Candida albicans cells lacking the Pil1 and Lsp1 proteins needed to form eisosome microdomains in the plasma membrane develop deep cell wall invaginations. Overproduction of the Sur7 membrane protein rescues the defects of pil1Δ lsp1Δ cells, demonstrating that a key role for eisosomes is to promote Sur7 regulation of plasma membrane function.
Abstract
The plasma membrane of the fungal pathogen Candida albicans forms a protective barrier that also mediates many processes needed for virulence, including cell wall synthesis, invasive hyphal morphogenesis, and nutrient uptake. Because compartmentalization of the plasma membrane is believed to coordinate these diverse activities, we examined plasma membrane microdomains termed eisosomes or membrane compartment of Can1 (MCC), which correspond to ∼200-nm-long furrows in the plasma membrane. A pil1∆ lsp1∆ mutant failed to form eisosomes and displayed strong defects in plasma membrane organization and morphogenesis, including extensive cell wall invaginations. Mutation of eisosome proteins Slm2, Pkh2, and Pkh3 did not cause similar cell wall defects, although pkh2∆ cells formed chains of furrows and pkh3∆ cells formed wider furrows, identifying novel roles for the Pkh protein kinases in regulating furrows. In contrast, the sur7∆ mutant formed cell wall invaginations similar to those for the pil1∆ lsp1∆ mutant even though it could form eisosomes and furrows. A PH-domain probe revealed that the regulatory lipid phosphatidylinositol 4,5-bisphosphate was enriched at sites of cell wall invaginations in both the sur7∆ and pil1∆ lsp1∆ cells, indicating that this contributes to the defects. The sur7∆ and pil1∆ lsp1∆ mutants displayed differential susceptibility to various types of stress, indicating that they affect overlapping but distinct functions. In support of this, many mutant phenotypes of the pil1∆ lsp1∆ cells were rescued by overexpressing SUR7. These results demonstrate that C. albicans eisosomes promote the ability of Sur7 to regulate plasma membrane organization.
INTRODUCTION
The plasma membrane is critically important as a protective barrier that also mediates a wide range of dynamic processes, including endocytosis, secretion, nutrient uptake, signal transduction, morphogenesis, and cell wall or extracellular matrix synthesis. Consistent with its complex function, the plasma membrane is composed of a diverse array of proteins and lipids. These various constituents are organized into specialized domains that bring together subsets of lipids and proteins by spontaneous segregation of membrane lipids, protein-mediated diffusion restrictions, and other mechanisms (Simons and Gerl, 2010; Ziolkowska et al., 2012). These features are believed to contribute to proper plasma membrane function and the homeostasis of the different lipids and proteins (Malinsky et al., 2013). However, the roles of plasma membrane organization are poorly understood due to the difficulties of studying hydrophobic membranes.
Recent studies with fungi have identified plasma membrane microdomains known as membrane compartment of Can1 (MCC), which are stable, protein-organized domains that can be visualized by microscopy (Malinsky et al., 2010, 2013; Douglas and Konopka, 2014). The MCC domains correspond to inward furrows in the plasma membrane (Stradalova et al., 2009), which are stabilized by a complex of cytosolic proteins termed the eisosome (Walther et al., 2006). In Saccharomyces cerevisiae, there are 50 of these punctate domains per cell, each of which is 300 nm long and 50 nm deep (Malinska et al., 2003; Walther et al., 2006). Outside the MCC, the plasma membrane is more dynamic—lateral diffusion of membrane proteins takes place in this region, and it also contains transient sites of endocytosis, secretion, and contact with the cortical ER (Malinska et al., 2004; Stradalova et al., 2012).
MCC/eisosome formation in S. cerevisiae is promoted by two homologous proteins, Pil1 and Lsp1, which assemble into long filaments on the cytoplasmic side of the plasma membrane (Karotki et al., 2011; Olivera-Couto et al., 2011; Ziolkowska et al., 2011). Pil1 and Lsp1 contain BAR domains, which are believed to bend the membrane to form the furrows (Olivera-Couto et al., 2011; Ziolkowska et al., 2011). At least 10 other proteins assemble onto the cytoplasmic surface of the eisosomes, including Seg1, a protein that facilitates eisosome formation (Moreira et al., 2012), the Pkh1/2 kinases, which regulate cell wall synthesis and stress responses (Walther et al., 2007), the Slm1/2 proteins, which regulate TORC2 and sphingolipid homeostasis (Berchtold et al., 2012), and quinone reductases, which were shown in C. albicans to promote resistance to oxidative stress (Li et al., 2015). Integral membrane proteins recruited into these domains include two distinct families of tetraspanner proteins related to Sur7 and Nce102 and several nutrient transporters, including the arginine transporter Can1 (Malinsky et al., 2010; Douglas and Konopka, 2014). Although the plasma membrane portion of these domains is known as the MCC and the cytoplasmic proteins that associate on the intracellular side are known as the eisosome, for simplicity we will often refer to both of these domains as eisosomes since most of our experiments focus on eisosome proteins.
Membrane furrows have been found in a wide range of fungi, lichens, and algae, but their function is not well understood (Lee et al., 2015). Therefore eisosomes in the human fungal pathogen Candida albicans were examined in this study because of the key role that the plasma membrane plays in virulence. Previous studies showed that deletion of SUR7 from C. albicans caused dramatic defects in plasma membrane organization and cell wall morphogenesis (Alvarez et al., 2008; Bernardo and Lee, 2010; Wang et al., 2011; Douglas et al., 2012). Although eisosomes still formed in the sur7∆ cells, there were extensive defects outside of the eisosomes, including mislocalization of actin and septins. The sur7∆ cells formed deep invaginations of cell wall growth and were more susceptible to compounds that perturb the cell wall (Alvarez et al., 2008; Wang et al., 2011; Douglas et al., 2012). To better understand the broad role of Sur7, we examined the effects of deleting the PIL1 and LSP1 genes, which are needed for eisosome formation. Similar but not identical defects were observed for sur7∆ and pil1∆ lsp1∆ mutants, indicating that they have overlapping but distinct functions. Consistent with this, some phenotypes of the pil1∆ lsp1∆ mutant could be partially rescued by overexpressing SUR7. These results provide new insights into membrane organization and identify a major role for C. albicans eisosomes in promoting Sur7 function.
RESULTS
Pil1 and Lsp1 play equal roles in eisosome formation
A previous study demonstrated that C. albicans Pil1 and Lsp1 localize to punctate eisosome domains (Reijnst et al., 2011). To elucidate their roles in C. albicans, we created PIL1 and LSP1 deletion mutant strains. Although C. albicans is diploid, for simplicity, the homozygous deletion strains will be referred to as pil1∆ and lsp1∆. Analysis of Lsp1-GFP in the pil1∆ mutant and Pil1-GFP in the lsp1∆ mutant revealed that both proteins formed punctate eisosome patches in the plasma membrane similar to the wild-type control strain (Figure 1A). The observation that Pil1 and Lsp1 have equal ability to form eisosomes was unexpected, since Lsp1 is not capable of forming eisosomes in the absence of Pil1 in S. cerevisiae or Ashbya gossypii (Walther et al., 2006; Seger et al., 2011), even though Pil1 and Lsp1 have very similar amino acid sequences (70% identical). Comparison of the amino acid sequences suggested that the functional differences may be due to six sites in Lsp1 from S. cerevisiae and A. gossypii that differed from residues that are conserved in the Pil1 proteins and in C. albicans Lsp1 (Supplemental Figure S1). These sites occur in the BAR-domain region (Ziolkowska et al., 2011), where they could affect the ability to interact with the plasma membrane. The amino acid comparison also suggested that C. albicans Lsp1 might be more closely related to S. cerevisiae Pil1. However, to avoid confusion, we will use the assignments made in the Candida Genome Database (Pil1 orf19.778/C1_04680W and Lsp1 orf19.3149/C2_06730W)
FIGURE 1:

Both Pil1 and Lsp1 promote eisosome formation in C. albicans. Lsp1, Pil1, and Sur7 tagged with GFP at their C-termini were visualized by fluorescence microscopy. (A) Lsp1-GFP showed typical punctate localization in both the wild-type and pil1∆ strains. Similar results were observed for Pil1-GFP in the wild-type and lsp1∆ strains. This indicates that both Lsp1 and Pil1 can promote eisosome formation. Top, the punctate eisosome patches on the top of the cell; bottom, eisosomes around the periphery in a midsection of the cell. (B) Localization of Lsp1-GFP and Sur7-GFP in the indicated strains, in which the different copies of PIL1 and LSP1 were progressively deleted. Sur7-GFP in the pil1∆ lsp1∆ strain, which lacked eisosomes, gave a lower signal. Therefore the Sur7-GFP signal was increased relative to the other images to visualize this sample. (C) Lsp1-GFP localization in wild-type and seg1∆ strains. Strains used were Pil1-GFP (YLD70-1), LSP1-GFP (YLD76-1), PIL1-GFP lsp1∆ (YHXW24-1), LSP1-GFP pil1∆ (YLD85-1), LSP1-GFP pil1∆ lsp1∆LSP1 (YHXW25-1), SUR7-GFP (YHXW4), SUR7-GFP pil1∆ (YHXW26-1), SUR7-GFP pil1∆ lsp1∆/LSP1 (YHXW27-1), SUR7-GFP pil1∆ lsp1∆ (YHXW28-1), and LSP1-GFP seg1∆ (YLD177-1). Bars, 5 μm.
