Abstract
Native electrospray ionization-mass spectrometry, with gas phase activation and solution compositions that partially release subcomplexes, can elucidate topologies of macromolecular assemblies. That so much complexity can be preserved in gas phase assemblies is remarkable, although a long-standing conundrum has been the differences between their gas and solution phase decompositions. Collision-induced dissociation of multimeric noncovalent complexes typically distributes products asymmetrically; i.e., by ejecting a single subunit bearing a large percentage of the excess charge. That unexpected behavior has been rationalized as one subunit “unfolding” to depart with more charge. We present an alternative explanation based on heterolytic ion-pair scission and rearrangement,
a mechanism that inherently partitions charge asymmetrically. Excessive barriers to dissociation are circumvented in this manner, when local charge rearrangements access a lower-barrier surface.
An implication of this ion pair consideration is that stability differences between high- and low-charge state ions usually attributed to Coulomb repulsion may, alternatively, be conveyed by attractive forces from ion pairs (salt bridges) stabilizing low-charge state ions. Should the number of ion pairs be roughly inversely related to charge, symmetric dissociations would be favored from highly charged complexes, as observed. Correlations between a gas phase protein’s size and charge reflect the quantity of restraining ion pairs. Collisionally-facilitated salt bridge rearrangement (SaBRe) may explain unusual size “contractions” seen for some activated, low charge state complexes. That some low-charged multimers preferentially cleave covalent bonds or shed small ions to disrupting noncovalent associations is also explained by greater ion pairing in low charge state complexes.
Keywords: electrospray ionization, noncovalent complexes, asymmetric dissociations, supercharging, salt bridges, ion pairs, tandem mass spectrometry
Graphical Abstract
Introduction
The discovery that electrospray ionization-mass spectrometry (ESI-MS) or native MS could preserve noncovalent associations [1,2] advanced structural biology by providing rapid, reliable assessments of protein interactions, subunit stoichiometry, and cofactor ligand identity from low picomole quantities of noncovalent assemblies [3–6]. Combined with solution compositions tailored to partially assemble or disassemble macromolecular complexes and subcomplexes, ESI-MS hints at the topology of complex assemblies [7–9], while tandem mass spectrometry (MS/MS) employing collision induced dissociation (CID) and electron capture dissociation (ECD) or electron transfer dissociation (ETD) reveal that ligand, cofactors and metal ions reside at or near their solution phase positions [10–13].
Two decades ago, a driving question was whether any of the elements of solution phase structure were retained in the gas phase. Although it was established that solution phase conformation, whether altered by pH or solvent, left an imprint on charge state distributions [14,15], other differences were not automatically assumed to survive. Equally charged ions created from different solution structures were interrogated by gas phase proton transfer [16–19], H/D exchange [20,21], and collision cross-section analysis in early attempts to answer this question [22,23]. Today’s landscape appears to be strikingly different, however. Rather than starting from assumptions that little, if any, structure may be preserved, crystal structure coordinates or reconstructions from cryo-electron microscopy now provide the starting point to fit structures to ion mobility-measured cross-sections [7,24–27].
A conundrum was exposed when the Smith laboratory [28] demonstrated that gas-phase avidin, concanavalin A, and hemoglobin tetramers dissociate by ejecting single monomers with charge densities (charge/mass ratios) significantly higher than their precursors. The observations were surprising because each gas phase complex dissociated by expelling a monomer, despite their known solution-phase assembly as “dimers of dimers.” Perhaps equally surprising was the unequal (asymmetric) partitioning of charge density [29–31]. CID of streptavidin 14+ tetramers released 7+ monomers and 7+ trimers (or 6+ monomers and 8+ trimers) [30], rather the expected 7+ dimers or even 3+/4+ monomers expected had charge density partitioned uniformly. Analogously, a hexameric “trimer of dimers” complex was observed decomposing to monomer plus pentamer with asymmetric charge partitioning [32]. Even specific, homomeric dimers of human galectin I and E. coli glyoxalase I and non-specific dimers of cytochrome c decompose to disparately charged monomers [33]. A striking example of an asymmetric dissociation is that of the 63+ α7β7β7α7 20S proteasome complex from Methanosarcina thermophila, decomposing to a monomer bearing 33% of the charge and a 27-mer with 67% remaining (Fig. S1) [34]. Interestingly, specific enolase dimers dissociate asymmetrically, while non-specific complexes dissociate symmetrically (Fig. 1).
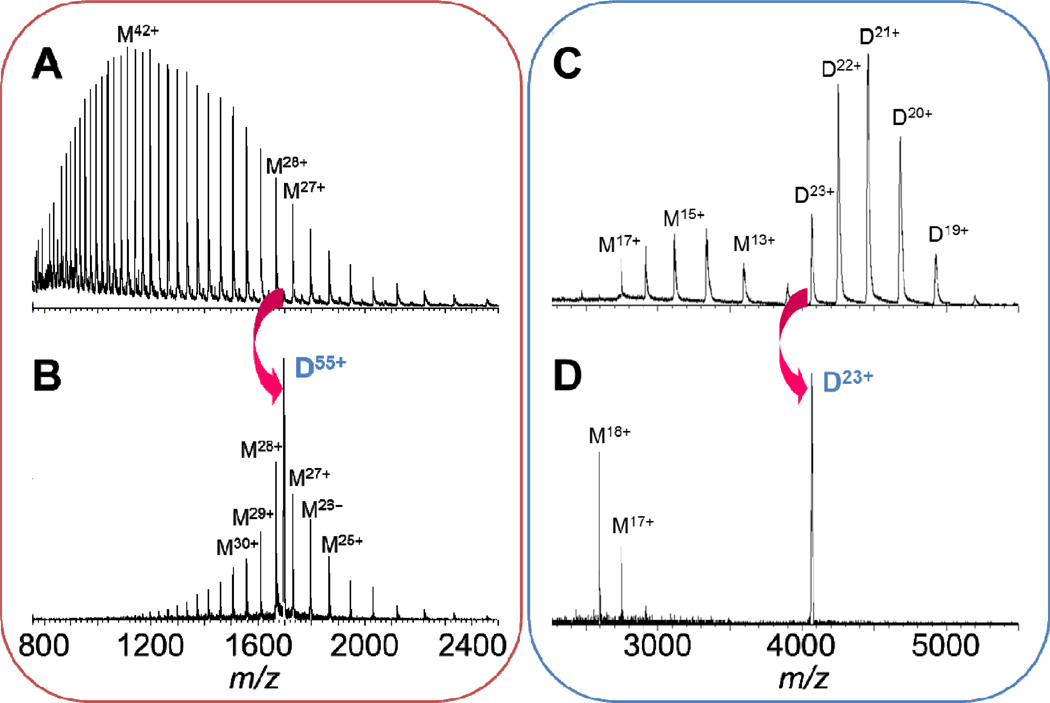
Figure 1.
ESI mass spectra of yeast enolase (46.7 kDa monomer; 93.5 kDa dimer) acquired with a quadrupole time-of-flight instrument. (A) Mass spectrum of enolase in 1:1 H2O/acetonitrile, 5% acetic acid (pH 2.5) and (B) CID mass spectrum of the nonspecific 55+ charged dimer (D, dimer), yielding a symmetric charge distribution of monomers (M, monomer). (C) Mass spectrum of enolase in 10 mM ammonium acetate and (D) CID mass spectrum of the specific 23+ charged dimer complex, distributing charge asymmetrically between monomers.
Light-Wahl, et al. [28] speculated that asymmetric charge partitioning could be driven Coulombically to unravel the tetramer, ejecting a charge-enriched monomer and leaving behind a compact trimer. Felitsyn, et al. [35] attributed the unusually high transition state entropies measured from dissociating Shiga-like toxin I pentamers to migrating protons that drove charge repulsion-induced denaturation. Jurchen, et al. [36] demonstrated that the observed proportion of symmetrically/asymmetrically dissociating homodimers depended on whether comprising subunits were intramolecularly cross-linked. That covalent linkages favored symmetric dissociations was taken to support the monomer unfolding mechanism proposed for asymmetric decompositions, because crosslinks were argued to reduce unfolding (i.e., decrease conformational flexibility) [36].
That rationale for asymmetric dissociations, in which one subunit unfolds withdrawing a larger portion of charge, has endured [37–39]. Nevertheless, it can be difficult to envision an activation process that would grossly change one monomer’s structure while leaving other subunits little-changed. What interaction could direct the energy extracted from tens to hundreds of collisions occurring anywhere on the molecule to drive the unfolding of a single subunit, even 5–10 nm away? Such energy flow differs from “classic” energy redistribution processes that would not favor one subunit over other identical subunits during energy randomization. Also hard to reconcile is a preference for unraveling a single, folded subunit over simply dislodging it [40]. These lingering questions encouraged us to seek alternative explanations for the asymmetric product distributions observed from dissociating noncovalent complexes.
Are Opposite Charges Common in Electrospray Ionization?
Grandori has advocated the consideration of opposite charges in ESI mechanisms [41–43]. The importance or ubiquity of opposing charge interactions (negatively charged sites within analytes dispersed by positive ion electrospray ionization (+ESI)) can be appreciated in the example of proteins modified chemically to add permanent positive charges [44]. When electrosprayed, these intrinsically-charged protein ions bear less charge overall than the number of fixed charges, even when sprayed from methanol/water, acid pH solutions. The absence of counter ion adducts in these “sub-charged” proteins established that at least one ion pair (e.g., −N(CH3)3+…−OOC−) was present in the gas phase protein ion. Despite these observations, it remains common to describe, for example, the 8+, 9+, and 10+ charge states of a protein as reflecting 8, 9, and 10 protonated basic sites, rather than as, e.g., 10 protonated basic sites and 2, 1, and 0 deprotonated acidic sites, even though it is rarely possible to distinguish between these compositions. Some of this bias may reflect an over-reliance on the intrinsic pKa values quoted for various amino acid residues and an under-appreciation of how they may be modulated by the local environment, e.g., altered by up to 5–6 pH units [45–47]. It also reflects assumptions about ion formation that model ESI as depositing charge onto completely uncharged analytes; i.e., with all residues in their neutral forms.
This bias is propagated in common explanations for the frequent observation that higher charge states dissociate at lower laboratory frame energies than low charge states. Reduced stability of higher charge states is typically attributed to like-charge repulsion. If we were to instead consider possible contributions from opposing charges, we might then suggest that stability differences could arise because lower charge states may be reinforced by a larger number of ionic bonds (salt bridges). Here we describe how many previously puzzling observations from ESI-MS and MS/MS analyses of proteins and protein complexes can be alternatively viewed by treating ion-pairing interactions and salt bridge rearrangements (SaBRe) as prevalent in gas phase protein ions.
Salt Bridges in Solution and in the Gas Phase are not Necessarily Identical
In solution, a salt bridge is defined as an attractive interaction between oppositely charged groups in which the donor and acceptor atoms are ≤ 4 Å apart [48]. Such hydrogen-bonded ion pairs can be formed when protonated lysines, arginines, or histidines, for example, donate protons to the carboxyl anions of well-positioned aspartic and glutamic acids. Conceivably, salt bridges existing in solution or newly formed could be transferred to the gas phase [49,50]. Moreover, attractive (and repulsive) interactions may be favored over distances greater than 4 Å in the gas phase, due to differences between solution and gas phase dielectric constants. This difference in interaction length, in concert with small structural rearrangements (perhaps only a few angstroms) arising from the activation inherent in transporting ions from atmospheric pressure to vacuum, suggests that the attractive interactions stabilizing electrosprayed complexes to dissociation are not necessarily limited to ion pairs annotated by NMR or X-ray crystallography. Consider if, in the “instant” that a protein is transferred to the gas phase by ESI, not all of its acidic residues are neutralized—that situation would be sufficient to spur new ion pairs to form. In that instant of transfer to the gas phase, charge can redistribute intramolecularly; e.g., it can migrate from protonated amines to carboxylate anions in a neutralization process, it can migrate to a region where it is stabilized by adjacent hydrogen-bonds, or it can interact with oppositely charged residues to form intramolecular salt-bridges. Some charge movement is to be expected upon transfer to the gas phase, where the basicities of carboxylate anions exceed those of neutral amines, inverting the basicity order from solution [42,43,51]. “Rattling” a protein ion via collisions may drive adjustments in residue positions that facilitate formation of additional or rearranged salt bridges in the gas phase.
