult body wall muscle cells from unc-89(su75) coimmunostained with antibodies to paramyosin and either UNC-98 or CSN-5. Note that in the merged images, the white areas (overlap of magenta and green) show that the paramyosin accumulations also contain UNC-98 and CSN-5. Scale bar, 10 μm.
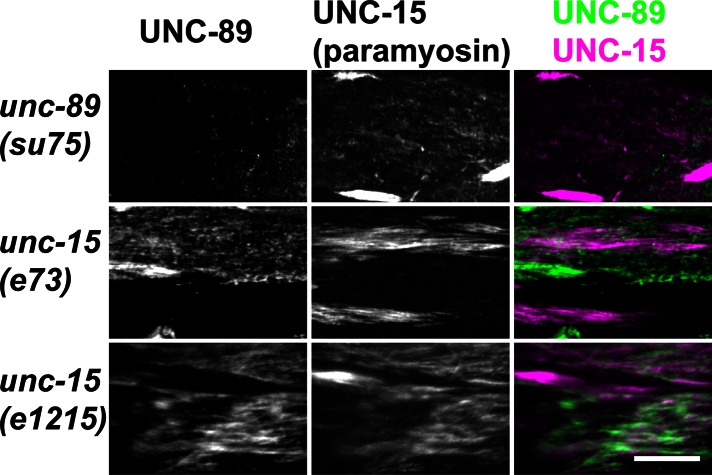
Next we reasoned that since the proper localization of paramyosin depended on interaction with UNC-89 (Figure 6A), then perhaps the proper localization of UNC-89 would depend on paramyosin. unc-15(e73) and unc-15(e1215) are missense mutations of unc-15 (Gengyo-Ando and Kagawa, 1991) and contain multifilament assemblages consisting of central paramyosin paracrystals and polar, thick filament–like structures (Epstein et al., 1987, 1993). As shown in Figure 8, immunostaining of the unc-15 mutants revealed clumping of both paramyosin and UNC-89. However, the clumps of the two proteins only partially colocalize. Moreover, as shown in Supplemental Figure S2, a wider, lower- magnification view, the accumulations of paramyosin in unc-89(su75) and either unc-15(e73) or unc-15(e1215) have different appearances. Overall there is less clumping of paramyosin in unc-89(su75) than there is in the two unc-15 missense alleles. One interpretation is that the loss of UNC-89 binding to paramyosin would only affect paramyosin incorporated into thick filaments, whereas the missense mutations of paramyosin would be more severe because they lead to self-aggregation or abnormal association with other proteins before paramyosin is incorporated into thick filaments.
FIGURE 8:
Paramyosin accumulations in unc-89(su75) and in two paramyosin missense mutants. Paramyosin paracrystals in unc-15(e73) and unc-15(e1215) show only partial colocalization with UNC-89. Each row contains images of the same portion of body wall muscle from the indicated nematode strains costained with anti-UNC-89 (green) and anti-UNC-15 (magenta); colocalization is indicated by white. A wider field view of the anti-paramyosin staining of these mutants is shown in Supplemental Figure S2. Scale bar, 10 μm.
A segment of UNC-89 containing PK2 (1/3IK-Ig-Fn-PK2) is sufficient for localization in the sarcomere
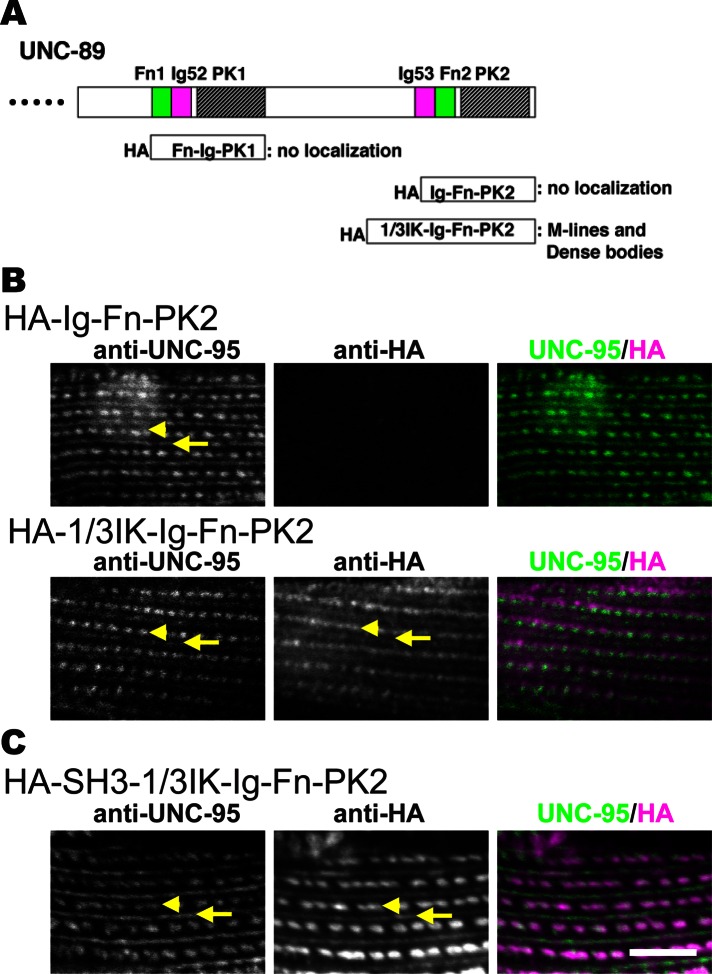
Because loss of UNC-89 isoforms containing the paramyosin-binding region (the SH3 domain) showed such a dramatic clumping of paramyosin, we wondered whether overexpression of the SH3 domain would also affect the organization of paramyosin. We first tried to overexpress the SH3 domain (UNC-89 residues 1–151, which contains the SH3 domain and flanking sequences) using a heat shock promoter in adults with either hemagglutinin (HA) or enhanced green fluorescent protein (EGFP) N-terminal tags. After heat shock at 30°C for 2 h, we made protein lysates and were able to detect either tagged protein by Western blot. After repeating this heat shock procedure, we incubated the worms at 20°C (their normal growing temperature) for 24 h to allow for sarcomere assembly in the presence of the SH3 domain. After fixation and immunostaining with anti-paramyosin and either anti-HA or anti-GFP, we found that paramyosin was normally localized, and we were not able to detect any HA-SH3 or EGFP-SH3 in the myofilament lattice. We suspected that HA-SH3 or EGFP-SH3 was not being incorporated into the sarcomere and consequently quickly degraded. We next considered whether we might have a better result if we were able to direct the localization of the overexpressed SH3 domain by attaching it to a protein that localizes to the native UNC-89 site, that is, the M-line. We reasoned that a good candidate for this would be UNC-89 itself. We already knew that the small isoforms of UNC-89, UNC-89-C and UNC-89-D, which consist of partial PK1-interkinase-Ig-Fn-PK2, localize to M-lines (Small et al., 2004). We expressed portions of this region (with HA tags) in transgenic nematodes to determine the minimum portion that is required to localize to M-lines. We heat shocked at 30°C for 2 h and then fixed and stained with anti-HA. As summarized in Figure 9A, neither Fn-Ig-PK1 nor Ig-Fn-PK2 localized in the sarcomere. However, adding the C-terminal one-third of the interkinase region N-terminal of Ig-Fn-PK2 as the segment “1/3IK-Ig-Fn-PK2” did allow it to localize to both M-lines and dense bodies (Figure 9, A and B, bottom row). Although we have never seen localization of UNC-89 to dense bodies using antibodies developed to three different regions of UNC-89 (Small et al., 2004), we have encountered examples of other proteins that show additional sites of localization when examined using transgenic overexpression. Examples include UNC-98 (Mercer et al., 2003) and UNC-96 (Mercer et al., 2006). In the case of UNC-96, antibodies localize only to M-lines, but when overexpressed with a GFP tag, they localize to M-lines and dense bodies (Mercer et al., 2006).
FIGURE 9:
A small portion of UNC-89, even with an added SH3 domain, can localize to sarcomeres. (A) Top, schematic of domains within the C-terminal portion of UNC-89. Below, portions tested with HA tags in transgenic animals for localization to the sarcomere. HA-1/3IK-Ig-Fn-PK2 localized to M-lines and dense bodies. (B) Localization of transgenically expressed portions of UNC-89. Each row shows costaining with anti-HA to localize the fragment and anti-UNC-95 to mark M-lines and dense bodies. As shown, HA-Ig-Fn-PK2 does not localize, but HA-1/3IK-Ig-Fn-PK2 does. (C) Localization of transgenically expressed chimeric protein HA-SH3-1/3IK-Ig-Fn-PK2. Note that inclusion of the SH3 domain still permits 1/3IK-Ig-Fn-PK2 to localize to M-lines and dense bodies. Arrows, M-lines; arrowheads, dense bodies. Scale bar, 10 μm.
