Abstract
Purpose
The majority of angiotensin-converting enzyme inhibitors (ACEIs) are synthesized as ester prodrugs that must be converted to their active forms in vivo in order to exert therapeutic effects. Hepatic carboxylesterase 1 (CES1) is the primary enzyme responsible for the bioactivation of ACEI prodrugs in humans. The genetic variant −816A>C (rs3785161) is a common variant located in the promoter region of the CES1P1 gene. Previous studies report conflicting results with regard to the association of this variant and therapeutic outcomes of CES1 substrate drugs. The purpose of this study was to determine the effect of the variant −816A>C on the activation of the ACEI prodrug trandolapril and the blood pressure (BP) lowering effect of trandolapril in hypertensive patients.
Methods
The −816A>C genotypes and CES1 expression and activity on trandolapril activation were determined in 100 individual human liver samples. Furthermore, the association of the −816A>C variant and the BP lowering effect of trandolapril was evaluated in hypertensive patients who participated in the INternational VErapamil SR Trandolapril Study (INVEST).
Results
Our in vitro study demonstrated that hepatic CES1 expression and activity did not differ among different −816A>C genotypes. Moreover, we were unable to identify a clinical association between BP lowering effects of trandolapril and −816A>C genotypes.
Conclusions
We conclude that the −816A>C variant is not associated with interindividual variability in CES1 expression, activity or therapeutic response to ACEI prodrugs.
Keywords: pharmacogenomics, carboxylesterase 1 (CES1), drug metabolism, angiotensin-converting enzyme inhibitors, trandolapril, hypertension
Introduction
Angiotensin-converting enzyme inhibitors (ACEIs) are commonly used for the treatment of hypertension, heart failure, and delaying progression of diabetic nephropathy. The majority of ACEIs are synthesized as ester prodrugs to improve otherwise poor oral bioavailability. We and others have found that the activation of ACEI prodrugs takes place primarily in the liver via carboxylesterase 1 (CES1)-catalyzed hydrolysis [1–4]. This bioactivation is essential for ACEI prodrugs to exert their intended therapeutic effects since the resulting acid metabolites are far more potent inhibitors of ACE activity relative to the intact parent compounds.
CES1 is the primary hepatic hydrolase responsible for metabolism of many important medications, endogenous substances, and environmental toxins. In humans, CES1 is encoded by the CES1 gene, located in chromosome 16. CES1P1 is a pseudogene in close proximity with the CES1 gene. The CES1P1 gene does not encode any functional protein due to a premature stop codon in exon 3. However, CES1P1VAR, a CES1P1 variation with a minor allele frequency (MAF) of approximately 30% in the general population, expresses functional CES1 protein, which is identical to that of the CES1 gene. CES1 and CES1P1 genes are highly polymorphic with numerous variants in both coding and non-coding regions. A single nucleotide polymorphism (SNP) −816A>C (rs3785161) within the promoter region of the CES1P1 gene was reported to be associated with a greater BP lowering effect of the ACEI prodrug imidapril in hypertensive patients [5], suggesting this SNP may be associated with higher level of CES1 expression. Furthermore, two clinical investigations were recently carried out evaluating the association between the −816A>C genotype and the antiplatelet activity of the CES1 substrate drug clopidogrel [6, 7]. However, the results from the two studies were contradictory with regard to potential effects of the SNP on the antiplatelet activity of clopidogrel. Thus, whether the −816A>C is a functional genetic variant associated with significantly altered CES1 expression and activity remains an open question.
In the present study, we assessed the potential impact of the variant −816A>C on CES1 expression and activity utilizing individual human liver samples. Moreover, the association between this SNP and antihypertensive effect of the ACEI prodrug trandolapril was evaluated in hypertensive patients who participated in the INternational VErapamil SR Trandolapril Study (INVEST).
Materials and Methods
Materials
A total of 100 individual normal human liver samples were obtained from the XenoTech LLC (Lenexa, KS) and the Cooperative Human Tissue Network (CHTN, Columbus, OH). Liver samples were obtained from 44 males and 56 females with ages ranging from 22 to 81 years old. The donors included 90 Caucasians, 6 African-Americans, 2 Hispanics, and 2 classified as ‘others’.
Trandolapril, trandolaprilat, and simvastatin acid were purchased from Toronto Research Chemicals Inc. (Toronto, Canada). Taq polymerase was obtained from New England Biolabs Inc. (Ipswich, MA). All other chemicals and agents were of the highest analytical grade commercially available.
INVEST-GENES study
The INternational VErapamil SR Trandolapril Study (INVEST) was an international, multicenter, parallel randomized controlled trial (clinicaltrials.gov identifier NCT00133692) that enrolled 22,576 hypertensive coronary artery disease (CAD) patients from 862 sites in 14 countries to compare a calcium channel blocker verapamil SR-based treatment strategy versus a beta blocker atenolol-based treatment strategy for the prevention of adverse cardiovascular outcomes [8, 9]. Briefly, participants were randomly assigned to one of the two treatment strategies and were followed with protocol visits every six weeks for the first six months and every six months until the last participant was enrolled. In order to achieve BP control, trandolapril and/or hydrochlorothiazide were added in a protocol-defined manner, and finally non-study antihypertensive drugs were included for BP control. At each visit, BP was measured twice with the patient in a seated position after a 5-minute period. The average of two seated cuff BP measurements was used as the BP at that visit. The BP response to trandolapril was calculated as (BP after trandospril treatment) – (BP before trandolapril use). Only patients with BP readings at these two visits were included in the BP response analysis.
