Abstract
Purpose
To evaluate the inter-study repeatability of multi-slice quantitative cardiovascular magnetic resonance myocardial blood flow (MBF), myocardial perfusion reserve (MPR), and extracellular volume (ECV). A unique saturation recovery self-gated acquisition was used for the perfusion scans.
Materials and Methods
An ungated golden angle radial turboFLASH pulse sequence was used to scan 10 subjects on two separate days on a 3T scanner. A single saturation pulse was followed by a set of 4 slices. Rest and hyperemia scans were acquired during free breathing. The images were reconstructed using an iterative algorithm with spatiotemporal constraints. The ungated images were retrospectively binned (self-gated) into near-systole and near-diastole. Deformable registration was performed to adjust for respiratory and residual cardiac motion, and the data was fit with a Fermi model to estimate the inter-study repeatability of quantitative self-gated MBF and MPR.
Results
The coefficient of variation (CoV) of the territorial MPR using the self-gated near-systole data was 18.6%. The self-gated near-diastole data gave less good CoV of MPR, equal to 46.2%. For MBFs, and using smaller (segmental) regions, the CoVs were 20.1% and 22.7% for the estimation of myocardial blood flow at stress and rest respectively using the self-gated near-systole data. The self-gated near-diastole data gave CoV=48.6% and 44.9% for stress and rest.
Conclusion
The self-gated free breathing technique for quantification of myocardial blood flow showed good repeatability for near-systole, with results comparable to published studies on inter-study repeatability of quantitative myocardial perfusion MRI using ECG-gating and breath-holds. Self-gated near-diastole data results were less repeatable.
Keywords: Inter-study repeatability, MRI myocardial perfusion, quantitative perfusion, ECG-gating, self-gated
INTRODUCTION
Magnetic resonance is a valuable tool for quantification of myocardial perfusion. Conventional studies for quantification of myocardial blood flow (MBF) acquire one or more slices to study the uptake and washout of a gadolinium based contrast agent (1). An ECG gating signal is used to ensure that images in a given slice are acquired during the same cardiac phase. However, ECG gating can suffer from problems such as the magneto-hemodynamic effect, signal distortion by magnetic field gradients, and the possibility of a poor gating signal due to patient size and shape (2).
Developments in MR reconstruction methods have made it possible to acquire multiple slices for myocardial perfusion assessment without the need for ECG gating (3). Multiple slices are acquired continuously between saturation pulses, at different cardiac phases. This ‘ungated’ data has been shown to be useful for visual assessment of coronary artery disease (3). Moreover, since data is acquired rapidly and continuously at a high temporal resolution, it is possible to split the acquired data into multiple cardiac phases. This classification of the data, for example into near-systolic and near-diastolic cardiac phases, can also be referred to as ‘self-gating’. A previous study by Likhite et al. reported that MBF estimates using a multi-slice self-gated approach were similar to those using an ECG-gated acquisition at rest (4). A comparison by Chen et al. in (5) showed similar results with a single slice acquisition for MBF estimates between ECG-gated and self-gated acquisition. Recently, Chen et al. extended their work to a multi-slice acquisition and reported that the self-gated acquisition gave MBF estimates comparable to those using an ECG-triggered acquisition (6).
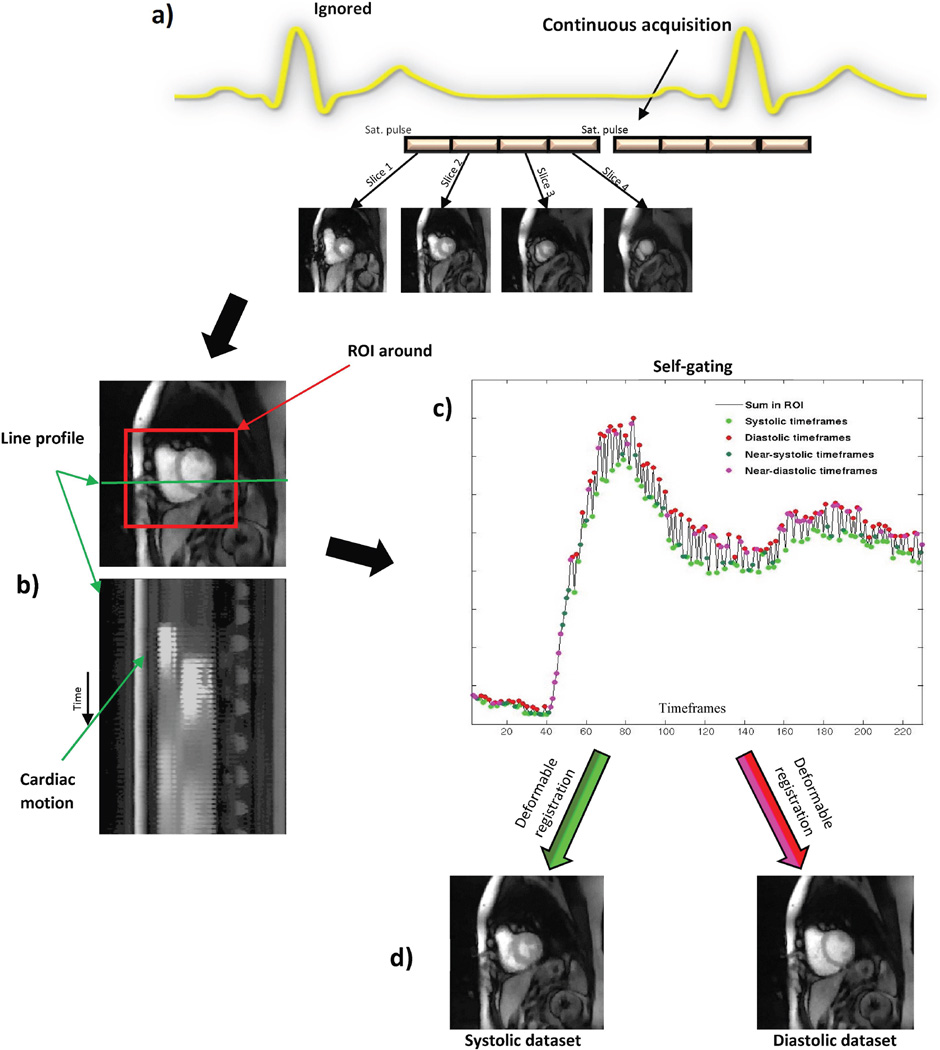
Figure 1a shows a schematic representation of the ungated/self-gated acquisition method. As seen from the figure, the scanner does not use the ECG as a trigger. The sequence acquires data continuously as a set of 4–5 slices after a saturation pulse; different images are acquired during different cardiac phases. By automatically binning this ungated data into two separate bins corresponding to near-systolic and near-diastolic cardiac phases, MBF estimates can be calculated (4).
