Abstract
This article describes data related to the research article titled “Functional characterization of importin α8 as a classical nuclear localization signal receptor” [1]. A GST pull-down assay showed that both importin α1 and α8, which are classical nuclear localization signal (cNLS) receptors, can form a dimer with importin α6, α7, or α8. Importin α8 has higher dimer-forming ability than importin α1. In addition, our data show that either importin α1 or importin α8 can form a heterodimer with importin α3, which exists in a preformed complex with cNLS substrates such as the conventional SV40TNLS or the p53 protein, resulting in the release of the cNLS substrates from importin α3.
Keywords: Nuclear transport, Importin α, Nuclear localization signal, Dimer
Specifications Table
| Subject area |
Biology |
| More specific subject area | Nucleocytoplasmic transport |
| Type of data | Figure |
| How data was acquired | GST pull-down, western blot |
| Data format | Raw image |
| Experimental factors | Bacterially expressed and purified recombinant proteins |
| Experimental features | Bound proteins precipitated by GST pull-down assay were subjected to SDS-PAGE and detected by immunoblotting |
| Data source location | Osaka, Japan |
| Data accessibility | Data are accessible in this article only |
Value of the data
-
•
These data are valuable to researchers interested in the molecular mechanisms by which the importin α-cNLS substrate complex dissociates in the nucleus.
-
•
These data show that importin α1 and α8 have substantial differences in dimer-forming ability, despite both proteins belonging to the same subfamily.
-
•
These data provide a new insight into the function of nuclear-localized importin αs.
1. Data
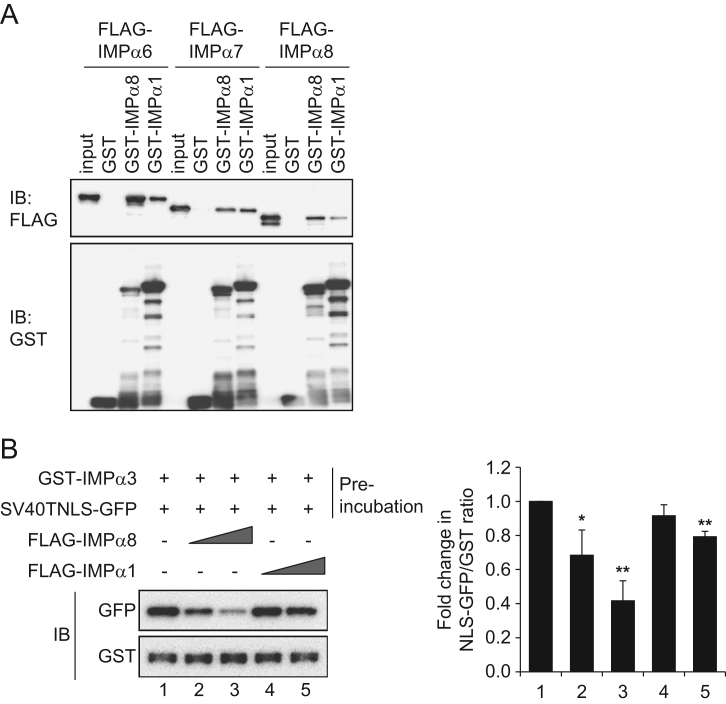
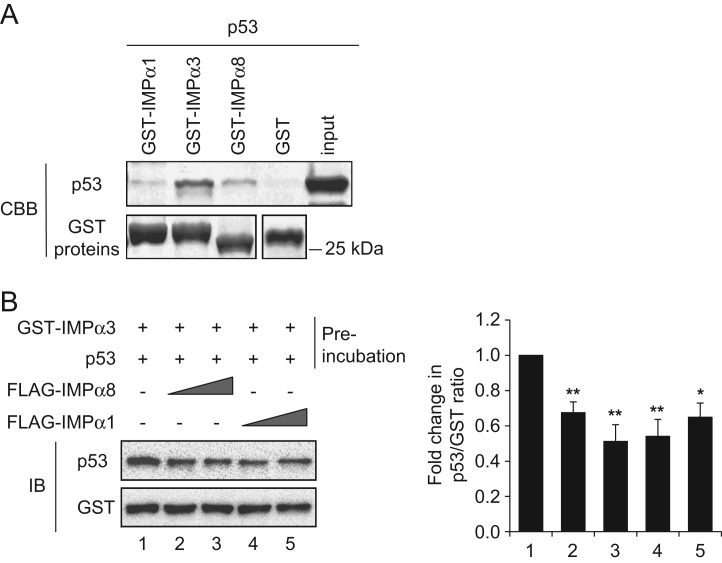
To examine whether importin αs can directly bind with other importin α subtypes, FLAG-tagged importin α6, α7, or α8 recombinant proteins were incubated with either GST-importin α8, or GST-importin α1, and analyzed by western blotting (Fig. 1A). To investigate the effect of heterodimerization of importin αs on its substrate binding, either an importin α3-SV40TNLS complex, or an importin α3-p53 complex was incubated with increasing amounts of FLAG-importin α8, or α1 recombinant proteins, and analyzed by western blotting (Figs. 1B and 2).
