Abstract
Didymella pinodes is the principal causal agent of ascochyta blight, one of the most important fungal diseases of pea (Pisum sativum) worldwide. Understanding its host specificity has crucial implications in epidemiology and management; however, this has not been clearly delineated yet. In this study we attempt to clarify the host range of D. pinodes and to compare it with that of other close Didymella spp. D. pinodes was very virulent on pea accessions, although differences in virulence were identified among isolates. On the contrary, studied isolates of D. fabae, D. rabiei, and D. lentil showed a reduced ability to infect pea not causing macroscopically visible symptoms on any of the pea accessions tested. D. pinodes isolates were also infective to some extend on almost all species tested including species such as Hedysarum coronarium, Lathyrus sativus, Lupinus albus, Medicago spp., Trifolium spp., Trigonella foenum-graecum, and Vicia articulata which were not mentioned before as hosts of D. pinodes. On the contrary, D. lentil and D. rabiei were more specific, infecting only lentil and chickpea, respectively. D. fabae was intermediate, infecting mainly faba bean, but also slightly other species such as Glycine max, Phaseolus vulgaris, Trifolium spp., Vicia sativa, and V. articulata. DNA sequence analysis of the nuclear ribosomal internal transcribed spacer region (ITS) was performed to confirm identity of the isolates studies and to determine phylogenetic relationship among the Didymella species, revealing the presence of two clearly distinct clades. Clade one was represented by two supported subclusters including D. fabae isolates as well as D. rabiei with D. lentil isolates. Clade two was the largest and included all the D. pinodes isolates as well as Phoma medicaginis var. pinodella. Genetic distance between D. pinodes and the other Didymella spp. isolates was not correlated with overall differences in pathogenicity. Based on evidences presented here, D. pinodes is not specialized on pea and its host range is larger than that of D. fabae, D. lentil, and D. rabiei. This has relevant implications in epidemiology and control as these species might act as alternative hosts for D. pinodes.
Keywords: pea, legume, ascochyta blight, dydimella pinodes, host range, disease management
Introduction
Cool season legumes play an important role in farming systems worldwide (Siddique et al., 2012). They provide important services to societies as they are important sources of oil, fiber, protein-rich food and feed while supplying nitrogen (N) to agro-ecosystems via their unique ability to fix atmospheric N2 in symbiosis with the soil bacteria rhizobia, increasing soil carbon content, and stimulating the productivity of the crops that follow (Jensen et al., 2012). Among them, field pea (Pisum sativum L.) is widely grown across cooler temperate zones of the world on about 6.2 m ha annually with total production generally ranging between 10 and 11 m tons (FAOSTAT, 2015).
Ascochyta blight diseases represent serious limitations to legume production worldwide (Rubiales and Fondevilla, 2012; Khan et al., 2013). Didymella fabae Jellis and Punith. (anamorph Ascochyta fabae Speg.), D. lentis Kaiser, Wang and Rogers (anamorph A. lentis Vassiljevsky) and D. rabiei (Kovachevski) v. Arx (anamorph A. rabiei (Pass) Labr.) are the causal agents of ascochyta blights on faba bean (Vicia faba L.), lentil (Lens culinaris Medik.), and chickpea (Cicer arietinum L.), respectively (Kaiser et al., 1997; Hernandez-Bello et al., 2006; Tivoli and Banniza, 2007). Yield losses caused by aschochyta blight are in order of 40% in lentil (Gossen and Derksen, 2003), but in severe cases losses higher than 90% have been reported in faba bean (Omri Benyoussef et al., 2012) and chickpea (Pande et al., 2005). In pea, this disease is caused by a complex of fungi formed by Ascochyta pisi Lib., Didymella pinodes (Berk and Blox) Petrak, Phoma medicaginis var. pinodella (L.K. Jones) Morgan-Jones and K.B. Burch and Phoma koolunga Davidson, Hartley, Priest, Krysinska-Kaczmarek, Herdina, McKay, and Scott (this last is, at the time, with limited presence in South and Western Australia; Tran et al., 2016). Of these, D. pinodes (formerly known as Mycosphaerella pinodes (Berk. and A. Bloxam) Vestergr., anamorph Ascochyta pinodes L.K. Jones) is the most predominant and damaging pathogen and under some conditions can cause yield losses up to 70% (Tivoli and Banniza, 2007).
D. pinodes remains an extremely difficult pathogen to control, primarily due to limited levels of host resistance available, and secondarily because fungicides are often uneconomic (Khan et al., 2013). Therefore, the main disease control strategy has been to avoid sowing close to infested field pea stubbles and/or to delay sowing of field pea crops for as long as possible in order to avoid the majority of ascospores, particularly those falling on emerging pea seedlings (Salam et al., 2011). Nevertheless, the late sowing is not an option in some countries due to the short crop season and this practice incurs unsustainable yield penalties in many instances (Khan et al., 2013). Other control measures involving crop rotation and intercropping have been also tested (Bailey et al., 2001; McDonald and Peck, 2009; Fernández-Aparicio et al., 2010) showing potential in disease reduction.
A better understanding of a pathogen's host range is critical to handle ascochyta blight and to break its cycle with more effectiveness, particularly in regions where pea is frequently grown and where the disease is endemic or where ascospores are an overriding primary source of initial infection. D. pinodes is known to be less specialized than other Didymella spp. (Sprague, 1929; Sattar, 1934; Le May et al., 2014), which increases the potential of this specie to survive. In fact, adjacent naturally infected alternative hosts could serve as important sources of inoculum to initiate disease epidemics on cultivated peas. So, the impact of alternative hosts on plant pathogen adaptation must be taken into account since they affect the survival of pathogen populations, and transmission opportunities to different components and ecological niches (wild/cultivated, cultivated/cultivated; Woolhouse et al., 2001), as recently showed for D. rabiei (Trapero-Casas and Kaiser, 2009). Nevertheless, despite its importance, the host range of D. pinodes on legume species other than Pisum spp. is poorly understood (Bretag, 2004; Taylor and Ford, 2007; Khan et al., 2013; Le May et al., 2014).
The aims of this study were therefore (i) to further refine the host range of D. pinodes within cultivated and wild legumes; (ii) to assess the susceptibility/resistance of different accessions within each of these legume species to nine isolates of D. pinodes from different geographical origin; (iii) to compare the host range of D. pinodes with that of other Didymella species; and (iv) to relate fungal isolates by ITS molecular markers.
Materials and methods
Fungal isolates
Nine isolates of D. pinodes, two isolates of D. fabae, one of D. lentil, and one of D. rabiei, all from IAS-CSIC fungal collection, were used in the experiments (information reported in Table 1). Local D. pinodes isolate Dp-CO-99, as well as isolates Dp-FR-88, Dp-PO-03 and Dp-JAP-03 have previously shown to differ in aggressiveness toward pea accessions (Fondevilla et al., 2005). All isolates were monoconidial and were preserved in sterile cellulose filter papers.
Table 1.
Codes of reference, specie definition, collecting site, year and GenBank accession relative to the fungus isolates used in the study.
| Fungal code | Fungal specie | Collecting site | Collecting year | GenBank n° |
|---|---|---|---|---|
| Dp-CO-99 | Didymella pinodes | Córdoba, Spain | 1999 | KR259388 |
| Dp-FR-88 | D. pinodes | Rennes, France | 2003 | KR259380 |
| Dp-PdT-03 | D. pinodes | Palmar de Troya, Spain | 2003 | KR259391 |
| Dp-PO-03 | D. pinodes | Wa̧sy, Poland | 2003 | KR259387 |
| Dp-JAP-03 | D. pinodes | Japan | 2003 | KR259392 |
| Dp-ANN-13 | D. pinodes | Annaba, Algeria | 2013 | KR259390 |
| Dp-M07-4 | D. pinodes | Perth, Australia | 2013 | KR259383 |
| Dp-Esc-13 | D. pinodes | Escacena del Campo, Spain | 2013 | KR259389 |
| Dp-KHM-13 | D. pinodes | Khemis Miliana, Algeria | 2013 | KR259386 |
| Df-AU04 | D. fabae | Gleisdorf, Austria | 2005 | KR259385 |
| Df-857 | D. fabae | France | 2005 | KR259384 |
| Dl-AL10 | D. lentil | Germany | 2010 | KR259381 |
| Dr-Pt04 | D. rabiei | Aleppo, Syria | 2010 | KR259382 |
| Ascochyta pisi | Pullman, USA | 2007 | DQ383954 | |
| D. pinodes | Canberra, Australia | 2009 | EU338435 | |
| Phoma koolunga | Canberra, Australia | 2009 | EU338427 | |
| P. medicaginis var. pinodella | Palampour, India | 2008 | FJ032641 |
Plant material
Disease responses were studied on accessions of 20 legumes species (Table 2): alfalfa (Medicago sativa L.), barrel medick (M. truncatula Gaertn.), button medick (M. orbicularis (L.) Bartal.), chickpea (Cicer arietinum L.), common bean (Phaseolus vulgaris L.), common vetch (Vicia sativa L.), faba bean (Vicia faba L.), fenugreek (Trigonella foenum-graecum L.), grass pea (Lathyrus sativus L.), lentil (Lens culinaris Medik.), oneflower vetch (Vicia articulata Hornem.), pea (Pisum sativum ssp. sativum L.), prinkly scorpion's tail (Scorpiurus muricatus L.), red clover (Trifolium pratense L.), soybean (Glycine max (L.), subterranean clover (T. subterraneum L.), sulla (Hedysarum coronarium L.), tawny pea (P. fulvum Sibth. & Sm.), white clover (T. repens L.), and white lupin (Lupinus albus L.). From 1 to 6 accessions per species were tested (Table 2).
Table 2.
