Abstract
Ischemic stress involves nutrient deprivation, hypoxia, acidosis, and altered levels of various ions and metabolites. Reperfusion, which abruptly alters these parameters, is a second stress to already stressed cells. Ischemic preconditioning, in which brief ischemia alternates with reperfusion to elicit a protective response to ischemia/reperfusion (I/R) injury, revealed the existence of a highly conserved, cell-autonomous, and nearly ubiquitous program. While we often assume that evolutionary selection is irrelevant with respect to myocardial infarctions—which generally occur long after reproduction—the program of ischemia tolerance may date back much further, to hibernating squirrels, turtles, and estivating frogs and snails (extremophiles), which must survive by entering a hypometabolic state. This relationship is further strengthened by the presence of similar signaling pathways and regulatory mechanisms such as mRNA localization and miRNA regulation. These parallels may offer new insights into the myocardial response to I/R injury. This review will explore some of the recent advances in our understanding of autophagy and mitochondrial turnover in the setting of I/R injury, and related findings drawn from research on hibernating extremophiles.
Introduction
Ischemic injury arises when blood flow is interrupted, thereby depriving the heart of oxygen and nutrients; the response of the heart is to enter a hypometabolic state. Additional injury occurs when oxygen-rich blood reperfuses the vulnerable myocardial tissue, triggering extensive mitochondrial production of reactive oxygen species and tissue injury.
Many interventions have been proposed to protect the heart against ischemia/reperfusion injury, among which conditioning is one of the best studied phenomena. Ischemic preconditioning (IPC), which consists of brief episodes of ischemia alternating with reperfusion, confers protection against a subsequent more sustained ischemia/reperfusion episode and reduces infarct size dramatically [1,2]. More recently, post-conditioning was suggested as yet another way to protect the heart which involves stuttering reperfusion after ischemia [3]. Finally in remote conditioning, episodic ischemia and reperfusion is administered in a remote organ, typically an extremity [4].
The hypometabolic state is also employed by extremophiles to withstand environmental hardship and is characterized by (1) signal transduction pathways that initiate and coordinate metabolic responses across cells and organs, accompanied by changes in gene expression; (2) reduction in the global metabolic rate, largely accomplished by re-prioritization of ATP utilization across the most energy-demanding processes including ion pumping and transcription/translation; (3) altered fuel utilization and handling of metabolic by-products; and (4) enhanced activity of defense mechanisms to stabilize macromolecules and support survival in the hypometabolic state.
There is another condition called hibernating myocardium in which patients experience a sustained reduction in regional blood flow, resulting in chronic ischemic stress. However, normalization of blood fails to fully restore cardiac contractility [5]. Comparing this response with the physiological model of hibernating animals in nature may yield insights that might lead to the development of novel therapies.
For the purpose of this review, we will focus largely on the signals and metabolic reprogramming necessary to achieve a hypometabolic state.
Signal transduction
Turtles can reduce their metabolic rate to as low as 10–20% of their normal aerobic rate during cold anoxic submersion, allowing them to meet their minimal energy requirements with anaerobic glycolysis [6,7]. Interestingly, adenosine, well-known to the cardiac research community as a key mediator of ischemic preconditioning, was also shown to be responsible for suppressing neuronal activity in the turtle brain during anoxia [8].
While phosphorylation mediated by protein kinases is the most common mechanism to regulate cell state such as metabolism, additional post-translational modifications play a role, including lysine acetylation [9] which has been reported in the preconditioned heart [10] as well as in extremophiles [11]. Transcriptional control mediated by HIF-1α is well-known and plays a key role in the establishing the hypometabolic state in extremophiles [12] as well as in cardioprotection [13]. In fact, the effect of preconditioning or remote conditioning was abrogated in heterozygous HIF-1α deficient mice suggesting its key role in cardioprotection [13,14].
Translational control—primarily suppression of CAP-dependent translation—is initiated by upstream kinase signaling. More recently, microRNAs have emerged as important players regulating the hypometabolic state in extremophiles and the preconditioned heart. These will be discussed in detail later in this review.
Hydrogen sulfide (H2S) is a global trigger and effector of the hypometabolic state, reducing oxygen consumption rapidly [15]. Endogenous production of H2S is accomplished by the translocation of cystathionine γ-lyase (CSE) to the mitochondria. At low concentrations (0.1–1 µM) and in the presence of oxygen, H2S enhanced mitochondrial bioenergetics by serving as an electron donor via sulfide quinone oxidoreductase to ubiquinone, whereas at higher concentrations (3–30 µM) it inhibited complex IV [16]. The inhibitory effect is magnified under hypoxic conditions [17]. H2S also activates HIF-1α, which mediates the transcriptional shift away from OXPHOS (oxidative phosphorylation) towards glycolysis.
ATP breakdown products (AMP, IMP (inosine monophosphate), and adenosine) all play a signaling role in establishing the hypometabolic state. AMP activates AMPK, which induces glucose uptake via Glut4 translocation to plasma membrane; the elevated AMP/ATP ratio also has allosteric effects on glycogen phosphorylase (regulates glycogen breakdown) and phosphofructokinase (rate-limiting enzyme in glycolysis). The role of AMPK may be nuanced, however, as studies in skeletal muscle suggest that AMPK is not essential for anaerobic metabolism of endogenous glycogen, but is important for the switch to OXPHOS, and contributes directly and indirectly to signaling for mitochondrial biogenesis [18]. AMPK activation is also an important trigger for activation of autophagy [19] and suppression of protein synthesis [20]. The importance of adenosine and AMPK activation in ischemic preconditioning are well-known [21]. In heart experiencing ischemic stress and energy depletion, AMP increases and activates glycogen phosphorylase. More importantly, it activates AMPK which results in the recruitment of glucose transporter (GLUT4) to the heart [22]. AMPK itself protects the heart through ATP-sensitive potassium channels [23]. Reduction in energy demand is achieved in part through activation of plasma membrane KATP channels [24–26]. The electrically active myocardium preserves Na+-pumping activity, but ATP-sensitive K+ channels open as ATP levels drop—in part a reflection of local ATP depletion by the Na+/K+ ATPase; in the extreme, this can result in cessation of electrical activity and thus contraction. However, under less severe conditions, activation of plasma membrane KATP channels shortens action potential duration and limits Ca+2 entry, thus attenuating OXPHOS and contractile activity. The activation of the mitochondrial KATP channel also contributes importantly to cardioprotection [27,28].
Another aspect of the ischemic stress response that may share parallels with hibernating or estivating animals includes shifts in fuel utilization. In the pig heart, ischemia or ischemia followed by 40min reperfusion resulted in upregulation of mRNA for glycolytic enzymes (hexokinase, phosphofructokinase, and glyceraldehyde-3-phosphate dehydrogenase), as well as glucose transporters (GLUT1 and GLUT4) in the reperfused heart, indicating a metabolic adaptation to ischemic stress. It is important to note that this metabolic adaptation towards glucose utilization persisted 8 weeks post-infarction [29]. Amino acids may be utilized nonoxidatively and generate fewer acidic byproducts [30]. Autophagy/lysosomal degradation will liberate amino acids that will provide metabolic support to the ischemic heart. Ischemic preconditioning is associated with decreased glycogen consumption, less lactate production, and higher ATP levels at the end of ischemia [31]. Upregulation of autophagy and a shift to amino acid metabolism might explain these findings. Fatty acid oxidation is not possible during hypoxia, and the ability to shift to alternate fuels, or metabolic flexibility, is a characteristic of a healthy, adaptable heart. Metabolic flexibility is diminished in Zucker obese (ZO) rats fed a Western diet for 7d, and is accompanied by contractile dysfunction [32]. It should come as no surprise, then, that ZO rats sustain larger infarcts than their lean counterparts, and show no infarct size reduction by ischemic preconditioning or pharmacologic preconditioning [33]. We have observed that ZO rats also exhibit impaired autophagy.
