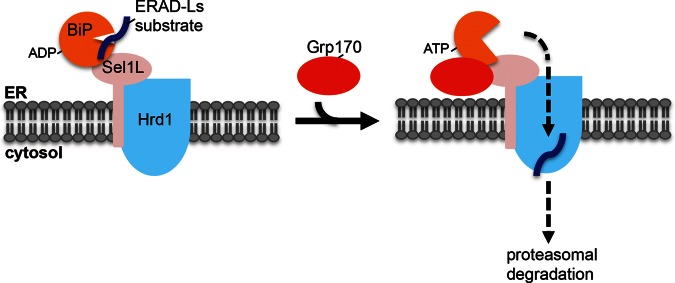
The Grp170 nucleotide exchange factor plays a critical role in the ERAD pathway. The positioning of this factor at the retrotranslocation site may afford a mechanism to couple client release from BiP and retrotranslocation.
Abstract
When a protein misfolds in the endoplasmic reticulum (ER), it retrotranslocates to the cytosol and is degraded by the proteasome via a pathway called ER-associated degradation (ERAD). To initiate ERAD, ADP-BiP is often recruited to the misfolded client, rendering it soluble and translocation competent. How the misfolded client is subsequently released from BiP so that it undergoes retrotranslocation, however, remains enigmatic. Here we demonstrate that the ER-resident nucleotide exchange factor (NEF) Grp170 plays an important role during ERAD of the misfolded glycosylated client null Hong Kong (NHK). As a NEF, Grp170 triggers nucleotide exchange of BiP to generate ATP-BiP. ATP-BiP disengages from NHK, enabling it to retrotranslocate to the cytosol. We demonstrate that Grp170 binds to Sel1L, an adapter of the transmembrane Hrd1 E3 ubiquitin ligase postulated to be the retrotranslocon, and links this interaction to Grp170’s function during ERAD. More broadly, Grp170 also promotes degradation of the nonglycosylated transthyretin (TTR) D18G misfolded client. Our findings thus establish a general function of Grp170 during ERAD and suggest that positioning this client-release factor at the retrotranslocation site may afford a mechanism to couple client release from BiP and retrotranslocation.
INTRODUCTION
The endoplasmic reticulum (ER) is endowed with robust folding machineries, which ensure that nascent polypeptide chains fold and mature properly before exiting this compartment. However, when a client misfolds, it is cleared from the ER via a protein quality control system called ER-associated degradation (ERAD; Smith et al., 2011; Brodsky, 2012; Olzmann et al., 2013; Christianson and Ye, 2014; Ruggiano et al., 2014). During initiation of ERAD, molecular chaperones recognize and bind to exposed hydrophobic regions of a misfolded client, preventing aggregation until the client is targeted to the ERAD membrane machinery (Hirsch et al., 2009; Claessen et al., 2012); this machinery’s central component is the Hrd1 E3 ubiquitin ligase, which is postulated to serve as the retrotranslocon (Carvalho et al., 2010; Stein et al., 2014). A misfolded client is typically captured by adapters of Hrd1, such as the transmembrane Sel1L protein and the luminal OS-9/XTP3-B lectins (Carvalho et al., 2006; Denic et al., 2006; Christianson et al., 2008; Hosokawa et al., 2008, 2009; Bernasconi et al., 2010), before being transferred to Hrd1. Once the client crosses the putative Hrd1 retrotranslocon and emerges into the cytosol, it is ubiquitinated by the E3 ligase, discharged into the cytosol, and finally degraded by the proteasome.
Within the ER lumen, the major Hsp70 chaperone BiP acts to prevent aggregation of a misfolded client by binding to its exposed hydrophobic patches until the client is delivered to the ERAD membrane machinery (Brodsky et al., 1999; Nishikawa et al., 2001; Kabani et al., 2003). BiP’s chaperone function is tightly regulated by its ATP-ADP binding states (Kampinga and Craig, 2010): ATP-BiP displays a low substrate-binding affinity, whereas ADP-BiP exhibits a high substrate-binding affinity. Cycling between these two opposing states is regulated by ER-resident J-proteins and nucleotide exchange factors (NEFs). Specifically, a J-protein stimulates BiP’s ATPase activity, converting ATP-BiP to ADP-BiP, which binds strongly to the substrate (Jin et al., 2008; Petrova et al., 2008). By contrast, a NEF induces an exchange of ADP for ATP to generate ATP-BiP. ATP-BiP in turn undergoes a conformational change that releases the substrate. Whereas several J-proteins have been implicated in ERAD (Nishikawa et al., 2001; Dong et al., 2008; Petrova et al., 2008; Hagiwara et al., 2011; Massey et al., 2011; Williams et al., 2013; Athanasiou et al., 2014), the role of NEFs in this quality control process is less clear.
Grp170 and Sil1 are two established ER-resident NEFs (Brodsky and Bracher, 2000; Kampinga and Craig, 2010; Behnke et al., 2015). Although Grp170 is also a member of the Hsp70 protein family, it represents an evolutionarily distinct form of canonical Hsp70 protein. One important distinction is that in addition to its recognized nucleotide exchange activity, Grp170 can also function as an ATP-independent chaperone/holdase—it binds to unfolded proteins (Behnke and Hendershot, 2014) or prevents protein aggregation (Easton et al., 2000; Park et al., 2003). In fact, the holdase but not the nucleotide exchange activity of the yeast Grp170 homologue LHS1, and Grp170, have been implicated in the ERAD of a membrane protein (Buck et al., 2013). However, in yeast, deletion of SIL1 (Travers et al., 2000) but not LHS1 (Buck et al., 2013) only moderately impaired the ERAD of the soluble misfolded substrate CPY*, suggesting that Sil1 may have a potential role in ERAD in yeast. However, whether this is due to Sil1’s nucleotide exchange activity or other undocumented functions remains unknown. Hence the precise source of nucleotide exchange activity required during ERAD has yet to be clearly established.
In this context, our laboratory recently reported that the DNA tumor virus SV40 hijacks Grp170’s nucleotide exchange activity to promote its ER-to-cytosol membrane transport, a critical infection step (Inoue and Tsai, 2015). Moreover, we also found that the nucleotide exchange activities of both Grp170 and Sil1 are used to drive ER membrane transport of the bacterially secreted cholera toxin (Williams et al., 2015). Because SV40 and cholera toxin are pathogens believed to disguise as misfolded proteins that co-opt elements of the ERAD pathway during entry, these findings raise the distinct possibility that Grp170 and Sil1 may play critical roles during ERAD of cellular misfolded clients.
In this study, we report that Grp170 but not Sil1 acts as a NEF to promote ERAD of the soluble cellular ERAD substrates glycosylated NHK and nonglycosylated TTR D18G. We further demonstrate that Grp170 binds to the Hrd1 adapter Sel1L and suggest that this interaction is critical for Grp170’s action during ERAD.
RESULTS
Grp170 is essential in degradation of the glycosylated NHK ERAD-Ls substrate
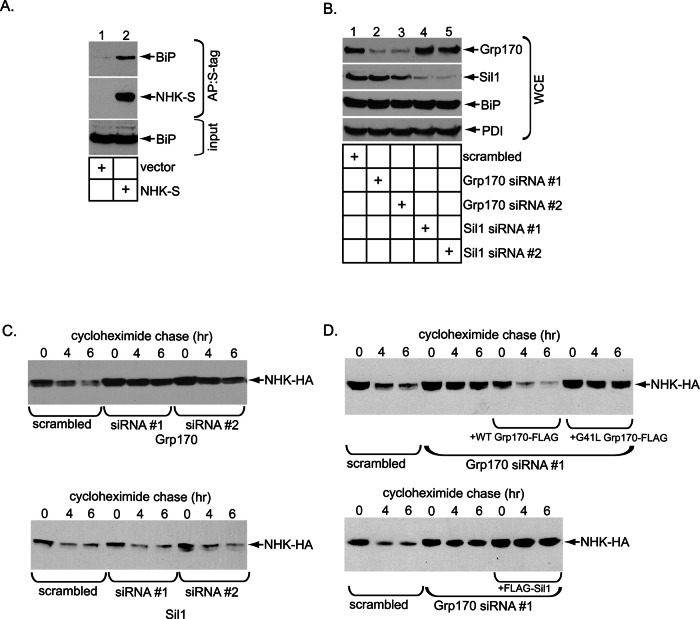
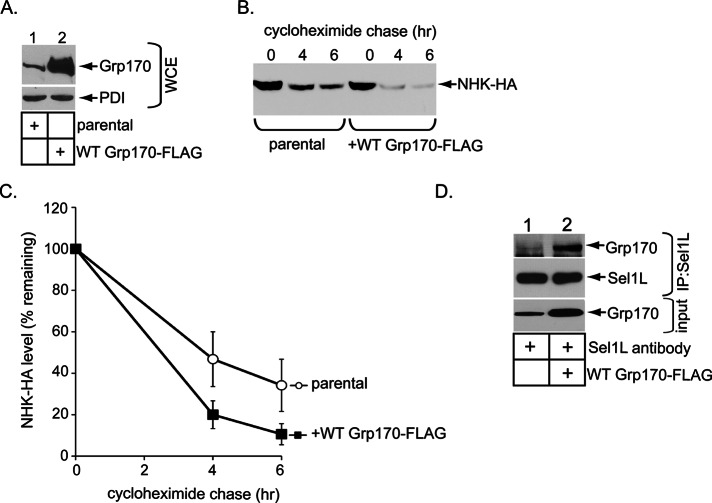
To ascertain whether Grp170 regulates retrotranslocation of cellular BiP-dependent ERAD clients, we chose a mutant form of α-1 antitrypsin called NHK as a model substrate. NHK is a soluble glycosylated ERAD client localized to the ER lumen and is therefore referred to as an ERAD-Ls substrate (Bernasconi et al., 2010). BiP was previously shown to interact with NHK (Hagiwara et al., 2011). To confirm this observation in our system, we lysed 293T cells transfected with either an empty vector or C-terminally S-tagged NHK (NHK-S) and subjected the resulting whole-cell extract (WCE) to affinity purification with S-protein–conjugated beads to precipitate NHK-S. Immunoblotting of the isolated proteins showed that NHK specifically coprecipitated BiP (Figure 1A, top, compare lane 2 to lane 1), demonstrating that BiP engages NHK.
