Abstract
The disaccharide trehalose is commonly considered to stimulate autophagy. Cell treatment with trehalose could decrease cytosolic aggregates of potentially pathogenic proteins, including mutant huntingtin, α-synuclein, and phosphorylated tau that are associated with neurodegenerative diseases. Here, we demonstrate that trehalose also alters the metabolism of the Alzheimer disease-related amyloid precursor protein (APP). Cell treatment with trehalose decreased the degradation of full-length APP and its C-terminal fragments. Trehalose also reduced the secretion of the amyloid-β peptide. Biochemical and cell biological experiments revealed that trehalose alters the subcellular distribution and decreases the degradation of APP C-terminal fragments in endolysosomal compartments. Trehalose also led to strong accumulation of the autophagic marker proteins LC3-II and p62, and decreased the proteolytic activation of the lysosomal hydrolase cathepsin D. The combined data indicate that trehalose decreases the lysosomal metabolism of APP by altering its endocytic vesicular transport.
Keywords: amyloid precursor protein (APP), amyloid-β (Aβ), autophagy, endosome, intracellular trafficking, lysosome, trehalose
Introduction
Alzheimer disease is characterized by the accumulation of extracellular amyloid plaques that contain aggregates of the amyloid β-peptide (Aβ).5 Aβ is derived from the amyloid precursor protein by sequential proteolytic processing by β- and γ-secretases (1, 2). The initial cleavage of amyloid precursor protein (APP) by β-secretase results in the secretion of the soluble ectodomain and the generation of a membrane-tethered C-terminal fragment (APP-CTFβ). Subsequently, APP-CTFβ can be cleaved by γ-secretase to liberate Aβ from cellular membranes (2). In an alternative pathway, APP can also be cleaved by α-secretase within the Aβ domain, thereby generating CTFα. The subsequent cleavage of APP-CTFα by γ-secretase results in secretion of a small peptide called p3 (2). It is important to note that APP is also metabolized by additional pathways, including proteasomal and lysosomal degradation (3–5).
The metabolism of APP is also regulated by macroautophagy (herein referred to as autophagy), a lysosome-dependent degradative pathway for long lived proteins, organelles, and nutrient recycling (6, 7). Autophagy starts with the formation of double-membrane vesicles, the so-called autophagosomes, which further fuse with lysosomes to become autolysosomes for degradation of autophagosome contents by lysosomal hydrolases (8). Autophagy is an essential prosurvival pathway induced by a variety of stress factors, including nutrient deprivation, growth factor withdrawal, oxidative stress, infection, and hypoxia (9). These factors contribute to the etiology of multiple diseases such as cancer, stroke, heart disease, and infection (10). Eukaryotic cells have a basal autophagic activity under normal physiological conditions. Cells deficient in autophagy show diffuse abnormal protein accumulation and mitochondrial disorganization (11, 12), suggesting that autophagy is important to maintain cellular homeostasis by eliminating protein aggregates and damaged organelles. Basal autophagy is particularly important and active in the liver and non-dividing cells such as neurons and myocytes (11, 13, 14). Dysfunction in autophagy is implicated in the pathogenesis of many human diseases, including different types of neurodegeneration (15, 16). Incompletely degraded autophagic vacuoles contain both APP and γ-secretase complex and could thus contribute to enhanced processing of APP into toxic Aβ (17–19).
Trehalose is a natural non-reducing disaccharide containing two d-glucose residues connected via an α,α-1,1 linkage. It functions not only as an energy source but is also used to protect cells against heat, cold, oxidation, or dehydration (20). Trehalose is considered to enhance autophagic activity (21). It has been shown to promote the cellular clearance of pathogenic proteins like mutant huntingtin, α-synuclein (21, 22), and phosphorylated tau (22–24) that are associated with Huntington, Parkinson, and Alzheimer diseases, respectively. However, the role of trehalose in the cellular metabolism of the Alzheimer disease-associated APP remains to be investigated.