Figure 1. Schematic of the ungated acquisition and self-gating.
a) Representation of ungated acquisition. The scanner ignores any ECG-triggering signals and acquires data continuously. b) Line profile through a slice of the ungated data shows the presence of cardiac motion. c) Self-gating of the ungated data into two discrete bins corresponding to near-systole and near-diastole. d) Every frame appears in one or the other of the near-systolic and near-diastolic datasets after deformable registration.
There have been relatively few studies looking at the reproducibility of quantitative MBF or myocardial perfusion reserve (MPR) (7–11). While the main aim of this study was to analyze the inter-study repeatability of quantitative MPR and MBF using a quantitative self-gated MR approach, we also studied the inter-study repeatability of myocardial fibrosis assessment by extracellular volume fraction (ECV) derived from modified look locker inversion recovery (MOLLI) T1 mapping (refer ‘Additional file 1’). Studies have reported good reproducibility of native and post contrast T1 derived from MOLLI T1 mapping (12–14). However, data on inter-study repeatability of ECV derived from MOLLI T1 mapping are limited (12, 15) and do not well-represent coronary artery disease and myocardial infarction populations.
MATERIALS AND METHODS
Overview
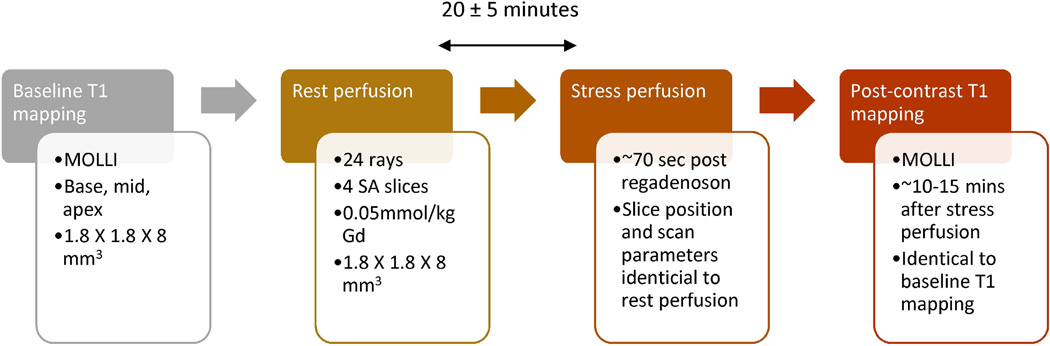
The study protocol involved acquisition of rest and stress perfusion data along with pre-contrast and post-contrast T1 mapping as shown in Figure 2. Pre-contrast MOLLI T1 mapping was performed first, followed by a rest perfusion scan. The rest perfusion scan was then followed by a stress perfusion scan. The rest and stress perfusion scans were separated by 20 ± 5 minutes. Post-contrast MOLLI T1 mapping with scan parameters and slices similar to pre-contrast T1 mapping was performed about 12 minutes after the contrast injection for stress perfusion. Details for the T1 mapping are provided in Additional File 1.
Figure 2. Summary of acquisition protocol.
The flow-chart describes the order and scan parameters for various parts of the study acquisition protocol.
Data acquisition
Ten subjects (48±12 years, 8 males, 2 females) were imaged on a Siemens 3T Verio scanner. Informed consent from the patients was obtained in accordance with the University of Utah Institutional Review Board. Table 1 shows a summary of the subjects involved in the study. Exclusion criteria included claustrophobia, pregnancy, unstable angina, pacemaker/defibrillator, severe hypotension, or glomerular filtration rate less than 30 ml/min/1.73 m2. Subjects were scanned on two separate days using a rest-stress protocol with no ECG gating. The separation between the two scans was 9.5 ± 4.5 days. Subjects did not have hemodynamic instability or adverse events during the interval between Scan 1 and Scan 2. The perfusion scans were performed using an ungated saturation recovery prepared TurboFLASH pulse sequence with golden angle radial acquisition. The acquisition parameters for the scans were 24 rays per image, TR=2.2 ms, TE=1.2 ms, flip angle=10°, resolution=1.8 × 1.8 × 8 mm3 voxels. Four short-axis (SA) slices were acquired after a single saturation pulse with a saturation recovery time of ~25 ms before the first slice. Gadoteridol (ProHance; Bracco Diagnostic, Princeton, NJ) 0.05 mmol/kg at rate of 5ml/sec was injected and ~240 frames were acquired over a minute with shallow breathing and no ECG gating. This was followed 20 ± 5 minutes later by an injection of regadenoson to induce hyperemia. Contrast was injected ~70 sec after regadenoson injection to ensure maximal stress (16) and the scan protocol was repeated to acquire four slices at stress. Slices were acquired from base to apex. The slices were positioned such that the slice 1 was as basal as possible without cutting through the valve plane. The same distance factor and slice thickness was used for all studies. Figure 2 shows a timeline of the acquisition protocol.
Table 1.
Summary of subjects in study
| Characteristics | # of subjects (n=10) |
|---|---|
| CAD | 5 (50%) |
| MI | 4 (40%) |
| Hypertension | 1 (10%) |
| Dyslipidemia | 5 (50%) |
| heart failure | 1 (10%) |
| AFIB | 1 (10%) |
| Smoking | 2 (20%) |
CAD-coronary artery disease, MI-myocardial infarction, AFIB- Atrial fibrillation
Reconstruction for perfusion scans
As seen from Figure 1a, the scanner acquired the perfusion data nearly continuously. An undersampled golden angle radial acquisition with 24 rays was used to acquire each image. The undersampled ungated radial data was reconstructed offline without manual intervention. The technique used a two step joint multi-coil spatio-temporal reconstruction with total variation constraints (17, 18). The cost function is shown in Eq. (1).
| (1) |
d is the k-space data from all coils, m is the combined coil image estimate, E is the encoding matrix that includes coil sensitivities (19). TVtime and TVspace are temporal and spatial total variation constraints with weights a1 and a2 respectively.