FIGURE 1:
Grp170 is essential in degradation of the glycosylated NHK ERAD-Ls substrate. (A) WCEs derived from 293T cells transfected with the indicated plasmid were incubated with S-protein–conjugated beads and the isolated proteins subjected to SDS–PAGE, followed by immunoblotting with anti–S-tag and BiP antibodies. Inputs were also analyzed by immunoblotting with the same antibodies. AP, affinity purification. (B) Cells expressing NHK-HA were transfected with the indicated siRNA and the resulting WCE analyzed by SDS–PAGE and immunoblotting with the indicated antibodies. (C) Top, cells expressing NHK-HA and transfected with scrambled or Grp170 siRNA #1 or #2 in B were treated with cycloheximide for the indicated time and harvested, and the resulting WCE was analyzed with an anti HA antibody. Bottom, as for the top, except that cells were transfected with scrambled or Sil1 siRNA #1 or #2. (D) Parental Flp-In T-REx 293 cells were transfected with either scrambled or Grp170 siRNA #1. Where indicated, cells transfected with Grp170 siRNA #1 also stably expressed either siRNA-resistant Grp170-FLAG or G41L Grp170-FLAG. After siRNA transfection, cells were transfected with NHK-HA, treated with cyclohexamide for the indicated time, and harvested, and the resulting WCE was analyzed as in C. Bottom, as for the top, except that cells stably expressing FLAG-Sil1 were used.
Because NHK interacts with BiP, we asked whether the two established NEFs in the ER, Grp170 and Sil1, might regulate retrotranslocation of NHK that leads to its proteasomal degradation. To this end, we knocked down Grp170 or Sil1 using two different small interfering RNAs (siRNAs) in cells expressing hemagglutinin (HA)-tagged NHK (NHK-HA) and examined NHK’s degradation rate using the standard cycloheximide chase approach. Effective Grp170 and Sil1 knockdowns were achieved in these cells using their respective siRNAs (Figure 1B, top, compare lanes 2 and 3 to lane 1, and second from top, compare lanes 4 and 5 to lane 1), without inducing significant BiP or PDI up-regulation (Figure 1B, bottom and next to bottom ). The lack of BiP and PDI up-regulation suggests that depleting Grp170 or Sil1 did not significantly induce ER stress, consistent with our recent reports demonstrating that silencing Grp170 or Sil1 did not trigger XBP1 splicing (Inoue and Tsai, 2015; Williams et al., 2015).
Accordingly, we probed the fate of NHK under these knockdown conditions. When compared with scrambled siRNA, Grp170 knockdown (using either siRNA) markedly blocked NHK degradation (Figure 1C, top; quantified in Supplemental Figure S1A, left), whereas Sil1 knockdown by either siRNA had no effect (Figure 1C, bottom; quantified in Supplemental Figure S1A, right). If Grp170’s nucleotide exchange activity but not its holdase activity is responsible for promoting NHK degradation, expression of the siRNA-resistant wild-type (WT) Grp170-FLAG but not the nucleotide exchange–defective G41L Grp170-FLAG (Inoue and Tsai, 2015) under the knockdown condition should restore NHK degradation. To test this, we constructed Flp-In T-REx 293 cells stably expressing the siRNA-resistant WT or G41L Grp170-FLAG under a tetracycline-inducible promoter and subjected them to the cyclohexamide chase experiment. When endogenous Grp170 was down-regulated by Grp170 siRNA #1 and the siRNA-resistant WT Grp170-FLAG was expressed at near-endogenous level (Supplemental Figure S1B, top, compare lane 3 to lane 1), the degradation rate of NHK was recovered to the same extent as the control (Figure 1D, top; quantified in Supplemental Figure S1C). By contrast, expressing the siRNA-resistant G41L Grp170-FLAG (Supplemental Figure S1B, top, compare lane 4 to lanes 3 and 1) had no effect on the degradation rate, with NHK-HA remaining stabilized (Figure 1D, top; quantified in Supplemental Figure S1C). These results are consistent with Grp170’s role in SV40 infection, in which expression of WT but not G41L Grp170 restored virus infection in Grp170-depleted cells (Inoue and Tsai, 2015). Using Flp-In T-REx 293 cells stably expressing FLAG-Sil1 under a tetracycline-inducible promoter, we also examined whether Sil1 overexpression might rescue NHK stabilization caused by Grp170 knockdown. Although the expression level of exogenous FLAG-Sil1 was at least fivefold more than that of endogenous Sil1 (Supplemental Figure S1D, middle row), it did not restore the NHK degradation rate (Figure 1D, bottom; quantified in Supplemental Figure S1E). These results are in line with the observation that silencing Sil1 did not affect NHK degradation (Figure 1C, bottom). Collectively these data indicate that the nucleotide exchange activity inherent in Grp170 is specifically used during retrotranslocation of the BiP-dependent ERAD substrate NHK.
Grp170’s nucleotide exchange activity triggers NHK release from BiP
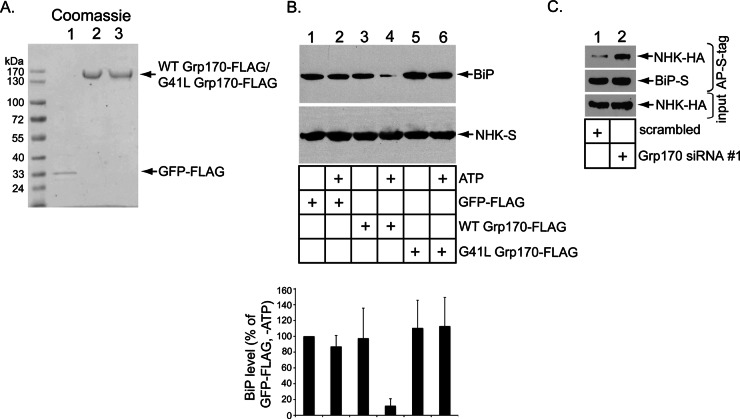
What might be a molecular explanation for why Grp170 plays a critical role during ERAD of NHK? The simplest explanation is that this NEF promotes release of NHK from BiP by catalyzing BiP nucleotide exchange. To directly test this, we asked whether adding recombinant Grp170 to the isolated BiP-NHK complex causes NHK to disengage from BiP. To this end, WT Grp170-FLAG, G41L Grp170-FLAG, and green fluorescent protein (GFP)–FLAG were expressed in and purified from 293T cells (Figure 2A). These recombinant proteins were then individually added to the isolated BiP-NHK-S complex in the presence or absence of ATP, followed by reisolation of this complex using S-protein–conjugated beads. We found that only addition of WT Grp170 in the presence of ATP caused release of BiP from NHK (Figure 2B, top, compare lane 4 to lanes 1–3 and 5–6; the BiP level is quantified in the graph). Thus WT but not the NEF-defective G41L Grp170 discharges NHK from BiP in an ATP-dependent manner. We conclude that Grp170 relies on its nucleotide exchange activity to release the misfolded client NHK from BiP; the strict dependence of this reaction on ATP is consistent with Grp170’s nucleotide exchange activity being ATP dependent (Steel et al., 2004; de Keyzer et al., 2009).
FIGURE 2:
Grp170’s nucleotide exchange activity triggers NHK release from BiP. (A) Coomassie staining of purified C-terminally FLAG-tagged GFP (GFP-FLAG), N-terminally FLAG-tagged WT Grp170 (WT Grp170-FLAG), and G41L Grp170 (G41L Grp170-FLAG) (B) Top, the BiP–NHK-S (10 nM) complex was incubated without ATP or with ATP (0.1 μM) for 10 min along with the indicated recombinant protein (125 nM). NHK-S was then reprecipitated and subjected to SDS–PAGE, followed by immunoblotting with anti–S-tag and BiP antibodies. Bottom, the BiP band intensity was quantified with ImageJ (National Institutes of Health, Bethesda, MD). Data represent mean ± SD of at least four independent experiments. (C) Cells expressing NHK-HA and BiP-S were transfected with scrambled or Grp170 #1 siRNA, the resulting WCEs were incubated with S-protein–conjugated beads, and the isolated proteins were subjected to SDS–PAGE, followed by immunoblotting with anti–S-tag and HA antibodies. Input WCEs were also analyzed by immunoblotting with an anti-HA antibody.