Overexpression of UNC-89’s SH3 domain results in mislocalization of paramyosin
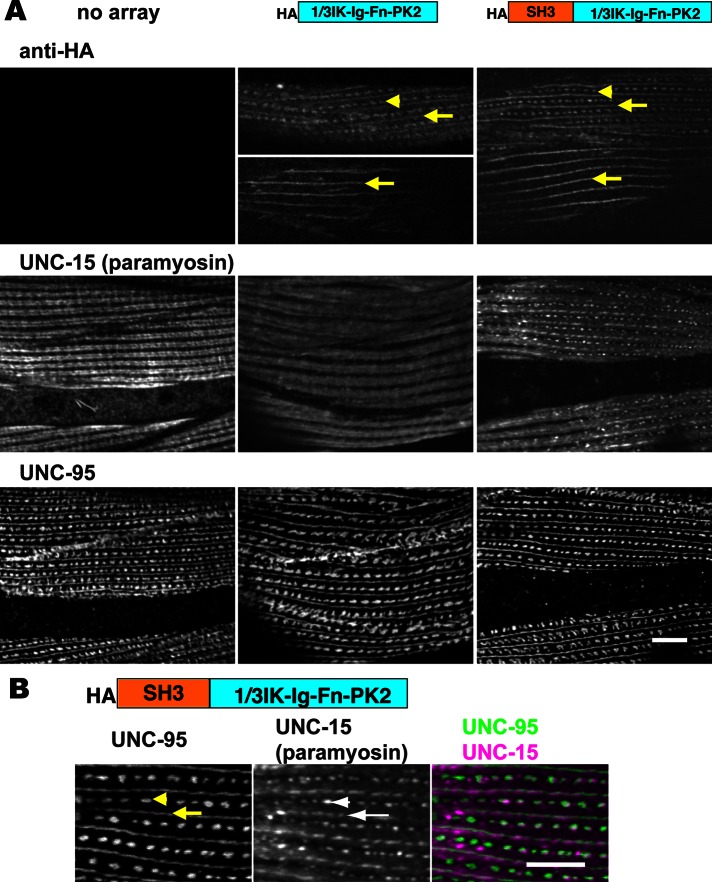
We then heat-shock expressed an artificial small UNC-89 molecule consisting of SH3-1/3IK-Ig-Fn-PK2 to direct SH3 to the M-line (again, we used UNC-89 residues 1–151, which contain the SH3 domain and flanking sequences.) As shown in Figure 9C, this chimeric protein also localizes to M-lines and dense bodies (after a 2-h heat shock and immediate fixation). However, when this chimeric protein was expressed with a 2-h heat shock followed by 24 h at 20°C, we did not detect a change in the localization of paramyosin (Supplemental Figure S3). In fact, after this protocol, we could not detect the chimeric protein by Western blot (Supplemental Figure S3A). Suspecting that if the protein was expressed it might be unstable, we heat shocked at 30°C for 2 h and then moved the worms to 20°C and made protein lysates from them immediately and at 2, 4, 6, 8, and 24 h later. As shown by Western blot (Supplemental Figure S3B), the protein levels decreased with time after heat shock and were undetectable at 24 h. Therefore we changed our heat shock protocol to a 30°C exposure continuously for 24 h. With this procedure, we found that both HA-1/3IK-Ig-Fn-PK2 and HA-SH3-1/3IK-Ig-Fn-PK2 were localized to M-lines and dense bodies and, in some muscle cells, just M-lines (Figure 10A, top). Transgenic lines expressing each of these chimeric proteins were immunostained with anti-paramyosin and anti-UNC-95 (a protein that localizes to the bases of both M-lines and dense bodies). As shown in Figure 10A, middle, neither heat shock on wild-type animals (“no array”) nor overexpression of HA-1/3IK-Ig-Fn-PK2 in transgenic animals affects the normal localization of paramyosin to A-bands. However, overexpression of HA-SH3-1/3IK-Ig-Fn-PK2 in transgenic animals results in mislocalization of paramyosin to M-lines and dense bodies. As shown in Figure 10B at higher magnification, overexpressed HA-SH3-1/3IK-Ig-Fn-PK2 clearly results in paramyosin being abnormally localized to M-lines (arrows) and dense bodies (arrowheads). The effect of overexpressing HA-SH3-1/3IK-Ig-Fn-PK2 is specific: the localization of UNC-95 is unaffected (Figure 10A, bottom, rightmost column), as are the localizations of the thick filament components MHC A, MHC B, and twitchin (Supplemental Figure S4). Therefore this experiment provides additional evidence that there is specific interaction between UNC-89’s SH3 domain with paramyosin in vivo.
FIGURE 10:
Overexpression of the UNC-89’s SH3 domain causes paramyosin to be mislocalized. (A) Immunolocalization of HA, paramyosin, and UNC-95 in body wall muscle cells of wild-type (no array) or transgenic animals expressing either HA-1/3IK-Ig-Fn-PK2 or HA-SH3-1/3IK-Ig-Fn-PK2 (as integrated arrays). Top row, anti-HA does not react to anything in wild-type muscle; in some animals or in some cells within the same animal, the fragments are localized to either both M-lines (arrow) and dense bodies (arrowheads) or only to M-lines (arrows). Middle row, in wild type (no array) or in animals overexpressing HA-1/3IK-Ig-Fn-PK2, paramyosin is localized to A-bands, with some reduced localization to M-lines; however, in animals overexpressing HA-SH3-1/3IK-Ig-Fn-PK2, paramyosin localizes in an abnormal pattern, to what appear to be M-lines and dense bodies. Bottom row, the overall organization of M-lines and dense bodies is normal in transgenic animals expressing either HA-1/3IK-Ig-Fn-PK2 or HA-SH3-1/3IK-Ig-Fn-PK2. (B) Coimmunostaining of anti-UNC-95 and anti-paramyosin antibodies in body wall muscle of transgenic nematodes overexpressing HA-SH3-1/3IK-Ig-Fn-PK2. UNC-95 is a marker for M-lines (arrows) and dense bodies (arrowheads). Note that paramyosin abnormally localizes to these structures. Scale bars, 10 μm.
Bioinformatics insight into the interaction between the SH3 domain of UNC-89 and paramyosin
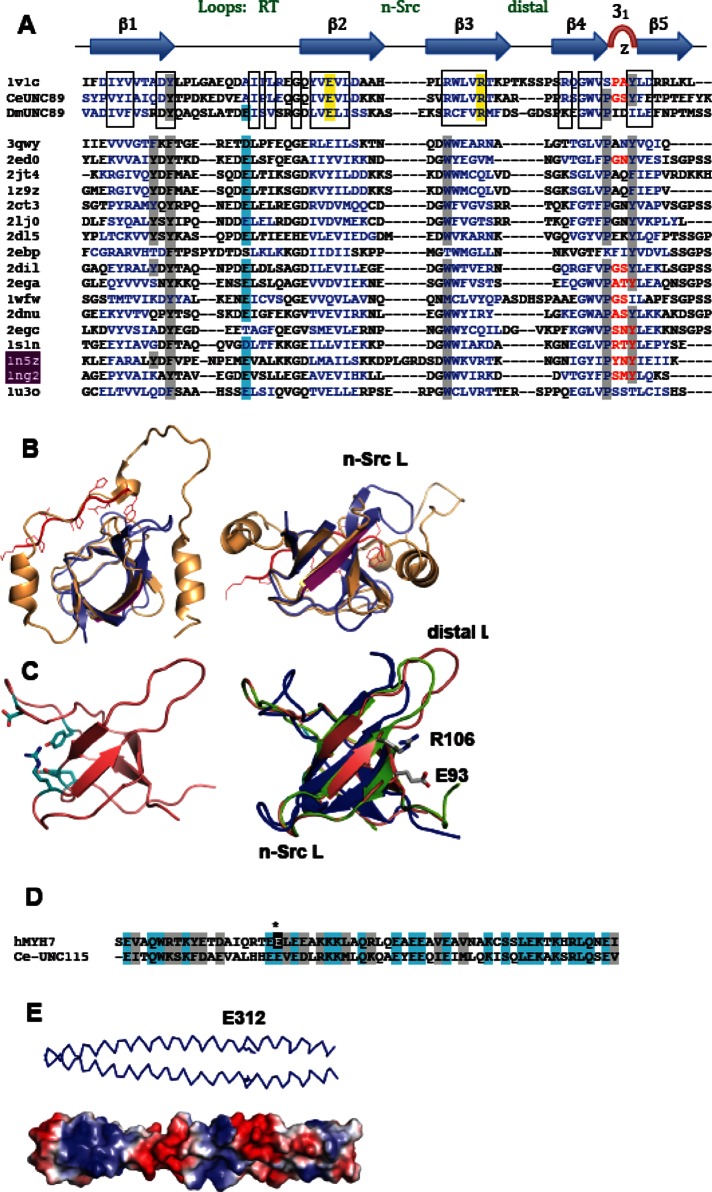
The interaction of UNC-89’s SH3 domain with paramyosin was unexpected, given that SH3 domains typically interact with proline-rich sequences. Paramyosin is almost entirely α-helical, with only three proline residues, all in the nonhelical, 30-residue-long N-terminus, which is not required for binding. The SH3 fold consists of a five-stranded β-barrel comprising three prominent loops (termed RT, n-Src, and distal loops) and a 310-helix turn (Figure 10A, top). The peptide-binding groove is located between the RT loop and the n-Src loop/310-turn. The groove is mostly hydrophobic and consists of three shallow pockets defined by conserved aromatic residues (Figure 11A, gray). The peptide ligand is typically proline rich, commonly containing a PxxP motif in extended, left-handed polyproline-2 conformation. Generally, proline residues in the peptide ligand are immediately preceded by hydrophobic residues forming two ΦP pairs in register (ΦPxΦP). Each ΦP dipeptide is accommodated in a hydrophobic pocket of the SH3. The third pocket includes in most cases a negatively charged residue (commonly from loop RT) that interacts with a basic residue in the ligand distal to the ΦPxΦP core. A number of nonconsensus SH3 target peptides have been identified that have atypical sequences (reviewed in Saksela and Permi, 2012), and cases are also known of totally divergent peptides that interact with an entirely different surface of the SH3 domain. For example, the peroxisomal Pex13p SH3 domain binds two different ligands, Pex5p and Pex14p, on opposite surfaces (Douangamath et al., 2002). The Pex14p ligand contains a conserved PxxP motif that docks into the canonical groove, and the Pex5p sequence is α-helical and binds onto strand β2 in the convex SH3 face. A further example is the autoinhibited SH3 domain of p47phox (Groemping et al., 2003). Its crystal structure shows how a C-terminal extension wraps around the SH3, docking into the canonical groove and following on to form a C-terminal helix that binds onto the convex domain face. This site is equivalent to that reported for the Pex13p-SH3/Pex5p helical interaction (Figure 11B), which suggests that this region of the SH3 fold might constitute a generally suitable enclave for the binding of helical peptides.
