Abstract
ZNF322A encoding a classical Cys2His2 zinc finger transcription factor was previously revealed as a potential oncogene in lung cancer patients. However, the oncogenic role of ZNF322A and its underlying mechanism in lung tumorigenesis remain elusive. Here we show ZNF322A protein overexpression in 123 Asian and 74 Caucasian lung cancer patients. Multivariate Cox regression analysis indicated that ZNF322A was an independent risk factor for a poor outcome in lung cancer, corroborating the Kaplan–Meier results that patients with ZNF322A protein overexpression had significantly poorer overall survival than other patients. Overexpression of ZNF322A promoted cell proliferation and soft agar growth by prolonging cell cycle in S phase in multiple lung cell lines, including the immortalized lung cell BEAS-2B. In addition, ZNF322A overexpression enhanced cell migration and invasion, whereas knockdown of ZNF322A reduced cell growth, invasion and metastasis abilities in vitro and in vivo. Quantitative proteomic analysis revealed potential ZNF322A-regulated downstream targets, including alpha-adducin (ADD1), cyclin D1 (CCND1), and p53. Using luciferase promoter activity assay combined with site-directed mutagenesis and sequential chromatin immunoprecipitation-PCR assay, we found that ZNF322A could form a complex with c-Jun and cooperatively activate ADD1 and CCND1 but repress p53 gene transcription by recruiting differential chromatin modifiers, such as histone deacetylase 3, in an AP-1 element dependent manner. Reconstitution experiments indicated that CCND1 and p53 were important to ZNF322A-mediated promotion of cell proliferation, whereas ADD1 was necessary for ZNF322A-mediated cell migration and invasion. Our results provide compelling evidence that ZNF322A overexpression transcriptionally dysregulates genes involved in cell growth and motility therefore contributes to lung tumorigenesis and poor prognosis.
Introduction
The Cys2His2 (C2H2) zinc finger (ZNF) proteins form the largest family of sequence-specific DNA-binding protein, which are encoded by 2% of human genes.1, 2 Accumulated lines of evidence have indicated that C2H2 ZNF proteins have critical roles in a wide spectrum of cellular processes, including differentiation, development, metabolism, apoptosis, autophagy and stemness maintenance.3, 4, 5, 6, 7 Recent studies revealed that aberrant expression of C2H2 ZNF proteins contributes to tumorigenesis in several cancers. For example, ZNF331 and ZNF545 act as tumor suppressors by regulating cell proliferation and migration in various cancers, including nasopharyngeal, esophageal, lung, gastric, colon and breast cancer.8, 9 In contrast, SALL4 and ZNF703 are overexpressed in gastric cancer and promote cell proliferation and migration.10, 11 Overexpression of SALL4 and ZFX confer self-renewal properties in gastric cancer and hepatocellular carcinoma, respectively.10, 12 Krüppel-like factor 4, can act as either oncogene or tumor suppressor gene depending on the cell context.13 These results indicate that C2H2 ZNF proteins show diverse functions in tumorigenesis. Therefore, it is important to elucidate the role of C2H2 ZNF proteins in tumorigenesis.
ZNF322A, a C2H2 ZNF protein, was first identified in human embryonic heart complementary DNA (cDNA) by Li et al.14 ZNF322A is expressed in nuclei and can activate artificial promoter containing SRE or AP-1 element implicating that ZNF322A may be a transcription activator but the downstream genes and biologic effects of ZNF322A remain unclear.14 In our previous study, the chromosome 6p22.1 where the ZNF322A gene is located, was found significantly amplified in both Asian and Caucasian lung cancer patients.15 However, the detailed tumorigenic effect and underlying mechanism of ZNF322A need further investigation.
To investigate the oncogenic role of ZNF322A in lung cancer, we conducted a series of clinical, cell, animal and proteomic studies. Clinically, ZNF322A was highly overexpressed in Asian and Caucasian lung cancer patients with poor prognosis. Overexpression of ZNF322A enhanced lung cancer cell growth, invasion and metastasis abilities in vitro and in vivo. Using proteomic analysis, we revealed ZNF322A could regulate proteins and associated network functions. Importantly, we found that ZNF322A cooperated with c-Jun to regulate expression of downstream genes including alpha-adducin (ADD1), cyclin D1 (CCND1) and p53 through AP-1 elements.
Results
ZNF322A overexpression correlates with poor prognosis in both Asian and Caucasian lung cancer patients
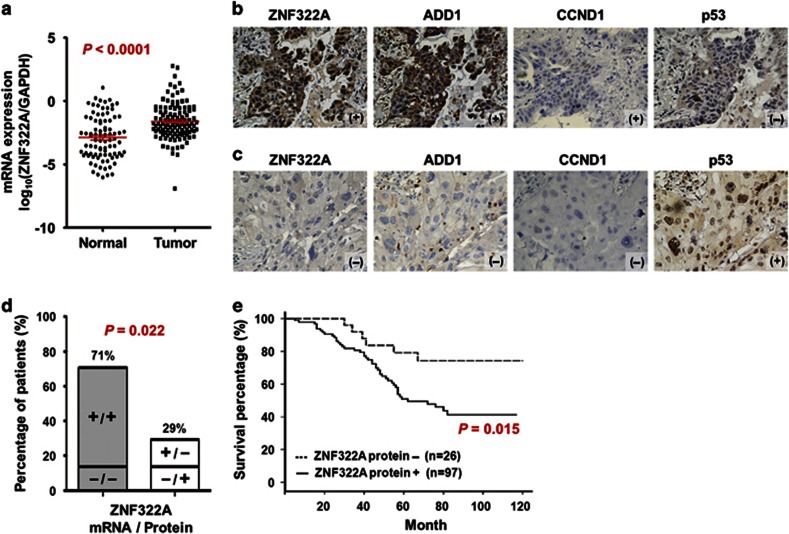
In this study, we conducted quantitative reverse transcription–PCR (qRT–PCR) analysis to examine ZNF322A mRNA expression in 123 Asian lung cancer patients. The mRNA expression level of ZNF322A in tumor tissues was significantly higher than that in the corresponding normal tissues (Figure 1a). In addition, ZNF322A protein was found to be overexpressed in 78.9% of these 123 Asian lung cancer patients using immunohistochemistry staining (Figures 1b and c, left; Supplementary Table S1). Moreover, a positive correlation between mRNA and protein expression was found (Figure 1d). Of note, immunohistochemistry staining of tissue arrays derived from 74 Caucasian lung cancer patients showed overexpression of ZNF322A protein in 72.9% of patients analyzed (Supplementary Figure S1A, Supplementary Table S1). ZNF322A mRNA expression in lung tumor samples of three published data sets16, 17, 18 deposited in Oncomine (https://www.oncomine.org) also showed significantly higher ZNF322A mRNA expression in tumor tissues compared with that in normal lung tissues (Supplementary Figure S2), suggesting that ZNF322A overexpression is common in lung cancers.
Figure 1.
Clinical significance of ZNF322A overexpression in lung cancer patients. (a) mRNA expression level of ZNF322A examined in 123 lung cancer patients. Dot plot demonstrates mRNA expression of log10 ratio between ZNF322A and GAPDH in tumor tissues and corresponding normal tissues. Red line indicates mean and s.e.m. (b and c) Representative immunohistochemistry images of ZNF322A, ADD1, CCND1 and p53 protein expression in tumor specimen of two lung cancer patients are shown. Patient 1 showed ZNF322A, ADD1, and CCND1 overexpression along with p53 downregulation (b), whereas patient 2 showed a reverse pattern (c). (d) Concordance analysis between mRNA and protein expression. + indicates increased mRNA expression and positive immunoreactivity, as opposed to − for reverse patterns. The percentage of the concordant group (gray columns) and discordant group (white columns) is indicated above. (e) Overall survival of lung cancer patients with ZNF322A overexpression (solid line) versus patients with ZNF322A normal expression (dotted line). P-values for comparison (a) and correlation analyzes (d) were determined using two-tailed Student's t-test and Pearson's χ2 test; and for survival analyzes (e) using log-rank test.