To corroborate this in vitro observation, we asked whether Grp170 regulates BiP-NHK interaction in intact cells. We reasoned that, if Grp170 catalyzes the release of NHK from BiP, depleting this NEF should trap NHK on BiP. Indeed, in Grp170-depleted cells, precipitation of S-tagged BiP (BiP-S) pulled down more NHK-HA than in control cells (Figure 2C, top, compare lane 2 to lane 1). This finding supports the idea that Grp170 is responsible for inducing NHK release from BiP, a step essential for NHK retrotranslocation leading to its proteasomal degradation.
Enzymatically active Grp170 binds to the Hrd1 adapter Sel1L
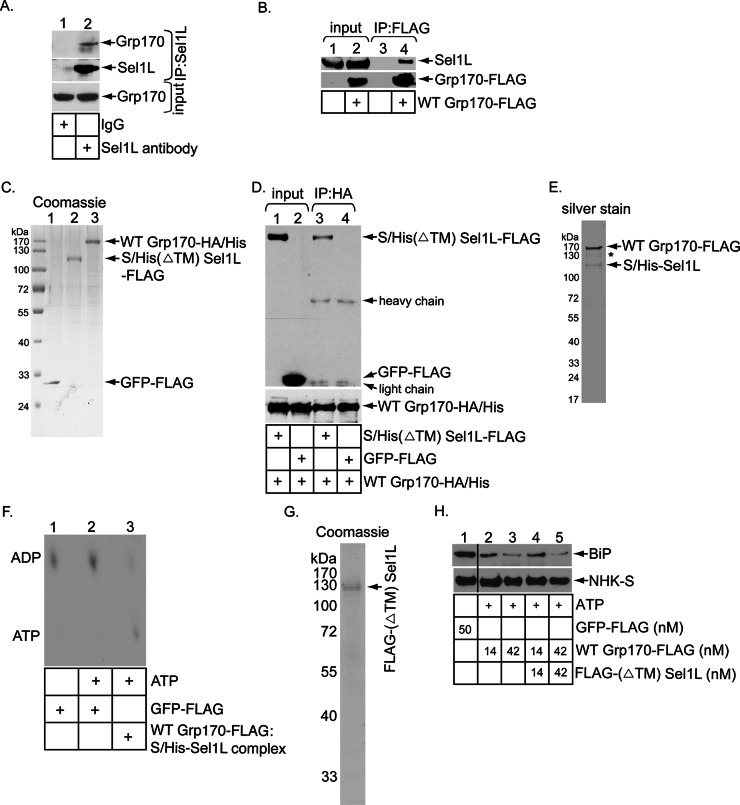
Because we previously found that an ER lumenal J-protein (i.e., ERdj5) complexes with the ERAD machinery via binding to Sel1L (Williams et al., 2013) and that it has also been reported that NHK is recruited to Sel1L in a J-protein–dependent manner (Ushioda et al., 2013), we asked whether Grp170 might be positioned to this machinery by interacting with Sel1L. Indeed, when endogenous Sel1L was immunoprecipitated, endogenous Grp170 was detected in the precipitated material (Figure 3A, top, compare lane 2 to lane 1). We then performed the reverse immunoprecipitation experiment. Because antibodies against endogenous Grp170 did not efficiently pull down Grp170, we used FLAG-tagged WT Grp170 (i.e., WT Grp170-FLAG) instead. When WT Grp170-FLAG was immunoprecipitated by FLAG antibody–conjugated agarose beads, endogenous Sel1L was precipitated (Figure 3B, top, compare lane 4 to lane 3). These results suggest that Grp170 is targeted to the ERAD membrane machinery by associating with Sel1L.
FIGURE 3:
Enzymatically active Grp170 binds to the Hrd1 adapter Sel1L. (A) Cells were incubated with the DSP cross-linker (1 mM) for 30 min, quenched, and semipermeabilized with digitonin, and the resulting pellet fraction was treated with Triton X-100. The resulting lysate was incubated with either a control immunoglobulin G or Sel1L-specific antibody, followed by protein G–agarose beads. The precipitated samples were analyzed by SDS–PAGE and immunoblotted with the indicated antibodies. (B) WCEs derived from Flp-In T-Rex-293 parental cells and cells stably expressing WT Grp170-FLAG were subjected to precipitation using FLAG antibody–conjugated agarose beads. The isolated proteins were analyzed by SDS–PAGE and immunoblotted with the indicated antibodies. Input WCEs were also analyzed by immunoblotting with the indicated antibodies. (C) Coomassie staining of purified C-terminally FLAG-tagged GFP (GFP-FLAG), C-terminally HA- and His-tagged WT Grp170 (WT Grp170-HA/His), and N-terminally S-/His-tagged, C-terminally FLAG-tagged transmembrane-deleted Sel1L [S/His(ΔTM) Sel1L-FLAG]. (D) WT Grp170-HA/His (150 nM) was incubated with either S/His(ΔTM) Sel1L-FLAG (150 nM) or GFP-FLAG (150 nM), and the samples were immunoprecipitated using an anti-HA antibody. The immunoprecipitates were subjected to SDS–PAGE, followed by immunoblotting with an anti-FLAG antibody. The input samples were also analyzed by immunoblotting with an anti-HA antibody. (E) Silver stain of the WT Grp170-FLAG:S/His-Sel1L complex. Asterisk denotes degraded WT Grp170-FLAG. (F) FLAG-BiP was incubated with [α-32P]ATP to form the radiolabeled ADP–BiP complex. The ADP–BiP complex was then incubated with GFP-FLAG or the isolated Grp170–Sel1L complex indicated in the absence or presence of unlabeled ATP. ADP release form BiP was analyzed by TLC. (G) Coomassie staining of purified, N-terminally FLAG-tagged, transmembrane-deleted Sel1L (FLAG-(ΔTM)Sel1L). (H) As in Figure 2B, except that the Grp170-FLAG was preincubated with or without FLAG-(ΔTM)Sel1L at 37ºC for 30 min before being added to the BiP-NHK-S complex. The black line indicates that intervening lanes in the same immunoblot have been spliced out.
Next we used purified recombinant proteins to evaluate whether Grp170 binds directly to Sel1L. To facilitate Sel1L purification, we deleted the transmembrane domain from S- and histidine (His)-tagged Sel1L (S/His-Sel1L) and fused a FLAG tag to its C-terminus, generating S/His(ΔTM) Sel1L-FLAG. To purify Grp170, we constructed C-terminally HA- and His-tagged WT Grp170 to create WT Grp170-HA/His. S/His(ΔTM) Sel1L-FLAG and Grp170-HA/His, as well as the control GFP-FLAG, were expressed in and purified from 293T cells (Figure 3C). Of importance, when Grp170-HA/His was incubated with either S/His(ΔTM) Sel1L-FLAG or GFP-FLAG and the samples subjected to precipitation with an HA antibody, only S/His(ΔTM) Sel1L-FLAG but not GFP-FLAG coprecipitated with Grp170-HA/His (Figure 3D, top, compare lanes 3 and 4). This result demonstrates that Grp170 binds directly to Sel1L.
To determine whether the Grp170 bound to Sel1L is enzymatically active, we isolated the Grp170-Sel1L complex using a tandem affinity purification approach and interrogated whether this complex harbors a nucleotide exchange activity. Cells were cotransfected with WT Grp170-FLAG and S/His-Sel1L. S/His-Sel1L was first isolated from the resulting cell extract by using a nickel column. After elution of S/His-Sel1L, the eluted material was subjected to precipitation by using FLAG antibody–conjugated agarose beads. The bound material was then eluted with a FLAG peptide in order to generate the WT Grp170-FLAG:S/His-Sel1L complex (Figure 3E). The S/His-Sel1L level appeared less than that of WT Grp170-FLAG, perhaps due to partial dissociation of the complex during purification. Next we used a nucleotide exchange assay (Inoue and Tsai, 2015) in which radiolabeled ADP-BiP was incubated with or without the WT Grp170-FLAG:S/His-Sel1L complex and assessed the extent of ADP release from BiP. To generate radiolabeled ADP-BiP, FLAG-BiP was preloaded with [α-32P]ATP and incubated to covert ATP-BiP into ADP-BiP. This sample was subjected to an initial round of spin column gel filtration to remove most of the excess radiolabeled ATP. Radiolabeled ADP-BiP was subsequently incubated with FLAG-GFP or WT Grp170-FLAG:S/His-Sel1L complex and then with unlabeled ATP to induce nucleotide exchange. The sample was then subjected to a second round of spin column gel filtration to remove the released nucleotides. Any radiolabeled ADP that stayed bound to BiP was detected by TLC.
Using this assay, we found that the WT Grp170-FLAG:S/His-Sel1L complex (but not the control GFP-FLAG) triggered release of ADP from radiolabeled ADP-BiP (Figure 3F, compare lane 3 to lane 2). Of note, a low radiolabeled ATP level was observed in the sample treated with the WT Grp170-FLAG:S/His-Sel1L complex. This was caused by the residual radiolabeled ATP (after the initial spin column gel filtration) that bound to Grp170, as reported previously (Inoue and Tsai, 2015). The nucleotide release activity in the WT Grp170-FLAG:S/His-Sel1L complex is due to WT Grp170 (but not contaminating factors) because the mutant G41L Grp170-FLAG:S/His-Sel1L complex (Supplemental Figure S2A) did not trigger ADP release from radiolabeled ADP-BiP (Supplemental Figure S2B). Thus enzymatically active Grp170 is recruited to Sel1L, where it presumably executes a nucleotide exchange reaction.
