RBM4 modulates alternative exon selection of Numb and up-regulates proneural Mash1 gene expression, possibly via specific Numb isoforms. RBM4 overexpression promotes neuronal cell differentiation. Moreover, RBM4 is essential for neurite outgrowth in primary cortical neurons by modulating specific Numb isoform expression.
Abstract
RBM4 participates in cell differentiation by regulating tissue-specific alternative pre-mRNA splicing. RBM4 also has been implicated in neurogenesis in the mouse embryonic brain. Using mouse embryonal carcinoma P19 cells as a neural differentiation model, we observed a temporal correlation between RBM4 expression and a change in splicing isoforms of Numb, a cell-fate determination gene. Knockdown of RBM4 affected the inclusion/exclusion of exons 3 and 9 of Numb in P19 cells. RBM4-deficient embryonic mouse brain also exhibited aberrant splicing of Numb pre-mRNA. Using a splicing reporter minigene assay, we demonstrated that RBM4 promoted exon 3 inclusion and exon 9 exclusion. Moreover, we found that RBM4 depletion reduced the expression of the proneural gene Mash1, and such reduction was reversed by an RBM4-induced Numb isoform containing exon 3 but lacking exon 9. Accordingly, induction of ectopic RBM4 expression in neuronal progenitor cells increased Mash1 expression and promoted cell differentiation. Finally, we found that RBM4 was also essential for neurite outgrowth from cortical neurons in vitro. Neurite outgrowth defects of RBM4-depleted neurons were rescued by RBM4-induced exon 9–lacking Numb isoforms. Therefore our findings indicate that RBM4 modulates exon selection of Numb to generate isoforms that promote neuronal cell differentiation and neurite outgrowth.
INTRODUCTION
A variety of biological processes, including cell differentiation and development, involve alternative pre-mRNA splicing. Splicing regulation is essentially governed by regulatory factors that bind to cis-elements in regulated exons or flanking introns and/or act through protein–protein interactions (Fedor, 2008; Coelho and Smith, 2014). RNA-binding motif protein 4 (RBM4) regulates alternative splicing of pre-mRNAs and is also involved in cell differentiation (Lin and Tarn, 2011; Lin et al., 2013, 2014). Mammalian genomes have two copies of Rbm4, namely Rbm4a and Rbm4b. Knockout of either Rbm4 reduces the size of embryonic pancreatic islets (Lin et al., 2013). Rbm4 knockout results in aberrant splicing patterns of several pancreatic factors and reduction in insulin gene expression in embryonic pancreas (Lin et al., 2013). These observations provide in vivo evidence that RBM4 is important for cell differentiation and development via its role in alternative splicing and gene expression.
The nervous system adopts alternative splicing for cell differentiation, morphogenesis, and even synapse formation and plasticity. A number of RNA-binding proteins, such as Nova, Rbfox, nSR100, and polypyrimidine tract-binding protein, are involved in splicing regulation during neuronal cell differentiation (Grabowski, 2011; Norris and Calarco, 2012; Zheng and Black, 2013). Genetic knockouts and cross-linking/immunoprecipitation/high-throughput sequencing experiments have revealed the details of splicing mechanisms involving these factors, as well as their roles in neural function and development (Grabowski, 2011; Norris and Calarco, 2012; Zheng and Black, 2013). For example, Nova2 knockout impairs the splicing isoform switch of Disabled-1 in the developing cerebral cortex and thus disrupts neuronal migration during lamina formation (Yano et al., 2010). Nova binds to YCAY nucleotide sequences (Y stands for pyrimidine) near regulated exons, and its binding shows position-dependent effects on splicing regulation (Licatalosi et al., 2008; Yano et al., 2010). The members of each splicing regulator family may exhibit different anatomical and temporal expression and therefore have different functions in brain. RBM4 has been detected in the cortical plate of embryonic mouse brain during gestation, and its level is reduced in fetal Down syndrome brain (Bernert et al., 2002), suggesting that RBM4 plays a role in neuronal differentiation and perhaps participates in pathogenesis.
In the present study, we find that Rbm4 knockout altered exon usage of Numb, a key cell-fate determination factor for neural development (Yan, 2010). During neurogenesis, Drosophila Numb plays a critical role in asymmetric cell division and specifies the fate of sibling neuron cells (Yan, 2010). Mammalian Numb has multiple functions during neural development, including maintaining neural progenitor cells through symmetric divisions, as well as promoting neuronal differentiation and even coordinating the polarization of migratory neurons (Li et al., 2003; Breunig and Rakic, 2011). Mammalian Numb genes express mRNA isoforms essentially via differential selection of two alternative exons (exons 3 and 9 in mouse Numb), generating protein isoforms containing different lengths of the phosphotyrosine-binding (PTB) domain and the proline-rich region (PRR) domain (Verdi et al., 1999). A switch in Numb isoforms is crucial for cortical development (Bani-Yaghoub et al., 2007). The long PRR (PRRL)–containing isoforms promote cell proliferation, whereas the short PRR (PRRS)–containing isoforms enhance differentiation in cultured neuronal cells (Verdi et al., 1999; Bani-Yaghoub et al., 2007). Numb is involved in Notch signaling, which maintains neuronal progenitor cells and inhibits their differentiation into neurons (Louvi and Artavanis-Tsakonas, 2006). In Drosophila, Numb promotes neuronal differentiation by antagonizing Notch signaling (Spana and Doe, 1996). Numb-PTBL isoforms negatively regulate Notch1 activity by promoting its ubiquitination and subsequent degradation (McGill and McGlade, 2003; McGill et al., 2009). Moreover, Numb isoforms function differently in neurite outgrowth and cell death in response to neurotrophic factors and changes in neuronal activity (Pedersen et al., 2002; Lu et al., 2009).