Subunit-subunit interfaces may actually be more susceptible to salt bridge rearrangements than other regions [40], especially when interfacial water molecules are lost. Increased susceptibility is implied by solution models of folding/association that suggest interfacial contacts are less highly optimized than intra-subunit folds. The folding models propose (1) an initial step in which nascent protein subunits fold to their native, lowest energy configurations followed by (2) association of folded subunits into complexes and finally, (3) relatively minor conformational adjustments to optimize binding. The latter adjustments are necessarily small, because the folded chains have only three rotational and three translational degrees of freedom remaining [40]. Consequently, those limitations leave many hydrophilic side chains and water molecules buried in subunit interfaces. Although these interfaces are less highly optimized than the structural folds within subunits, they are stabilized in vivo by bound water molecules. ESI mass spectra often present some residual water when analytes are introduced with little activation [52–55], but once these water molecules are detached, the interfaces must be stabilized by other means. Coulomb attraction can be exploited as a gas phase stabilizing force if local rearrangements permit some opposite charge alignment across subunits.
In principle, counter ions; e.g., ammonium or acetate, may also stabilize proteins in the gas phase. Counter ions are frequently associated with charge sites on gas phase proteins, but can be dislodged by collisional activation. Whether they are dislodged as ions or neutrals is governed by their gas phase basicities/acidities compared to those of the associated protein sites; e.g., the charge stripping observed from collisionally activated 29+ and 21+ glycogen phosphorylase B kinase hexamers [56] is consistent with release of NH4+ and/or (CH3CH2)3NH+. Charge stripping can free opposing charge sites to reposition and form new interactions, while the loss of neutralized counter ions can eliminate opposite charge sites. Activated counter ions could, in principle, migrate to re-pair to new sites, but dissociation is expected to be more common.
The ramifications of a variable number of ionic interactions, either ones persisting from solution or created by gas phase rearrangements (over as little as a few angstroms), and potentially spanning subunits, leads to the question: Have we underestimated the contributions from ion pairing to gas phase protein structures, particularly for the non-denaturing conditions employed in noncovalent (native) complex studies? Could the strong inverse correlation observed between a gas phase protein’s “compactness” and its charge [37,57], particularly following gas-phase collisions, reflect an inverse relationship between charge state and number of salt bridges? Might mechanisms other than “maintained protein folds (conformations)” yield gas phase collision cross-sections resembling those calculated from crystal structures? Perhaps the gas phase retention of a few structural relationships known from solution simply reflects their reinforcement by a few salt bridge “staples”.
How Important is Coulomb Repulsion in Ionized Complexes?
Coulomb’s law (equation 1) describes the electrostatic repulsion between two point charges of like polarity and magnitude (q1 and q2), located a distance of r12 apart.
| (1) |
where ε corresponds to the effective dielectric of the medium. For highly charged ions, the individual electrostatic interactions for each charge must be summed; a general equation expressing the overall Coulomb repulsion, (EREP), has been presented [58,59]
| (2) |
From expression 2, it can be seen that evaluating overall Coulomb energy requires knowledge of charge locations.
Redirecting our focus to noncovalent complexes, we might ponder whether partitioning charge asymmetrically from dissociating complexes; e.g., homodimers, reduces electrostatic repulsion more than symmetric allocations. That is, one might naively consider that decomposing a 16+ precursor to 11+ and 5+ products would reduce the dissociation barrier by 14% compared to decomposition to 8+ and 8+ (5 · 11 = 55 versus 8 · 8 = 64). Equation 1 reminds us, however, that treating the energetics of electrostatic interactions in this manner would not be accurate because the individual charges are localized; rij values must be considered. Nevertheless, it is clear that reducing any Coulomb barrier to generating products can be important for facilitating tandem mass spectrometry analyses of noncovalent complexes [35,60,61].
On a potential energy surface, barrier height is determined in part by the subset of charge interactions that vary in rij (equation 1) as the reaction coordinate is traversed. Gronert [59] noted that multiply protonated ions with charge separations > 10 Å are expected to display reactivities akin to singly charged ions; hence, only charges within 10 Å or less of other charges could contribute significantly to the barrier. The observation [62] that charging in ESI-MS requires at least three uncharged residues separating charge-bearing ones is consistent with a 10 Å distance, given an average length of 3.8 Å per amino acid [63]. This 10 Å limit is an important consideration when attributing observations to Coulomb repulsion. Although it is tempting to attribute charge state-dependent differences in stability to excessive repulsive forces within higher charge state species, it is not the overall magnitude of charge that is important, but the distances between individual charges (sum of pairwise interactions as in equation 1). Even if charge sites are known or assumed (enabling the pairwise interactions to be summed), the significance of the calculated value lies in comparison to the total internal energy of a protein dimer possessing, perhaps, 25000 degrees of freedom [64]. Similarly, it is sometimes assumed that the maximum number of charges placed on an analyte by “native” ESI is limited by Coulomb repulsion or surface charging, such that any increase in charge (whether achieved by adding a supercharging reagent or by other means) beyond that obtained by spraying analyte in aqueous, millimolar ammonium acetate must reflect unfolding because only by extending over more space could that additional charge be accommodated on the protein [36]. To make such arguments requires knowing the spatial arrangement of the charge sites. A related assumption made by some is that successively higher protein charge states produced by ESI will necessarily possess gas phase structures (e.g., as revealed by ion mobility cross-sections) that are successively less and less like the native solution phase structure (less and less “folded”), either because their unfolded nature in solution allowed them to assume more charge during the ESI process or because assuming more charge irrevocably led to Coulomb repulsion-induced unfolding. However, simple calculations (supplemental material) demonstrate that Coulomb repulsion’s importance in limiting charging is far from obvious; e.g., to distribute 16 positive charges at least 10 Å apart on the surface of a folded holo-myoglobin-sized sphere could cover less than ¼ of the available surface area, yet +ESI of native myoglobin typically deposits only 8–10 charges. Under some conditions it should clearly be feasible to distribute additional charges onto native structures, such that none is within 10 Å of another.
Heterolytic versus Homolytic Ion Pair Cleavages can Account for Asymmetric versus Symmetric Charge Allocations
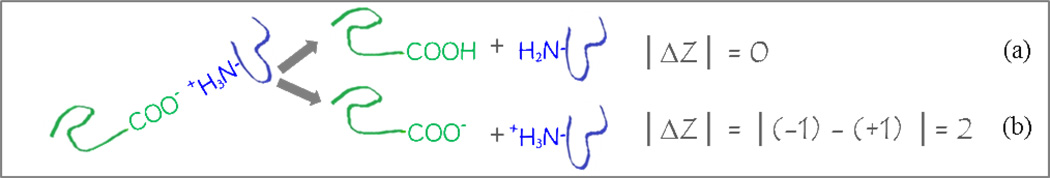
If not by gross rearrangement (unfolding) of one subunit’s structure, is there another mechanism to explain how charge could be allocated asymmetrically among dissociating subunits of a homodimer? We believe there is. Returning to the concept of salt bridges, consider the products and the difference in charge between them (│ΔZ│) for dissociation by two different pathways of a carboxylic acid/amine pair spanning two protein monomers. Scheme 1 illustrates homolytic (a) and heterolytic (b) cleavages of an ion pair to yield neutral (a), or oppositely charged (b) products. Note that for the dimer precursor, the presence or absence of an ion pair on the left hand side of Scheme 1 would not impact its charge state distribution (CSD) or overall charge, because the opposite charges cancel. However, the heterolytic cleavage, (b), inherently distributes charge asymmetrically because disrupting a salt bridge yields a charge difference | ΔZ | = 2 between the two products. Hence, heterolytically cleaving existing ion pairs in gas phase noncovalent complexes provides a mechanism to asymmetrically distribute charge to the products, but is heterolytic cleavage energetically feasible?
Scheme 1.
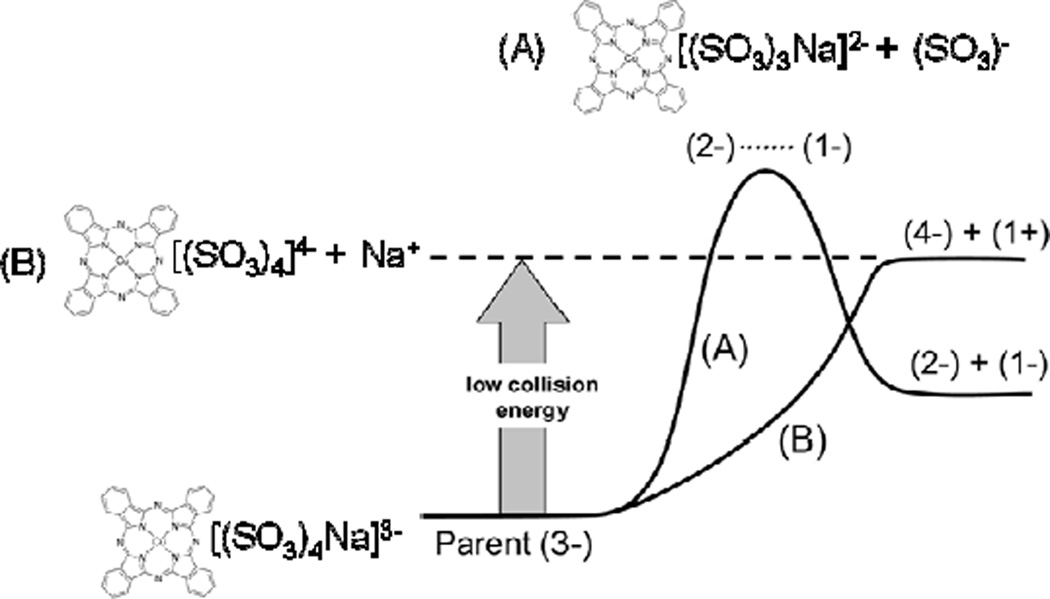
A key precedent for the heterolytic cleavage of a multiply charged ion exists in the dissociation of [Cu phthalocyanine (SO3)4Na]3−, for which the thermodynamically-favored dissociation to singly- and doubly-charged anions is only observed at high collision energies (Fig. 2). At low energies the dissociation is blocked by a substantial electrostatic kinetic barrier. Instead, barrierless dissociation to a quadruply charged anion and singly charged cation prevails [65]. This demonstration, performed on a triple quadrupole mass spectrometer, establishes that heterolytic and homolytic ion pair cleavages can be observed by ESI-MS/MS.
Figure 2.
Illustration of the homolytic and heterolytic dissociation channels for triply-charged [Cu phthalocyanine (SO3)4Na]3−. Dissociation along thermodynamically favored channel A (homolytic) is accessible only at high collision energies, due to its electrostatic kinetic barrier. Low collision energy dissociations access the more endothermic, barrierless channel B. (Modified from J. Am. Soc. Mass Spectrom., Vol. 19, S. Hashemi, M. J. Y. Jarvis, and D. K. Bohme, 375–379 (2008), with permission from American Society for Mass Spectrometry.)
Why wouldn’t the proton transfer/homolytic pathway (neutralization) be so favorable that the amount of heterolytic cleavage would be negligible? After all, for a simple gas phase species such as NH4+ ‥ −OOCCH3, it is easy to conclude that a proton would ultimately migrate to create NH3 and CH3COOH, the favored products based on relative gas phase proton affinities.
If we look at the accumulated evidence regarding the energetics of gas phase peptides, amino acids, and small amino acid clusters [66–71], we find that the most stable neutral structures are not always nonionic. Although glycine zwitterions are predicted to be unstable by ~84 kJ/mol, zwitterionic arginine is expected to be only 4–12 kJ/mol less stable than the nonionic form, and adding a methyl to one side chain yields a zwitterion that is more stable than nonionic methyl-arginine [72]. The most stable gas phase structure for neutral arginine dimer has two salt bridges. Neutral Arg3 assembles zwitterionic arginines, maintaining 12 hydrogen bonds between the guanidinium and carboxylate groups of adjacent molecules [68]. The additional Coulomb energy from these salt bridges more than compensates for the energetic cost of generating zwitterionic arginine. Moreover, despite the instability of glycine zwitterions, anionic glycine dimers, [Gly2-H]−, predominantly form salt bridge structures, stabilized by the attractive Coulomb interaction between opposite charges and by enhanced hydrogen bonding networks that outweigh the energetic cost associated with proton transfer [66]. If ground state structures for these very small, gas phase species are salt-bridged, then it is likely that the considerable self-solvation available within large proteins could support many low-energy salt-bridged configurations. In the complex molecular environment of a large protein, the synergistic interaction of salt bridge arrays, hydrogen bonding, and other charges can modulate the favorability of heterolytic versus homolytic cleavage. Moreover, instances in which one charged cleavage product re-pairs to a different charge site (e.g., changing from an inter-subunit bridge to an intra-subunit bridge, see Scheme 2) are especially favorable, because they circumvent the energy demands of charge-separation.