Although Lsp1–green fluorescent protein (GFP) formed punctate patches in the pil1∆ mutant, there was ∼24% reduction in the number of patches (Figure 1B; 15 cells; p < 0.001). This was further exacerbated in a pil1∆ strain that contained only one copy of LSP1 (pil1∆/∆ LSP1-GFP/lsp1∆), which displayed ∼57% reduction in eisosome patches (15 cells, p < 0.001). Sur7-GFP showed expected localization to punctate domains in wild type, but slight abnormalities in its localization were detected in the pil1∆ strain (Figure 1B). Further deletion of one copy of LSP1 from the pil1∆ strain resulted in atypical Sur7-GFP patches that were much larger than the punctate eisosome patches. Of interest, Sur7-GFP could still be detected at the plasma membrane in the pil1∆ lsp1∆ strain, which lacks eisosomes, although the size, distribution, and intensities of the Sur7-GFP patches varied (Figure 1B). Similar results have been reported for S. cerevisiae Sur7-GFP in a pil1∆ strain that lacks eisosomes (Walther et al., 2006; Stradalova et al., 2012). The fact that Sur7-GFP was still detectable at the plasma membrane has significance for conclusions to be made later regarding its role in the absence of Pil1 and Lsp1 (see Discussion).
A C. albicans seg1∆ mutant was also analyzed because the S. cerevisiae and A. gossypii Seg1 localizes to eisosomes and plays a role in promoting eisosome formation (Seger et al., 2011; Moreira et al., 2012). In contrast, a C. albicans seg1∆ mutant showed at most a slight delay in forming eisosomes in small buds (see arrow in Figure 1C). Seg1 may be less critical for eisosome formation in C. albicans, since both Pil1 and Lsp1 appear to be fully functional.
Similar but not identical cell wall invaginations are present in sur7∆ and pil1∆ lsp1∆ mutants
The C. albicans sur7∆ mutant forms extensive invaginations of the cell wall that accumulated with the increased age of the cell (Alvarez et al., 2008; Wang et al., 2011). Similar invaginations were seen for the pil1∆ lsp1∆ mutant using calcofluor white to stain the cell wall (Figure 2A). The sur7∆ and pil1∆ lsp1∆ mutants were also similar, in that the cells were often present in clumps, indicating a defect in cytokinesis (Figure 2A). These defects were not detected in the pil1∆ or lsp1∆ single mutants or in a pil1∆ lsp1∆ strain that was complemented with either LSP1 or PIL1. Higher-resolution images obtained with transmission electron microscopy (TEM) revealed that, as seen previously for sur7∆ cells (Alvarez et al., 2008), the pil1∆ lsp1∆ cells contained torus-shaped structures in the middle of the cell that represent a cross section of tubes of cell wall material (marked with white arrows in Figure 2B). However, the mutants differed in the types of invaginations that were present at the periphery of the cells (marked with black arrows on the outside of the cell in Figure 2B). The sur7∆ mutant predominantly contained pointed invaginations of cell wall at the periphery, whereas the pil1∆ lsp1∆ mutant had fewer spiky invaginations and instead contained large, rounded invaginations similar to those observed in a S. cerevisiae pil1∆ mutant (Walther et al., 2006). Thus the sur7∆ and pil1∆ lsp1∆ mutants display similar but not identical cell wall defects.
FIGURE 2:
The pil1∆ lsp1∆ mutant forms large invaginations of the cell wall. (A) The cells were stained with calcofluor white, and then cell wall structures were photographed by fluorescence microscopy. Note that the pil1∆ and lsp1∆ mutants displayed very little altered cell wall, whereas the pil1∆ lsp1∆ double mutant displayed cell wall invaginations comparable to those of the sur7∆ mutant. White bar, 5 μm. (B) Cell sections detected by transmission electron microscopy. Note the presence of thicker cell walls and invaginations of cell wall in both the pil1∆ lsp1∆ double mutant and the sur7∆ mutant. Black arrows on the outer edges of the cells point to regions where there are spiky invaginations in the sur7∆ mutant and the large, round cell wall invagination in the pil1∆ lsp1∆ mutant. The white arrows point to the cell wall tubes. Black bar, 1 μm. The strains used were wild type (DIC185), lsp1∆ (YLD77-11-24-1), pil1∆ (YLD73-9-2-1), pil1∆ lsp1∆ (YHXW21-1), sur7∆ (YJA11), pil1∆ lsp1∆ with a copy of LSP1 reintegrated (YHXW23-1), pil1∆ lsp1∆ with a copy of PIL1 integrated (YHXW22-1), and seg1∆ (YLD182-40-14-3).
The pil1∆ lsp1∆ mutant displays defects in actin organization and morphogenesis
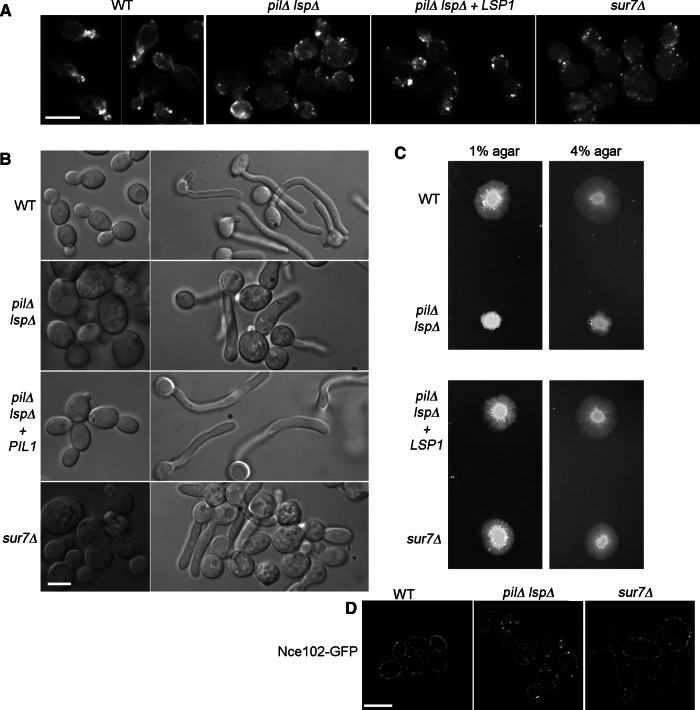
The abnormal cell morphology suggested that the pil1∆ lsp1∆ mutant cells have a defect in actin polarization. To examine this, we stained cells with the actin-binding agent rhodamine-phalloidin (Figure 3A). Wild-type cells showed the expected localization of actin patches at the cortical regions of the bud and actin filaments leading back into the mother cell. In contrast, actin was depolarized in the pil1∆ lsp1∆ mutant, as actin patches could be detected in both the bud and the mother cell, and there were fewer detectable actin filaments. This phenotype is similar to the sur7∆ mutant (Figure 3A). Introduction of one copy of LSP1 partially rescued the phenotype of the pil1∆ lsp1∆ cells, as there were still some abnormalities.
FIGURE 3:
The pil1∆ lsp1∆ mutant displays defects in actin localization and hyphal morphogenesis. (A) The indicated cells types were stained with rhodamine-phalloidin to detect actin. Note the lack of polarized actin patches and fewer actin cables in the pil1∆ lsp1∆ cells and the sur7∆ cells. (B) Left, budding cells. Right, cells in synthetic medium that were treated with 15% serum for 1.5 h at 37°C to induce hyphal formation. Although the pil1∆ lsp1∆ mutant could be induced to form elongated cells, they did not form typical hyphae with long, parallel walls. (C) Cells were spotted onto solid-medium plates containing 4% bovine serum and then either 1% agar (left) or 4% agar (right). The higher agar concentration weakly rescued the invasive growth defect of the pil1∆ lsp1∆ mutant. (D) Nce102-GFP localized to punctate patches in the wild-type and sur7∆ strains, which form eisosomes. In contrast, Nce102-GFP localized evenly in the plasma membrane in the pil1∆ lsp1∆ strain, consistent with the absence of eisosomes. The Nce102-GFP signal seen in the center of the cells is due to the plasma membrane that is associated with the cell wall invaginations and does not represent a punctate localization of Nce102-GFP in the pil1∆ lsp1∆ or sur7∆ cells. Strains used were wild type (DIC185), pil1∆ lsp1∆ (YHXW21-1) sur7∆ (YJA11), and pil1∆ lsp1∆ with a copy of LSP1 reintegrated (YHXW23-1), NCE102-GFP (YHXW15), NCE102-GFP pil1∆ lsp1∆ (YHXW29-1), and NCE102-GFP sur7∆ (YHXW30-1). Bars, 5 μm.