The genetic substudy of INVEST, INVEST-GENES, collected DNA samples from 5,979 participants residing in the United States including Puerto Rico. Participants provided written informed consent to participate in INVEST and INVEST-GENES. The study was approved by an ethics committee for all participating study sites, and was conducted in accordance with the Declaration of Helsinki and the U.S. Code of Federal Regulations for Protection of Human Subjects. This analysis included 486 patients with analyzable data for BP response to trandolapril treatment.
Genotyping of the −816A>C variant
For the clinical study, genomic DNA was isolated from mouthwash buccal cell samples collected from participants using a commercially available kit (PureGene, Gentra Systems Inc., Minneapolis, MN). Genotyping analysis was carried out on TaqMan® 7900 HT SNP genotyping platform using Taqman® allele discrimination assays from Applied Biosystems (Life Technologies, Carlsbad, CA). The custom designed probes (6FAM-CATCACCCCTACTGCMGBNFQ, and VIC-CATCACACCTACTGCT-MGBNFQ), and PCR primers (Forward: 5’-CCTTAATTTGGTGATTTCACATTGC-3’; Reverse: 5’-CAAGACATGGTTCAGCTTCTCAAG-3’) were purchased from Applied Biosystems (Life Technologies, Carlsbad, CA), and used in 5 µl reactions in 384-well plates according to the manufacturer's recommendations.
For human liver samples, total genomic DNA was extracted from 100 individuals with Pure Link™ Genomic DNA Mini Kit (Life technology, Austin, TX) according to the manufacturer’s instructions. To determine the −816A>C genotypes, the promoter region of CES1P1 was amplified using the primers and thermocycling conditions summarized in Supplemental Tables 1 and 2. The PCR products were purified with the Pure Link™ Quick PCR Purification Kit (Life technologies, Austin, TX), then subjected to Sanger sequencing analysis utilizing the same PCR primers.
Western blot study of CES1 expression in human livers
Human liver s9 fraction (HLS9) samples were prepared according to a standard procedure [10]. Levels of CES1 expression in individual HLS9 samples were determined by an established western blot assay [2, 10]. CES1 expression of each sample was semi-quantified by comparing the density of CES1 bands with the bands obtained from various amounts of pooled HLS9 samples.
Hydrolysis of trandolapril in individual human liver s9 fractions
An in vitro incubation study was performed to assess CES1 activity on trandolapril hydrolysis (i.e. bioactivation) in individual human liver samples. Trandolapril (200 µM) and human HLS9 (0.2 mg/ml) were freshly prepared in phosphate buffered saline (PBS, pH7.4) from stock solutions, and hydrolysis was initiated by mixing equal volumes of trandolapril and HLS9 fraction work solutions. After incubation at 37 °C for10 min, the reaction was terminated by adding 4-fold volume of acetonitrile containing the analytic internal standard, simvastatin acid (20 ng/ml). Samples were then briefly vortexed, and centrifuged at 13,200 rpm at 4°C for 20 min to remove precipitated proteins. Ten µl of the supernatant was injected into an HPLC-MS/MS system for the analysis of the concentrations of trandolaprilat, the active acid metabolite of trandolapril formed via CES1-mediated hydrolysis.
HPLC-MS/MS assay for trandolaprilat
An HPLC-MS/MS assay was developed to determine concentrations of the trandolapril active metabolite, trandolaprilat, based on a previously published method with some modifications [11]. The HPLC-MS/MS analytical system consisted of a Shimadzu Prominence HPLC system and an Applied Biosystems API 4000 QTRAP® mass spectrometer. The liquid chromatographic separation was performed on a reverse phase column (Restek, Ultra II® C18, 2.0 × 150 mm, 5 micron) with gradient elution at a constant flow rate of 0.3 ml/min at 40°C. The mobile phase consisted of 2 mM ammonium acetate and 0.2% formic acid in water (A) and 2 mM ammonium acetate and 0.2% formic acid in methanol (B). The linear gradient was run as the follows: 0 min, 50% B; 3 – 5 min, ramped to 95% B; 5.5 – 8.5 min, returned to 50% B. The entire acquisition time was 8.5 min. The MS was operated in an electrospray negative ionization mode using multiple reaction monitoring (MRM) with the m/z transitions of 429.0>168.0, 401.0>168.0, and 435.3>319.0, for trandolapril, trandolaprilat, and simvastatin acid (I.S.), respectively. The assay was validated for accuracy and precision using blank matrix spiked with three concentrations (0.03, 3, 10 µM) of trandolaprilat. Accuracy and precision were within 98.0% to 100.9% and 0.3% to 1.0%, respectively.
Statistical analysis
The continuous variables are presented as mean and standard deviation (SD) and categorical variables are presented as numbers and percentages as appropriate. Hardy-Weinberg Equilibrium (HWE) was tested within each race group using chi-square test with 1 degree of freedom. The BP response to trandolapril therapy was calculated as the changes of BP following trandolapril treatment (i.e. BP after trandolapril therapy - BP before trandolapril therapy). The association between the −816A>C genotypes and BP response was evaluated using linear regression within each race/ethnicity group, adjusting for age, gender, trandolapril dose and baseline BP. For in vitro experiments, data are presented as mean ± SD of three independent experiments. One-way analysis of variance (ANOVA) was utilized to assess the effect of the −816A>C genotypes on the expression and activity of CES1 in human liver samples. The correlation between CES1 expression and activity on trandolapril activation was assessed with Pearson correlation analysis. All statistical analysis was performed in SAS 9.4 (Cary, NC).