Scheme 2.
Excising salt bridges would likely enable formerly constrained, compact structures to rearrange, but expansion (unfolding) is not required to generate the product charge asymmetry. Conceivably, the low charge density product of a heterolytic cleavage could rearrange its now unpaired, opposing charges to form new salt bridges, recouping the energy lost in separating the opposite charges that spanned subunits, reinforcing its structure against activation-induced deformation, or even securing it into a more compact form, accessed by activation. In contrast, the high charge density product, depleted in opposing charges/ion pairs could be deformed more easily if too few salt bridge reinforcements remain to stabilize it.
In this view, even if a 16+ homodimer’s dissociation initially expelled equal-sized 12+ and 4+ products, subsequently measured cross-sections for the former could easily exceed those for the latter if (1) the 12+ has fewer stabilizing salt bridges (vide infra), and (2) its higher charge ensures that it receives more activation during the mobility measurement.
How Strong is the Evidence Supporting a Single Monomer’s Unfolding versus Heterolytic Ion Pair Scission/Rearrangement?
The opposing charge/salt bridge model finds support in recent CID studies on assemblies that yielded little evidence of unfolding (expansion) in collision cross-sections (Ω) of the monomeric products released from asymmetrically-dissociating noncovalent complexes [73–75]. During dissociation of 24+ [3(CP2:TR) + 3CP2] a capsid intermediate for MS2 phage, the expelled coat protein (CP) monomer withdrew 25% of the charge while accounting for only 8% of the total mass. Despite an apparent tripling of charge, the expelled monomer’s cross-section measured only 15% larger than that calculated for folded units within an intact ribonucleotide complex [73]. As collision cross-sections are proportional to apparent surface area, the measurement demonstrates that, at most, 15% extra surface area is required to accommodate 300% of the subunit’s initial charge, reiterating the previous points about the charging not necessarily being Coulomb-limited in ESI and “native” MS. Similarly, monomers expelled from collisionally-activated 800 kDa GroEL 14-mers exited the assembly with almost 50% of the total charge packed in a subunit cross-section only 21% larger than the value calculated from complex-bound subunits; i.e., 26 charges added to the monomer’s average 4.7+ charge [73]. Only 21% more surface area to accommodate 600% more charge? These results are inconsistent with the hypothesis that a single monomer expands to accommodate excess charge in a Coulomb repulsion-driven process, because the charge is actually concentrating in these expelled subunits!
It has been argued that charge partitioning, which is asymmetric with respect to the mass of ions, is symmetric with respect to the surface area of the ions; i.e., the surface area charge density is constant over the two product ions and is decreased versus the precursor, making the whole process Coulombically favorable [76]. The examples above [73], in which surface area increases of only 15 and 21% accommodated 300 and 600% more charge, respectively, clearly disagree with the proposed constant charge density.
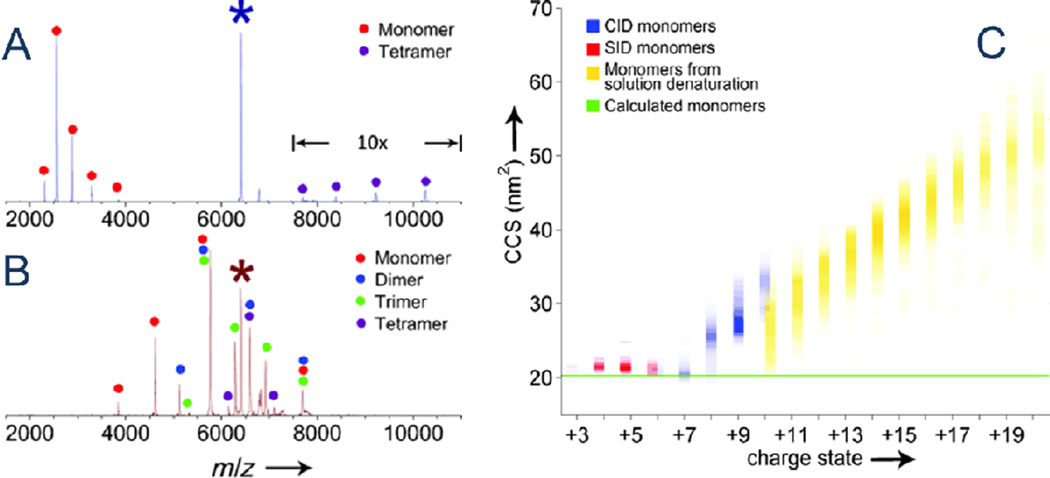
A third example that displays little monomer expansion while partitioning charge asymmetrically is CID of C-reactive protein (CRP) pentamers. Monomers (6+ and 7+) expelled from decomposing 18+ precursors were found to be compact with collision cross-sections (CCS) similar to calculated values and to all monomers (3+ - 6+) released by surface induced dissociation (SID) [74] (Fig. 3C). SID dissociates mass-selected ions by colliding them against a surface, imparting more collision energy to analyte than CID (for the same lab frame energy), and potentially accessing higher energy dissociation pathways. The single collision activation event drives unimolecular dissociation. Because SID of many noncovalent complexes yields products more similar in charge density than those from CID, these “symmetric” dissociations are thought to proceed without monomer unfolding. The dramatic differences in charge partitioning between asymmetric CID and symmetric SID are apparent in Fig. 3A and B. Nevertheless, Fig. 3C significantly reveals that 6+ CRP monomers, whether generated by symmetric (SID) or asymmetric dissociation (CID) pathways, bear equal cross-sections. Those cross-sections also match that for CID-released 7+ monomer. This equality in cross-sections raises the question of whether highly charged CID product cross-sections are larger than those of most SID products because the former unfolded in accordance with the dissociation mechanism, or because they carry more charge (and hence are subjected to more activation by the measurement). Control CCS measurements for equally-charged, supposedly folded subunits are rarely available for comparison. It could be argued that it may be impossible for some subunits to remain compact if they carry as much charge as is borne by asymmetric dissociation products, but proving that the dissociation mechanism expands or unfolds the product requires establishing that the larger cross-sections do not reflect charge state-dependent activation from the mobility measurement.
Figure 3.
Tandem mass spectra of 18+ CRP pentamer in CID (A) and SID (B). Product ions are labeled with colored circles and precursor ions with asterisks. (C) Collision cross-section profile of CRP monomer product ions for CID (blue, +6-+10) and SID (red, +3-+6) from a native CRP pentamer precursor, as well as solution denatured CRP monomers (yellow, +10-+20) over different charge states. The green line is the calculated CCS for the CRP monomer clipped from the crystal structure. Color depth of the spots is proportional to the square root of the relative abundance of the species. Monomer 3+-7+ product ions are compact and agree with monomer CCSs calculated from the crystal structure. (Reprinted from Figs. 1 and 2 in Angew. Chem. Int. Ed., Vol. 51, M. Zhou, S. Dagan, and V. H. Wysocki, 4336–4339 (2012), with permission.)
Interestingly, two additional CID/SID comparisons yielded similar CCS values for identically charged monomer products from symmetric and asymmetric decompositions [74]. Transthyretin (TTR) tetramers (15+), subjected to SID and CID, yielded 3+-7+ and 5+-9+ products, respectively. Cross-sections for TTR 5+, 6+ and 7+ SID products were little-different from those produced by CID. Similarly, serum amyloid P pentamers (24+) dissociated to 4+-7+ (SID) and 7+-13+ (CID) monomers, with products in the single charge state common to both SID and CID dissociation, 7+, possessing identical cross-sections. Additionally, the CID-produced 7+ cross-section was consistent with the size calculated from the crystal structure. These observations were attributed to the correlation between charge state and gas phase structure [74], but the CCS measurements described above are clearly inconsistent with a single monomer’s unfolding event driving asymmetric dissociation.
Building a Homolytic versus Heterolytic Ion Pair Cleavage Model
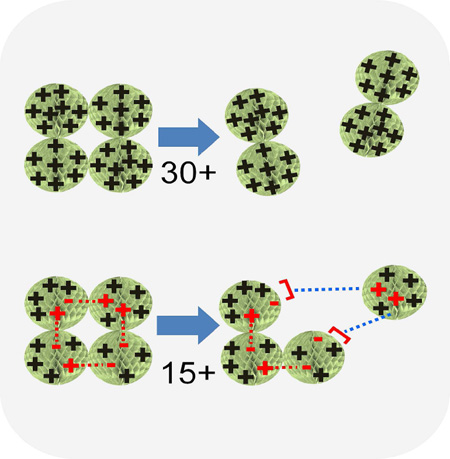
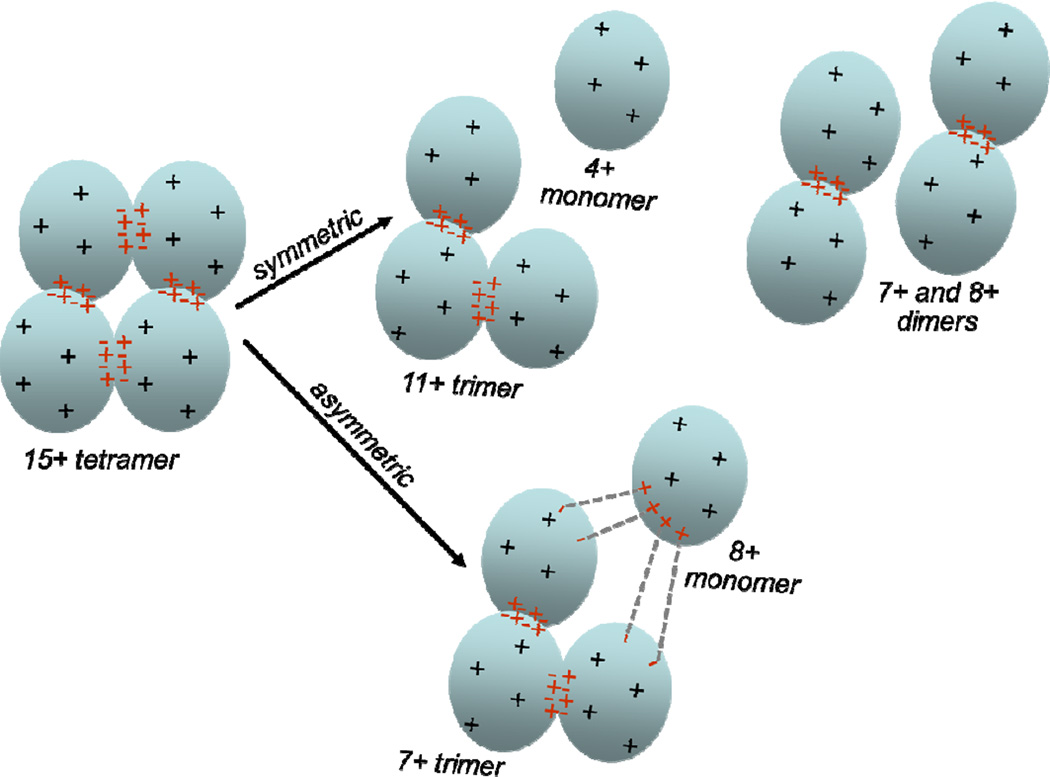
A 15+ ion could have 30 side chains carrying positive charges and 15 carrying negative charges, it could have 15 positively charged side chains with no negatively charged ones, or it could have any combination of positive and negative charges that sum to 15+. Coulomb barriers to dissociation (due to like-charge repulsion) could exist in either arrangement, depending on charge locations. If ion pairs are present, we envision potential rearrangements and homolytic and/or heterolytic scissions proceeding through an activated complex until a distribution (potential energy surface) with a sufficiently low barrier to dissociation can be accessed. Breaking salt bridges within an undissociated multimer would not change its overall charge, but could alter its allocation amongst the products (Fig. 4). Although the low barrier pathway (Fig. 5) would not yield the lowest enthalpy products, less activation energy would be required. In contrast to the unfolded subunit view of dissociation, where a single subunit’s structure undergoes an enormous change, the separating ion pair view does not require that the ejected subunit’s structure differ largely from that of the subunits maintaining association, other than in the disposition of cleaved ion pairs.
Figure 4.
Cartoon of a multimer dissociating symmetrically vs. asymmetrically with ion pairs. The asymmetric process is drawn assuming that only heterolytic ion pair cleavages create the charge asymmetry. Should charge migration also contribute, the number of heterolytically cleaved ion pairs spanning interfaces would be reduced.