Several splicing regulators can modulate Numb exon selection. The neuron-specific splicing factor Rbfox3 represses the inclusion exon 12 (equivalent to rodent exon 9) of chick Numb, resulting in PRRS isoforms that are critical for neuronal differentiation and spinal cord development (Kim et al., 2013). Aberrant alternative splicing of Numb is also implicated in tumorigenesis (Bechara et al., 2013; Zong et al., 2014). The RNA-binding protein QKI suppresses exon 12 inclusion of Numb, producing isoforms that inhibit cell proliferation. Down-regulation of QKI-5 in lung cancer promotes tumorigenesis (Zong et al., 2014). A similar scenario was observed in another Numb splicing regulator, RBM10, whose mutations in lung cancer also contribute to tumor progression (Bechara et al., 2013). Because the isoform switch of Numb is important for neuronal development and cancer pathogenesis, it is necessary to understand its underlying mechanism.
Our preliminary observation that Rbm4 knockout resulted in abnormal Numb isoform ratios in embryonic brain prompted us to characterize the role of RBM4 in regulating Numb splicing and neuronal differentiation via Numb-mediated pathways.
RESULTS
RBM4 expression and the splicing switch of neuronal transcripts during neuronal differentiation
The fact that RBM4 is expressed in mouse embryonic brain (Brooks et al., 2009) prompted us to assess the role of RBM4 in brain development. However, Rbm4 single-gene knockout did not exhibit visible defects in brain (unpublished data). We then performed Golgi staining of newborn neurons but did not observe any significant difference in the number of primary dendrites and axonal length between wild-type and knockout mice (Supplemental Figure S1). Nevertheless, we cultured cortical neurons isolated from Rbm4a−/− embryos. The growth rate of Rbm4a-knockout neurites was slightly slower than that of wild type at 1 d in vitro (DIV1) but had no significant difference from the wild type at a later time (DIV3; Supplemental Figure S2). We reasoned that two Rbm4 genes may functionally compensate for each other.
We then used mouse embryonal carcinoma P19 cells to examine RBM4 expression during neuronal differentiation. We treated the cells with retinoic acid (RA) to form embryoid bodies. After RA treatment, we cultured cells in a neurobasal medium in the presence of a proliferation inhibitor (see Materials and Methods). On induction of differentiation, the cells exhibited protruding processes and expressed the neuronal marker MAP2 (Figure 1). Rbm4b mRNA level slightly increased until day 2 and then decreased, whereas Rbm4a abundance was essentially constant (Figure 1). Immunoblotting using anti-RBM4 showed that the level of total RBM4 was transiently up-regulated and also peaked at day 2 (Figure 1).
FIGURE 1:

The expression pattern of RBM4 and neuronal transcript isoforms during neuronal differentiation of P19 cells. P19 cells (2 × 106) were cultured in the differentiation medium containing 1 μM RA to allow embryoid body formation (undifferentiated; UD). Four days later, 1 × 106 cells were reseeded into a six-well plate, and the media were changed to the neurobasal medium containing B27 supplement (see Materials and Methods) to allow neuronal differentiation (D0). After 2 d (D2), cells were treated with 10 μM cytosine β-d-arabinofuranoside hydrochloride (AraC). Cells were harvested on UD, D0, D2, D4, and D6. Total RNA was isolated and analyzed by RT-PCR using primers complementary to the indicated transcripts and isoforms as listed in Supplemental Table S1. Total cell lysate proteins were analyzed by immunoblotting using antibodies against RBM4 and GAPDH.
We previously demonstrated that RBM4 binds preferentially to CU-rich elements (Lin and Tarn, 2011). Therefore we selected a set of previously reported neuronal transcripts that exhibit a differentiation-associated change in splicing pattern and contain CU-rich sequences near regulated exons (Salomonis et al., 2008). We then examined their splicing patterns in P19 cells. All examined transcripts (CLCB, Smap1, Kif1b, PKM, and Numb) exhibited a splicing switch during P19 cell differentiation (Figure 1). Kif1b is a direct target of RBM4 (Lu et al., 2013). Our result thus showed that the switch of Kif1bβ3 to Kif1bβ, which involves exon 25 inclusion, indeed correlated well with the up-regulation of RBM4 (Figure 1). Among these genes, Numb is important for neuronal development (Verdi et al., 1999; Bani-Yaghoub et al., 2007). Mouse Numb isoform expression during neuronal development essentially involves differential selection of exon 3 (E3) and E9 (Bani-Yaghoub et al., 2007). Induction of neuronal differentiation immediately resulted in E3 inclusion and E9 skipping (day 0), which coincided with the increase in total RBM4 protein (Figure 1). We thus inferred that RBM4 might promote E3 inclusion and E9 skipping. Nevertheless, a splicing suppressor might counteract the effect of RBM4 on E3 at and after D2, resulting in E3 skipping (Figure 1).
RBM4 modulates alternative splicing of Numb
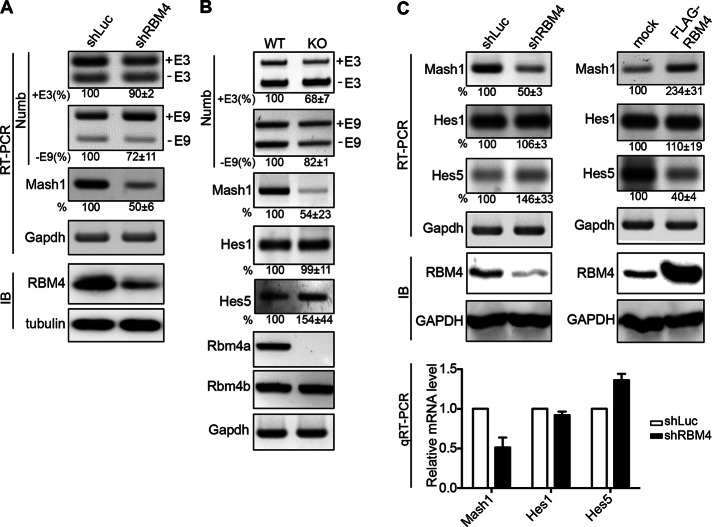
To evaluate whether RBM4 is involved in Numb pre-mRNA splicing, we attempted to deplete RBM4 in P19 cells using short hairpin RNA (shRNA) targeting RBM4 (shRBM4). We transfected cells with a vector expressing shRBM4. RBM4 protein level was reduced by ∼70% (Figure 2A). Under this condition, both E3 inclusion and E9 skipping were suppressed (∼10 and ∼30%, respectively; Figure 2A). In addition, we examined Numb isoform expression in Rbm4a-knockout mice. RNA was extracted from mouse whole brain at embryonic day 13.5 (E13.5) and subjected to reverse transcription-PCR (RT-PCR). The absence of Rbm4a mRNA confirmed Rbm4a knockout. Compared with wild-type littermates, E3 inclusion and E9 skipping were reduced by ∼30 and ∼20%, respectively, in Rbm4a-knockout embryonic brain (Figure 2B). These data further suggested that RBM4 modulates Numb isoform expression.