Autophagy
Autophagy is the highly conserved catabolic process in which cytoplasmic components are engulfed by a unique cup-shaped membrane called the isolation membrane or phagophore. The Atg5/Atg12-Atg16L complex and lipidated LC3 facilitate membrane elongation, which eventually closes upon itself to form a double-membrane structure called the autophagosome containing engulfed cytoplasmic cargo. The autophagosomes fuse with lysosomes which deliver hydrolytic enzymes able to degrade the cytoplasmic components [34] in the degrading structures called autophagolysosomes or autolysosomes. Autophagy is required under basal conditions to maintain cellular homeostasis. It is especially important for long-lived cells such as neurons and cardiomyocytes, in order to remove damaged organelles or toxic protein aggregates [35]. Autophagy also serves to eliminate excess organelles which are no longer in demand, and to degrade non-essential components during nutrient deprivation such as starvation, ischemia, or hibernation.
Mitophagy and biogenesis
Removal of mitochondria by this process is called mitophagy. The best-studied pathway for mitophagy involves the PINK1/Parkin pathway, in which mitochondrial depolarization results in accumulation of PINK1, which phosphorylates ubiquitin to activate Parkin, and phosphorylates other targets that may serve as Parkin receptors. Other mechanisms certainly exist to accomplish mitochondrial turnover, including “general” autophagy, Bnip3 and Nix, and others (including TRAF2, optineurin and Bcl2-L-13)[36–42], as well as mitochondrial protein quality control proteases in matrix and intermembrane space, and the ubiquitin-proteasome system for many outer membrane proteins. Several of these alternative pathways synergize with Parkin, suggesting that Parkin plays a central coordinating role. However, selective elimination of mitochondria that are unable to maintain a threshold membrane potential or to support functional protein import is expertly accomplished by the PINK1-Parkin pathway, allowing the cell to eliminate only the least functional organelles.
Parkin plays a role in cardioprotection but may also contribute to mitochondrial homeostasis in the heart. The Parkin knockout mouse described by Kubli et al.[43,44], exhibited mitochondrial abnormalities seen by EM at 3 and 6 months of age, although crude measures of cardiac function revealed no abnormality out to 12mo of age. Respirometry (performed on mitochondria from hearts of 3mo mice) revealed no abnormalities, but studies were not performed in older mice despite the observation that mitochondrial morphology was increasingly abnormal. Parkin deficiency exacerbated ischemia/reperfusion injury, yet in cardiomyocytes from wild type animals, hypoxia did not result in upregulation of Parkin expression. This suggests that endogenous, basally-expressed Parkin plays an important role in the acute response to injury. It further indicates that there must be enough Parkin present under basal conditions to mount a useful response. This is consistent with our study which showed that Parkin was essential for cardioprotection mediated by ischemic preconditioning[45]. Ischemic preconditioning is generally regarded to be independent of de novo transcription; thus the protective role of Parkin in that context must also be due to preexisting Parkin. Some have discounted a role for Parkin in physiologic homeostasis because it can be difficult to find Parkin-decorated mitochondria under basal conditions[46]. Consider the numbers: rat cardiomyocytes have ~10,000 mitochondria per cell[47–50]; based on a half-life of 15 days[51], then one mitochondrion is replaced every 4 minutes. According to Twig et al.[52], a hypo-polarized mitochondrion may persist for several hours before it is engulfed by an autophagosome (in the INS1 cell line; persistence in cardiomyocytes may be shorter). Even if Parkin-decorated mitochondria persist for 2 hr before autophagic elimination, there will be only 120 decorated mitochondria out of a population of 10,000, making their detection under homeostatic conditions quite challenging. There are no dirty tables in a well-run restaurant if the busboys are doing their job properly. Their role becomes glaringly evident when a busload of tourists arrive at once, dine, and depart together, leaving behind a sea of dirty tables now swarming with busboys (like mitochondria decorated with Parkin after ischemic stress). But it would be false logic to suggest that the busboys only play a role when tour buses arrive.
Mitochondria respond to the energy demand of the cell by producing ATP production through oxidative phosphorylation (OXPHOS) complexes. During hypoxia, mitochondria may produce reactive oxygen species (ROS) and may lose membrane potential. In some settings, the F0F1 ATPase may run in reverse, hydrolyzing ATP in order to restore membrane potential, thereby further depleting ATP supplies in the already-stressed cell. In response to ROS or mitochondrial depolarization, mitophagy is activated to remove the damaged mitochondria to limit further tissue injury [53]. In fact, it has been shown that ischemic preconditioning activates mitophagy as a cell protective mechanism [45]. When dysfunctional mitochondria with low membrane potential are cleared via the PINK1/Parkin pathway, new mitochondria with functional OXPHOS components can be made to restore cell homeostasis [54]. Autophagy and mitophagy are also activated during hibernation [55], whereas protein synthesis is largely suppressed.
During myocardial ischemia, protein synthesis and degradation rates are slowed, but resume after reperfusion [56]. AMPK activation suppresses protein synthesis through eEF2 kinase, which in turn phosphorylated and inactivated eEF2 (eukaryotic elongation factor-2) to suppress protein synthesis [57]. A growing body of work puts eEF2 and its regulatory kinase at the helm for control of protein synthesis in response to nutrient availability and workload [58]. Global reduction in protein turnover may mask dramatic changes in specific subsets of proteins. CAP-dependent translation is suppressed by sequestration of eIF4E by its binding protein (4E-BP1) during ischemia [59]. Hypoxia is associated with an increase in translation of mRNAs with an internal ribosomal entry site (IRES) and a decrease of CAP-dependent translation. Recently, this was shown to be regulated by PINK1 which activates 4E-BP1 to switch on IRES-dependent translation [60]. An important IRES target is HIF-1α, which directs a metabolic shift from OXPHOS to glycolysis through transcriptional regulation.
Interestingly, HIF-1α also directs the upregulation of Bnip3, which mediates mitophagy; PINK1 also plays a key role in mitophagy. While proteasomal protein degradation is an energy-demanding process, glucose, amino acids, and fatty acids are liberated from the breakdown of glycogen, proteins, and membranes delivered to the lysosome as autophagic cargo, and thus may provide metabolic support under conditions of stress. Ischemia impairs autophagic flux, but it is upregulated during reperfusion [61], and ischemic preconditioning is dependent on upregulation of autophagic flux [45] and more specifically, Parkin-dependent mitophagy [62] HIF-1α is an important mediator of ischemic preconditioning [63] and is also important in hibernating mammals [64] Proteomic analysis of hibernating squirrels reveals a role for autophagy regulated through increased lysine acetylation associated with Sirt3 downregulation [65]. In contrast, Sirt3 plays a protective role in the ischemic heart [66] its functions in the heart may be more complex [67].
Regulation of autophagy
Autophagy is increasingly recognized to be a protective pathway in both extremophiles [65,68] and the ischemic heart [45] This pathway serves to eliminate damaged mitochondria that may generate reactive oxygen species, aggregates of misfolded proteins, and other unwanted cytoplasmic elements, while at the same time liberating metabolic substrates for fuel utilization. The importance of autophagy in the heart has been covered extensively in many reviews [54,69,70].