FIGURE 11:
Predicted molecular features of UNC-89-SH3 and UNC-15(294-376). (A) Sequence alignment of SH3 domains of known structure most similar to UNC-89-SH3. PDB codes are given; 1v1c is from human obscurin. Hydrophobic residues (gray) and negatively charged residue (blue) classically found in the PxxP binding groove are highlighted. SH3, known to bind helical peptides in their convex surface, are in red; 1n5z is Pex13p-SH3, and 1ng2 is p47phox-SH3. Conserved groups across UNC-89-SH3 from human, nematode, and fly are boxed. Conserved residues that are potential mediators of interactions at the convex face are in yellow. (B) Two perpendicular views of the crystal structures of Pex13p-SH3 (1n5z; blue) and p47phox-SH3 (1ng2; orange). Pex13p-SH3 is in complex with a canonical PxxP-containing peptide from Pex14p (red); the binding site of the helical peptide from Pex15p is magenta (strand β2). p47phox contains the SH3-surrounding inhibitory sequence. (C) Left, NMR structure of human obscurin’s SH3 (PDB 1V1C) displaying residues in the PxxP binding region. Right, 3D models of the SH3 domains from UNC-89 from C. elegans (blue) and D. melanogaster (green) superimposed on human obscurin’s SH3. The conserved charged residues in the convex region are shown. (D) Alignment of skip-2–containing sequences from UNC-15 and human cardiac myosin hMYH7 (E312 is marked and sequence identity highlighted). (E) A 3D model of UNC-15 showing the skip 2 residue, E312.
The structure of the SH3 domain from human obscurin has been elucidated using nuclear magnetic resonance (NMR; Protein Data Bank [PDB] ID 1CVC; Figure 11C, left). It shares 54 and 48% sequence similarity with the SH3 domains of UNC-89 from C. elegans and Drosophila melanogaster, respectively. This modest conservation contrasts with the high levels of sequence identity found within the respective animal classes. We calculated three-dimensional (3D) models of the SH3 domains of UNC-89 from C. elegans and D. melanogaster (Figure 11C, right). The most conserved region across the three domains is the β-sheet, whereas the RT, n-Src, and distal loops vary notably in composition (Figure 11A). The distal loop is the most divergent region of the fold, varying also in length. The three domains show the presence of a somewhat degenerated PxxP binding groove, where the classical tryptophan residue in strand β3 is replaced by an arginine that protrudes from the binding groove. A BLAST analysis revealed that lysine is also found in this position across sequences from vertebrates, insects, and nematodes, although nematodes often contain a serine instead (asparagine is only rarely observed). The “specificity” glutamate in loop RT (blue in Figure 11A) is identifiable only in the fly, possibly having shifted positions in the human and worm domains. This variability in the PxxP binding groove might point to a nonconsensus ligand sequence for this site or, potentially, a lack of binding functionality. On the other hand, the exposed surface of strands β2 and β3 is highly conserved across animal groups. In particular, the charged residues E93 and R106 (C. elegans numbering) are present in all sequences from vertebrates, insects, and nematodes examined here. This region binds helical motifs in Pex13p SH3 and p47phox. It will be of interest to explore whether in UNC-89 (obscurin) it might support the interaction with helical electrostatic ligands such as paramyosin identified in this study.
The sequences of the rod portion of myosin heavy chains and paramyosin share very high conservation and both consist of heptad repeats that induce their folding into coiled-coils. Simultaneously, the sequences have a longer, 28-residue repeat (38 copies) that results in charged residues being displayed on the surface of the coiled-coil in a periodic manner. This charge distribution directs the staggered parallel assembly of myosin rods into a thick filament (McLachlan and Karn, 1982; Kagawa et al., 1989). There are four widely spaced “skip residues” in the rod portion of myosin, which introduce a discontinuity in the phasing of the heptads that is believed to result in the deformation of the coiled-coil. Each skip is located at the end of a 28- residue repeat following position c of a heptad. The crystal structure of regions of human cardiac myosin containing each of the skip residues confirmed that the skips result in local relaxation of the coiled-coil (Taylor et al., 2015). The biological significance of each skip residue was tested by introducing tagged myosin into cardiomyocytes in which each of the skip residues had been deleted; deletion of skip 3 resulted in misincorporation by the presence of myosin aggregates; deletion of skip 4 resulted in lack of antiparallel assembly of myosin rods in the bare zone. Of interest, deletion of skip 1 or skip 2 had no obvious phenotype, and these were speculated to play other, as-yet-unidentified sarcomeric roles (Taylor et al., 2015). The SH3-binding segment of paramyosin residues 294–376 shares high similarity (64%) with the region of myosin MYH7 containing skip 2 (Figure 11D). The glutamate residue that causes this skip in MYH7 (E1385) is also present in UNC-15 (E312; Figure 11D). Our assignment of E312 as skip 2 in UNC-15 is in agreement with an earlier analysis (Kagawa et al., 1989). It is conceivable that UNC-89’s SH3 might specifically recognize the quaternary structure of UNC-15 at this locus due to the unwinding distortion that this skip introduces in its coiled-coil.
DISCUSSION
Our results demonstrate that paramyosin is a new binding partner for the giant muscle polypeptide UNC-89 in C. elegans muscle. The UNC-89/paramyosin interaction was initially discovered by screening a yeast two-hybrid library with the SH3-DH-PH region of UNC-89 as bait. In vitro binding assays using purified recombinant proteins showed that minimally this interaction requires the SH3 domain (residues 58–131) of UNC-89 and residues 294–376 of paramyosin (Figure 12A). Biolayer interferometry showed that the two protein segments bind with KD = 1.1 μM with a relatively slow off-rate. Evidence that the UNC-89/paramyosin interaction occurs in vivo includes both mutant and transgenic analysis. Paramyosin forms accumulations or aggregates in unc-89 mutants that lack expression of the large SH3 domain–containing isoforms; these paramyosin aggregates are not found in normal muscle or unc-89 mutants that do express the SH3 domain–containing isoforms. Paramyosin accumulations can be found in as early as L1 larvae. These results suggest that the interaction of UNC-89 with paramyosin is important during sarcomere assembly (during the L1 stage) and perhaps during sarcomere maintenance via turnover, as there is no new sarcomere assembly during the adult stage (Mackenzie et al., 1978). Paramyosin accumulations in unc-89 mutants were also shown to contain a trio of interacting proteins found previously in paramyosin accumulations in unc-98 and unc-96 mutants: UNC-96 and UNC-98, two nematode-specific M-line proteins, and CSN-5, a highly conserved component of COP9 signalosomes, which regulates ubiquitin-mediated proteolysis. After identifying a minimal region of UNC-89 that can localize to M-lines and dense bodies when expressed in transgenic animals, we used it to direct the localization of the SH3 domain to these structures. This chimeric artificial protein (SH3-minimal localizing fragment) is able to direct paramyosin to M-lines and dense bodies, providing further evidence that the SH3 domain of UNC-89 can interact with paramyosin in vivo. The interaction of UNC-89’s SH3 domain with paramyosin was surprising, given that SH3 domains usually interact with proline- rich sequences that are absent in coiled-coil proteins like paramyosin. To provide some insight into how UNC-89’s SH3 domain binds to paramyosin, we made a homology model of UNC-89’s SH3 domain from both C. elegans and Drosophila. The models suggested a somewhat degenerated PxxP binding groove, which may suggest lack of binding ability to this type of sequence, as well as conservation of exposed surfaces of strands β2 and β3, which may be significant for binding paramyosin, as this region has been shown to bind α-helical motifs in Pex13p SH3 and p47phox. We also found that the region of paramyosin that binds to UNC-89’s SH3 contains a skip residue and has strong homology (and conservation of the skip-causing glutamate) to skip 2 of human cardiac myosin MYH7, which was shown recently by crystallography (Taylor et al., 2015) to result in relaxation of the coiled-coil. We speculate that UNC-89’s SH3 domain might recognize and bind to the surface structure created by the unwinding distortion of the coiled-coil.