FIGURE 2:
RBM4 knockdown impairs Numb exon selection and reduces Mash1 expression. (A) P19 cells were transfected with plasmid expressing RBM4-targeting shRNA (shRBM4) or luciferase-targeting shRNA (shLuc), followed by treatment with 1 μM RA for 2 d and subsequently in the neural basal medium for 2 d. RNA and protein were respectively analyzed by RT-PCR and immunoblotting using specific primers and antibodies as indicated. E3 inclusion (+E3) and E9 skipping (−E9) percentages were calculated by the ratio of +E3 (or −E9) to total in individual lanes (control was set as 100%). Relative Mash1 level is indicated. The averages and SEs were obtained from three independent experiments; the same applies hereafter. (B) Total RNA was isolated from E13.5 brain of Rbm4a-knockout and wild-type littermate embryos and analyzed by RT-PCR using gene specific primers. Splicing efficiency calculation was as in A. Relative Mash1, Hes1, and Hes5 levels were measured from three pairs of mouse embryos. The averages and SEs are indicted at the bottom of the individual gels; the wild type was set as 100%. (C) P19 cells were transiently transfected with the vector expressing shRBM4 (left) or FLAG-RBM4 (right); shLuc and empty vector were used as control in their respective transfections. RT-PCR, immunoblotting and calculations of Mash1, Hes1, and Hes5 levels were essentially as in B. RNA from the shRNA transfectants was also analyzed by quantitative RT-PCR; bar graph shows averages with SEs of three independent experiments.
To confirm the effect of RBM4 on the utilization of both alternative exons of Numb, we generated Numb minigene reporters containing either E3 or E9 (Figure 3A). Overexpression of human RBM4 promoted E3 inclusion in both P19 and HEK293T cells (Figure 3B). RBM4 overexpression caused E9 skipping in P19 cells, but its effect was less apparent in HEK293T cells (Figure 3B). We reasoned that the E9-skipped transcript predominated in HEK293T cells, so that its level could not be further enhanced by RBM4 overexpression. Finally, using immunoprecipitation and RT-PCR, we observed that RBM4 associated with the Numb transcript but not Mash1, which encodes a transcription factor essential for neural differentiation (Figure 3C). Together these results suggest that RBM4 might directly regulate the selection of both functionally important exons of Numb.
FIGURE 3:
RBM4 regulates Numb E3 and E9 usage in a minigene assay. (A) Schematic representation of the Numb E3 and E9 minigene constructs. Primers used in RT-PCR (B) are indicated by arrows, and their sequences are listed in Supplemental Table S1. (B) The minigene assay was performed in P19 cells (left) and HEK293T cells (right). Cells were transiently transfected with the Numb E3 or E9 minigene plasmid and empty or RBM4 expression vector. Thirty hours posttransfection, total RNA was isolated and analyzed by RT-PCR. Relative E3 inclusion (+E3) and E9 skipping (−E9) efficiencies in P19 cells were calculated as in Figure 2A. (C) Transfection of P19 cells was as in B. Cell lysates were subjected to immunoprecipitation using anti-FLAG M2 beads. Input and bound RNAs were analyzed by RT-PCR using primers complementary to Mash1 or Numb (spanning the region of E8 and the downstream intron; Supplemental Table S1). We used 2 and 5% of the inputs in RT-PCR and immunoblotting, respectively.
RBM4 modulates the expression of neuronal differentiation genes
We next evaluated whether RBM4 also influences the expression of Notch target genes. Compared with the wild type, the levels of the hairy and Enhancer-of-split 1 (Hes1) mRNAs in E13.5 brain of Rbm4-deficint mice were not significantly altered, whereas the Hes5 mRNA level was increased by ∼50% (Figure 2B). A similar result was observed in RBM4-knockdown P19 cells, in which Hes5 mRNA level was elevated but Hes1 remained essentially unchanged (Figure 2C, left). Overexpression of RBM4 reduced the level of Hes5 by ∼60% in P19 cells, whereas the Hes1 level was only minimally changed (Figure 2C, right). Therefore RBM4 had a more prominent effect on Hes5 expression and had little or no effect on Hes1.
We next examined the level of Mash1, a downstream target of Hes proteins (Kageyama et al., 2010). Mash1 mRNA level correlated with that of RBM4 protein level during P19 cell differentiation (Figure 1) and was significantly down-regulated in RBM4-depleted P19 cells and Rbm4a-knockout brain (Figure 2, A–C, left). The expression of Mash1 was up-regulated in P19 cells upon RBM4 overexpression (Figure 2C, right), suggesting that RBM4 promotes Mash1 expression.
RBM4 modulates the expression of Mash1 via specific Numb isoforms
Because Numb-PRRS isoforms can induce Mash1 expression (Bani-Yaghoub et al., 2007), we examined whether any Numb isoform could rescue Mash1 expression in RBM4-knockdown cells. We transfected P19 cells with the RBM4-targeting shRNA vector twice with a 2-d interval. We added an expression vector encoding FLAG-tagged RBM4 or each of the FLAG-tagged Numb isoforms (Figure 4A) during the second transfection. Immunoblotting using anti-FLAG antibody showed appropriate expression of RBM4 and Numb isoforms in transfected cells (Figure 4B, lane 3, RBM4; lanes 6–9, Numb isoforms). RT-PCR revealed that RBM4 could restore Mash1 expression (lane 3). Among Numb isoforms, the PRRS isoforms, particularly Numb2, rescued Mash1 more efficiently (lanes 7 and 9), whereas Numb3 (PTBS-PRRL) failed (lane 8). Because RBM4 promoted E9 skipping and might thus increase the relative proportion of PRRS isoforms, our results support a role for RBM4 in promoting Mash1 expression, perhaps via Numb-mediated pathways.