Considering the involvement of autophagy in different cellular functions and implications for defective autophagy in pathological conditions, regulation of this process is of great significance. Signaling pathways modulate autophagy via protein-protein interaction and post-translational modifications including phosphorylation [71,72]. Rapid induction of autophagy occurs mainly through post-translational modification of proteins involved in this process. However, prolonged induction of autophagy leads to activation of transcription factors including TFEB [73] which regulate the expression of key autophagy components or regulators such as DRAM1 [74,75].
mRNA localization and translational control represent another level of gene regulation which has advantages over transcriptional control because of the speed and lower energy requirement if transcription can be bypassed. Hibernation offers an interesting example of stress-induced mRNA localization in gene regulation. Some eukaryotes have evolved mechanisms including metabolic depression and upregulation of anti-apoptotic genes to tolerate a variety of stresses such as nutrient deprivation, oxidative damage, and I/R which occur during torpor/arousal cycles of hibernation and estivation [76]. Anoxia changes the translation capability of several mRNAs in kidney, liver, heart and red muscles of turtle [77]. Moreover, large-scale mRNA sequencing studies have shown no variation in mRNA level of the genes involved in gluconeogenesis, amino acid metabolism or antioxidant response during torpor/arousal cycles, yet an increase in protein content of these genes has been reported. This discrepancy has been explained by storage of mRNAs during torpor cycle and their immediate translation upon arousal [78]. Moreover, loss of polysomes and stabilization of mRNAs during hibernation has been shown previously [79]. Those mRNA transcripts which exit polysomes can be targeted by de-adenylating enzymes and assemble in translationally repressed mRNP (mRNA/protein) complexes which form distinct foci in the cytoplasm called processing bodies [80]. It is attractive to speculate that this same phenomenon is activated in the myocardium during I/R.
P-bodies
Processing bodies or P-bodies are distinct structures composed of enzymes and proteins involved in mRNA degradation such as decapping enzyme Dcp1/2, the activator of decapping Dhh1p/Rck1/p54, Pat1, Lsm1-7, Edc3, and exonuclease Xrn1. They also require mRNAs in their structure as treating purified P-bodies with RNAse A disrupts them [81]. Several studies indicate that mRNAs in P-bodies are destined for degradation. As mentioned before, decapping factors are concentrated in P-bodies. Moreover, inhibition of mRNAs from entering P-bodies by trapping them in polysomes results in a reduction in the size of P-bodies, whereas inhibiting the exonuclease activity leads to the formation of larger P-bodies [82].
P-bodies are not merely for mRNA decay. In fact, several studies have shown that silent intact mRNAs can exit P-bodies to re-enter the translation machinery (Fig. 1). In a study published by Parker and colleagues in 2005, it was shown that removal of glucose resulted in the rapid loss of polysomes and accumulation of reporter mRNAs and decapping enzymes in P-bodies. Interestingly, resupplying glucose led to loss of Dcp2p, Dhh1p, and reporter mRNAs from P-bodies. To confirm that mRNAs were entering translation machinery after exiting P-bodies, they analyzed the pattern of mRNA in sucrose gradients before and during glucose deprivation and after glucose restoration. The association of mRNAs with polysomes was observed in the log phase. They shifted to the non-translating fraction (P-bodies) during starvation, and re-entered the translation machinery after re-addition of glucose [80]. A recent study demonstrated translational regulation of the autophagy gene, Ambra1, in hypoxic conditions. Ambra1 mRNA localized to cytoplasmic P-bodies upon CoCl2 treatment, resulting in its translational suppression concurrent with activation of apoptosis [83]. This study supports the notion that ischemic preconditioning might involve translational controls.
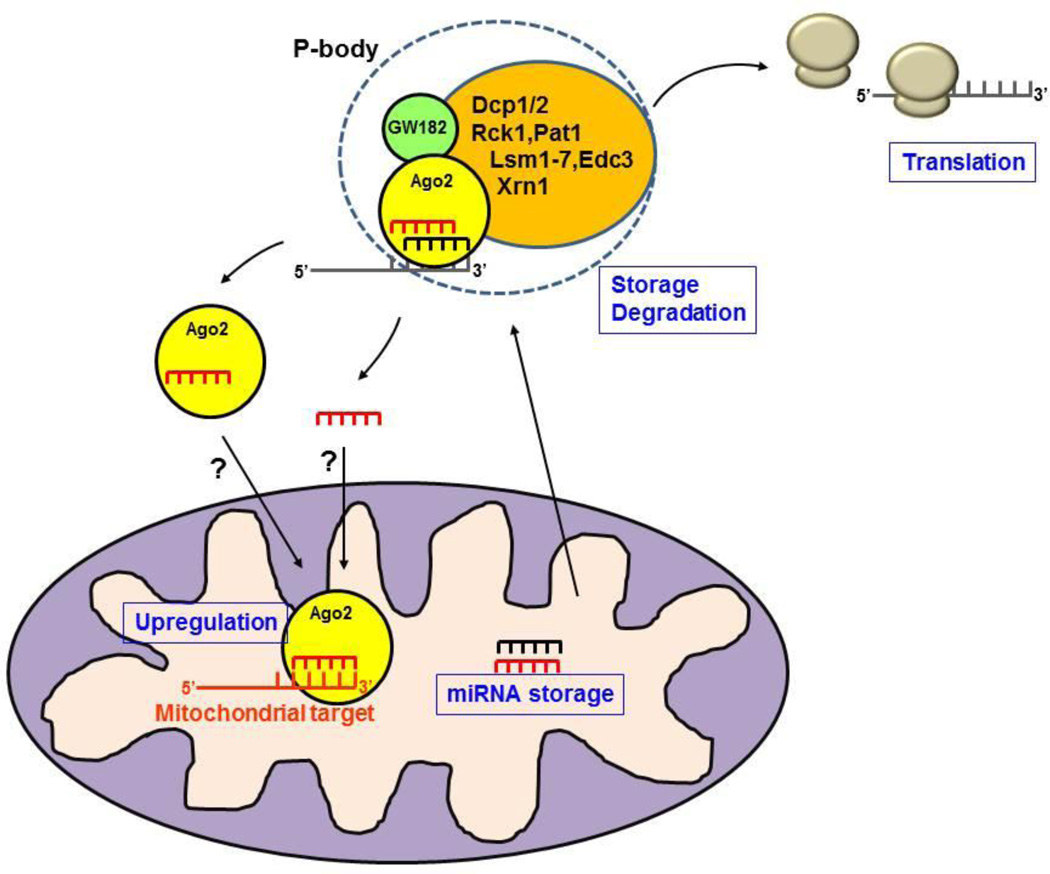
Fig. 1. Interaction between P-bodies, miRNAs, and mitochondria.
The conventional P-body comprises Ago2, GW182, RNA, and processing enzymes. mRNAs can be degraded in P-bodies or stored and released later to form polysomes and resume translation. P-bodies interact with mitochondria, and Ago2 and miRNAs are present in mitochondrial matrix. It is not known yet whether Ago2 delivers miRNAs to the Ago2 pool inside the mitochondria or whether it enters mitochondria with its miRNA cargo. The miRNAs may be stored in mitochondrial matrix for later release to the cytosol, or may regulate mitochondrial mRNA translation. In the absence of GW182, Ago2 and miRNAs can enhance translation.