Fig. 1.
Western blot analysis indicates that importin α subtypes have the potential to form heterodimers with other importin αs. (A) GST-importin α8 (IMPα8) and GST-importin α1 (IMPα1) were incubated with FLAG-importin α6 (IMPα6), FLAG-importin α7 (IMPα7), or FLAG-importin α8 (IMPα8) recombinant proteins. Bound proteins were detected by anti-FLAG antibody or anti-GST antibody, respectively. FLAG-tagged importin αs (0.625 pmol) were loaded as an input. (B) GST-importin α3 (IMPα3) immobilized on GSH beads was preincubated with SV40TNLS-GFP (Preincubation), and then an equal or 10 times higher amount of FLAG-importin α8 or FLAG-importin α1 was added. Left panels: representative immunoblot (IB) images of the NLS-GFP and GST-importin α3 bands. Right panels: relative fold changes in the NLS-GFP/GST ratio in the presence of either importin α8 or α1, which were normalized to the control condition (without FLAG-importin αs). The results are from three independent experiments and have been presented as mean ± SEM. The numbers 1–5 correspond to the lane numbers in the left panels. **p<0.01, *p<0.05; Student׳s t-test.
Fig. 2.
Either importin α8 or α1 dissociates the importin α3-p53 complex by dimer formation with importin α3. (A) p53 preferentially binds to importin α3 rather than importin α1 or α8. Pull-down assays were performed with the p53 recombinant protein and with GST-importin α1 (IMPα1), GST-importin α3 (IMPα3), or GST-importin α8 (IMPα8) immobilized on GSH beads. After incubation at 4 °C for 1 h, the beads were washed and bound proteins were analyzed by SDS-PAGE. Proteins are detected by Coomassie staining. p53 (10 pmol) was loaded as an input control. (B) GST-importin α3 (IMPα3) immobilized on GSH beads was preincubated with a five times higher amount of p53 to form a complex (Preincubation). Then, increasing amounts of FLAG-importin α8 or FLAG-importin α1 (equal or 10 times higher amount than GST-IMPα3) were added. Bound proteins were analyzed by western blotting using the antibodies indicated. Left panels: representative immunoblot images of the p53 and GST-importin α3 bands. Right panels: relative fold changes in the p53/GST ratio in the presence of either FLAG-importin α8 or FLAG-importin α1, which were normalized to the control condition (without FLAG-importin αs). The results are from three independent experiments and have been presented as mean ± SEM. The numbers 1–5 correspond to the lane numbers described in the left panels. **p<0.01, *p<0.05; Student׳s t-test.