Response of legume species to isolates of D. pinodes in seedling stage, measured 10 days after inoculation under controlled conditions.
| Legume host | Code | Dp-M07-4 | Dp-Esc-13 | Dp-KHM-13 | Dp-ANN-13 | Dp-JAP-03 | Dp-PO-03 | Dp-CO-99 | Dp-PdT-03 | Dp-FR-88 | Dl-AL10 | Dr-Pt04 | Df-AU04 | Df-857 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DRA | DSB | DR | DS | DR | DS | DR | DS | DR | DS | DR | DS | DR | DS | DR | DS | DR | DS | DR | DS | DR | DS | DR | DS | DR | DS | ||
| PEAS | |||||||||||||||||||||||||||
| Pisum sativum | Messire3 | 5 | 80bc* | 5 | 77b | 5 | 63b | 5 | 63ab | 4.7 | 66bc | 5 | 50a | 5 | 79ab | 5 | 50a | 5 | 50a | 0 | 0 | 0 | 0 | 2 | 2a | 0 | 0 |
| P. sativum | J20 | 5 | 87ab | 5 | 93a | 5 | 98a | 5 | 80ab | 5 | 100a | 5 | 62a | 5 | 87a | 5 | 43a | 4 | 35a | 0 | 0 | 0 | 0 | 0 | 0b | 0 | 0 |
| P. sativum | J4 | 5 | 95a | 5 | 90ab | 5 | 75ab | 5 | 83ab | 5 | 100a | 4.3 | 40a | 5 | 57bc | 5 | 25a | 4 | 15b | 0 | 0 | 0 | 0 | 0 | 0b | 0 | 0 |
| P. sativum | 6NIL | 5 | 83ab | 5 | 93a | 5 | 82ab | 5 | 92a | 5 | 77b | 5 | 70a | 5 | 47c | 5 | 25a | 5 | 30a | 0 | 0 | 0 | 0 | 0 | 0b | 0 | 0 |
| P. fulvum | IFPI3260 | 4 | 67c | 3.3 | 57c | 4.7 | 60b | 4 | 51.7b | 4 | 42c | 4.7 | 63a | 1.3 | 7d | 3 | 25a | 3 | 22b | 0 | 0 | 0 | 0 | 0 | 0b | 0 | 0 |
| AVERAGE | 82A | 81A | 76A | 74A | 78A | 57A | 54A | 30A | 26A | 0B | 0B | 0.3C | 0E | ||||||||||||||
| WHITE LUPIN | |||||||||||||||||||||||||||
| Lupinus albus | Giza 21 | – | – | 5 | 95a | – | – | 5 | 40a | 4 | 7b | 5 | 47a | 4 | 15a | – | – | 3 | 13a | – | – | 0 | 0 | 0 | 0 | 0 | 0 |
| L. albus | Lup344 | 3 | 20a | 5 | 100a | 5 | 47a | 5 | 50a | 5 | 40ab | 4.7 | 43a | 5 | 25a | 3.3 | 40a | 3 | 5a | – | – | 0 | 0 | 0 | 0 | 0 | 0 |
| L. albus | Lup354 | 4 | 35a | 5 | 22b | 5 | 37ab | 5 | 50a | 5 | 67a | 4 | 40a | 5 | 20a | 3 | 9b | 3 | 12a | 3.3 | 20a | 0 | 0 | 0 | 0 | 0 | 0 |
| L. albus | Lup394 | 4.3 | 20a | 5 | 25b | 5 | 26b | 5 | 50a | 5 | 40ab | 4.7 | 44a | 5 | 25a | 5 | 46a | 3.3 | 15a | 0 | 0b | 0 | 0 | 0 | 0 | 0 | 0 |
| AVERAGE | 25CDE | 53B | 37B | 47B | 38B | 43B | 21.5BC | 33A | 9B | 10B | 0B | 0C | 0E | ||||||||||||||
| CLOVERS | |||||||||||||||||||||||||||
| Trifolium pratense | E075 | 4.7 | 47a | 4.3 | 30a | 4.7 | 37a | 5 | 58a | 4 | 40a | 4.7 | 38a | 3 | 20a | 4.7 | 30a | 3 | 10a | 2.7 | 12a | 0 | 0 | 3 | 13a | 3 | 8a |
| T. subterraneum | E085 | 4 | 20a | 3.3 | 20a | 4 | 20ab | 4 | 37a | 5 | 32a | 3.5 | 20a | 2 | 17a | 3 | 20a | 3 | 3a | 0 | 0b | 0 | 0 | 0 | 0b | 2.3 | 6a |
| T. repens | Anteria1 | 3.7 | 26a | 4.3 | 30a | 4.3 | 17b | 4.7 | 30a | 4.2 | 38a | 3.3 | 15a | 2 | 16a | 5 | 32a | 3 | 8a | 0 | 0b | 0 | 0 | 0 | 0b | 2 | 7a |
| AVERAGE | 31BCD | 27CDE | 24BC | 42BC | 36B | 25BC | 17BC | 27A | 7B | 4B | 0B | 4BC | 8CD | ||||||||||||||
| MEDICKS | |||||||||||||||||||||||||||
| Medicago truncatula | Parabinga1 | – | – | 3 | 20ab | 2.7 | 10a | 2.3 | 35a | 5 | 40b | 2 | 5b | 2.7 | 25a | 2 | 5a | 1.3 | 5a | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| M. truncatula | Paraggio1 | 2 | 20a | 2 | 12b | 4 | 20a | 2.3 | 27a | 3.7 | 37b | 2 | 5b | 2 | 30a | – | – | 2 | 6a | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| M. truncatula | M2634 | 1.7 | 18a | 3.7 | 53a | 2.7 | 10a | 2 | 30a | 4.5 | 80a | 2.3 | 30a | 2 | 37a | 2 | 5a | 1.3 | 11a | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| M. orbicularis | M2644 | 2 | 27a | – | – | 2.7 | 7a | 3 | 15a | 4.3 | 37b | – | – | 2.7 | 35a | 2 | 10a | – | – | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| M. orbicularis | M2814 | – | – | 4.3 | 47a | 3 | 10a | 3.3 | 21a | 4.5 | 60ab | 4.7 | 32a | – | – | 2 | 8a | 3 | 15a | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| AVERAGE | 22DEF | 36BCD | 12CD | 25CD | 38B | 23C | 34B | 7BC | 9B | 0B | 0B | 0C | 0E | ||||||||||||||
| ONEFLOWER VETCH | |||||||||||||||||||||||||||
| Vicia articulata | BGE0133766 | 4 | 37a | 3 | 37a | 4 | 27a | 4.3 | 45ab | 4 | 23a | 2.3 | 23a | – | – | 4.3 | 40a | 2.7 | 8a | 0 | 0 | 0 | 0 | 3 | 8a | 3 | 4a |
| V. articulata | BGE0139846 | 4 | 43a | 4.3 | 60a | 3.7 | 30a | 4 | 43ab | 4 | 53a | 3 | 25a | 3 | 30a | 3.7 | 28a | 3 | 11a | 0 | 0 | 0 | 0 | 0 | 0b | 0 | 0a |
| V. articulata | BGE0139856 | 4 | 40a | 3.7 | 63a | 4.3 | 33a | 4.3 | 53a | 5 | 53a | 3.7 | 33a | 2 | 15a | 4.3 | 30a | 3 | 8a | 0 | 0 | 2.3 | 3a | 0 | 0b | 2.3 | 3a |
| V. articulata | BGE0188246 | 4 | 43a | 4 | 60a | 4.3 | 32a | 4 | 28b | 4 | 50a | 3 | 35a | 3 | 20a | 4.7 | 33a | 2.7 | 7a | 0 | 0 | 0 | 0 | 0 | 0b | 0 | 0a |
| AVERAGE | 41BC | 55B | 30B | 43BC | 48B | 29BC | 22BC | 35A | 9B | 0B | 0.8B | 2BC | 2DE | ||||||||||||||
| CHICKPEA | |||||||||||||||||||||||||||
| Cicer arietinum | ILC722 | 3 | 15a | 4 | 13a | 2.7 | 6a | 4.3 | 8a | 1 | 1a | 4.3 | 30a | 0.7 | 2a | 4.3 | 7a | 1.3 | 3a | 0 | 0b | 0.3 | 5b | 0 | 0 | 0 | 0 |
| C. arietinum | M382 | 3.3 | 10a | 3.4 | 15a | 3.7 | 5a | 4.7 | 8a | 3.3 | 10a | 5 | 30a | 4.5 | 25b | 5 | 15a | 1.3 | 2a | 0 | 0b | 4.7 | 40a | 0 | 0 | 0 | 0 |
| C. arietinum | AS182 | 1.7 | 9a | 3 | 8a | 3 | 4a | – | – | 3 | 12a | 4.7 | 21a | 3 | 5a | 4.3 | 7a | 1 | 3a | 3 | 30a | 5 | 37a | 0 | 0 | 0 | 0 |
| C. arietinum | AS192 | 2.7 | 10a | 2 | 13a | 3 | 5a | 5 | 5a | 3 | 12a | 5 | 28a | 3 | 7a | 5 | 8a | – | – | 0 | 0b | 3.8 | 35a | 0 | 0 | 0 | 0 |
| C. arietinum | AS232 | – | – | 3 | 10a | – | – | 5 | 7a | 2.7 | 5a | – | – | – | – | 5 | 10a | 1 | 1a | 0 | 0b | 2.6 | 40b | 0 | 0 | 0 | 0 |
| AVERAGE | 10EF | 12EF | 5D | 7DE | 8D | 28BC | 8C | 10BC | 2B | 6B | 31.4A | 0C | 0E | ||||||||||||||
| LENTIL | |||||||||||||||||||||||||||
| Lens culinaris | S172 | 4 | 29a | 4.3 | 62a | 4 | 47a | 4.7 | 53a | 5 | 48ab | 3.7 | 35a | – | – | 3.7 | 15a | 2 | 17a | 4.3 | 24a | 0 | 0 | 0 | 0 | 3 | 6a |
| L. culinaris | S232 | 4 | 57a | 3.7 | 60a | 4.7 | 47a | 5 | 67a | 5 | 60a | 2.7 | 25a | 2.3 | 10a | 4 | 33a | 2 | 8ab | 4.3 | 28a | 0 | 0 | 0 | 0 | 1.5 | 3ab |
| L. culinaris | R52 | 4 | 30a | 4.7 | 57a | 4 | 27b | 4.7 | 37a | 4 | 50ab | 3.3 | 30a | 3 | 22a | 4 | 17a | 2 | 4b | 4 | 45a | 0 | 0 | 0 | 0 | 0 | 0b |
| L. culinaris | R172 | 4 | 43a | 4 | 50a | 4.5 | 42ab | 4.7 | 45a | 4 | 32b | 3.7 | 20a | 1.3 | 7a | 3.3 | 7b | 0.7 | 2b | 4 | 48a | 0 | 0 | 0 | 0 | 1.3 | 1b |
| AVERAGE | 40BCD | 58B | 40B | 49B | 46B | 27BC | 13BC | 18AB | 6B | 35A | 0B | 0C | 3CDE | ||||||||||||||
| SOYBEAN | |||||||||||||||||||||||||||
| Glycine max | PI081005 | 1.7 | 8 | 0 | 0 | 3 | 10 | 3.3 | 12 | 3 | 30 | 1 | 1 | 2 | 17 | 2 | 5 | 3 | 4 | 0 | 0 | 3 | 4 | 3.3 | 17 | 4 | 30 |
| AVERAGE | 8EF | 0F | 10CD | 12DE | 30BC | 1D | 17BC | 5BC | 4B | 0B | 4AB | 17A | 30A | ||||||||||||||
| Host specie | Code | Dp-M07-4 | Dp-Esc-13 | Dp-KHM-13 | Dp-ANN-13 | Dp-JAP-03 | Dp-PO-03 | Dp-CO-99 | Dp-PdT-03 | Dp-FR-88 | Dl-AL10 | Dr-Pt04 | Df-AU04 | Df-857 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DRA | DSB | DR | DS | DR | DS | DR | DS | DR | DS | DR | DS | DR | DS | DR | DS | DR | DS | DR | DS | DR | DS | DR | DS | DR | DS | ||
| COMMON VETCH | |||||||||||||||||||||||||||
| Vicia sativa | 31514 | 3 | 33a | 2 | 15a | 3.3 | 30a | 3 | 13a | 2 | 6a | 3 | 20a | 1.3 | 10a | 2.7 | 10a | 3 | 3a | 0 | 0b | 1 | 2a | 4 | 5a | 3.7 | 8a |
| V. sativa | 31544 | 3 | 37a | 2.3 | 28a | 3.3 | 35a | 3 | 23a | 2.3 | 18a | 3 | 20a | 2 | 10a | 2.7 | 8a | 3 | 11a | 1.7 | 7ab | 1.7 | 5a | 4 | 10a | 4 | 6a |
| V. sativa | 31554 | 3.7 | 33a | 2.3 | 27a | 3 | 35a | 3 | 15a | 2 | 17a | 2.3 | 20a | 1.7 | 10a | 2.3 | 13a | 1 | 3a | 0.9 | 2b | 0.5 | 2a | 4 | 8a | 4 | 8a |
