The last few years have seen tremendous advances in understanding the genomic landscape, clonal architecture and evolution of the chronic lymphocytic leukaemia (CLL) genome (Braggio et al, 2012; Quesada et al, 2012; Schuh et al, 2012; Landau et al, 2013). Significant efforts are already underway in understand the clinical implications of some novel “driver” genes, including SF3B1, NOTCH1, MYD88 and BIRC3 (Rossi et al, 2012, 2013). Despite the advance in the characterization of these genes, there is high genetic heterogeneity in CLL with several additional genes recurrently affected where no further analyses have been provided.
Furthermore, most of the studies were performed in heterogeneous cohorts collected at different disease stages from patients subjected to multiple therapeutic approaches. Here we focused on the characterization of a homogeneous cohort of untreated CLL with active disease prior to the inclusion in a PCR (pentostatin/cyclophosphamide/rituximab) trial (Kay et al, 2007). Detailed clinical information is available in the Supplementary material. We analysed 12 cases by whole exome sequencing (WES), and additional 36 cases by targeted deep sequencing (TDS). In two cases, we further analysed sequential samples collected at relapse by WES and TDS. Clonal B-cells were enriched using the EasySep Human CD19+ Cell Enrichment Kit with an average purity of 91% cells after enrichment (range 66–99%). T-cells were enriched using a CD3-Positive Selection Kit and subsequently used as germline samples. For WES, a paired-end library was generated as per Illumina protocol followed by exome capture using SureSelect 50 Mb Enrichment kit (Agilent, Santa Clara, CA, USA) and 100 bp paired-end libraries were sequenced in HiSeq2000. Somatic single nucleotide variants and insertions/ deletions were called by SomaticSniper and GATK Somatic Indel Detector, respectively. Pair-wise analyses were performed by comparing respective tumour samples with germline samples. Somatic variants were functionally annotated with snpEFF, PolyPhen-2 and MutationTaster. Targeted sequencing was performed in 24 genes that have been found recurrently mutated in CLL (Supplementary Table SI) using IonTorrent PGM sequencer (Life Technologies, Carlsbad, CA, USA) as per manufacturer’s protocol. Somatic variants were identified using the variant caller of IonTorrent suite. Protein expression was assessed by Western blotting. Overall survival was measured from date of initial disease presentation to the date of death or last follow-up using Kaplan–Meier. Significance (P < 0·05) was estimated by log-rank test.
One-third of cases (17 of 48) had mutated IGHV, 37% were ZAP70-positive and 37% were CD38-positive (Supplementary Table SII). Patients were followed-up for a median of 40 months after first-line therapy with 48% of cases showing disease progression after therapy (median of 17 months between treatment and progression). WES and TDS were performed in 12 (102-fold coverage) and 36 (720-fold coverage) cases, respectively. Copy-number abnormalities (CNAs) were screened in all 48 cases by array-based comparative genomic hybridization.