The two step process referred to means that first a preliminary reconstruction was done by minimizing C in Eq. (1) using all of the acquired ungated data. A coil sensitivity map for each coil was computed using all of the acquired data by grouping 250 rays from adjacent time frames and averaging the resulting images and then using the method described in (20). This preliminary reconstruction was only used to perform self-gating (described below), and 25 iterations of gradient descent to minimize C were sufficient in this first step to generate reasonable quality images for this purpose. After self-gating (described below), k-space data frames were grouped/binned into near-systole and near-diastole. Final reconstruction (using Eq. (1)) of the image sets in each of the two bins was then performed separately. Binning the k-space data frames reduced inter-frame cardiac motion and increased the temporal correlations leading to more effective application of the temporal TV constraint and better image quality. Reconstructed images from each bin were then grouped together to form the original ungated series for subsequent processing.
Self-gating
The final reconstructed dataset contained images acquired at different cardiac phases due to the absence of ECG gating. Figure 1b shows a line profile through a slice of one such ungated dataset from this study.
Because of the presence of cardiac motion, it was difficult to use such ungated data for quantification of MBF. Hence self-gating - splitting the ungated dataset into near-systolic and near-diastolic datasets as described in (4), was done. The self-gating used the sums in a region of interest (2). This process gave a time varying 1D signal as shown in Figure 1c. The 1D signal had an amplitude that changes between timeframes. Since the data was acquired rapidly without any ECG-triggering, the neighboring timeframes did not need to be in the same cardiac phase. This change in amplitude of the 1D signal between successive timeframes was due to the change in size of blood pool during different cardiac phases. During systole, the size of the myocardium and the blood pool was small, hence the smaller value for the sum in the region. The size of the blood pool was larger during diastole and gave a larger value for the sum in the region. The images that corresponded to the peaks in the 1D signal were classified as diastolic time frames and those corresponding to troughs were classified as systolic time frames. The remaining images were classified into the systolic or diastolic bin depending on which was closest to the summed signal intensity. Thus all of the acquired images were used. The process included automatic location of the heart and was completely automatic (see (4)).
Deformable registration
After the ungated data was binned into near-systole and near-diastole, there was residual cardiac motion and respiratory motion in each of the datasets. To correct for these motions, deformable registration using Advanced Normalization Tools (ANTs) (21) and fitted model images as reference images (22) was done. The model images were generated by fitting each voxel’s time course to the convolution of an input function and a decaying exponential. Deformable registration was done with the following set of ANTs parameters: step size for transformation model = 0.15, sigma (deformation field) = 2, sigma (similarity field) = 2. This self-gating and deformable registration was similar to methods that we used previously (4) to quantify MBF. In that previous work, it was noted that the model images used as reference images sometimes had a patchy appearance. These artificial patches in the model images sometimes interfered with the performance of deformable registration. During this study, we found that two iterations of deformable registration along with model generation gave improved results. During the first iteration, the model images were generated using the original self-gated perfusion dataset. These model images were used as reference images to perform deformable registration using ANTS. The motion corrected images thus obtained after deformable registration were free from breathing motion but still had some residual cardiac motion. These motion corrected intermediate images were used to generate a new set of model images. The new model images looked sharper and did not show the presence of artificial patches. These new sharper model images were again used as reference images with ANTs to perform deformable registration on the input data. More than two iterations of the process did not produce noticeable improvements. The entire process of deformable registration was completely automatic.
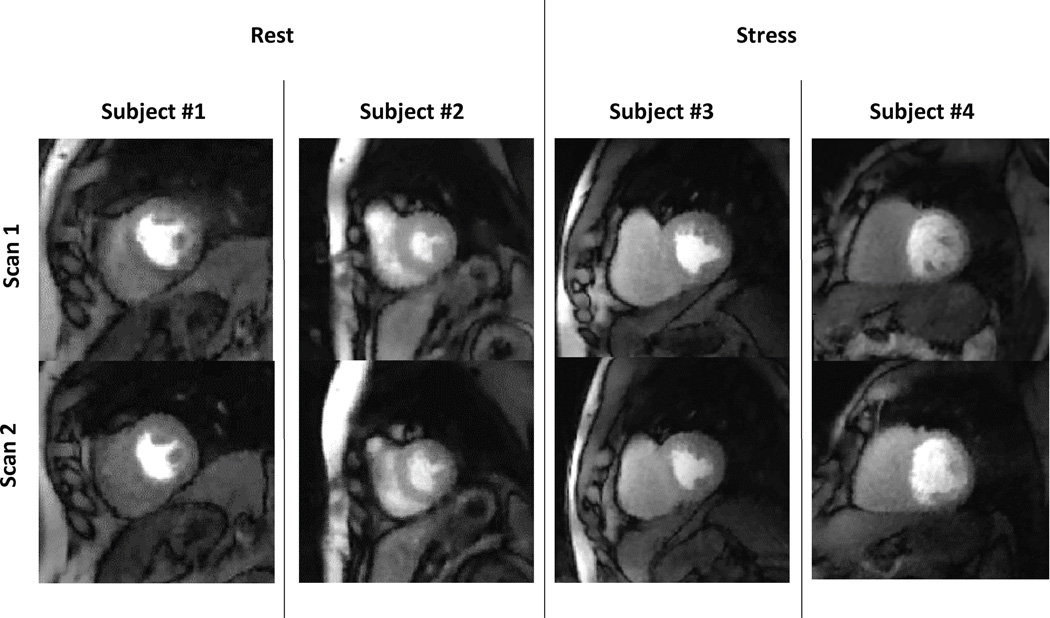
The use of self-gating along with deformable registration gave near-systole and near-diastole datasets from a single scan, as shown in Figure 1d. These two datasets were processed individually to quantify MBF at near-systole and near-diastole. Figure 3 shows the same slice from scan 1 and from scan 2 for four of the subjects in the study.
Figure 3. Data comparison between scan 1 and scan 2.
The figure compares two matching slices from rest and two matching slices from stress from scan1 and scan 2 for four volunteers.
Quantification of MBF
The quantification process was done using custom software based on (23) built in MATLAB® (The Mathworks, Inc., Natick, MA).
Tissue curves and arterial input function (AIF)
After registration, segmentation was done manually by drawing endocardial and epicardial contours on a single time frame of the images. The segmentation was done conservatively to minimize error in estimation of MBF (24). These contours were then copied to the remaining images in the time series. The copied contours were checked and could be shifted manually for each frame if it gave a better match. The segmented myocardium was divided into six circumferential regions. This process was repeated for all of the slices in the dataset. The segmentation was done by a single observer (4 years experience) to avoid inter-observer variability.