| V. sativa | 31564 | 3 | 25a | 2 | 23a | 3.7 | 36a | 3 | 23a | 2 | 12a | 2.7 | 13a | 1.3 | 8a | 2 | 8a | 0.3 | 1b | 2 | 10a | 0 | 0a | 4 | 5a | 4 | 8a |
| AVERAGE | 32BCD | 23DEF | 34B | 18DE | 14CD | 18CD | 10C | 10BC | 5B | 4B | 2B | 7B | 7C | ||||||||||||||
| GRASS PEA | |||||||||||||||||||||||||||
| Lathyrus sativus | ILAT13 | 4 | 33a | 4.7 | 40a | 4 | 37a | – | – | 4.7 | 40a | – | – | – | – | 4.3 | 32a | 2.3 | 4a | 0 | 0b | 0 | 0 | 0 | 0b | 0 | 0 |
| L. sativus | ILAT103 | 3.7 | 50a | 4.3 | 57a | 3.7 | 17a | 5 | 63a | 4.7 | 37a | 4.3 | 48a | 2.3 | 17a | 4 | 20a | 3.5 | 12a | 1 | 2a | 0 | 0 | 4 | 5ab | 0 | 0 |
| L. sativus | ILAT163 | 4 | 43a | 5 | 53a | 4 | 28a | 5 | 50a | 4.3 | 50a | 3.7 | 33a | 1.7 | 10a | 3.7 | 17a | 3 | 7a | 0 | 0b | 0 | 0 | 4 | 10a | 0 | 0 |
| L. sativus | ILAT183 | 4.3 | 46a | 4 | 37a | 4.3 | 37a | 5 | 55a | 4.7 | 43a | 4.7 | 48a | 1 | 7a | 4.3 | 23a | 2.4 | 5a | – | – | 0 | 0 | 0 | 0b | 0 | 0 |
| L. sativus | BGE0171846 | 4.3 | 42a | 5 | 70a | 2 | 27a | 5 | 47a | 4.7 | 45a | 4 | 37a | 0 | 0b | 4 | 30a | 3.7 | 11a | – | – | 0 | 0 | – | – | – | – |
| AVERAGE | 43B | 51B | 29B | 54B | 36B | 42B | 13AB | 26A | 8B | 0.6B | 0B | 3BC | 0E | ||||||||||||||
| SULLA | |||||||||||||||||||||||||||
| Hedysarum coronarium | Sparacia1 | 2.7 | 20a | 3.3 | 58a | 4 | 43a | 2.3 | 8 | 4.7 | 47a | 3 | 20b | 2 | 15a | 1.7 | 9b | 0.7 | 1a | 0 | 0b | 0 | 0 | 0 | 0 | 0 | 0 |
| H. coronarium | Grimaldi1 | 1.5 | 10a | 2.7 | 33a | 4 | 45a | – | – | 5 | 60a | 3 | 42a | 1 | 5b | 1.7 | 5b | 1.3 | 10a | 0 | 0b | 0 | 0 | 0 | 0 | 0 | 0 |
DR, Disease rating following 0–5 scale defined by Roger and Tivoli (1996).
DS, final disease severity (%) measured under controlled conditions.
Commercial varieties are named.
Belonging to IAS-CSIC collection.
Provided by ICARDA (Syria).
Collected by authors.
Provided by USDA (USA).
Provided by CRF-INIA (Spain).
Data followed with different letters, per column (lower letter types) and host species (capital letter type), are significantly different (LSD test, P = 0.01). − Not determined.
To ensure experiments with a uniform plant development stage, seeds were scarified by nicking with a razor blade and then germinated for 48 h on wet filter paper in a Petri dish at 4°C. The Petri dishes were then transferred to 20°C for 5–7 days. Germinated seeds were planted into plastic pots (6 × 6 × 10 cm) filled with a 1:1 mixture of sand and peat in a rust-free growth chamber. Plants were pre-germinated and sown at 3 days intervals in order to be able to select seedlings at the same growing stage at the time of inoculation. There were three independent replicates per fungal isolate, arranged in a complete randomized design. Each replicate consisted of 3 pots with 5 plants each per accession. Experiments were repeated three times. Pea cv. Messire was included in each replication as a common susceptible check. Plants were grown in a growth chamber at 20°C, under a photoperiod of 14/10 h day/night regime, with 148 μmol/m2s irradiance at plant canopy for 3 weeks, until the plants reached the 4–5-leaf stage.
Plant inoculation
Plants with 4-5 leaves were inoculated as described by Fondevilla et al. (2005) with some modifications. Inoculum was prepared by multiplying spores of each isolate on PDA (Potato Dextrose Agar) medium with chloramphenicol (60 mg/l PDA) and ampicillin (50 mg/l PDA) at 20°C with 16 h light/8 h dark photoperiod. Spore suspensions were prepared by flooding the surface of 10-day-old cultures with sterile distilled water, gently scraping the colony with a glass rod and filtering the suspension through two layers of sterile cheesecloth. Concentration of pycnidiospores was determined with a haemocytometer and adjusted to 106 spores/ml. Tween 20 (VWR) was added as wetting agent (two drops per 500 ml pycnidiospore suspension). The pycnidiospore suspensions were sprayed at the 4–5-leaf stage using a handheld sprayer at a rate of 1 ml per plant. After inoculation, plants were covered with a polyethylene sheet during the first 24 h in darkness, and high humidity was ensured by ultrasonic humidifiers operating for 15 min every 2 h. Later on, the polyethylene cover was removed and plants were maintained 9 more days in a growth chamber (under conditions described above). Every 2 days, water was added to the trays to maintain high relative humidity (95–100%).
Disease assessment
Plant response to infection was visually assessed 10 days after inoculation using two separate assessments. Disease severity (DS) was assessed by a visual estimation of the percent of diseased tissue per plant (Fondevilla et al., 2005). In addition, disease rating (DR) was visually assessed on leaves over the first, second and third nodes of each plant using a 0–5 scale defined by Roger and Tivoli (1996) were 0 = no lesions; 1 = a few scattered flecks; 2 = numerous flecks; 3 = 10–15% of the leaf area necrotic and appearance of coalescent necrosis; 4 = 50% of the leaf area dehydrated or necrotic; 5 = 75–100% of the leaf area dehydrated or necrotic. DR was then calculated as the average of values scored per node. Accessions displaying an average DR > 3 combined with DS > 35% were considered as highly susceptible, accessions displaying an average DR > 3 combined with DS values lower than 35% were considered as susceptible, accessions showing an average DR included between 2 and 3 combined with DS values < 35% were considered as moderately resistant and, finally, accessions displaying DR < 2 combined with DS values < 10% were considered as highly resistant.
DNA extraction and its amplification
Monoconidial cultures of the 13 isolates were grown in Petri dishes using PDA medium as described above. Mycelium was collected by flooding the surface of 5-day-old cultures with sterile distilled water (2 ml per Petri dishes), gently scraping the colony with a glass rod and filtering the suspension through two layers of sterile cheesecloth. Three Petri dishes per isolate were used, in order to ensure sufficient amount of fungal material. Suspension was centrifuged at maximum speed (14,000 rpm) and pellet was collected. DNA was extracted from ground mycelium using the DNeasy plant minikit (Qiagen, Ltd.). DNA concentration was determined using an ND-1000 spectrophotometer (NanoDrop Technologies) and adjusted to 20 ng μl/1 for PCR. Primers ITS1 and ITS2 were used to amplify the nuclear ribosomal internal transcribed spacer (ITS) region ITS1-5.8S-ITS2 following the protocol described by White et al. (1990). PCR products were extracted with a sterile scalpel and purified using the QIAquick Gel Extraction kit (Qiagen®) following the protocol of the manufacturer. The purified products were cloned using the pGEM-T Easy Vector Systems kit (Promega, Madison, WI, USA) following Barilli et al. (2011) protocol. Sequencing was carried out on an ABI 3730 XL sequencer (Applied Biosystems, Foster City, CA, USA) at the DNA Sequencing Service, STAB VIDA GENOMICS LAB, Caparica, Portugal. For each isolate, two clones were sequenced. Both forward and reverse strands were sequenced for each clone. ITS sequences were submitted to GenBank.
In addition to this, sequences from Ascochyta pisi, Didymella pinodes, Phoma koolunga, and P. medicaginis var. pinodella (Table 1) retrieved from GenBank (http://www.ncbi.nih.gov; Davidson et al., 2007; Peever et al., 2007) were included in the analysis.