As expected for cells with an actin defect, the pil1∆ lsp1∆ mutant did not efficiently form highly polarized hyphal cells after induction with serum (Figure 3B). Some cells could form elongated pseudohyphae, whereas other cells failed to initiate filamentous growth. A similar phenotype was detected for sur7∆ cells. This variable ability of different cells to undergo filamentous growth may be due to varying degrees of abnormal cell wall morphogenesis present in the cells at the time they were induced (Figure 2).
Although some of the pil1∆ lsp1∆ cells formed elongated pseudohyphae in liquid culture, the mutant cells were strongly defective in invasive growth into 1% agar–containing serum (Figure 3C). This defect was stronger than that observed for the sur7∆ mutant (Figure 3C). The ability of the pil1∆ lsp1∆ mutant to undergo invasive growth was slightly improved when assayed for ability to invade into a more rigid matrix of 4% agar. This was interesting because a mutant that lacks the MCC/eisosome domain protein Nce102 displays a similar phenotype. An nce102∆ mutant is defective in invading 1% agar but invades well into denser, 4% agar (Douglas et al., 2013), indicating that a separate pathway activated by the surrounding matrix can compensate for the nce102∆ defect. Analysis of Nce102-GFP in the pil1∆ lsp1∆ mutant showed that it localized efficiently at the plasma membrane, although it was evenly distributed and was not present in the punctate patches seen for the wild type or the sur7∆ mutant (Figure 3D). This raises the possibility that the abnormal Nce102 localization contributes to the pil1∆ lsp1∆ phenotype. The failure of the Nce102-GFP to localize to punctate clusters in the pil1∆ lsp1∆ mutant is also significant because it confirms that the eisosomes are defective in this pil1∆ lsp1∆ mutant.
Differential effects of inhibitors on pil1∆ lsp1∆ and sur7∆ mutants
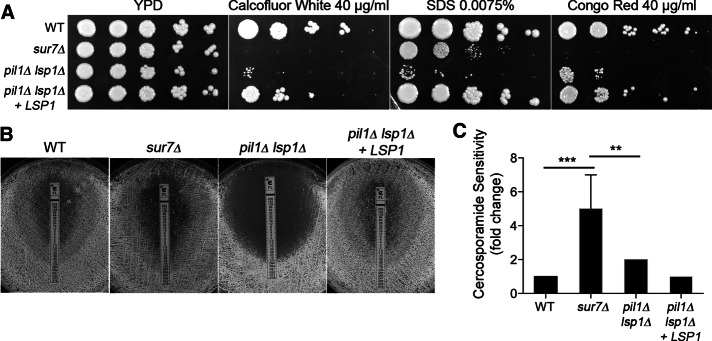
The unusual cell wall invaginations prompted us to examine whether the pil1∆ lsp1∆ mutant was sensitive to agents that exacerbate cell wall defects. The pil1∆ lsp1∆ mutant was similar to sur7∆ cells in being more sensitive than wild-type cells to calcofluor white, Congo red, and SDS (Figure 4A) and the ergosterol synthesis inhibitor fluconazole (Figure 4B). However, it was significant that the two mutants displayed differential sensitivity to these inhibitors. Of the two mutants, sur7∆ was more sensitive to calcofluor white and Congo red, which bind to the cell wall. In contrast, the pil1∆ lsp1∆ mutant was more sensitive to the detergent SDS, and it showed a clearer zone of growth inhibition surrounding an Etest strip containing a gradient of fluconazole, an antifungal drug that blocks ergosterol synthesis (Figure 4B). Control studies confirmed that these phenotypes were complemented by reintroduction of a copy of LSP1 into the pil1∆ lsp1∆ mutant cells. The greater sensitivity of the pil1∆ lsp1∆ mutant to fluconazole is consistent with studies showing that certain cell wall mutants, such as those affecting the Pkc1 pathway, are more susceptible to fluconazole (LaFayette et al., 2010). However, the pil1∆ lsp1∆ mutant was not more sensitive to cercosporamide, an inhibitor of fungal Pkc1 (Sussman et al., 2004), whereas the sur7∆ mutant was fivefold more sensitive to cercosporamide (Figure 4C). Taken together, these data demonstrate that pil1∆ lsp1∆ and sur7∆ have similar but not identical defects in cell wall function.
FIGURE 4:
The pil1∆ lsp1∆ and sur7∆ mutants show differential sensitivity to cell wall–perturbing agents. (A) Dilutions of cells were spotted onto solid-medium agar plates containing the indicated perturbing agent. The plates were incubated for 2 d at 37°C and then photographed. (B) The sensitivity to fluconazole was assayed by plating a lawn of cells and then placing an E strip containing a gradient of fluconazole on top. Plates were incubated for 2 d at 37°C and then photographed. (C) Sensitivity to cercosporamide was determined by growing cells in the presence of different concentrations of this Pkc1 inhibitor at 37°C for 2 d. Strains used were wild type (DIC185), pil1∆ lsp1∆ (YHXW21-1), sur7∆ (YJA11), and pil1∆ lsp1∆ with a copy of LSP1 reintegrated (YHXW23-1). **p < 0.01, ***p < 0.001.
Slm2 does not play an important role in C. albicans cell wall morphogenesis
To investigate the basis for the cell wall defects of the pil1∆ lsp1∆ and sur7∆ mutants, we analyzed the C. albicans orthologues of the S. cerevisiae Slm1 and Slm2 proteins, which localize to eisosomes and influence morphogenesis (Berchtold et al., 2012). Two C. albicans proteins showed significant homology to the Slm proteins. The Orf19.3505 protein (C6_02060W) had greater similarity to Slm1/2 from S. cerevisiae (∼40% identity over 425 amino acids) than Orf19.4043 (C5_05440C; ∼30% identity over 300 amino acids). When fused to GFP, Orf19.3035 colocalized with Lsp1–red fluorescent protein (RFP) in punctate eisosome patches in the plasma membrane (Figure 5A), as expected for the Slm proteins (Kamble et al., 2011; Berchtold et al., 2012). We will refer to Orf19.3505 as Slm2, since it shows higher amino acid similarity to Slm2 than to Slm1 from S. cerevisiae. In contrast, Orf19.4043 showed better similarity to Ask10 and Rgc1 from S. cerevisiae, and Orf19.4043-GFP localized to the cytoplasm (Figure 5A), as do Ask10 and Rgc1 under these conditions (Huh et al., 2003; Beese et al., 2009). Thus, consistent with C. albicans not having undergone the ancient genome duplication that S. cerevisiae did, C. albicans appears to have just one orthologue of each.
FIGURE 5:

The C. albicans orthologue of S. cerevisiae Slm1/2 localizes to eisosomes but is not needed for proper cell wall morphogenesis. (A) Orf19.3505-GFP (Slm2-GFP) colocalized with Lsp1-RFP, indicating that it is present in eisosomes, consistent with it being the C. albicans orthologue of S. cerevisiae Slm1 and Slm2 proteins. In contrast, Orf19.4043-GFP displayed a diffuse cytoplasmic localization, consistent with it being the orthologue of the S. cerevisiae Ask10 and Rgc1 proteins. (B) In the sur7∆ mutant, Orf19.3035-GFP (Slm2-GFP) localized to punctate clusters, although there were fewer patches than in a wild-type strain. In contrast, Orf19.3035-GFP showed a very different abnormal localization in the eisosome mutant pil1∆ lsp1∆. The predominant pattern was two major spots at opposite poles of the cells. (C) Left, cells lacking or overexpressing ORF19.3035 (SLM2) do not display cell wall or morphogenesis defects. Cells were stained with calcofluor white. Right, the cells could be induced to form hyphae by treatment with 15% serum for 1.5 h. Strains used were ORF19.3035-GFP LSP1-RFP (YHXW32-1), ORF19.4043-GFP LSP1-RFP (YHXW31-1), ORF19.3505-GFP sur7∆ (YHXW33-1), ORF19.3505 pil1lsp1 (YHXW34-1), orf19.3505∆ (YHXW36-1), and ORF19.3505 overexpressed (YHXW37-1). Bars, 5 μm.
In pil1∆ lsp1∆ cells, Slm2-GFP was not in punctate patches and instead was usually present in large patches near the ends of the cells (Figure 5B). A small fraction of cells showed a variety of different localization patterns that are likely due to Slm2-GFP associated with the plasma membrane surrounding the abnormal cell wall invaginations. In contrast, sur7∆ cells contained Slm2-GFP in the expected punctate eisosome patches, although there were fewer patches than in wild-type cells.