The tissue curves were recorded by averaging the signal intensity (SI) in each of the six circumferential segments of the myocardium. The AIF was obtained from the most basal self-gated near-diastolic slice. The most basal slice had the shortest saturation recovery time and thus the lowest signal saturation. The AIF was obtained automatically by averaging a subset of pixels inside the endocardial border. Only the values lying between 85%–95% of the maximum were averaged. For this work, the most basal slice was only used to obtain the AIF and the remaining 3 slices were used to obtain the tissue SI curves. Assuming the prescribed flip angle (α = 10°), Eq. (2) was used to obtain the T1.
| (2) |
where SRT was the saturation recovery time for the slice, and M0 was the total magnetization. The pulse sequence acquired five proton density frames at the beginning of each perfusion scan. The average of these proton density frames were used to estimate the total magnetization M0.
Using the T1 values calculated by Eq. (2), the AIF and the tissue SI curves were converted to [Gd] (gadolinium concentration) assuming fast exchange of water (25), precontrast T1 of 1660 ms for tissue at 3T (26), and a longitudinal relaxivity of 3.7mmol−1 sec−1 for gadoteridol (27). The conversion to [Gd] was done to account for any signal saturation in the AIF or tissue curves. This simplified the use of the basal AIF for all of the slices, since use of delta signal intensity curves (signal intensity curves with pre-contrast signal subtracted off) requires scaling between slices. Since the saturation recovery time was short for the first slice (~25ms), this method of obtaining the AIF from the first slice was similar to the dual sequence approach (28). The accuracy of the AIF obtained as described above was studied earlier (4) (Refer to ‘Additional file 3: Mini-website in (4)) and found to give AIFs comparable to a dual bolus AIF (29).
Model fitting
The AIF and tissue curves converted to gadolinium concentration were truncated automatically to only include the first pass of the contrast agent. The automatic truncation of tissue curves and the AIF was done by considering the zero crossing of the first derivative of the tissue curves and the AIF as described in (30). These truncated curves were then fit to a Fermi model by constrained deconvolution and the myocardial blood flow (MBF) was reported. The Fermi model was:
| (2) |
Here G and k represented the influx and efflux parameter for the contrast agent (CA). Δt represented the delay between LV blood pool enhancement and myocardial tissue enhancement. τ controlled the duration of the plateau portion of the Fermi function. Myocardial blood flow was reported as hF (t = 0), after the data was adjusted for time delay between blood and tissue enhancement (31, 32).
The resting MBF values were normalized by the rate pressure product (RPP) to account for differences in heart rate and blood pressure. The normalization was done as follows: Corrected MBF = MBF × (10000/individual RPP). The individual RPP was calculated as the product of the systolic blood pressure and the resting heart rate. A comparison of the non-normalized resting MBF with the normalized resting MBF values was also performed. A summary of those results is included in ‘Additional file 2’.
The same processing steps were followed for all other slices. The segmental MBFs from stress and rest were reported and the ratio of stress MBF to rest MBF was evaluated to give the segmental myocardial perfusion reserve (MPR). The segmental MPR values estimated were combined and assigned to the three coronary artery territories as described in (33) to give territorial MPR. The MPRs, as well as rest and stress MBFs estimated from the two different scans were compared to evaluate the inter-study repeatability.
The estimation of ECV using MOLLI T1 mapping is explained in ‘Additional file 1’.
STATISTICAL ANALYSIS
Patient characteristics were summarized as N (%) for the risk factors and comorbidity status.
A total of 180 segmental MBF values were obtained from each of the self-gated rest (systole), self-gated rest (diastole), self-gated stress (systole) and self-gated stress (diastole), at each visit. Segmental MPR was calculated for these MBF values and then computed for the three coronary artery territories as described in (33). This resulted in 3 territorial MPR values per subject. The normality of the MPR and MBF values was checked using the boxplot (34). The inter-study reproducibility was represented by the coefficient of variation (CoV), calculated as the standard deviation of the within-subject difference between the two scans relative to the mean of the two scans and expressed as a percent. The statistical analyses were performed using SAS (SAS Inst. Inc., Cary, NC, USA) version 9.4. Bland-Altman analysis was performed to assess the agreement between studies using the technique for data with repeated measures (35).
In addition, global flow values and perfusion reserve were calculated as an average over all of the segments and slices for each subject. These values were used to calculate intra-class correlation coefficient (ICC).
RESULTS
There was no statistically significant difference of heart rate between scans at baseline between the two visits: 66±12 vs 65±11, p=0.75.
A CoV of 18.6% indicating good inter-study repeatability was obtained for territorial MPR estimates using the self-gated near-systole dataset. Less good inter-study repeatability of CoV=46.2% was seen for the territorial MPR estimates from self-gated near-diastole data. Table 2 shows the mean rest (corrected for RPP) and stress MBF values calculated during scan 1 and scan 2 along with the segmental CoV and global ICC. A CoV of 22.7% and 20.1% were obtained for segmental MBF estimated at rest and stress respectively using the self-gated systole datasets. A CoV of 44.9% and 49.0% was obtained for the segmental MBF estimated at rest and stress respectively using the self-gated diastole datasets.
Table 2.
Summary of MPR and MBF values for scan 1 and scan 2 using self-gated near-systole and self-gated near-diastole.