MicroRNA
In metazoans, P-bodies contain the components of microRNA machinery, including Ago proteins, Rck, and GW182 [81,84]. MicroRNAs are 18–25 nucleotide long non-coding RNAs which regulate gene expression post-transcriptionally, mainly through interaction with 3’ UTR of the target gene. MicroRNAs are produced as long non-coding transcripts, pre-mature miRNAs, which undergo several processing steps to yield mature miRNAs. These mature miRNAs then pair with RISC (RNA-induced Silencing Complex) to bind the target gene and repress its expression through translational inhibition or mRNA degradation. miRNAs regulate many cellular processes including cell proliferation, differentiation, apoptosis, and autophagy [85].
Mitochondrial microRNAs
miRNAs are known to function mainly in the cytoplasm but several studies revealed that they are also present in mitochondria, where they affect different aspects of mitochondrial function (Table 1). 15 miRNAs were detected in highly purified mitochondria isolated from rat liver by means of miRNA microarray. Functional enrichment analysis showed that these miRNAs could potentially affect apoptosis, cell death and cell cycle [86]. In a different study, 13 miRNAs were found to be enriched in the mitochondrial fraction, of which three mapped to the mitochondrial genome [87]. Not only mature miRNAs were detected in the mitochondria, but also pre-miRNAs were detected by in situ hybridization and confirmed by qRT-PCR [88]. Other components of miRNA machinery such as Ago2 were also found in the mitochondria, localized to the inner membrane. They were capable of binding several mitochondrial-encoded transcripts [87,89,90].
Table1. Mitochondrial miRNAs.
List of published miRNAs identified in mitochondria, the method of detection, their mRNA targets, tissue of origin, global effect if known, and the citation.
| miRNA/ Pre miRNA |
Technique | Target | Tissue/Cell Type |
Effect | Reference |
|---|---|---|---|---|---|
| 33 Pre/25 miR |
In silico analysis | ND6, ND4L, ND4, COX1, ND1 | HeLa cells | Myogenesis, inflammation, fibrosis, oncogenic and oncosuppressor activities | [88]* |
| 46 miRNA | qRT-PCR | ||||
| miR 365 pre-miR 302a pre-Let7b |
In situ hybridization | ||||
| 15 miRNA | miRNA microarray |
COX3* |
Rat liver-derived mitochondria | Cell cycle Cell division (in silico prediction) | [86] |
| miR 130a,b miR 140 miR 320 miR 494 |
TaqMan miRNA assay | ||||
| 13 miRNA: miR 1974 miR 1977 miR 1978 |
Microarray | ND1, ND4, ND5, ND6, COX1, COX2* | Mitochondria isolated from HeLa cells | ATP synthesis coupled electron transport, cell cycle, translation initiation | [63] |
| miR 494 miR 1275 miR 1974 |
qRT-PCR | ||||
| miR-1 | qRT-PCR | COX1, ND1 | Mitochondria isolated from C2C12 myoblast and myotubes | Boost ATP production in muscle | [89] |
| miR-1192 miR-883 |
Microarray | COX1 | Mitochondria isolated from adult rat heart | Complex IV remodeling Increased ROS generation | [96] |
| miR 181-c | qRT-PCR, in situ hybridization | ||||
| MiR 181-c | In vivo miRNA overexpression | COX1 | Mouse heart | Complex IV dysfunction Increased ROS generation and oxygen consumption | [97] |
| 20 miRNA: miR-122 miR-805 miR-690 |
miRNA microarray stemloop miRNA qPCR assay | WARS, TFAM, BCL2, Pter, Gli3, Cftr, Mal, Clock, Hlf, Lif, sfrs8, Lcor, LBR, CD28, KRAS | Mouse liver mitochondria | Mitochondria dysfunction | [90]* |
| miR-705 miR-202- 5p miR-134 | |||||
| 35 miRNAs | Deep sequencing | Mitochondria isolated from HeLa and HEK293 | [103] | ||
| Let7b,g miR-122 miR-1819 miR-221 miR-320 |
qRT-PCR |
(indicates that targets were identified by in silico analysis).
Using cells transfected with Rck fused to RFP and mitochondrially targeted GFP, Huang et al. showed that the interaction between P-bodies and mitochondria is highly dynamic with each individual contact lasting about 18 seconds [91]. This association was decreased to some extent after vinblastine treatment, revealing the importance of an intact microtubule network for this association. Interestingly, the interactions were independent of translational state since disrupting polysomes did not alter the contacts. Depolarizing mitochondria by CCCP also didn’t affect the association, although it decreased the silencing by siRNA and miRNA machinery. More detailed analysis revealed that CCCP treatment resulted in depletion of Ago2 from P-bodies but not Rck or Ge-1. It seems that there is cross-talk between mitochondria and P-bodies which is important for the function of P-bodies including silencing [91]. However, it is still unknown whether this association might also deliver miRNAs or previously sequestered mRNAs to the mitochondria.
Although the main function of miRNAs is inhibiting translation, it has been shown that they can exert the opposite role under certain circumstances. The switch from inhibition to activation capacity of miRNAs was first demonstrated in quiescent cells where under serum starvation conditions, miR-363-3 was recruited to the TNFα 3’UTR, upregulating its translation [92].
A recent study by Zhang et al. reported that muscle-specific miR-1 enhances translation of mitochondrial transcripts during muscle differentiation. It was observed that protein levels of ND1 and COX1 increased during differentiation without a significant increase in mRNA levels or mtDNA copy number. Furthermore, they detected Ago2 in isolated mitochondria and mitoplasts which increased following differentiation. They also confirmed its association with mitochondrial transcripts by immunoprecipitation experiments [89]. It has been shown that miRNAs can enhance translation by binding to an mRNA target lacking cap and poly-A tail, if Ago2 is detached from GW182. These criteria are all present in mitochondrial transcripts as they all resemble prokaryotic transcripts lacking 5’cap and 3’polyA tail. Notably, GW182 was not detected inside the mitochondria. These data revealed the dual nature of miRNA pathways in response to the energy demand of the cell during differentiation by increasing mitochondrial proteins while suppressing cytoplasmic targets [89] (Fig. 1).
MicroRNA in heart
Considering the contractile function and high energy demand of heart, mitochondrial function and regulation should be kept intact. In fact, numerous miRNAs have been implicated in heart disease, a topic which is thoroughly covered in another review [93]. For example, miR-21 is involved in hypertrophy [94] while miR-126 is responsible for neo-angiogenesis after myocardial ischemia [95].
Mitochondria isolated from adult rat hearts also contained microRNAs, with miR-181c highly enriched in mitochondrial fractions [96]. Localization of miR-181c to the mitochondria was further confirmed by qRT-PCR and in situ hybridization analysis. This microRNA was immunoprecipitated with Ago2 along with COX1 transcript, implicating it as the primary target of miR-181c. Overexpressing miR-181c in the cells resulted in a decrease of COX1 protein while at the same time COX2 mRNA and protein increased. This could affect the proper assembly of complex IV, possibly explaining the increase in ROS production with miR-181c overexpression. To study the in vivo functional implication of miR-181c, nanovectors were used to deliver it to the mice. Overexpression of miR-181c resulted in heart failure. COX1 mRNA and protein levels decreased, confirming the in vitro results. Unlike the in vitro results, however, COX2 and COX3 proteins were decreased as well as COX1 [97].
MicroRNA in hibernation
Discordant results obtained from transcriptome and proteome analysis of hibernating animals suggests the presence of a post-transcriptional regulatory mechanism with miRNAs being a potential candidate [98,99]. In fact, differential expression of miRNAs in organs of hibernating animals has been shown [100,101]. For example, anti-apoptotic miR-21 was increased in the kidney during torpor while miR-24 was downregulated in heart and skeletal muscles. Interestingly, protein levels of Dicer (one of the enzymes involved in processing miRNAs) were increased 2.7 fold in the heart [100]. In a different study, increased levels of miR-29a, miR-152, miR-195, miR-223, and miR-486 were observed in the liver of hibernating squirrels while miR-378 was downregulated in skeletal muscle. Interestingly, the protein levels of FAS (fatty acid synthase), target of miR-195 were decreased in the liver [102].