Statistical analysis
Disease responses
All isolate x species combinations (including several accessions per species) were arranged in a completely randomized design in a controlled condition growth chamber. For the whole data set, only final disease severity values were included in the statistical analysis. Disease severity was first analyzed by taking into account differences in pathogenicity between the 13 Didymella spp. isolates according to the species evaluated (by averaging disease severity among accessions within each species).
Disease severity was assessed for every Didymella spp. isolate between accessions within each species. The whole experiment was repeated three times. Before performing analyses of variance, the normality and equality of variances were checked using Shapiro–Wilk's (Shapiro and Wilk, 1965) and Bartlett's tests (Little and Hills, 1978) respectively. When necessary, DS percentage data were transformed to angles (y = arcsine (x/100)) and again checked before applying analysis of variance. Differences between isolates, species, or accessions within species were compared by analysis of variance (ANOVA) followed by a least significant difference (LSD) test, with values of P < 0.01 considered significant. Statistical analyses were performed with Statistix software (version 8.0; Analytical Software, Tallahassee, USA).
Disease rating (DR) was visually estimated as the mean disease score over the first, second and third leaves of each accession within each specie.
The entire data set was analyzed by Principal Component Analysis (PCA) using the web-based software PAST (Hammer et al., 2001), available at http://nhm2.uio.no/norlex/past/download.html, with the following settings: covariance matrix type, four principal components, 1-fold change threshold for clusters, and 0.3 correlation thresholds for clusters. PCA results were represented as a biplot, with accessions more susceptible to a specific Didymella spp. isolate (according to both DS and DR) located in the same area of the graph.
ITS sequence analysis
Sequences were aligned and adjusted manually with Mega version 6 (Tamura et al., 2013) using the penalties of 15 for gap opening and 6.66 for gap extension. Estimates of genetic similarity (GS) were calculated for all possible pairs of genotypes according to Rho similarity coefficient (Posada and Crandall, 1998).
The evolutionary history was inferred using the unweighted pair-group method with arithmetic average (UPGMA; Sneath and Sokal, 1973). The evolutionary distances were computed using the Maximum Composite Likelihood method (Tamura et al., 2004) and a dendrograms was constructed.
The trees were rooted using P. koolunga as outgroup. The scores between 50 and 74 bootstrap percentages (BS) were defined as weak support, scores between 75 and 89% BS as moderate support and scores > 90% BS as strong support. A likelihood ratchet employs multiple sequential truncated searches on different starting trees created by fast algorithmic searches on reweighed data, in the hope of exploring a larger pro- portion of tree space, analogous to the parsimony ratchet (Nixon, 1999). We ran 200 iterations with the general time reversible likelihood model of evolution with gamma distribution (GTR+G) and uniformly reweighing 15% of the data-set per iteration. Bootstrap support values from 1000 replicates were calculated using the heuristic search with random addition-sequence with 10 replicates limited to 10,000 tree rearrangements (branch swaps) imposed separately for each addition-sequence replicate (rearlimit = 10,000; limitperrep = yes). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances are reported in the units of the number of base substitutions per site. The rate variation among sites was modeled with a gamma distribution (shape parameter = 1). All positions containing gaps and missing data were excluded in analyses.
Results
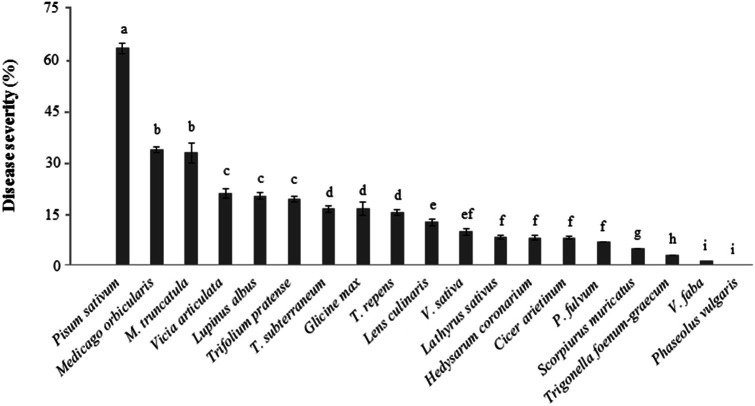
The local Didymella pinodes isolate Dp-CO-99 caused different disease rating (DR) (Table 2) as well as significantly different disease severity (DS) values on the tested legume species (P < 0.01; Figure 1). The highest levels of susceptibility were found in P. sativum (DR = 5; DS = 67%) confirming expectations (Fondevilla et al., 2005), followed by L. albus (DR = 4.7; DS > 20%), Trifolium spp., Medicago spp., V. articulata, C. arietinum, and L. culinaris (2 ≤ DR < 3; DS > 15%). Some infection was also observed on G. max, V. sativa, L. sativus, H. coronarium, P. fulvum, S. muricatus, V. faba, and T. foenum-graecum although at the level of resistance (DR < 2; DS < 20%). P. vulgaris did not showed any symptoms of fungal infection (DS and DR = 0; Figure 1).
Figure 1.
Percentage of disease severity (DS%) measured on foliar organs of different legume species in response to inoculation with D. pinodes isolate Dp-CO-99 under controlled conditions. Averages per species are presented. The bars indicate the standard deviation; different letters indicate significant differences (P = 0.01).
Results from cross inoculations performed with different Didymella spp. showed that the legume species under study displayed differential resistance/susceptibility to each isolate as indicated by significant specie x isolate interactions in ANOVA (P < 0.01; Table 2). Statistical analysis showed a significant effect of legume species (sum of squares = 353,064, P < 0.001), fungal isolates (sum of squares = 125,118, P < 0.001), and their interaction (sum of squares = 75,346, P < 0.001), indicating that not all D. pinodes isolates displayed the same infection pattern toward the legume species involved in this study.
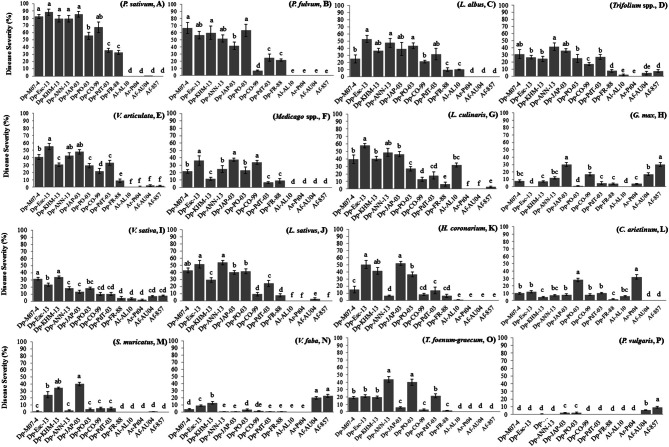
P. sativum accessions showed DR values = 4 against all D. pinodes tested (Table 2), although level of infection varied greatly (DS from 15 to 100%). Isolates Dp-M07-4 (DS 80–95%), Dp-Esc-13 (DS 77–93.3%), Dp-JAP-03 (DS 66–100%), and Dp-KHM-13 (DS 63–98%) were the most aggressive on cultivated peas (Table 2, Figure 2A). P. fulvum was generally more resistant than P. sativum, with DR ranging from 1.3 to 4.7 and DS from 7 to 67%. In particular, accession IFPI3260 confirmed here its high resistance against Dp-CO-99 (DR = 1.3, DS = 6.7; Figure 2B; Fondevilla et al., 2005). In addition, P. fulvum was also moderately resistant to isolates Dp-FR-88 and Dp-Esc-13 (DR = 3; DS < 25%). As for P. sativum, accession IFPI3260 was immune to other Didymella spp. isolates tested (Table 2).
Figure 2.
Disease severity (%) measured on whole plants of different legume species after infection by isolates of Didymella spp. under controlled conditions. Averages per species are presented: (A) pea (Pisum sativum), (B) tawny pea (P. fulvum), (C) white lupin (Lupinus albus), (D) clovers (Trifolium pratense, T. subterraneum, T. repens), (E) oneflower vetch (V. articulata), (F) medicks (Medicago orbicularis, M. truncatula), (G) lentil (Lens culinaris), (H) soybean (Glycine max), (I) common vetch (V. sativa), (J) grass pea (Lathyrus sativus), (K) sulla (Hedysarum coronarium), (L) chickpea (Cicer arietinum), (M) prinkly scorpion's tail (Scorpiorus muricatus), (N) faba bean (Vicia faba), (O) fenugreek (Trigonella foenum-graecum), (P) common bean (Phaseolus vulgaris). The bars indicate the standard deviation. Different letters indicate significant differences (P = 0.01).
Accessions from L. albus were also susceptible to D. pinodes (DR = 3), showing level of infection that varied depending on the isolate tested (average DS = 34%, range 9–100%). Isolates Dp-Esc-13 and Dp-ANN-13 were the most virulent (DR = 5; DS > 40%; Table 2, Figure 2C). By contrary, L. albus was resistant to both D. rabiei and D. fabae, while only accession Lup35 was moderately infected by D. lentil (Table 2).
Trifolium spp. showed responses to D. pinodes infections that were from moderately resistant to susceptible (averages ranging between DR 2.5–4.6 and DS 7–42%; Table 2). Isolate Dp-ANN-13 was the most virulent (DR > 4.7; DS > 30%) while Dp-CO-99 and Dp-FR-88 the lesser (DR < 3.7; DS < 20%; Table 2, Figure 2D). Accessions studied were not infected by D. rabiei, whereas T. pratense was slightly infected by D. lentil and D. fabae (Table 2).
V. articulata accessions were from highly susceptible to moderate resistant against D. pinodes inoculations (averages ranging between DR 2.6–4.3 and DS 9–55%), being differences significant among accessions and isolates (P < 0.01) (Table 2, Figure 2E). V. articulata was immune to D. lentil, whereas only certain accessions were slightly infected by D. rabiei or D. fabae (DR from 2.3 to 3.7, DS < 8%).
Similarly, Medicago spp. accessions showed from resistance to susceptibility to D. pinodes infections. Nevertheless, differences between plant species were not consistent (Table 2, Figure 2F). Isolate Dp-JAP-03 was the most virulent on all Medicago accessions studied inciting DR ranging from 3.7 to 5 and DS ranging from 37 to 80%. Medicago accessions were not affected by any other Didymella spp.