Of interest, a C. albicans slm2∆ mutant grew well, which contrasts with the fact that an S. cerevisiae mutant lacking both of the redundant SLM1 and SLM2 genes is not viable (Berchtold et al., 2012). Furthermore, there were no detectable abnormalities in budding for the C. albicans slm2∆ cells or in the ability to form hyphae when induced with serum (Figure 5C). Overexpression of SLM2 by introduction of an extra copy under control of the strong ADH1 promoter also did not have obvious effects on morphogenesis and did not cause formation of cell wall invaginations. Thus, although Slm2-GFP localizes to eisosomes, it does not appear to play a significant role in morphogenesis and therefore seems unlikely to contribute to the defects observed for the pil1∆ lsp1∆ and sur7∆ mutants.
pkh mutants form larger eisosomes but not cell wall invaginations
The Pkh kinases were next investigated for a role in the pil1∆ lsp1∆ phenotype since in S. cerevisiae, the Pkh1 and Pkh2 paralogues localize to eisosomes and regulate Pkc1 and other stress responses (Roelants et al., 2002). The more distantly related Pkh3 kinase has not been studied well but appears to carry out similar functions (Inagaki et al., 1999). C. albicans contains only one gene related to PKH1 and PKH2 (C1_12410C or orf19.5224). We previously referred to this gene as PKH1 (Douglas et al., 2013), but will now refer to it as PKH2 to conform to the Candida Genome Database. The C. albicans pkh2∆ mutant displayed increased sensitivity to Congo red and SDS, and these phenotypes were exacerbated by further deletion of one copy of PKH3 (pkh2∆/∆ pkh3∆/PKH3; Douglas et al., 2013). We were unable to isolate a pkh2∆ pkh3∆ double mutant, perhaps because the combined defect makes cells sick or is lethal.
The role of the Pkh kinases in S. cerevisiae eisosome formation is controversial; some studies indicated they promote eisosome formation, and other studies reported that they promote eisosome disassembly (Douglas and Konopka, 2014). Of interest, the C. albicans pkh2∆/∆ pkh3∆/PKH3 mutant showed abundant eisosomes (Figure 6A). In addition, filaments of Lsp1-GFP were detected in Z-stack images from the middle of the cell away from the plasma membrane, indicating a role for the Pkh kinases in preventing polymerization of Pil1 and Lsp1 in the cytoplasm. Analysis of the top view of the cells showed that the pkh2∆/∆ pkh3∆/PKH3 mutant cells displayed an extended meshwork of Lsp1-GFP rather than the isolated punctate patches seen in wild-type cells (Figure 6B). The pkh2∆ mutant and to a lesser degree the pkh3∆ mutant also showed enlarged eisosome structures. This indicates that the Pkh kinases are not required for eisosome assembly in C. albicans and are instead needed to prevent assembly or promote disassembly.
FIGURE 6:
The Pkh2 and Pkh3 kinases prevent formation of elongated eisosomes. (A) Localization of Lsp1-GFP in wild-type control and pkh2∆/∆ pkh3∆/PKH3 cells. The full diploid genotype is listed for this strain to designate that it lacks PKH2 and is heterozygous for PKH3. Cells were analyzed by fluorescence microscopy, and representative midsections are shown. Right, an enlarged midsection for the pkh2∆/∆ pkh3∆/PKH3 strain, revealing intracellular filaments of Lsp1-GFP. (B) Top view of cells, showing the pattern of Lsp1-GFP in the indicated wild-type or pkh mutant strain. Deletion of the PKH genes resulted in formation of elongated patches of Lsp1-GFP. (C) Cell walls were detected by staining with calcofluor white. (D) Dilutions of the indicated cells were spotted onto agar plates to test the effect on growth of different inhibitory compounds. These data revealed a different pattern of sensitivity to these agents for the pil1∆ lsp1∆ mutant and the pkh2∆/∆ pkh3∆/PKH3 mutant. The top row of assays was carried out with rich YPD medium. The bottom row of spot assays was carried out with SD medium (synthetic medium with dextrose). (E) Plasma membrane furrows were analyzed in the indicated strains by freeze-etching methods and TEM. Strains used for A and B were LSP1-GFP (YLD76-1), LSP1-GFP pkh2∆ (YLD173-2), LSP1-GFP pkh3∆ (YLD178-1), and LSP1-GFP pkh2∆/∆ pkh3∆/PKH3 (YLD174-4). Strains used for C–E were wild-type control (DIC185), sur7∆ (YJA11), pil1∆ lsp1∆ (YHXW21-1), pkh2∆/∆ pkh3∆PKH3 (YLD146-6-54-1), and pkh2∆/∆ pkh3∆PKH3 plus PKH2 (YLD203-3-3). Bars, 5 μm (A, C), 1 μm (B), 250 nm (E).
The effects of mutating the Pkh kinases on membrane furrow formation were analyzed in cells processed by freeze-etching and imaged by TEM (samples shown in Figure 6E, and whole cells shown in Supplement Figures S2–S5). As expected, wild-type cells formed typical membrane furrows, which were absent in pil1∆ lsp1∆ cells. Both the sur7∆ and nce102∆ cells contained furrows, although on average they were slightly longer than in the wild-type cells (Supplemental Figure S6). The pkh3∆ mutant cells formed unusual furrows that were wider. In contrast, the pkh2∆ and the pkh2∆/∆ pkh3∆/PKH3 mutants formed long chains of furrows (Figure 6E). This suggests that extensive filament formation of Lsp1-GFP seen in pkh2∆/∆ pkh3∆/PKH3 cells (Figure 6A) induces chains of furrows rather than longer furrows that have been seen in other species (Kabeche et al., 2011; Lee et al., 2015).
In spite of the altered eisosomes, the cell wall morphology of the pkh2∆/∆ pkh3∆/PKH3 mutant cells was normal, as we did not detect invaginations after staining with calcofluor white (Figure 6C). To determine whether a defect in Pkh kinase activity might contribute to the other phenotypes in the sur7∆ or pil1∆ lsp1∆ mutants, we tested cells for sensitivity to agents that exacerbate cell wall defects. The results showed that there was no direct correlation between phenotypes of the pkh2∆/∆ pkh3∆/PKH3 mutant and sur7∆ or pil1∆ lsp1∆ cells. The pkh2∆/∆ pkh3∆/PKH3 mutant was less sensitive to either calcofluor white or SDS than are the sur7∆ and pil1∆ lsp1∆ cells (Figure 6D). Conversely, the pkh2∆/∆ pkh3∆/PKH3 cells were more sensitive to caffeine than were the sur7∆ or pil1∆ lsp1∆ cells. The response to rapamycin distinguished all three mutants (Figure 6D). The pkh2∆/∆ pkh3∆/PKH3 cells were slightly sensitive to rapamycin, the sur7∆ cells were very sensitive to rapamycin, and the pil1∆ lsp1∆ cells were slightly resistant (note the better growth of pil1∆ lsp1∆ cells in columns 3 and 4 of Figure 6D). These differences indicate that the defects caused by the absence of the Pkh kinases do not correlate with the phenotypes of the sur7∆ and pil1∆ lsp1∆ mutants. These results also further demonstrate that the sur7∆ and pil1∆ lsp1∆ mutants have distinct phenotypes.
PI4,5P2 clusters in the plasma membrane at sites of cell wall invaginations
Studies on the role of the Inp51, a phosphatidylinositol 4,5-bisphosphate (PI4,5P2) phosphatase, showed that an inp51∆ mutant that has elevated levels of the lipid PI4,5P2 formed invaginations of cell wall growth that are similar to those seen in the sur7∆ and the pil1∆ lsp1∆ mutant cells (Badrane et al., 2012). PI4,5P2 is an important regulatory lipid that binds to many different signaling and morphogenesis proteins (Sun et al., 2013). Therefore we examined the localization of PI4,5P2 in the plasma membrane using a previously described fusion between the human PLCδ1 PH domain and RFP (Badrane et al., 2012). As expected, the PH-RFP probe localized to the plasma membrane in wild-type cells (Figure 7A). In contrast, both the sur7∆ and the pil1∆ lsp1∆ cells showed abnormal clusters of PH-RFP at sites of invaginations of the plasma membrane. These results indicate that eisosomes regulate the distribution of PI4,5P2 in the plasma membrane. Eisosomes in S. cerevisiae and Schizosaccharomyces pombe have been implicated in regulating PI4,5P2 levels and distribution into patches in response to stress (Murphy et al., 2011; Frohlich et al., 2014; Kabeche et al., 2014, 2015).
FIGURE 7:
A PH-domain probe indicates that sites of cell wall invaginations correlate with increased PI4,5P2. (A) Indicated cell types carrying the Cdc10 septin fused to GFP (Cdc10-GFP) and the PLCδ1 PH domain fused to RFP (PH-RFP) were analyzed by fluorescence microscopy. Note the similar localization of these PI4,5P2-binding proteins. (B) Cell walls were stained with calcofluor white to detect abnormalities. The fraction of cells displaying strongly abnormal cell wall was higher in the sur7∆ and pil1∆ lsp1∆ mutants than in the inp51∆ mutant, which lacks this PI4,5P2 phosphatase. Bottom, representative samples of cells that were scored as wild type, minor defect, or major defect. The contrast was adjusted when scoring samples to make minor defects more obvious. (C) Dilutions of cells were spotted onto rich YPD medium in the absence and presence of 35 μg/ml calcofluor white to perturb the cell wall. Two independent inp51∆ mutants. Strains used were CDC10-GFP PH-RFP (YHXW47-1), CDC10-GFP PH-RFP sur7∆ (YHXW41-1), CDC10-GFP PH-RFP pil1∆ lsp1∆ (YHXW39-1), WT control (DIC185), inp51∆-1 (YXH42-1), inp51∆-2 (YXH42-2), sur7∆ (YJA11), and pil1∆ lsp1∆ (YHXW21-1). Bars, 5 μm.