Perspective
The response to ischemic stress is orchestrated on multiple levels: transcriptional, translational, and post-translational. The coordination of immediate responses (post-translational modifications, changes in subcellular localization), mid-range responses (suppression of translation of some mRNAs and activation of others, mediated in part by miRNAs), and longer-term responses (transcription, induction of new regulatory miRNAs and lncRNAs) positions the cell to respond not only to the acute challenge but to alter its homeostatic functions to adapt to the changed environment in the longer term. Hibernating squirrels suppress protein translation and activate autophagy during entry into hibernation. They upregulate mitochondrial biogenesis and new protein synthesis at the end of torpor, when energy supplies are at a minimum, in order to support thermogenesis and shivering to raise body temperature. This response is necessary, for the squirrel must emerge, find food, and mate if its genes are to be passed on. Hibernation and emergence from torpor are orchestrated by post-translational modifications and miRNAs; these may serve to suppress RNA translation but may also thriftily store the RNAs in P-bodies, allowing their rapid and energy-efficient return to the translational machinery prior to emerging from torpor. If this evolutionarily conserved program is activated in the ischemic myocardium, we might also expect to see upregulation of mitochondrial biogenesis at some point even though ischemia persists. We might speculate that ischemic preconditioning would amplify the response, allowing more mRNAs to be produced and stored, allowing for a more rapid recovery of function at the end of ischemia. Further work is needed to better translate the language of extremophile hibernation to cardioprotective mechanisms, as well as to understand how these responses might be counterproductive in certain settings such as chronic mild ischemia or the diabetic heart.
Highlights.
Lost in Translation
The cell in preparation for a rainy day
May see fit to store some RNA.
Messages encoding oxphos ‘zymes
That may come in handy in sunnier times.
To survive the storm of nutrient loss
Or preparing to withstand the winter frost
Hibernators conserve their energy
And meet their needs with autophagy.
But in fuel nadir and near energy-less,
They nevertheless start mito-biogenesis
Expecting that they’ll soon emerge
From winter burrow or feel reperfusion surge.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Murry CE, Jennings RB, Reimer KA. Preconditioning with ischemia: a delay of lethal cell injury in ischemic myocardium. Circulation. 1986;74:1124–1136. doi: 10.1161/01.cir.74.5.1124. [DOI] [PubMed] [Google Scholar]
- 2.Heusch G. Molecular basis of cardioprotection: signal transduction in ischemic pre-, post-, and remote conditioning. Circ Res. 2015;116:674–699. doi: 10.1161/CIRCRESAHA.116.305348. [DOI] [PubMed] [Google Scholar]
- 3.Zhao Z-Q, Corvera JS, Halkos ME, Kerendi F, Wang N-P, Guyton Ra, et al. Inhibition of myocardial injury by ischemic postconditioning during reperfusion: comparison with ischemic preconditioning. Am J Physiol Heart Circ Physiol. 2003;285:H579–H588. doi: 10.1152/ajpheart.01064.2002. [DOI] [PubMed] [Google Scholar]
- 4.Przyklenk K, Bauer B, Ovize M, Kloner Ra, Whittaker P. Regional ischemic “preconditioning” protects remote virgin myocardium from subsequent sustained coronary occlusion. Circulation. 1993;87:893–899. doi: 10.1161/01.cir.87.3.893. [DOI] [PubMed] [Google Scholar]
- 5.Colbert RW, Holley CT, Stone LH, Crampton M, Adabag S, Garcia S, et al. The Recovery of Hibernating Hearts Lies on a Spectrum: from Bears in Nature to Patients with Coronary Artery Disease. J Cardiovasc Transl Res. 2015;8(4):244–252. doi: 10.1007/s12265-015-9625-5. [DOI] [PubMed] [Google Scholar]
- 6.Jackson DC. Living without oxygen: Lessons from the freshwater turtle. Comp Biochem Physiol - A Mol Integr Physiol. 2000;125:299–315. doi: 10.1016/s1095-6433(00)00160-4. [DOI] [PubMed] [Google Scholar]
- 7.Hochachka PPW, Somero GN. Biochemical Adaptation: Mechanism and Process in Physiological Evolution. 2002 [Google Scholar]
- 8.Lutz PL, Nilsson GE. Vertebrate brains at the pilot light. Respir. Physiol. Neurobiol. 2004;141:285–296. doi: 10.1016/j.resp.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 9.Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, et al. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nadtochiy SM, Redman E, Rahman I, Brookes PS. Lysine deacetylation in ischaemic preconditioning: The role of SIRT1. Cardiovasc Res. 2011;89:643–649. doi: 10.1093/cvr/cvq287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang J, Biggar KK, Storey KB. Regulation of p53 by reversible post-transcriptional and post-translational mechanisms in liver and skeletal muscle of an anoxia tolerant turtle, Trachemys scripta elegans. Gene. 2013;513:147–155. doi: 10.1016/j.gene.2012.10.049. [DOI] [PubMed] [Google Scholar]
- 12.Zhu X, Smith MA, Perry G, Wang Y, Ross AP, Zhao HW, et al. MAPKs are differentially modulated in arctic ground squirrels during hibernation. J Neurosci Res. 2005;80:862–868. doi: 10.1002/jnr.20526. [DOI] [PubMed] [Google Scholar]
- 13.Cai Z, Luo W, Zhan H, Semenza GL. Hypoxia-inducible factor 1 is required for remote ischemic preconditioning of the heart. Proc Natl Acad Sci U S A. 2013;110:17462–17467. doi: 10.1073/pnas.1317158110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cai Z, Zhong H, Bosch-Marce M, Fox-Talbot K, Wang L, Wei C, et al. Complete loss of ischaemic preconditioning-induced cardioprotection in mice with partial deficiency of HIF-1 alpha. Cardiovasc Res. 2008;77:463–470. doi: 10.1093/cvr/cvm035. [DOI] [PubMed] [Google Scholar]
- 15.Blackstone E, Morrison M, Roth MB. H2S induces a suspended animation-like state in mice. Science. 2005;308:518. doi: 10.1126/science.1108581. [DOI] [PubMed] [Google Scholar]
- 16.Módis K, Coletta C, Erdélyi K, Papapetropoulos A, Szabo C. Intramitochondrial hydrogen sulfide production by 3-mercaptopyruvate sulfurtransferase maintains mitochondrial electron flow and supports cellular bioenergetics. FASEB J. 2013;27:601–611. doi: 10.1096/fj.12-216507. [DOI] [PubMed] [Google Scholar]
- 17.Matallo J, Vogt J, McCook O, Wachter U, Tillmans F, Groeger M, et al. Sulfide-Inhibition of Mitochondrial Respiration at Very Low Oxygen Concentrations. Nitric Oxide. 2014:1–6. doi: 10.1016/j.niox.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hardie DG, Sakamoto K. AMPK: a key sensor of fuel and energy status in skeletal muscle. Physiology (Bethesda) 2006;21:48–60. doi: 10.1152/physiol.00044.2005. [DOI] [PubMed] [Google Scholar]
- 19.Shang L, Wang X. AMPK and mTOR coordinate the regulation of Ulk1 and mammalian autophagy initiation. Autophagy. 2011;7:924–926. doi: 10.4161/auto.7.8.15860. [DOI] [PubMed] [Google Scholar]