2. Experimental design, materials and methods
2.1. Plasmid construction
The N-terminus FLAG-tagged cDNAs encoding full-length human importin α6 (KPNA5, NM_012316) or full-length human-importin α7 (KPNA6, NM_002269) were amplified from either HEK293 cells or MRK-nu-1 cells (JCRB Cell Bank, Osaka, Japan) by PCR using the following primers: importin α6 Forward: 5′-CCCGAATTCCGCCATGGACTACAAGGACGACGACGACAAGATGGATGCCATGGCTAGTCC-3′ and importin α6 Reverse: 5′-CCCGCGGCCGCCTCGAGTTAAAGTTGAAATCCATCC-3′ or importin α7 Forward: 5′-CCCGAATTCCGCCATGGACTACAAGGACGACGACGACAAGATGGAGACCATGGCGAGC-3′ and importin α7 Reverse: 5′-CCCGCGGCCGCCTCGAGTTATAGCTGGAAGCCCTCC-3′. The PCR program was as follows: 2 min at 94 °C followed by 40 cycles of 30 s at 98 °C and 15 min at 68 °C. The PCR products were digested with EcoRI and NotI, and then subcloned into the pGEX6P3 plasmid (GE Healthcare, Tokyo, Japan). The construct integrity of pGEX6P3/FLAG-h-importin α6 and pGEX6P3/FLAG-h-importin α7 was confirmed by DNA sequencing using the following primers: pGEX 5′ sequencing primer: 5′-GGGCTGGCAAGCCACGTTTGGTG-3′, pGEX 3′ sequencing primer: 5′-CCGGGAGCTGCATGTGTCAGAGG-3′, importin α6 sequencing primer: 5′-GCATCTGGAACTTTTCTGCATACC-3′, or importin α7 sequencing primer: 5′-GTACATTACAGTTTGAAGCTGCCT-3′.
The human-importin α8 (KPNA7, NM_001145715) cDNA with the FLAG-tag at the N-terminus was amplified by PCR from the pcDNA5/3xFLAG-h-importin α8 plasmid [1]. The primers were as follows: importin α8 Forward: 5′-CCCGAATTCCGCCATGGACTACAAGGACGACGACGACAAGATGCCGACCTTAGATGCTCC-3′ and importin α8 Reverse: 5′-CCCGCGGCCGCCTCGAGCTATTTTTTTGCTAAGC-3′. The PCR program was the same as that described above. The PCR products were inserted into EcoRI and NotI sites of pGEX6P3, and then sequenced using either the pGEX 5′ or pGEX 3′ sequencing primer and the importin α8 sequencing primer: 5′-CAACATCGCTTCAGGGACTTCG-3′.
The construct encoding SV40 large T antigen NLS (PPKKKRKVED, pGEX6P2-SV40TNLS-GFP) was subcloned from pGEX2T-SV40TNLS-GFP [2]. The plasmids pGEX6P3/FLAG-human-importin α1 (KPNA2), pGEX6P2-mouse-importin α2 (KPNA2, which we referred to as m-importin α1), and pGEX2T-human-importin α3 (KPNA4, Qip1) were obtained as described previously [1], [3], [4].
The human cDNA encoding the tumor protein p53 (NM_000546) was amplified from MCF7 cells by PCR performed using the following primers: p53 Forward: 5′-CACGGATCCATGGAGGAGCCGCAGTCAGATC-3′ and p53 Reverse: 5′-GGACTCGAGTCAGTCTGAGTCAGGCCCTTCTG-3′. The PCR program was as follows: one cycle of 2 min at 94 °C; 40 cycles of 15 s at 94 °C, 30 sec at 64 °C, and 1 min 20 s at 68 °C; and one cycle of 10 min at 68 °C. The PCR product was subcloned into the BamHI and XhoI sites of pGEX6P1, and then verified by sequencing.