FIGURE 12:
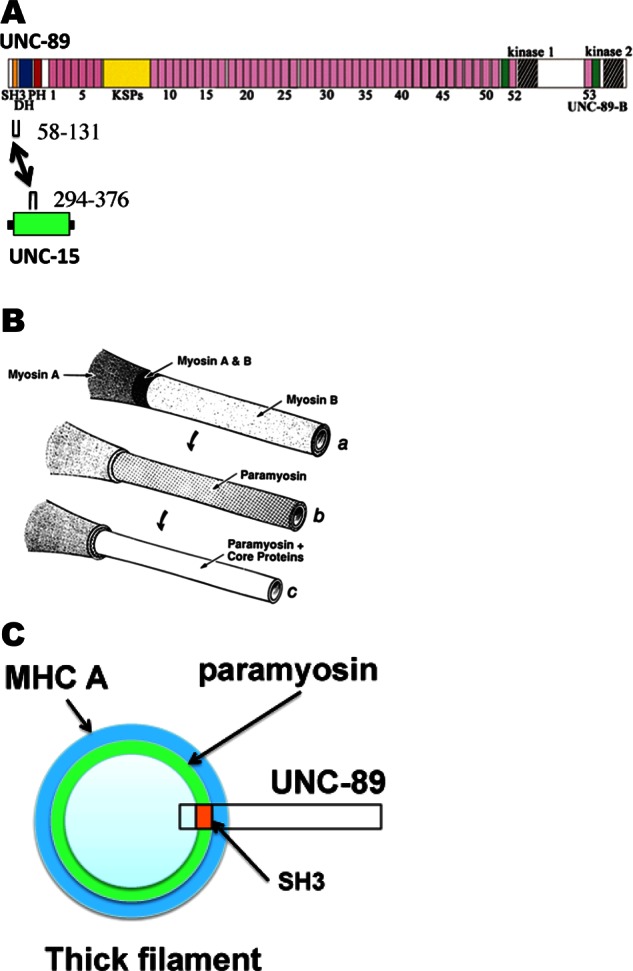
Summary of results and model for how UNC-89 associates with thick filaments at the M-line. (A) The SH3 domain of UNC-89 near its C-terminus interacts with a small portion (residues 294–376) of the α-helical coiled-coil region of UNC-15 (paramyosin). In UNC-89, the color scheme used to designate domains is the same as in Figure 1. In UNC-15, green denotes predicted coiled-coil, and black indicates the nonhelical tail pieces. (B) Model of nematode thick filament from Deitiker and Epstein (1993; reprinted, with permission). There are three concentric layers: an outer layer consisting of the myosins MHC A and MHC B; an intermediate layer consisting of paramyosin; and a core consisting of paramyosin and core proteins, later shown to be filagenins. (C) Speculation as to how the SH3 domain of UNC-89 interacts with paramyosin in thick filaments. A cross section of the thick filament near the M-line based on the model shown in B. The green annulus denotes where paramyosin is located (both intermediate and core layers). It is speculated that gaps or crevices along the myosin surface allow penetration of UNC-89 so that its SH3 domain can interact with paramyosin. The major portion of UNC-89 is thus likely to project from the thick filament surface and interact with other proteins at the M-line.
Of interest, the novel function of UNC-89’s SH3 domain is a second example in which UNC-89 likely uses “classical” domains for “nonclassical” functions. PH domains usually bind to acid phospholipids and function in membrane targeting. However, the PH domain of UNC-89 does not bind to inositol-1,4,5-triphosphate, and its NMR structure shows that it has a closed conformation, and its presumed phospholipid-binding pocket is lined with negative charges, which would make it incapable of binding to acidic phospholipids (Blomberg et al., 2000). Although Blomberg et al. (2000) speculated that UNC-89’s PH domain might alternatively function in protein–protein interactions, such a role has not yet been found by experiment.
The optical biosensing experiments indicated fast on-rate and a slow off-rate between UNC-89’s SH3 and the segment of paramyosin. This is compatible with the robust signals we obtained using far Western assays. The measured KD ≈ 1.1 μM of the interaction is also compatible with reports of the binding of the myosin rod or paramyosin with several other proteins. For example, the mammalian protein myosin-binding protein C binds through its C-terminal Ig domain to myosin, and, when measured for binding by cosedimentation to light meromyosin (LMM) filaments, had KD ≈ 3.5 μM (Flashman et al., 2007). We reported that for C. elegans muscle, the N-terminal portion of the M-line protein UNC-98 binds to purified myosin by enzyme-linked immunosorbent assay (ELISA) with KD ≈ 0.30 μM (Miller et al., 2006). In contrast, we also reported that full-length UNC-98 and UNC-96 bind to fragments of paramyosin by ELISA with higher affinity—56 and 49 nM, respectively (Miller et al., 2008).
Based on gradual solubilization of purified thick filaments with buffers of increasing salt concentrations (Epstein et al., 1985), the model for the nematode thick filament is that it is a series of concentric layers—an outer layer consisting of myosins MHC A and B, an intermediate layer of paramyosin, and an inner layer of paramyosin and the filagenins (Deitkiker and Epstein, 1993; Liu et al., 1998; Figure 12B). Analysis of electron microscope (EM) images of isolated cores indicated that the core consists of seven subfilaments of paramyosin coupled to form a tube by the filagenins (Epstein et al., 1995). In vertebrate muscles, the M-line is observable by EM to be a structure in which the shafts of thick filaments are cross-linked at their surfaces by a series of struts and additional filaments (Knappeis and Carlsen, 1968; Luther and Squire, 1978). Presumably, similar cross-linking of thick filaments occurs in C. elegans. UNC-89 is localized exclusively to the M-line, from its base, where it is anchored at the muscle cell membrane, continuing throughout the depth of the myofilament lattice (Warner et al., 2013; Reedy, Hoppe, and Benian, unpublished data). Because paramyosin seems buried under an outer layer of the myosins, it is difficult to understand how interaction of UNC-89 with paramyosin can occur at the M-line. Both myosin rods and paramyosin dimers have alternating zones of positive and negative charges along their surface that support their assembly into filaments primarily by electrostatic interactions (McLachlan and Karn, 1982; Kagawa et al., 1989). It is possible that the distortions introduced by skip residues in the helical rod bundles of paramyosin and myosin are such that these proteins do not form tight smooth layers but that bulges and/or crevices exist in their surface topography. These discontinuities would permit access of UNC-89’s SH3 domain to the otherwise internal paramyosin layer (Figure 12C). Not mutually exclusive with this possibility is that during sarcomere assembly, paramyosin and UNC-89 associate before they are incorporated into thick filaments and M-lines. Consistent with this idea, during embryonic muscle development, paramyosin can be detected in 420-min embryos (Epstein et al., 1993), during which time thick filament precursors are being assembled, and this is the same time at which staining with MH42, the monoclonal later shown to react with UNC-89 (Benian et al., 1996), is first detected (Hresko et al., 1994). Although additional sarcomeres are not being added in adult muscle, presumably there is removal of damaged proteins and their replacement with newly synthesized ones. One way to interpret the paramyosin accumulations found in unc-89(su75) and unc-89(r452) is that in the absence of UNC-89, which is capable of binding to paramyosin, paramyosin forms abnormal aggregates, and this reflects a paramyosin pool that was otherwise destined to form replacement thick filaments.
A related question is that, if UNC-89 interacts with paramyosin that is distributed throughout the long axis of thick filaments, how is UNC-89 restricted to M-lines? The answer likely arises from other binding partners of UNC-89 that emanate from the cell membrane base of the M-lines, which begins with the transmembrane protein integrin. The cytoplasmic tail of integrin exists in a complex with UNC-112 (kindlin), PAT-4 (ILK), PAT-6 (α-parvin), and UNC-97 (PINCH; Qadota and Benian, 2010). UNC-97 interacts with LIM-9 (FHL), which in turn interacts with UNC-89 via its Ig52-Fn1-PK1 and 1/3 interkinase region (Xiong et al., 2009). PAT-6 (α-parvin) interacts with CPNA-1, which in turn interacts with UNC-89 via its Ig1-3 domains (Warner et al., 2013).