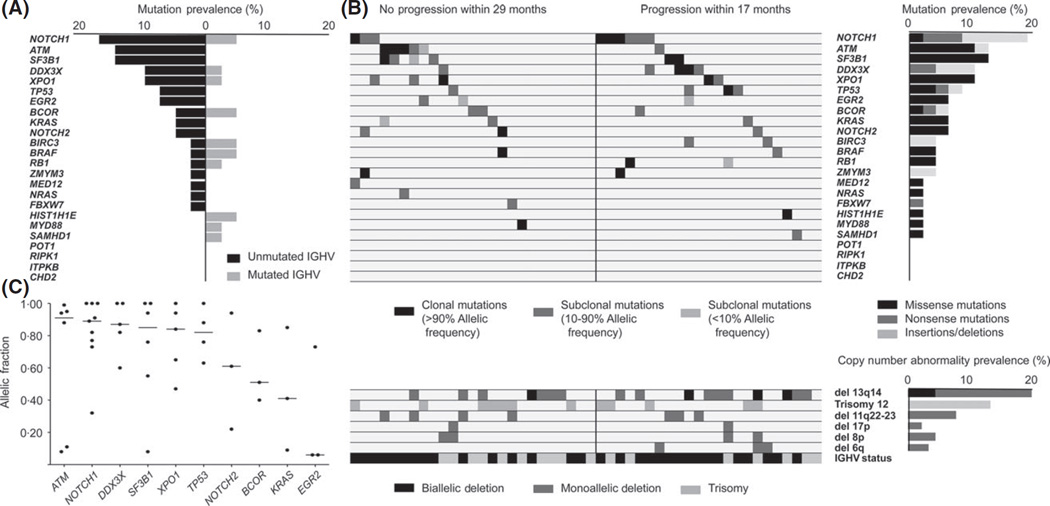
NOTCH1 was the most commonly mutated gene (19%), followed by ATM, SF3B1 (12% each), XPO1, DDX3X (10% each) and TP53 (8%). All mutations were predicted to be damaging by PolyPhen, SIFT and MutationTaster. The most common CNAs were -13q14 (50%), +12 (23%), -11q22 (17%), -8p11-p12 and -17p13 (6% each). In 39 cases (81%), at least one of the 24 genes of interest was mutated and mutations and/or CNAs were found in 45 cases (94%). The complete list of mutated genes and CNAs are included in Supplementary Tables SIII and SIV, respectively.
The incidence of mutations in driver genes were 3 × higher in unmutated IGHV (U-CLL) compared to mutated IGHV (M-CLL) (Fig 1A). ATM, SF3B1, EGR2, KRAS and BIRC3 were found mutated only in U-CLL. Additionally, the incidence of NOTCH1, DDX3X and XPO1 mutations in U-CLL was double that of M-CLL.
Fig 1.
Genomic landscape in a cohort of untreated active CLL. (A) Prevalence of mutations in 24 genes in unmutated (U-CLL) and mutated (M-CLL) IGHV CLL cases. (B) Upper left panel: Heatmap of mutations in CLL cohort. Horizontal rows depict mutations in a particular gene and vertical columns represent individual patients. Black, dark grey and light grey closed boxes show the mutation status as clonal (more than 90%), subclonal (present in 10–90%) and small subclonal (present in <10%) allelic fractions, respectively. Solid black vertical line differentiates the cases with no progression after therapy (left side) and cases that progressed after therapy (right side). Upper right panel: Mutation prevalence. Black, dark grey and light grey closed boxes represent the percentage of missense mutations, nonsense mutations and frameshift indels, respectively. Lower left panel: Copy-number aberrations in the six most commonly affected chromosomal regions. Horizontal rows depict particular aberration and vertical columns represent individual patients. Lower right panel: Copy number abnormality prevalence. For lower panel (left and right), black, dark grey and light grey closed boxes represent biallellic deletion, monoallellic deletion and monoallelic gain, respectively. (C) Clonal status of mutated genes. Horizontal axis represents genes mutated in three or more cases and vertical axis represents allelic frequency distribution mutation in genes in cohort. Each dot represents a case and horizontal solid black line is the median allelic fraction affected per mutations in the particular gene.
Considering only the cases that progressed after therapy (n = 23), the most commonly mutated genes were NOTCH1 (30%), followed by DDX3X, (17%), TP53 (13%), BIRC3, SF3B1 and XPO1 (9% each)(Fig 1B). The clonal status of mutations varied from clonal to subclonal presence in different cases (Fig 1C).
Overall, we identified a higher prevalence of mutations in NOTCH1, XPO1 and DDX3X in our cohort. Eight of nine mutations in NOTCH1 were either nonsense or frameshift indels in the PEST domain and p.P2514Rfs*4 was the most common mutation (5/9 cases). XPO1 was mutated in five cases and the most frequent mutation was found in codon 571 (4/5 cases). The remaining mutation was identified in the HEAT domain (p.D624G).