To determine whether high ZNF322A protein expression is associated with poor patient outcome, we performed univariate and multivariate Cox regression analysis in 123 Asian lung cancer patients. The univariate Cox regression analysis revealed that patients with ZNF322A overexpression, smoking habit, late stage or larger tumor size status had poor outcome (P=0.020, hazard ratio=2.75, 95% confidence interval=1.17–6.46 for ZNF322A overexpression; Table 1). Notably, multivariate Cox regression analysis showed that overexpression of ZNF322A protein correlated with a relative risk of death of 3.26 (P=0.016), even after adjusting for the smoking habit, late stage and larger tumor size status (Table 1), suggesting that ZNF322A overexpression was an independent risk factor of poor outcome. To further define the prognostic effects of ZNF322A overexpression in lung cancer patients, survival curves were generated using the Kaplan–Meier method. Patients with ZNF322A protein overexpression showed significantly poor overall survival than other patients (Figure 1e). Caucasian patients with ZNF322A protein overexpression in our cohort of tissue arrays also correlated with poor prognosis (Supplementary Figure S1B). In summary, our clinical studies indicated that ZNF322A was significantly overexpressed both in Asian and Caucasian lung cancer patients with poor prognosis.
Table 1. Cox regression analysis of risk factors for cancer-related death in lung cancer patients.
| Characteristics |
Univariate analysis |
Multivariate analysis |
||
|---|---|---|---|---|
| HR (95% CI) | P-valuea | HR (95% CI) | P-valuea | |
| ZNF322A protein expression | ||||
| Normal expression | 1.00 | 1.00 | ||
| Overexpression | 2.75 (1.17–6.46) | 0.020 | 3.26 (1.25–8.49) | 0.016 |
| Age | ||||
| <65 year-old | 1.00 | —b | ||
| ⩾65 year-old | 0.81 (0.48–1.38) | 0.433 | —b | —b |
| Gender | ||||
| Female | 1.00 | —b | ||
| Male | 1.48 (0.85–2.57) | 0.165 | —b | —b |
| Smoking habit | ||||
| Non-smoker | 1.00 | 1.00 | ||
| Smoker | 1.87 (1.02–3.42) | 0.044 | 1.93 (1.01–3.68) | 0.046 |
| Type | ||||
| SCC | 1.00 | —b | ||
| ADC | 0.96 (0.53–1.75) | 0.897 | —b | —b |
| Stage | ||||
| Stage I–II | 1.00 | 1.00 | ||
| Stage III–IV | 2.17 (1.25–3.77) | 0.006 | 1.54 (0.80–2.96) | 0.194 |
| T stage | ||||
| Stage 1–2 | 1.00 | 1.00 | ||
| Stage 3–4 | 2.21 (1.03–4.74) | 0.042 | 1.44 (0.45–4.57) | 0.539 |
| N stage | ||||
| N0 | 1.00 | —b | ||
| ⩾N1 | 1.46 (0.84–2.52) | 0.180 | —b | —b |
| M stage | ||||
| M0 | 1.00 | —b | ||
| ⩾M1 | 1.94 (0.69–5.44) | 0.209 | —b | —b |
Abbreviations: ADC, adenocarcinoma; CI, confidence interval; HR, hazard ratio; SCC, squamous cell carcinoma.
Bold values indicate statistical significance (P<0.05).
The variables without significant HR in the univariate analysis were not included in the multivariate analysis.
ZNF322A overexpression promotes cell proliferation and motility in lung cancer cells
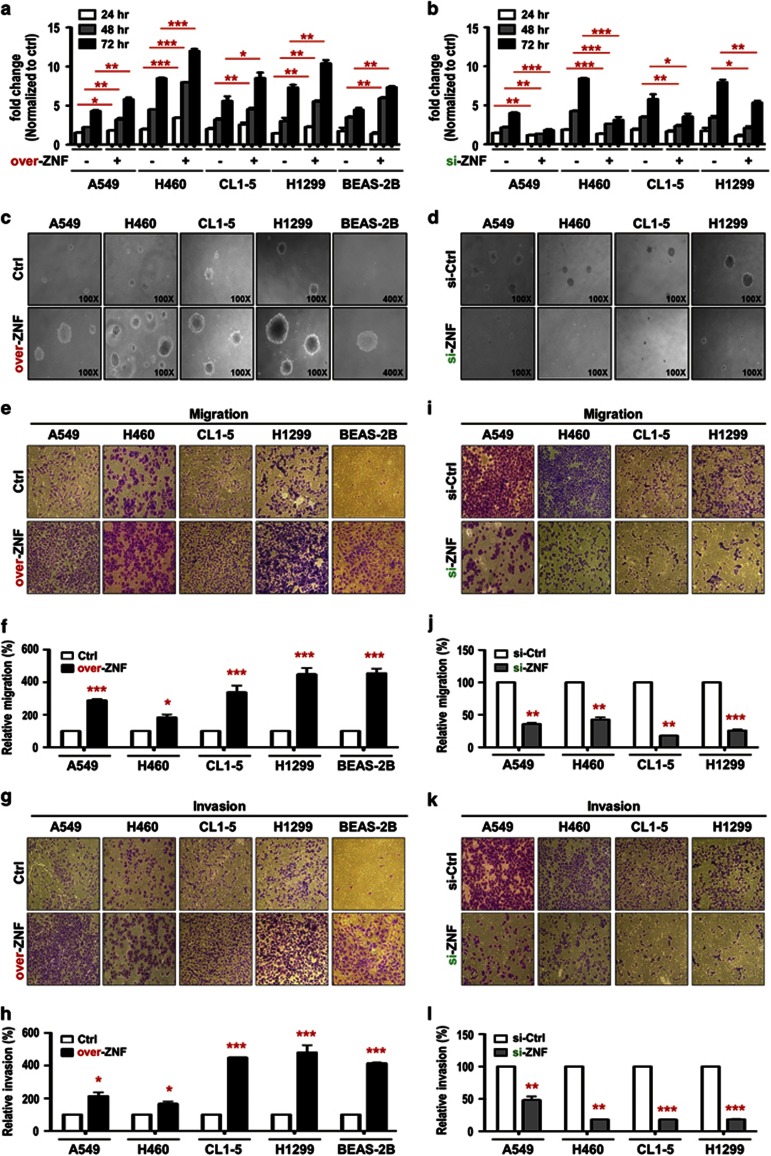
Our clinical data suggest that ZNF322A may function as an oncogene. Therefore, we examined ZNF322A expression by immunoblotting and found high ZNF322A expression in various lung cancer cell lines compared with that in the MRC5 normal lung cell line and the BEAS-2B immortalized lung cell line (Supplementary Figure S3). Cell proliferation was significantly increased following ectopic overexpression of ZNF322A in A549, H460, CL1-5 and H1299 lung cancer cells (Figure 2a), whereas ZNF322A knockdown inhibited cell proliferation (Figure 2b). Soft agar growth assay further supported that ZNF322A overexpression promoted anchorage-independent cell growth (Figure 2c), whereas ZNF322A silencing inhibited colony formation of various lung cancer cells (Figure 2d). Notably, ZNF322A overexpression increased cell proliferation rate and soft agar growth in BEAS-2B, the immortalized bronchial cells (Figures 2a and c). Ectopic ZNF322A expressing cells displayed a higher proportion of cells in S phase (Supplementary Figures S4A and B), whereas knockdown of ZNF322A showed less proportion of cells in the S phase determined by flow cytometry (Supplementary Figure S4C). Representative immunoblots for ZNF322A overexpression or silencing in all functional studies were shown in Supplementary Figure S5.
Figure 2.