FIGURE 4:
Numb rescues Mash1 expression in RBM4-knockdown P19 cells in an isoform-specific manner. (A) Schematic of four Numb isoforms; the different lengths of the PTB and PRR domains result from differential usage of E3 and E9, respectively. (B) P19 cells were transfected by the shLuc or shRBM4 vector twice in a 2-d interval. At the second transfection, the mock vector (lanes 2 and 5) or the vector encoding FLAG-RBM4 (lane 3) or FLAG-Numb1-4 (lanes 6–9) was added. Cells were harvested 2 d after transfection. Total RNA was isolated for RT-PCR (Mash1 and GAPDH). Total protein was subjected to immunoblotting using anti–α-tubulin or anti-RBM4, which detected both endogenous and overexpressed RBM4. Right, bar graph showing quantitation of the RT-PCR signal intensities; average values were derived from three experiments. Statistical analysis between treatment groups was performed with Student’s t test. *p < 0.05.
Overexpression of RBM4 promotes neuronal differentiation
The result that RBM4 overexpression up-regulated Mash1 prompted us to evaluate whether RBM4 promotes neuronal differentiation. We used a neural stem/progenitor cell line, KT98, derived from transgenic mice with SV40 T-antigen–induced brain tumors bearing a stable green fluorescent protein transgene under the control of the brain-specific FGF1 promoter (Hsu et al., 2009). We established a stable clone that could express FLAG-tagged RBM4 upon doxycycline induction and then selected GFP-positive cells with self-renewal and multipotency properties using fluorescence-activated cell sorting. Doxycycline treatment induced RBM4 expression (Figure 5A). Under this condition, the level of Mash1 was elevated by ∼30%, consistent with that observed in P19 cells (Figure 2C). On RBM4 induction, Numb E3 inclusion increased, albeit minimally (∼10%), but E9 skipping was not enhanced, possibly because it predominated in untreated cells (Figure 5A). Furthermore, to examine whether RBM4 could promote neuronal differentiation, we cultured cells in medium containing growth factors to allow neurosphere formation, followed by differentiation. Doxycycline treatment increased neurite length by 50% (Figure 5B), suggesting that RBM4 overexpression promoted neuronal differentiation.
FIGURE 5:

RBM4 enhances Mash1 expression and promotes differentiation of neuronal progenitor KT98 cells. (A) KT98-RBM4 cells that contained F1B-driven GFP and RBM4 transgene were cultured as described in Materials and Methods. RBM4 overexpression from the transgene was induced by 2 μg/ml doxycycline for 7 d. Total RNA was isolated from cells for RT-PCR. Mash1 and GAPDH were visualized by agarose gel electrophoresis; Numb isoforms were detected by Southern blotting using 32P-radiolabeled primers. Relative exon inclusion or exclusion efficiencies were calculated as in Figure 2A. Bar graph shows quantitative RT-PCR of the level of Mash1 mRNA in mock- or doxycycline-treated cells from three experiments. (B) KT98-RBM4 cells were induced into neurospheres and followed by differentiation induction in medium containing B27 and growth factors (see Materials and Methods). Doxycycline (2 μg/ml) was added at the first day of differentiation induction. The averages of neurite length were measured from >100 neurite-bearing cells. *p < 0.05.
RBM4 is involved in neurite outgrowth
Previous reports indicated that Numb also controls neurite outgrowth of neuroblastic PC-12 cells (Lu et al., 2009). Rbm4a−/− cortical neurons exhibited poor neurite outgrowth compared with the wild type when cultured for a short period of time (Supplemental Figure S2). We therefore examined whether Numb isoform expression changed during neuronal cell development. We isolated cortical neurons from E18.5 rat embryos and subjected them to in vitro culture for up to 9 d. MAP2 expression increased throughout the culture period (Figure 6A). Although Rbm4 mRNAs were up-regulated, total RBM4 protein level did not increase (Figure 6A). We observed a gradual increase in E3 inclusion and E9 skipping of Numb (Figure 6A). To test whether RBM4 is involved in neurite elongation, we cotransfected the expression vectors for shRBM4 and GFP into cortical neurons at DIV1. Using GFP as indicator of shRNA transfection, we observed that the neurite length of RBM4-knockdown neurons was reduced by ∼40% compared with mock-treated cells at DIV3 (Figure 6B). Next we cotransfected an RBM4 or Numb isoform expression vector with the shRNA and GFP vectors and found that RBM4 and either of the PRRS isoforms (Numb2 or 4) could rescue neurite outgrowth of RBM4-depleted cortical neurons (Figure 6B). The activity of the PRRS isoforms in neurite elongation was coincident with E9 skipping, which prevailed at later stages of neuronal development (Figures 1 and 6). Therefore our data suggested that RBM4 may regulate Numb isoform expression, which in turn influences downstream pathways linked to neurite outgrowth.
FIGURE 6:
RBM4-induced Numb isoforms restore neurite outgrowth in RBM4-depleted cortical neurons. (A) Cortical neurons from E18.5 rat embryos were cultured in vitro and harvested at DIV1, 5, and 9. RNA and protein were separately isolated for RT-PCR and immunoblotting analysis. (B) The shLuc or shRBM4 expression vector was cotransfected with either the FLAG-RBM4 or FLAG-Numb expression vector at DIV1, and images were collected at DIV3. Representative images. Bar graph shows the number of neurite-bearing cells from four selected fields; >100 cells were quantified. *p < 0.05.
DISCUSSION
The present study shows for the first time that a single pre-mRNA splicing regulator, RBM4, contributes to the selection of two alternative exons of Numb. RBM4-mediated Numb isoform expression is important for neuronal differentiation and neurite outgrowth.