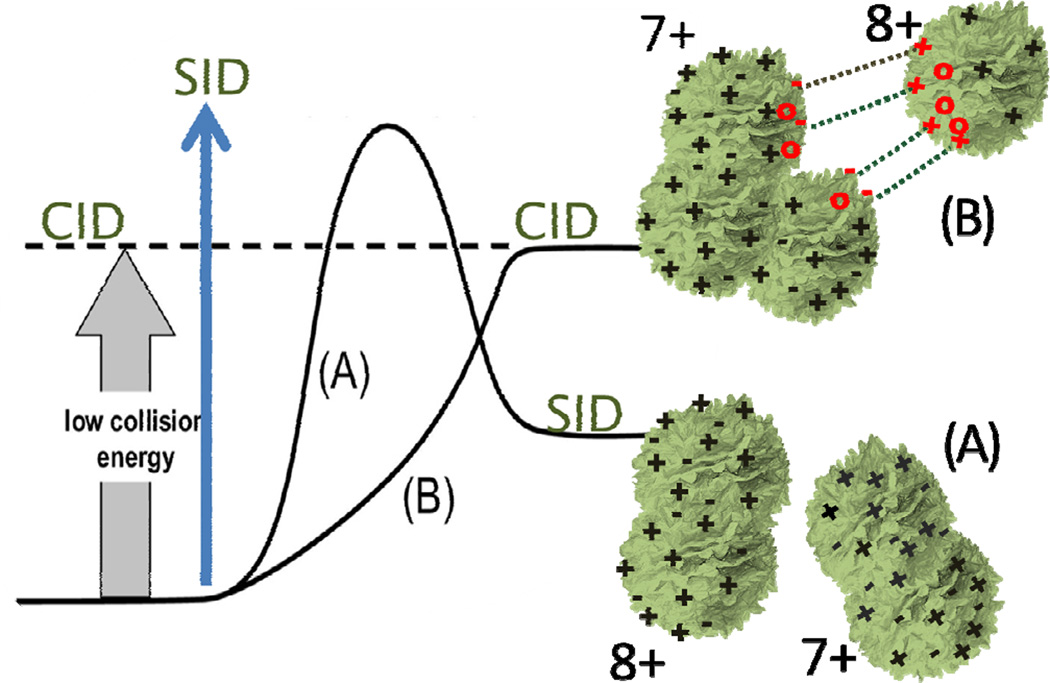
Figure 5.
Illustration of homolytic (A) and heterolytic (B) dissociation channels for a 15+ tetramer. Dissociation along thermodynamically favored channel A (homolytic) is accessible only at high collision energies, due to its electrostatic kinetic barrier. Low collision energy dissociations access the more endothermic, barrierless channel B.
Heterolytically cleaving an ion pair (separating point charges) presents no activation barrier in the reaction coordinate, in contrast to the repulsive barriers encountered when like charges are separated. It does, however, require significant energy input, rationalizing the higher enthalpy associated with this pathway.
That a low energy process (e.g., CID) allocates mass and charge asymmetrically from non-protein alkyl trimethyl ammonium bromide clusters (ATA+Br−)n (equation 3), or from inorganic Cs3(CsI)693+ and Cs4(CsI)924+, while SID allocates them symmetrically [39,77] implies that unfolding/charge migration is not essential to partition charge asymmetrically. These are simply clusters after all, not subject to the complex landscape of protein folding. The long distance charge migration invoked for decomposing protein complexes hardly seems plausible when extended to Br− and/or ATA+ charge carriers. The results of equation 3 are, however, consistent with the lower activation barrier presented by heterolytic ion pair cleavage processes.
| (3) |
Previously, strong support for the subunit unfolding/proton migration view was inferred from data showing diminished charge asymmetry when non-specific complexes constrained by intra-subunit crosslinks dissociated [36,78]. However reducing conformational flexibility and eliminating potential charge sites in crosslinking reactions obviously reduces an assembly’s ability to rearrange ion pairs and hydrogen bonds during activation and alters its dissociation threshold, suggesting that the crosslinking-diminished charge asymmetry can be explained equally well by alterations in the preference for homolytic versus heterolytic ion pair cleavage, or by a combination of both ideas.
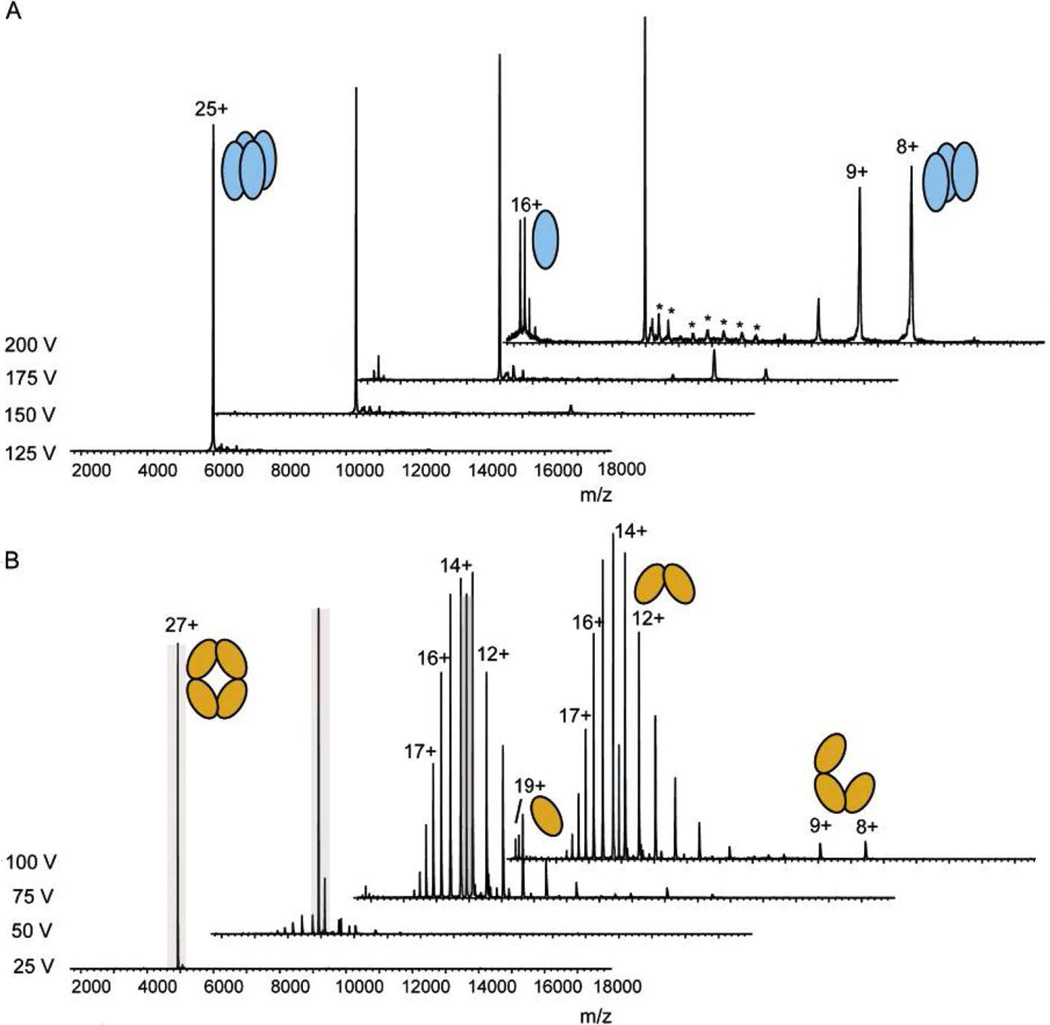
What can be learned from energy dependences? Transthyretin (TTR) 15+ homotetramer complexes dissociated by CID and SID have been compared [39,79]. CID at a 1350 eV collision energy yielded monomers and trimers, with the most intense monomer carrying 8 positive charges, or 53% of the total charge, while SID (at energies yielding a similar amount of surviving precursor), typically released monomers carrying ~4 positive charges, a roughly equal (symmetric) allocation of charge. Only at 450 eV (dissociation threshold voltage) was SID observed to allocate charge asymmetrically, releasing 8+ monomers [79]. CID dissociations of TTR 9+-15+ primarily distributed charge asymmetrically, even from the low charge precursors where covalent cleavage was competitive with disassembly of the noncovalent complex [80]. Only the highest precursor charge state, 15+, displayed some contributions from a symmetric CID pathway—release of 7+ and 8+ dimers [80]. The SID observations are understandable in that by depositing energy with a favorable center-of-mass in a single collision, SID accesses high barrier symmetric dissociation pathways unsurmountable by CID (Fig. 5). For products to be generated by CID, which deposits energy less efficiently and over multiple collisions, a pathway avoiding high barriers appears essential. In instances where barriers are excessive and unavoidable, covalent bond cleavage can take precedence over the disruption of noncovalent interactions, as observed for TTR charge states ≤ 9+ [80]. This latter instance is likely when many ion pairs span subunits. If the number of ion pairs is inversely related to charge state, then covalent bond cleavage is most likely from low charge state complexes, whereas symmetric dissociation might be accessible to CID from high charge state ions. Indeed, force field and massive density functional calculations by Marchese et al. [42] strengthen this supposition, predicting that low charge states should be zwitterionic, but high charge states might not.
We propose that a few ion pairs spanning subunit interfaces simply cleave heterolytically to access the alternative pathway needed for asymmetric dissociation in CID. (See Fig. 4.) “Pre-activation” or heating ions by elevating atmosphere/vacuum interface voltages can facilitate ion pair rearrangement and proton transfer/neutralization. Evidence of these rearrangements exists in the mobility-selected SID spectra of pre-activated TTR [39]. In that study, TTR tetramers (30 or 117 V cone voltage) were delivered to the ion mobility spectrometer (IMS) for mobility separation prior to SID. Drift time distributions for low cone voltage TTR 15+ (“native-like”) consisted of a single, sharp peak at ~10 msec, while those from TTR 15+ at high cone voltage spanned 10–20 msec with 3 peaks, attributed to differential unfolding. SID of the expanded tetramers was found to release remarkable amounts of trimer and complementary highly charged monomer (+7 to +9), interpreted to indicate that one of the monomers in the complex had been partially unfolded, promoting its SID detachment from the complex as a highly charged monomer.
We suggest an alternate interpretation. Differing mobilities could reflect size differences for various ion pair and charge site arrangements in pre-activated tetramers. Increased size can arise from expansion of all or a subset of the subunits. Some expanded precursors could possess structures with more ion pairs placed at dimer/dimer or monomer/trimer interfaces than would be present in the unactivated tetramer. (Note that relocating salt bridges to interfaces does not preclude increases in collision cross-section during pre-activation, because other regions in the complex would correspondingly lose their restraints against expansion.) Akin to Figs. 4 and 5 it can be imagined that SID of tetramers having ion pairs pre-aligned across subunit interfaces could promote release of monomers, dimers, and trimers by heterolytic cleavages to yield high and low charge/mass products.
There are two reasons to prefer a mechanism invoking ion pair rearrangements over one invoking the unfolding of a single subunit. The first is that “pre-activated” TTR tetramers were found to be more resistant to collisional dissociation (as revealed by precursor survival yields), than unactivated, “native-like” tetramers, despite the former’s generally larger collision cross-sections [39]. Pre-activated precursors were similarly more resistant to SID than unactivated precursors. It is unclear how one supposedly unfolding, extended subunit within an otherwise folded complex would be “reinforced” against detachment. In contrast, a mechanism reinforcing interfaces by adding subunit-spanning salt bridges does predict that pre-activated complexes could display increased stability to dissociation. A second factor disfavoring the single-subunit unfolding mechanism is that SID spectra from pre-activated precursors displayed intense dimer and trimer peaks absent or of much lower intensity in SID and CID spectra of unactivated precursors. The striking differences were noted, but not explained in the initial study [39]. It is not clear how a mechanism altering only one subunit could drive so many changes in dimer and trimer abundances and charge state distributions. The differences appear more consistent with a mechanism rearranging charge and ion pairs throughout the complex.
The presence of opposing charges within ESI analytes may also explain a puzzling size compaction, i.e., smaller collision cross-sections, observed when ring-like complexes, complexes with central cavities, or lower charge state complexes are collisionally activated [57,73,74,81,82,56]. The reduced cross-sections for such complexes are highly correlated to charge state, with the largest changes obtained by activating the lowest charge states at the same lab-frame energy [57]. If we consider a model where the lowest charge states of protein CSDs represent species carrying more opposing charges (i.e., ion pairs), then we can imagine a process wherein gas phase collisional activation facilitates formation of rearranged and/or additional salt bridges to “staple” subunits separated in solution by the central cavity. Higher charge states are less likely to collapse, because they have fewer ion pairs available to “fasten” the rearranged complex.