The Grp170–Sel1L interaction might stimulate Grp170’s nucleotide exchange activity, thereby enhancing substrate release from BiP. However, in a titration experiment, we found that inclusion of the recombinant FLAG-tagged Sel1L (ΔTM) protein (Figure 3G) did not enhance the concentration-dependent, Grp170-mediated NHK release from BiP (Figure 3H), indicating that Sel1L did not stimulate Grp170’s nucleotide exchange function. These results suggest that Sel1L serves as a scaffold to recruit enzymatically active Grp170 proximal to the retrotranslocation site.
Depleting Grp170 traps the NHK–BiP complex on Sel1L
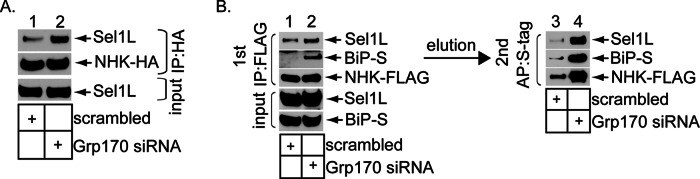
We performed three independent experiments to evaluate the functional link between Grp170’s interaction with Sel1L and its role during ERAD. First, we envisioned that when BiP captures a misfolded client in the ER lumen, it targets the client to the ERAD membrane machinery, in part due to BiP’s ability to bind to Sel1L (Denic et al., 2006; Hosokawa et al., 2008). Our present study suggests that Grp170 is also recruited to this machinery via association with Sel1L. This would potentially allow Grp170 to trigger client release from BiP at the retrotranslocation site. In this scenario, not only should depleting Grp170 prevent client release from BiP (as observed in Figure 2C), but the resulting client-BiP complex should now be trapped on Sel1L. To initially test whether downregulating Grp170 traps NHK on Sel1L, we immunoprecipitated NHK-HA from control or Grp170-depleted cells, and analyzed the precipitate for the presence of endogenous Sel1L. Indeed, immunoprecipitation of NHK-HA pulled down an increased level of Sel1L when Grp170 was depleted (Figure 4A, top, compare lane 2 to lane 1), indicating that NHK is trapped on Sel1L in the absence of Grp170. To further evaluate whether it is the NHK-BiP complex (and not simply NHK) that becomes trapped on Sel1L in Grp170’s absence, we performed a sequential pulldown experiment. In the first precipitation, NHK-FLAG was pulled down and the precipitated material eluted (with a FLAG peptide). Next the eluted material was subjected to a second round of precipitation by S-affinity purification to isolate BiP-S from the eluted sample, and the precipitated material was subjected to immunoblotting. We found low BiP (Figure 4B, middle, lane 3), Sel1L (Figure 4B, top, lane 3) and NHK (Figure 4B, bottom, lane 3) levels using this method. Thus the NHK precipitated material contains BiP: when this pool of BiP was precipitated, it also pulled down Sel1L. These findings suggest that the NHK-BiP complex associates with Sel1L. Of importance, depleting Grp170 significantly increased the level of BiP-S that can be isolated from the initial NHK precipitated/eluted material (Figure 4B, middle, compare lane 4 to lane 3), consistent with the observation that downregulating Grp170 stimulated NHK–BiP interaction (Figure 2C). Not surprisingly, precipitating this increased BiP level also led to enhanced coprecipitation of Sel1L (Figure 4B, top, compare lane 4 to lane 3) and NHK (Figure 4B, bottom, compare lane 4 to lane 3). Collectively these data demonstrate that in the absence of Grp170, the NHK–BiP complex is trapped on Sel1L. These results also support the notion that Grp170 executes its role proximal to the Sel1L–Hrd1 complex, in line with the finding that Grp170 is physically localized to Sel1L (Figure 3).
FIGURE 4:
Depleting Grp170 traps the NHK-BiP complex on Sel1L. (A) Cells expressing NHK-HA were transfected with scrambled siRNA or Grp170 siRNA #1, treated with cycloheximide and MG132 for 4 h, and harvested. After lysis, the resulting WCEs were subjected to immunoprecipitation using an anti-HA antibody. The resulting precipitated proteins were analyzed with SDS–PAGE, followed by immunoblotting with anti–FLAG-tag and S-tag antibodies. Input WCEs were also analyzed with an anti-Sel1L antibody. (B) Cells expressing NHK-FLAG and BiP-S were transfected with scrambled siRNA or Grp170 siRNA #1, treated with cycloheximide and MG132 for 4 h, and harvested. The resulting WCEs were subjected to immunoprecipitation using FLAG antibody–conjugated beads. The bound proteins were eluted with FLAG peptide and separated with SDS–PAGE, followed by immunoblotting with the indicated antibodies. Input WCEs were also analyzed. In addition, the eluted protein samples were further incubated with the S-protein–conjugated beads to pull down BiP-S, and the isolated proteins were analyzed by immunoblotting with the indicated antibodies.
Grp170 overexpression stimulates NHK degradation and increases Grp170–Sel1L interaction
Second, using Flp-In T-REx 293 parental cells or cells stably expressing WT Grp170-FLAG (Figure 5A, panel, lanes 1 and 2), we found that overexpressing WT Grp170-FLAG stimulated NHK degradation (Figure 5, B, quantified in C). In these cells, pull-down studies revealed that more Grp170 was recruited to Sel1L when WT Grp170-FLAG was overexpressed (Figure 5D, top, compare lane 2 to lane 1). The correlation between enhanced NHK degradation and increased Grp170 binding to Sel1L further strengthens the importance of the Grp170–Sel1L interaction during Grp170’s function in ERAD.
FIGURE 5:
Grp170 overexpression stimulates NHK degradation and increases the Grp170–Se11L interaction. (A) Flp-In T-REx 293 cells or cells overexpressing WT Grp170-FLAG were transfected with NHK-HA, and the resulting WCEs were analyzed by the indicated antibodies. (B) Cells in A were treated with cycloheximide for the indicated time and harvested, and the resulting WCE was analyzed with an anti-HA antibody. (C) The NHK-HA band intensity in B was quantified with ImageJ. Data represent the mean ± SD of at least three independent experiments. (D) Flp-In T-REx 293 cells or cells overexpressing WT Grp170-FLAG were processed as in Figure 3A.
Grp170 overexpression cannot bypass Sel1L depletion during NHK degradation
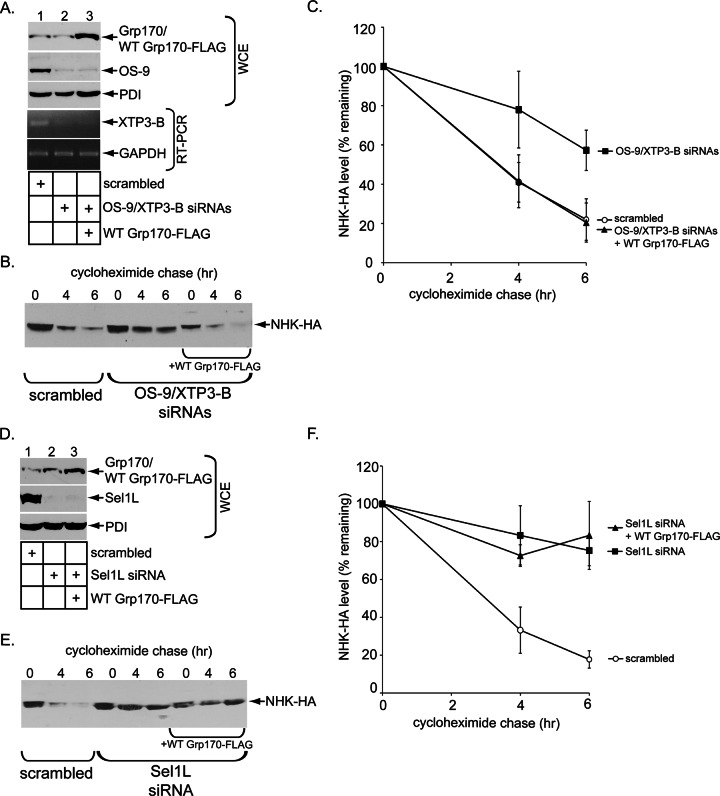
Third, we asked whether overexpressing Grp170 might bypass Sel1L’s requirement during NHK degradation. As a control, we initially found that whereas the simultaneous depletion of the OS-9/XTP3-B lectins (Figure 6A, lane 2) impaired NHK degradation (Figure 6, B, quantified in C), overexpressing Grp170 under this condition (Figure 6A, lane 3) restored degradation of NHK (Figure 6, B, quantified in C). When Sel1L was knocked down (Figure 6D, lane 2), NHK degradation was similarly disrupted (Figure 6, E, quantified in F). However, overexpressing Grp170 in the Sel1L-depleted cells did not rescue NHK degradation (Figure 6, E, quantified in F). These experiments suggest a stringent requirement for Sel1L during the Grp170-dependent ERAD pathway, consistent with the idea that Grp170’s interaction with Sel1L is crucial to this NEF’s mechanism of action.