RBM4 regulates alternative splicing of Numb
Numb was initially identified as a Notch antagonist, and its many other functions were discovered later (Gulino et al., 2010). For example, Numb plays a role in membrane trafficking via association with clathrin, and it also regulates actin dynamics (Gulino et al., 2010). In neurons, Numb is critical for neurogenesis and can also regulate neurite outgrowth and synapse formation of mature neurons (Nishimura et al., 2003; Lu et al., 2009). However, the biochemical properties and biological functions of each Numb isoform have not been completely and clearly assigned. Neuronal differentiation involves differential selection of E3 and E9, whereas E9 inclusion has been implicated in cancer progression (Bechara et al., 2013; Kim et al., 2013; Zong et al., 2014). RBM4 is the first splicing factor shown to determine E3 selection. RBM4 regulates exon selection generally by binding CU-rich elements (Lin and Tarn, 2011). However, deletion or mutation of several CU-rich motifs near Numb E3 did not affect RBM4-mediated exon selection (unpublished data). In addition, RBM4 suppresses E9 inclusion, as reported for several other splicing regulators (Bechara et al., 2013; Kim et al., 2013; Zong et al., 2014). Rbfox3 and RBM10 bind to intronic sequences immediately upstream of E9, whereas QKI-5 binds to E9 to prevent the core splicing factor SF1 from binding to the branch-site sequence of the upstream intron (Bechara et al., 2013; Kim et al., 2013; Zong et al., 2014). Although we found that RBM4 associates with Numb transcripts (Figure 3), its exact binding site(s) are still to be determined. Our present results imply a role for RBM4-mediated Numb isoform expression in neuronal cell differentiation. It is also worth noting that RBM4-mediated suppression of Numb E9 inclusion may contribute to tumor suppression (Bechara et al., 2013; Zong et al., 2014). Thus further exploration of the mechanism underlying RBM4-mediated splicing regulation of Numb is warranted.
Potential role for RBM4 in regulating Notch signaling
RBM4-regulated Numb splicing may possibly influence cellular Notch signaling activity. Notch signaling activates the expression of Hes1/5 and subsequently leads to Mash1 suppression during neurogenesis (Kageyama et al., 2010). The Numb-Notch signaling pathway controls asymmetric division during neural development of Drosophila, whereas mammalian Notch functions to maintain neural stem cells and determine specific cell fates (Faigle and Song, 2013). Moreover, mammalian Numb generates isoforms with differential activities in modulating Notch protein processing. Numb-PTBS isoforms are relatively weak inhibitors of Notch (Bani-Yaghoub et al., 2007). Notch activation–induced Hes1/5 expression suppresses Mash1 and another neuronal transcription factor, neurogenin 2 (Kageyama et al., 2010). We observed that Rbm4a knockout or RBM4 depletion increased Hes5 level, and RBM4 overexpression reduced Hes5 (Figure 2). However, the effect of RBM4 on Hes1 was negligible. We assumed that this is because Hes1 expression oscillates through negative feedback (Kageyama et al., 2010), so that we might have been unable to detect a significant change in Hes1 expression in our experimental assessment. Nevertheless, RBM4 knockdown and overexpression, respectively, suppressed and induced Mash1 in P19 cells (Figure 2). RBM4 overexpression also induced Mash1 in KT98 cells (Figure 5), confirming a close correlation between RBM4 and Mash1 expression. Moreover, we observed that RBM4-induced Numb isoforms exhibited a higher potential for induction of Mash1 expression and neurite outgrowth (Figures 4 and 6), which is consistent with the ability of Numb-PRRS to promote Mash1 expression and neuronal differentiation (Bani-Yaghoub et al., 2007). Nevertheless, why RBM4 has a differential effect on Hes1/5 expression (Figure 2) remains to be answered. Together our results suggest that RBM4 modulates Mash1 expression via the Numb-Notch-Hes pathway, but at present we cannot exclude other possible mechanisms.
Roles of RBM4 in neuronal differentiation and neurite outgrowth
Numb isoforms have overlapping but distinct roles in neuronal differentiation (Verdi et al., 1999; Bani-Yaghoub et al., 2007). It is not fully clear how individual isoforms function and how the balance between these isoforms is developmentally regulated. E3 was included in RA-induced embryo bodies but subsequently skipped upon induction of differentiation (Figure 1). Such transient inclusion of E3 in Numb (PTBL) causes Notch inhibition and thereby initiates neuronal differentiation (Kageyama et al., 2010). Although RBM4 overexpression or depletion affected Numb isoform expression to different extents in different cell types, RBM4-induced E3 inclusion might contribute to Mash1 expression—a possible consequence of Notch inhibition—at early differentiation stages. Indeed, we observed that RBM4-induced Numb2 (PTBL-PRRS) had a greater ability to restore Mash1 expression in RBM4-depleted cells (Figure 4). Therefore RBM4 induces Mash1 expression, at least in part, by regulating the Numb isoform switch, which subsequently promotes neuronal differentiation. However, the possibility remains that RBM4-induced Mash1 results from combined effects of different pathways and is influenced by different cellular contexts.
Notch and Numb are also involved in neurite outgrowth. Numb may function by modulating Notch activity (Berezovska et al., 1999) or endocytosis and recycling at the growth cone via its association with neuronal cell adhesion factors (Nishimura et al., 2003). Indeed, Numb-PTBS isoforms promote neurite outgrowth in response to neurotrophic factors or neuronal activation (Lu et al., 2009). However, our observation that Numb2 and Numb4 reversed neurite outgrowth in RBM4-depleted neurons (Figure 6) suggests that the PRR domain may determine neurite outgrowth during cell differentiation. This result coincides with the observed gradual and perhaps irreversible E9 skipping during neuronal differentiation and maturation (Figures 1 and 6). Together our in vitro data suggest that specific Numb isoforms are involved in RBM4-mediated neuronal differentiation and neurite outgrowth. However, whether RBM4-regulated Numb isoforms modulate neurite outgrowth by inhibiting Notch activity or via Notch-independent pathways remains to be answered.