Results
The −816A>C genotypes were not associated with the BP lowering effect of trandolapril in INVEST participants
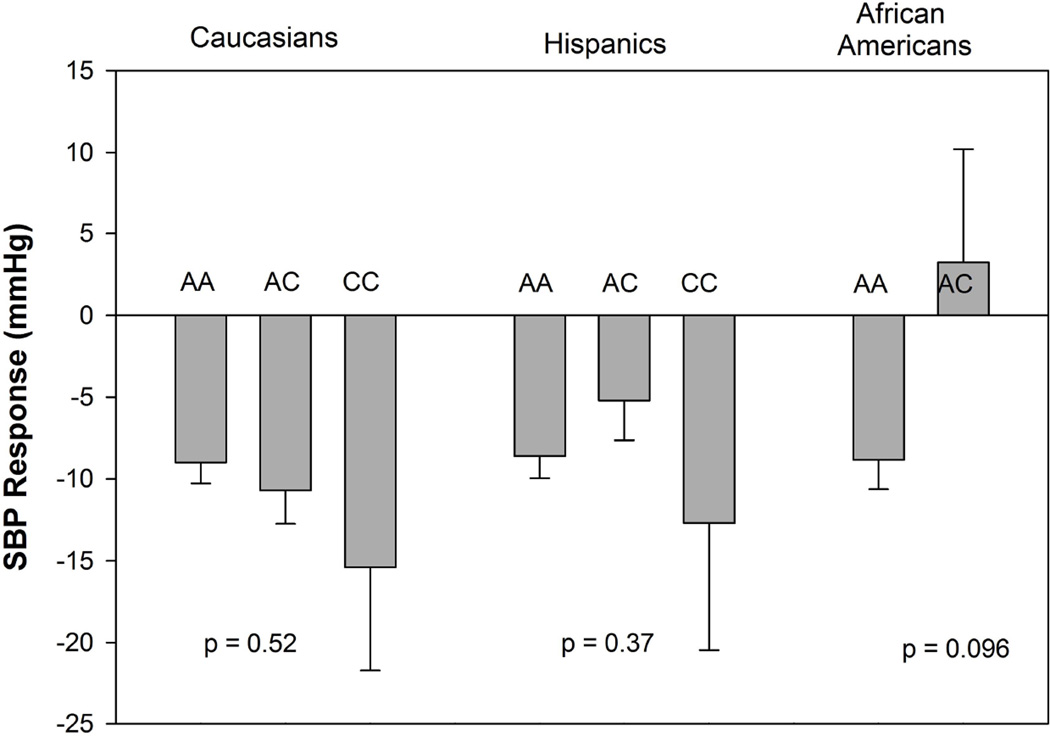
Baseline demographics of the 486 INVEST-GENES patient samples with analyzable genotype and BP response to trandolapril are shown in Table 1. This subset of participants were largely comparable to the overall INVEST participants, except for higher baseline blood pressure and higher percentage of black and Hispanics and lower percentage of white participants. The minor allele frequency (MAF) of −816 A>C was 16.6% in Caucasians, 13.5% in Hispanics and 3.3% in African Americans, respectively. The genotype frequencies of this SNP in all race groups did not deviate from the Hardy Weinberg Equilibrium (Table 2). We were unable to identify any evidence that this SNP was associated with a systolic BP (SBP) response to trandolapril (Figure 1). The p values were 0.52 in Caucasians, 0.37 in Hispanics and 0.096 in African Americans, respectively.
Table 1.
Baseline demographics of the 486 INVEST-GENES participants
| Characteristics | This study (n=486) | INVEST (n=22,576) | p |
|---|---|---|---|
| Age, mean (SD), years | 66 (10) | 66 (10) | 0.42 |
| Women | 285 (59%) | 11770 (52%) | 0.23 |
| Baseline BP, mean (SD), mmHg | |||
| Systolic | 151 (18) | 148 (18) | 0.0001 |
| Diastolic | 87 (10) | 87 (12) | 0.0015 |
| Race/ethnicity | 0.0006 | ||
| White | 214 (44%) | 10925 (48%) | |
| Black | 77 (16%) | 3029 (13%) | |
| Hispanic | 189 (39%) | 8045 (36%) | |
| Other/multiracial | 6 (1%) | 577 (3%) | |
| BMI, mean (SD), kg/m2 | 30 (6) | 29 (7) | 0.04 |
| Smoking history | 189 (39%) | 2265 (41%) | 0.31 |
| Past Medical History | |||
| Myocardial infarction | 117 (24%) | 7218 (32%) | 0.65 |
| Angina pectoris | 348 (72%) | 15045 (67%) | 0.19 |
| Stroke/TIA | 38 (8%) | 1629 (7%) | 0.46 |
| Left ventricular hypertrophy | 88 (18%) | 4948 (22%) | 0.06 |
| Heart failure (class I-III) | 15 (3%) | 1256 (6%) | 0.73 |
| Peripheral vascular disease | 53 (11%) | 2699 (12%) | 0.91 |
| Diabetes | 137 (28%) | 6401 (28%) | 0.997 |
| Hypercholesterolemia | 254 (52%) | 12448 (55%) | 0.34 |
| Renal impairment | 8 (2%) | 424 (2%) | 0.9 |
Table 2.