- 20.Shaw RJ. LKB1 and AMP-activated protein kinase control of mTOR signalling and growth. Acta Physiol. 2009;196:65–80. doi: 10.1111/j.1748-1716.2009.01972.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Young LH. AMP-activated protein kinase conducts the ischemic stress response orchestra. Circulation. 2008;117:832–840. doi: 10.1161/CIRCULATIONAHA.107.713115. [DOI] [PubMed] [Google Scholar]
- 22.Russell RR, Li J, Coven DL, Pypaert M, Zechner C, Palmeri M, et al. AMP-activated protein kinase mediates ischemic glucose uptake and prevents postischemic cardiac dysfunction, apoptosis, and injury. J Clin Invest. 2004;114:495–503. doi: 10.1172/JCI19297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sukhodub A, Jovanovic S, Du Q, Budas G, Clelland AK, Shen M, et al. AMP-activated protein kinase mediates preconditioning in cardiomyocytes by regulating activity and trafficking of sarcolemmal ATP-sensitive K(+) channels. JCell Physiol. 2007;210:224–236. doi: 10.1002/jcp.20862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bian J-S, Yong QC, Pan T-T, Feng Z-N, Ali MY, Zhou S, et al. Role of hydrogen sulfide in the cardioprotection caused by ischemic preconditioning in the rat heart and cardiac myocytes. J Pharmacol Exp Ther. 2006;316:670–678. doi: 10.1124/jpet.105.092023. [DOI] [PubMed] [Google Scholar]
- 25.Johansen D, Ytrehus K, Baxter GF. Exogenous hydrogen sulfide (H2S) protects against regional myocardial ischemia-reperfusion injury--Evidence for a role of K ATP channels. Basic Res Cardiol. 2006;101:53–60. doi: 10.1007/s00395-005-0569-9. [DOI] [PubMed] [Google Scholar]
- 26.Pan TT, Feng ZN, Lee SW, Moore PK, Bian JS. Endogenous hydrogen sulfide contributes to the cardioprotection by metabolic inhibition preconditioning in the rat ventricular myocytes. J Mol Cell Cardiol. 2006;40:119–130. doi: 10.1016/j.yjmcc.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 27.Garlid KD, Paucek P, Yarov-Yarovoy V, Murray HN, Darbenzio RB, D’Alonzo AJ, et al. Cardioprotective effect of diazoxide and its interaction with mitochondrial ATP-sensitive K+ channels. Possible mechanism of cardioprotection. Circ Res. 1997;81:1072–1082. doi: 10.1161/01.res.81.6.1072. [DOI] [PubMed] [Google Scholar]
- 28.Haruna T, Horie M, Kouchi I, Nawada R, Tsuchiya K, Akao M, et al. Coordinate Interaction Between ATP-Sensitive K+ Channel and Na+,K+-ATPase Modulates Ischemic Preconditioning. Circulation. 1998;98:2905–2910. doi: 10.1161/01.cir.98.25.2905. [DOI] [PubMed] [Google Scholar]
- 29.Remondino A, Rosenblatt-Velin N, Montessuit C, Tardy I, Papageorgiou I, Dorsaz PA, et al. Altered expression of proteins of metabolic regulation during remodeling of the left ventricle after myocardial infarction. J Mol Cell Cardiol. 2000;32:2025–2034. doi: 10.1006/jmcc.2000.1234. [DOI] [PubMed] [Google Scholar]
- 30.Drake KJ, Sidorov VY, McGuinness OP, Wasserman DH, Wikswo JP. Amino acids as metabolic substrates during cardiac ischemia. Exp Biol Med (Maywood) 2012;237:1369–1378. doi: 10.1258/ebm.2012.012025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Finegan BA, Lopaschuk GD, Gandhi M, Clanachan AS. Ischemic preconditioning inhibits glycolysis and proton production in isolated working rat hearts. Am J Physiol. 1995;269:H1767–H1775. doi: 10.1152/ajpheart.1995.269.5.H1767. [DOI] [PubMed] [Google Scholar]
- 32.Burgmaier M, Sen S, Philip F, Wilson CR, Miller CC, Young ME, et al. Metabolic adaptation follows contractile dysfunction in the heart of obese Zucker rats fed a high-fat “Western” diet. Obesity (Silver Spring) 2010;18:1895–1901. doi: 10.1038/oby.2009.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katakam PVG, Jordan JE, Snipes JA, Tulbert CD, Miller AW, Busija DW. Myocardial preconditioning against ischemia-reperfusion injury is abolished in Zucker obese rats with insulin resistance. Am J Physiol Regul Integr Comp Physiol. 2007;292:R920–R926. doi: 10.1152/ajpregu.00520.2006. [DOI] [PubMed] [Google Scholar]
- 34.Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 35.Iwata J, Ezaki J, Komatsu M, Yokota S, Ueno T, Tanida I, et al. Excess peroxisomes are degraded by autophagic machinery in mammals. J Biol Chem. 2006;281:4035–4041. doi: 10.1074/jbc.M512283200. [DOI] [PubMed] [Google Scholar]
- 36.Yang K-C, Ma X, Liu H, Murphy J, Barger PM, Mann DL, et al. Tumor Necrosis Factor Receptor-Associated Factor 2 Mediates Mitochondrial Autophagy. Circ Hear Fail. 2014;8:175–187. doi: 10.1161/CIRCHEARTFAILURE.114.001635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ding WX, Ni HM, Li M, Liao Y, Chen X, Stolz DB, et al. Nix is critical to two distinct phases of mitophagy, reactive oxygen species-mediated autophagy induction and Parkin-ubiquitin-p62-mediated mitochondrial priming. J Biol Chem. 2010;285:27879–27890. doi: 10.1074/jbc.M110.119537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee Y, Lee H-Y, Hanna Ra, Gustafsson aB. Mitochondrial autophagy by Bnip3 involves Drp1-mediated mitochondrial fission and recruitment of Parkin in cardiac myocytes. AJP Hear Circ Physiol. 2011;301:H1924–H1931. doi: 10.1152/ajpheart.00368.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu L, Feng D, Chen G, Chen M, Zheng Q, Song P, et al. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat Cell Biol. 2012;14:177–185. doi: 10.1038/ncb2422. [DOI] [PubMed] [Google Scholar]
- 40.Wong YC, Holzbaur EL. Optineurin is an autophagy receptor for damaged mitochondria in parkin-mediated mitophagy that is disrupted by an ALS-linked mutation. Proc Natl Acad Sci U S A. 2014;111:E4439–E4448. doi: 10.1073/pnas.1405752111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murakawa T, Yamaguchi O, Hashimoto A, Hikoso S, Takeda T, Oka T, et al. Bcl-2-like protein 13 is a mammalian Atg32 homologue that mediates mitophagy and mitochondrial fragmentation. Nat Commun. 2015;6:7527. doi: 10.1038/ncomms8527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carreira RS, Lee Y, Ghochani M, Gustafsson ÅB, Gottlieb Ra. Cyclophilin D is required for mitochondrial removal by autophagy in cardiac cells. Autophagy. 2010;6:462–472. doi: 10.4161/auto.6.4.11553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kubli Da, Zhang X, Lee Y, Hanna Ra, Quinsay MN, Nguyen CK, et al. Parkin protein deficiency exacerbates cardiac injury and reduces survival following myocardial infarction. J Biol Chem. 2013;288:915–926. doi: 10.1074/jbc.M112.411363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kubli Da, Quinsay MN, Gustafsson AB. Parkin deficiency results in accumulation of abnormal mitochondria in aging myocytes. Commun Integr Biol. 2013;6:e24511. doi: 10.4161/cib.24511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang C, Andres AM, Ratliff EP, Hernandez G, Lee P, Gottlieb RA. Preconditioning involves selective mitophagy mediated by parkin and p62/SQSTM1. PLoS One. 2011;6(6):e20975. doi: 10.1371/journal.pone.0020975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Song M, Gong G, Burelle Y, Gustafsson AB, Kitsis RN, Matkovich SJ, et al. Interdependence of Parkin-Mediated Mitophagy and Mitochondrial Fission in Adult Mouse Hearts. 2015;117:346–351. doi: 10.1161/CIRCRESAHA.117.306859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Page E, McCallister LP, Power B. Stereological measurements of cardiac ultrastructures implicated in excitation-contraction coupling. Proc Natl Acad Sci U S A. 1971;68:1465–1466. doi: 10.1073/pnas.68.7.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mcdonald K, Chu C, Francis G, Judd D, Carlyle W, Toher C, et al. The effect of delayed reperfusion following infarction in the rat on structural changes in viable myocardium. 1997;36:347–353. doi: 10.1016/s0008-6363(97)00201-0. [DOI] [PubMed] [Google Scholar]