| Scan 1 | Scan 2 | Segmental CoV |
Global ICC |
|||
|---|---|---|---|---|---|---|
| MPR | Mean ± SD | IQR | Mean ± SD | IQR | ||
| Systole | 2.6 ± 0.7 | 2.2–2.9 | 2.5 ± 0.8 | 1.8–3.2 | 25% | 0.88 |
| Diastole | 2.9 ± 0.9 | 2.4–3.4 | 3 ± 1.2 | 2.2–3.6 | 47.2% | 0.3 |
| Stress MBF (ml/min/g) | ||||||
| Systole | 1.7 ± 0.5 | 1.4–2.1 | 1.7 ± 0.6 | 1.2–2 | 20.1% | 0.90 |
| Diastole | 1.8 ± 0.7 | 1.2–2.2 | 2 ± 0.9 | 1.3–2.4 | 48.6% | 0.54 |
| Rest MBF (ml/min/g) | ||||||
| Systole | 0.7 ± 0.2 | 0.5–0.8 | 0.7 ± 0.2 | 0.5–0.8 | 22.7% | 0.77 |
| Diastole | 0.7 ± 0.3 | 0.5–0.8 | 0.7 ± 0.3 | 0.5–0.8 | 44.9% | 0.56 |
SD- standard deviation, IQR- interquartile range, CoV- Coefficient of variation, ICC- Intraclass correlation coefficient
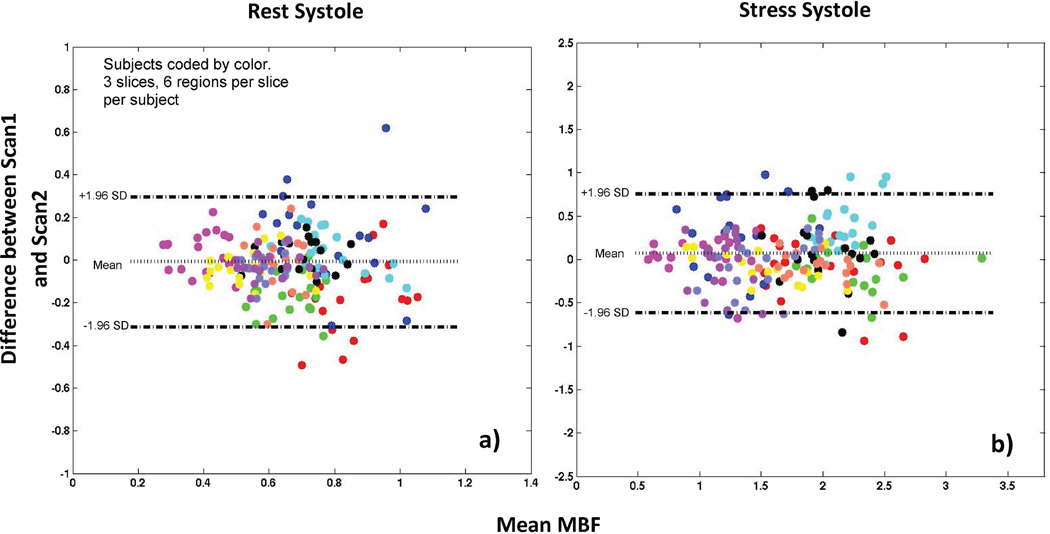
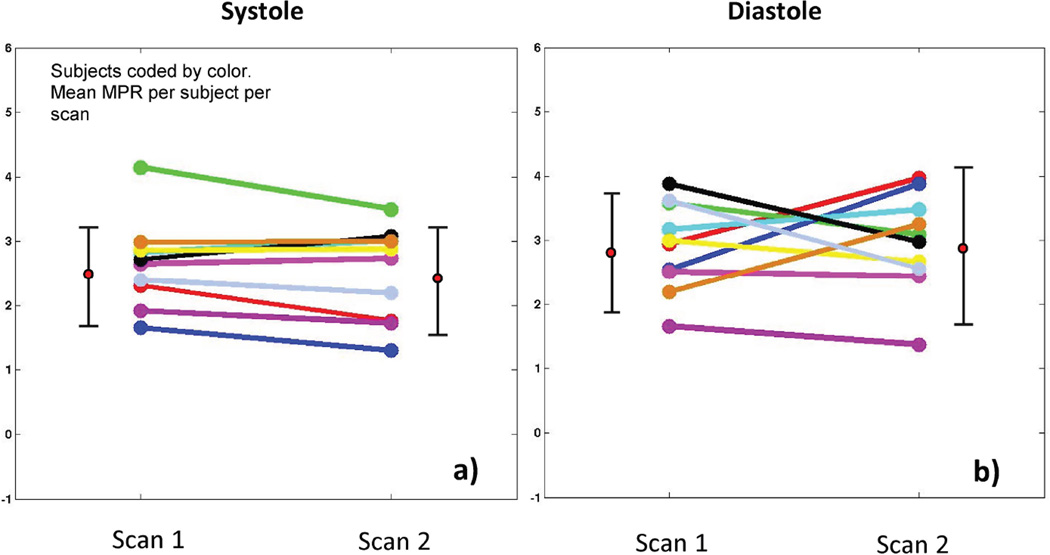
Figure 4 shows Bland-Altman plots comparing MBF estimates between scan1 and scan 2. Figure 5 shows a plot comparing the myocardial perfusion reserve (MPR) between scan 1 and scan 2.
Figure 4. Bland-Altman plot for self-gated near-systole data.
The Bland-Altman plot shows an absence of bias between estimated MBF values at scan 1 and scan 2 during a) rest and b) stress
Figure 5. Comparison of MPR values between scan 1 and scan 2.
The plot compares the mean MPR values of scan 1 and scan 2 for all ten subjects using a) self-gated near-systole b) self-gated near-diastole.
For the MOLLI ECV measurements, a CoV of 6.8% was obtained for inter-study repeatability. The section detailing the results on inter-study repeatability of ECV measurement using MOLLI T1 mapping is included in ‘Additional file 1’.
DISCUSSION
The main aim of this study was to evaluate the inter-study reproducibility of MPR estimation using the self-gated technique described in (4). One of the major advantages of MR is the absence of ionizing radiation. This makes MR suitable for longitudinal studies. Dynamic PET is arguably more standard for quantifying myocardial perfusion (36). However, the radiation dose for serial measurements can be prohibitive. Dynamic CT techniques are being developed for estimation of perfusion index (37), although the radiation dose for even a single study is a major drawback. The average dose for rest and stress perfusion for quantification of MBF using CT perfusion was 18mSv in (38).
Poor ECG gating has been a source of problems for quantitative perfusion MR. A missed trigger or corrupted ECG gating signal, especially during the uptake of contrast, can lead to errors in the quantification of perfusion. The added complexity of making adjustments to get a good ECG signal can cost valuable time. The self-gated technique for quantification of perfusion (4) is simple and does not require any timing adjustments. The self-gated technique can be run as a ‘push-button’ technique with minimal operator interaction. These advantages may be useful for longitudinal examinations, but a major question that this study investigated is to what degree free-breathing and cardiac motion impact the repeatability of the blood flow estimates.