Genotype and allele frequency of the −816 A>C variant in the INVEST-GENES samples and the 100 human liver samples
| INVEST GENES participants (n = 486) | Liver samples (n = 100) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Race/Ethnicity | AA | AC | CC | MAF (%) | P (HWE) | AA | AC | CC | MAF (%) | P (HWE) |
| Caucasian | 149 | 59 | 6 | 16.6 | 0.96 | 62 | 26 | 2 | 16.7 | 0.7 |
| African American | 72 | 5 | 0 | 3.25 | 0.77 | 6 | 0 | 0 | NA | NA |
| Hispanic | 142 | 43 | 4 | 13.5 | 0.73 | 2 | 0 | 0 | NA | NA |
| Others | 4 | 2 | 0 | 16.7 | 0.62 | 1 | 0 | 1 | NA | 0.15 |
Figure 1.
Systolic blood pressure response to trandolapril by CES1 −816 A>C genotype in each race/ethnicity group. The p values were adjusted for age, gender, baseline systolic blood pressure and dose. Bars: adjusted blood pressure response; error bars: standard error of the mean.
The −816A>C variant did not affect CES1 expression and activity on trandolapril activation in human livers
The promoter region of the CES1P1 gene was sequenced to determine the −816A>C genotypes in individual human liver samples. The MAF of the −816A>C was 16.7% within the Caucasian subjects genotyped (Table 1), which is consistent with the data from the INVEST GENES study. MAFs were not determined in other racial groups due to small sample sizes.
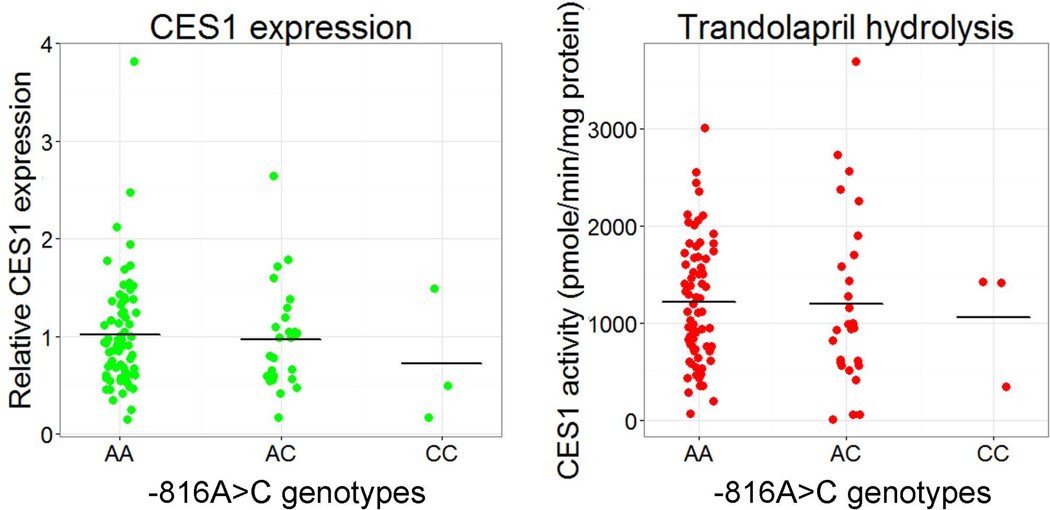
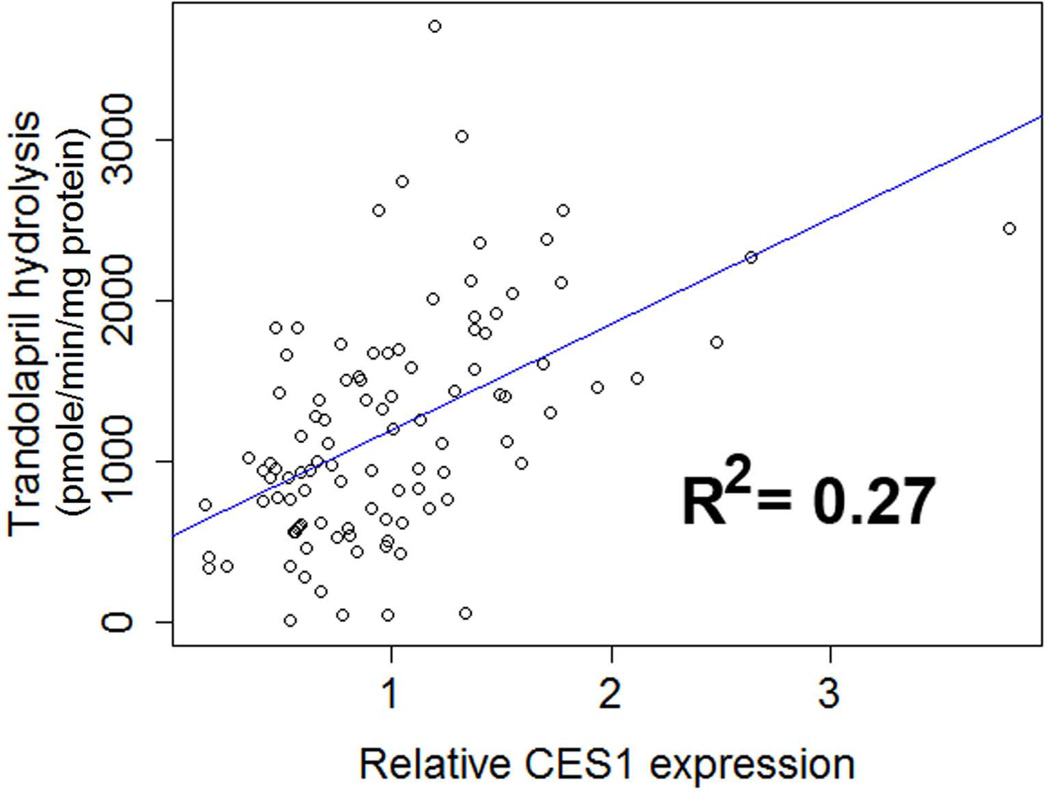
Western blotting study demonstrated that the protein expression of CES1 in the human liver samples were comparable among the three −816A>C genotypes (i.e. AA, AC, and CC, P = 0.51, Figure 2A). Additionally, the genotypes were not associated with CES1 activity on trandolapril hydrolysis (P = 0.57), as measured by the formation of the active metabolite trandolaprilat (Figure 2B). The correlation analysis of CES1 expression and activity on trandolapril activation showed that the activity was significantly correlated to CES1 expression levels (P<0.001, R2=0.27, Figure 3).
Figure 2.
Effect of the CES1P1 variant −816A>C on CES1 expression (A) and activity on trandolapril activation (B) in human liver samples (n=100). The data were categorized into three groups based on the −816A>C genotypes (i.e. AA, AC, CC). Horizontal bars represent mean values in each group.
Figure 3.
Correlation analysis of CES1 expression and CES1 activity on trandolapril hydrolysis in human liver samples (n=100).
Discussion
It has been well documented that pharmacokinetics and therapeutic outcomes of ACEIs varies significantly among individual patients [12–17]. Given that the majority of ACEIs are CES1-activated ester prodrugs, the functional status of CES1 could be an important contributing factor to interindividual variability in the pharmacokinetics and pharmacodynamics of ACEIs. A clinical study conducted by Geshi and associates suggested that the −816A>C, a common CES1P1 promoter variant, was associated with greater BP lowering effect of the ACEI prodrug imidapril in Japanese hypertensive patients [5]. This finding led to the assumption that this SNP might be associated with the enhanced CES1 expression, and consequently increases imidapril activation and antihypertensive effects. The clinical significance of the variant −816A>C was further investigated by two recently published clopidogrel clinical studies. One of the studies reported that platelet activity in patients treated with clopidogrel was higher in the −816C carriers relative to that in non-carriers, suggesting that CES1 activity is higher in patients carrying the minor C allele, an observation in agreement with the previous imidapril study. However, a second clopidogrel study published in the same year indicated that platelet activity was lower in the C allele carriers than those with the AA genotype, suggesting that the C allele is associated with decreased CES1 function, an observation contradictory to the findings from the two aforementioned studies.