ZNF322A overexpression promotes proliferation, soft agar growth, migration and invasion abilities in lung cells. (a and b) ZNF322A overexpression (over-ZNF) promoted cell proliferation in A549, H460, CL1-5, H1299 and BEAS-2B cells, whereas ZNF322A knockdown (si-ZNF) suppressed cell proliferation. (c and d) ZNF322A overexpression promoted colony formation ability of A549, H460, CL1-5, H1299 and BEAS-2B cells, whereas ZNF322A knockdown decreased colony formation ability. (e-h) ZNF322A overexpression promoted cell migration and invasion abilities in A549, H460, CL1-5, H1299 and BEAS-2B cells. (i-l) ZNF322A knockdown decreased cell migration and invasion abilities. P-values determined using two-tailed Student's t-test. Data represent mean±s.e.m. (n=3). *P<0.05; **P<0.01; ***P<0.001.
We further investigated whether ZNF322A affected lung cancer cell motility in vitro by transwell migration and invasion assays. ZNF322A overexpression significantly increased cell migration and invasion abilities in A549, H460, CL1-5 and H1299 lung cancer cells and even in the BEAS-2B bronchial cells (Figures 2e–h) at 12 h without affecting cell proliferation (data not shown), whereas knockdown of ZNF322A significantly decreased cell migration and invasion (Figures 2i–l). These data suggest that ZNF322A has an important role in lung tumorigenesis by promoting cell proliferation, anchorage-independent growth, migration and invasion in various lung cell lines, including the immortalized bronchial cells BEAS-2B.
ZNF322A overexpression accelerates tumor growth and tumor metastasis in vivo
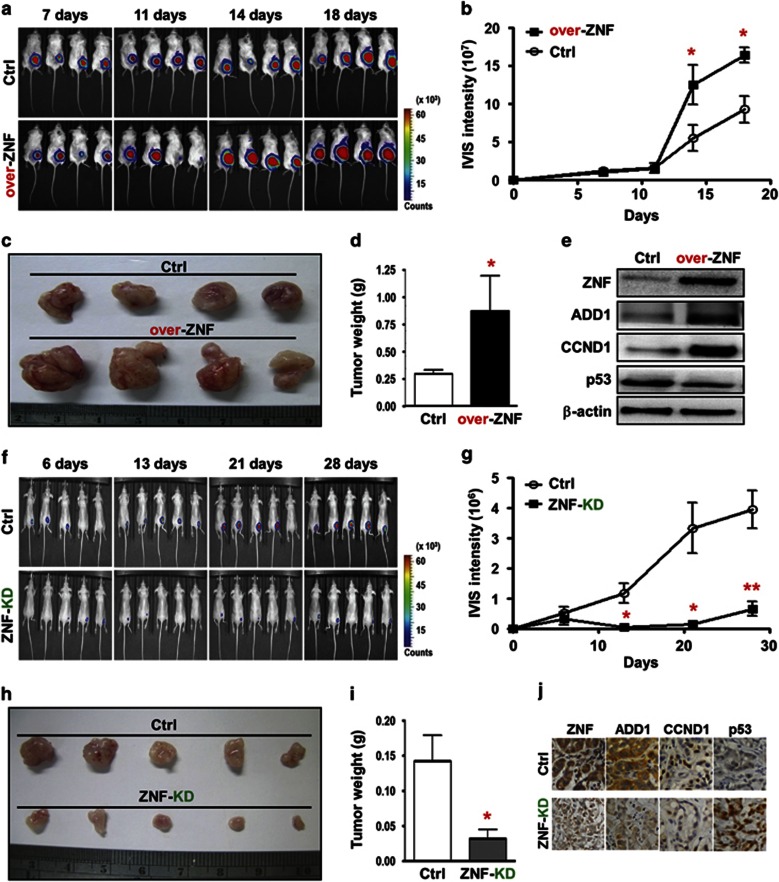
According to our cell-based results, we speculated that ZNF322A overexpression could promote tumor growth and facilitate tumor metastases in animals. To explore the effect of ZNF322A on tumor growth in vivo, severe combined immunodeficient mice were subcutaneously injected with H460-Luc cells transfected with empty vector or ZNF322A expression vector. IVIS spectrum in vivo imaging and intensity analysis demonstrated that mice injected with ZNF322A overexpressing H460-Luc cells showed significantly stronger luciferase signals on day 14 and day 18 compared with control group (Figures 3a and b). In addition, ZNF322A overexpression significantly increased tumor size and tumor weight compared with control (Figures 3c and d). Immunoblotting confirmed ZNF322A overexpression in xenografts collected on day 18 (Figure 3e). In the reciprocal experiment, A549-Luc cells with ZNF322A knockdown were injected into BALB/c nude mice and observed for tumor growth. Tumor growth, tumor size and tumor weight were significantly suppressed in ZNF322A knockdown group (Figures 3f–i). Immunohistochemistry confirmed that ZNF322A was silenced in xenografts collected on day 28 (Figure 3j).
Figure 3.
ZNF322A overexpression accelerates tumor growth in vivo. (a–d) H460-Luc cells transfected with empty vector (Ctrl) or ZNF322A expression vector (over-ZNF) were subcutaneously injected into severe combined immunodeficient mice. Luciferase intensity of tumor images (a) and quantitative IVIS intensity (b) were obtained on indicated days. Xenograft images (c) and quantitative weight (d) of tumor taken from mice on day 18 are shown. (e) Xenografts were subjected to tissue-immunoblotting to examine expression level of the indicated proteins. (f–i) Control (Ctrl) or ZNF322A knockdown A549-Luc cells were subcutaneously injected into BALB/c nude mice. Luciferase intensity of tumor images (f) and quantitative IVIS intensity (g) were obtained on indicated days. Xenograft images (h) and quantitative weight (i) of tumor taken from mice at day 28 are shown. (j) Xenografts were subjected to immunohistochemistry to examine expression of the indicated proteins. Data are mean±s.e.m. (n=5 mice per group). *P<0.05; **P<0.01.
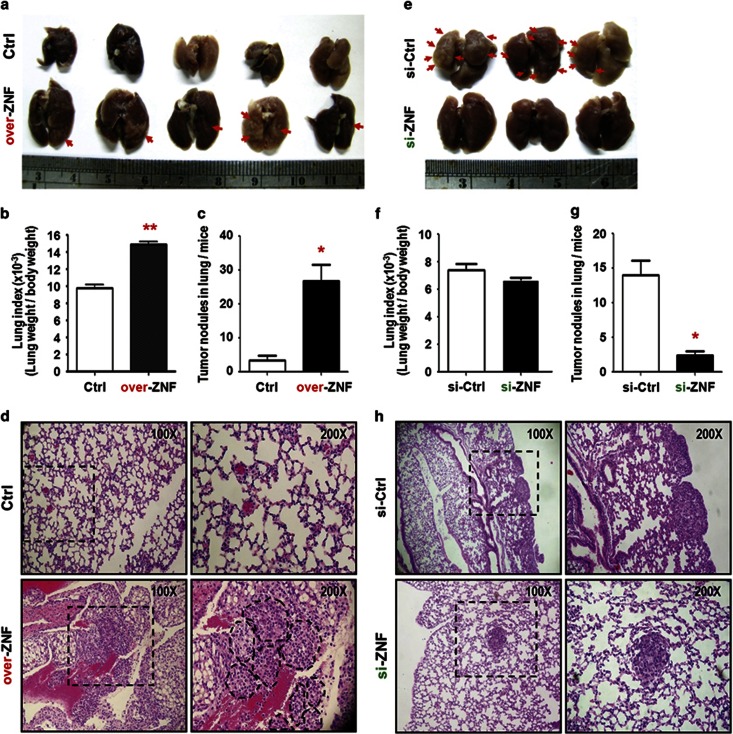
To examine if ZNF322A promotes tumor metastasis in vivo, experimental metastasis animal models were performed. A549 control cells or ZNF322A overexpressing cells were intravenously injected into severe combined immunodeficient mice via the tail vein. Mice were killed at the fourth week and lung tissues were collected. The group of mice injected with ZNF322A overexpressing cells bore more tumor nodules in lung tissue than the control group (Figure 4a). Lung index (lung weight/body weight) and tumor nodules were significantly higher in ZNF322A overexpression group (Figures 4b and c). Hematoxylin and eosin staining showed significantly more and bigger metastasized tumor nodules in ZNF322A overexpression group (Figure 4d). Conversely, ZNF322A knockdown suppressed lung metastasis, lung index and tumor nodules (Figures 4e–h). These in vivo results corroborated with in vitro data that ZNF322A overexpression promoted tumor growth and tumor metastasis.