In this study, we analyzed the role of trehalose in the cellular metabolism of APP. Interestingly, trehalose strongly decreased the degradation of APP and its CTFs. By biochemical and cell biological approaches, we demonstrate that trehalose interferes with endocytic vesicular trafficking as indicated by decreased processing of cathepsin D (Cat D), redistribution of lysosome-associated protein (LAMP)2, and accumulation of the autophagy-related proteins microtubule-associated protein 1 light chain 3 (LC3) and p62. The combined data demonstrate that trehalose exerts complex effects on vesicular trafficking, thereby decreasing the metabolism of APP and the secretion of Aβ.
Experimental Procedures
Antibodies, Constructs, and Reagents
APP-CT (C1/6.1) antibody has been described (25). Human cathepsin D-CT antibody was a generous gift from Dr. S. Höning (University of Cologne, Cologne, Germany). Anti-LAMP2 antibody was purchased from the Iowa Hybridoma Bank. Other antibodies were purchased from the indicated companies: anti-EEA1 and anti-LC3 (MBL International Corp.; Nanotools); anti-beclin-1, anti-mechanistic target of rapamycin (mTOR), and anti-phospho-mTOR (Ser-2448) antibodies (Cell Signaling Technology); 6E10 antibody (Biolegend); anti-Rab9 and anti-calnexin (Santa Cruz Biotechnology); anti-Rab7 (Abcam); anti-tau (BD Biosciences); anti-α-synuclein (Rockland); anti-β-actin, anti-TGN46, and anti-mouse, anti-rabbit, and anti-goat IgG-peroxidase (Sigma-Aldrich); and anti-mouse, anti-rabbit, and anti-goat Alexa Flour 488 and 546 antibodies (Invitrogen).
The mCherry-GFP-LC3 construct has been described previously (26) and was generously provided by Dr. Jörg Höhfeld (University of Bonn, Bonn, Germany). Trehalose, maltose, glucose, and 4′,6-diamidino-2-phenyindole (DAPI) were obtained from Sigma-Aldrich. Bafilomycin A1 and chloroquine were purchased from Enzo Life Sciences. Cycloheximide was obtained from Sigma-Aldrich. DMEM, Earle's balanced salt solution (EBSS), fetal calf serum (FCS), penicillin, streptomycin, ampicillin, and puromycin were purchased from Invitrogen.
Cell Lines and Cell Culture
Human neuroglioma H4, human neuroblastoma SH-SY5Y, human embryonic kidney 293 (HEK-293), and human hepatocellular carcinoma cells (Hep-G2) were obtained from ATCC. Mouse embryonic fibroblasts wild type (MEF-WT) and presenilin 1/2 double knock-out (MEF-PSdKO) were a generous gift from Dr. De Strooper (Vlaams Instituut voor Biotechnologie, Flanders, Belgium) and were described previously (27). MEF-WT and MEF-Atg5KO were obtained from the RIKEN Cell Bank.
Cells were cultured in DMEM supplemented with 10% FCS and 1% penicillin/streptomycin. H4-mCherry-GFP-LC3 stable clones were generated from transfection of H4 cells with mCherry-GFP-LC3 constructs. The protocol was performed with Lipofectamine (Invitrogen) according to the manufacturer's instructions. Clones were selected with 25 μg/ml puromycin.
Immunocytochemistry
Cells were processed for immunocytochemistry as described previously (28). Briefly, cells were cultured on poly-l-lysine-coated coverslips and fixed with 4% paraformaldehyde for 20 min. Subsequently, cells were kept in blocking solution (90% PBS, 10% FCS, 0.1% Triton X-100) at room temperature for 1 h, incubated with primary antibodies at 4 °C overnight or at room temperature for 1–2 h, and then incubated with secondary antibodies for 1 h at room temperature. Cells were imaged with a Carl Zeiss Axio Imager 2 ApoTome fluorescence microscope for optical sectioning. Signal intensity and co-localization were analyzed with AxioVision software. For quantification, randomly selected images (n = 10) were used. Signals were quantified in two randomly chosen boxes (150 μm2) per cell. Values represent means ± standard deviation (S.D.).