To elucidate potential impact of the −816A>C variant on hepatic CES1 function, we determined CES1 expression and its activity on trandolapril activation in individual normal human liver samples (n=100). No significant differences of CES1 expression were observed among the three −816A>C genotypes (A/A, A/C, and C/C). Consistent with the expression profiles, CES1 activity toward trandolapril bioactivation was not found to be associated with the −816A>C genotypes. Furthermore, BP lowering effects of trandolapril did not significantly differ between the A/A, A/C, and C/C genotypes, which is in full agreement with the in vitro human liver study. Taken together, these results suggest that the −816A>C variant is unlikely to be a contributing factor to varied CES1 function and interindividual variability in response to trandolapril or any number of other medications predominantly metabolized by CES1.
Substantial variations in CES1 expression and activity were observed in the 100 individual human liver samples assayed, which is likely attributable to both genetic and non-genetic factors. To date, over 1000 SNPs have been identified within the loci of the CES1 and CES1P1 genes. The first documented clinically significant CES1 nonsynonymous variant G143E (rs71647871) was originally discovered in our laboratory from a study subject who participated in a clinical pharmacokinetic study. The MAFs of the G143E were found to be 3.7%, 4.3%, 2.0%, and 0% in European, African American, Hispanic, and Asian populations, respectively. The G143E is a loss-of-function variant for the metabolism of several CES1 substrates including methylphenidate, clopidogrel, and many ACEI prodrugs while the variant exhibits significantly decreased activity for catalyzing hydrolysis (activation) of the antivirus prodrug oseltamivir [2, 4, 18–20]. Several clinical studies have demonstrated that pharmacokinetics and/or pharmacodynamics of CES1 substrate medications were significantly altered in the subjects carrying the G143E variant. Besides the G143E variant, more than 200 nonsynonymous CES1 variants have been reported in various SNP databases (http://exac.broadinstitute.org). Most of these SNPs are rare variants with MAFs less than 0.1%, and the functionality of these variants largely remains undetermined. In addition to nonsynonymous variants, many SNPs were discovered in regulatory regions of the CES1 and CES1P1 genes. For example,, the −75T>G (rs3815583) is a common SNP located in 5’-untranslated region (5’-UTR) of CES1, and was associated with appetite reduction in patients with attention-deficit/hyperactivity disorder treated with the CES1 substrate methylphenidate [21]. However, it remains unknown as to whether this SNP is associated with altered CES1 expression and catalytic function. In addition to genetic variants, various environmental factors, such as many medications serving as CES1 inhibitors, may affect CES1 activity, and consequently alter pharmacokinetics and pharmacodynamics of medications metabolized by CES1 [10, 22–29].