Response of L. culinaris accessions to D. pinodes varied greatly, depending on the isolate tested (averages ranging between DR 0.7–5 and DS 4–67%) (Table 2, Figure 2G). As for peas, isolates Dp-Esc-13, Dp-JAP-03, and Dp-M07-4 were highly virulent on all accessions tested (DR > 3.7, DS > 40%; Table 2). By contrary, lentils were less damaged by isolate Dp-FR-88 (DR ≤ 2, DS < 10%). As expected, all accessions tested were susceptible to D. lentil, with no significant differences between them (DR ≥ 4, DS ≥ 20%). By contrary, D. rabiei did not cause any symptoms on lentils and D. fabae was only slightly infective (DR < 3, DS ≤ 6%; Table 2).
Accession PI08100 from G. max showed from moderate to high resistance against D. pinodes infections (Figure 2H), being isolate Dp-JAP-03 the most virulent (DR = 3, DS = 30%). By contrary, no symptoms were found on PI08100 after Dp-PO-03 and Dp-Esc-13 inoculations. This accession was immune to D. lentil, slightly infected by D. rabiei (DR = 3, DS = 4%) and susceptible to D. fabae (DR > 3.3, DS > 17%; Table 2).
Similarly, responses from V. sativa varied greatly, being resistant to isolates Dp-CO-99 and Dp-FR-88 (averages DR < 1.8 and DS < 10%) and susceptible to Dp-KHM-13 (DR > 3, DS > 30%), with no significantly difference among accessions (Table 2, Figure 2I). V. sativa showed a fully compatible interaction with both D. fabae isolates in spite of a reduced severity (DS < 10%). Nevertheless, both D. rabiei and D. lentil caused foliar symptoms at reduced rates (DR < 2, DS ≤ 10; Table 2).
Except for local isolate Dp-CO-99, studied L. sativus accessions were moderately or highly susceptible to all D. pinodes isolates studied, being isolates Dp-Esc-13 and Dp-ANN-13 the most virulent (DR > 4, DS > 37%; Table 2, Figure 2J). Accessions from L. sativus were immune or highly resistant to infection from D. rabiei, D. lentil, and D. fabae isolates (Table 2).
Responses of C. arietinum varied greatly depending both on the D. pinodes isolate employed as well as the accession tested (Figure 2L, Table 2), but infection was always reduced compared to pea accessions. Accessions showed DR from low to high, depending on the isolate, but always with low DS (<30 %). Isolate Dp-Po-03 was the most virulent on chickpea (DR > 4.3), while all accessions were resistant to isolate Dp-FR-88 (DR < 1.3, DS < 3%). Chickpea was resistant to both D. fabae isolates, while accession AS18 showed moderate susceptibility to Dl-AL10 infection. Chickpea showed a fully compatible interaction with D. rabiei isolate studied (Dr-Pt04) although significant differences between accessions were found (Table 2).
H. coronarium, S. muricatus, and T. foenum-graecum showed differential responses to D. pinodes inoculations depending principally on the isolate tested (P < 0.01; Figures 2K,M–O, respectively). In general, accessions showed symptoms that were significantly reduced comparing with P. sativum, also if some exceptions were found (e.g., H. coronarium and DP-JAP-03 or T. foenum-graecum and Dp-PO-03 with DR ≥ 4 and DS ≥ 30%; Table 2). With the exception of isolate Dp-KHM-13, V. faba was highly resistant against almost all D. pinodes studied (DR ≤ 2 and DS ≤ 10%; Table 2). Accessions belonging to H. coronarium, S. muricatus, and T. foenum-graecum were highly resistant or immune to infection with other Didymella spp. V. faba was highly susceptible to both D. fabae isolates with no significant differences among accessions, while no symptoms were found after Dr-Pt04 and Dl-AL10 inoculations (Table 2).
Finally, P. vulgaris was highly resistant to all Didymella spp. isolates since no or limited symptoms were foundflentils were less damaged by isolate on all accessions tested (DR ≤ 1.3, DS ≤ 2%) with exception of D. fabae that caused compatible interactions (DR ≥ 3.3) although with reduced DS values (Table 2, Figure 2P).
Among the isolates tested, Dp-KHM-13 was the most virulent being common bean the unique legume specie tested that was immune, while Dp-FR-88 was the lesser damaging isolate (Table 3). Isolate Dl-AL10 (D. lentil) was only virulent on L. culinaris accessions, while isolate Dr-Pt04 (D. rabiei) showed symptoms on C. arietinum and, although limited, on G. max. Finally, G. max, P. vulgaris, T. pratense, V. sativa, and V. faba were susceptible to isolates from D. fabae (Table 3).
Table 3.
Disease reaction of fourteen isolates of Didymella spp. from different geographical origin on fifteen leguminous species, performed under controlled growing conditions.
| Host specie | N | Dp-M07-4 | Dp-Esc-13 | Dp-KHM-13 | Dp-ANN-13 | Dp-JAP-03 | Dp-PO-03 | Dp-CO-99 | Dp-PdT-03 | Dp-FR-88 | Dl-AL10 | Dr-Pt04 | Df-AU04 | Df-857 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pisum sativum | 4 | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | – | − | − | − |
| P. fulvum | 1 | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | – | + | + | – | – | – | – |
| Lupinus albus | 4 | +++/+* | ++++/+++ | ++++/+++ | ++++ | ++++/+++ | ++++ | +++ | ++++/+ | +/+++ | −/+++ | – | – | – |
| Trifolium pretense | 1 | ++++ | +++ | ++++ | ++++ | ++++ | ++++ | + | +++ | + | + | – | + | + |
| T. repens | 1 | +++ | +++ | +++ | +++ | ++++ | +++ | + | +++ | + | – | – | – | + |
| T. subterraneum | 1 | +++ | +++ | +++ | ++++ | +++ | +++ | + | + | + | – | – | – | + |
| Lathyrus sativus | 5 | ++++/+++ | ++++ | +++/++++ | ++++ | ++++ | ++++/+++ | −/+ | +++ | +/+++ | – | – | −/+++ | – |
| Lens culinaris | 4 | ++++/+++ | ++++ | ++++/+++ | ++++ | ++++/+++ | +++/+ | +/- | +++ | −/+ | ++++/+++ | – | – | −/+ |
| Vicia articulate | 4 | ++++ | ++++ | +++ | ++++/+++ | ++++/+++ | +++/+ | + | +++/++++ | + | – | −/+ | −/+ | −/+ |
| Trigonella foenum-graecum | 3 | +++ | +++/+ | +++/+ | ++++/+++ | – | +++/++++ | – | −/+ | – | – | – | – | – |
| Medicago orbicularis | 2 | + | ++++ | + | +++ | ++++ | +++ | + | + | + | – | – | – | – |
| M. truncatula | 3 | + | +/+++ | +/+++ | + | ++++ | + | + | – | −/+ | – | – | – | – |
| Cicer arietinum | 5 | −/+++ | +++/+ | +/+++ | +++ | −/+++ | +++ | −/++++ | +++ | – | −/+ | ++++/− | – | – |
| Hedysarum coronarium | 4 | +/- | ++++/+ | ++++ | + | ++++ | ++++/+ | −/+ | −/+ | – | – | – | – | – |
| Scorpiurus muricatus | 4 | – | −/+++ | −/++++ | – | ++++/+ | – | – | – | – | – | – | – | – |
| Vicia sativa | 4 | +++/++++ | + | +++/++++ | + | + | + | – | + | −/+ | – | – | +++ | +++ |
| Glycine max | 1 | – | – | + | +++ | +++ | – | + | – | + | – | + | +++ | +++ |
| Vicia faba | 4 | – | −/+ | + | – | – | – | – | – | – | – | – | +++ | +++ |
| Phaseolus vulgaris | 4 | – | – | – | – | – | – | – | – | – | – | – | +++ | +++ |
Results correspond to the combination of Disease Rate (based on a 0–5 scale, Roger and Tivoli 1996) and the % of the whole plant covered by symptoms (% Disease Severity, DS %) as follows:
++++, highly susceptible (DR ≥ 3 and DS > 35%); +++, susceptible (DR ≥ 3 and DS < 35%); +, moderately resistant (2 < DR < 3 and DS < 35%); –,highly resistant (DR < 2 and DS < 10%).
When several classifications are reported means that genotypes from the same specie showed different responses to the same fungal isolate. Classification reported in first position is the most common.
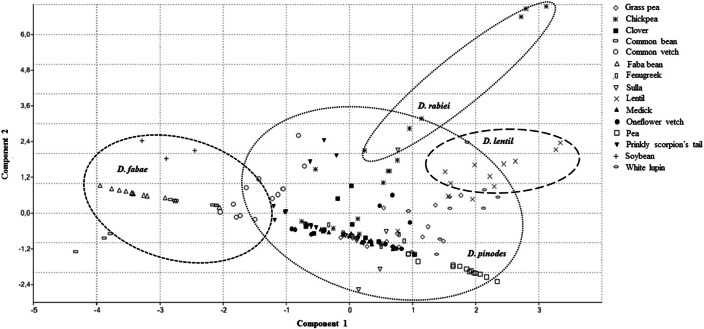
Principal component analysis (PCA) showed that two principal axes gave eigenvalues greater than 1, while the other axis all had eigenvalues lesser than 1 (Table 4). Hence, the first two principal components were considered important and contribute the most in the distribution of variation existing among the isolates. The component 1 had an eigenvalue of 2.8034, accounted for 40.62% of the overall variance in the data set (Table 4). Component 2 had an eigenvalue of 2.2101 and accounted for 31.1% of the total variance. Hence, the two principal components contributed for 71.69% of the total variability (Table 4). The first pc was more related to the level of aggressiveness expressed by D. pinodes, D. lentil, and D. rabiei isolates, while the second pc contributed for those expressed by D. fabae isolates to all cultivars tested (Figure 3). On the other hand, we can also appreciate certain host specificity between the legumes and fungal isolate species. The scattered diagram showed a major distance between isolates belonging to D. fabae and D. rabiei with the rest that were studied (Figure 3).
Table 4.
Principal components for disease rating (DR) and disease severity (DS) values of 13 isolates of Didymella spp.
| Component 1 | Component 2 | |
|---|---|---|
| Eigenvalues | 2.8034 | 2.2101 |
| Proportion of variance | 40.623 | 31.067 |
| Cumulative variance | 40.623 | 71.690 |
Figure 3.
Scattered diagram generated by principal component analysis (PCA) showing associations between Disease Severity and Disease Rating response performed by 13 isolates of Didymella spp. on 15 leguminous species. A short distance between plant accessions and fungal isolate in the component space is indicative in susceptibility of the plant/pathogen interaction.