Cell Lysis, Isolation of Cellular Membranes, and Western Immunoblotting
For lysis, cells were incubated in radioimmune precipitation assay buffer (150 mm NaCl, 10 mm Tris, pH 8.0, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 5 mm EDTA) supplemented with 1% phosphatase inhibitor and 1% proteinase inhibitor (Roche Applied Science) for 15 min. Particulate materials were removed by centrifugation at 13,200 rpm for 15 min, and supernatants were used for further analyses.
To isolate cellular membranes, cells were incubated in hypotonic buffer D (10 mm Tris-HCl, pH 7.5, 10 mm NaCl, 0.1 mm EGTA, 25 mm glycerol 2-phosphate, 1 mm DTT) supplemented with 1% proteinase inhibitor on ice for 10 min. Cells were then homogenized using 21-gauge needles and centrifuged at 1000 rpm for 10 min to pellet nuclei. The supernatant was then centrifuged at 13,200 rpm for 30 min to obtain the membrane fraction as a pellet.
Membrane fractions or cellular lysates were separated by sodium dodecylsulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes (Schleicher & Schüll). Membranes were blocked in 5% nonfat milk for 1 h and incubated with primary antibodies overnight and appropriate secondary antibodies for 1 h. Proteins were detected by enhanced chemiluminescence (ECL) using ECL reagent (GE Healthcare) and an ECL imaging station (ChemiDoc XRS, Bio-Rad). Quantification of signals was done by the Quantity One software package (Bio-Rad).
Subcellular Fractionation
For subcellular fractionation, cells were subjected to a hypotonic shock as described above. Postnuclear fractions (3800 rpm, 5 min) were loaded onto a discontinuous gradient (30, 20, 17.5, 15, 12.5, 10, 7.5, 5, and 2.5% OptiPrepTM; Sigma Aldrich; 1.2 ml each) and separated at 100,000 × g at 4 °C for 8 h without using the brake. Single fractions were collected at a volume of 1 ml, and proteins were precipitated with trichloroacetic acid.
Measurement of Aβ Variants
Cells were grown on 6-well culture plates until 70% confluence in DMEM as described above. For collection of Aβ, 750 μl of fresh medium was added overnight. Conditioned media were cleared by centrifugation. Cells were briefly washed and lysed in radioimmune precipitation assay buffer. Both cell lysates and conditioned media were analyzed by electrochemiluminescence technology (MesoScale Discovery) for Aβ38, Aβ40, and Aβ42 according to the manufacturer's protocol.
Metabolic Radiolabeling and Pulse-Chase Experiments
Cells were grown in Petri dishes until 80% confluence. Cells were washed with PBS and incubated in serum and methionine-free medium for 30 min. Cells were then incubated with 20 μCi of 35S-radiolabeled methionine/cysteine for 30 min to pulse label newly synthesized proteins. After pulse labeling, cells were washed with label free-medium containing 10% serum and a 5-fold excess of unlabeled methionine and chased for the indicated time periods. Cells were then lysed, and proteins were separated by SDS-PAGE. After transfer to PVDF membrane, radiolabeled proteins were quantified by phosphorimaging.
In Vitro γ-Secretase Assay
The in vitro γ-secretase assay was carried out as described previously (18, 29). Briefly, cells were lysed in hypotonic buffer D, and membranes were isolated as described above. The membrane pellet was then resuspended in citrate buffer (150 mm sodium citrate, distilled H2O, pH 6.4 adjusted with citric acid) supplemented with 1% protease/phosphatase inhibitors and incubated in the absence or presence of γ-secretase modulators at 37 °C for 2 h. Samples were centrifuged at 13,200 rpm for 1 h. The resulting pellets and supernatants were separated by SDS-PAGE, and proteins were detected by Western immunoblotting.
Data Analysis and Statistics
Statistical analyses were carried out by calculation of S.D. and Student's t test. Significance is indicated by asterisks as follows: * for p < 0.05, ** for p < 0.01, *** for p < 0.001, and n.s. for not significant.