The highly homologous CES1 and CES1P1 genes are located in the same chromosome region, suggesting the two genes were originated by gene duplication during an evolutionary process. It is interesting that the pseudogene CES1P1 variation CES1P1VAR is a functional gene encoding functional CES1 protein which is identical to the protein product of CES1. Therefore, it has been suspected that the CES1P1/CES1P1VAR genotypes may affect CES1 expression. However, our in vitro human liver study has revealed that the CES1P1/CES1P1VAR genotypes are not associated with CES1 protein expression levels in human livers [4], which is likely due to that the transcription efficiency of CES1P1/CES1P1VAR is only 2% of the CES1 gene, thus overall contribution of CES1P1VAR gene to CES1 expression appears negligible. The −816A>C is a variant residing in the promoter region of the CES1P1/CES1P1VAR gene. A previous study suggested that the −816A>C may be associated with higher transcription activity based on an in vitro reporter assay [30]. However, the present study demonstrated that the −816A>C genotypes do not significantly influence hepatic CES1 expression and activity, which is consistent with the fact that the SNP is in almost complete linkage with the nonfunctional pseudogene CES1P1 [31], and the transcription efficiency of the CES1P1/CES1P1VAR in the liver is extremely low relative to the CES1 gene [32]. We did not pursue the study of other CES1 variants due to the limited DNA materials from our clinical samples.
In summary, our study has provided both in vitro and in vivo evidence supporting our conclusion that the 816A>C variant is unlikely to be a contributing factor to interindividual variability in CES1 function nor variability in response to the treatment of ACEI prodrugs and other medications metabolized by CES1. Further study is warranted to elucidate the function of numerous, and as yet, unstudied CES1 genetic polymorphisms which may enable the greater use of CES1 variants as biomarkers for individualizing pharmacotherapy involving CES1 substrate drugs.
Supplementary Material
Acknowledgments
Research reported in this publication was supported in part by the National Institute on Aging (R21AG048500) (Hao-Jie Zhu), the National Center for Advancing Translational Sciences of the National Institutes of Health under Award Number 2UL1TR000433 (Hao-Jie Zhu) and the American Association of Colleges of Pharmacy (AACP) 2015 New Investigator Award (Hao-Jie Zhu).
References
- 1.Takai S, Matsuda A, Usami Y, Adachi T, Sugiyama T, Katagiri Y, Tatematsu M, Hirano K. Hydrolytic profile for ester- or amide-linkage by carboxylesterases pI 5.3 and 4.5 from human liver. Biol Pharm Bull. 1997;20(8):869–873. doi: 10.1248/bpb.20.869. [DOI] [PubMed] [Google Scholar]
- 2.Zhu HJ, Appel DI, Johnson JA, Chavin KD, Markowitz JS. Role of carboxylesterase 1 and impact of natural genetic variants on the hydrolysis of trandolapril. Biochem Pharmacol. 2009;77(7):1266–1272. doi: 10.1016/j.bcp.2008.12.017. DOI S0006-2952(08)00908-8 [pii] 10.1016/j.bcp.2008.12.017. [DOI] [PubMed] [Google Scholar]
- 3.Thomsen R, Rasmussen HB, Linnet K. In Vitro Drug Metabolism by Human Carboxylesterase 1: Focus on Angiotensin-converting Enzyme Inhibitors. Drug Metab Dispos. 2013 doi: 10.1124/dmd.113.053512. [DOI] [PubMed] [Google Scholar]
- 4.Wang X, Wang G, Shi J, Aa J, Comas R, Liang Y, Zhu HJ. CES1 genetic variation affects the activation of angiotensin-converting enzyme inhibitors. Pharmacogenomics J. 2015 doi: 10.1038/tpj.2015.42. [pii] DOI 10.1038/tpj.2015.42 tpj201542 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Geshi E, Kimura T, Yoshimura M, Suzuki H, Koba S, Sakai T, Saito T, Koga A, Muramatsu M, Katagiri T. A single nucleotide polymorphism in the carboxylesterase gene is associated with the responsiveness to imidapril medication and the promoter activity. Hypertens Res. 2005;28(9):719–725. doi: 10.1291/hypres.28.719. [DOI] [PubMed] [Google Scholar]
- 6.Xie C, Ding X, Gao J, Wang H, Hang Y, Zhang H, Zhang J, Jiang B, Miao L. The effects of CES1A2 A(−816)C and CYP2C19 loss-of-function polymorphisms on clopidogrel response variability among Chinese patients with coronary heart disease. Pharmacogenet Genomics. 2014 doi: 10.1097/FPC.0000000000000035. DOI 10.1097/FPC.0000000000000035. [DOI] [PubMed] [Google Scholar]