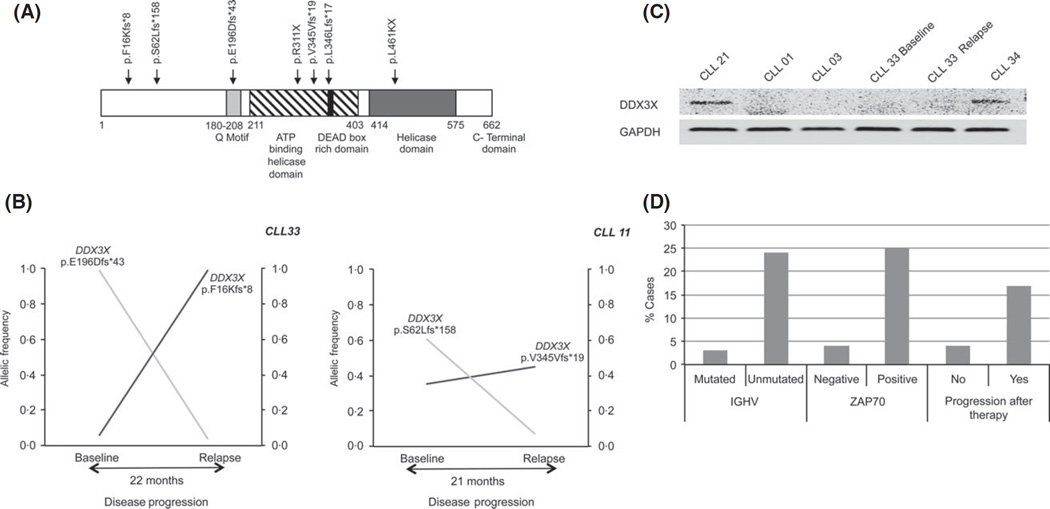
An interesting finding was the identification of DDX3X mutations in 10% of cases. DDX3X is located on chromosome X and was preferentially mutated in males (4/5 cases). All identified mutations have a truncating effect, either nonsense mutations or frameshift indels (Fig 2A). Moreover, we identified two independent truncating mutations in two of five cases. Analysis of sequential samples from these two cases showed that these mutations waxed and waned differently between time points, confirming their presence into different subclones (Fig 2B). Next, we confirmed loss of protein expression in cases harbouring DDX3X mutations (Fig 2C). Moreover, we recently found DDX3X mutations in premalignant monoclonal B-cell lymphocytosis (Ojha et al, 2014).
Fig 2.
Characterization of somatic mutations in DDX3X. (A) Location and type of somatic mutations found in DDX3X. (B) Two independent DDX3X mutations affecting different subclones were found in two cases. Allelic frequency of each mutation in sequential samples is denoted by black and grey lines. X-axis denotes disease progression in months. Y-axis denotes allelic frequency. (C) Protein expression level of DDX3X and GAPDH in cases with DDX3X mutation (CLL01, CLL03, CLL33 Baseline and Relapse) and controls with wild type DDX3X (CLL21 and CLL34). (D) Association of DDX3X mutation and clinical variables. Vertical axis shows % cases with DDX3X mutation.
Our analysis suggested an association between DDX3X inactivation and clinically unfavourable features and poor outcome. Thus, DDX3X mutations were preferentially found in U-CLL (24% cases with mutations) compared with M-CLL (3%), and ZAP70-positive (25%) compared with ZAP70-negative (4%). Furthermore, DDX3X mutations were more frequently found in cases that relapsed after therapy (17%) than cases with stable disease (4%)(Fig 2D). Finally, cases with mutated DDX3X showed a trend to shorter survival, even though the differences were not statistically significant (P = 0·15).