Future studies need to clarify how splicing factors collectively regulate the balance between various Numb isoforms and how Numb isoforms coordinately function in neural development.
MATERIALS AND METHODS
Plasmids
The expression vectors for FLAG-tagged human and mouse RBM4a and RBM4 mutant have been described (Lu et al., 2013). Plasmid pAS4.1w-hRBM4-IRES-RFP-Ppuro-aOn was constructed by inserting the open reading frame encoding human RBM4 (hRBM4) and the fragment IRES-RFP (internal ribosome entry site followed by red fluorescent protein, derived from pTagRFP-N; Evrogen, Moscow, Russia) into the multiple cloning sites of vector pAS4.1w-Ppuro-aOn, which was purchased from the National RNAi Core Facility, Academia Sinica. The shRNA vectors for targeting RBM4 (pLKO.1-shRBM4-puro) or luciferase (as control; pLKO.1-shLuc-puro) were also purchased from the National RNAi Core Facility. The plasmids expressing four Numb isoforms (Verdi et al., 1999) were constructed by inserting each of the FLAG-tagged Numb isoforms into vector pcDNA3.1: Numb1 (PTBLPRRL), Numb2 (PTBLPRRS), Numb3 (PTBSPRRL), and Numb4 (PTBSPRRS).
To construct the mouse Numb E3 and E9 minigenes, the PCR-amplified DNA fragments as follows were ligated and inserted into vector pCH110. The Numb-E3 minigene contained a 375–base pair exon 2–intron fragment (nucleotides [nt] 83831622–83831996 of chromosome 12, genome assembly GRCm38.p3, accession number NC_000078.6), a 633–base pair intron–exon 3–intron fragment (nt 83824959–83825591), and a 375–base pair intron–exon 4 fragment (nt 83811286–83811660). The Numb-E9 minigene contained a 450–base pair exon–intron 8 fragment (nt 83799208–83799657), a 747–base pair intron–exon 9–intron fragment (nt 83796897-83797643), and a 550–base pair intron–exon 10 fragment (nt 83795905–83796454).
RNA extraction from embryonic mouse brain
Total RNA from E13.5 Rbm4a knockout (Lin et al., 2013) and wild-type littermate brains was extracted using TRIzol reagent (Life Technologies, Carlsbad, CA) following manufacturer’s instructions and used for RT-PCR.
Culture and neuronal differentiation of P19 cells
The mouse embryonal carcinoma cell line P19 was maintained in DMEM supplemented with 7.5% newborn calf serum, 2.5% fetal bovine serum (FBS), 2 mM l-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin (all from Life Technologies).
For differentiation of P19 cells, 2 × 106 cells/ml were first suspension cultured in bacterial-grade dishes containing the differentiation medium to induce embryoid body formation; the differentiation medium was composed of DMEM, 3.75% FBS, 1.25% newborn calf serum, a mixture of penicillin, streptomycin, and l-glutamine (as described), and 1 μM all-trans RA (Sigma-Aldrich, St. Louis, MO). Subsequently, 1 × 106 cells were placed in neurobasal medium (Life Technologies) containing B27 supplement minus vitamin A (Life Technologies) and 10 μM DNA synthesis inhibitor cytosine β-d-arabinofuranoside hydrochloride (Sigma-Aldrich). The medium was replaced every 2 d during the induction of cell differentiation.
Culture and neuronal differentiation of KT98 cells
KT98 cells derived from F1B-Tag transgenic mouse line 98 (Hsu et al., 2009) were maintained in DMEM/F12 (Life Technologies) supplemented with 10% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin. F1B-driven GFP expression was used as an indicator for isolating a population of KT98 cells with higher self-renewal and multipotent capacities (Hsu et al., 2009). To establish stable clones for inducible expression of RBM4 (KT98-RBM4), GFP-positive KT98 cells were transfected with plasmid pAS4.1w-hRBM4-IRES-RFP-Ppuro-aOn. Transfectants were selected with 3 μg/ml puromycin (Sigma-Aldrich) for 4 d, followed by induction with 2 μg/ml doxycycline (Sigma-Aldrich) for 7 d. GFP and RFP double-positive KT98 cells were sorted using a FACSAria III flow cytometer (BD Biosciences, Franklin Lakes, NJ) each week for 4 wk.
For neuronal differentiation of KT98-RBM4 cells, neurospheres were initially induced in DMEM/F12 supplemented with B27, 20 ng/ml epidermal growth factor (EGF), 20 ng/ml fibroblast growth factor 2 (FGF2), and 2 μg/ml heparin for 7 d, followed by culture in DMEM/F12 containing 2% FBS, 10 ng/ml platelet-derived growth factor (PDGF), 50 ng/ml brain-derived neurotrophic factor (BDNF), and 50 ng/ml glial cell–derived neurotrophic factor for 7–9 d (Hsu et al., 2009). Differentiated cells were verified as described (Hsu et al., 2009).
Primary rat cortical neuron culture
Cortical neurons were isolated from rat embryos (E18.5) and seeded at 1 × 105 cells/well in 24-well plates with coverslips that had been precoated with 1 mg/ml poly-d-lysine (Merck Millipore, Billerica, MA). Cells were cultured in the neurobasal medium containing 2% B27, 0.5 mM l-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin. Neurons were transfected with the RBM4-targeting shRNA (shRBM4) vector alone or together with the human RBM4 or mouse Numb expression vector. Morphological analysis of neuronal cells was carried out as described (Chen et al., 2012).
Splicing assays
In general, 2 × 105 P19 or HEK293T cells were seeded in each well of a six-well plate 1 d before transfection, followed by 2 or 4 μg of pcDNA-FLAG hRBM4 overexpression plasmid or shRNA-expressing plasmids using Lipofectamine 2000 (Life Technologies) according to manufacturer’s instructions. For minigene reporter assays, cells were cotransfected with 0.5 or 1 μg of E3 or E9 minigene vector. Transfected cells were harvested at 30 h posttransfection. Total RNA was extracted, followed by RT-PCR.