Table 3 summarizes the results from previously published results on repeatability of perfusion cardiovascular MRI (8–11). The studies listed in Table 3 all made use of ECG-gated, breath-held acquisitions. Three of the four studies reported results from a single mid-ventricular slice. The current study was free-breathing and self-gated. Four slices were acquired. The most basal slice, with the lowest SRT, was used only to obtain the AIF. The other three slices were used to quantify MBF.
Table 3.
Summary of prior works on repeatability of cardiovascular MRI perfusion
| Prior work | Study protocol | Results |
|---|---|---|
| Elkington et al. (8), 2005 |
|
Regional Coefficient of variation
|
| Morton et al. (9), 2012 |
|
Territorial coefficient of variations
|
| Larghat et al. (10), 2013 |
|
Global coefficient of variations
|
| Jerosch-Herold et al. (11), 2008 |
|
Regional coefficient of repeatability (1.96 X Coefficient of variation)
|
One advantage of using the self-gated technique was the ability to acquire systolic or near-systolic and near-diastolic perfusion images in the same scan. Depending on heart rate and how the data was binned, it would be possible to obtain truly systolic and diastolic phases by using only subsets of the data (although there would be fewer time frames). As described in earlier sections, we acquired ungated data and retrospectively self-gated all of the data into two discrete bins corresponding to near-systole and near-diastole. The high temporal frame rate during the acquisition made it possible to split the data into two bins, although this was a somewhat arbitrary split in that some of the data was between systole and diastole. These in-between frames could be discarded or the data could be binned into for example four cardiac phases.
Prior ECG-gated studies (39, 40) have shown that there is no significant difference in MBF between systolic and diastolic phases at rest. However, at stress, diastolic flows were found to be higher than systolic flows in healthy subjects (39, 40). Using the self-gated near-systole and self-gated near-diastole datasets for quantification of MBF, it was seen that the MBF values were similar between the cardiac phases during rest. At stress, the MBF values estimated using the near-diastole dataset were higher than the MBF values estimated using the near-systole dataset. These results are in line with the previous studies (39, 40). However, during this study, the self-gating did not use purely systolic or diastolic data frames, and the diastolic tissue curves were noisy due to the low wall thickness of the myocardium during diastole. Imperfect processing and registration also affected near-diastole images more due to the thin wall. This made the quantification of MBF challenging at diastole, and the large spread of values from near-diastole make it prudent to have little confidence in the diastolic results. Further studies are needed to evaluate the differences between systolic and diastolic perfusion obtained using self-gated techniques, including methods that may self-gate or bin the data differently than done here. For example, images could be required to be closer to true diastole in order to be binned/gated into the diastolic phase.
One of the limitations of the study is the number of subjects. Ten subjects were imaged at rest and stress twice at an interval of ~10 days. Other inter-study repeatability studies of perfusion have also been relatively small, with 11 to 30 subjects (Table 3), likely in part due to the challenges with performing repeated stress studies. The results here are comparable to the previously published ECG gated breath-held studies (8, 9).
Segmentation of the myocardium is an aspect of this study that requires further study. Biglands et al. observed in (24) that errors in segmentation of the myocardium can lead to errors in the estimation of MBF for ECG-gated studies. With a self-gated approach, the problem of segmentation becomes more challenging especially during diastole. For this study, a conservative approach for segmentation was followed as suggested in (24) to minimize the error in the estimation of MBF.
While MBFs and MPR from the Fermi model have been validated by microspheres (41), it is still not clear if these values reflect perfusion or if extraction or permeability also needs to be considered. Even if the MBFs reported here and in (8–11) more correctly reflect a flow index, measuring their repeatability is important.
In this study, multiple slices were acquired after a single saturation pulse. As a result, the SRT was different for different slices. However the process of conversion of signal intensity to gadolinium concentration as described by eq. (2) accounts for these differences in SRT, although SNR and contrast will vary between slices. This may produce a difference in precision of estimated flow values between slices. To study the effect of differences in SRT on perfusion values, the perfusion values for both of the scans of all subjects were combined with slice number as the variable. The results are presented in Table 4. The MBFs and MPRs for different slices (SRT range – ~25ms to ~190ms) had a similar mean and std. dev for segmental flows.
Table 4.
Summary of MBF and MPR values for different slices using self-gated near-systole dataset.
| Slice 2 | Slice 3 | Slice 4 | |
|---|---|---|---|
| Rest (ml/min/g) | 0.7 ± 0.1 | 0.7 ± 0.2 | 0.7 ± 0.2 |
| Stress (ml/min/g) | 1.8 ± 0.5 | 1.7 ± 0.5 | 1.7 ± 0.5 |
| MPR | 2.6 ± 0.7 | 2.6 ± 0.8 | 2.5 ± 0.7 |
Using eq. (2) to convert signal to gadolinium concentration may have inaccuracies. For example, a saturation pulse efficiency of 100% and a flip angle equal to the prescribed value were assumed. A study by Broadbent et al. (42) examined the sensitivity of signal correction techniques to changes in these parameters. Their work found that the proton density weighted signal correction technique as used in this paper was robust to variations in saturation efficiency and flip angle. As well, T2* effects were not modeled in eq. (2), although there have been reports of mild effects on the AIF (43). However, the process of conversion of AIF signal intensity to gadolinium concentration was validated (Refer to ‘Additional file 3: Mini-website in (4)). In that study of resting perfusion, the higher dose AIF was converted to gadolinium concentration and compared to a lower dose dual bolus AIF. The match between the dual bolus AIF, which should have little T2* effect, and the higher dose AIF converted to gadolinium concentration using eq. (2) suggests a negligible effect of T2* on peak AIF with the doses and acquisition parameters used here.
Another important finding of this study is the repeatability of quantification with regadenoson. It is an open question if an injection of regadenoson, which results in relatively short-lived hyperemia, is as repeatable for quantitation as is adenosine infusion that keeps hyperemia stable during the study. The results of the current study imply that when gadolinium is given approximately 70 seconds after the regadenoson, that this does give repeatable stress levels. The previous vasodilator studies with adenosine or dipyridamole reported in Table 3 reported similar or not as good repeatability.
In conclusion, the self-gated near-systole free breathing acquisition method had a CoV of 18.6% for territorial MPR estimates. This measure of inter-study repeatability was comparable or better than published CoVs from studies that used ECG-gated breath-held acquisitions. However, the near-diastolic datasets as processed here had a CoV of 46.2% for territorial MPR, which was worse than reported in the ECG-gated breath-held studies. The self-gated MBF values were analyzed with smaller (6 segments per slice) regions and gave a comparable or lower CoV than the published ECG-gated studies when near-systole data was used. Segmental near-diastole data were not as repeatable. The ease of use and insensitivity to ECG related problems makes self-gated myocardial perfusion a viable tool for longitudinal studies.