- 7.Zou J-J, Chen S-L, Fan H-W, Tan J, He B-S, Xie H-G. The CES1A −816C as a genetic marker to predict greater platelet clopidogrel response in patients with percutaneous coronary intervention. Journal of Cardiovascular Pharmacology. 2014;63(2):178–183. doi: 10.1097/FJC.0000000000000037. [DOI] [PubMed] [Google Scholar]
- 8.Pepine CJ, Handberg-Thurmond E, Marks RG, Conlon M, Cooper-DeHoff R, Volkers P, Zellig P. Rationale and design of the International Verapamil SR/Trandolapril Study (INVEST): an Internet-based randomized trial in coronary artery disease patients with hypertension. J Am Coll Cardiol. 1998;32(5):1228–1237. doi: 10.1016/s0735-1097(98)00423-9. [DOI] [PubMed] [Google Scholar]
- 9.Pepine CJ, Handberg EM, Cooper-DeHoff RM, Marks RG, Kowey P, Messerli FH, Mancia G, Cangiano JL, Garcia-Barreto D, Keltai M, Erdine S, Bristol HA, Kolb HR, Bakris GL, Cohen JD, Parmley WW. A calcium antagonist vs a non-calcium antagonist hypertension treatment strategy for patients with coronary artery disease. The International Verapamil-Trandolapril Study (INVEST): a randomized controlled trial. JAMA. 2003;290(21):2805–2816. doi: 10.1001/jama.290.21.2805. [DOI] [PubMed] [Google Scholar]
- 10.Zhu HJ, Appel DI, Jiang Y, Markowitz JS. Age- and sex-related expression and activity of carboxylesterase 1 and 2 in mouse and human liver. Drug Metab Dispos. 2009;37(9):1819–1825. doi: 10.1124/dmd.109.028209. DOI dmd.109.028209 [pii] 10.1124/dmd.109.028209. [DOI] [PubMed] [Google Scholar]
- 11.Nirogi RV, Kandikere VN, Shrivastava W, Mudigonda K. Quantification of trandolapril and its metabolite trandolaprilat in human plasma by liquid chromatography/tandem mass spectrometry using solid-phase extraction. Rapid Commun Mass Spectrom. 2006;20(24):3709–3716. doi: 10.1002/rcm.2794. [DOI] [PubMed] [Google Scholar]
- 12.Tsoukas G, Anand S, Yang K. Dose-dependent antihypertensive efficacy and tolerability of perindopril in a large, observational, 12-week, general practice-based study. Am J Cardiovasc Drugs. 2011;11(1):45–55. doi: 10.2165/11587000-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 13.Malacco E, Omboni S, Volpe M, Auteri A, Zanchetti A. Antihypertensive efficacy and safety of olmesartan medoxomil and ramipril in elderly patients with mild to moderate essential hypertension: the ESPORT study. J Hypertens. 2010;28(11):2342–2350. doi: 10.1097/HJH.0b013e32833e116b. [DOI] [PubMed] [Google Scholar]
- 14.Ionescu DD. Antihypertensive efficacy of perindopril 5–10 mg/day in primary health care: an open-label, prospective, observational study. Clin Drug Investig. 2009;29(12):767–776. doi: 10.2165/11319700-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 15.Mugellini A, Dobovisek J, Planinc D, Cremonesi G, Fogari R. Efficacy and safety of delapril plus manidipine compared with enalapril plus hydrochlorothiazide in mild to moderate essential hypertension: results of a randomized trial. Clin Ther. 2004;26(9):1419–1426. doi: 10.1016/j.clinthera.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 16.Kostis JB, Packer M, Black HR, Schmieder R, Henry D, Levy E. Omapatrilat and enalapril in patients with hypertension: the Omapatrilat Cardiovascular Treatment vs. Enalapril (OCTAVE) trial. Am J Hypertens. 2004;17(2):103–111. doi: 10.1016/j.amjhyper.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 17.Poirier L, Bourgeois J, Lefebvre J, Archambault F, Lacourciere Y. ACE Inhibitors as First-Line Treatment Agents: A Comparative Study of Trandolapril and Enalapril on Casual and Ambulatory Blood Pressures. Am J Ther. 1995;2(3):159–164. [PubMed] [Google Scholar]
- 18.Zhu HJ, Wang X, Gawronski BE, Brinda BJ, Angiolillo DJ, Markowitz JS. Carboxylesterase 1 as a determinant of clopidogrel metabolism and activation. J Pharmacol Exp Ther. 2013;344(3):665–672. doi: 10.1124/jpet.112.201640. jpet.112.201640 [pii] [DOI] [PubMed] [Google Scholar]
- 19.Zhu HJ, Patrick KS, Yuan HJ, Wang JS, Donovan JL, DeVane CL, Malcolm R, Johnson JA, Youngblood GL, Sweet DH, Langaee TY, Markowitz JS. Two CES1 gene mutations lead to dysfunctional carboxylesterase 1 activity in man: clinical significance and molecular basis. Am J Hum Genet. 2008;82(6):1241–1248. doi: 10.1016/j.ajhg.2008.04.015. DOI S0002-9297(08)00275-9 [pii] 10.1016/j.ajhg.2008.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu HJ, Markowitz JS. Activation of the antiviral prodrug oseltamivir is impaired by two newly identified carboxylesterase 1 variants. Drug Metab Dispos. 2009;37(2):264–267. doi: 10.1124/dmd.108.024943. DOI dmd.108.024943 [pii] 10.1124/dmd.108.024943. [DOI] [PubMed] [Google Scholar]