Figure 4.
ZNF322A overexpression accelerates tumor metastasis in vivo. (a) Representative lung images taken from mice at 4th week are shown. A549 cells transfected with empty vector (Ctrl) or ZNF322A expression vector (over-ZNF) were injected into severe combined immunodeficient mice via the tail vein. (b and c) Lung index (lung weight/body weigh) (b) and quantitative tumor nodules (c) show bigger tumor mass and more lung nodules in over-ZNF group compared with Ctrl groups. (d) Hematoxylin and eosin staining of tumor micrometastasis are shown. (Original magnification: × 100). A twofold enlarged image is also shown with tumor nodules indicated by dotted lines. (e-h) Control (Ctrl) or ZNF322A knockdown A549 cells were injected into mice via the tail vein. Representative lung images (e), lung index (f), quantitative tumor nodules (g) and heamatoxylin and eosin staining (h) are shown. Data are mean±s.e.m. (n=5 mice per group). *P<0.05; **P<0.01.
Quantitative proteomics identifies ZNF322A-regulated proteins
To identify ZNF322A downstream proteins, we used isobaric tags for relative and absolute quantization and identified 1108 proteins regulated by ZNF322A (Supplementary Table S2). Most of them were involved in signal transduction and protein phosphorylation, cell motility, embryonic development, vesicle-mediated transport, generation of energy and chromatin organization (Supplementary Figure S6A). Notably, gene ontology term ‘cancer' ranked as the top 1 associated network function and disease (Supplementary Figure S6B). Supplementary Figure S7 shows 10 of the significantly differentially expressed ZNF322A-regulated proteins in isobaric tags for relative and absolute quantization data, which have been verified by immunoblotting.
ZNF322A positively regulates ADD1 and CCND1 expression but negatively regulates p53 expression at transcription level
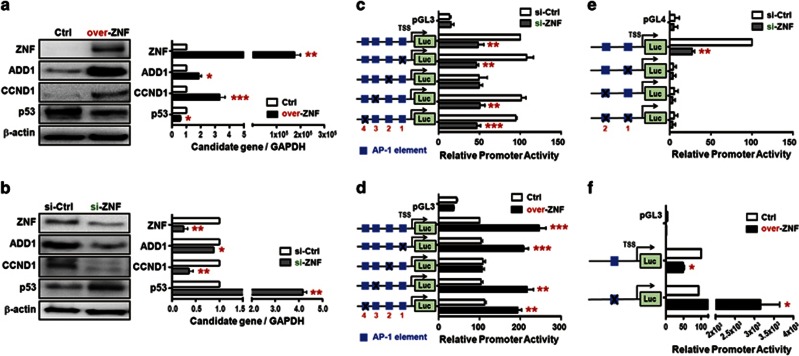
As ZNF322A has been suggested to regulate expression of AP-1 containing genes, we further integrated literature searches and our proteomic data set followed by transcription factor-binding site prediction to identify putative ZNF322A transcriptional targets. Three AP-1 containing genes including ADD1, CCND1 and p53 were revealed. To determine whether ZNF322A regulated the expression of ADD1, CCND1 and p53, immunoblotting and qRT–PCR analyzes were performed on cells with ZNF322A overexpression or knockdown. Importantly, ZNF322A overexpression significantly upregulated the mRNA and protein expression of ADD1 and CCND1 but downregulated the expression of p53 (Figure 5a), whereas knockdown of ZNF322A significantly decreased the expression of ADD1 and CCND1 but increased the expression of p53 (Figure 5b).
Figure 5.
ZNF322A upregulates ADD1 and CCND1 expression but downregulates p53 expression at transcriptional level. (a) A549 cell transfected with empty vector (Ctrl) or ZNF322A expression vector (over-ZNF) were harvested and subjected to immunoblotting (left) and qRT–PCR (right) analyzes as indicated. (b) A549 cell transfected with si-control (si-Ctrl) or si-ZNF322A (si-ZNF) oligo were detected for protein (left) and mRNA (right) expression. (c–f) ZNF322A transcriptionally regulates downstream gene through AP-1 elements. Dual luciferase activity assays were performed using ADD1 promoter (c and d), CCND1 promoter (e) or p53 promoter (f) containing wild-type AP-1 (blue box) or mutant AP-1 element (marked with X symbol). Data represent promoter activity in A549 cells expressing si-ZNF (gray bar) compared with si-Ctrl or over-ZNF (black bar) compared with Ctrl. TSS, transcription start site. Data are mean±s.e.m. (n=3). *P<0.05; **P<0.01; ***P<0.001.
To further clarify whether ZNF322A regulated expression of downstream genes through AP-1 elements at the transcriptional level, luciferase promoter activity assay combined with site-directed mutagenesis were performed. As shown in Figure 5c, knockdown of ZNF322A decreased promoter activity of the wild-type ADD1 promoter (containing four AP-1 sites) or promoters containing either mutation at first, third or fourth AP-1 element next to the transcription start site, whereas ZNF322A did not affect the ADD1 promoter activity with mutation at the second AP-1 element. Consistently, the second AP-1 element was required for ZNF322A-mediated activation as evident from the ZNF322A overexpression assays (Figure 5d). Similar results were obtained in another lung cell model (Supplementary Figure S8). ZNF322A knockdown reduced the activity of wild-type CCND1 promoter (containing two AP-1 elements). However, ZNF322A did not affect the activity of the CCND1 promoters containing mutation at either one of the AP-1 elements (Figure 5e). In addition, ZNF322A overexpression significantly suppressed activity of the p53 promoter in the presence of wild-type AP-1 element, whereas mutation of AP-1 element reversed the suppressive effect of ZNF322A on the p53 promoter (Figure 5f). Altogether, these results demonstrated that ZNF322A transcriptionally regulated downstream genes expression through AP-1 element.
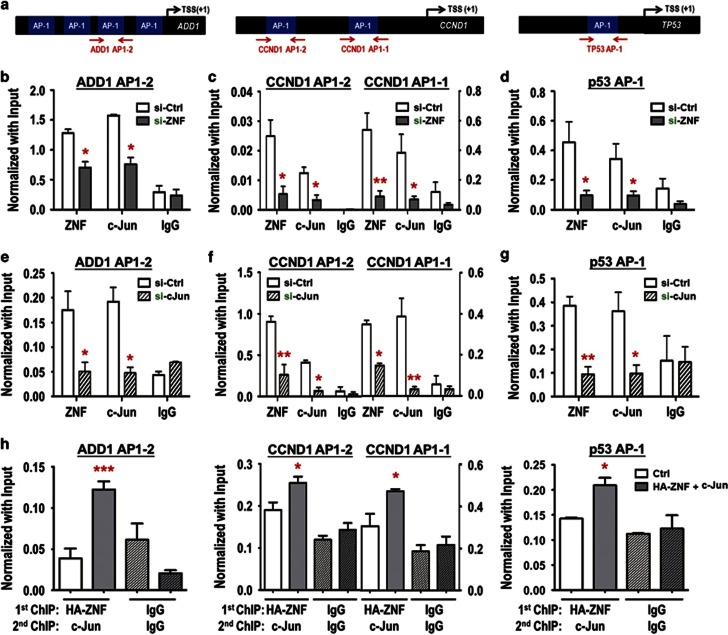
ZNF322A and c-Jun cooperatively bind to AP-1 elements on the ADD1, CCND1 and p53 promoters
c-Jun is a well-known member of the AP-1 transcription factor family that binds to AP-1 element,19 this prompted us to investigate the interplay between c-Jun and ZNF322A on promoters containing AP-1 element. Chromatin immunoprecipitation (ChIP)-qPCR assays of the ADD1, CCND1 and p53 promoters (Figure 6a) were performed using antibodies against ZNF322A and c-Jun in A549 cells upon ZNF322A knockdown (Figures 6b–d) or c-Jun knockdown (Figures 6e–g). In the context of all three promoters, ZNF322A knockdown not only decreased ZNF322A binding but also significantly attenuated c-Jun targeting to the AP-1 elements (Figures 6b–d). In addition, c-Jun knockdown led to loss of ZNF322A binding (Figures 6e–g), suggesting that ZNF322A and c-Jun are both essential for binding at the AP-1 elements of the targeted genes. Indeed, immunoprecipitation-western blot results showed that ZNF322A and c-Jun formed a protein complex at both exogenous and endogenous level (Supplementary Figure S9). Consistently, re-ChIP results further supported the co-occupancy of ZNF322A and c-Jun at ADD1, CCND1 and p53 promoters (Figure 6h). These results strongly suggest that ZNF322A and c-Jun work cooperatively to regulate transcription of the targeted genes.