2.2. Recombinant protein purification
Recombinant proteins fused to GST were purified as follows: The expression vectors were transformed into Escherichia coli Rosetta, and then the cells were grown at 37 °C in LB medium containing 50 μg/mL ampicillin. Expression was induced by addition of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), followed by incubation at 20 °C for 12 h. The bacteria were lysed in lysis buffer (50 mM Tris–HCl at pH 8.3, 500 mM NaCl, 1 mM EDTA, 2 mM dithiothreitol (DTT), 0.2 mM phenylmethylsulfonyl fluoride (PMSF), 1 μg/mL aprotinin (Nacalai Tesque, Kyoto, Japan), 1 μg/mL pepstatin (Peptide Institute, Osaka, Japan), and 1 μg/mL leupeptin (Peptide Institute)) by freeze–thawing twice and passing through a French press (model: FA-078, SLM Instruments, Rochester, NY, USA). The cell lysates were sonicated using a Sonifier 250 (Branson, Danbury, CT, USA), and centrifuged at 20,400g at 4 °C for 30 min. The resultant supernatant was incubated with glutathione-Sepharose 4B beads (GSH beads, GE Healthcare, Tokyo, Japan) at 4 °C for 12 h. After the GSH beads were washed five times with lysis buffer, GST-tagged proteins were eluted with elution buffer (20 mM glutathione, 100 mM Tris–HCl at pH 8.3, 100 mM NaCl, 1 mM EDTA, 2 mM DTT, and 1 μg/mL each of aprotinin, pepstatin, and leupeptin). Cleavage of GST from the GST-fused protein was performed using PreScission protease (GE Healthcare, Piscataway, NJ, USA) with 10 units/mg of fusion protein at 4 °C for 12 h in cleavage buffer (50 mM Tri-HCl at pH 7.0, 150 mM NaCl, 1 mM EDTA, 1 mM DTT, and 1 µg/mL each of aprotinin, leupeptin, and pepstatin). Finally, the purified proteins were dialyzed against dialysis buffer (20 mM HEPES at pH 7.3, 110 mM CH3COOK, 2 mM DTT, and 1 μg/mL each of aprotinin, pepstatin, and leupeptin) and concentrated by ultrafiltration using Amicon Ultra centrifugal filter units (Merck Millipore, Tullagreen, Ireland).
2.3. GST pull-down assay
Fig. 1A: Bacterially produced FLAG-h-importin α6, α7, and α8 recombinant proteins (100 pmol each) were incubated with GST, GST-h-importin α8 (KPNA7), or GST-m-importin α1 (KPNA2) immobilized on GSH beads in 200 μL of transport buffer (TB; 20 mM HEPES at pH 7.3, 110 mM potassium acetate, 2 mM magnesium acetate, 1 mM EGTA, 1 mM DTT, 500 μM PMSF, and 1 μg/mL each of aprotinin, pepstatin, and leupeptin) with 0.1% Triton X-100 at 4 °C for 1 h. After washing five times with TB containing 0.1% Triton X-100, the beads were suspended in sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer (50 mM Tris–HCl at pH 6.8, 34.7 mM SDS, 50% glycerol, 25% β-mercaptoethanol, and bromophenol blue). Bound proteins were analyzed by western blotting with specific antibodies described.
Fig. 1B: GST-h-importin α3 (50 pmol) immobilized on GSH beads was incubated with the SV40TNLS substrate (SV40TNLS-GFP, 50 pmol) at 4 °C for 1 h. After washing the beads to remove unbound proteins, either 50 pmol or 500 pmol of FLAG-h-importin α8 or FLAG-h-importin α1 was mixed with the importin α3-SV40TNLS complex at 4 °C for 1 h. The beads were then washed five times with TB containing 0.1% Triton X-100 and suspended in SDS-PAGE loading buffer. Bound proteins were analyzed by western blotting with specific antibodies described.
Fig. 2A: GST-h-importin α1, α3, or α8 (50 pmol each) immobilized on GSH beads were incubated with the p53 protein (50 pmol) in 200 μL TB containing 0.1% Triton X-100 at 4 °C for 1 h. After washing the beads to remove unbound proteins, bound proteins were subjected to 10% SDS-PAGE and stained with Coomassie Brilliant Blue (CBB).
Fig. 2B: GST-h-importin α3 (50 pmol) immobilized on GSH beads was incubated with the p53 protein (250 pmol) in 200 μL TB containing 0.1% Triton X-100 at 4 °C for 1 h. After washing the beads, either 50 pmol or 500 pmol of FLAG-h-importin α8 or FLAG-h-importin α1 was added to the importin α3-p53 complex at 4 °C for 1 h. The beads were then washed five times with TB containing 0.1% Triton X-100, and bound proteins were analyzed by western blotting with the specific antibodies described below.