Supplementary Material
Acknowledgments
Grant support:
This publication was made possible by Grant Number R01 HL113224 from the NIH. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH
REFERENCES
- 1.Jerosch-Herold M. Quantification of myocardial perfusion by cardiovascular magnetic resonance. J Cardiovasc Magn Reson. 2010;12:57. doi: 10.1186/1532-429X-12-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Larson AC, White RD, Laub G, McVeigh ER, Li D, Simonetti OP. Self-gated cardiac cine MRI. Magn Reson Med. 2004;51(1):93–102. doi: 10.1002/mrm.10664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harrison A, Adluru G, Damal K, et al. Rapid ungated myocardial perfusion cardiovascular magnetic resonance: preliminary diagnostic accuracy. J Cardiovasc Magn Reson. 2013;15:26. doi: 10.1186/1532-429X-15-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Likhite D, Adluru G, Hu N, McGann C, DiBella E. Quantification of myocardial perfusion with self-gated cardiovascular magnetic resonance. J Cardiovasc Magn Reson. 2015;17(1):14. doi: 10.1186/s12968-015-0109-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen D, Sharif B, Dharmakumar R, et al. Quantification of myocardial blood flow using non-ECG-triggered MR imaging. Magn Reson Med. 2014 doi: 10.1002/mrm.25451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen D, Sharif B, Bi X, et al. Quantification of myocardial blood flow using non-electrocardiogram-triggered MRI with three-slice coverage. Magn Reson Med. 2015 doi: 10.1002/mrm.25787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muhling OM, Dickson ME, Zenovich A, et al. Quantitative magnetic resonance first-pass perfusion analysis: inter- and intraobserver agreement. J Cardiovasc Magn Reson. 2001;3(3):247–256. doi: 10.1081/jcmr-100107473. [DOI] [PubMed] [Google Scholar]
- 8.Elkington AG, Gatehouse PD, Ablitt NA, Yang GZ, Firmin DN, Pennell DJ. Interstudy reproducibility of quantitative perfusion cardiovascular magnetic resonance. J Cardiovasc Magn Reson. 2005;7(5):815–822. doi: 10.1080/10976640500287877. [DOI] [PubMed] [Google Scholar]
- 9.Morton G, Jogiya R, Plein S, Schuster A, Chiribiri A, Nagel E. Quantitative cardiovascular magnetic resonance perfusion imaging: inter-study reproducibility. Eur Heart J Cardiovasc Imaging. 2012;13(11):954–960. doi: 10.1093/ehjci/jes103. [DOI] [PubMed] [Google Scholar]
- 10.Larghat AM, Maredia N, Biglands J, et al. Reproducibility of first-pass cardiovascular magnetic resonance myocardial perfusion. J Magn Reson Imaging. 2013;37(4):865–874. doi: 10.1002/jmri.23889. [DOI] [PubMed] [Google Scholar]
- 11.Jerosch-Herold M, Vazquez G, Wang L, Jacobs DR, Jr, Folsom AR. Variability of myocardial blood flow measurements by magnetic resonance imaging in the multi-ethnic study of atherosclerosis. Invest Radiol. 2008;43(3):155–161. doi: 10.1097/RLI.0b013e31815abebd. [DOI] [PubMed] [Google Scholar]
- 12.Singh A, Horsfield MA, Bekele S, Khan JN, Greiser A, McCann GP. Myocardial T1 and extracellular volume fraction measurement in asymptomatic patients with aortic stenosis: reproducibility and comparison with age-matched controls. Eur Heart J Cardiovasc Imaging. 2015;16(7):763–770. doi: 10.1093/ehjci/jev007. [DOI] [PubMed] [Google Scholar]
- 13.Pica S, Sado DM, Maestrini V, et al. Reproducibility of native myocardial T1 mapping in the assessment of Fabry disease and its role in early detection of cardiac involvement by cardiovascular magnetic resonance. J Cardiovasc Magn Reson. 2014;16:99. doi: 10.1186/s12968-014-0099-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Messroghli DR, Plein S, Higgins DM, et al. Human myocardium: single-breath-hold MR T1 mapping with high spatial resolution--reproducibility study. Radiology. 2006;238(3):1004–1012. doi: 10.1148/radiol.2382041903. [DOI] [PubMed] [Google Scholar]
- 15.Roujol S, Weingartner S, Foppa M, et al. Accuracy, precision, and reproducibility of four T1 mapping sequences: a head-to-head comparison of MOLLI, ShMOLLI, SASHA, and SAPPHIRE. Radiology. 2014;272(3):683–689. doi: 10.1148/radiol.14140296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vasu S, Bandettini WP, Hsu LY, et al. Regadenoson and adenosine are equivalent vasodilators and are superior than dipyridamole- a study of first pass quantitative perfusion cardiovascular magnetic resonance. J Cardiovasc Magn Reson. 2013;15:85. doi: 10.1186/1532-429X-15-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adluru G, McGann C, Speier P, Kholmovski EG, Shaaban A, Dibella EV. Acquisition and reconstruction of undersampled radial data for myocardial perfusion magnetic resonance imaging. J Magn Reson Imaging. 2009;29(2):466–473. doi: 10.1002/jmri.21585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rudin LI, Osher S, Fatemi E. Nonlinear total variation based noise removal algorithms. Physica D: Nonlinear Phenomena. 1992;60(1–4):259–268. [Google Scholar]
- 19.Block KT, Uecker M, Frahm J. Undersampled radial MRI with multiple coils Iterative image reconstruction using a total variation constraint. Magn Reson Med. 2007;57(6):1086–1098. doi: 10.1002/mrm.21236. [DOI] [PubMed] [Google Scholar]
- 20.Walsh DO, Gmitro AF, Marcellin MW. Adaptive reconstruction of phased array MR imagery. Magn Reson Med. 2000;43(5):682–690. doi: 10.1002/(sici)1522-2594(200005)43:5<682::aid-mrm10>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 21.Avants B. Advanced Normalization Tools (ANTS) Release 1.5. 2011 [Google Scholar]