Let’s consider the collisional dissociation of the torroidal complex HSP16.9, a dodecamer composed of stacked hexamer rings. Collisional dissociation of the 32+ ion of this homo-oligomer asymmetrically releases monomers and undecamers with CSDs centered on ~14+ and ~18+, respectively [83]. If charge on an intact 12-mer is uniformly distributed initially, then each monomer would bear 2–3 positive charges on average (Fig. S2A). Upon collisional activation, the monomer unfolding hypothesis would have each of 11 subunits transfer almost 40% of its charge to a departing 12th subunit (Fig. S2A, right hand side), in a process coordinated across the 95 Å x 55 Å assembly [84]. The ion pair hypothesis (Fig. S2B, illustrating only half of the complex) considers each monomer in the stacked rings to link to at least 3 other monomers by ion pairs (e.g., one subunit on each side and one on the adjacent ring). Without long range transfers of 12 charges, a departing monomer would need to heterolytically cleave four ion pairs per contacting unit to accumulate 12 additional charges. Ion pairing requires a departing monomer to simply flick off linkages to adjacent subunits, enabling it to step around barriers, in contrast to the current paradigm that postulates coordinated charge movement across tens or hundreds of angstroms.
Evidence for the SaBRe Ion Pairing Model
SaBRe ion pairing predicts that sequence mutations which alter the number of basic or acidic amino acids can alter the extent of asymmetry in charge partitioning by altering the number of heterolytic ion pair cleavages. The direction of that change (e.g., increasing the number of positive charges deposited on an expelled monomer) should be inverted for the opposite polarity (i.e., to decrease the negative charge deposited on expelled monomers in negative ion mode). These predictions contrast with the behavior anticipated by the unfolded monomer hypothesis, where charge partitioning would, instead, be governed by the surface area of the unfolded monomer. The veracity of SaBRe predictions is evident in work by Sinelnikov, et al. [85] comparing charge states for monomers released in positive and negative ion mode from dissociating Shiga toxin pentamers Stx1, Stx2, and mutants, which demonstrated that charge asymmetry is, indeed, sequence and polarity dependent. Those results are discussed in detail in the supplemental material.
Sequence mutations can lead to different CCSs for equally-charged mutant and wild type (WT) complexes. Beyond these differences, ΔΩ, SaBRe predicts that mutations which weaken or reinforce subunit interfaces (i.e., by eliminating or introducing an inter-subunit salt bridge) will lead the differences to increase in magnitude with increasing charge. That is, differences in resistance to deformation become more prominent at increasing lab frame energies and, at constant voltage, ΔΩ will increase with Z. This trend was displayed by hemoglobin tetramers (αh βh )2, from normal (HbA) and sickle cell (HbS) variants [86], as described in the supplemental material. The HbS, HbA ΔΩ became larger with increasing charge, consistent the loss of one potential salt bridge per HbS β-subunit.
Recent studies explored the stabilization of complexes to CID provided by divalent cations, demonstrating that bound Ca2+ or Mg2+ ions reinforce hemoglobin tetramers with multidentate ion bridges [87–89]. The stabilization was proposed to arise from intra-subunit divalent metal ion “staples” that inhibited unfolding. Folded subunits were to thus retain enough of their initial non-covalent bonds and higher order structure to remain associated in complexes [89]. This proposal and an alternative based on salt bridges are detailed in the supplemental material. The alternative argues that divalent metal and “organic” staples (ion paired-amino acid side chains) bind and rearrange inter-subunit to stabilize complexes against dissociation. Both models rationalize the observation that asymmetrically ejected monomers tend to carry less than the average metal load.
How can Ligand Association be Maintained Despite Subunit “Unfolding”?
The release of charge-enriched subunits that still maintain their ligand associations [90] presents another conundrum for the idea that in order for highly charge-enriched subunits to be released, they must be unfolded. One association that persists, despite “unfolding” is the cobalt ligand associated with the a-subunits of toyocamycin nitrile hydratase, as mentioned previously [75]. A more complex example is from CID of the RNA polymerase II (517 kDa) complex [8]. The 48+ dodecamer was shown to dissociate asymmetrically by eliminating not only highly charged Rpb4 and Rpb7 subunits, but also the 44.5 kDa Rpb4/7 heterodimer. Rpb4 and Rpb7 are known to associate in vivo, but it is quite remarkable that this apparently specific heterodimer can be ejected from RNA polymerase II with 24–26 positive charges. Despite 50% of the charge being ejected onto 9% of the mass, the Rpb4/7 association is preserved [90].
Ion pairs easily rationalize these persistent interactions—they are maintained by Coulombic attraction. Collisions could slightly reposition an anionic ligand to a nearby protonated site, such that ligand remained associated despite ejection of the subunit to which it was previously bound. In positive ion mode, relocating an anion ligand away from a soon-to-be expelled subunit and onto a positive charge site elsewhere decrements by 1 the surviving aggregate’s overall charge, while incrementing by 1 that of the released subunit.
The CID behavior of streptavidin homotetramers (S4) complexed with four biotin (B) or four 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-biotinyl (Btl) ligands [90] provides interesting data for testing hypotheses. (S4 + 4B)16+ was observed to decompose primarily by releasing neutral B, although subunit loss (e.g., S7+) constituted a minor pathway at higher collision energies. In contrast, expulsion of free ligand was not observed for related (S4 + 4Btl)16+ complexes. Instead, subunit loss (e.g., S7+) comprised the major dissociation pathway. Expelled (S + Btl)x+ products were also weakly observed [90], comprising another example in which noncovalent associations are preserved despite release by asymmetric dissociations. Why is B ligand dislodged from protonated streptavidin complexes, but not Btl? If unfolding is required for asymmetric charge partitioning, why would a ligand remain associated with the highly charged, expelled subunit?
We consider that although biotin and Btl are both acidic (Btl is a phosphate diester, while biotin has a valeric acid tail); phosphoric acids are more acidic than carboxylic acids. Hence, the Btl ligand’s higher gas phase acidity would favor heterolytic cleavage/rearrangement of its salt bridges with protonated subunits, making (S4 + 4Btl)n+ complexes more likely to dissociate by expelling highly charged subunits than by releasing neutral or positively charged Btl. Btl’s higher propensity to form ion pairs (or cleave heterolytically) also explains why asymmetric dissociation by subunit expulsion is enhanced by 85% for (S4 + 4Btl)n+, compared to (S4 + 4B)n+ [90]. When a subunit-ligand ion pair cleaves heterolytically, the departing subunit should be enriched by one charge; thus, if ion pairing is more important in (S4 + 4Btl)n+ than in (S4 + 4B)n+, Sx+ expelled from the former should bear more charge on average than Sx+ expelled from the latter. Indeed, the average charge state of Sx+ product ions released from (S4 + 4Btl)n+ was found to be 7.1 ± 0.1 (for +15) and 7.3 ± 0.1 (for +16) versus 6.8 ± 0.1 (+15) and 6.9 ± 0.1 (+16) for release from (S4 + 4B)n+ [90].
The dominance of electrostatic interactions is reiterated in CID of RNase S/CTP complexes [91], which break a covalent bond to eject CMP, yet maintain the noncovalent diphosphate/S-protein association.
Collision-induced dissociation studies of cholera toxin B pentamers (CTB5) complexed to saccharide GM1 yielded three particularly interesting observations [90]. In positive ion mode, (CTB5+5GM1)15+ decomposed primarily by expelling a CTB subunit with 4–7 positive charges, a result considered surprising because the native structure would require ligand release before or during subunit detachment. It was argued that bound ligand must migrate to a different subunit in the activated complex, enabling the decomposing complex to retain ligand while expelling a subunit. At higher collision energies, small amounts of (CTB + GM1)5+ or (CTB + GM1)6+ complexes were observed to detach from the precursor, equally surprising because the subunit/ligand association was preserved, despite asymmetric charge partitioning. That observation was interpreted as indicating that monomer unfolding did not preclude ligand binding, but the inability to collisionally dislodge free ligand is curious.
The stability of the associated ligand implies that an electrostatic interaction may be stabilizing it. Pentasaccharide GM1 contains a sialic acid that could pair with a basic site on a CTB subunit. As mentioned earlier, small amounts of activation can rearrange local structure and salt bridges to an extent that, while not necessarily altering a protein’s collision cross-section, would not leave it “natively folded” either, rationalizing the release of a subunit while ligand was retained by the stripped complex. Ion pairs could also explain how ligand could remain associated with the expelled, highly charged subunit, and the inability to dislodge free ligand. This hypothesis predicts different behavior for deprotonated (negative ion mode) complexes, namely that collisions should readily detach the sialylated ligand as an anion, consistent with observations.
These subtle product distribution differences arising when positive or negative charges are incorporated into mutants [85] or ligands [90] highlight the importance of Coulomb attraction in complexes and the ion pairs that lie at the center of charge partitioning asymmetry.
How Symmetric and Asymmetric Dissociations Differ
If heterolytically cleaved ion pairs define asymmetric dissociations, then we might suspect that asymmetric dissociations accessible to CID would require more energy to accomplish than the handful of CID-accessible symmetric dissociations. We find that this prediction is verified in existing data [83], comparing asymmetrically dissociating HSP16.9 (described above), to symmetrically dissociating stable protein 1 (SP-1), a related dodecamer. Those measurements demonstrated that the intact HSP16.9 32+ ion was seven times more resistant to dissociation than the SP-1 27+ ion (based on the activation needed to dissociate 50% of the complexes; Elab=11413 for HSP16.9 vs. 1552 eV for SP-1, where Elab = charge x voltage). It is similarly verified for the fragile tetrameric 2-keto-3-deoxyarabinonate, which cleaves symmetrically at only 50 V, in contrast to functionally-related arabinose dehydrogenase, dissociating asymmetrically at > 3 times the Elab energy (Fig. 6) [92]. Calcineurin, a heterodimer assuming more than one gas phase conformation and displaying both symmetric and asymmetric products, requires a slightly lower Elab threshold to dissociate symmetrically [93]. Similarly, dissociating non-specific 11+ cytochrome c dimers (100 mM NH4OAc) produces a highly symmetric distribution of complementary ions at low collision energies, but reveals a second, asymmetric pathway at higher energies [36].
Figure 6.
Tetrameric 2-keto-3-deoxyarabinonate, a complex with limited inter-subunit contacts, cleaves symmetrically at only 50 V, while functionally-related arabinose dehydrogenase, a tetramer with extensive contacts, dissociates at higher voltages and yields asymmetric product distributions. (A) Tandem mass spectra of arabinose dehydrogenase at acceleration voltages ranging from 50 to 200 V after selection of the 25+ ion of the tetrameric species. (B) Tandem mass spectra of 2-keto-3-deoxyarabinonate dehydratase at acceleration voltages ranging from 10 to 100 V after selection of the 27+ ion of the tetrameric species. The gray column indicates the precursor ion of 2-keto-3-deoxyarabinonate dehydratase. At high collision energies, some covalent fragmentation reactions took place. The stars indicate these fragments. (Reprinted with permission from Van den Heuvel, et al., Anal. Chem. 78, 7473–7483 (2006), copyright 2006 American Chemical Society.)
We might also guess that the number of ion pairs would be inversely correlated to charge state. That prediction is consistent with observations from complexes displaying both symmetric and asymmetric dissociation pathways, for which asymmetric pathways dominate in the low charge state precursors [36,39]. An example of this behavior is present in cytochrome c dimers sprayed from 1:1 water/methanol, 2% acetic acid. Decomposing 11+ and 13+ homodimers decompose primarily by allocating charge asymmetrically, while 17+ and 19+ dimers dissociate symmetrically; 15+ dimers dissociate along both pathways. Likewise, a pathway by which transthyretin tetramers symmetrically dissociate into dimers is observed for TTR 15+, but not for lower charge states [80], and 30+ SAP pentamers release low charged monomers and dimers (symmetric-type products), but 25+ pentamers do not, instead dissociating asymmetrically to expel highly charged monomers [57].