2.4. Antibodies
The following antibodies were used for western blotting: anti-FLAG M2 antibody (F1804, 0.1 μg/mL, Sigma-Aldrich, St. Louis, MO, USA), anti-GST antibody (sc-138, 0.04 μg/mL, Santa Cruz Biotechnology, Texas, USA), anti-GFP antibody (M048-3, 0.1 μg/mL, MBL, Nagoya, Japan), anti-p53 (FL-393) antibody (sc-6243, 0.04 μg/mL, Santa Cruz Biotechnology), and horseradish peroxidase (HRP)-conjugated anti-mouse or anti-rabbit IgG secondary antibodies (1: 10,000 dilution, Jackson ImmunoResearch Lab., West Grove, PA, USA)
2.5. Western blotting
Samples were loaded on a 10% SDS-PAGE gel, and the separated proteins in the gel were transferred onto an Immobilon-P membrane (PVDF membrane; Merck Millipore, Darmstadt, Germany) using a semi-dry transfer blotting system (Trans-Blot Turbo Transfer System, BioRad Laboratories, Inc., Hercules, CA, USA). The membrane was blocked with blocking buffer consisting of 3% skim milk in Tris-buffered saline (TBS; TAKARA BIO, Shiga, Japan) with 0.05% Tween (TBS-T) for 1 h. The membrane was probed with primary antibodies diluted in Can Get Signal Immunoreaction Enhancer Solution 1 (TOYOBO, Osaka, Japan) at 4 °C overnight, and then incubated with the HRP-conjugated secondary antibody diluted in Can Get Signal Immunoreaction Enhancer Solution 2 (TOYOBO) at room temperature (RT) for 1 h. After the membrane was washed with TBS-T, it was developed with Chemi-Lumi One L or Super (Nacalai Tesque). The intensity of each western blot signal was quantified by Image J software (http://rsbweb.nih.gov/ij/).
Acknowledgments
We thank Hitomi Inoue for the technical support provided for this study. We also thank Yoshinari Yasuda for constructing the pGEX6P1-p53 plasmid. This work was supported in part by JSPS Grant-in-Aid for Scientific Research (C) (No. 15K07068) to Y.M. and Grant-in-Aid for Scientific Research on Innovative Areas (No. 25116008) to M.O.
Footnotes
Supplementary data associated with this article can be found in the online version at 10.1016/j.dib.2016.03.080.
Contributor Information
Yoichi Miyamoto, Email: ymiyamoto@nibiohn.go.jp.
Masahiro Oka, Email: moka@nibiohn.go.jp.
Appendix A. Supplementary material
Supplementary material
References
- 1.Kimoto C., Moriyama T., Tsujii A., Igarashi Y., Obuse C., Miyamoto Y., Oka M., Yoneda Y. Functional characterization of importin α8 as a classical nuclear localization signal receptor. Biochim. Biophys. Acta. 2015;1853:2676–2683. doi: 10.1016/j.bbamcr.2015.07.017. [DOI] [PubMed] [Google Scholar]
- 2.Miyamoto Y., Baker M.A., Whiley P.A., Arjomand A., Ludeman J., Wong C., Jans D.A., Loveland K.L. Towards delineation of a developmental α-importome in the mammalian male germline. Biochim. Biophys. Acta. 2013;1833:731–742. doi: 10.1016/j.bbamcr.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 3.Sekimoto T., Imamoto N., Nakajima K., Hirano T., Yoneda Y. Extracellular signal-dependent nuclear import of Stat1 is mediated by nuclear pore-targeting complex formation with NPI-1, but not Rch1. EMBO J. 1997;16:7067–7077. doi: 10.1093/emboj/16.23.7067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miyamoto Y., Imamoto N., Sekimoto T., Tachibana T., Seki T., Tada S., Enomoto T., Yoneda Y. Differential modes of nuclear localization signal (NLS) recognition by three distinct classes of NLS receptors. J. Biol. Chem. 1997;272:26375–26381. doi: 10.1074/jbc.272.42.26375. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material