- 22.Adluru G, DiBella EV, Schabel MC. Model-based registration for dynamic cardiac perfusion MRI. J Magn Reson Imaging. 2006;24(5):1062–1070. doi: 10.1002/jmri.20756. [DOI] [PubMed] [Google Scholar]
- 23.Pack NA, Vijayakumar S, Kim TH, McGann CJ, DiBella EVR, editors. A semi-automatic software package for analysis of dynamic contrast-enhanced MRI myocardial perfusion studies. Computers in Cardiology. 2009 Sep 13–16;2009 2009. [Google Scholar]
- 24.Biglands J, Magee D, Boyle R, Larghat A, Plein S, Radjenovic A. Evaluation of the effect of myocardial segmentation errors on myocardial blood flow estimates from DCE-MRI. Phys Med Biol. 2011;56(8):2423–2443. doi: 10.1088/0031-9155/56/8/007. [DOI] [PubMed] [Google Scholar]
- 25.Donahue KM, Weisskoff RM, Burstein D. Water diffusion and exchange as they influence contrast enhancement. J Magn Reson Imaging. 1997;7(1):102–110. doi: 10.1002/jmri.1880070114. [DOI] [PubMed] [Google Scholar]
- 26.Sharma P, Socolow J, Patel S, Pettigrew RI, Oshinski JN. Effect of Gd-DTPA-BMA on blood and myocardial T1 at 1.5T and 3T in humans. J Magn Reson Imaging. 2006;23(3):323–330. doi: 10.1002/jmri.20504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanal E, Maravilla K, Rowley HA. Gadolinium contrast agents for CNS imaging: current concepts and clinical evidence. AJNR Am J Neuroradiol. 2014;35(12):2215–2226. doi: 10.3174/ajnr.A3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gatehouse PD, Elkington AG, Ablitt NA, Yang GZ, Pennell DJ, Firmin DN. Accurate assessment of the arterial input function during high-dose myocardial perfusion cardiovascular magnetic resonance. J Magn Reson Imaging. 2004;20(1):39–45. doi: 10.1002/jmri.20054. [DOI] [PubMed] [Google Scholar]
- 29.Kershaw LE, Cheng HL. A general dual-bolus approach for quantitative DCE-MRI. Magn Reson Imaging. 2011;29(2):160–166. doi: 10.1016/j.mri.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 30.Larghat A, Biglands J, Maredia N, et al. Endocardial and epicardial myocardial perfusion determined by semi-quantitative and quantitative myocardial perfusion magnetic resonance. Int J Cardiovasc Imaging. 2012;28(6):1499–1511. doi: 10.1007/s10554-011-9982-3. [DOI] [PubMed] [Google Scholar]
- 31.Jerosch-Herold M, Wilke N, Stillman AE. Magnetic resonance quantification of the myocardial perfusion reserve with a Fermi function model for constrained deconvolution. Med Phys. 1998;25(1):73–84. doi: 10.1118/1.598163. [DOI] [PubMed] [Google Scholar]
- 32.Clough AV, al-Tinawi A, Linehan JH, Dawson CA. Regional transit time estimation from image residue curves. Ann Biomed Eng. 1994;22(2):128–143. doi: 10.1007/BF02390371. [DOI] [PubMed] [Google Scholar]
- 33.Cerqueira MD, Weissman NJ, Dilsizian V, et al. Standardized myocardial segmentation and nomenclature for tomographic imaging of the heart. A statement for healthcare professionals from the Cardiac Imaging Committee of the Council on Clinical Cardiology of the American Heart Association. Circulation. 2002;105(4):539–542. doi: 10.1161/hc0402.102975. [DOI] [PubMed] [Google Scholar]
- 34.Zar JH. Biostatistical Analysis. 4th Edition ed. Prentice Hall; 1998. Biostatistical Analysis. [Google Scholar]
- 35.Bland JM, Altman DG. Agreement between methods of measurement with multiple observations per individual. J Biopharm Stat. 2007;17(4):571–582. doi: 10.1080/10543400701329422. [DOI] [PubMed] [Google Scholar]
- 36.Kaufmann PA, Camici PG. Myocardial blood flow measurement by PET: technical aspects and clinical applications. J Nucl Med. 2005;46(1):75–88. [PubMed] [Google Scholar]
- 37.Christian TF, Frankish ML, Sisemoore JH, et al. Myocardial perfusion imaging with first-pass computed tomographic imaging: Measurement of coronary flow reserve in an animal model of regional hyperemia. J Nucl Cardiol. 2010;17(4):625–630. doi: 10.1007/s12350-010-9206-6. [DOI] [PubMed] [Google Scholar]
- 38.Ho KT, Chua KC, Klotz E, Panknin C. Stress and rest dynamic myocardial perfusion imaging by evaluation of complete time-attenuation curves with dual-source CT. JACC Cardiovasc Imaging. 2010;3(8):811–820. doi: 10.1016/j.jcmg.2010.05.009. [DOI] [PubMed] [Google Scholar]
- 39.Motwani M, Kidambi A, Uddin A, Sourbron S, Greenwood JP, Plein S. Quantification of myocardial blood flow with cardiovascular magnetic resonance throughout the cardiac cycle. J Cardiovasc Magn Reson. 2015;17(1):4. doi: 10.1186/s12968-015-0107-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Radjenovic A, Biglands JD, Larghat A, et al. Estimates of systolic and diastolic myocardial blood flow by dynamic contrast-enhanced MRI. Magn Reson Med. 2010;64(6):1696–1703. doi: 10.1002/mrm.22538. [DOI] [PubMed] [Google Scholar]
- 41.Schuster A, Zarinabad N, Ishida M, et al. Quantitative assessment of magnetic resonance derived myocardial perfusion measurements using advanced techniques: microsphere validation in an explanted pig heart system. J Cardiovasc Magn Reson. 2014;16:82. doi: 10.1186/s12968-014-0082-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Broadbent DA, Biglands JD, Ripley DP, et al. Sensitivity of quantitative myocardial dynamic contrast-enhanced MRI to saturation pulse efficiency, noise and t measurement error: Comparison of nonlinearity correction methods. Magn Reson Med. 2015 doi: 10.1002/mrm.25726. [DOI] [PubMed] [Google Scholar]
- 43.Kellman P, Aletras AH, Hsu LY, McVeigh ER, Arai AE. T2* measurement during first-pass contrast-enhanced cardiac perfusion imaging. Magn Reson Med. 2006;56(5):1132–1134. doi: 10.1002/mrm.21061. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.