The prediction that the extent of ion pairing is inversely correlated to charge state is also consistent with observations that the “compaction” of activated cavity-containing and other complexes is most pronounced for low charge state ions, as observed for SAP pentamers and TRAP oligomers [57,73]. Ion pair rearrangements provide “adhesive” to secure the compacted oligomers produced by collisional activation and deformation. When asymmetric dissociations expel a charge-enriched monomer by heterolytically cleaving salt bridges, the stripped complex is enriched in opposite charges, potentially available to restrain it to a “collapsed” volume. A striking example of contraction in a stripped complex is the 45% discrepancy between the collision cross-section measured for GroEL 13-mer (following expulsion of just one of the original fourteen subunits) and the value calculated from its crystal structure [73].
Additional evidence of a correlation between ion pairs and charge state is that the laboratory frame energy required to dissociate SP-1 and textilotoxin complexes decreases with increasing charge state [80,83,94]. Traditionally, Coulomb repulsion has been invoked to explain a decrease in dissociation energy with increasing charge, but Coulomb attraction from additional ion pairs in low charge states should not be ignored. In fact, imparting greater ion pair stabilization to low charge state complexes explains why some lowly charged multimers tend to cleave covalent bonds or shed small ions (charge-strip) in preference or in competition to disrupting their noncovalent association [80,95]. For example, the same CID energy that expels charge-enriched monomers from 25+ serum amyloid P pentamers, also releases [(SAP)5 – y40]22+, [(SAP)5 – y40]21+, and [(SAP)5 – y30]23+, along with their complementary ions, revealing that covalent bond cleavage competes with asymmetric dissociation for this charge state. Moreover, [(SAP)5 – 2y40]19+ product ions were also observed, challenging theory to explain how a pentameric complex could remain associated despite enduring two covalent bond cleavages! Interestingly, SID of 25+ pentamers yielded symmetric dissociation products, but no covalent backbone cleavages. In CID of TTR, backbone cleavages are preferred over subunit release for 9+ tetramers and comprise the sole dissociation pathway of 8+ complexes [80]. Supplemental Figure S3 summarizes relationships between charge state, ion pairs, and dissociation pathways. Preferential covalent bond cleavage has also been noted for noncovalent complexes between DNA and basic peptides [96], where electrostatic interactions are also important.
Further Ramifications of Opposite Charges, Salt Bridges and Rearrangements
These ideas about the influence of opposite charges and salt bridges on dissociating noncovalent complexes can also explain the resistance of certain protein structural regions to cleavage by electron capture and electron transfer excitation. It may well be that some of the increased sequence coverage and efficiency observed in dissociations of higher charge states with electron-based dissociation methods reflect a reduction in the ion pairs that inhibit release of products. Hydrogen bonds are generally blamed for limitations in cleavage product release; but ionic hydrogen bonds (salt bridges and polydentate proton-binding interactions) constitute the strongest of these associations.
We do not wish to completely neglect the stabilization provided to gas phase protein complexes by other ionic hydrogen bonds; e.g., a proton hydrogen-bonded to two groups, akin to a proton-bound dimer. Meot-Ner noted that they provide up to 35 kcal/mol of stabilization—up to a third of the stabilization from covalent bonds [97]. Similarly, migrating protons can account for some of the asymmetry observed when some noncovalent complexes decompose; contributions from these processes should not be completely excluded. However, the desire to explain collected observations including why some low-charged noncovalent complexes cleave covalent bonds in preference to noncovalent ones, and why sequence mutations alter the charge borne by ejected monomers in a polarity-dependent fashion require some departure from today’s unfolding monomer paradigm.
In reconsidering the importance of protein folds to gas phase dissociation dynamics, we should also revisit evidence for their retention in the gas phase. If salt bridges are actually providing much of the stabilization against distortion observed in CCSs of gas phase proteins and protein complexes, must the solution phase arrangement of hydrogen bonds and van der Waals interactions (the conformation) necessarily also be retained? Does correspondence between the CCSs measured for proteins delivered by native ESI and the CCSs calculated from crystal structure coordinates necessarily require that structural arrangements (folds) are preserved in the gas phase? Perhaps it does not, as discussed in the supplemental material.
Our ideas about ion pairs and opposing charges suggest that charge-manipulated ions (supercharged or charge-reduced) differ subtly from equally charged ions produced directly by ESI, providing predictions by which our theories can be tested, as detailed in the supplemental materials, along with experimental evidence from the literature that upholds the predictions. One notable experiment is the decomposition of non-specific cytochrome c 13+ dimers produced either directly or by charge stripping 17+ dimers. Jurchen, et al. [78] found that while the charge-stripped dimers partitioned charge symmetrically, dimers produced directly by ESI released asymmetrically-charged products, consistent with the hypothesis that the latter is capable of more ion pairs. Alternate mechanisms would have predicted no difference in the dissociations or the opposite behavior: increased asymmetry from decomposing charge-stripped dimers, reflecting Coulomb repulsion from an initially higher charge state.
It is tempting to extend these ideas to other macromolecules, but caution is recommended. Although tandem MS of duplex DNA and DNA-drug complexes show some similarities to protein complexes; e.g., preference for covalent over noncovalent cleavage from low charge state precursors [98], the molecules differ in ionizing groups and structure, changing the importance of various interactions. In long DNA strands, highly charged precursors allocated charge asymmetrically, but lower charge states distributed it symmetrically [99], a contrast with the behavior of protein complexes.
Conclusions
It is worth pondering whether ion pairing contributions to gas phase protein structures have been underestimated in the past, particularly for the non-denaturing conditions employed in noncovalent complex studies. The strong correlation observed between a gas phase protein’s “compactness” and its charge [37,57], particularly following gas-phase collisions, could reflect a relationship between charge state and number of salt bridges constraining structure. Lower charge states may be stabilized by a larger number of salt bridges. Although charge repulsion has traditionally been credited for this correlation, the short distances spanned by salt bridges would clearly also influence stability.
The existing paradigm for multimer dissociation argues that one monomer unfolds to accommodate excess charge, but little evidence of that unfolding has been found in the collision cross-sections of monomeric products released from asymmetric dissociations when potential size alterations from charge state-dependent ion activation are excluded. We suggest that local charge rearrangements, homolytic, and/or heterolytic scissions of ion pairs proceed throughout the activated complex until the system can cross to a low-barrier potential energy surface cleaving (or appearing to cleave) ion pairs heterolytically to decompose. Heterolytic scission of ion pairs inherently distributes charge asymmetrically. Although such a cleavage may enable a compact structure to expand, unfolding of tertiary structure (assumedly transferred from solution) is not required for product ion charge asymmetry.
If the number of ion pairs within a complex is inversely related to its overall charge, then symmetric dissociations are more likely from higher charge states, asymmetric distributions from medium-low charge states, and intra-subunit covalent bond cleavages from the lowest-charged species (where subunit dissociation requires more free energy than even cleaving covalent bonds). For a given charge state, collisional activation that deposits energy slowly is more likely to yield asymmetric product distributions, while the rapid energy deposition of SID should favor symmetric distributions (kinetic energy exceeding the barrier height). Rare examples displaying symmetric product distributions from CID (without supercharging) are likely to reflect complexes that are weakly bound in the gas phase (few ion pairs).
A consideration of ion pairs in protein structures should not be limited to those annotated by NMR or X-ray crystallography, because salt bridge influence spans longer distances in the gas phase than in solution. Differences between gas and solution phase basicity also imply that sites ionized in solution are not necessarily the same as in the gas phase, and vice versa. Activating proteins by gas phase collisions may facilitate the formation of additional or rearranged salt bridges, not necessarily changing conformation globally and without changing the charge state of an ion or complex. Collisional activation-facilitated ion pairing or re-pairing could explain the puzzling “contraction” observed in some ion mobility-based measurements of cross-sections for protein complexes.
Ion pairing considerations may have ramifications for previous and future molecular modeling results where, e.g., all of the acidic residues in a protein ion dispersed by positive ESI may have been simulated as charge-neutral. It is important that models capture dynamic adjustments in charge location during activation, along with their effect on the basicity and acidity of sites around them.
Supplementary Material
Acknowledgments
Support from the US National Institutes of Health (R01GM103479) and the US Department of Energy (UCLA Institute for Genomics and Proteomics; DE-FC03-02ER63421) are acknowledged.
References
- 1.Ganem B, Li Y-T, Henion JD. Detection of Noncovalent Receptor-Ligand Complexes by Mass Spectrometry. J. Am. Chem. Soc. 1991;113:6294–6296. [Google Scholar]
- 2.Ganem B, Li Y-T, Henion JD. Observation of Noncovalent Enzyme-Substrate and Enzyme-Product Complexes by Ion-Spray Mass Spectrometry. J. Am. Chem. Soc. 1991;113:7818–7819. [Google Scholar]
- 3.Katta V, Chait BT. Observation of the Heme-Globin Complex in Native Myoglobin by Electrospray-Ionization Mass Spectrometry. J. Am. Chem. Soc. 1991;113:8534–8535. [Google Scholar]
- 4.Baca M, Kent SBH. Direct Observation of a Ternary Complex between the Dimeric Enzyme HIV-1 Protease and a Substrate-Based Inhibitor. J. Am. Chem. Soc. 1992;114:3992–3993. [Google Scholar]
- 5.Smith RD, Light-Wahl KJ, Winger BE, Loo JA. Preservation of Noncovalent Associations in Electrospray Ionization-Mass Spectrometry: Multiply Charged Polypeptide and Protein Dimers. Organic Mass Spectrometry. 1992;27:811–821. [Google Scholar]