Figure 6.
ZNF322A and c-Jun bind cooperatively to AP-1 elements on ADD1, CCND1 and p53 promoters. (a) Promoter map for ADD1, CCND1 and p53 genes. AP-1 elements are indicated as blue boxes and regions examined for ChIP–qPCR are marked by red arrows. (b–g) A549 cells transfected with si-ZNF322A (si-ZNF) oligo (b-d) or si-c-Jun (si-c Jun) oligo (e–g) were subjected to ChIP–qPCR analysis. (h) Re-ChIP assays were performed in A549 cells expressing control vector (white bar) or ectopically expressing ZNF322A and c-Jun (gray bar). qPCR products of the target gene relative to input are indicated on the y axis and antibodies for the proteins analyzed are on the x axis. IgG serves as a negative control. Data are mean±s.e.m. (n=3). *P<0.05; **P<0.01; ***P<0.001.
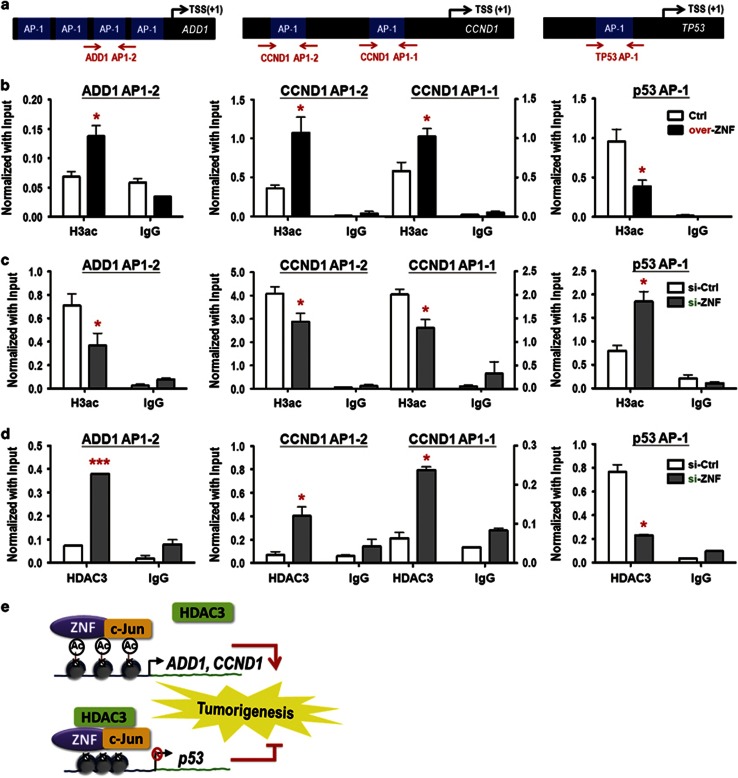
ZNF322A modulates chromatin structure of ADD1, CCND1 and p53 promoters by recruiting differential chromatin modifiers
Previous studies showed that AP-1 transcription factor could positively or negatively regulate downstream gene expression by recruiting different chromatin modifiers, such as p300 co-activator or SMRT/NcoR repressive complex to modulate chromatin structures.20, 21, 22 Therefore, we examined the level of acetylated histone 3, an active mark for opened chromatin structures, around AP-1 elements on ZNF322A downstream genes (Figure 7a). The ChIP–qPCR results showed that acetylated histone 3 was accumulated around AP-1 elements on ADD1 and CCND1 promoters but not on p53 promoter upon ZNF322A overexpression (Figure 7b), whereas ZNF322A knockdown had a reverse effect (Figure 7c). Moreover, silencing ZNF322A increased binding of histone deacetylase 3 (HDAC3), a component of SMRT/NcoR repressive complex, to AP-1 element on ADD1 and CCND1 promoters. In contrast, silencing ZNF322A decreased HDAC3 occupancy on AP-1 elements on p53 promoter (Figure 7d). These results implicated that ZNF322A alone or together with c-Jun may recruit HDAC3 in the context of p53 promoter, but not in the ADD1 and CCND1 promoters, to suppress p53 expression (Figure 7e).
Figure 7.
ZNF322A modulates the chromatin structure of ADD1, CCND1 and p53 promoter regions. (a) Promoter map for ADD1, CCND1 and p53 genes. (b and c) A549 cells transfected with ZNF322A expression vector (over-ZNF) (b) or si-ZNF322A oligo (si-ZNF) (c) were subjected to ChIP–qPCR analyzes for acetylated histone 3 (H3ac) open chromatin mark. (d) A549 cells transfected with si-ZNF oligo were subjected to ChIP–qPCR analysis for histone deacetylase 3 (HDAC3) repressive protein. Data are mean±s.e.m. (n=3). *P<0.05; ***P<0.001. (e) Schematic representation of transcriptional induction of ZNF322A at ADD1 and CCND1 and repression at p53, and subsequent promotion of cell proliferation, tumor metastasis and poor prognosis.
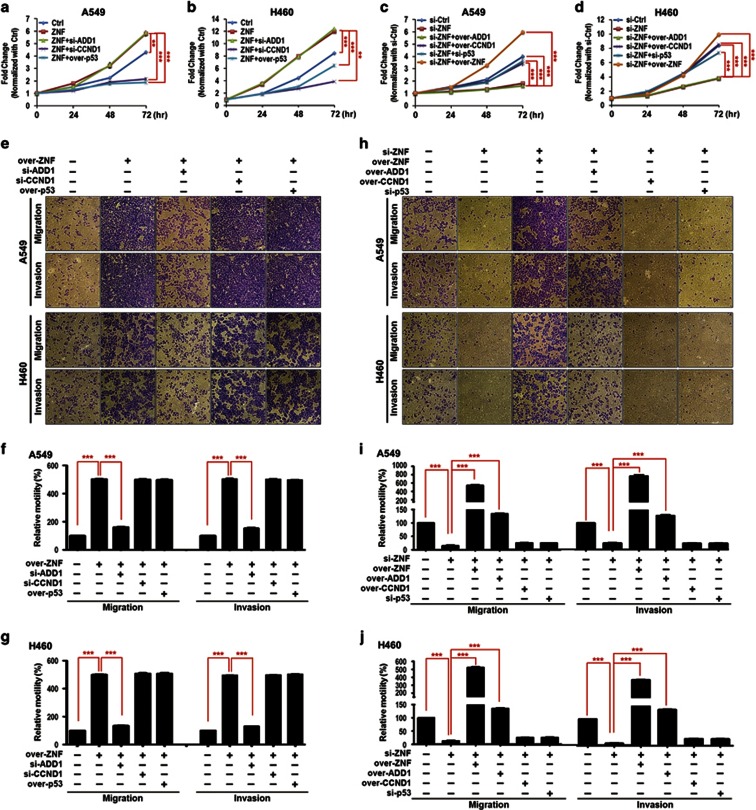
ZNF322A exerts its oncogenic effects in lung cancer via regulation of ADD1, CCND1 and p53
To confirm the specificity of the ZNF322A expression vector and si-RNA and the causal relationship between ZNF322A and its downstream targets in lung tumorigenesis, we performed reconstitution experiments by treating A549 and H460 cells with ZNF322A expression vector alone or together with si-ADD1 oligo, si-CCND1 oligo or p53 expression vector. On the other hand, A549 and H460 cells were treated with si-ZNF322A oligo alone or together with ZNF322A, ADD1 or CCND1 expression vector or si-p53 oligo. Treated cells were then subjected to proliferation, migration and invasion assays. As shown in Figure 8, overexpression of ZNF322A increased cell proliferation rate, which was abolished by silencing CCND1 or overexpressing p53 (Figures 8a and b). In the reciprocal experiment, decreased cell proliferation by ZNF322A silencing was rescued by overexpression of ZNF322A and CCND1 or knockdown of p53. Reconstitution of ADD1 expression, however, did not reverse the effect of ZNF322A on cell proliferation (Figures 8c and d). Notably, knockdown of ADD1 suppressed migration and invasion abilities, which were promoted by ZNF322A overexpression in A549 and H460 cells (Figures 8e–g). Reconstitution of ZNF322A and ADD1 expression restored the migration and invasion abilities in ZNF322A silenced cells (Figures 8h–j). These results suggested that CCND1 and p53 are important to ZNF322A-mediated promotion of cell proliferation, whereas ADD1 is necessary for ZNF322A-mediated cell migration and invasion.