Badrane et al. (2012) also found that septins were mislocalized to ectopic sites in the inp51∆ mutant (similar to what was observed in sur7∆ cells; Alvarez et al., 2008). We therefore examined a GFP-tagged version of the Cdc10 septin and found that it was mislocalized to sites away from the bud neck in pil1∆ lsp1∆ cells, similar to the sur7∆ cells (Figure 7A). In both of these mutants, Cdc10-GFP was detected near regions of membrane invagination that contained enriched levels of PH-RFP. However, some regions enriched in PH-RFP contained low or nondetectable levels of Cdc10-GFP. Septin proteins may be recruited to these regions because it was shown in S. cerevisiae that the binding to PI4,5P2 promotes assembly of septin filaments (Bertin et al., 2010).
Comparison of the cell wall defects indicated that the phenotypes of the sur7∆ and pil1∆ lsp1∆ cells are not solely due to abnormal regulation of Inp51. Calcofluor white staining revealed that the sur7∆ and pil1∆ lsp1∆ cells had more extensive cell wall invaginations than the inp51∆ cells (Figure 7B). Whereas only 12% of the inp51∆ cells showed major cell wall invaginations, >40% of the pil1∆ lsp1∆ cells and 60% of the sur7∆ cells did so. Furthermore, the inp51∆ mutant did not show increased sensitivity to calcofluor white under conditions in which sur7∆ and the pil1∆ lsp1∆ cells are strongly inhibited (Figure 7C). Thus the phenotypes of sur7∆ and the pil1∆ lsp1∆ cells must be due, at least in part, to effects on other pathways that regulate PI4,5P2 and not just Inp51.
Overexpression of SUR7 partially rescues phenotypes of pil1∆ lsp1∆ mutant cells
The similar but distinct phenotypes of the sur7∆ and the pil1∆ lsp1∆ cells suggested that Sur7 and the eisosome proteins Pil1 and Lsp1 have independent functions. For example, as described earlier, sur7∆ cells were more sensitive to compounds that affect the cell wall (e.g., cercosporamide and calcofluor white) and to rapamycin, whereas pil1∆ lsp1∆ cells were more susceptible to fluconazole and had a stronger defect in invasive hyphal growth. To examine this relationship further, we tried to construct a sur7∆ pil1∆ lsp1∆ triple mutant but were not successful. It was rare to find a mutant with one copy of SUR7 deleted in the pil1∆ lsp1∆ mutant under conditions in which they were plentiful in the wild-type background, and deletion of a second copy of SUR7 showed that there was still a wild-type SUR7 present. Owing to these difficulties, we instead examined the effects of overexpressing SUR7 in the pil1∆ lsp1∆ mutant.
Of interest, overexpressing SUR7 by integrating an extra copy of SUR7 in the pil1∆ lsp1∆ cells caused a significant improvement in resistance to SDS, calcofluor white, and Congo red (Figure 8A). Overexpression of SUR7 also decreased the clumps of large cells that represent the more extreme phenotype of pil1∆ lsp1∆ cells (Figure 8B). Overexpression of SUR7 caused a reduction in the number of cells with obvious cell wall abnormalities from 40.4% in the pil1∆ lsp1∆ cells to 18.4% for pil1∆ lsp1∆ cells with an extra copy of SUR7 (Figure 8C). Integration of two extra copies of SUR7 (2xSUR7) in the pil1∆ lsp1∆ cells further improved the phenotypes but did not fully restore cells to a wild-type phenotype.
FIGURE 8:
Overexpression of SUR7 partially suppresses the cell wall phenotypes of pil1∆ lsp1∆ cells. Wild type, pil1∆ lsp1∆, and versions of pil1∆ lsp1∆ carrying one extra copy of SUR7 integrated into the genome (+1xSUR7) or two extra copies of SUR7 (+2xSUR7) were analyzed. (A) Spot assays showing the growth of the strains indicated on the left to the compounds listed above. YPD indicates the rich nutrient medium, which was used as the base medium for all of the assays. (B) Wild-type and indicated mutant cells were stained with calcofluor white, and then cell wall structures were photographed by fluorescence microscopy. (C) Relative proportions of cells with cell wall invaginations detected by calcofluor white staining as described in Figure 7B. (D) Relative sensitivity to fluconazole for the indicated strains. Cells were spread on an agar plate, and then filter disks containing 0.25, 0.5, or 1 μg fluconazole were placed on top and the plates incubated for 2 d at 37°C. Strains used were wild-type control (DIC185), pil1∆ lsp1∆ (YHXW21-1), pil1∆ lsp1∆ +1xSUR7 (YHXW45-1), and pil1∆ lsp1∆ +2xSUR7 (YHXW46-1). Bar, 5 μm.
In contrast to the foregoing results, overexpression of SUR7 only caused a minor change in the susceptibility of pil1∆ lsp1∆ cells to a low dose of fluconazole and did not cause a significant difference at higher doses (Figure 8D). This is consistent with SUR7 playing a minor role in resistance to fluconazole compared with Pil1 and Lsp1 (Figure 4B).
DISCUSSION
C. albicans eisosomes were examined to better define the roles of plasma membrane compartmentalization in this important human fungal pathogen. A pil1∆ lsp1∆ mutant that lacks eisosomes displayed strong defects in plasma membrane organization, cell wall morphogenesis, and invasive hyphal growth that were similar to sur7∆ cells that do make eisosomes (Alvarez et al., 2008). Further comparison of sur7∆ and pil1∆ lsp1∆ phenotypes indicated that one key function for eisosomes in C. albicans is to promote the ability of Sur7 to regulate the distribution of PI4,5P2 and cell wall morphogenesis. The pil1∆ lsp1∆ mutant also had distinct properties, such as stronger defects in invasive hyphal growth and increased sensitivity to fluconazole, indicating that eisosomes have other functions independent of Sur7.
Eisosome variation among species
Analysis of C. albicans mutants lacking different eisosome proteins revealed important similarities and differences with other fungi. A major difference is that Sur7 has important roles in C. albicans, whereas its deletion causes relatively minor phenotypes in S. cerevisiae (Young et al., 2002; Alvarez et al., 2008). In addition, Pil1 and Lsp1 have equal roles in C. albicans, whereas Lsp1 cannot promote eisosome formation on its own in S. cerevisiae or A. gossypii (Walther et al., 2006; Seger et al., 2011). This may help to explain why seg1∆ had a minor effect in C. albicans, since its role in promoting eisosome formation may not be as important when Lsp1 is fully functional. Although the role of the Pkh kinases in regulating S. cerevisiae eisosome formation is controversial (Douglas and Konopka, 2014), in C. albicans, the Pkh kinases were not needed for eisosome formation. Of interest, the pkh2∆ and pkh3∆ phenotypes indicated that the Pkh kinases are important for regulating the placement and width of eisosomes, which represents new roles for these kinases. It was also unexpected that a C. albicans strain lacking Slm2 had no obvious phenotypes (Figure 5), whereas production of either Slm1 or Slm2 is required in S. cerevisiae for morphogenesis and viability (Berchtold et al., 2012). Similarly, Nce102 does not appear to have a significant influence on eisosome assembly or furrow formation in C. albicans as it does in S. cerevisiae, S. pombe, and Aspergillus nidulans (Frohlich et al., 2009; Stradalova et al., 2009; Kabeche et al., 2011; Douglas et al., 2013). These species-specific differences suggest that eisosome proteins have overlapping functions that enable some proteins to take on stronger roles in different species. This might also help to explain why eisosome proteins are not as highly conserved as is the presence of plasma membrane furrows, which are detected in a broad range of fungi, algae, and lichens (Lee et al., 2015).
An important role for C. albicans Pil1 and Lsp1 is to promote Sur7 function
The fact that Sur7 plays a key role in C. albicans makes it a good model system for determining the function of this MCC/eisosome component. Sur7 is an integral membrane protein that is predicted to span the plasma membrane four times. Of interest, the first extracellular loop of Sur7 contains an amino acid motif similar to one found at the same position in the claudin proteins, which are a family of tetraspan proteins that promote the formation of specialized membrane domains at sites of tight junctions in mammalian cells (Alvarez et al., 2008). Claudin proteins mediate contacts across tight junctions that restrict the passage of chemicals or microorganisms between cells (Furuse and Tsukita, 2006). Sur7 could play a similar type of structural role in eisosomes, perhaps mediating contact across furrows or forming a lateral scaffold.
To better define Sur7 function, we compared a sur7∆ mutant that makes eisosomes to a pil1∆ lsp1∆ mutant that lacks eisosomes. Both mutants showed abnormal cell wall invaginations (Figure 2), mislocalized actin (Figure 3), and increased sensitivity to agents that affect cell wall function (Figure 4). However, there were significant differences. For example, the sur7∆ mutant was more sensitive than the pil1∆ lsp1∆ mutant to agents that affect the cell wall, including Congo red, calcofluor white, and the Pkc1 inhibitor cercosporamide (Figure 4). In contrast, the pil1∆ lsp1∆ mutant was more sensitive than the sur7∆ mutant to agents that affect the plasma membrane, such as SDS and the ergosterol synthesis inhibitor fluconazole (Figure 4). The sur7∆ cells were also distinct, in that they were more susceptible to caffeine and rapamycin (Figure 6), whereas the pil1∆ lsp1∆ mutant had a stronger defect in invasive hyphal growth (Figure 3). These differences indicate that Sur7 and Pil1/Lsp1 have overlapping but distinct functions.