- 49.Fawcett DW, McNutt NS. The Ultrastructure of the Cat Myocardium: I. Ventricular Papillary Muscle. J Cell Biol. 1969;42:1–45. doi: 10.1083/jcb.42.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nouette-Gaulain K, Malgat M, Rocher C, Savineau JP, Marthan R, Mazat JP, et al. Time course of differential mitochondrial energy metabolism adaptation to chronic hypoxia in right and left ventricles. Cardiovasc Res. 2005;66:132–140. doi: 10.1016/j.cardiores.2004.12.023. [DOI] [PubMed] [Google Scholar]
- 51.Kim T-Y, Wang D, Kim aK, Lau E, Lin aJ, Liem Da, et al. Metabolic Labeling Reveals Proteome Dynamics of Mouse Mitochondria. Mol Cell Proteomics. 2012:1–27. doi: 10.1074/mcp.M112.021162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Twig G, Elorza A, Molina AJa, Mohamed H, Wikstrom JD, Walzer G, et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Twig G, Shirihai OS. The interplay between mitochondrial dynamics and mitophagy. Antioxid Redox Signal. 2011;14:1939–1951. doi: 10.1089/ars.2010.3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Andres AM, Stotland A, Queliconi BB, Gottlieb Ra. A time to reap, a time to sow: mitophagy and biogenesis in cardiac pathophysiology. J Mol Cell Cardiol. 2015;78:62–72. doi: 10.1016/j.yjmcc.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Talaei F. Modulation of mTOR and autophagy in hibernating hamster lung and the application of the potential mechanism to improve the recellularization process of decellularized lung scaffolds. J Regen Med Tissue Eng. 2014;3:1. [Google Scholar]
- 56.Williams EH, Kao RL, Morgan HE. Protein degradation and synthesis during recovery from myocardial ischemia. Am J Physiol. 1981;240:E268–E273. doi: 10.1152/ajpendo.1981.240.3.E268. [DOI] [PubMed] [Google Scholar]
- 57.Horman S, Beauloye C, Vertommen D, Vanoverschelde JL, Hue L, Rider MH. Myocardial Ischemia and Increased Heart Work Modulate the Phosphorylation State of Eukaryotic Elongation Factor-2. J Biol Chem. 2003;278:41970–41976. doi: 10.1074/jbc.M302403200. [DOI] [PubMed] [Google Scholar]
- 58.Kaul G, Pattan G, Rafeequi T. Eukaryotic elongation factor-2 (eEF2): Its regulation and peptide chain elongation. Cell Biochem Funct. 2011;29:227–234. doi: 10.1002/cbf.1740. [DOI] [PubMed] [Google Scholar]
- 59.Connolly EP, Thuillier V, Rouy D, Bouétard G, Schneider RJ. Inhibition of Cap-initiation complexes linked to a novel mechanism of eIF4G depletion in acute myocardial ischemia. Cell Death Differ. 2006;13:1586–1594. doi: 10.1038/sj.cdd.4401854. [DOI] [PubMed] [Google Scholar]
- 60.Lin W, Wadlington NL, Chen L, Zhuang X, Brorson JR, Kang UJ. Loss of PINK1 attenuates HIF-1α induction by preventing 4E-BP1-dependent switch in protein translation under hypoxia. J Neurosci. 2014;34:3079–3089. doi: 10.1523/JNEUROSCI.2286-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xie H, Xu Q, Jia J, Ao G, Sun Y, Hu L, et al. Hydrogen sulfide protects against myocardial ischemia and reperfusion injury by activating AMP-activated protein kinase to restore autophagic flux. Biochem Biophys Res Commun. 2015;458:632–638. doi: 10.1016/j.bbrc.2015.02.017. [DOI] [PubMed] [Google Scholar]
- 62.Andres AM, Hernandez G, Lee P, Huang C, Ratliff EP, Sin J, et al. Mitophagy is Required for Acute Cardioprotection by Simvastatin. Antioxid Redox Signal. 2014;21(14):1960–1973. doi: 10.1089/ars.2013.5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Semenza GL. Hypoxia-inducible factor 1: Regulator of mitochondrial metabolism and mediator of ischemic preconditioning. Biochim Biophys Acta - Mol Cell Res. 2011;1813:1263–1268. doi: 10.1016/j.bbamcr.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Maistrovski Y, Biggar KK, Storey KB. HIF-1α regulation in mammalian hibernators: Role of non-coding RNA in HIF-1α control during torpor in ground squirrels and bats. J Comp Physiol B Biochem Syst Environ Physiol. 2012;182:849–859. doi: 10.1007/s00360-012-0662-y. [DOI] [PubMed] [Google Scholar]
- 65.Hindle AG, Grabek KR, Epperson LE, Karimpour-Fard A, Martin SL. Metabolic changes associated with the long winter fast dominate the liver proteome in 13-lined ground squirrels. Physiol Genomics. 2014;46:348–361. doi: 10.1152/physiolgenomics.00190.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Porter G, Urciuoli WR, Brookes PS, Nadtochiy SM. SIRT3 deficiency exacerbates ischemia-reperfusion injury: implication for aged hearts. Am J Physiol Heart Circ Physiol. 2014;306(12):H1602–H1609. doi: 10.1152/ajpheart.00027.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lombard DB, Zwaans BMM. SIRT3: As simple as it seems? Gerontology. 2013;60:56–64. doi: 10.1159/000354382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Garbarino VR, Orr ME, Rodriguez Ka, Buffenstein R. Mechanisms of oxidative stress resistance in the brain: Lessons learned from hypoxia tolerant extremophilic vertebrates. Arch Biochem Biophys. 2015;576:8–16. doi: 10.1016/j.abb.2015.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gustafsson AB, Gottlieb Ra. Autophagy in ischemic heart disease. Circ Res. 2009;104:150–158. doi: 10.1161/CIRCRESAHA.108.187427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gottlieb RA, Mentzer RM. Autophagy during cardiac stress: joys and frustrations of autophagy. Annu Rev Physiol. 2010;72:45–59. doi: 10.1146/annurev-physiol-021909-135757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Maiuri MC, Le Toumelin G, Criollo A, Rain J-C, Gautier F, Juin P, et al. Functional and physical interaction between Bcl-X(L) and a BH3-like domain in Beclin-1. EMBO J. 2007;26:2527–2539. doi: 10.1038/sj.emboj.7601689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xiucui M, Godar RJ, Liu H, Diwan A. Enhancing lysosome biogenesis attenuates BNIP3- induced cardiomyocyte death. Autophagy. 2012;8:297–309. doi: 10.4161/auto.18658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pietrocola F, Izzo V, Niso-Santano M, Vacchelli E, Galluzzi L, Maiuri MC, et al. Regulation of autophagy by stress-responsive transcription factors. Semin Cancer Biol. 2013;23:310–322. doi: 10.1016/j.semcancer.2013.05.008. [DOI] [PubMed] [Google Scholar]