Figure 8.
Reconstituted ZNF322A, ADD1, CCND1 and p53 expression reverses the oncogenic effects of ZNF322A in lung cancer cells. (a and b) Knockdown of CCND1 (si-CCND1) and overexpression of p53 (over-p53) reduced cell proliferation in ZNF322A overexpressing A549 and H460 cells. (c and d) Reconstitution of ZNF322A (over-ZNF) and CCND1 (over-CCND1) and knockdown of p53 (si-p53) restored cell proliferation in ZNF322A silenced (si-ZNF) A549 and H460 cells. (e–g) Knockdown of ADD1 (si-ADD1) reversed the oncogenic effects of ZNF322A on migration and invasion abilities in vitro. (h–j) Reconstituted ZNF322A (over-ZNF) and ADD1 (over-ADD1) expression restored cell migration and invasion abilities in ZNF322A silenced lung cancer cells. Cells were transfected with indicated expression vectors with or without siRNAs. P-values determined using two-tailed Student's t-test. Data are mean±s.e.m. (n=3). **P<0.01; ***P<0.001.
Discussion
Here we provide compelling evidence to demonstrate the oncogenic role of ZNF322A in lung tumorigenesis and the underlying mechanism. Our data show that enforced expression of ZNF322A conferred anchorage-independent growth and transwell cell motility to multiple lung cancer cell lines, including A549, H460, CL1-5 and H1299 and the immortalized bronchial epithelial cell line BEAS-2B. In addition, overexpression of ZNF322A promoted tumor growth and metastasis in vivo. ZNF322A cooperated with c-Jun to regulate downstream gene expression, including ADD1, CCND1 and p53, through the AP-1 elements. ZNF322A knockdown showed opposite effects on gene expression and cancer behaviors compared with ZNF322A overexpression. Moreover, ZNF322A was significantly overexpressed in lung cancer patients and was associated with poor overall survival. Collectively, our results suggest that ZNF322A is a potential oncogenic transcription factor in lung cancer.
Some ZNF proteins work as transcriptional repressors by disrupting pre-initiation complex assembly or recruiting corepressors.23, 24, 25 For example, ZNF217 has been found to suppress downstream gene expression by interacting with corepressors, including CoREST, lysine demethylase 1, histone deacetylase 2 and C-terminal binding protein.23 Some ZNF proteins, on the other hand, work as transcriptional activators by interacting with coactivators, including CBP/p300 and C/EBP.26, 27 Interestingly, our study shows that ZNF322A functions as an oncoprotein to activate transcription of oncogenic downstream genes, ADD1 and CCND1, but repress tumor suppressor downstream gene, p53, by differentially recruiting HDAC3 (Figure 7). Further investigation to identify ZNF322A interacting chromatin modifiers, especially coactivators, will help to unveil the underlying mechanism of differential regulation on downstream genes by ZNF322A.
ZNF322A downstream gene ADD1 encodes α-adducin, which has an important role in the assembly of membrane skeleton by tethering actin and spectrin.28, 29, 30 Studies have shown that phosphorylation of adducins diminishes their association with actin and spectrin, and therefore reduces cell migration ability.31, 32 However, transcriptional regulation of α-adducin expression remains unclear. Here we provide the first evidence that ZNF322A could transcriptionally upregulate ADD1 expression in an AP-1 element-dependent manner. Of note, our results indicated that ZNF322A and c-Jun cooperatively regulate ADD1 expression through the second AP-1 element but not the other three AP-1 elements (Figures 5c and d), suggesting the selectivity of ZNF322A targeting on downstream genes. The differential binding affinity of ZNF322A/c-Jun to these four AP-1 elements may diverge in the context of the DNA sequences flanking the elements, which may change its interaction with other transcription factors. Further studies will be pursued to validate the responsive DNA elements of ZNF322A and its interacting transcriptional regulators in more ZNF322A target genes.
ZNF322A downstream genes CCND1 and p53 have been shown to regulate cell cycle progression at G1/S transition.33 A previous study showed that AP-1 could transcriptionally activate CCND1 expression leading to breast cancer cell growth through the distal AP-1 element.34 In this study, we show that both the proximal and distal AP-1 elements are essential for CCND1 expression under ZNF322A/c-Jun complex regulation in lung cancer model (Figure 5e). Intriguingly, although c-Jun is a transcription activator for genes containing AP-1 elements, one previous study demonstrated that c-Jun represses p53 transcription by targeting to the variant AP-1 element on the p53 promoter.35 On the basis of our data, there is a possibility that ZNF322A may help c-Jun negatively regulate p53 expression by recruiting corepressors to target the AP-1 site (Figure 7d). Of note, although wild-type p53 promoter activity was suppressed by ZNF322A overexpression, p53 promoter activity with mutant AP-1 element was significantly enhanced (Figure 5f). In our ongoing studies, we identified bona fide ZNF322A binding elements by integrating ChIP-sequencing and RNA-sequencing data and found that ZNF322A possessed different DNA binding elements on its positively and negatively regulated genes. p53 promoter analysis showed that top 1 ZNF322A positively regulated element was significantly enriched at upstream of the AP-1 element examined (data not shown). Therefore, we hypothesize that ectopically expressed ZNF322A will target to its bona fide positive element on p53 promoter region and enhance p53 promoter activity when the nearby AP-1 element is mutated. Further studies will be performed to elucidate the underlying mechanism of differential regulation of ZNF322A on p53 promoter, focusing on the differential binding elements and interacting partners.
To further confirm the relationship between ZNF322A, ADD1, CCND1 and p53, we compared their protein expression patterns in tumor sample from patients. Indeed, patients with ZNF322A overexpression showed coordinated overexpression of ADD1 and CCND1 along with downregulation of p53 protein expression, whereas other patients with decreased ZNF322A expression showed low expression of ADD1 and CCND1 and a positive immunostaining for p53 (Figures 1b and c). Similar expression patterns were observed for tumor xenografts from animal studies (Figures 3e and j). Tumor suppressor p53 gene has been found to be mutated in multiple cancers.36 Studies also show that some mutations of the p53 gene may lead to p53 stabilization and thus overexpression in cancers.37, 38 To clarify whether the inverse correlation between ZNF322A and p53 expression resulted from transcriptional repression of p53 by ZNF322A or from p53 mutation, we performed cDNA-sequencing analyzes of exons 5–11 of p53 gene in the two lung cancer samples analyzed in Figures 1b and c and some patients whose cDNA were available in the concordant group presented in Figure 1d. Sequencing results revealed that the p53 in these patients were wild-type in the region analyzed (Supplementary Figure S10). Notably, analyzes of ADD1, CCND1 and p53 mRNA levels using tumor specimens from lung cancer patients revealed that the expression of ZNF322A positively correlated with ADD1 and CCND1, while negatively correlated with p53 (Supplementary Figure S11). Together, these results suggest that ZNF322A exerts its oncogenic role through positively regulating ADD1 and CCND1 expression but negatively regulating p53 expression at the transcription level.