FIGURE 6:
Grp170 overexpression cannot bypass Sel1L depletion during NHK degradation. (A) After gene knockdown using the indicated siRNAs, cells were further transfected with NHK-HA and either a vector control or WT Grp170-FLAG, and the cells were subjected to cell lysis or total RNA extraction. The resulting WCEs were analyzed by immunoblotting with the indicated antibodies (top three panels), and the resulting total RNA samples were subjected to RT-PCR using primer sets specific for XTP3-B or GAPDH (bottom two panels). XTP3-B mRNA but not its protein level was examined due to the poor quality of the XTP3-B antibody. (B) Cells in A were treated with cyclohexamide for the indicated time and harvested, and the resulting WCE was analyzed by immunoblotting with an anti-HA antibody. (C) The NHK-HA band intensity in B was quantified as in Figure 5C. (D) As in A, except that an siRNA against Sel1L was used. (E) Cells in D were subjected to a cyclohexamide chase analysis as in B. (F) The NHK-HA band intensity in E was quantified as in Figure 5C.
Grp170 promotes degradation of a nonglycosylated ERAD-Ls substrate
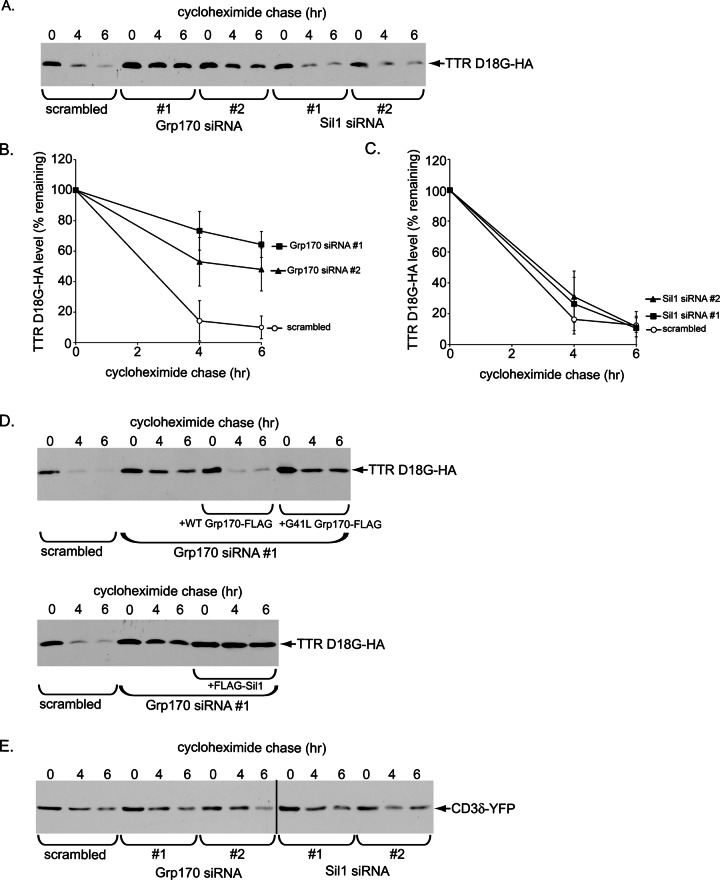
Because NHK is a glycosylated ERAD-Ls substrate, we tested whether the mutant nonglycosylated ERAD-Ls substrate TTR D18G (Christianson et al., 2012) might also require Grp170. Similar to the fate of NHK, depleting Grp170 (using either siRNA) blocked TTR D18G degradation (Figure 7, A, quantified in B), whereas knocking down Sil1 (using either siRNA) exerted no effect (Figure 6, A, quantified in C). Furthermore, in a knockdown-rescue experiment, reexpressing (siRNA-resistant) WT but not the NEF-defective G41L Grp170 mutant in Grp170-depleted cells restored TTR D18G degradation (Figure 7D, top), indicating that Grp170’s nucleotide exchange activity is essential in driving this client’s degradation. By contrast, overexpressing Sil1 in Grp170-depleted cells did not rescue degradation of TTR D18G (Figure 7D, bottom), in line with the observation that down-regulating Sil1 did not affect ERAD of this client (Figure 7A). Of importance, degradation of the Sel1L-independent ERAD-Lm client CD3δ (Bernasconi et al., 2010; Ninagawa et al., 2011), a membrane protein, was unaffected by the loss of Grp170 (or Sil1; Figure 7E), suggesting that Grp170 is dispensable for clients that do not require Sel1L. These experiments indicate that Grp170 can act more generally against glycosylated and nonglycosylated misfolded clients during the ERAD process.
FIGURE 7:
Grp170 promotes degradation of a nonglycosylated ERAD-Ls substrate. (A) Cells expressing TTR D18G-HA and transfected with scrambled,Grp170 siRNA (#1 or #2), or Sil1 siRNA (#1 or #2) were treated with cycloheximide for the indicated time and harvested, and the resulting WCE analyzed with an anti-HA antibody. (B, C) The TTR D18G-HA band intensity in A was quantified as in Figure 5C. (D) Top, parental Flp-In T-REx 293 cells were transfected with either scrambled or Grp170 siRNA #1. Where indicated, cells transfected with Grp170 siRNA #1 also stably expressed either siRNA-resistant WT Grp170-FLAG or G41L Grp170-FLAG. After siRNA transfection, cells were transfected with TTR D18G-HA, treated with cyclohexamide for the indicated time, and harvested, and the resulting WCE was analyzed as in A. Bottom, as for the top, except that cells stably expressing FLAG-Sil1 were used. (E) As in A, except that CD3d-YFP was used.
DISCUSSION
Our study identified Grp170 as the NEF that induces release of misfolded cellular ERAD-Ls clients from BiP, enabling them to undergo retrotranslocation leading to their proteasomal degradation (Figure 8). In addition, we found that enzymatically active Grp170 is recruited to the ERAD membrane machinery via binding to Sel1L, where it likely executes its NEF function. We will address these two points separately.
FIGURE 8.
Model depicting the role of the Grp170 nucleotide exchange factor during ERAD. During retrotranslocation of an ERAD-Ls client, the BiP–client complex is delivered to the Sel1L–Hrd1 membrane complex. When Grp170 is recruited to Sel1L, this nucleotide exchange factor triggers client release from BiP by converting ADP-BiP to ATP-BiP. This reaction enables the client to cross the postulated Hrd1 retrotranslocon, where it is extracted into the cytosol and targeted to the proteasome for degradation. An alternative scenario is possible in which Grp170 is first recruited to Sel1L, where it awaits the BiP–client complex.
The Grp170 nucleotide exchange activity promotes degradation of ERAD-Ls substrates
Using a cycloheximide chase approach in the context of a knockdown-rescue strategy in cells, our analyses pinpoint the nucleotide exchange activity of Grp170 but not Sil1 as crucial in promoting ERAD of NHK and TTR D18G. Additional cell-based studies revealed that depleting Grp170 prevented NHK release from BiP, indicating that Grp170 uses its nucleotide exchange function to disengage the client from BiP. These observations are further supported by in vitro analyses demonstrating that recombinant Grp170 is capable of inducing NHK release from BiP. What might account for the selective use of Grp170 but not Sil1 in regulating ERAD of NHK and TTR D18G? Because Grp170 and Sil1 bind to substrate-free BiP with a similar efficiency in vitro (Inoue and Tsai, 2015), it is unlikely that their affinity for BiP per se dictates the specificity. This is in agreement with observations that both NEFs convert substrate-free ADP-BiP to ATP-BiP with similar efficiency (Steel et al., 2004; Weitzmann et al., 2007). Instead, because BiP has been reported to reciprocally stimulate Grp170’s ATPase activity (Steel et al., 2004), this reaction may increase Grp170’s affinity for substrates. Grp170 also harbors a unique holdase domain that interacts with hydrophobic proteins (Park et al., 2003), which is absent in Sil1. These properties found in Grp170 might support a more stable Grp170-client-BiP complex required for the subsequent Grp170-dependent client release step. Our proposed role of Grp170 during ERAD is hinted at by previous observations that Grp170/Lhs1p in yeast (Travers et al., 2000) and in mammalian cells (Christianson et al., 2012) is up-regulated during ER stress, a condition in which maximal ERAD efficiency is essential to clear the buildup of misfolded ER clients. Of interest, SV40 also selectively uses Grp170 but not Sil1 to undergo ER membrane penetration (Inoue and Tsai, 2015), whereas cholera toxin is more promiscuous, using both NEFs during its ER membrane translocation (Williams et al., 2015). Further experiments are needed to clarify why cholera toxin, in contrast to NHK/TTR D18G and SV40, can use either NEF during its cellular entry.
Grp170 interaction with Sel1L: a potential mechanism to couple substrate release from BiP and retrotranslocation?
How do BiP-dependent ERAD substrates avoid repeated futile cycles of reengaging BiP once they are released from BiP? A possible scenario to overcome this nonproductive cycle is to juxtapose Grp170 to the membrane-bound ERAD machinery. In this scenario, substrates that are released from BiP would be efficiently transported across the ER membrane without interference from BiP. This is reminiscent of the cytosolic Bag1 NEF, which couples release of substrates from cytosolic Hsp70 family proteins and ubiquitin-dependent proteasomal degradation by binding to the proteasome itself (Luders et al., 2000).