Given that Drosophila thick filaments also contain paramyosin, we suggest that Drosophila UNC-89 (Small et al., 2004; Katzemich et al., 2015) might also associate with paramyosin through its SH3 domain. Although vertebrate muscle does not contain paramyosin, perhaps it is possible for the SH3 domain of vertebrate obscurin to interact with myosin. An obscurin–myosin interaction seems unlikely, however, for the following reasons. In obscurin-A isoforms (Fukuzawa et al., 2005), there are up to 55 Ig and Fn3 domains separating the M-band targeting N-terminal Ig1-3 (via titin–myomesin interactions; Fukuzawa et al., 2008) from the obscurin SH3 domain, and the C-terminus of obscurin-A interacts with small ankyrin 1 at the sarcoplasmic reticulum (Bagnato et al., 2003; Kontrogianni-Konstantopoulos et al., 2003). Myosin and the SH3 are therefore separated by ∼200 nm, and a direct interaction seems unlikely. Nevertheless, it is possible that obscurin’s SH3 domain interacts with another coiled-coiled protein.
MATERIALS AND METHODS
Screening of yeast two-hybrid library
Yeast two-hybrid screening of the C. elegans cDNA library RB2 was performed as described (Miller et al., 2006) with some modifications. We first tried to screen the library with this previous protocol using the bait plasmid pGBDU-UNC-89 SH3-DH-PH (plasmid described in Warner et al., 2013). However, after shuffling off the bait plasmid, the yeast host strain harboring only prey plasmids did not grow well in selective media, meaning that isolation of prey plasmids from yeast cells was practically impossible. We changed the bait plasmid to pGBKT7-UNC-89 SH3-DH-PH, a bait plasmid with kanamycin resistance as the bacterial marker (pGBKT7; Takara Clontech, Mountain View, CA). To make pGBKT7-UNC-89 SH3-DH-PH, we cloned the NcoI/XhoI fragment insert from pGBDU-UNC-89 SH3-DH-PH into NcoI/SalI-digested pGBKT7. Because of the kanamycin selection for the bait, it was not necessary to remove the bait plasmid from the yeast cells of the two-hybrid positive clones in order to grow and isolate the ampicillin-resistant prey plasmids.
Domain mapping by yeast two-hybrid assay
Twenty prey clones containing various UNC-15 regions were originally isolated from cDNA library screening. UNC-89 bait fragments lacking SH3, DH, or PH domain were amplified by two-step PCR using GK1-8 primers (Supplemental Table S1) and confirmed by DNA sequencing. Using those prey and bait clones, we detected interaction by our standard yeast two-hybrid method (Miller et al., 2006).
Domain mapping by far Western assay
Segments of paramyosin spanning residues 221–873, 221–446, 294–446, 294–376, and 365–446 were expressed as an MBP fusion protein in Escherichia coli. cDNAs encoding UNC-15– corresponding regions were PCR amplified using primers GK10-19 (Supplemental Table S1), and its sequence was confirmed to be error-free. Plasmids expressing MBP fusion with UNC-15 (1–693, 1–446, 437–709, 699–873) were described previously (Miller et al., 2008). Portions of UNC-89 (1–500, 1–70/125–500, 1–151, 58–131) were cloned into pGEX-KK-1 for expression of the corresponding GST fusion proteins, beginning with cDNA PCR fragments generated using primers (GK1, 2, 5, 6, 9, 20, 21) indicated in Supplemental Table S1. MBP, GST, and fusion proteins were purified as described previously (Benian et al., 1993; Mercer et al., 2006). Supplemental Figure S5 shows SDS–PAGE of 2 μg each of the 15 expressed recombinant proteins. Far Western assays were conducted as follows: 2 μg of MBP and 4 μg of MBP-UNC-15 (221–873) or 2 μg of MBP-UNC-15 (1–693, 1–446, 437–709, 699–873, 221–446, 294–446, 221–373, 294–376, 365–446) were resolved by SDS–PAGE, transferred to nitrocellulose membrane, and blocked overnight at room temperature in 5% milk and Tris-buffered saline/Tween 20 (TBS-T). Portions of this blot containing MBP and MBP-UNC-15 were separately incubated with 1 μg/ml GST or GST-UNC-89 (1–500, 1–70/125–500, 1–151, 58–131) at room temperature for 1 h, washed multiple times in TBS-T, reacted with anti-GST conjugated to horseradish peroxidase (at 1:3000 dilution; A7340; Sigma-Aldrich, St. Louis, MO), washed, and then reactions visualized by enhanced chemiluminescence (ECL; Pierce, Thermo Fisher Scientific, Waltham, MA).
Biolayer interferometry
BLI experiments were performed using a FortéBio (Menlo Park, CA) Octet QK using anti-GST sensors. Assays were done in 96-well plates at 25°C. We used 200-μl volumes in each well. Ligand GST-SH3 (UNC-89 (58–131)) was loaded onto sensors for 600 s and followed by baseline measurements in binding buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 10% glycerol) for 300 s. Association was measured by dipping sensors into solutions of analyte protein and was followed by moving sensors to buffer only to monitor dissociation. For qualitative experiments, all analyte concentrations were 7.2 μM. A serial dilution of MBP-UNC-15 (294–376) was made for full characterization. Binding was fit to a global 1:1 association-then-dissociation model using GraphPad Prism 5.02.
C. elegans strains
Standard growth conditions for C. elegans were used (Brenner, 1974). Wild-type nematodes were the N2 (Bristol) strain. Strain HE75 unc-89(su75) was provided by Henry Epstein (University of Texas Medical Branch at Galveston, Galveston, TX). unc-89(tm752) was obtained from Shohei Mitani (Tokyo Women’s Medical University School of Medicine, Tokyo, Japan). CB1460 unc-89(e1460) was obtained from Robert Waterston (University of Washington, Seattle, WA). Strain TR625 unc-89(r452) was obtained from Phil Anderson (University of Wisconsin, Madison, WI). These unc-89 mutants were outcrossed to wild type three or four times. The unc-15 alleles e73 and e1215,and N2 (Bristol) were obtained from the Caenorhabditis Genetics Center (University of Minnesota, Minneapolis, MN).
Immunolocalization in adult body-wall muscle
Adult and L1 larva nematodes were fixed and immunostained according to the method described by Nonet et al. (1993) and described in further detail by Wilson et al. (2012a). The following primary antibodies were used at 1:200 dilution: anti-UNC-89 (rabbit polyclonal EU30; Benian et al., 1996), anti-paramyosin (mouse monoclonal 5-23; Miller et al., 1983), anti–MHC A (mouse monoclonal 5-6; Miller et al., 1983), anti–MHC B (mouse monoclonal 5-8; Miller et al., 1983), anti–UNC-95 (rabbit polyclonal Benian-13; Qadota et al., 2007), anti-twitchin (rabbit polyclonal I I II; Benian et al., 1996), and anti-HA (mouse monoclonal; H3663; Sigma-Aldrich). The following antibodies were used at 1:100 dilution: anti-UNC-98 (rabbit polyclonal EU131; Mercer et al., 2003), and anti–CSN-5 (rabbit polyclonal; Miller et al., 2009). Secondary antibodies, also used at 1:200 dilution, included anti-rabbit Alexa 488 (Invitrogen, Thermo Fisher Scientific) and anti-mouse Alexa 594 (Invitrogen). Images were captured at room temperature with a Zeiss confocal system (LSM510) equipped with an Axiovert 100M microscope and an Apochromat ×63/1.4 numerical aperture oil immersion objective in ×2.5 zoom mode. The color balances of the images were adjusted by using Photoshop (Adobe, San Jose, CA).
Western blots
We used the procedure of Hannak et al. (2002) to prepare total protein lysates from nematodes. Western blots to detect the large isoforms of UNC-89 were carried out on wild type and the unc-89 alleles su75, r452, e1460, and tm752. Briefly, equal amounts of total protein from these strains were separated by 4.5% polyacrylamide–SDS Laemmli gels, transferred to nitrocellulose membrane, and reacted with affinity-purified rabbit EU30 anti–UNC-89 antibody (generated to Ig3-6) at 1:500 dilution. As a loading control, equal amounts of total protein from these strains were separated by 10% SDS–PAGE, transferred to nitrocellulose membrane, and reacted with the mouse monoclonal MH4 antibody at 1:100, which detects an intermediate filament protein expressed in the hypodermis (Francis and Waterston, 1985). To detect expression of HA-SH3-1/3IK-Ig-Fn-PK2, Hannak extracts were separated by 10% SDS–PAGE, transferred to membrane, and reacted with mouse monoclonal to HA (H3663; Sigma-Aldrich) at 1:200 dilution.