- 21.Bruxel EM, Salatino-Oliveira A, Genro JP, Zeni CP, Polanczyk GV, Chazan R, Rohde LA, Hutz MH. Association of a carboxylesterase 1 polymorphism with appetite reduction in children and adolescents with attention-deficit/hyperactivity disorder treated with methylphenidate. Pharmacogenomics J. 2012 doi: 10.1038/tpj.2012.25. tpj201225 [pii] [DOI] [PubMed] [Google Scholar]
- 22.Shi D, Yang D, Prinssen EP, Davies BE, Yan B. Surge in expression of carboxylesterase 1 during the post-neonatal stage enables a rapid gain of the capacity to activate the anti-influenza prodrug oseltamivir. J Infect Dis. 2011;203(7):937–942. doi: 10.1093/infdis/jiq145. DOI jiq145 [pii] 10.1093/infdis/jiq145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang D, Pearce RE, Wang X, Gaedigk R, Wan YJ, Yan B. Human carboxylesterases HCE1 and HCE2: ontogenic expression, inter-individual variability and differential hydrolysis of oseltamivir, aspirin, deltamethrin and permethrin. Biochem Pharmacol. 2009;77(2):238–247. doi: 10.1016/j.bcp.2008.10.005. DOI S0006-2952(08)00716-8 [pii] 10.1016/j.bcp.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu HJ, Appel DI, Peterson YK, Wang ZC, Markowitz JS. Identification of selected therapeutic agents as inhibitors of carboxylesterase 1: Potential sources of metabolic drug interactions. Toxicology. 2010;270(2–3):59–65. doi: 10.1016/j.tox.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 25.Hatfield MJ, Potter PM. Carboxylesterase inhibitors. Expert Opin Ther Pat. 2011 doi: 10.1517/13543776.2011.586339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi D, Yang J, Yang D, LeCluyse EL, Black C, You L, Akhlaghi F, Yan B. Anti-influenza prodrug oseltamivir is activated by carboxylesterase human carboxylesterase 1, and the activation is inhibited by antiplatelet agent clopidogrel. J Pharmacol Exp Ther. 2006;319(3):1477–1484. doi: 10.1124/jpet.106.111807. [DOI] [PubMed] [Google Scholar]
- 27.Rhoades JA, Peterson YK, Zhu HJ, Appel DI, Peloquin CA, Markowitz JS. Prediction and In Vitro Evaluation of Selected Protease Inhibitor Antiviral Drugs as Inhibitors of Carboxylesterase 1: A Potential Source of Drug-Drug Interactions. Pharm Res. 2011 doi: 10.1007/s11095-011-0637-9. [DOI] [PubMed] [Google Scholar]
- 28.Kristensen KE, Zhu HJ, Wang X, Gislason GH, Torp-Pedersen C, Rasmussen HB, Markowitz JS, Hansen PR. Clopidogrel bioactivation and risk of bleeding in patients cotreated with angiotensin-converting enzyme inhibitors after myocardial infarction: a proof-of-concept study. Clin Pharmacol Ther. 2014;96(6):713–722. doi: 10.1038/clpt.2014.183. [DOI] [PubMed] [Google Scholar]
- 29.Tarkiainen EK, Holmberg MT, Tornio A, Neuvonen M, Neuvonen PJ, Backman JT, Niemi M. Carboxylesterase 1 c.428G>A single nucleotide variation increases the antiplatelet effects of clopidogrel by reducing its hydrolysis in humans. Clin Pharmacol Ther. 2015;97(6):650–658. doi: 10.1002/cpt.101. [DOI] [PubMed] [Google Scholar]
- 30.Yoshimura M, Kimura T, Ishii M, Ishii K, Matsuura T, Geshi E, Hosokawa M, Muramatsu M. Functional polymorphisms in carboxylesterase1A2 (CES1A2) gene involves specific protein 1 (Sp1) binding sites. Biochem Biophys Res Commun. 2008;369(3):939–942. doi: 10.1016/j.bbrc.2008.02.120. [DOI] [PubMed] [Google Scholar]
- 31.Sai K, Saito Y, Tatewaki N, Hosokawa M, Kaniwa N, Nishimaki-Mogami T, Naito M, Sawada J, Shirao K, Hamaguchi T, Yamamoto N, Kunitoh H, Tamura T, Yamada Y, Ohe Y, Yoshida T, Minami H, Ohtsu A, Matsumura Y, Saijo N, Okuda H. Association of carboxylesterase 1A genotypes with irinotecan pharmacokinetics in Japanese cancer patients. Br J Clin Pharmacol. 2010;70(2):222–233. doi: 10.1111/j.1365-2125.2010.03695.x. DOI BCP3695 [pii] 10.1111/j.1365-2125.2010.03695.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fukami T, Nakajima M, Maruichi T, Takahashi S, Takamiya M, Aoki Y, McLeod HL, Yokoi T. Structure and characterization of human carboxylesterase 1A1, 1A2, and 1A3 genes. Pharmacogenet Genomics. 2008;18(10):911–920. doi: 10.1097/FPC.0b013e32830b0c5e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.