This postulated scenario led us to identify Sel1L of the ERAD membrane complex as a Grp170-binding partner. This interaction may not be surprising because Grp170’s N-terminal domain is similar to BiP, which has already been documented to bind to Sel1L (Denic et al., 2006; Hosokawa et al., 2009), although it is unclear whether BiP and Grp170 bind to the same site on Sel1L (see later discussion). Because Sel1L can engage a misfolded substrate (Mueller et al., 2006), this raises the possibility that Grp170 may be a substrate rather than a binding partner of Sel1L. To rule out this possibility, we isolated the Grp170–Sel1L complex and found that enzymatically active Grp170 associated with Sel1L, indicating that Grp170 bound to Sel1L is not misfolded and is likely a binding partner instead. At steady state, the Grp170–Sel1L interaction is likely weak or transient because only a low level of interaction between the two endogenous components was detected. This might explain why Grp170 was not identified as a Sel1L-interacting protein in a previous proteomics study (Christianson et al., 2012). Despite this, we hypothesize that the Grp170–Sel1L interaction is crucial for Grp170’s function during retrotranslocation of misfolded clients. This proposal is supported by the findings that the NHK–BiP complex is trapped on Sel1L in the absence of Grp170 and that overexpressing Grp170 stimulated NHK degradation, which is concomitant with increased localization of Grp170 to Sel1L. In addition, Grp170 overexpression can only bypass the loss of OS-9/XTB3-B but not Sel1L during NHK degradation, demonstrating a strict Sel1L requirement during the Grp170-dependent ERAD pathway. We do not know why overexpressing Grp170 compensates for the loss of OS-9 and XTB3-B. Because these lectins also bind to Sel1L (Christianson et al., 2008; Hosokawa et al., 2008, 2009), one possibility is that they help to stabilize the client–BiP complex interaction with Sel1L. When these lectins are depleted, the client–BiP complex would dissociate from Sel1L, thereby preventing the client from being released into the retrotranslocation channel. With increased localization of Grp170 to Sel1L, the client might be efficiently released from BiP and delivered successfully into the channel before dissociating from Sel1L. We note that the previous reports that ERAD of NHK (Bernasconi et al., 2010) and TTR D18G (Christianson et al., 2012) rely on the Sel1LxHrd1 complex are in complete agreement with our present study demonstrating that retrotranslocation of these misfolded clients also depends on Grp170, which is physically linked to Sel1L, and by extension Hrd1.
Given that our results suggest that Grp170 localization to Sel1L is critical for promoting efficient NHK degradation, we were surprised to find that overexpressing Grp170 stimulated degradation of this misfolded substrate since a significant fraction of the overexpressed Grp170 is expected to not bind to Sel1L and instead remain as a free pool. This free Grp170 is likely to lead to nonproductive NHK release from BiP at a site distal to the retrotranslocation site. To reconcile this observation, it is possible that by inducing NHK release from BiP, the free pool of Grp170 might in fact promote the released NHK to exit the futile chaperone cycle and enter into the productive cycle in which NHK is captured preferentially by BiP located at the retrotranslocation site. In this scenario, a J-protein—which stimulates BiP to capture its substrates—would need to be positioned at the retrotranslocation site. This possibility is consistent with our previous report demonstrating that the J-protein ERdj5 binds to Sel1L (Williams et al., 2013). In addition, there is also evidence that the J-protein ERdj4 interacts with Derlin-1 (Lai et al., 2012), a membrane component of the core Hrd1-Sel1L retrotranslocation machinery.
In summary, our cumulative findings lead us to propose the following model describing how Grp170 participates in ERAD, as depicted in Figure 8. Because BiP/Kar2p binds to Sel1L (Denic et al., 2006; Hosokawa et al., 2008), we envision that the client–BiP complex is first recruited to Sel1L. Subsequent recruitment of Grp170 to Sel1L enables this NEF to trigger client release from BiP at the retrotranslocation site, thus coupling client release with retrotranslocation. However, it is possible that the sequence of events is reversed: Grp170 is first recruited to Sel1L, where it awaits the client–BiP complex. These two possibilities may not be mutually exclusive, with one possibility potentially favored over the other, depending on changes in cellular conditions. In either of these scenarios, a single Sel1L should accommodate both Grp170 and the client–BiP complex. If so, this would suggest that Grp170 and BiP are targeted to different sites on Sel1L in order to avoid competition. Because Sel1L harbors multiple Sel1L repeats similar to tetratricopeptide (TPR) protein–protein interaction domains, it is conceivable that a different combination of these TPR domains is used to engage Grp170 and BiP. Future investigations are clearly needed to clarify the precise nature of the Grp170–versus BiP–Sel1L interactions.
MATERIALS AND METHODS
Materials
We used polyclonal OS-9, S-tag, and Sel1L and monoclonal PDI antibodies from Abcam (Cambridge, MA), a monoclonal BiP antibody from BD (Franklin, NJ), monoclonal Grp170 and polyclonal Sil1 antibodies from GeneTex (Los Angeles, CA), polyclonal Sel1L antibodies and monoclonal FLAG antibody from Sigma-Aldrich (St. Louis, MO), monoclonal GFP and polyclonal HA antibodies from Protein Tech Group (Chicago, IL), and monoclonal HA antibody from Covance (Princeton, NJ). FLAG M2 antibody–conjugated agarose beads and FLAG and 3X FLAG peptides were purchased from Sigma-Aldrich, and S-protein conjugated agarose beads and digitonin were from EMD Millipore Chemicals (San Diego, CA). The 12CA5 HA monoclonal antibody was isolated from the hybridoma culture supernatant.
DNA constructs
The plasmids encoding HA-tagged NHK and TTR D18G were generous gifts from R. Kopito (Stanford University, Stanford, CA). To generate the S tagged–protein expression vectors, each cDNA was inserted into pCDNA3.1(−) in frame with the S tag sequence by standard cloning methods. To generate S/His-Sel1L, Sel1L cDNA was amplified by PCR and replaced with the Sel1L (1–372) coding sequence in the plasmid expressing S/His-Sel1L (1–372) (Williams et al., 2013). Note that only Sel1L coding sequence lacking amino acids 114–134 was successfully obtained without mutation during cloning, likely due to the full-length Sel1L sequence being unstable or toxic to Escherichia coli. Because these omitted sequences partially overlap with the fibronectin domain that was reported to be dispensable for ERAD (Ninagawa et al., 2011), we used Sel1L sequence lacking the amino acids 114–134 as wild type in this study. To generate S/His(ΔTM) Sel1L-FLAG or FLAG-(ΔTM) Sel1L, the coding sequence was amplified by PCR with a primer containing the FLAG-tag sequence, and the resulting PCR product was subcloned in pCDNA3.1 (−).
siRNA-mediated gene knockdown
The target sequences of the siRNAs used in this study were as follows:
Grp170 #1 siRNA: (5′-GCUCAAUAAGGCCAAGUUUTT-3′)
Grp170 #2 siRNA: (5′-GCCUUUAAAGUGAAGCCAUTT-3′)
Sil1 #1 siRNA: (5′-GCUGAUCAACAAGUUCAAUTT-3′)
Sil1 #2 siRNA; (5′-GCGCUCUUUGAUCUUGAAUTT-3′)
Sel1L siRNA: (5′-GCUCAGUAGUACAGAGAAUUU-3′)
OS-9.1 siRNA: (5′-AUCCCUGAGUUGUUGAGCCCAAU-3′)
OS-9.2 siRNA: (5′-UAACAAACUGGACAGCAGCGUUUCC-3′)
XTP3-B siRNA: (5′-GGACAAGGAUAGUGGGAAAUU-3′)
Duplex siRNA (10 nM) was reverse transfected into cells using Lipofectamine RNAiMAX (Thermo Fisher Scientific, Waltham, MA) according to the manufacturer’s protocol. Allstar negative control siRNA (Qiagen, Hilden, Germany) was used as a scrambled siRNA. The efficacy of the siRNA against XTP3-B was confirmed by reverse transcription (RT)-PCR using the primer set forward, 5′-CACTGCCAGGATCTCCATTT-3′, and reverse, 5′-CAGTGAGCACTGGCTGAGAG-3′. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA level was analyzed by using the primer set forward, 5′-GATTCCACCCATGGCAAATTC-3′, and reverse, 5′-GTCATGAGTCCTTCCACGATAC-3′, and its level served as a loading control.
Cycloheximide chase experiment
Cells were reverse transfected with duplex siRNA (10 or 30 nM) using Lipofectamine RNAiMAX (Thermo Fisher Scientific) according to the manufacturer’s protocol and plated. At 24 h after siRNA transfection, cells were further transfected with the indicated ERAD substrates using polyethylenimine (PEI; Polysciences, Warrington, PA). At 24 h after DNA transfection, cells were incubated in the presence of 100 μg/ml cycloheximide, harvested at the indicated time, and lysed in a buffer containing 50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES; pH 7.5), 150 mM NaCl, 1% Triton X-100, and 1 mM phenylmethylsulfonyl fluoride (PMSF). The resulting WCEs were subjected to SDS–PAGE, followed by immunoblotting with the appropriate antibodies.