Determination of mutation sites in unc-89(su75) and unc-89(r452)
Genomic DNA from each strain was prepared and provided to the Emory Integrated Genomics Core (EIGC) for Illumina sequencing. The Genomic Services Laboratory at the HudsonAlpha Institute for Biotechnology (Huntsville, AL) performed quality control and constructed standard Illumina sequencing libraries following the manufacturer’s instructions. Sequencing was performed on an Illumina Hiseq version 3 with 100–base pair paired-end reads. Raw data were returned to the EIGC for bioinformatics analysis. Raw reads were mapped against the C. elegans reference genome (WS190/ce6) with the PEMapper software package. PEMapper maps short reads to a reference genome by first decomposing those reads into k-mers (16-mers in this case) and then performing a hash-based “rough mapping” of each read. After roughly mapping the read, the fine-scale position of the read was determined by a local Smith–Waterman alignment. Genotypes were determined using PECaller. Intuitively, we envision the six channels of data (number of A, C, G, T, deletion, and insertion reads) as being multinomially sampled, with some probability of drawing a read from each of the channels, but the probability varies from experiment to experiment and is drawn from a Dirichlet distribution. PECaller combines data across samples in order to identify the genotype with the highest likelihood at each sequenced base. Both PEMapper and PECaller are part of a custom software package developed at Emory University for mapping and identifying variant sites from Illumina raw sequencing data (D. J. Cutler and M. E. Zwick, personal communication). Our analysis then focused on the ∼60-kb unc-89 gene. PECaller identified a single homozygous mutation in the coding sequencing in each sequenced mutant strain.
Construction and expression of UNC-89 fragments in transgenic nematodes
HA-tagged UNC-89 fragments were expressed under the control of two different types of heat shock promoters (pPD49.78 and pPD49.83). UNC-89 1/3IK-Ig-Fn-PK2, UNC-89 SH3-1/3IK-Ig-Fn-PK2, and UNC-89 SH3 were amplified by PCR using GK22-26 (Supplemental Table S1) and cloned into the HA-tagged vector, pKS-HA(Nhex2), and their DNA sequences were confirmed. UNC-89 SH3 was also cloned into pEGFP-C1 vector. NheI-digested fragments of HA-UNC-89 Fn-Ig-PK1 (Qadota et al., 2008b), HA-UNC-89 Ig-Fn-PK2 (Qadota et al., 2008b), HA-UNC-89 1/3IK-Ig-Fn-PK2, HA-UNC-89 SH3-1/3IK-Ig-Fn-PK2, HA-UNC-89 SH3, and EGFP-UNC-89 SH3 were cloned into NheI-digested pPD49.78 and pPD49.83, resulting in pPD49.78/83-HA-UNC-89 Fn-Ig-PK1, pPD49.78/83-HA-UNC-89 Ig-Fn-PK2, pPD49.78/83-HA-UNC-89 1/3IK-Ig-Fn-PK2, pPD49.78/83-HA-UNC-89 SH3-1/3IK-Ig-Fn-PK2, pPD49.78/83-HA-UNC-89 SH3, and pPD49.78/83-EGFP-UNC-89 SH3. pPD49.78/83-HA-UNC-89 (Fn-Ig-PK1, Ig-Fn-PK2, 1/3IK-Ig-Fn-PK2, SH3-1/3IK-Ig-Fn-PK2, SH3) were mixed with pTG96 (SUR-5::NLS::GFP) as a transformation marker (Yochem et al., 1998) and injected into wild-type N2 worms. pPD49.78/83-EGFP-UNC-89 SH3 were mixed with pRF4 (rol-6 dominant) as a transformation marker (Mello et al., 1991) and injected into wild-type N2 worms. Transgenic lines with extrachromosomal arrays containing pPD49.78/83-HA-UNC-89 Fn-Ig-PK1, HA-UNC-89 Ig-Fn-PK2 (called sfEx59), HA-UNC-89 1/3IK-Ig-Fn-PK2 (called sfEx52), HA-UNC-89 SH3-1/3IK-Ig-Fn-PK2 (called sfEx51), HA-UNC-89 SH3, and pTG96 were established by picking up GFP- positive worms using a GFP dissection microscope. Transgenic lines with extrachromosomal arrays containing pPD49.78/83-EGFP-UNC-89 SH3 and pRF4 were established by picking up Rol worms.
Integration of extrachromosomal arrays
The extrachromosomal arrays containing pPD49.78/83-HA-UNC-89 (1/3IK-Ig-Fn-PK2 and SH3-1/3IK-Ig-Fn-PK2) and pTG96 were integrated into the genome by ultraviolet irradiation (Mitani, 1995) with some modifications (P. Barrett, personal communication). The resulting integrated nematode lines are called sfIs11,for HA-UNC-89 SH3-1/3IK-Ig-Fn-PK2 and sfIs13, for HA-UNC-89 1/3IK-Ig-Fn-PK2.
Overexpression experiments
To examine the localization of HA-tagged UNC-89 fragments (Figure 8), expression of the HA- tagged UNC-89 proteins was induced by incubation of the transgenic worms at 30°C for 2 h (heat shock). Heat-shocked transgenic worms were fixed using the method described previously (Nonet et al., 1993) and stained by anti-GFP to verify the existence of extrachromosomal array (at 1:200 dilution; Life Technologies, Thermo Fisher Scientific; A11122), anti–UNC-95 (to identify dense bodies and M-lines in muscle cells), and anti-HA (to determine the localization of HA-tagged UNC-89 fragments). To investigate the effect of expression of HA-UNC-89 1/3IK-Ig-Fn-PK2 and HA-UNC-89 SH3-1/3IK-Ig-Fn-PK2 on muscle organization (Figure 9), wild-type worms and integrated worms were exposed at 30°C for 24 h. Worms were fixed using the method described previously (Nonet et al., 1993) and stained by anti-paramyosin (to examine the localization of paramyosin), anti-UNC-95 (to identify dense bodies and M-lines in muscle cells), anti-HA (to determine the localization of HA-tagged UNC-89 fragments), and anti-MHC A, anti-MHC B, and anti-twitchin (to determine any effects on these thick filament proteins). We prepared worm lysates from transgenic worms with or without heat shock and examined the expression of HA-tagged UNC-89 proteins by Western blot, reacting with anti-HA (1:200 dilution; H3663; Sigma-Aldrich).
Sequence analysis and homology modeling of SH3 domains and paramyosin
Various 3D models of the SH3 domains of UNC-89 from C. elegans and D. melanogaster were calculated using the Phyre2 (Kelley et al., 2015) and ModWeb v.r166 (https://modbase.compbio.ucsf.edu/modweb) servers, as well as a local installation of MODELLER v9.15 (https://salilab.org/modeller/). The latter used as template various combinations of SH3 domain structures (listed in Figure 11A), with sequence similarity being identified and evaluated using BLAST (http://blast.ncbi.nlm.nih.gov/) against the Protein Data Bank (www.pdb.org). Visual inspection of 3D models was in PyMOL v1.7.4 (www.pymol.org), which revealed good consensus in the modeling of the β-barrel but typical variability in the loop regions. We calculated 3D models of UNC-15(294–376) using Phyre2 with PDB ID 4XA3 as template, as well as by manual mutagenesis in silico in PyMOL.
General sequence searches and alignments of SH3 domains of UNC-89/obscurin from vertebrates, insects, and nematodes were carried out independently per each taxon using BLAST. Other sequence alignments and comparisons used T-coffee (www.tcoffee.org/).
Supplementary Material
Acknowledgments
We thank Henry Epstein (deceased), Shohei Mitani, Robert Waterston, and Phil Anderson for various nematode strains. We also thank Robert Barstead for the C. elegans yeast two-hybrid library RB2, Andrew Fire for pPD49.78 and pPD49.83, and Victor Faundez for plasmid pGBKT7. Some of the nematode strains used in this work were provided by the Caenorhabditis Genetics Center (funded by the National Institutes of Health, Center for Research Resources). This study was supported in part by National Institutes of Health Grant R01AR064307 and a pilot grant from Merial Limited to G.M.B. This study was also supported in part by the Emory Integrated Genomics Core, which is subsidized by the Emory University School of Medicine and is one of the Emory Integrated Core Facilities.
Abbreviations used:
- GFP
green fluorescent protein
- GST
glutathione S-transferase
- MBP
maltose-binding protein.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-09-0675) on March 23, 2016.