RT-PCR and immunoprecipitation
For RT-PCR, 2 μg of total RNA was treated with RQ1 DNase (Promega, Madison, WI), followed by RT using oligo d(T) primer and the SuperScript III kit (Life Technologies). The resulting cDNA was then analyzed by PCR using corresponding specific primers. RT-PCR products were visualized by agarose gel electrophoresis or further detected with Southern blotting using 32P-labeled primers as described (Lin and Tarn, 2011). Band intensities were analyzed using ImageJ, 1.47 version (imagej.nih.gov/ij). Quantitative RT-PCR was performed using the SensiFAST SYBR No-ROX kit (Bioline, Taunton, MA) according to the manufacturer’s instructions. Supplemental Table S1 lists the primers used.
For immunoprecipitation, 2 × 106 P19 cells were transfected for 2 d with 24 μg of the pcDNA-flag-hRBM4a or control plasmid as described. After harvest, cells were lysed in hypotonic lysis buffer (10 mM Tris-HCl, pH 7.5, 10 mM NaCl, 10 mM EDTA, 0.5% Triton X-100) containing protease inhibitor cocktail (Roche, Indianapolis, IN), followed by immunoprecipitation using anti-FLAG M2 beads as described (Lin et al., 2013). RNA was subjected to RT-PCR using primers specific to mouse Numb exon 8 (Supplemental Table S1).
Immunoblotting
Immunoblotting was performed as described (Lin and Tarn, 2011). We used affinity-purified polyclonal antibodies against RBM4 (Lin and Tarn, 2011), α-tubulin (GenScript, Piscataway, NJ), glyceraldehyde 3-phosphate dehydrogenase (GAPDH; Proteintech, Rosemont, IL), and FLAG (Sigma-Aldrich).
Statistical analysis
The Student’s t-test was performed to determine the significance of differences between treatment groups in Figures 4–6. Error bars represent SEM. p < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We are grateful to Wen-Cheng Chiang for initiating this study, Ming-Shiang Tsai for knockout-mouse maintenance, and Simon Chang for mouse brain analysis. We also thank the Core Facility of the Institute of Biomedical Sciences, Academia Sinica, for technical assistance. This work was funded by Grant 103-2311-B-001-027-MY3 from the Ministry of Science and Technology, Taiwan.
Abbreviations used:
- AraC
cytosine β-d-arabinofuranoside hydrochloride
- BDNF
brain-derived neurotrophic factor
- EGF
epidermal growth factor
- FGF2
fibroblast growth factor 2
- PDGF
platelet-derived growth factor
- RA
retinoic acid.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-11-0798) on March 23, 2016.
REFERENCES
- Bani-Yaghoub M, Kubu CJ, Cowling R, Rochira J, Nikopoulos GN, Bellum S, Verdi JM. A switch in numb isoforms is a critical step in cortical development. Dev Dyn. 2007;236:696–705. doi: 10.1002/dvdy.21072. [DOI] [PubMed] [Google Scholar]
- Bechara EG, Sebestyén E, Bernardis I, Eyras E, Valcárcel J. RBM5, 6, and 10 differentially regulate NUMB alternative splicing to control cancer cell proliferation. Mol Cell. 2013;52:720–733. doi: 10.1016/j.molcel.2013.11.010. [DOI] [PubMed] [Google Scholar]
- Berezovska O, McLean P, Knowles R, Frosh M, Lu FM, Lux SE, Hyman BT. Notch1 inhibits neurite outgrowth in postmitotic primary neurons. Neuroscience. 1999;93:433–439. doi: 10.1016/s0306-4522(99)00157-8. [DOI] [PubMed] [Google Scholar]
- Bernert G, Fountoulakis M, Lubec G. Manifold decreased protein levels of matrin 3, reduced motor protein HMP and hlark in fetal Down’s syndrome brain. Proteomics. 2002;2:1752–1757. doi: 10.1002/1615-9861(200212)2:12<1752::AID-PROT1752>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Breunig JJ, Rakic P. Coordinating migratory neuron polarization by numbing communication. Dev Cell. 2011;20:578–580. doi: 10.1016/j.devcel.2011.05.002. [DOI] [PubMed] [Google Scholar]
- Brooks YS, Wang G, Yang Z, Smith KK, Bieberich E, Ko L. Functional pre- mRNA trans-splicing of coactivator CoAA and corepressor RBM4 during stem/progenitor cell differentiation. J Biol Chem. 2009;284:18033–18046. doi: 10.1074/jbc.M109.006999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HH, Yu HI, Chiang WC, Lin YD, Shia BC, Tarn WY. hnRNP Q regulates Cdc42 mediated neuronal morphogenesis. Mol Cell Biol. 2012;32:2224–2238. doi: 10.1128/MCB.06550-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho MB, Smith C. Regulation of alternative pre-mRNA splicing. Methods Mol Biol. 2014;1126:55–82. doi: 10.1007/978-1-62703-980-2_5. [DOI] [PubMed] [Google Scholar]
- Faigle R, Song H. Signaling mechanisms regulating adult neural stem cells and neurogenesis. Biochim Biophys Acta. 2013;1830:2435–2448. doi: 10.1016/j.bbagen.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedor MJ. Alternative splicing minireview series: combinatorial control facilitates splicing regulation of gene expression and enhances genome diversity. J Biol Chem. 2008;283:1209–1210. doi: 10.1074/jbc.R700046200. [DOI] [PubMed] [Google Scholar]
- Grabowski Alternative splicing takes shape during neuronal development. Curr Opin Genet Dev. 2011;21:388–394. doi: 10.1016/j.gde.2011.03.005. [DOI] [PubMed] [Google Scholar]
- Gulino A, Marcotullio LD, Screpanti I. The multiple functions of Numb. Exp Cell Res. 2010;316:900–906. doi: 10.1016/j.yexcr.2009.11.017. [DOI] [PubMed] [Google Scholar]