The weaker signal of Sur7-GFP in the plasma membrane of pil1∆ lsp1∆ cells led us to examine the effects of overexpressing SUR7. The results showed that SUR7 overexpression in the pil1∆ lsp1∆ mutant increased resistance to the cell wall inhibitors Congo red and calcofluor white and improved cell wall morphology (Figure 8). The ability of SUR7 overexpression to suppress pil1∆ lsp1∆ defects indicates that Sur7 can function in the absence of Pil1 and Lsp1. This indicates that an important role of eisosomes is to promote Sur7 function, most likely by stabilizing it in the membrane. Consistent with this, Sur7 is one of the most stable proteins in S. cerevisiae (Thayer et al., 2014). Eisosomes also likely have other roles in the cell, as SUR7 overexpression did not rescue the fluconazole sensitivity of the pil1∆ lsp1∆ cells (Figure 8D).
Eisosomes regulate morphogenesis
Eisosomes regulate cell wall growth in a manner distinct from the new cell wall synthesis at the tips of buds and hyphae. Eisosomes take time to form and are not present at these sites of new morphogenesis (Moreira et al., 2009; Reijnst et al., 2011). Consistent with this, sur7∆ daughter cells look relatively normal, but the older, mother cells grow larger and form cell wall invaginations (Wang et al., 2011). Similar results were observed for the pil1∆ lsp1∆ cells. The phenotypes of a C. albicans inp51∆ mutant first suggested that altered regulation of PI4,5P2 might be involved in forming the cell wall invaginations. Mutation of the PI4,5P2 phosphatase INP51 in C. albicans caused cell wall invaginations and mislocalization of septins similar to what was observed for the sur7∆ and pil1∆ lsp1∆ mutants (Alvarez et al., 2008; Badrane et al., 2012). In S. cerevisiae, an inp51∆ inp52 mutant lacking two PI4,5P2 phosphatases also forms abnormal invaginations (Singer-Kruger et al., 1998). PI4,5P2 plays important roles by influencing actin localization and recruiting morphogenesis proteins to the plasma membrane (Strahl and Thorner, 2007; Vernay et al., 2012). A PH-RFP probe that binds PI4,5P2 showed that sites of altered cell wall growth were associated with elevated levels of PI4,5P2 in the sur7∆ and pil1∆ lsp1∆ mutants (Figure 7) similar to what was reported for the inp51∆ mutant (Badrane et al., 2012). Consistent with this, septin proteins that bind PI4,5P2 (Bertin et al., 2010) and cell wall morphogenesis proteins also localized at these sites. This suggests that PI4,5P2 inappropriately recruits morphogenesis proteins that promote cell wall invaginations.
Patches of PI4,5P2 were detected in the plasma membrane of S. pombe and S. cerevisiae in response to osmotic stress, which might be related to the PI4,5P2 patches seen in C. albicans sur7∆ and pil1∆ lsp1∆ mutants (Kabeche et al., 2015). These stress-induced PI4,5P2 patches were not associated with cell wall invaginations, perhaps because they are transient. S. cerevisiae and S. pombe eisosomes have been linked to negative regulation of PI4,5P2 levels (Frohlich et al., 2014; Kabeche et al., 2014, 2015). An S. cerevisiae pil1∆ mutant that fails to recruit Inp51 to the plasma membrane displayed elevated PI4,5P2 levels (Frohlich et al., 2014). However, C. albicans Sur7 does not exclusively regulate Inp51, since the phenotype of inp51∆ cells is weaker than that of sur7∆ or pil1∆ lsp1∆ cells (Figure 7). Eisosomes in C. albicans must either regulate additional PI4,5P2 phosphatases or interfere with PI4,5P2 levels in some other way. Of interest, since PI4,5P2 is important for eisosome assembly (Olivera-Couto et al., 2011), there could be a feedback loop in which increased PI4,5P2 levels promote eisosome assembly, which in turn negatively regulates PI4,5P2 levels.
Eisosome proteins promote fungal virulence
The pil1∆ lsp1∆ mutant defects provide additional evidence that eisosomes are important for C. albicans virulence functions. Previous studies showed that a sur7∆ mutant has a very strong virulence defect in a mouse model of disseminated candidiasis (Alvarez et al., 2008). An nce102∆ mutant has decreased virulence, which is likely due to its defect in invasive hyphal growth (Douglas et al., 2013). A family of four quinone reductase proteins that localize to eisosomes in C. albicans promotes a novel antioxidant pathway that is needed for virulence (Li et al., 2015). Furthermore, the pil1∆ lsp1∆ mutant shows increased susceptibility to fluconazole (Figure 4). Thus eisosome proteins represent interesting new drug targets. Further studies on eisosomes will contribute to developing new therapeutic approaches and improving the efficacy of current antifungal drugs.
MATERIALS AND METHODS
Strains and media
The C. albicans strains are described in Supplemental Table S1. Cells were grown in either rich yeast extract/peptone/dextrose (YPD) medium or complete synthetic medium containing yeast nitrogen base, dextrose, amino acids, and uridine (Sherman, 1991). Deletion mutants lacking LSP1, PIL1, SEG1, ORF19.3505 (SLM2), PKH2, or PKH3 were constructed in C. albicans strain BWP17 essentially as described previously (Wilson et al., 1999). PCR primers containing ∼70 base pairs of sequence homologous to the sequences flanking the open reading frame of the targeted gene were used to amplify either the ARG4 or HIS1 selectable marker genes. Proper integration of the deletion cassettes was verified by PCR using combinations of primers that flanked the deleted gene and primers that annealed within the deletion cassette, as well as primers that were used to confirm the absence of the deleted gene. The ura3 auxotrophy was corrected by transforming cells with an IRO1-URA3 fragment that was released from plasmid pBSK-URA by digestion with NotI and PstI. Alternatively, URA3 was used to select for reintegration of a copy of the corresponding wild-type gene to create complemented strains. Complementing plasmids were constructed by inserting a C. albicans gene containing the open reading frame plus sequences ∼1 kb upstream and 300 base pairs downstream between the SacI and SacII sites of pDDB57. Plasmids were cut within the promoter region and transformed into the appropriate deletion mutant to create the complementing strain. The PIL1 complementing plasmid was cut with SnaBI, the LSP1 complementing plasmid with BsaBI, the PKH2 complementing plasmid with SnaBI, and the SUR7-URA3 and SUR7-NAT1 complementing plasmids with BsrGI. To overexpress SUR7, the SUR7-URA3 plasmid carrying the open reading frame and ∼1 kb upstream sequence was cut in the promoter region with BsrGI and then integrated into the genome to create strain HXWY45-1. SUR7 overexpression strain HXWY46-1 was created by integrating the SUR7-NAT1 plasmid in a similar manner, using selection for nourseothricin resistance.
A pil1∆ lsp1∆ double mutant was constructed by deleting one copy of PIL1 with ARG4 and one copy of LSP1 with HIS1, as described. The second copy of each allele was then deleted using the SAT flipper (Reuss et al., 2004). In brief, PCR primers containing ∼70 base pairs of homology to PIL1 or LSP1 were used to amplify the SAT Flipper cassette, which contains a nourseothricin resistance gene. After obtaining a deletion of the second allele of PIL1, we grew the cells on maltose medium to stimulate recombination to pop out the SAT flipper cassette and leave behind one copy of the FRT recombination site. This process was then repeated to delete the remaining copy of the LSP1 gene and create the lsp1::HIS1/lsp1::FRT pil1::ARG4/pil1::FRT strain, which for brevity is referred to as the pil1∆ lsp1 mutant.
The GFP variant was fused to the 3′ ends of the open reading frames for LSP1, PIL1, SEG1, SUR7, ORF19.3505 (SLM2), ORF19.4043, and NCE102 as described previously (Zhang and Konopka, 2010; Douglas et al., 2013). The correct integration was confirmed by PCR analysis and production of a fluorescent protein, which was detected by microscopy. The human PLCδ1 PH domain fused to RFP was introduced near the RP10 locus as described previously (Badrane et al., 2012).
Fluorescence microscopy
Strains that produce fluorescent fusion proteins were grown overnight and analyzed by microscopy using a Zeiss (Jena, Germany) Axiovert 200 M microscope equipped with an AxioCam HRm camera and Zeiss AxioVision software for deconvoluting images. Actin localization was examined by phalloidin staining. Cells were grown to log phase in YPD medium at 30°C, fixed in 5% formaldehyde for 75 min at 30°C, incubated at room temperature for 30 min in 0.1 M potassium phosphate buffer (pH 7.5) containing 0.1% Triton X-100, and then washed twice in 0.1 M potassium phosphate buffer (pH 7.5). The cells were then stained by addition of 5 U of rhodamine-phalloidin (Invitrogen, Carlsbad, CA), incubated overnight at 4°C, washed, and then examined by fluorescence microscopy. Cell-wall chitin was stained by incubating cells with 40 μg/ml calcofluor white (Fluorescent Brightener 28; Sigma-Aldrich, St. Louis, MO) as described previously (Pringle, 1991). Calcofluor white does not cross the plasma membrane, which prevents it from efficiently staining the invaginations of cell wall growth in the mutant cells. To better visualize the cell wall invaginations, we first permeabilized cells with methanol and acetone and then stained them with calcofluor white.