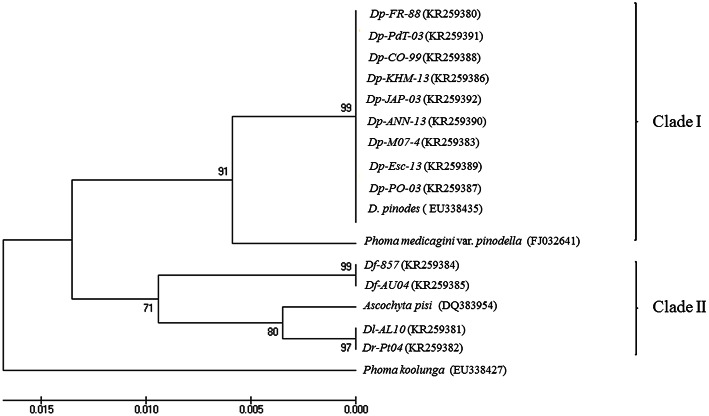
ITS analysis by MEGA6 originates an optimal tree with the sum of branch length = 0.06595538. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches (Figure 4). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances are reported in the units of the number of base substitutions per site. The rate variation among sites was modeled with a gamma distribution (shape parameter = 1). The analysis involved 17 nucleotide sequences. All positions containing gaps and missing data were eliminated. There were a total of 437 positions in the final dataset.
Figure 4.
UPGMA dendrograms of 13 samples of Didymella spp. based on Dice distance for Internal Transcribed Spacer regions analysis.
From the dendrogram generated, using UPGMA with the genetic distance coefficient, the 17 isolates could be classified into two main clusters that clearly separate all isolates belonging to D. pinodes from the others (Figure 4). Cluster 1 (bootstrap support [BS] = 91 from Maximum Composite Likelihood analysis) included all isolates from D. pinodes used for the study as well as the D. pinodes isolate from GenBank. D. pinodes isolates showed to be monophyletic since they were included in a unique well-supported branch ([BS] = 99). The isolate of P. medicaginis var. pinodella was also included in this clade although it was divergent and on a branch apart from the rest of the isolates included.
Clade II ([BS] = 71) comprised two isolates of D. fabae, one isolate from D. lentil, one isolate from D. rabiei and one isolate from A. pisi. D. fabae isolates showed to be monophyletic since they were included in a unique well-supported branch ([BS] = 99). By contrary, isolates from D. lentil and D. rabiei clustered together in other strongly supported branch ([BS] = 97) where A. pisi was apart ([BS] = 80). Finally, isolate from P. koolunga did not fit with any other isolates.
Discussion
Cool season legumes play an important role for human food and animal feed throughout the world. These crops are attacked by numerous aerial fungal pathogens that cause considerable losses in quality and quantity (Tivoli et al., 2006; Muehlbauer and Chen, 2007). The major necrotrophic fungal diseases are ascochyta blight on various grain legumes and Didymella pinodes was reported as the principal agent causing aschochyta blight on peas (Tivoli et al., 2006; Khan et al., 2013).
The aim of this current study was to analyse variations in the susceptibility of different legume species to D. pinodes compared to other Didymella spp., as well as to characterize the disease response of different cultivars within different legume species toward several D. pinodes isolates under controlled conditions. The results demonstrated that D. pinodes is able to cause disease in a number of legume species, that D. pinodes isolates from different geographical origin are differentially aggressive toward the legume species, and that cultivars within each legume species responded differentially to D. pinodes.
Infection of several host species is common in agrosystems leading to change in epidemic characteristics and pathogenicity. As a result, these processes will modify the survival of pathogen populations and their transmission (Woolhouse et al., 2001). In fact, variation in disease response can be significant at both the host species level as well as the host cultivar level, as was recently shown (Moussart et al., 2008; Le May et al., 2014). In the current study, cultivars from 20 different legume species were used to characterize the behavior of D. pinodes isolates sampled from pea. Visible symptoms caused by D. pinodes isolates were observed on all the legume species examined in this study, excepted with common bean. Large differences in susceptibility to D. pinodes were observed among the infected hosts, with Pisum spp. being the most susceptible, followed by L. sativus, L. culinaris, L. albus, Medicago spp., Trifolium spp., T. foenum-graecum, and V. articulata. In contrast to other Didymella species, D. pinodes appears to have the widest host range, since only accessions from lentil and chickpea were severely infected by D. lentil and D. rabiei, respectively, while D. fabae infected principally beans (common bean, faba bean, and soybean) and common vetch. Results for D. lentil and D. rabiei agreed with previous studies which demonstrated that artificial inoculations with Ascochyta fungi in the greenhouse and/or growth chambers are host-specific (Kaiser et al., 1997; Khan et al., 1999; Hernandez-Bello et al., 2006; Peever et al., 2007). In fact, it was previously found that D. fabae, D. lentil, and D. rabiei only diseased their respective hosts, while no visible symptoms were observed on any of the plant species other than faba bean, lentil and chickpea (Kaiser et al., 1997; Trapero-Casas and Kaiser, 2009). Nevertheless, for D. rabiei, Trapero-Casas and Kaiser (2009) also found that the fungus was able to survive on other leguminous or weeds, even though it did not show any visible symptoms and that this phenomenon could serve as secondary reservoirs in the absence of the natural host. In our study, isolates from D. fabae were highly virulent on faba bean but were also able to slightly infect other beans and vetch.
Regarding D. pinodes virulence, the results obtained with pea genotypes with very low levels of partial resistance were similar to those obtained by Fondevilla et al. (2005) and Le May et al. (2014) with common vetch and clover. All genotypes studied from P. sativum showed high susceptibility to all isolates tested, while accession IFPI3260 from P. fulvum (tawny pea) displayed a certain degree of partial resistance. These results confirms that only incomplete resistance is available for cultivated pea, while the highest levels of resistance are available in related Pisum species. In fact, sources of resistance to D. pinodes were recently found in accessions belonging to P. fulvum, P. sativum ssp. syriacum, and P. sativum ssp. elatius (Zhang et al., 2003; Fondevilla et al., 2005; Carrillo et al., 2013). Accession IFPI3260 showed from moderate to high resistance against 4 out of 9 D. pinodes isolates tested under controlled conditions. This accession was previously identified also as an important source of resistance against pea powdery mildew (Erysiphe pisi DC) and pea rust (Uromyces pisi (Pers.) Wint) (Fondevilla et al., 2007; Barilli et al., 2009) and is included in our department plant breeding programme.
Lathyrus has been reported as a resistant leguminous to D. pinodes infection firstly by Weimer (1947) who studied accessions belonging to L. tingitanus, L. sativus and L. hirsutus, followed by another relevant report (Gurung et al., 2002) which confirmed resistance of L. sativus, and added L. ochrus and L. clymenum as species with high degree of resistance. Nevertheless, all accessions from L. sativus used in our study resulted to be highly susceptible to all D. pinodes isolates tested under controlled conditions. Susceptibility in white lupin (L. albus), lentil (L. culinaris), fenugreek (T. foenum-graecum) and oneflower vetch (V. articulata) is described here for the first time, expanding the current knowledge of D. pinodes's host range.
The almost complete absence of symptoms in common bean (P. vulgaris) against several D. pinodes isolates may indicate that this species is a non-host species or that the fungus had invaded the host tissues internally although no visible symptoms were observed. This has been previously found for D. rabiei, which was recovered consistently from inoculated tissue of pea without causing any visible symptoms (Trapero-Casas and Kaiser, 2009). Future histological studies will be necessary to clarify this fact.
Unlike common bean, common vetch (V. sativa), faba bean (V. faba), and soybean (G. max) may be defined as a host plant under conditions of high inoculum pressure, but all genotypes studied displayed a very high level of partial resistance against the set of fungal isolates tested. As the conditions used in this study were very favorable for disease development on plants, the results would require confirmation by testing under different infection conditions such as in the field since growth habit, canopy morphology, lodging and precocity can affect D. pinodes development (Khan et al., 2013) and plant susceptibility since it was reported that plant symptoms were more severe at plant maturity than at the seedling stage (Zhang et al., 2003). In addition, plant seasonality might also be another factor that influenced plant susceptibility in the field. Common vetch and faba bean are cool season legumes, whereas common bean and soybean are summer crops. Influences of mean temperatures and humidity on host plant susceptibility during crop development needs to be further investigated, as on Didymella spp. the temperatures before and after the fungal infection period affected disease development and symptom expression (Trapero-Casas and Kaiser, 1992; Roger et al., 1999; Frenkel et al., 2008).
The use of faba bean has been previously tested in pea intercropped field as an alternative control measure to limit aschochyta blight (Fernández-Aparicio et al., 2010), leading to a fungal reduction by up to 60%. Introduction of species as common bean, common vetch, faba bean, soybean in pea rotation or intercropped may be tested in relation with a reduction of aerial spores during the cropping season and the survival of the pathogen into the soil residues by chlamydospore and sclerotium production. In fact it has been previously reported that introduction of plants with modified characteristics than pea imposes a non-host barrier, and as a consequence, less conidia are surviving and successfully transported to new developing host tissue (Zhang et al., 2003; McDonald and Peck, 2009; Fernández-Aparicio et al., 2010).
The existence of susceptible, partially and highly resistant genotypes within the same species (as in medicks, sulla, fenugreek, chickpea, prinkly scorpion's tail) suggest that the reaction may therefore be described as cultivar specific since the fungal ability to infect these other species depends on the susceptibility of the cultivar chosen (Moussart et al., 2008). C. arietinum accessions showed different degrees of susceptibility depending on the accession and the isolate tested, nevertheless cv. ILC72 was one of the lesser diseased after D. pinodes inoculation. ILC72 is a D. rabiei resistant line from ICARDA which showed a degree of resistance in the field and in controlled environments (Muehlbauer and Chen, 2007), as confirmed here. This accession has been thoroughly used in breeding programmes worldwide, as well in studies of the genetic of resistance to aschochyta blight (Cobos et al., 2006; Muehlbauer and Chen, 2007; Madrid et al., 2014). Susceptibility found here to certain D. pinodes isolates in cultivars belonging to H. coronarium, Medicago spp., S. muricatus and T. foenum-graecum is also described here for the first time. The susceptibility of these pasture legume species need to be tracked under field conditions before to become a serious agricultural problem. Thus, for each legume species, it should be interesting to enlarge the set of genotypes tested to make possible the identification of resistant genotypes.