- Hsu YC, Lee DC, Chen SL, Liao WC, Lin JW, Chiu WT, Chiu IM. Brain-specific 1B promoter of FGF1 gene facilitates the isolation of neural stem/progenitor cells with self-renewal and multipotent capacities. Dev Dyn. 2009;238:302–314. doi: 10.1002/dvdy.21753. [DOI] [PubMed] [Google Scholar]
- Kageyama R, Niwa Y, Shimojo H, Kobayashi T, Ohtsuka T. Ultradian oscillations in Notch signaling regulate dynamic biological events. Curr Top Dev Biol. 2010;92:311–331. doi: 10.1016/S0070-2153(10)92010-3. [DOI] [PubMed] [Google Scholar]
- Kim KK, Nam J, Mukouyama YS, Kawamoto S. Rbfox3-regulated alternative splicing of Numb promotes neuronal differentiation during development. J Cell Biol. 2013;200:443–458. doi: 10.1083/jcb.201206146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HS, Wang D, Shen Q, Schonemann MD, Gorski JA, Jones KR, Temple S, Jan LY, Jan YN. Inactivation of Numb and Numb like in embryonic dorsal forebrain impairs neurogenesis and disrupts cortical morphogenesis. Neuron. 2003;40:1105–1118. doi: 10.1016/s0896-6273(03)00755-4. [DOI] [PubMed] [Google Scholar]
- Lin JC, Tarn WY. RBM4 down-regulates PTB and antagonizes its activity in muscle cell-specific alternative splicing. J Cell Biol. 2011;193:509–520. doi: 10.1083/jcb.201007131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JC, Tarn WY, Hsieh WK. Emerging role for RNA binding motif protein 4 in the development of brown adipocytes. Biochim Biophys Acta. 2014;1843:769–779. doi: 10.1016/j.bbamcr.2013.12.018. [DOI] [PubMed] [Google Scholar]
- Lin JC, Yan YT, Hsieh WK, Peng PJ, Su CH, Tarn WY. RBM4 promotes pancreas cell differentiation and insulin expression. Mol Cell Biol. 2013;33:319–327. doi: 10.1128/MCB.01266-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louvi A, Artavanis-Tsakonas S. Notch signalling in vertebrate neural development. Nat Rev Neurosci. 2006;7:93–102. doi: 10.1038/nrn1847. [DOI] [PubMed] [Google Scholar]
- Lu CB, Fu W, Xu X, Mattson MP. Numb-mediated neurite outgrowth is isoform-dependent, and requires activation of voltage-dependent calcium channels. Neurosci. 2009;161:403–412. doi: 10.1016/j.neuroscience.2009.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu CC, Chen TH, Wu JR, Chen HH, Yu HY, Tarn WY. Phylogenetic and molecular characterization of the splicing factor RBM4. PLoS One. 2013;8:e59092. doi: 10.1371/journal.pone.0059092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGill MA, Dho SE, Weinmaster G, McGlade CJ. Numb regulates post-endocytic trafficking and degradation of Notch1. J Biol Chem. 2009;284:26427–26438. doi: 10.1074/jbc.M109.014845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGill MA, McGlade CJ. Mammalian numb proteins promote Notch1 receptor ubiquitination and degradation of the Notch1 intracellular domain. J Biol Chem. 2003;278:23196–23203. doi: 10.1074/jbc.M302827200. [DOI] [PubMed] [Google Scholar]
- Nishimura T, Fukata Y, Kato K, Yamaguchi T, Matsuura Y, Kamiguchi H, Kaibuchi K. CRMP-2 regulates polarized Numb-mediated endocytosis for axon growth. Nat Cell Biol. 2003;5:819–26. doi: 10.1038/ncb1039. [DOI] [PubMed] [Google Scholar]
- Norris AD, Calarco JA. Emerging roles of alternative pre-mRNA splicing regulation in neuronal development and function. Front Neurosci. 2012;6:122. doi: 10.3389/fnins.2012.00122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen WA, Chan SL, Zhu H, Abdur-Rahman LA, Verdi JM, Mattson MP. Numb isoforms containing a short PTB domain promote neurotrophic factor-induced differentiation and neurotrophic factor withdrawal-induced death of PC12 Cells. J Neurochem. 2002;82:976–986. doi: 10.1046/j.1471-4159.2002.01036.x. [DOI] [PubMed] [Google Scholar]
- Salomonis N, Nelson B, Vranizan K, Pico AR, Hanspers K, Kuchinsky A, Ta L, Mercola M, Conklin BR. Alternative splicing in the differentiation of human embryonic stem cells into cardiac precursors. PLoS Comput Biol. 2008;5:e1000553. doi: 10.1371/journal.pcbi.1000553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spana EP, Doe CQ. Numb antagonize Notch signaling to specify sibling neuron cell fates. Neuron. 1996;17:21–26. doi: 10.1016/s0896-6273(00)80277-9. [DOI] [PubMed] [Google Scholar]
- Verdi JM, Bashirullah A, Goldhawk DE, Kubu CJ, Jamali M, Meakin SO, Lipshitz HD. Distinct human Numb isoforms regulate differentiation vs. proliferation in the neuronal lineage. Proc Natl Acad Sci USA. 1999;96:10472–10476. doi: 10.1073/pnas.96.18.10472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan B. Numb—from flies to humans. Brain Dev. 2010;32:293–298. doi: 10.1016/j.braindev.2009.03.008. [DOI] [PubMed] [Google Scholar]
- Yano M, Hayakawa-Yano Y, Mele A, Darnell RB. Nova2 regulates neuronal migration through an RNA switch in disabled-1 signaling. Neuron. 2010;66:848–858. doi: 10.1016/j.neuron.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S, Black DL. Alternative pre-mRNA splicing in neurons: growing up and extending its reach. Trends Genet. 2013;29:442–448. doi: 10.1016/j.tig.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong FY, Fu X, Wei WJ, Luo YG, Heiner M, Cao LJ, Fang Z, Fang R, Lu D, Ji H, et al. The RNA-binding protein QKI suppresses cancer-associated aberrant splicing. PLoS Genet. 2014;10:e1004289. doi: 10.1371/journal.pgen.1004289. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.