DDX3X is an ATP-dependent RNA helicase involved in several steps of RNA processing pathways, including transcription, translation initiation, splicing and mRNA export. Recent studies have identified DDX3X as a tumour suppressor gene in medulloblastoma and documented its role in signal transduction through pathogenic WNT/b-catenin signalling (Pugh et al, 2012). Furthermore, synergistic DDX3X and TP53 transcriptional suppression of CDKN1A expression has been documented in non-small cell lung carcinoma (Wu et al, 2013).
We recognize the limitation of the relatively small number of cases in this study; nevertheless, this is a unique study with homogeneous cohort of CLL patients with active disease needing initial therapy. We identified recurrent inactivating DDX3X mutations, especially in cases with unfavourable clinical markers. Furthermore, the truncated nature of all mutations, the presence of multiple mutations in different subclones and the association with unfavourable clinical markers implicate DDX3X as a bona fide tumour suppressor gene in CLL.
Supplementary Material
Acknowledgments
This work was supported by the Henry Predolin Foundation, the Marriott Specialized Workforce Development Awards in Individualized Medicine, the Fraternal Order of Eagles and grant NIH-CA95241.
PCR trial was funded with research support by Hospira. T. S. has received research funding from Hospira, Genentech, Flaxo-Smith-Kline, Jannsen, Celgene and Cephalon. N.K. is on the data safety monitoring committee for Gilead and Celgene and has received research support from Phamacyclics. R.F. has received a patent for the prognostication of Multiple Myeloma based on genetic categorization of the disease and has received consulting fees from Medtronic, Otsuka, Celgene, Genzyme, BMS, Lilly, Onyx, Binding Site, Millennium and AMGEN.
Footnotes
Conflict of interest
The remaining authors have no conflicts of interest to disclose.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Data S1. Materials and methods.
Table SI. List of recurrently mutated genes in CLL sequenced by targeted deep sequencing.
Table SII. Clinical information for 48 PCR-CLL cases.
Table SIII. List of mutations identified.
Table SIV. Copy number abnormalities found in the cohort by aCGH.
References
- Braggio E, Kay NE, VanWier S, Tschumper RC, Smoley S, Eckel-Passow JE, Sassoon T, Barrett M, Van Dyke DL, Byrd JC, Jelinek DF, Shanafelt TD, Fonseca R. Longitudinal genome-wide analysis of patients with chronic lymphocytic leukemia reveals complex evolution of clonal architecture at disease progression and at the time of relapse. Leukemia. 2012;26:1698–1701. doi: 10.1038/leu.2012.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay NE, Geyer SM, Call TG, Shanafelt TD, Zent CS, Jelinek DF, Tschumper R, Bone ND, Dewald GW, Lin TS, Heerema NA, Smith L, Grever MR, Byrd JC. Combination chemoimmunotherapy with pentostatin, cyclophosphamide, and rituximab shows significant clinical activity with low accompanying toxicity in previously untreated B chronic lymphocytic leukemia. Blood. 2007;109:405–411. doi: 10.1182/blood-2006-07-033274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landau DA, Carter SL, Stojanov P, McKenna A, Stevenson K, Lawrence MS, Sougnez C, Stewart C, Sivachenko A, Wang L, Wan Y, Zhang W, Shukla SA, Vartanov A, Fernandes SM, Saksena G, Cibulskis K, Tesar B, Gabriel S, Hacohen N, Meyerson M, Lander ES, Neuberg D, Brown JR, Getz G, Wu CJ. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell. 2013;152:714–726. doi: 10.1016/j.cell.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojha J, Secreto C, Rabe K, da Silva JA, Tschumper R, Van Dyke D, Slager S, Fonseca R, Shanafelt T, Kay N, Braggio E. Monoclonal B-cell lymphocytosis is characterized by mutations in CLL putative driver genes and clonal heterogeneity clonal competition many years prior to disease progression. Leukemia. 2014 doi: 10.1038/leu.2014.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugh TJ, Weeraratne SD, Archer TC, Pomeranz Krummel DA, Auclair D, Bochicchio J, Carneiro MO, Carter SL, Cibulskis K, Erlich RL, Greulich H, Lawrence MS, Lennon NJ, McKenna A, Meldrim J, Ramos AH, Ross MG, Russ C, Shefler E, Sivachenko A, Sogoloff B, Stojanov P, Tamayo P, Mesirov JP, Amani V, Teider N, Sengupta S, Francois JP, Northcott PA, Taylor MD, Yu F, Crabtree GR, Kautzman AG, Gabriel SB, Getz G, Jager N, Jones DT, Lichter P, Pfister SM, Roberts TM, Meyerson M, Pomeroy SL, Cho YJ. Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature. 2012;488:106–110. doi: 10.1038/nature11329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada V, Conde L, Villamor N, Ordonez GR, Jares P, Bassaganyas L, Ramsay AJ, Bea S, Pinyol M, Martinez-Trillos A, Lopez-Guerra M, Colomer D, Navarro A, Baumann T, Aymerich M, Rozman M, Delgado J, Gine E, Hernandez JM, Gonzalez-Diaz M, Puente DA, Velasco G, Freije JM, Tubio JM, Royo R, Gelpi JL, Orozco M, Pisano DG, Zamora J, Vazquez M, Valencia A, Himmelbauer H, Bayes M, Heath S, Gut M, Gut I, Estivill X, Lopez-Guillermo A, Puente XS, Campo E, Lopez-Otin C. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nature Genetics. 2012;44:47–52. doi: 10.1038/ng.1032. [DOI] [PubMed] [Google Scholar]
- Rossi D, Fangazio M, Rasi S, Vaisitti T, Monti S, Cresta S, Chiaretti S, Del Giudice I, Fabbri G, Bruscaggin A, Spina V, Deambrogi C, Marinelli M, Fama R, Greco M, Daniele G, Forconi F, Gattei V, Bertoni F, Deaglio S, Pasqualucci L, Guarini A, Dalla-Favera R, Foa R, Gaidano G. Disruption of BIRC3 associates with fludarabine chemorefractoriness in TP53 wild-type chronic lymphocytic leukemia. Blood. 2012;119:2854–2862. doi: 10.1182/blood-2011-12-395673. [DOI] [PubMed] [Google Scholar]
- Rossi D, Rasi S, Spina V, Bruscaggin A, Monti S, Ciardullo C, Deambrogi C, Khiabanian H, Serra R, Bertoni F, Forconi F, Laurenti L, Marasca R, Dal-Bo M, Rossi FM, Bulian P, Nomdedeu J, Del Poeta G, Gattei V, Pasqualucci L, Rabadan R, Foa R, Dalla-Favera R, Gaidano G. Integrated mutational and cytogenetic analysis identifies new prognostic subgroups in chronic lymphocytic leukemia. Blood. 2013;121:1403–1412. doi: 10.1182/blood-2012-09-458265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh A, Becq J, Humphray S, Alexa A, Burns A, Clifford R, Feller SM, Grocock R, Henderson S, Khrebtukova I, Kingsbury Z, Luo S, McBride D, Murray L, Menju T, Timbs A, Ross M, Taylor J, Bentley D. Monitoring chronic lymphocytic leukemia progression by whole genome sequencing reveals heterogeneous clonal evolution patterns. Blood. 2012;120:4191–4196. doi: 10.1182/blood-2012-05-433540. [DOI] [PubMed] [Google Scholar]
- Wu DW, Lee MC, Wang J, Chen CY, Cheng YW, Lee H. DDX3 loss by p53 inactivation promotes tumor malignancy via the MDM2/Slug/E-cadherin pathway and poor patient outcome in non-small-cell lung cancer. Oncogene. 2013;33:1515–1526. doi: 10.1038/onc.2013.107. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.