REFERENCES
- Abdiche Y, Malashock D, Pinkerton A, Pons J. Determining kinetics and affinities of protein interactions using a parallel real-time label-free biosensor, the Octet. Anal Biochem. 2008;377:209–217. doi: 10.1016/j.ab.2008.03.035. [DOI] [PubMed] [Google Scholar]
- Bagnato P, Barone V, Giacomello E, Rossi D, Sorrentino V. Binding of an ankyrin-1 isoform to obscuring suggests a molecular link between the sarcoplasmic reticulum and myofibrils in striated muscles. J Cell Biol. 2003;160:245–253. doi: 10.1083/jcb.200208109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benian GM, Ayme-Southgate A, Tinley TL. The genetics and molecular biology of the titin/connectin-like proteins of invertebrates. Rev Physiol Biochem Pharmacol. 1999;138:235–268. doi: 10.1007/BFb0119629. [DOI] [PubMed] [Google Scholar]
- Benian GM, L’Hernault SW, Morris ME. Additional sequence complexity in the muscle gene, unc-22, and its encoded protein, twitchin, of Caenorhabditis elegans. Genetics. 1993;134:1097–1104. doi: 10.1093/genetics/134.4.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benian GM, Mayans O. Titin and obscurin: giants holding hands and discovery of a new Ig domain subset. J Mol Biol. 2015;427:707–714. doi: 10.1016/j.jmb.2014.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benian GM, Tinley TL, Tang X, Borodovsky M. The Caenorhabditis elegans gene unc-89, required for muscle M-line assembly, encodes a giant modular protein composed of Ig and signal transduction domains. J Cell Biol. 1996;132:835–848. doi: 10.1083/jcb.132.5.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomberg N, Baraldi E, Sattler M, Saraste M, Nilges M. Structure of a PH domain from the C. elegans muscle protein UNC-89 suggests a novel function. Stucture. 2000;8:1079–1087. doi: 10.1016/s0969-2126(00)00509-8. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen C, Szent-Gyorgyi AG, Kendrick-Jones J. Paramyosin and the filaments of molluscan “catch” muscles. I. Paramyosin: structure and assembly. J Mol Biol. 1971;56:223–237. doi: 10.1016/0022-2836(71)90461-x. [DOI] [PubMed] [Google Scholar]
- Concepcion J, Witte K, Wartchow C, Choo S, Yao D, Persson H, Wei J, Li P, Heidecker B, Ma W, et al. Label-free detection of biomolecular interactions using biolayer interferometry for kinetic characterization. Comb Chem High Throughput Screen. 2009;12:791–800. doi: 10.2174/138620709789104915. [DOI] [PubMed] [Google Scholar]
- Deitiker PR, Epstein HF. Thick filament substructures in C. elegans: evidence for two populations of paramyosin. J Cell Biol. 1993;123:303–311. doi: 10.1083/jcb.123.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douangamath A, Filipp FV, Klein AT, Barnett P, Zou P, Voorn-Brouwer T, Vega MC, Mayans OM, Sattler M, Distel B, et al. Topography for independent binding of alpha-helical and PPII-helical ligands to a peroxisomal SH3 domain. Mol Cell. 2002;10:1007–1017. doi: 10.1016/s1097-2765(02)00749-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein HF, Casey DL, Ortiz I. Myosin and paramyosin of Caenorhabditis elegans embryos assemble into nascent structures distinct from thick filaments and multi-filament assemblages. J Cell Biol. 1993;122:845–858. doi: 10.1083/jcb.122.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein HF, Lu GY, Deitiker PR, Oritz I, Schmid MF. Preliminary three-dimensional model for nematode thick filament core. J Struct Biol. 1995;115:163–174. doi: 10.1006/jsbi.1995.1041. [DOI] [PubMed] [Google Scholar]
- Epstein HF, Miller DM, Ortiz I, Berliner GC. Myosin and paramyosin are organized about a newly identified core structure. J Cell Biol. 1985;100:904–915. doi: 10.1083/jcb.100.3.904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein HF, Ortiz I, Berliner GC. Assemblages of multiple thick filaments in nematode mutants. J Muscle Res Cell Motil. 1987;8:527–536. doi: 10.1007/BF01567911. [DOI] [PubMed] [Google Scholar]
- Ferrara TM, Flaherty DB, Benian GM. Titin/connectin-related proteins in C. elegans: a review and new findings. J Muscle Res Cell Motil. 2005;26:435–447. doi: 10.1007/s10974-005-9027-4. [DOI] [PubMed] [Google Scholar]
- Flashman E, Watkins H, Redwood C. Localization of the binding site of the C-terminal domain of cardiac myosin-binding protein-C on the myosin rod. Biochem J. 2007;401:97–102. doi: 10.1042/BJ20060500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford-Speelman DL, Roche JA, Bowman AL, Bloch RJ. The rho-guanine nucleotide exchange domain of obscurin activates rhoA signaling in skeletal muscle. Mol Biol Cell. 2009;20:3905–3917. doi: 10.1091/mbc.E08-10-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis GR, Waterston RH. Muscle organization in Caenorhabditis elegans: localization of proteins implicated in thin filament attachment and I-band organization. J Cell Biol. 1985;101:1532–1549. doi: 10.1083/jcb.101.4.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuzawa A, Idowu S, Gautel M. Complete human gene structure of obscurin: implications for isoform generation by differential splicing. J Muscle Res Cell Motil. 2005;26:427–434. doi: 10.1007/s10974-005-9025-6. [DOI] [PubMed] [Google Scholar]
- Fukuzawa A, Lange S, Holt MR, Vihola A, Carmignac V, Ferreiro A, Udd AB, Gautel M. Interactions with titin and myomesin target obscurin and its small homologue, obscurin-like 1, to the sarcomeric M-band: implications for hereditary myopathies. J Cell Sci. 2008;121:1841–1851. doi: 10.1242/jcs.028019. [DOI] [PubMed] [Google Scholar]
- Gengyo-Ando K, Kagawa H. Single charge change on the helical surface of the paramyosin rod dramatically disrupts thick filament assembly in Caenorhabditis elegans. J Mol Biol. 1991;219:429–441. doi: 10.1016/0022-2836(91)90184-8. [DOI] [PubMed] [Google Scholar]
- Groemping Y, Lapouge K, Smerdon SJ, Rittinger K. Molecular basis of phosphorylation-induced activation of the NADPH oxidase. Cell. 2003;113:343–355. doi: 10.1016/s0092-8674(03)00314-3. [DOI] [PubMed] [Google Scholar]
- Hannak E, Oegema K, Kirkham M, Gonczy P, Habermann B, Hyman AA. The kinetically dominant assembly pathway for centrosomal asters in Caenorhabditis elegans is gamma-tubulin dependent. J Cell Biol. 2002;157:591–602. doi: 10.1083/jcb.200202047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hresko MC, Williams BD, Waterston RH. Assembly of body wall muscle and muscle cell attachment structures in Caenorhabditis elegans. J Cell Biol. 1994;124:491–506. doi: 10.1083/jcb.124.4.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu LY, Kontrogianni-Konstantopoulos A. The kinase domains of obscurin interact with intercellular adhesion proteins. FASEB J. 2013;27:2001–2012. doi: 10.1096/fj.12-221317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagawa H, Gengyo K, McLachlan AD, Brenner S, Karn J. Paramyosin gene (unc-15) of Caenorhabditis elegans. Molecular cloning, nucleotide sequence and models for thick filament structure. J Mol Biol. 1989;207:311–333. doi: 10.1016/0022-2836(89)90257-x. [DOI] [PubMed] [Google Scholar]
- Katzemich A, West RJ, Fukuzawa A, Sweeney ST, Gautel M, Sparrow J, Bullard B. Binding partners of the kinase domains in Drosophila obscurin and their effect on the structure of the flight muscle. J Cell Sci. 2015;128:3386–3397. doi: 10.1242/jcs.170639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knappeis GG, Carlsen F. The ultrastructure of the M line in skeletal muscle. J Cell Biol. 1968;38:202–211. doi: 10.1083/jcb.38.1.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontrogianni-Konstantopoulos A, Ackermann MA, Bowman AL, Yap SV, Bloch RJ. Muscle giants: molecular scaffolds in sarcomerogenesis. Physiol Rev. 2009;89:1217–1267. doi: 10.1152/physrev.00017.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontrogianni-Konstantopoulos A, Jones EM, Van Rossum DB, Bloch RJ. Obscurin is a ligand for small ankyrin 1 in skeletal muscle. Mol Biol Cell. 2003;14:1138–1148. doi: 10.1091/mbc.E02-07-0411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange S, Ouyang K, Meyer G, Cui L, Cheng H, Lieber RL, Chen J. Obscurin determines the architecture of the longitudinal sarcoplasmic reticulum. J Cell Sci. 2009;122:2640–2650. doi: 10.1242/jcs.046193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange S, Perera S, Teh P, Chen J. Obscurin and KCTD6 regulate cullin-depenent small ankyrin-1 (sAnk1.5) protein turnover. Mol Biol Cell. 2012;23:2490–2504. doi: 10.1091/mbc.E12-01-0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecroisey C, Brouilly N, Qadota H, Mariol MC, Rochette NC, Martin E, Benian GM, Ségalat L, Mounier N, Gieseler K. ZYX-1, the unique zyxin protein of Caenorhabditis elegans, is involved in dystrophin-dependent muscle degeneration. Mol Biol Cell. 2013;24:1232–1249. doi: 10.1091/mbc.E12-09-0679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine RJC, Elfvin M, Dewey MM, Walcott B. Paramyosin in invertebrate muscles II. Content in relation to structure and function. J Cell Biol. 1976;71:273–279. doi: 10.1083/jcb.71.1.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linke WA, Hamdani N. Gigantic business: titin properties and function through thick and thin. Circ Res. 2014;114:1052–1068. doi: 10.1161/CIRCRESAHA.114.301286. [DOI] [PubMed] [Google Scholar]