Precipitation of S-tagged proteins with S-protein–conjugated beads
The 293T cells transfected with the indicated plasmid were harvested and lysed in a buffer containing 50 mM HEPES (pH 7.5), 150 mM NaCl, 1% Triton X-100, and 1 mM PMSF. After centrifugation, the resulting lysates were incubated with S-protein–conjugated beads (EMB Millipore, Darmstadt, Germany) at 4°C for 2 h. The bound proteins were washed, eluted by SDS sample buffer, and subjected to SDS–PAGE, followed by immunoblotting with the appropriate antibodies. For sequential FLAG/S affinity purification, 293T cells transfected with the indicated siRNAs and DNA constructs were harvested and lysed in a buffer containing 50 mM HEPES (pH 7.5), 150 mM NaCl, 1% Triton X-100, and 1 mM PMSF. After centrifugation, the resulting lysates were incubated with FLAG antibody–conjugated beads at 4°C for 2 h. The bound proteins were washed and eluted with a buffer containing 50 mM HEPES (pH 7.5), 150 mM NaCl, 1% Triton X-100, and 0.1 mg/ml FLAG peptide. The eluted material was then incubated with S-protein–conjugated beads.
Purification of recombinant proteins
All recombinant proteins used in this study were expressed in and isolated from HEK 293T cells transfected with the indicated DNA constructs. FLAG-tagged recombinant proteins were purified as described previously (Inoue and Tsai, 2015). To purify S/His(ΔTM) Sel1L-FLAG, 293T cells transfected with the DNA construct were harvested at 24 h posttransfection, semipermeabilized in a buffer containing 50 mM HEPES (pH 7.5), 150 mM NaCl, 0.02% digitonin, and 1 mM PMSF, and centrifuged at 16,100 × g for 10 min. The resulting pellet fraction was further lysed with a buffer containing 50 mM HEPES (pH 7.5), 150 mM NaCl, 1% Triton X-100, and 1 mM PMSF and centrifuged at 16,100 × g for 10 min. The supernatant fraction was incubated with 2 mM ATP and 2 mM MgCl2 for 30 min, mixed with 0.5 M imidazole solution and 5 M NaCl to generate a final 30 mM imidazole and 500 mM NaCl sample solution, and applied to a HisTrap HP column (GE HealthCare, Chicago, IL) in a fast-performance liquid chromatography system (Bio-Rad, Hercules, CA). After the column was extensively washed with a buffer containing 50 mM HEPES (pH 7.5), 500 mM NaCl, 0.1% Triton X-100, and 30 mM imidazole, bound proteins were eluted with a 30–500 mM imidazole gradient. The peak fractions of S/His(ΔTM) Sel1L-FLAG were pooled and incubated with FLAG M2 agarose beads. The S/His(ΔTM) Sel1L-FLAG bound to beads was extensively washed with a buffer containing 50 mM HEPES (pH 7.5), 150 mM NaCl, and 0.1% Triton X-100 and eluted with 0.1 mg/ml FLAG peptide. For purification of the Grp170-FLAG:S/His-Sel1L complex, cells expressing S/His-Sel1L and Grp170-FLAG were processed as described for the purification of S/His(ΔTM) Sel1L-FLAG, except that a 20–500 mM imidazole gradient was used. The peak fractions of S/His (ΔTM) Sel1L were pooled and incubated with FLAG M2 agarose beads. The beads were extensively washed with a buffer containing 20 mM HEPES (pH 7.5), 50 mM KCl, and 0.1% Triton X-100 and incubated with 2 mM ATP and 2 mM MgCl2. After the beads were washed with the buffer, the Grp170-FLAG:S/His-Sel1L complex was eluted with 0.1 mg/ml 3xFLAG peptide. To purify FLAG-(ΔTM) Sel1, 293T cells transfected with the DNA construct were processed as described for the purification of S/His(ΔTM) Sel1L-FLAG, except that the resulting cell lysate was directly incubated with FLAG M2 agarose beads. After extensive washing of the beads, FLAG-(ΔTM) Sel1 was eluted with 0.1 mg/ml FLAG peptide.
In vitro release of NHK from BiP
NHK-S was isolated from the DNA-transfected 293T cells using S-protein–conjugated beads. The NHK-S bound beads were suspended in a buffer containing 20 mM HEPES (pH 7.5), 50 mM KCl, and 0.1% Triton X-100 and incubated with the indicated recombinant proteins in the presence or absence of ATP at 30 ºC for 10 min. After incubation, the beads were washed extensively, and the bound proteins were eluted with SDS sample buffer and separated by SDS–PAGE, followed by immunoblotting with anti–S-tag and BiP antibodies.
In vitro binding assay
Recombinant proteins were mixed and incubated at 37ºC for 30 min and subjected to immunoprecipitation with an anti-HA antibody using Protein G magnetic beads. The immune complexes were washed, eluted by SDS sample buffer, and subjected to SDS–PAGE, followed by immunoblotting with the appropriate antibodies.
Generating stable cell lines
Flp-In T-Rex-293 cells (Thermo Fisher Scientific) were cotransfected with pOG44 and either pCDNA5/FRT/TO encoding RNA interference–resistant WT, mutant Grp170-FLAG, or FLAG-Sil1 using Lipofectamine 2000 (Life Technologies). At 24 h posttransfection, cells were split and cultured in DMEM medium plus 100 μg/ml hygromycin and 5 μg/ml blasticidin for 10–15 d. Hygromycin-resistant colonies were cloned.
Nucleotide exchange assay
The detailed method of this assay was described previously (Inoue and Tsai, 2015). To test the nucleotide exchange activity of the Grp170-Sel1L complex, FLAG-BiP (3 μM) was first incubated with 50 μCi of [α-32P]ATP (3000 Ci/mmol) in a final volume of 11 μl (50 μCi is equivalent to 1.5 μM ATP in this reaction) at 37 ºC for 2 h to generate [α-32P]ADP FLAG-BiP, and the reaction was subjected to an initial spin gel filtration column to remove the free nucleotides. Despite this step to remove free nucleotides, a small residual amount of [α-32P]ATP was still present in the resulting [α-32P]ADP FLAG-BiP sample. Next the Grp170-Sel1 complex (20 nM) was added to [α-32P]ADP FLAG-BiP (20 nM) at 23ºC for 20 min and then further incubated with cold ATP (4 nM) and MgCl2 (2 mM) for 1 min to release [α-32P]ADP from BiP. Note that the residual free [α-32P]ATP in the [α-32P]ADP FLAG-BiP sample can be incorporated into WT Grp170 during this incubation step. After removal of the free nucleotides, the reaction was spotted onto a PEI cellulose plate and analyzed via TLC.
Supplementary Material
Acknowledgments
We thank Kaiyu He and Corey Cunningham for critical review of the manuscript. B.T. is funded by the National Institutes of Health (RO1 AI064296-08). This work is also partially supported by the Protein Folding Disease Initiative at the University of Michigan Medical School.
Abbreviations used:
- AP
affinity purification
- ER
endoplasmic reticulum
- ERAD
endoplasmic reticulum-associated degradation
- GFP
green fluorescent protein
- HA
hemagglutinin
- IP
immunoprecipitation
- MW
molecular weight
- NEF
nucleotide exchange factor
- NHK
null Hong Kong
- PEI
polyethylenimine
- PMSF
phenylmethylsulfonyl fluoride
- siRNA
small interfering RNA
- TM
transmembrane domain
- TTR
transthyretin
- WCE
whole-cell extract
- WT
wild type
- YFP
yellow fluorescent protein.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-01-0033) on March 30, 2016.