- 6.Ganguly AK, Pramanik BN, Tsarbopoulos A, Covey TR, Huang E, Fuhrman SA. Mass Spectrometric Detection of the Noncovalent GDP-Bound Conformational State of the Human H-ras Protein. J. Am. Chem. Soc. 1992;114:6559–6560. [Google Scholar]
- 7.Zhou M, Politis A, Davies RB, Liko I, Wu K-J, Stewart AG, Stock D, Robinson CV. Ion Mobility-Mass Spectrometry of a Rotary ATPase Reveals ATP-Induced Reduction in Conformational Flexibility. Nature Chem. 2014;6:208–215. doi: 10.1038/nchem.1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lorenzen K, Vannini A, Cramer P, Heck AJR. Structural Biology of RNA Polymerase III: Mass Spectrometry Elucidates Subcomplex Architecture. Structure. 2007;15:1237–1245. doi: 10.1016/j.str.2007.07.016. [DOI] [PubMed] [Google Scholar]
- 9.Sharon M, Mao H, Erba EB, Stephens E, Zheng N, Robinson CV. Symmetrical Modularity of the COP9 Signalosome Complex Suggests its Multifunctionality. Structure. 2009;17:31–40. doi: 10.1016/j.str.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 10.Xie Y, Zhang J, Yin S, Loo JA. Top-Down ESI-ECD-FT-ICR Mass Spectrometry Localizes Noncovalent Protein-Ligand Binding Sites. J. Am. Chem. Soc. 2006;128:14432–14433. doi: 10.1021/ja063197p. [DOI] [PubMed] [Google Scholar]
- 11.Yin S, Loo JA. Elucidating the Site of Protein-ATP Binding by Top-Down Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2010;21(6):899–907. doi: 10.1016/j.jasms.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 12.Yin S, Loo JA. Top-Down Mass Spectrometry of Supercharged Native Protein-Ligand Complexes. Int. J. Mass Spectrom. 2011;300:118–122. doi: 10.1016/j.ijms.2010.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Enyenihi AA, Yang H, Ytterberg AJ, Lyutvinskiy Y, Zubarev RA. Heme Binding in Gas-Phase Holo-Myoglobin Cations: Distal Becomes Proximal? J. Am. Soc. Mass Spectrom. 2011;22:1763–1770. doi: 10.1007/s13361-011-0182-0. [DOI] [PubMed] [Google Scholar]
- 14.Chowdhury SK, Katta V, Chait BT. Probing Conformational Changes in Proteins by Mass Spectrometry. J. Am. Chem. Soc. 1990;112:9012–9013. [Google Scholar]
- 15.Loo JA, Ogorzalek Loo RR, Udseth HR, Edmonds CG, Smith RD. Solvent-induced Conformational Changes of Polypeptides Probed by Electrospray Ionization-Mass Spectrometry. Rapid Commun. Mass Spectrom. 1991;5(3):101–105. doi: 10.1002/rcm.1290050303. [DOI] [PubMed] [Google Scholar]
- 16.Ogorzalek Loo RR, Loo JA, Udseth HR, Fulton JL, Smith RD. Protein Structural Effects in Gas Phase Ion/Molecule Reactions with Diethylamine. Rapid Commun. Mass Spectrometry. 1992;6:159–165. doi: 10.1002/rcm.1290060302. [DOI] [PubMed] [Google Scholar]
- 17.Ogorzalek Loo RR, Smith RD. Investigation of the Gas-Phase Structure of Electrosprayed Proteins using Ion-Molecule Reactions. J. Am. Soc. Mass Spectrom. 1994;5:207–220. doi: 10.1016/1044-0305(94)85011-9. [DOI] [PubMed] [Google Scholar]
- 18.Ogorzalek Loo RR, Smith RD. Proton Transfer Reactions of Multiply Charged Peptide and Protein Cations and Anions. J. Mass Spectrom. 1995;30:339–347. [Google Scholar]
- 19.Ogorzalek Loo RR, Winger BE, Smith RD. Proton Transfer Reaction Studies of Multiply Charged Proteins in a High Mass-to-Charge Ratio Quadrupole Mass Spectrometer. J. Am. Soc. Mass Spectrom. 1994;5:1064–1071. doi: 10.1016/1044-0305(94)85067-4. [DOI] [PubMed] [Google Scholar]
- 20.Winger BE, Light-Wahl KJ, Rockwood AL, Smith RD. Probing Qualitative Conformation Differences of Multiply Protonated Gas-Phase Proteins via Hydrogen/Deuterium Isotopic Exchange with Water-d2. J. Am. Chem. Soc. 1992;114:5897–5898. [Google Scholar]
- 21.Suckau D, Shi Y, Beu SC, Senko MW, Quinn JP, Wampler FM, III, McLafferty FW. Coexisting Stable Conformations of Gaseous Protein Ions. Proc. Natl. Acad. Sci. USA. 1993;90:790–793. doi: 10.1073/pnas.90.3.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Covey T, Douglas DJ. Collision Cross Sections for Protein Ions. J. Am. Soc. Mass Spectrom. 1993;4:616–623. doi: 10.1016/1044-0305(93)85025-S. [DOI] [PubMed] [Google Scholar]
- 23.Clemmer DE, Hudgins RR, Jarrold MF. Naked Protein Conformations: Cytochrome c in the Gas Phase. J. Am. Chem. Soc. 1995;117:10141–10142. [Google Scholar]
- 24.Chen SH, Russell DH. How Closely Related Are Conformations of Protein Ions Sampled by IM-MS to Native Solution Structures? J. Am. Soc. Mass Spectrom. 2015;26:1433–1443. doi: 10.1007/s13361-015-1191-1. [DOI] [PubMed] [Google Scholar]
- 25.Ruotolo BT, Benesch JLP, Sandercock AM, Hyung S-J, Robinson CV. Ion Mobility-Mass Spectrometry Analysis of Large Protein Complexes. Nature Prot. 2008;3:1139–1152. doi: 10.1038/nprot.2008.78. [DOI] [PubMed] [Google Scholar]
- 26.Sharon M, Robinson CV. The Role of Mass Spectrometry in Structure Elucidation of Dynamic Protein Complexes. Annu. Rev. Biochem. 2007;76:167–193. doi: 10.1146/annurev.biochem.76.061005.090816. [DOI] [PubMed] [Google Scholar]
- 27.Uetrecht C, Rose RJ, van Duijn E, Lorenzen K, Heck AJ. Ion Mobility Mass Spectrometry of Proteins and Protein Assemblies. Chem. Soc. Rev. 2010;39:1633–1655. doi: 10.1039/b914002f. [DOI] [PubMed] [Google Scholar]
- 28.Light-Wahl KJ, Schwartz BL, Smith RD. Observation of the noncovalent quaternary associations of proteins by electrospray ionization mass spectrometry. J. Am. Chem. Soc. 1994;116(12):5271–5278. [Google Scholar]
- 29.Schwartz BL, Light-Wahl KJ, Smith RD. Observation of Noncovalent Complexes to the Avidin Tetramer by Electrospray Ionization Mass Spectrometry. J. Am. Soc. Mass Spectrom. 1994;5:201–204. doi: 10.1016/1044-0305(94)85034-8. [DOI] [PubMed] [Google Scholar]
- 30.Schwartz BL, Bruce JE, Anderson GA, Hofstadler SA, Rockwood AL, Smith RD, Chilkoti A, Stayton PS. Dissociation of Tetrameric Ions of Noncovalent Streptavidin Complexes Formed by Electrospray Ionization. J. Am. Soc. Mass Spectrom. 1995;6:459–465. doi: 10.1016/1044-0305(95)00191-F. [DOI] [PubMed] [Google Scholar]
- 31.Loo JA. Structure Determination of Protein Noncovalent Complexes by Mass Spectrometry. ASMS Conference on Mass Spectrometry and Allied Topics; Chicago, Il. 2001. [Google Scholar]
- 32.Fitzgerald MC, Chernushevich I, Standing KG, Whitman CP, Kent SB. Probing the Oligomeric Structure of an Enzyme by Electrospray Ionization Time-of-Flight Mass Spectrometry. Proc Nat. Acad. Sci. USA. 1996;93:6851–6856. doi: 10.1073/pnas.93.14.6851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Versluis C, van der Staaij A, Stokvis E, Heck AJR, de Craene B. Metastable ion formation and disparate charge separation in the gas-phase dissection of protein assemblies studied by orthogonal time-of-flight mass spectrometry. J. Am. Soc. Mass Spectrom. 2001;12:329–336. doi: 10.1016/S1044-0305(00)00227-0. [DOI] [PubMed] [Google Scholar]
- 34.Loo JA, Berhane B, Kaddis CS, Wooding KM, Xie Y, Kaufman SL, Chernushevich IV. Electrospray Ionization Mass Spectrometry and Ion Mobility Analysis of the 20S Proteasome Complex. J. Am. Soc. Mass Spectrom. 2005;16:998–1008. doi: 10.1016/j.jasms.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 35.Felitsyn N, Kitova EN, Klassen JS. Thermal Decomposition of a Gaseous Multiprotein Complex Studied by Blackbody Infrared Radiative Dissociation. Investigating the Origin of the Asymmetric Dissociation Behavior. Anal. Chem. 2001;73(19):4647–4661. doi: 10.1021/ac0103975. [DOI] [PubMed] [Google Scholar]
- 36.Jurchen JC, Williams ER. Origin of Asymmetric Charge Partitioning in the Dissociation of Gas-Phase Protein Homodimers. J. Am. Chem. Soc. 2003;125:2817–2826. doi: 10.1021/ja0211508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hall Z, Robinson CV. Do Charge State Signatures Guarantee Protein Conformations? J. Am. Soc. Mass Spectrom. 2012;23:1161–1168. doi: 10.1007/s13361-012-0393-z. [DOI] [PubMed] [Google Scholar]
- 38.van Duijn E. Current Limitations in Native Mass Spectrometry Based Structural Biology. J. Am. Soc. Mass Spectrom. 2010;21:971–978. doi: 10.1016/j.jasms.2009.12.010. [DOI] [PubMed] [Google Scholar]
- 39.Zhou M, Huang C, Wysocki VH. Surface-Induced Dissociation of Ion Mobility-Separated Noncovalent Complexes in a Quadrupole/Time-of-Flight Mass Spectrometer. Anal. Chem. 2012;84:6016–6023. doi: 10.1021/ac300810u. [DOI] [PubMed] [Google Scholar]
- 40.Xu D, Tsai C-J, Nussinov R. Hydrogen Bonds and Salt Bridges Across Protein-Protein Interfaces. Protein Eng. 1997;10:999–1012. doi: 10.1093/protein/10.9.999. [DOI] [PubMed] [Google Scholar]
- 41.Šamalikova M, Grandori R. Role of Opposite Charges in Protein Electrospray Ionization Mass Spectrometry. J. Mass Spectrom. 2003;38:941–947. doi: 10.1002/jms.507. [DOI] [PubMed] [Google Scholar]
- 42.Marchese R, Grandori R, Carloni P, Raugei S. On the Zwitterionic Nature of Gas-Phase Peptides and Protein Ions. PLOS Comput. Biol. 2010;6:e1000775. doi: 10.1371/journal.pcbi.1000775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grandori R. Origin of the Conformation Dependence of Protein Charge-State Distributions in Electrospray Ionization Mass Spectrometry. J. Mass Spectrom. 2003;38:11–15. doi: 10.1002/jms.390. [DOI] [PubMed] [Google Scholar]
- 44.Krusemark CJ, Frey BL, Belshaw PJ, Smith LM. Modifying the Charge State Distribution of Proteins in Electrospray Ionization Mass Spectrometry by Chemical Derivatization. J. Am. Soc. Mass Spectrom. 2009;20:1617–1625. doi: 10.1016/j.jasms.2009.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gitlin I, Carbeck JD, Whitesides GM. Why Are Proteins Charged? Networks of Charge-Charge Interactions in Proteins Measured by Charge Ladders and Capillary Electrophoresis. Angew. Chem. Int. Ed. 2006;45:3022–3060. doi: 10.1002/anie.200502530. [DOI] [PubMed] [Google Scholar]
- 46.Mathews CK, van Holde KE. Biochemistry. Redwood City, CA: Benjamin/Cummings; 1990. [Google Scholar]
- 47.Pace CN, Grimsley GR, Scholtz JM. Protein Ionizable Groups: pK Values and Their Contribution to Protein Stability and Solubility. J. Biol. Chem. 2009;284:13285–13289. doi: 10.1074/jbc.R800080200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barlow DJ, Thornton JM. Ion-Pairs in Proteins. J. Mol. Biol. 1983;168:867–885. doi: 10.1016/s0022-2836(83)80079-5. [DOI] [PubMed] [Google Scholar]
- 49.Breuker K, Bruschweiler S, Tollinger M. Electrostatic Stabilization of a Native Protein Structure in the Gas Phase. Angew. Chem. Int. Ed. 2011;50:873–877. doi: 10.1002/anie.201005112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Z, Browne SJ, Vachet RW. Exploring Salt Bridge Sructures of Gas-Phase Protein Ions using Multiple stages of Electron Transfer and Collision Induced Dissociation. J. Am. Soc. Mass Spectrom. 2014 doi: 10.1007/s13361-013-0821-8. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Prakash H, Mazumdar S. Direct Correlation of the Crystal Structure of Proteins with the Maximum Positive and Negative Charge States of Gaseous Protein Ions Produced by Electrospray Ionization. J. Am. Soc. Mass Spectrom. 2005;16:1409–1421. doi: 10.1016/j.jasms.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 52.Chowdhury SK, Katta V, Chait BT. An Electrospray-ionization Mass Spectrometer with New Features. Rapid Commun. Mass Spectrom. 1990;4(3):81–87. doi: 10.1002/rcm.1290040305. [DOI] [PubMed] [Google Scholar]
- 53.Rodriguez-Cruz SE, Klassen JS, Williams ER. Hydration of Gas-Phase Gramicidin S (M + 2H)2+ Ions Formed by Electrospray: The Transition From Solution to Gas-Phase Structure. J. Am. Soc. Mass Spectrom. 1997;8:565–568. [Google Scholar]