In conclusion, we have uncovered an oncogenic role of ZNF322A in multiple lung cancer cell lines and their derived xenografts. Mechanistically, ZNF322A transcriptionally activated oncogenes, such as ADD1 and CCND1, and transcriptionally suppressed tumor suppressor genes, such as p53, by targeting to the AP-1 element on the promoter. Clinically, lung cancer patients with ZNF322A overexpression significantly correlated with poor prognosis. These results collectively show that ZNF322A is an oncogenic transcription factor involved in lung tumorigenesis. Therapeutic strategies such as efficient delivery of ZNF322A interference RNAs or treatment with targeted inhibitors of ZNF322A downstream genes could facilitate the development of cancer therapy for lung cancer.
Materials and methods
Study population
We recruited 123 Asian lung cancer patients from National Cheng Kung University Hospital, Taiwan and 74 Caucasian lung cancer patients from The University of Chicago Comprehensive Cancer Center, USA after obtaining appropriate institutional review board permission and informed consent from the patients. Paraffin blocks of tumors were collected for immunohistochemistry. The detailed clinicopathological characteristics of the enrolled patients are listed in Supplementary Table S1.
qRT–PCR assay
For RNA expression assay, total RNA was extracted using Trizol reagent (Invitrogen, Carlsbad, CA, USA). qRT–PCR was used to analyze the expression of ZNF322A, ADD1, CCND1 and p53 in patient samples and cell lines. Target gene expression levels were normalized based on GAPDH expression levels. Primers used for qRT–PCR analysis are described in Supplementary Table S3.
Immunohistochemistry assay
Antibodies used and their experimental conditions are listed in Supplementary Table S4. Staining was scored as +++ if >75% tumor cells were immunostaining-positive; ++ for 50–75% + for 25–50% +/– for 10–25% cells and− if <10% were positive. The protein expression level was graded as ‘overexpression' if the score were ++ and +++.
Cell lines and culture conditions
Human lung cells A549, H460, H1299 and BEAS-2B were purchased from ATCC. Human lung cell line CL1-5 was obtained from Dr Pan-Chyr Yang (National Taiwan University, Taipei, Taiwan). Cells were cultured in accordance with ATCC recommended culture conditions.
Immunoblotting analysis
Samples containing equal amounts of protein (50 μg) were separated on a 10% SDS–PAGE and electroblotted onto Immobilon-P membranes (Millipore, Bedford, MA, USA). Immunoblotting was performed using the conditions described in Supplementary Table S4.
Plasmid, RNAi and transfection
Plasmids used in the study are listed in Supplementary Table S5. siGENOME SMARTpool siRNAs against ZNF322A and c-Jun were purchased from Dharmacon (Lafayette, CO, USA). siRNAs against ADD1, CCND1 and p53 were purchased from Invitrogen. Plasmids and RNAi transfections were carried out with Lipofetamine 2000 (Invitrogen).
Cell proliferation and anchorage-independent growth assays
Cell transfected with expression plasmid for ZNF322A, ADD1, CCND1 or p53, and oligo for si-ZNF322A, si-ADD1, si-CCND1 or si-p53 were seeded in 6 cm dishes and cultured for 24, 48 and 72 h. Cells were then stained with 0.4% Trypan Blue solution (Sigma-Aldrich, St Louis, MO, USA) to determine the cell number.
Anchorage-independent colony formation assay was carried out by soft agar growth of 3 × 103 cells for 12–17 days. Colonies consisting of >30 cells were counted.
Transwell migration and invasion assay
For transwell migration and invasion assays, 2 × 105 cells were seeded onto the upper chamber with or without Matrigel (Sigma-Aldrich) and cultured for 12 h. Cells attached on the reverse side of the membrane were then fixed and stained. Seven random views were photographed and quantified.
Tumor growth and experimental metastasis assays in vivo
Six-week-old male mice were obtained from National Cheng Kung University Laboratory Animal Center and raised in pathogen-free conditions after obtaining appropriate institutional review board permission. For tumor growth analysis, ZNF322A was overexpressed in H460-Luc cells, and silenced in A549-Luc cells. H460-Luc and A549-Luc cells (5 × 106 cells) were implanted subcutaneously into severe combined immunodeficient mice and BALB/c nude mice, respectively. These mice were then given endotoxin-free luciferase substrate (Promega, Madison, WI, USA) and photographed using IVIS Spectrum Noninvasive Quantitative Molecular Imaging System (Xenogen Corp., Alameda, CA, USA). Mice were killed 18–28 days after injection, and then tumors were excised and weighted. For experimental metastasis assay, 1 × 106 A549 cells, with ZNF322A overexpressed or silenced, were injected into the tail vein of severe combined immunodeficient mice. Mice were killed after 6 weeks of implantation and lung tissues were resected, fixed and embedded in paraffin for histologic hematoxylin and eosin staining, immunoblotting and immunohistochemistry analyzes.
Site-directed mutagenesis and dual luciferase promoter assay
Mutations of AP-1 elements within ADD1, CCND1 and p53 promoter regions were generated by site-directed mutagenesis using indicated wild-type promoter vectors and specific primers listed in Supplementary Table S3. Dual luciferase reporter assay kit (Promega) was used to determine gene promoter activity according to the protocols provided by the manufacturer. Data were represented as the mean of ratio of firefly luciferase to Renilla luciferase activity.
ChIP–qPCR assay
Chromatin was immunoprecipitated for 16 h at 4 °C using anti-ZNF322A, anti-c-Jun, anti-HA, anti-acetylated histone 3, anti-HDAC3 or normal IgG listed in Supplementary Table S4. The DNA samples recovered from ChIP were analyzed by quantitative PCR using specific primers (Supplementary Table S3).
Statistical analysis
Pearson's χ2 test was used to compare the correlation of ZNF322A mRNA and protein expression level in lung cancer patients. Overall survival curves were calculated according to the Kaplan–Meier method. Cox regression comparison was performed to analyze the relative risk for patient poor outcome. Two-tailed Student's t-test was used in cell and animal studies. Each experiment was repeated at least three times and represented as mean±s.e.m. P<0.05 was considered to be statistically significant.