Electron microscopy
TEM was used to analyze cell wall invaginations in cells that were fixed with 3% EM-grade glutaraldehyde in 1× sodium cacodylate buffer (pH 7.4) at room temperature for 1 h. The fixed cells were resuspended in a 4% permanganate solution, washed, and then stained with uranyl acetate. The samples were then dehydrated by resuspending the cells in a graded series of acetone, washed with acetonitrile, and then embedded in Epon resin. A Reichert-Jung UltracutE ultramicrotome was used to cut ultrathin sections (80 nm), which were placed on Formvar-coated slot copper grids. The sections were then viewed with a Tecnai12 BioTwinG2 electron microscope (FEI, Hillsboro, OR), and an XR-60 CCD Digital Camera System (Advanced Microscopy Techniques, Woburn, MA) was used to acquire digital images. Sample preparation and TEM analyses were carried out at the Central Microscopy Imaging Center at Stony Brook University.
For analysis of membrane furrows, cells from an overnight culture were harvested by centrifugation (1 min at 1500 × g) and washed in 50 mM potassium phosphate buffer (pH 5.5). A 2-μl aliquot of the concentrated cell suspension was loaded onto a gold carrier and frozen rapidly in liquid nitrogen. The sample was cut with a cold knife (≤−185°C), etched for 4 min (−97°C; pressure ≤1.3105 Pa) in a CFE-50 freeze-etch unit (Cressington, Watford, United Kingdom), shadowed (1-nm Pt/C, 45°C; 10-nm C, 90°C), and cleaned in fresh 70% H2SO4 for 16 h (Rachel et al., 2002). Air-dried sample surface replicas were examined in an FEI Morgagni 268(D) transmission electron microscope at 80 kV. Images were captured with a Megaview II (Olympus Corp., Münster, Germany) charge-coupled device camera.
Growth inhibition assays
Spot assays were used to test the sensitivity of cells to SDS, Congo red, calcofluor white, rapamycin, and caffeine. A 10-fold dilution series of cells was prepared, and then 3 μl of each dilution was spotted on the surface of YPD plates containing the indicated concentration of chemical. The plates were incubated at 37°C for 2 d and then photographed to record the extent of growth. The sensitivity of cells to cercosporamide was assayed by testing the ability of cells to grow in a twofold dilution series of different concentrations of cercosporamide in 96-well plates at 37°C for 2 d. The extent of growth in each well was then quantified using a spectrophotometer. The sensitivity of cells to fluconazole was assayed using Etest strips (AB Biodisk North America, Piscataway, NJ). We spread 1 × 106 cells on an agar plate containing RPMI 1640 medium (Sigma-Aldrich), 0.165 M 3-(N-morpholino)propanesulfonic acid, pH 7, and 2% dextrose. An Etest strip was applied to the surface of each agar plate; the plates were incubated at 37°C for 48 h and then photographed. Similar studies were also carried out by placing filter disks carrying different amounts of fluconazole on the lawns of cells spread on an agar plate.
Supplementary Material
Acknowledgments
We thank Scott Filler for providing the pBSK-URA plasmid and Cornelius Clancy and Hassan Badrane for providing the PH domain–RFP fusion plasmid. This work was supported by a Public Health Service grant from the National Institute of Allergy and Infectious Diseases (AI-47837; to J.B.K.) and by the Czech Science Foundation (project 15-10641S; to P.V. and J.M.).
Abbreviations used:
- GFP
green fluorescent protein
- MCC
membrane compartment of Can1
- MCP
membrane compartment of Pma1
- PI4,5P2
phosphatidylinositol 4,5-bisphosphate
- RFP
red fluorescent protein
- TEM
transmission electron microscopy.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-01-0065) on March 23, 2016.
REFERENCES
- Alvarez FJ, Douglas LM, Rosebrock A, Konopka JB. The Sur7 protein regulates plasma membrane organization and prevents intracellular cell wall growth in Candida albicans. Mol Biol Cell. 2008;19:5214–5225. doi: 10.1091/mbc.E08-05-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badrane H, Nguyen MH, Blankenship JR, Cheng S, Hao B, Mitchell AP, Clancy CJ. Rapid redistribution of phosphatidylinositol-(4,5)-bisphosphate and septins during the Candida albicans response to caspofungin. Antimicrob Agents Chemother. 2012;56:4614–4624. doi: 10.1128/AAC.00112-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beese SE, Negishi T, Levin DE. Identification of positive regulators of the yeast fps1 glycerol channel. PLoS Genet. 2009;5:e1000738. doi: 10.1371/journal.pgen.1000738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berchtold D, Piccolis M, Chiaruttini N, Riezman I, Riezman H, Roux A, Walther TC, Loewith R. Plasma membrane stress induces relocalization of Slm proteins and activation of TORC2 to promote sphingolipid synthesis. Nat Cell Biol. 2012;14:542–547. doi: 10.1038/ncb2480. [DOI] [PubMed] [Google Scholar]
- Bernardo SM, Lee SA. Candida albicans SUR7 contributes to secretion, biofilm formation, and macrophage killing. BMC Microbiol. 2010;10:133. doi: 10.1186/1471-2180-10-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertin A, McMurray MA, Thai L, Garcia G, 3rd, Votin V, Grob P, Allyn T, Thorner J, Nogales E. Phosphatidylinositol-4,5-bisphosphate promotes budding yeast septin filament assembly and organization. J Mol Biol. 2010;404:711–731. doi: 10.1016/j.jmb.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas LM, Konopka JB. Fungal membrane organization: the eisosome concept. Annu Rev Microbiol. 2014;68:377–393. doi: 10.1146/annurev-micro-091313-103507. [DOI] [PubMed] [Google Scholar]
- Douglas LM, Wang HX, Keppler-Ross S, Dean N, Konopka JB. Sur7 promotes plasma membrane organization and is needed for resistance to stressful conditions and to the invasive growth and virulence of Candida albicans. MBio. 2012;3:e00254-00211. doi: 10.1128/mBio.00254-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas LM, Wang HX, Konopka JB. The MARVEL domain protein Nce102 regulates actin organization and invasive growth of Candida albicans. MBio. 2013;4:e00723-13. doi: 10.1128/mBio.00723-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohlich F, Christiano R, Olson DK, Alcazar-Roman A, DeCamilli P, Walther TC. A role for eisosomes in maintenance of plasma membrane phosphoinositide levels. Mol Biol Cell. 2014;25:2797–2806. doi: 10.1091/mbc.E13-11-0639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohlich F, Moreira K, Aguilar PS, Hubner NC, Mann M, Walter P, Walther TC. A genome-wide screen for genes affecting eisosomes reveals Nce102 function in sphingolipid signaling. J Cell Biol. 2009;185:1227–1242. doi: 10.1083/jcb.200811081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse M, Tsukita S. Claudins in occluding junctions of humans and flies. Trends Cell Biol. 2006;16:181–188. doi: 10.1016/j.tcb.2006.02.006. [DOI] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O’Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Inagaki M, Schmelzle T, Yamaguchi K, Irie K, Hall MN, Matsumoto K. PDK1 homologs activate the Pkc1-mitogen-activated protein kinase pathway in yeast. Mol Cell Biol. 1999;19:8344–8352. doi: 10.1128/mcb.19.12.8344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeche R, Baldissard S, Hammond J, Howard L, Moseley JB. The filament-forming protein Pil1 assembles linear eisosomes in fission yeast. Mol Biol Cell. 2011;22:4059–4067. doi: 10.1091/mbc.E11-07-0605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeche R, Madrid M, Cansado J, Moseley JB. Eisosomes regulate PI(4,5)P2 cortical clusters and mitogen-activated protein (MAP) kinase signaling upon osmotic stress. J Biol Chem. 2015;290:25960–25973. doi: 10.1074/jbc.M115.674192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeche R, Roguev A, Krogan NJ, Moseley JB. A Pil1-Sle1-Syj1-Tax4 functional pathway links eisosomes with PI(4,5)P2 regulation. J Cell Sci. 2014;127:1318–1326. doi: 10.1242/jcs.143545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamble C, Jain S, Murphy E, Kim K. Requirements of Slm proteins for proper eisosome organization, endocytic trafficking and recycling in the yeast Saccharomyces cerevisiae. J Biosci. 2011;36:79–96. doi: 10.1007/s12038-011-9018-0. [DOI] [PubMed] [Google Scholar]