- 75.Yu M, Jiang Y, Feng Q, Ouyang Y, Gan J. DRAM1 Protects Neuroblastoma Cells from Oxygen-Glucose Deprivation/Reperfusion-Induced Injury via Autophagy. Int J Mol Sci. 2014;15:19253–19264. doi: 10.3390/ijms151019253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rouble AN, Hefler J, Mamady H, Storey KB, Tessier SN. Anti-apoptotic signaling as a cytoprotective mechanism in mammalian hibernation. PeerJ. 2013;1:e29. doi: 10.7717/peerj.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Douglas DN, Giband M, Altosaar I, Storey KB. Anoxia induces changes in translatable mRNA populations in turtle organs: a possible adaptive strategy for anaerobiosis. J Comp Physiol B. 1994;164:405–414. doi: 10.1007/BF00302557. [DOI] [PubMed] [Google Scholar]
- 78.Yan J, Barnes BM, Kohl F, Marr TG. Modulation of gene expression in hibernating arctic ground squirrels. Physiol Genomics. 2008;32:170–181. doi: 10.1152/physiolgenomics.00075.2007. [DOI] [PubMed] [Google Scholar]
- 79.Knight JE, Narus EN, Martin SL, Jacobson A, Barnes BM, Boyer BB. mRNA stability and polysome loss in hibernating Arctic ground squirrels (Spermophilus parryii) Mol Cell Biol. 2000;20:6374–6379. doi: 10.1128/mcb.20.17.6374-6379.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Brengues M, Teixeira D, Parker R. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science. 2005;310:486–489. doi: 10.1126/science.1115791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Parker R, Sheth U. P Bodies and the Control of mRNA Translation and Degradation. Mol Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 82.Sheth U, Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pourpirali S, Valacca C, Merlo P, Rizza S, D’Amico S, Cecconi F. Prolonged Pseudohypoxia Targets Ambra1 mRNA to P-Bodies for Translational Repression. PLoS One. 2015;10:e0129750. doi: 10.1371/journal.pone.0129750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sen GL, Blau HM. Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nat Cell Biol. 2005;7:633–636. doi: 10.1038/ncb1265. [DOI] [PubMed] [Google Scholar]
- 85.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 86.Kren BT, Wong PY-P, Sarver A, Zhang X, Zeng Y, Steer CJ. MicroRNAs identified in highly purified liver-derived mitochondria may play a role in apoptosis. RNA Biol. 2009;6:65–72. doi: 10.4161/rna.6.1.7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bandiera S, Rüberg S, Girard M, Cagnard N, Hanein S, Chrétien D, et al. Nuclear outsourcing of RNA interference components to human mitochondria. PLoS One. 2011;6(6):e20746. doi: 10.1371/journal.pone.0020746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Barrey E, Saint-Auret G, Bonnamy B, Damas D, Boyer O, Gidrol X. Pre-microRNA and mature microRNA in human mitochondria. PLoS One. 2011;6(5):e20220. doi: 10.1371/journal.pone.0020220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang X, Zuo X, Yang B, Li Z, Xue Y, Zhou Y, et al. MicroRNA directly enhances mitochondrial translation during muscle differentiation. Cell. 2014;158:607–619. doi: 10.1016/j.cell.2014.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bian Z, Li L-M, Tang R, Hou D-X, Chen X, Zhang C-Y, et al. Identification of mouse liver mitochondria-associated miRNAs and their potential biological functions. Cell Res. 2010;20:1076–1078. doi: 10.1038/cr.2010.119. [DOI] [PubMed] [Google Scholar]
- 91.Huang L, Mollet S, Souquere S, Le Roy F, Ernoult-Lange M, Pierron G, et al. Mitochondria associate with P-bodies and modulate microRNA-mediated RNA interference. J Biol Chem. 2011;286:24219–24220. doi: 10.1074/jbc.M111.240259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 93.Van Rooij E, Marshall WS, Olson EN. Toward microRNA-based therapeutics for heart disease: The sense in antisense. Circ Res. 2008;103:919–928. doi: 10.1161/CIRCRESAHA.108.183426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cheng Y, Ji R, Yue J, Yang J, Liu X, Chen H, et al. MicroRNAs are aberrantly expressed in hypertrophic heart: do they play a role in cardiac hypertrophy? Am J Pathol. 2007;170:1831–1840. doi: 10.2353/ajpath.2007.061170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD, et al. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci U S A. 2006;103:18255–18260. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Das S, Ferlito M, Kent Oa, Fox-Talbot K, Wang R, Liu D, et al. Nuclear miRNA regulates the mitochondrial genome in the heart. Circ Res. 2012;110:1596–1603. doi: 10.1161/CIRCRESAHA.112.267732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Das S, Bedja D, Campbell N, Dunkerly B, Chenna V, Maitra A, et al. miR-181c regulates the mitochondrial genome, bioenergetics, and propensity for heart failure in vivo. PLoS One. 2014;9:1–9. doi: 10.1371/journal.pone.0096820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yan J, Burman A, Nichols C, Alila L, Showe LC, Showe MK, et al. Detection of differential gene expression in brown adipose tissue of hibernating arctic ground squirrels with mouse microarrays. Physiol Genomics. 2006;25:346–353. doi: 10.1152/physiolgenomics.00260.2005. [DOI] [PubMed] [Google Scholar]
- 99.Shao C, Liu Y, Ruan H, Li Y, Wang H, Kohl F, et al. Shotgun proteomics analysis of hibernating arctic ground squirrels. Mol Cell Proteomics. 2010;9:313–326. doi: 10.1074/mcp.M900260-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Morin PJ, Dubuc A, Storey KB. Differential expression of microRNA species in organs of hibernating ground squirrels: A role in translational suppression during torpor. Biochim Biophys Acta - Gene Regul Mech. 2008;1779:628–633. doi: 10.1016/j.bbagrm.2008.07.011. [DOI] [PubMed] [Google Scholar]
- 101.Liu Y, Hu W, Wang H, Lu M, Shao C, Menzel C, et al. Genomic analysis of miRNAs in an extreme mammalian hibernator, the Arctic ground squirrel. Physiol Genomics. 2010;42A:39–51. doi: 10.1152/physiolgenomics.00054.2010. [DOI] [PubMed] [Google Scholar]
- 102.Lang-Ouellette D, Morin P., Jr Differential expression of miRNAs with metabolic implications in hibernating thirteen-lined ground squirrels, Ictidomys tridecemlineatus. Mol Cell Biochem. 2014;394:291–298. doi: 10.1007/s11010-014-2105-4. [DOI] [PubMed] [Google Scholar]
- 103.Sripada L, Tomar D, Prajapati P, Singh R, Singh AK, Singh R. Systematic Analysis of Small RNAs Associated with Human Mitochondria by Deep Sequencing: Detailed Analysis of Mitochondrial Associated miRNA. PLoS One. 2012;7(9):e44873. doi: 10.1371/journal.pone.0044873. [DOI] [PMC free article] [PubMed] [Google Scholar]