In terms of pathogenicity, results on peas showed that the local isolate Dp-Co-99 was not always the most aggressive. In fact, disease severity measured on the primary host plants showed that isolates Dp-M07-4, Dp-Esc-13, Dp-KHM-13, and Dp-ANN-13 (from Perth, Australia, Escacena del Campo, Spain and both Khemis Miliana and Annaba from Algeria, respectively) were significantly more aggressive, hence dangerous if introduced in other fields. Migration of invasive organisms might lead to selective emergence of adapted isolates in novel geographic regions and on specific host genotypes (Leo et al., 2015). The evolutionary potential of pathogens may be increased and subsequently adapt to overcome host resistances (Linde et al., 2009). Available resistance to D. pinodes is partial and governed by multiple quantitative resistance loci (Rubiales and Fondevilla, 2012). Pathogen aggressiveness could incur a gradual evolution and adaptation that may lead to an “erosion” of resistance, especially if a monoculture farming system is applied (Gandon, 2002).
D. pinodes is a teleomorph of A. pinodes that reproduces asexually by pycnidia containing splash-dispersed pycnospores (Roger and Tivoli, 1996), and sexually by perithecia releasing wind-dispersed ascospores (Tivoli and Banniza, 2007). With the presence of sexual reproduction, new combination of genes could arise in the field, from one growing season to the next (Ali et al., 1994). The existence of pathotypes between D. pinodes isolates is still a matter of concern since there are numerous reports analyzing differential reaction of fungal isolate collection on various hosts leading to ambiguous conclusions (Ali et al., 1978; Zhang et al., 2003; Setti et al., 2009, 2011). Here, despite their large geographical distance (Africa, Australia and Europe), we found a similarity between the host range pattern and the low genetic variability between the D. pinodes isolates used for the study. Both results from D. pinodes host range as well as molecular ITS analysis indicate a lack of pathotypes within the fungal collection used here.
Conclusions
Knowledge of the host range is important to determine whether other crops could be affected. Understanding of population diversity and identification of pathogenic variation within plant species will assist in the management of ascochyta blight diseases.
If common bean is a non-host to D. pinodes as our results suggest, the use of this specie may have positive effect on soil infestation and subsequent disease development. Conversely, the use of grass pea, clover, lentil, oneflower vetch, white lupin might considerably increase the inoculum potential of the soil, having a deleterious effect on the subsequent pea crop. Ascospores produced in pseudothecia on overwintered debris of alternative hosts may serve as important sources of primary inoculum and/or inoculum necessary for secondary infections later in the growing season, as other aschochyta species did (Trapero-Casas et al., 1996; Trapero-Casas and Kaiser, 2009). Infected alternative hosts also may aid in the pathogen's survival from one growing season to the next, as do pea debris and infected seeds (Kaiser, 1990, 1992, 1997).
The use of chickpea, medick, sulla or fenugreek cultivars with qualitative resistance could be considered, but studies on the risk of resistance breakdown are required. As well, it would be important to determine if and which species could act as bridging hosts allowing for the crossing of D. pinodes isolates from one legume with those from another, as demonstrated with Ascochyta spp. by Hernandez-Bello et al. (2006) for A. pisi and A. fabae isolates. This is especially important in light of the plasticity of D. pinodes which is highly adaptable under the influence of biotic and abiotic factors (Le May et al., 2014).
Author contributions
EB designed experiments. Together with MC carried out the experimental work. EB carried out most of the data analysis and contributed to the writing of the manuscript. DR contributed to the interpretation of results and writing of the manuscript. MC also contributed to critical reading.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by Spanish project AGL2014-52871-R. EB was granted by a contract founded by the Spanish JAEdoc program and MC by the Spanish Juan de la Cierva program. Drs. Tivoli B., Boros L., Kiba A., Bauniza B., Bani M., and Lichtenzveig J. are acknowledged for providing D. pinodes isolates described in Table 1.
References
- Ali S. M., Nitschke L. F., Dube A. J., Krause M. R., Cameron B. (1978). Selection of pea lines for resistance to pathotypes of Ascochyta pinodes, Ascochyta pisi and Phoma medicaginis var. pinodella. Aust. J. Agr. Res. 29, 841–849. 10.1071/AR9780841 [DOI] [Google Scholar]
- Ali S. M., Sharma B., Ambrose M. J. (1994). Current status and future strategy in breeding pea to improve resistance to biotic and abiotic stresses. Euphytica 73, 115–126. 10.1007/BF00027188 [DOI] [Google Scholar]
- Bailey K. L., Gossen B. D., Lafond G. R., Watson P. R., Derksen D. A. (2001). Effect of tillage and crop rotation on root and foliar diseases of wheat and pea in Saskatchewan from 1991 to 1998: univariate and multivariate analyses. Can. J. Plant Sci. 81, 789–803. 10.4141/P00-152 [DOI] [Google Scholar]
- Barilli E., Sillero J. C., Moral A., Rubiales D. (2009). Characterization of resistance response of pea (Pisum spp.) against rust (Uromyces pisi). Plant Breed. 128, 665–670. 10.1111/j.1439-0523.2008.01622.x [DOI] [Google Scholar]
- Barilli E., Satovic Z., Sillero J. C., Rubiales D., Torres A. M. (2011). Phylogenetic analysis of Uromyces species infecting grain and forage legumes by sequence analysis of nuclear ribosomal Internal Transcribed Spacer region. J. Phytopathol. 159, 137–145. 10.1111/j.1439-0434.2010.01736.x [DOI] [Google Scholar]
- Bretag T. W. (2004). Review: Ascochyta Blight in Field Peas. Horsham, VIC: Victorian Department of Primary Industries. [Google Scholar]
- Carrillo E., Rubiales D., Pérez-de-Luque A., Fondevilla S. (2013). Characterization of mechanisms of resistance against Didymella pinodes in Pisum spp. Eur. J. Plant Pathol. 135, 761–769. 10.1007/s10658-012-0116-0 [DOI] [Google Scholar]
- Cobos M. J., Rubio J., Strange R. N., Moreno M. T., Gil J., Millan T. (2006). A new QTL for Ascochyta blight resistance in a RIL population derived from an interspecific cross in chickpea. Euphytica 149, 105–111. 10.1007/s10681-005-9058-3 [DOI] [Google Scholar]
- Davidson J. A., Hartley D., Priest M., Krysinska-Kaczmarek M., Herdina H., McKay A., et al. (2007). A new species of Phoma causes Ascochyta blight symtoms on field peas (Pisum sativum) in South Australia. Mycologia 101, 120–128. 10.3852/07-199 [DOI] [PubMed] [Google Scholar]
- FAOSTAT (2015). FAOSTAT, Production, Crops. Food and Agriculture Organization of the United Nations. Available online at: http://faostat.fao.org/site/567/DesktopDefault.aspx?PageID=567#ancor (Accessed 30.09.15).
- Fernández-Aparicio M., Amri M., Kharrat M., Rubiales D. (2010). Intercropping reduces Mycosphaerella pinodes severity and delays upward progress on the pea plant. Crop Prot. 29, 744–750. 10.1016/j.cropro.2010.02.013 [DOI] [Google Scholar]
- Fondevilla S., Avila C. M., Cubero J. L., Rubiales D. (2005). Response to Mycosphaerella pinodes in a germplasm collection of Pisum spp. Plant Breeding 124, 313–315. 10.1111/j.1439-0523.2005.01104.x [DOI] [Google Scholar]
- Fondevilla S., Cubero J. I., Rubiales D. (2007). Inheritance of resistance to Mycosphaerella pinodes in two wild accessions of Pisum. Eur. J. Plant Pathol. 119, 53–58. 10.1007/s10658-007-9146-4 [DOI] [Google Scholar]
- Frenkel O., Sherman A., Abbo S., Shtienberg D. (2008). Different ecological affinities and aggressiveness patterns among Didymella rabiei isolates from sympatric domesticated chickpea and wild Cicer judaicum. Phytopathology 98, 600–608. 10.1094/PHYTO-98-5-0600 [DOI] [PubMed] [Google Scholar]
- Gandon S. (2002). Local adaptation and the geometry of host-parasite coevolution. Ecol. Lett. 5, 246–256. 10.1046/j.1461-0248.2002.00305.x [DOI] [Google Scholar]
- Gossen B. D., Derksen D. A. (2003). Impact of tillage and crop rotation on Aschochyta blight (Ascochyta lentis) of lentil. Can. J. Plant Sci. 83, 411–415. 10.4141/P02-088 [DOI] [Google Scholar]
- Gurung A. M., Pang E. C. K., Taylor P. W. J. (2002). Examination of Pisum and Lathyrus species as sources of Ascochyta blight resistance for field pea (Pisum sativum). Australas. Plant Path. 31, 41–45. 10.1071/AP01069 [DOI] [Google Scholar]
- Hammer Ø., Harper D. A. T., Ryan P. D. (2001). PAST: Paleontological Statistics Software Package for Education and Data Analysis. Palaeontologia Electronica 4. 9. Avaliable online at: http://palaeo-electronica.org/2001_1/past/issue1_01.htm
- Hernandez-Bello M. A., Chilvers M. I., Akamatsu H., Peever T. L. (2006). Host specificity of Ascochyta spp. infecting legumes of the Viciae and Cicerae tribes and pathogenicity of an interspecific hybrid. Phytopathology 96, 1148–1156. 10.1094/PHYTO-96-1148 [DOI] [PubMed] [Google Scholar]
- Jensen E. S., Peoples M. B., Boddey R. M., Gresshoff P. M., Henrik H. N., Alves B. J. R., et al. (2012). Legumes for mitigation of climate change and the provision of feedstock for biofuels and biorefineries. A review. Agron. Sustain. Dev. 32, 329–364. 10.1007/s13593-011-0056-7 [DOI] [Google Scholar]