- Karotki L, Huiskonen JT, Stefan CJ, Ziolkowska NE, Roth R, Surma MA, Krogan NJ, Emr SD, Heuser J, Grunewald K, et al. Eisosome proteins assemble into a membrane scaffold. J Cell Biol. 2011;195:889–902. doi: 10.1083/jcb.201104040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFayette SL, Collins C, Zaas AK, Schell WA, Betancourt-Quiroz M, Gunatilaka AA, Perfect JR, Cowen LE. PKC signaling regulates drug resistance of the fungal pathogen Candida albicans via circuitry comprised of Mkc1, calcineurin, and Hsp90. PLoS Pathog. 2010;6:e1001069. doi: 10.1371/journal.ppat.1001069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Heuser JE, Roth R, Goodenough U. Eisosome ultrastructure and evolution in fungi, microalgae and lichens. Eukaryot Cell. 2015;14:1017–1042. doi: 10.1128/EC.00106-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Naseem S, Sharma S, Konopka JB. Flavodoxin-like proteins protect Candida albicans from oxidative stress and promote virulence. PLoS Pathog. 2015;11:e1005147. doi: 10.1371/journal.ppat.1005147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinska K, Malinsky J, Opekarova M, Tanner W. Visualization of protein compartmentation within the plasma membrane of living yeast cells. Mol Biol Cell. 2003;14:4427–4436. doi: 10.1091/mbc.E03-04-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinska K, Malinsky J, Opekarova M, Tanner W. Distribution of Can1p into stable domains reflects lateral protein segregation within the plasma membrane of living S. cerevisiae cells. J Cell Sci. 2004;117:6031–6041. doi: 10.1242/jcs.01493. [DOI] [PubMed] [Google Scholar]
- Malinsky J, Opekarova M, Grossmann G, Tanner W. Membrane microdomains, rafts, and detergent-resistant membranes in plants and fungi. Annu Rev Plant Biol. 2013;64:501–529. doi: 10.1146/annurev-arplant-050312-120103. [DOI] [PubMed] [Google Scholar]
- Malinsky J, Opekarova M, Tanner W. The lateral compartmentation of the yeast plasma membrane. Yeast. 2010;27:473–478. doi: 10.1002/yea.1772. [DOI] [PubMed] [Google Scholar]
- Moreira KE, Schuck S, Schrul B, Frohlich F, Moseley JB, Walther TC, Walter P. Seg1 controls eisosome assembly and shape. J Cell Biol. 2012;198:405–420. doi: 10.1083/jcb.201202097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira KE, Walther TC, Aguilar PS, Walter P. Pil1 controls eisosome biogenesis. Mol Biol Cell. 2009;20:809–818. doi: 10.1091/mbc.E08-03-0313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy ER, Boxberger J, Colvin R, Lee SJ, Zahn G, Loor F, Kim K. Pil1, an eisosome organizer, plays an important role in the recruitment of synaptojanins and amphiphysins to facilitate receptor-mediated endocytosis in yeast. Eur J Cell Biol. 2011;90:825–833. doi: 10.1016/j.ejcb.2011.06.006. [DOI] [PubMed] [Google Scholar]
- Olivera-Couto A, Grana M, Harispe L, Aguilar PS. The eisosome core is composed of BAR domain proteins. Mol Biol Cell. 2011;22:2360–2372. doi: 10.1091/mbc.E10-12-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle JR ( Staining of bud scars and other cell wall chitin with calcofluor. Methods Enzymol. 1991;194:732–735. doi: 10.1016/0076-6879(91)94055-h. [DOI] [PubMed] [Google Scholar]
- Rachel R, Wyschkony I, Riehl S, Huber H. The ultrastructure of Ignicoccus: evidence for a novel outer membrane and for intracellular vesicle budding in an archaeon. Archaea. 2002;1:9–18. doi: 10.1155/2002/307480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijnst P, Walther A, Wendland J. Dual-colour fluorescence microscopy using yEmCherry-/GFP-tagging of eisosome components Pil1 and Lsp1 in Candida albicans. Yeast. 2011;28:331–338. doi: 10.1002/yea.1841. [DOI] [PubMed] [Google Scholar]
- Reuss O, Vik A, Kolter R, Morschhauser J. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene. 2004;341:119–127. doi: 10.1016/j.gene.2004.06.021. [DOI] [PubMed] [Google Scholar]
- Roelants FM, Torrance PD, Bezman N, Thorner J. Pkh1 and pkh2 differentially phosphorylate and activate ypk1 and ykr2 and define protein kinase modules required for maintenance of cell wall integrity. Mol Biol Cell. 2002;13:3005–3028. doi: 10.1091/mbc.E02-04-0201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seger S, Rischatsch R, Philippsen P. Formation and stability of eisosomes in the filamentous fungus Ashbya gossypii. J Cell Sci. 2011;124:1629–1634. doi: 10.1242/jcs.082487. [DOI] [PubMed] [Google Scholar]
- Sherman F ( Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- Simons K, Gerl MJ. Revitalizing membrane rafts: new tools and insights. Nat Rev Mol Cell Biol. 2010;11:688–699. doi: 10.1038/nrm2977. [DOI] [PubMed] [Google Scholar]
- Singer-Kruger B, Nemoto Y, Daniell L, Ferro-Novick S, De Camilli P. Synaptojanin family members are implicated in endocytic membrane traffic in yeast. J Cell Sci. 1998;111:3347–3356. doi: 10.1242/jcs.111.22.3347. [DOI] [PubMed] [Google Scholar]
- Stradalova V, Blazikova M, Grossmann G, Opekarova M, Tanner W, Malinsky J. Distribution of cortical endoplasmic reticulum determines positioning of endocytic events in yeast plasma membrane. PloS One. 2012;7:e35132. doi: 10.1371/journal.pone.0035132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stradalova V, Stahlschmidt W, Grossmann G, Blazikova M, Rachel R, Tanner W, Malinsky J. Furrow-like invaginations of the yeast plasma membrane correspond to membrane compartment of Can1. J Cell Sci. 2009;122:2887–2894. doi: 10.1242/jcs.051227. [DOI] [PubMed] [Google Scholar]
- Strahl T, Thorner J. Synthesis and function of membrane phosphoinositides in budding yeast, Saccharomyces cerevisiae. Biochim Biophys Acta. 2007;1771:353–404. doi: 10.1016/j.bbalip.2007.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Thapa N, Hedman AC, Anderson RA. Phosphatidylinositol 4,5-bisphosphate: targeted production and signaling. Bioessays. 2013;35:513–522. doi: 10.1002/bies.201200171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman A, Huss K, Chio LC, Heidler S, Shaw M, Ma D, Zhu G, Campbell RM, Park TS, Kulanthaivel P, et al. Discovery of cercosporamide, a known antifungal natural product, as a selective Pkc1 kinase inhibitor through high-throughput screening. Eukaryot Cell. 2004;3:932–943. doi: 10.1128/EC.3.4.932-943.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thayer NH, Leverich CK, Fitzgibbon MP, Nelson ZW, Henderson KA, Gafken PR, Hsu JJ, Gottschling DE. Identification of long-lived proteins retained in cells undergoing repeated asymmetric divisions. Proc Natl Acad Sci USA. 2014;111:14019–14026. doi: 10.1073/pnas.1416079111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernay A, Schaub S, Guillas I, Bassilana M, Arkowitz RA. A steep phosphoinositide bis-phosphate gradient forms during fungal filamentous growth. J Cell Biol. 2012;198:711–730. doi: 10.1083/jcb.201203099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther TC, Aguilar PS, Frohlich F, Chu F, Moreira K, Burlingame AL, Walter P. Pkh-kinases control eisosome assembly and organization. EMBO J. 2007;26:4946–4955. doi: 10.1038/sj.emboj.7601933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther TC, Brickner JH, Aguilar PS, Bernales S, Pantoja C, Walter P. Eisosomes mark static sites of endocytosis. Nature. 2006;439:998–1003. doi: 10.1038/nature04472. [DOI] [PubMed] [Google Scholar]
- Wang HX, Douglas LM, Aimanianda V, Latge JP, Konopka JB. The Candida albicans Sur7 protein is needed for proper synthesis of the fibrillar component of the cell wall that confers strength. Eukaryot Cell. 2011;10:72–80. doi: 10.1128/EC.00167-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RB, Davis D, Mitchell AP. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J Bacteriol. 1999;181:1868–1874. doi: 10.1128/jb.181.6.1868-1874.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young ME, Karpova TS, Brugger B, Moschenross DM, Wang GK, Schneiter R, Wieland FT, Cooper JA. The Sur7p family defines novel cortical domains in Saccharomyces cerevisiae, affects sphingolipid metabolism, and is involved in sporulation. Mol Cell Biol. 2002;22:927–934. doi: 10.1128/MCB.22.3.927-934.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Konopka JB. A photostable green fluorescent protein variant for analysis of protein localization in Candida albicans. Eukaryot Cell. 2010;9:224–226. doi: 10.1128/EC.00327-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziolkowska NE, Christiano R, Walther TC. Organized living: formation mechanisms and functions of plasma membrane domains in yeast. Trends Cell Biol. 2012;22:151–158. doi: 10.1016/j.tcb.2011.12.002. [DOI] [PubMed] [Google Scholar]
- Ziolkowska NE, Karotki L, Rehman M, Huiskonen JT, Walther TC. Eisosome-driven plasma membrane organization is mediated by BAR domains. Nat Struct Mol Biol. 2011;18:854–856. doi: 10.1038/nsmb.2080. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.