REFERENCES
- Athanasiou D, Bevilacqua D, Aguila M, McCulley C, Kanuga N, Iwawaki T, Paul Chapple J, Cheetham ME. The co-chaperone and reductase ERdj5 facilitates rod opsin biogenesis and quality control. Hum Mol Genet. 2014;23:6594–6606. doi: 10.1093/hmg/ddu385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnke J, Feige MJ, Hendershot LM. BiP and its nucleotide exchange factors Grp170 and Sil1: mechanisms of action and biological functions. J Mol Biol. 2015;427:1589–1608. doi: 10.1016/j.jmb.2015.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnke J, Hendershot LM. The large Hsp70 Grp170 binds to unfolded protein substrates in vivo with a regulation distinct from conventional Hsp70s. J Biol Chem. 2014;289:2899–2907. doi: 10.1074/jbc.M113.507491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernasconi R, Galli C, Calanca V, Nakajima T, Molinari M. Stringent requirement for HRD1, SEL1L, and OS-9/XTP3-B for disposal of ERAD-LS substrates. J Cell Biol. 2010;188:223–235. doi: 10.1083/jcb.200910042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky JL. Cleaning up: ER-associated degradation to the rescue. Cell. 2012;151:1163–1167. doi: 10.1016/j.cell.2012.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky JL, Bracher A. Nucleotide exchange factors for Hsp70 molecular chaperones. 2000. In: Madame Curie Bioscience Database [Internet]. Available at www.ncbi.nlm.nih.gov/books/NBK5987/ [DOI] [PMC free article] [PubMed]
- Brodsky JL, Werner ED, Dubas ME, Goeckeler JL, Kruse KB, McCracken AA. The requirement for molecular chaperones during endoplasmic reticulum-associated protein degradation demonstrates that protein export and import are mechanistically distinct. J Biol Chem. 1999;274:3453–3460. doi: 10.1074/jbc.274.6.3453. [DOI] [PubMed] [Google Scholar]
- Buck TM, Plavchak L, Roy A, Donnelly BF, Kashlan OB, Kleyman TR, Subramanya AR, Brodsky JL. The Lhs1/GRP170 chaperones facilitate the endoplasmic reticulum associated degradation of the epithelial sodium channel. J Biol Chem. 2013;288:18366–13680. doi: 10.1074/jbc.M113.469882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho P, Goder V, Rapoport TA. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell. 2006;126:361–373. doi: 10.1016/j.cell.2006.05.043. [DOI] [PubMed] [Google Scholar]
- Carvalho P, Stanley AM, Rapoport TA. Retrotranslocation of a misfolded luminal ER protein by the ubiquitin-ligase Hrd1p. Cell. 2010;143:579–591. doi: 10.1016/j.cell.2010.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson JC, Olzmann JA, Shaler TA, Sowa ME, Bennett EJ, Richter CM, Tyler RE, Greenblatt EJ, Harper JW, Kopito RR. Defining human ERAD networks through an integrative mapping strategy. Nat Cell Biol. 2012;14:93–105. doi: 10.1038/ncb2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson JC, Shaler TA, Tyler RE, Kopito RR. OS-9 and GRP94 deliver mutant alpha1-antitrypsin to the Hrd1-SEL1L ubiquitin ligase complex for ERAD. Nat Cell Biol. 2008;10:272–282. doi: 10.1038/ncb1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson JC, Ye Y. Cleaning up in the endoplasmic reticulum: ubiquitin in charge. Nat Struct Mol Biol. 2014;21:325–335. doi: 10.1038/nsmb.2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claessen JH, Kundrat L, Ploegh HL. Protein quality control in the ER: balancing the ubiquitin checkbook. Trends Cell Biol. 2012;22:22–32. doi: 10.1016/j.tcb.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Keyzer J, Steel GJ, Hale SJ, Humphries D, Stirling CJ. Nucleotide binding by Lhs1p is essential for its nucleotide exchange activity and for function in vivo. J Biol Chem. 2009;284:31564–31571. doi: 10.1074/jbc.M109.055160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denic V, Quan EM, Weissman JS. A luminal surveillance complex that selects misfolded glycoproteins for ER-associated degradation. Cell. 2006;126:349–359. doi: 10.1016/j.cell.2006.05.045. [DOI] [PubMed] [Google Scholar]
- Dong M, Bridges JP, Apsley K, Xu Y, Weaver TE. ERdj4 and ERdj5 are required for endoplasmic reticulum-associated protein degradation of misfolded surfactant protein C. Mol Biol Cell. 2008;19:2620–2630. doi: 10.1091/mbc.E07-07-0674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easton DP, Kaneko Y, Subjeck JR. The hsp110 and Grp1 70 stress proteins: newly recognized relatives of the Hsp70s. Cell Stress Chaperones. 2000;5:276–290. doi: 10.1379/1466-1268(2000)005<0276:thagsp>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagiwara M, Maegawa K, Suzuki M, Ushioda R, Araki K, Matsumoto Y, Hoseki J, Nagata K, Inaba K. Structural basis of an ERAD pathway mediated by the ER-resident protein disulfide reductase ERdj5. Mol Cell. 2011;41:432–444. doi: 10.1016/j.molcel.2011.01.021. [DOI] [PubMed] [Google Scholar]
- Hirsch C, Gauss R, Horn SC, Neuber O, Sommer T. The ubiquitylation machinery of the endoplasmic reticulum. Nature. 2009;458:453–460. doi: 10.1038/nature07962. [DOI] [PubMed] [Google Scholar]
- Hosokawa N, Kamiya Y, Kamiya D, Kato K, Nagata K. Human OS-9, a lectin required for glycoprotein endoplasmic reticulum-associated degradation, recognizes mannose-trimmed N-glycans. J Biol Chem. 2009;284:17061–17068. doi: 10.1074/jbc.M809725200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N, Wada I, Nagasawa K, Moriyama T, Okawa K, Nagata K. Human XTP3-B forms an endoplasmic reticulum quality control scaffold with the HRD1-SEL1L ubiquitin ligase complex and BiP. J Biol Chem. 2008;283:20914–20924. doi: 10.1074/jbc.M709336200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T, Tsai B. A nucleotide exchange factor promotes endoplasmic reticulum-to-cytosol membrane penetration of the nonenveloped virus simian virus 40. J Virol. 2015;89:4069–4079. doi: 10.1128/JVI.03552-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Awad W, Petrova K, Hendershot LM. Regulated release of ERdj3 from unfolded proteins by BiP. EMBO J. 2008;27:2873–2882. doi: 10.1038/emboj.2008.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabani M, Kelley SS, Morrow MW, Montgomery DL, Sivendran R, Rose MD, Gierasch LM, Brodsky JL. Dependence of endoplasmic reticulum-associated degradation on the peptide binding domain and concentration of BiP. Mol Biol Cell. 2003;14:3437–3448. doi: 10.1091/mbc.E02-12-0847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampinga HH, Craig EA. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol. 2010;11:579–592. doi: 10.1038/nrm2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CW, Otero JH, Hendershot LM, Snapp E. ERdj4 protein is a soluble endoplasmic reticulum (ER) DnaJ family protein that interacts with ER-associated degradation machinery. J Biol Chem. 2012;287:7969–7978. doi: 10.1074/jbc.M111.311290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luders J, Demand J, Hohfeld J. The ubiquitin-related BAG-1 provides a link between the molecular chaperones Hsc70/Hsp70 and the proteasome. J Biol Chem. 2000;275:4613–4617. doi: 10.1074/jbc.275.7.4613. [DOI] [PubMed] [Google Scholar]
- Massey S, Burress H, Taylor M, Nemec KN, Ray S, Haslam DB, Teter K. Structural and functional interactions between the cholera toxin A1 subunit and ERdj3/HEDJ, a chaperone of the endoplasmic reticulum. Infect Immun. 2011;79:4739–4747. doi: 10.1128/IAI.05503-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller B, Lilley BN, Ploegh HL. SEL1L, the homologue of yeast Hrd3p, is involved in protein dislocation from the mammalian ER. J Cell Biol. 2006;175:261–270. doi: 10.1083/jcb.200605196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninagawa S, Okada T, Takeda S, Mori K. SEL1L is required for endoplasmic reticulum-associated degradation of misfolded luminal proteins but not transmembrane proteins in chicken DT40 cell line. Cell Struct Funct. 2011;36:187–195. doi: 10.1247/csf.11018. [DOI] [PubMed] [Google Scholar]
- Nishikawa SI, Fewell SW, Kato Y, Brodsky JL, Endo T. Molecular chaperones in the yeast endoplasmic reticulum maintain the solubility of proteins for retrotranslocation and degradation. J Cell Biol. 2001;153:1061–1070. doi: 10.1083/jcb.153.5.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olzmann JA, Kopito RR, Christianson JC. The mammalian endoplasmic reticulum-associated degradation system. Cold Spring Harb Perspect Biol. 2013;5 doi: 10.1101/cshperspect.a013185. a013185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Easton DP, Chen X, MacDonald IJ, Wang XY, Subjeck JR. The chaperoning properties of mouse grp170, a member of the third family of hsp70 related proteins. Biochemistry. 2003;42:14893–14902. doi: 10.1021/bi030122e. [DOI] [PubMed] [Google Scholar]
- Petrova K, Oyadomari S, Hendershot LM, Ron D. Regulated association of misfolded endoplasmic reticulum lumenal proteins with P58/DNAJc3. EMBO J. 2008;27:2862–2872. doi: 10.1038/emboj.2008.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggiano A, Foresti O, Carvalho P. Quality control: ER-associated degradation: protein quality control and beyond. J Cell Biol. 2014;204:869–879. doi: 10.1083/jcb.201312042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MH, Ploegh HL, Weissman JS. Road to ruin: targeting proteins for degradation in the endoplasmic reticulum. Science. 2011;334:1086–1090. doi: 10.1126/science.1209235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steel GJ, Fullerton DM, Tyson JR, Stirling CJ. Coordinated activation of Hsp70 chaperones. Science. 2004;303:98–101. doi: 10.1126/science.1092287. [DOI] [PubMed] [Google Scholar]
- Stein A, Ruggiano A, Carvalho P, Rapoport TA. Key steps in ERAD of luminal ER proteins reconstituted with purified components. Cell. 2014;158:1375–1388. doi: 10.1016/j.cell.2014.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–258. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- Ushioda R, Hoseki J, Nagata K. Glycosylation-independent ERAD pathway serves as a backup system under ER stress. Mol Biol Cell. 2013;24:3155–3163. doi: 10.1091/mbc.E13-03-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitzmann A, Baldes C, Dudek J, Zimmermann R. The heat shock protein 70 molecular chaperone network in the pancreatic endoplasmic reticulum—a quantitative approach. FEBS J. 2007;274:5175–5187. doi: 10.1111/j.1742-4658.2007.06039.x. [DOI] [PubMed] [Google Scholar]
- Williams JM, Inoue T, Banks L, Tsai B. The ERdj5-Sel1L complex facilitates cholera toxin retrotranslocation. Mol Biol Cell. 2013;24:785–795. doi: 10.1091/mbc.E12-07-0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JM, Inoue T, Chen G, Tsai B. The nucleotide exchange factors Grp170 and Sil1 induce cholera toxin release from BiP to enable retrotranslocation. Mol Biol Cell. 2015;26:2181–2189. doi: 10.1091/mbc.E15-01-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.