- Liu F, Bauer CC, Ortiz I, Cook RG, Schmid MF, Epstein HF. β-filagenin, a newly identified protein coassembling with myosin and paramyosin in Caenorhabditis elegans. J Cell Biol. 1998;140:347–353. doi: 10.1083/jcb.140.2.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Ortiz I, Hutagalung A, Bauer CC, Cook RG, Epstein HF. Differential assembly of alpha- and gamma-filagenins into thick filaments in Caenorhabditis elegans. J Cell Sci. 2000;113:4001–4012. doi: 10.1242/jcs.113.22.4001. [DOI] [PubMed] [Google Scholar]
- Luther P, Squire J. Three-dimensional structure of the vertebrate muscle M-region. J Mol Biol. 1978;125:313–324. doi: 10.1016/0022-2836(78)90405-9. [DOI] [PubMed] [Google Scholar]
- Mackenzie JM, Garcea RL, Zengel JM, Epstein HF. Muscle development in Caenorhabditis elegans: mutants exhibiting retarded sarcomere construction. Cell. 1978;15:751–762. doi: 10.1016/0092-8674(78)90261-1. [DOI] [PubMed] [Google Scholar]
- McLachlan AD, Karn J. Periodic charge distributions in the myosin rod amino acid sequence match cross-bridge spacings in muscle. Nature. 1982;299:226–231. doi: 10.1038/299226a0. [DOI] [PubMed] [Google Scholar]
- Meissner B, Warner A, Wong K, Dube N, Lorch A, McKay SJ, Khattra J, Rogalski T, Somasiri A, Chaudhry I, et al. An integrated strategy to study muscle development and myofilament structure in Caenorhabditis elegans. PLoS Genet. 2009;5:e1000537. doi: 10.1371/journal.pgen.1000537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer KB, Flaherty DB, Miller RK, Qadota H, Tinley TL, Moerman DG, Benian GM. Caenorhabditis elegans UNC-98, a C2H2 Zn finger protein, is a novel partner of UNC-97/PINCH in muscle adhesion complexes. Mol Biol Cell. 2003;14:2492–2507. doi: 10.1091/mbc.E02-10-0676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer KB, Miller RK, Tinley TL, Sheth S, Qadota H, Benian GM. Caenorhabditis elegans UNC-96 is a new component of M-lines that interacts with UNC-98 and paramyosin and is required in adult muscle for assembly and/or maintenance of thick filaments. Mol Biol Cell. 2006;17:3832–3847. doi: 10.1091/mbc.E06-02-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DM, Ortiz I, Berliner GC, Epstein HF. Differential localization of two myosins within nematode thick filaments. Cell. 1983;34:477–490. doi: 10.1016/0092-8674(83)90381-1. [DOI] [PubMed] [Google Scholar]
- Miller RK, Qadota H, Landsverk ML, Mercer KB, Epstein HF, Benian GM. UNC-98 links an integrin-associated complex to thick filaments in Caenorhabditis elegans muscle. J Cell Biol. 2006;175:853–859. doi: 10.1083/jcb.200608043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RK, Qadota H, Mercer KB, Gernert KM, Benian GM. UNC-98 and UNC-96 interact with paramyosin to promote its incorporation into thick filaments of C. elegans. Mol Biol Cell. 2008;19:1529–1539. doi: 10.1091/mbc.E07-07-0723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RK, Qadota H, Stark TJ, Mercer KB, Wortham TS, Anyanful A, Benian GM. CSN-5, a component of the COP9 signalosome complex, regulates the levels of UNC-96 and UNC-98, two components of M-lines in Caenorhabditis elegans muscle. Mol Biol Cell. 2009;20:3608–3616. doi: 10.1091/mbc.E09-03-0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitani S. Genetic regulation of mec-3 gene expression implicated in the specification of the mechanosensory neuron cell types in Caenorhabditis elegans. Dev Growth Differ. 1995;37:551–557. doi: 10.1046/j.1440-169X.1995.t01-4-00010.x. [DOI] [PubMed] [Google Scholar]
- Muller SA, Haner M, Ortiz I, Aebi U, Epstein HF. STEM analysis of Caenorhabditis elegans muscle thick filaments: evidence for microdifferentiated substructures. J Mol Biol. 2001;305:1035–1044. doi: 10.1006/jmbi.2000.4363. [DOI] [PubMed] [Google Scholar]
- Nahabedian JF, Qadota H, Stirman JN, Lu H, Benian GM. Bending amplitude—a new quantitative assay of C. elegans locomotion: identification of phenotypes for mutants in genes encoding muscle focal adhesion components. Methods. 2012;56:95–102. doi: 10.1016/j.ymeth.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonet ML, Grundahl K, Meyer BJ, Rand JB. Synaptic function is impaired but not eliminated in C. elegans mutants lacking synaptotagmin. Cell. 1993;73:1291–1305. doi: 10.1016/0092-8674(93)90357-v. [DOI] [PubMed] [Google Scholar]
- Qadota H, Benian GM. Molecular structure of sarcomere-to-membrane attachment at M-lines in C. elegans muscle. J Biomed Biotechnol. 2010;2010:864749. doi: 10.1155/2010/864749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qadota H, Blangy A, Xiong G, Benian GM. The DH-PH region of the giant protein UNC-89 activates RHO-1 GTPase in Caenorhabditis elegans body wall muscle. J Mol Biol. 2008a;383:747–752. doi: 10.1016/j.jmb.2008.08.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qadota H, McGaha LA, Mercer KB, Stark TJ, Ferrara TM, Benian GM. A novel protein phosphatase is a binding partner for the protein kinase domains of UNC-89 (Obscurin) in Caenorhabditis elegans. Mol Biol Cell. 2008b;19:2424–2432. doi: 10.1091/mbc.E08-01-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qadota H, Mercer KB, Miller RK, Kaibuchi K, Benian GM. Two LIM domain proteins and UNC-96 link UNC-97/pinch to myosin thick filaments in Caenorhabditis elegans muscle. Mol Biol Cell. 2007;18:4317–4326. doi: 10.1091/mbc.E07-03-0278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saksela K, Permi P. SH3 domain ligand binding: What’s the consensus and where’s the specificity. FEBS Lett. 2012;586:2609–2614. doi: 10.1016/j.febslet.2012.04.042. [DOI] [PubMed] [Google Scholar]
- Small TM, Gernert KM, Flaherty DB, Mercer KB, Borodovsky M, Benian GM. Three new isoforms of Caenorhabditis elegans UNC-89 containing MLCK-like protein kinase domains. J Mol Biol. 2004;342:91–108. doi: 10.1016/j.jmb.2004.07.006. [DOI] [PubMed] [Google Scholar]
- Spooner PM, Bonner J, Maricq AV, Benian GM, Norman KR. Large isoforms of UNC-89 (obscurin) are required for muscle cell architecture and optimal calcium release in Caenorhabditis elegans. PLoS One. 2012;(7):e40182. doi: 10.1371/journal.pone.0040182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor KC, Buvoli M, Korkmaz EN, Buvoli A, Zheng Y, Heinze NT, Cui Q, Leinwand LA, Rayment I. Skip residues modulate the structural properties of the myosin rod and guide thick filament assembly. Proc Natl Acad Sci USA. 2015;112:E3806–E3815. doi: 10.1073/pnas.1505813112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner A, Xiong G, Qadota H, Rogalski T, Vogl AW, Moerman DG, Benian GM. CPNA-1, a copine domain protein, is located at integrin adhesion sites, and is required for myofilament stability in C. elegans. Mol Biol Cell. 2013;24:601–616. doi: 10.1091/mbc.E12-06-0478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston RH, Fishpool RM, Brenner S. Mutants affecting paramyosin in Caenorhabditis elegans. J Mol Biol. 1977;117:679–697. doi: 10.1016/0022-2836(77)90064-x. [DOI] [PubMed] [Google Scholar]
- Waterston RH, Thomson JN, Brenner S. Mutants with altered muscle structure in C. elegans. Dev Biol. 1980;77:271–302. doi: 10.1016/0012-1606(80)90475-3. [DOI] [PubMed] [Google Scholar]
- Wilson KJ, Qadota H, Benian GM. Immunofluorescent localization of proteins in Caenorhabditis elegans muscle. Methods Mol Biol. 2012a;798:171–181. doi: 10.1007/978-1-61779-343-1_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson KJ, Qadota H, Mains PE, Benian GM. UNC-89 (obscurin) binds to MEL-26, a BTB-domain protein, and affects the function of MEI-1 (katanin) in striated muscle of Caenorhabditis elegans. Mol Biol Cell. 2012b;23:2623–2634. doi: 10.1091/mbc.E12-01-0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong G, Qadota H, Mercer KB, McGaha LA, Oberhauser AF, Benian GM. A LIM-9 (FHL)/SCPL-1 (SCP) complex interacts with the C-terminal protein kinase regions of UNC-89 (obscurin) in Caenorhabditis elegans muscle. J Mol Biol. 2009;386:976–988. doi: 10.1016/j.jmb.2009.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yochem J, Gu T, Han M. A new marker for mosaic analysis in Caenorhabditis elegans indicates a fusion between hyp6 and hyp7, two major components of the hypodermis. Genetics. 1998;149:1323–1334. doi: 10.1093/genetics/149.3.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.