- Kaiser W. J. (1990). Host range of the Ascochyta blight pathogen of chickpea. Phytopathology 80, 889–890. [Google Scholar]
- Kaiser W. J. (1992). Fungi associated with the seeds of commercial lentils from the U.S. Pacific Northwest. Plant Dis. 76, 605–610. 10.1094/PD-76-0605 [DOI] [Google Scholar]
- Kaiser W. J. (1997). Inter- and intranational spread of aschochyta pathogens of chickpea, faba bean, and lentil. Can. J. Plant Pathol. 19, 215–224. 10.1080/07060669709500556 [DOI] [Google Scholar]
- Kaiser W. J., Wang B. C., Rogers J. D. (1997). Ascochyta fabae and A. lentis: Host specificity, teleomorphs (Didymella), hybrid analysis, and taxonomic status. Plant Dis. 81, 809–816. 10.1094/PDIS.1997.81.7.809 [DOI] [PubMed] [Google Scholar]
- Khan M. S. A., Ramsey M. D., Scott E. S. (1999). Host range studies with an Australian isolate of Ascochyta rabiei. Aus. J. Agric. Res. 28, 170–173. 10.1071/ap99028 [DOI] [Google Scholar]
- Khan T. N., Timmerman-Vaughan G. M., Rubiales D., Warkentin T. D., Siddique K. H. M., Erskine W., et al. (2013). Didymella pinodes and its management in field pea: challenges and opportunities. Field Crops Res. 148, 61–77. 10.1016/j.fcr.2013.04.003 [DOI] [Google Scholar]
- Le May C., Guibert M., Baranger A., Tivoli B. (2014). A wide range of cultivated legume species act as alternative hosts for the pea aschochyta blight fungus, Didymella pinodes. Plant Pathol. 63, 877–887. 10.1111/ppa.12154 [DOI] [Google Scholar]
- Leo A. E., Ford R., Linde C. C. (2015). Genetic homogeneity of a recently introduced pathogen of chickpea, Ascochyta rabiei, to Australia. Biol. Invasions 17, 609–623. 10.1007/s10530-014-0752-8 [DOI] [Google Scholar]
- Linde C. C., Zala M., McDonald B. A. (2009). Molecular evidence for recent founder populations and human-mediated migration in the barley scald pathogen Rhynchosporium secalis. Mol. Phylogenet. Evol. 51, 454–464. 10.1016/j.ympev.2009.03.002 [DOI] [PubMed] [Google Scholar]
- Little T. M., Hills F. J. (1978). Agricultural Experimentation: Design and Analysis. 350 New York, NY: Wiley. [Google Scholar]
- Madrid E., Barilli E., Gil J., Huguet T., Gentzbittel L., Rubiales D. (2014). Detection of partial resistance quantitative trait loci against Didymella pinodes in Medicago truncatula. Mol. Breed. 33, 589–599. 10.1007/s11032-013-9976-z [DOI] [Google Scholar]
- McDonald G. K., Peck D. (2009). Effects of crop rotation, residue retention and sowing time on the incidence and survival of Ascochyta blight and its effect on grain yield of field peas (Pisum sativum L.). Field Crop Res. 111, 11–21. 10.1016/j.fcr.2008.10.001 [DOI] [Google Scholar]
- Moussart A., Even M. N., Tivoli B. (2008). Reaction of genotypes from several species of grain and forage legumes to infection with a French pea isolate of the oomycete Aphanomyces euteiches. Eur. J. Plant Pathol. 122, 321–333. 10.1007/s10658-008-9297-y [DOI] [Google Scholar]
- Muehlbauer F. J., Chen W. (2007). Resistance to Aschochyta blights of cool season food legumes. Eur. J. Plant Pathol. 119, 135–141. 10.1007/s10658-007-9180-2 [DOI] [Google Scholar]
- Nixon K. C. (1999). The Parsimony Ratchet, a new method for rapid parsimony analysis. Cladistics 15, 407–414. 10.1111/j.1096-0031.1999.tb00277.x [DOI] [PubMed] [Google Scholar]
- Omri Benyoussef N., Le May C., Mlayeh O., Kharrat M. (2012). First report of Didymella fabae, teleomorph of Ascochyta fabae, on faba bean crop debris in Tunisia. Phytopathol. Mediterr. 51, 369–373. [Google Scholar]
- Pande S., Siddique K. H. M., Kishore G. K., Bayaa B., Gaur P. M., Gowda C. L. L., et al. (2005). Ascochyta blight of chickpea (Cicer arietinum L.): a review of biology, pathogenicity, and disease management. Aust. J. Agr. Res. 56, 317–332. 10.1071/AR04143 [DOI] [Google Scholar]
- Peever T. L., Barve M. P., Stone L. J. (2007). Evolutionary relationships among Ascochyta species infecting wild and cultivated hosts in the legume tribes Cicereae and Vicieae. Mycologia 99, 59–77. 10.3852/mycologia.99.1.59 [DOI] [PubMed] [Google Scholar]
- Posada D., Crandall K. A. (1998). Modeltest: testing the model of DNA substitution. Bioinformatics 14, 817–818. 10.1093/bioinformatics/14.9.817 [DOI] [PubMed] [Google Scholar]
- Roger C., Tivoli B. (1996). Spatio-temporal development of pycnidia and perithecia and dissemination of spores of Mycosphaerella pinodes on pea (Pisum sativum). Plant Pathol. 45, 518–528. 10.1046/j.1365-3059.1996.d01-139.x [DOI] [Google Scholar]
- Roger C., Tivoli B., Huber L. (1999). Effects of interrupted wet periods and different temperatures on the development of Ascochyta blight caused by Mycosphaerella pinodes on pea (Pisum sativum) seedlings. Plant Pathol. 48, 10–18. 10.1046/j.1365-3059.1999.00311.x [DOI] [Google Scholar]
- Rubiales D., Fondevilla S. (2012). Future prospects for Aschochyta blight resistance breeding in cool season food legumes. Front. Plant Sci. 3:27. 10.3389/fpls.2012.00027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salam M. U., Galloway J., MacLeod W. J., Davidson J. A., Seymour M., Pritchard I., et al. (2011). Blackspot manager model predicts the maturity and release of ascospores in relation to Ascochyta blight on field pea. Australas. Plant Path. 40, 621–631. 10.1007/s13313-011-0035-0 [DOI] [Google Scholar]
- Sattar A. (1934). A comparative study of the fungi associated with blight diseases of certain cultivated leguminous plants. T. Br. Mycol. Soc. 18, 276–301. 10.1016/S0007-1536(34)80013-7 [DOI] [Google Scholar]
- Shapiro S. S., Wilk M. B. (1965). An analysis of variance test for normality (complete samples). Biometrika 52, 591–611. 10.1093/biomet/52.3-4.591 [DOI] [Google Scholar]
- Setti B., Bencheikh M., Henni J., Neema C. (2009). Comparative aggressiveness of Mycosphaerella pinodes on peas from different regions in western Algeria. Phytopathol. Mediterr. 48, 195–204. 10.14601/Phytopathol_Mediterr-2787 [DOI] [Google Scholar]
- Setti B., Bencheikh M., Henni J., Neema C. (2011). Morphological and virulence variation among isolates of Mycosphaerella pinodes the causal agent of pea leaf blight. Afr. J. Agric. Res. 6, 1067–1075. [Google Scholar]
- Siddique K. M., Johansen C., Turner N., Jeuffroy M.-H., Hashem A., Sakar D., et al. (2012). Innovations in agronomy for food legumes. A review. Agron. Sust. Devel. 32, 45–64. 10.1007/s13593-011-0021-5 [DOI] [Google Scholar]
- Sneath P. H. A., Sokal R. R. (1973). Numerical taxonomy: the principles and practice of numerical classification. Syst. Zool. 24, 263–268. [Google Scholar]
- Sprague R. (1929). Host range and life history studies of some leguminous ascochytae. Phytopathology 19, 917–932. [Google Scholar]
- Tamura K., Nei M., Kumar S. (2004). Prospects for inferring very large phylogenies by using the neighbour-joining method. Proc. Natl. Acad. Sci. U.S.A. 101, 11030–11035. 10.1073/pnas.0404206101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor P. W. J., Ford R. (2007). Diagnostics, genetic diversity and pathogenic variation of aschochyta blight of cool season food and feed legumes. Eur. J. Plant Pathol. 119, 127–133. 10.1007/s10658-007-9177-x [DOI] [Google Scholar]
- Tivoli B., Baranger A., Avila C. M., Banniza S., Barbetti M., Chen W., et al. (2006). Screening techniques and sources of resistance to foliar diseases caused by major necrotrophic fungi in grain legumes. Euphytica 147, 223–253. 10.1007/s10681-006-3131-4 [DOI] [Google Scholar]
- Tivoli B., Banniza S. (2007). Comparison of the epidemiology of Ascochyta blights on grain legumes. Eur. J. Plant Pathol. 119, 59–76. 10.1007/s10658-007-9117-9 [DOI] [Google Scholar]
- Tran H. S., You M. P., Khan T. N., Barbetti M. J. (2016). Pea black spot disease complex on field pea: dissecting the roles of the different pathogens in causing epicotyl and root disease. Eur. J. Plant Pathol. 144, 595–605. 10.1007/s10658-015-0798-1 [DOI] [Google Scholar]
- Trapero-Casas A., Kaiser W. J. (1992). Influence of temperature, wetness period, plant age, and inoculum concentration on infection and development of Ascochyta blight of chickpea. Phytopathology 82, 589–596. 10.1094/Phyto-82-589 [DOI] [Google Scholar]
- Trapero-Casas A., Kaiser W. J. (2009). Alternative hosts and plant tissues for the survival, sporulation and spread of the Ascochyta blight pathogen of chickpea. Eur. J. Plant Pathol. 125, 573–587. 10.1007/s10658-009-9507-2 [DOI] [Google Scholar]
- Trapero-Casas A., Navas-Cortés J. A., Jiménez-Díaz R. M. (1996). Airborne ascospores of Didymella rabiei as a major primary inoculum for Ascochyta blight epidemics in chickpea crops in southern Spain. Eur. J. Plant Pathol. 102, 237–245. 10.1007/BF01877962 [DOI] [Google Scholar]
- Weimer J. L. (1947). Resistance of Lathyrus spp. and Pisum spp. to Ascochyta pinodella and Mycosphaerella pinodes. J. Agric. Res. 75, 181–190. [Google Scholar]
- White T. J., Bruns T., Lee S., Taylor J. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, in PCR Protocols: a Guide to Methods and Applications, eds Innis M. A., Gelfand D. H., Sninsky J. J., White T. J. (San Diego, CA: Academic Press; ), 315–322. [Google Scholar]
- Woolhouse M. E. J., Taylor L. H., Haydon D. T. (2001). Population biology of multihost pathogens. Science 292, 1109–1112. 10.1126/science.1059026 [DOI] [PubMed] [Google Scholar]
- Zhang J. X., Fernando W. G. D., Xue A. G. (2003). Virulence and genetic variability among isolates of Mycosphaerella pinodes. Plant Dis. 87, 1376–1383. 10.1094/PDIS.2003.87.11.1376 [DOI] [PubMed] [Google Scholar]