Acknowledgments
This work was supported by the Taiwan Ministry of Science and Technology Grant (103-2627-B-006-007; 103-2627-B-002-002), Taiwan Ministry of Health and Welfare Grant (MOHW103-TD-B-111-06) and the Aim for the Top University Project Grant. We are grateful for the support from the Human Biobank, Research Center of Clinical Medicine, National Cheng Kung University Hospital. Accession numbers: Data from proteomics was deposited in the ProteomeXchange Consortium with accession number PXD001123.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
Supplementary Material
References
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J et al. Initial sequencing and analysis of the human genome. Nature 2001; 409: 860–921. [DOI] [PubMed] [Google Scholar]
- Tupler R, Perini G, Green MR. Expressing the human genome. Nature 2001; 409: 832–833. [DOI] [PubMed] [Google Scholar]
- Wagner S, Hess MA, Ormonde-Hanson P, Malandro J, Hu H, Chen M et al. A broad role for the zinc finger protein ZNF202 in human metabolism. J Biol Chem 2000; 275: 15685–15690. [DOI] [PubMed] [Google Scholar]
- Jheon AH, Ganss B, Cheifetz S, Sodek J. Characterization of a novel KRAB-C2H2 zinc finger transcription factor involved in bone development. J Biol Chem 2001; 276: 18282–18289. [DOI] [PubMed] [Google Scholar]
- Tian C, Xing G, Xie P, Lu K, Nie J, Wang J et al. KRAB-type zinc-finger protein Apak specifically regulates p53-dependent apoptosis. Nat Cell Biol 2009; 11: 580–591. [DOI] [PubMed] [Google Scholar]
- Chauhan S, Goodwin JG, Chauhan S, Manyam G, Wang J, Kamat AM et al. ZKSCAN3 is a master transcriptional repressor of autophagy. Mol Cell 2013; 50: 16–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z, Yan Y, Zhang P, Andrianakos R, Hasegawa K, Lyu J et al. Klf4 interacts directly with Oct4 and Sox2 to promote reprogramming. Stem Cells 2009; 27: 2969–2978. [DOI] [PubMed] [Google Scholar]
- Yu J, Liang QY, Wang J, Cheng Y, Wang S, Poon TC et al. Zinc-finger protein 331, a novel putative tumor suppressor, suppresses growth and invasiveness of gastric cancer. Oncogene 2013; 32: 307–317. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Liang P, Geng H, Wang Z, Li L, Cheng SH et al. A novel 19q13 nucleolar zinc finger protein suppresses tumor cell growth through inhibiting ribosome biogenesis and inducing apoptosis but is frequently silenced in multiple carcinomas. Mol Cancer Res 2012; 10: 925–936. [DOI] [PubMed] [Google Scholar]
- Zhang L, Xu Z, Xu X, Zhang B, Wu H, Wang M et al. SALL4, a novel marker for human gastric carcinogenesis and metastasis. Oncogene 2014; 33: 5491–5500. [DOI] [PubMed] [Google Scholar]
- Yang G, Ma F, Zhong M, Fang L, Peng Y, Xin X et al. ZNF703 acts as an oncogene that promotes progression in gastric cancer. Oncol Rep 2014; 31: 1877–1882. [DOI] [PubMed] [Google Scholar]
- Lai KP, Chen J, He M, Ching AK, Lau C, Lai PB et al. Overexpression of ZFX confers self-renewal and chemoresistance properties in hepatocellular carcinoma. Int J Cancer 2014; 135: 1790–1799. [DOI] [PubMed] [Google Scholar]
- Evans PM, Liu C. Roles of Krüpel-like factor 4 in normal homeostasis, cancer and stem cells. Acta Biochim Biophys Sin 2008; 40: 554–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Wang Y, Zhang C, Yuan W, Wang J, Zhu C et al. ZNF322, a novel human C2H2 Krüppel-like zinc-finger protein, regulates transcriptional activation in MAPK signaling pathways. Biochem Biophys Res Commun 2004; 325: 1383–1392. [DOI] [PubMed] [Google Scholar]
- Lo FY, Chang JW, Chang IS, Chen YJ, Hsu HS, Huang SF et al. The database of chromosome imbalance regions and genes resided in lung cancer from Asian and Caucasian identified by array-comparative genomic hybridization. BMC Cancer 2012; 12: 235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou J, Aerts J, den Hamer B, van Ijcken W, den Bakker M, Riegman P et al. Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PLoS One 2010; 5: e10312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selamat SA, Chung BS, Girard L, Zhang W, Zhang Y, Campan M et al. Genome-scale analysis of DNA methylation in lung adenocarcinoma and integration with mRNA expression. Genome Res 2012; 22: 1197–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okayama H, Kohno T, Ishii Y, Shimada Y, Shiraishi K, Iwakawa R et al. Identification of genes upregulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res 2012; 72: 100–111. [DOI] [PubMed] [Google Scholar]
- Eferl R, Wagner EF. AP-1: a double-edged sword in tumorigenesis. Nat Rev Cancer 2003; 3: 859–868. [DOI] [PubMed] [Google Scholar]
- Kim MK, Shin JM, Eun HC, Chung JH. The role of p300 histone acetyltransferase in UV-induced histone modifications and MMP-1 gene transcription. PLoS One 2009; 4: e4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa S, Lozach J, Jepsen K, Sawka-Verhelle D, Perissi V, Sasik R et al. A nuclear receptor corepressor transcriptional checkpoint controlling activator protein 1-dependent gene networks required for macrophage activation. Proc Natl Acad Sci USA 2004; 101: 14461–14466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinenov Y, Kerppola TK. Close encounters of many kinds: Fos-Jun interactions that mediate transcription regulatory specificity. Oncogene 2001; 20: 2438–2452. [DOI] [PubMed] [Google Scholar]
- Cowger JJ, Zhao Q, Isovic M, Torchia J. Biochemical characterization of the zinc-finger protein 217 transcriptional repressor complex: identification of a ZNF217 consensus recognition sequence. Oncogene 2007; 26: 3378–3386. [DOI] [PubMed] [Google Scholar]
- Gocke CB, Yu H. ZNF198 stabilizes the LSD1-CoREST-HDAC1 complex on chromatin through its MYM-type zinc fingers. PLoS One 2008; 3: e3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frietze S, O'Geen H, Blahnik KR, Jin VX, Farnham PJ. ZNF274 recruits the histone methyltransferase SETDB1 to the 3' ends of ZNF genes. PLoS One 2010; 5: e15082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon BN, Kim MK, Yoon JH, Kim MY, An H, Noh HJ et al. Two ZNF509 (ZBTB49) isoforms induce cell-cycle arrest by activating transcription of p21/CDKN1A and RB upon exposure to genotoxic stress. Nucleic Acids Res 2015; 42: 11447–11461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meruvu S, Hugendubler L, Mueller E. Regulation of adipocyte differentiation by the zinc finger protein ZNF638. J Biol Chem 2011; 286: 26516–26523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner K, Bennett K. Modulation of spectrin-actin assembly by erythrocyte adducin. Nature 1987; 328: 359–362. [DOI] [PubMed] [Google Scholar]
- Bennett V, Gardner K, Steiner JP. Brain adducin: a protein kinase C substrate that may mediate site-directed assembly at the spectrin-actin junction. J Biol Chem 1998; 263: 5860–5869. [PubMed] [Google Scholar]
- Li X, Matsuoka Y, Bennett V. Adducin preferentially recruits spectrin to the fast growing ends of actin filaments in a complex requiring the MARCKS-related domain and a newly defined oligomerization domain. J Biol Chem 1998; 273: 19329–19338. [DOI] [PubMed] [Google Scholar]
- Matsuoka Y, Li X, Bennett V. Adducin is an in vivo substrate for protein kinase C: phosphorylation in the MARCKS-related domain inhibits activity in promoting spectrin-actin complexes and occurs in many cells, including dendritic spines of neurons. J Cell Biol 1998; 142: 485–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CL, Hsieh YT, Chen HC. Phosphorylaiton of adducin by protein kinase Cδ promotes cell motility. J Cell Sci 2007; 120: 1157–1167. [DOI] [PubMed] [Google Scholar]
- Bartek J, Lukas J. Mammalian G1- and S-phase checkpoints in response to DNA damage. Curr Opin Cell Biol 2001; 13: 738–747. [DOI] [PubMed] [Google Scholar]
- Shen Q, Uray IP, Li Y, Krisko TI, Strecker TE, Kim HT et al. The AP-1 transcription factor regulates breast cancer cell growth via cyclins and E2F factors. Oncogene 2007; 27: 366–377. [DOI] [PubMed] [Google Scholar]
- Schreiber M, Kolbus A, Piu F, Szabowski A, Möhle-Steinlein U, Tian J et al. Control of cell cycle progression by c-Jun is p53 dependent. Genes Dev 1999; 13: 607–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C et al. Mutational landscape and significance across 12 major cancer types. Nature 2013; 502: 333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oren M, Rotter V. Mutant p53 gain-of-function in cancer. Cold Spring Harb Perspect Biol 2010; 2: a001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Marchenko ND, Schulz R, Fischer V, Velasco-Hernandez T, Talos F et al. Functional inactivation of endogenous MDM2 and CHIP by HSP90 causes aberrant stabilization of mutant p53 in human cancer cells. Mol Cancer Res 2011; 9: 577–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.