- 54.Chung E, Henriques D, Renzoni D, Zvelebil M, Bradshaw JM, Waksman G, Robinson CV, Ladbury JE. Mass Spectrometric and Thermodynamic Studies Reveal the Role of Water Molecules in Complexes formed between SH2 Domains and Tyrosyl Phosphopeptides. Structure. 1998;6:1141–1151. doi: 10.1016/s0969-2126(98)00115-4. [DOI] [PubMed] [Google Scholar]
- 55.Chung EW, Henriques DA, Renzoni D, Morton CJ, Mulhern TD, Pitkeathly MC, Ladbury JE, Robinson CV. Probing the Nature of Interactions in SH2 Binding Interfaces-Evidence from Electrospray Ionization Mass Spectrometry Protein. Science. 1999;8:1962–1970. doi: 10.1110/ps.8.10.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ma X, Zhou M, Wysocki VH. Surface Induced Dissociation Yields Quaternary Substructure of Refractory Noncovalent Phosphorylase B and Glutamate Dehydrogenase Complexes. J. Am. Soc. Mass Spectrom. 2014;25:368–379. doi: 10.1007/s13361-013-0790-y. [DOI] [PubMed] [Google Scholar]
- 57.Hall Z, Politis A, Bush MF, Smith LJ, Robinson CV. Charge-State Dependent Compaction and Dissociation of Protein Complexes: Insights from Ion Mobility and Molecular Dynamics. J. Am. Chem. Soc. 2012;34:3429–3438. doi: 10.1021/ja2096859. [DOI] [PubMed] [Google Scholar]
- 58.Williams ER. Proton Transfer Reactivity of Large Multiply Charged Ions. J. Mass Spectrom. 1996;31:831–842. doi: 10.1002/(SICI)1096-9888(199608)31:8<831::AID-JMS392>3.0.CO;2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gronert S. Determining the Gas-Phase Properties and Reactivities of Multiply Charged Ions. J. Mass Spectrom. 1999;34:787–796. doi: 10.1002/(SICI)1096-9888(199908)34:8<787::AID-JMS847>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 60.Felitsyn N, Kitova EN, Klassen JS. Thermal Dissociation of the Protein Homodimer Ecotin in the Gas Phase. J. Am. Soc. Mass Spectrom. 2002;13:1432–1442. doi: 10.1016/S1044-0305(02)00647-5. [DOI] [PubMed] [Google Scholar]
- 61.Mauk MR, Mauk AG, Chen YL, Douglas DJ. Tandem Mass Spectrometry of Protein–Protein Complexes: Cytochrome c–Cytochrome b5. J. Am. Soc. Mass Spectrom. 2002;13:59–71. doi: 10.1016/S1044-0305(01)00331-2. [DOI] [PubMed] [Google Scholar]
- 62.Douglass KA, Venter AR. Predicting the Highest Intensity Ion in Multiple Charging Envelopes Observed for Denatured Proteins during Electrospray Ionization Mass Spectrometry by Inspection of the Amino Acid Sequence. Anal. Chem. 2013;85:8212–8218. doi: 10.1021/ac401245r. [DOI] [PubMed] [Google Scholar]
- 63.Stirnemann G, Kang S-g, Zhou R, Berne BJ. How Force Unfolding Differs from Chemical Denaturation. Proc Nat. Acad. Sci. USA. 2014;111:3413–3418. doi: 10.1073/pnas.1400752111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Drahos L, Vekey K. Determination of the Thermal Energy and its Distribution in Peptides. J. Am. Soc. Mass Spectrom. 1999;10:323–328. [Google Scholar]
- 65.Hashemi S, Jarvis MJY, Bohme DK. Gas-Phase Observation of the Heterolytic Dissociation of Negative Ions into Counter Ions: Dissociation of [Cu phthalocyanine (SO3)4Na]3− . J. Am. Soc. Mass Spectrom. 2008;19:375–379. doi: 10.1016/j.jasms.2007.11.014. [DOI] [PubMed] [Google Scholar]
- 66.Heiles S, Cooper RJ, Berden G, Oomens J, Williams ER. Hydrogen Bond Mediated Stabilization of the Salt Bridge Structure for the Glycine Dimer Anion. Phys. Chem. Chem. Phys. 2015;17:30642. doi: 10.1039/c5cp06120b. [DOI] [PubMed] [Google Scholar]
- 67.Jarrold MF. Peptides and Proteins in the Vapor Phase. Annu. Rev. Phys. Chem. 2000;51:179–207. doi: 10.1146/annurev.physchem.51.1.179. [DOI] [PubMed] [Google Scholar]
- 68.Julian RR, Beauchamp JL, Goddard III WA. Cooperative Salt Bridge Stabilization of Gas-Phase Zwitterions in Neutral Arginine Clusters. J. Phys. Chem. A. 2002;106:32–34. [Google Scholar]
- 69.Polfer NC, Oomens J. Vibrational Spectroscopy of Bare and Solvated Ionic Complexes of Biological Relevance. Mass Spectrom Rev. 2009;28:468–494. doi: 10.1002/mas.20215. [DOI] [PubMed] [Google Scholar]
- 70.Zhang Z, Browne SJ, Vachet RW. Exploring Salt Bridge Sructures of Gas-Phase Protein Ions using Multiple stages of Electron Transfer and Collision Induced Dissociation. J. Am. Soc. Mass Spectrom. 2014;25:604–613. doi: 10.1007/s13361-013-0821-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Patrick AL, Polfer N. H2SO4 and SO3 Transfer Reactions in a Sulfopeptide-Basic Peptide Complex. Anal. Chem. 2015;87:9551–9554. doi: 10.1021/acs.analchem.5b02479. [DOI] [PubMed] [Google Scholar]
- 72.Julian RR, Jarrold MF. Gas Phase Zwitterions in the Absence of a Net Charge. J. Phys. Chem. A. 2004;108:10861–10864. [Google Scholar]
- 73.Knapman TW, Morton VL, Stonehouse NJ, Stockley PG, Ashcroft AE. Determining the Topology of Virus Assembly Intermediates Using Ion Mobility Spectrometry-Mass Spectrometry. Rapid Commun. Mass Spectrom. 2010;24:3033–3042. doi: 10.1002/rcm.4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhou M, Dagan S, Wysocki VH. Protein Subunits Released by Surface Collisions of Noncovalent Complexes: Nativelike Compact Structures Revealed by Ion Mobility Mass Spectrometry. Angew. Chem. Int. Ed. 2012;51:4336–4339. doi: 10.1002/anie.201108700. [DOI] [PubMed] [Google Scholar]
- 75.Blackwell AE, Dodds ED, Bandarian B, Wysocki V. Revealing the Quaternary Structure of a Heterogeneous Noncovalent Protein Complex through Surface-Induced Dissociation. Anal. Chem. 2011;83:2862–2865. doi: 10.1021/ac200452b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Benesch JLP, Aqulina JA, Ruotolo BT, Sobott F, Robinson CV. Tandem Mass Spectrometry Reveals the Quaternary Organization of Macromolecular Assemblies. Chemistry & Biology. 2006;13:597–605. doi: 10.1016/j.chembiol.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 77.Zhang Y, Zhou M, Ma X, Wysocki VH. Comparison between Surface-Induced Dissociation (SID) and Collision-Induced Dissociation (CID) of Ion Mobility (IM)-Separated Detergent Clusters. 61st ASMS Conference on Mass Spectrometry and Allied Topics; May 20-24 2013; Minneapolis, MN. p. TP012. [Google Scholar]
- 78.Jurchen JC, Garcia DE, Williams ER. Further Studies on the Origins of Asymmetric Charge Partitioning in Protein Homodimers. J. Am. Soc. Mass Spectrom. 2004;15:1408–1415. doi: 10.1016/j.jasms.2004.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Beardsley RL, Jones CMSA, Galhena AS, Wysocki VH. Noncovalent Protein Tetramers and Pentamers with “n” Charges Yield Monomers with n/4 and n/5 Charges. Anal. Chem. 2009;81:1347–1356. doi: 10.1021/ac801883k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pagel K, Hyung SJ, Ruotolo BT, Robinson CV. Alternate Dissociation Pathways Identified in Charge-Reduced Protein Complex Ions. Anal. Chem. 2010;82:5363–5372. doi: 10.1021/ac101121r. [DOI] [PubMed] [Google Scholar]
- 81.Benesch JLP. Collisional Activation of Protein Complexes: Picking Up the Pieces. J. Am. Soc. Mass Spectrom. 2009;20:341–348. doi: 10.1016/j.jasms.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 82.Hogan CJ, Ruotolo BT, Robinson CV, Fernández de la Mora J. Tandem Differential Mobility Analysis-Mass Spectrometry Reveals Partial Gas-Phase Collapse of the GroEL Complex. J. Phys. Chem. B. 2011;115:3614–3621. doi: 10.1021/jp109172k. [DOI] [PubMed] [Google Scholar]
- 83.Erba EB, Ruotolo BT, Barsky D, Robinson CV. Ion Mobility-Mass Spectrometry Reveals the Influence of Subunit Packing and Charge on the Dissociation of Multiprotein Complexes. Anal. Chem. 2010;82:9702–9710. doi: 10.1021/ac101778e. [DOI] [PubMed] [Google Scholar]
- 84.van Montfort RLM, Basha E, Friedrich KL, Slingsby C, Vierling E. Crystal Structure and Assembly of a Eukaryotic Small Heat Shock Protein. Nature Structural Biology. 2001;8:1025–1030. doi: 10.1038/nsb722. [DOI] [PubMed] [Google Scholar]
- 85.Sinelnikov I, Kitova EN, Klassen JS, Armstrong GD. Effects of Single Amino Acid Substitution on the Dissociation of Multiply Charged Multiprotein Complexes in the Gas Phase. J. Am. Soc. Mass Spectrom. 2007;18:688–692. doi: 10.1016/j.jasms.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 86.Scarff CA, Patel VJ, Thalassinos K, Scrivens JH. Probing Hemoglobin Structure by Means of Traveling-Wave Ion Mobility Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2009;20:625–631. doi: 10.1016/j.jasms.2008.11.023. [DOI] [PubMed] [Google Scholar]
- 87.Han L, Hyung S-J, Ruotolo BT. Bound Cations Significantly Stabilize the Structure of Multiprotein Complexes in the Gas Phase. Angew. Chem. Int. Ed. 2012;51:5692–5695. doi: 10.1002/anie.201109127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Han L, Ruotolo BT. Traveling-Wave Ion Mobility-Mass Spectrometry Reveals Additional Mechanistic Details in the Stabilization of Protein Complex Ions Through Tuned Salt Additives. Int. J. Ion Mobil. Spectrom. 2013;16:41–50. doi: 10.1007/s12127-013-0121-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liu J, Konermann L. Cation-Induced Stabilization of Protein Complexes in the Gas Phase: Mechanistic Insights From Hemoglobin Dissociation Studies. J. Am. Soc. Mass Spectrom. 2014;25:595–603. doi: 10.1007/s13361-013-0814-7. [DOI] [PubMed] [Google Scholar]
- 90.Zhang Y, Deng L, Kitova EN, Klassen JS. Dissociation of Multisubunit Protein- Ligand Complexes in the Gas Phase. Evidence for Ligand Migration. J. Am. Soc. Mass Spectrom. 2013;24:1573–1583. doi: 10.1007/s13361-013-0712-z. [DOI] [PubMed] [Google Scholar]
- 91.Sheng Yin YX, Loo Joseph A. Mass Spectrometry of Protein-Ligand Complexes: Enhanced Gas-Phase Stability of Ribonuclease-Nucleotide Complexes. J. Am. Soc. Mass Spectrom. 2008;19:1199–1208. doi: 10.1016/j.jasms.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.van den Heuvel RHH, van Duijn E, Mazon H, Synowsky SA, Lorenzen K, Versluis C, Brouns SJJ, Langridge D, van der Oost J, Hoyes J, Heck AJR. Improving the Performance of a Quadrupole Time-of-Flight Instrument for Macromolecular Mass Spectrometry. Anal. Chem. 2006;78:7473–7483. doi: 10.1021/ac061039a. [DOI] [PubMed] [Google Scholar]
- 93.Kükrer B, Barbu IM, Copps J, Hogan P, Taylor SS, van Duijn E, Heck AJR. Conformational Isomers of Calcineurin Follow Distinct Dissociation Pathways. J. Am. Soc. Mass Spectrom. 2012;23:1534–1543. doi: 10.1007/s13361-012-0441-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Aquilina JA. The Major Toxin from the Australian Common Brown Snake is a Hexamer with Unusual Gas-Phase Dissociation Properties. Proteins: Structure, Function, and Bioinformatics. 2009;75:478–485. doi: 10.1002/prot.22259. [DOI] [PubMed] [Google Scholar]
- 95.Wysocki VH, Jones CM, Galhena AS, Blackwell AE. Surface-Induced Dissociation Shows Potential to Be More Informative Than Collision-Induced Dissociation for Structural Studies of Large Systems. J. Am. Soc. Mass Spectrom. 2008;19:903–913. doi: 10.1016/j.jasms.2008.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Alves S, Woods A, Delvolvé A, Tabet JC. Influence of Salt Bridge Interactions on the Gas-Phase Stability of DNA/Peptide Complexes. Int. J. Mass Spectrom. 2008;278:122–128. [Google Scholar]
- 97.Meot-Ner (Mautner) M. Update 1 of: Strong Ionic Hydrogen Bonds. Chem. Rev. 2012;112(10):PR22–PR103. doi: 10.1021/cr200430n. [DOI] [PubMed] [Google Scholar]
- 98.Keller KM, Zhang J, Oehlers L, Brodbelt JS. Influence of Initial Charge State on Fragmentation Patterns for Noncovalent Drug/DNA Duplex Complexes. J. Mass Spectrom. 2005;40:1362–1371. doi: 10.1002/jms.927. [DOI] [PubMed] [Google Scholar]
- 99.Madsen JA, Brodbelt JS. Asymmetric Charge Partitioning upon Dissociation of DNA Duplexes. J. Am. Soc. Mass Spectrom. 2010;21:1144–1150. doi: 10.1016/j.jasms.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.