Results
Trehalose Impairs the Metabolism of APP and Decreases the Secretion of Aβ
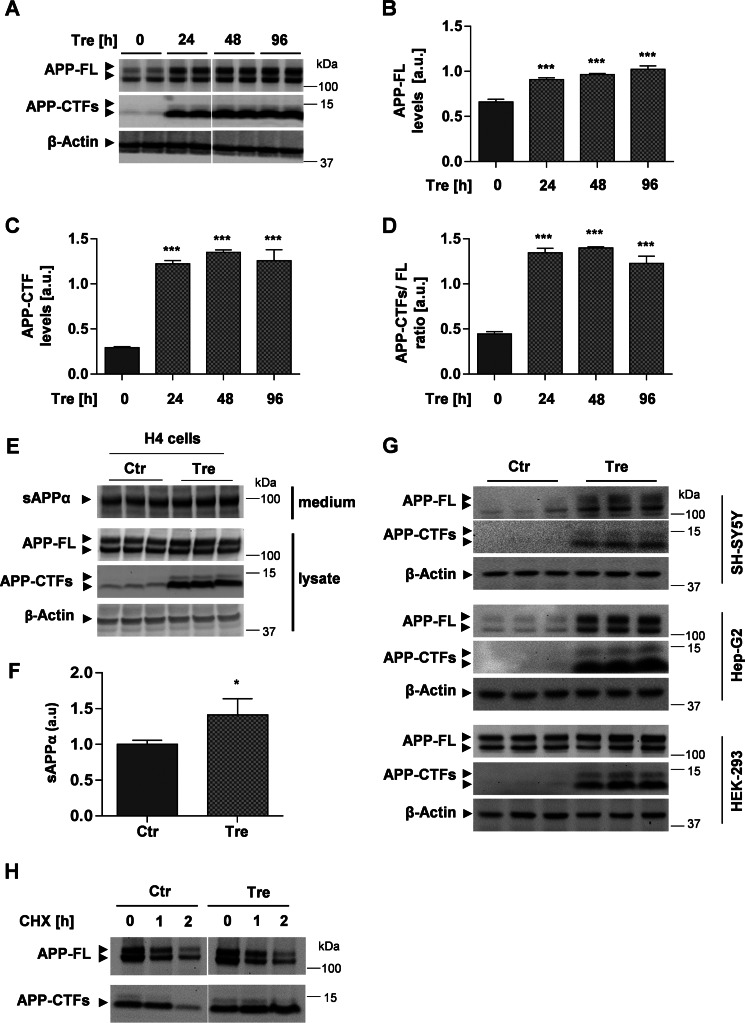
The degradation of APP and its CTFs involves autophagic and lysosomal pathways (17–19, 25, 26). To assess the effect of trehalose on APP metabolism, human neuroglioma H4 cells that endogenously express APP were incubated with trehalose for different time periods, and APP levels were analyzed by Western immunoblotting. Cell treatment with trehalose strongly increased levels of APP full length (FL) (Fig. 1, A and B) and, even more pronounced, of APP-CTFs (Fig. 1, A, C, and D). Trehalose also significantly increased the secretion of soluble APP derived from α-secretory processing into conditioned media (Fig. 1, E and F), suggesting that the increased levels of cellular APP-FL were not caused by inhibition of APP secretion.
FIGURE 1.
Trehalose induces accumulation of APP and APP-CTFs. A, Western immunoblot of APP and APP-CTFs. H4 cells were incubated in the presence or absence of 100 mm trehalose (Tre) for the indicated time periods. Cell lysates were subjected to SDS-PAGE and Western immunoblotting. β-Actin was used as a loading control. B–D, quantification of APP-FL (B), APP-CTFs (C), and the ratio APP-CTFs/FL (D) by ECL imaging. Values represent means ± S.D. of six biological replicates. E, H4 cells were incubated in the presence or absence of 100 mm trehalose (Tre) for 24 h. Soluble APPα in conditioned media and APP-FL and APP-CTFs in cell lysates were analyzed by Western immunoblotting. F, quantification of soluble APPα in media by ECL imaging. Values represent means ± S.D. of three biological replicates. G, the indicated cell types (SH-SY5Y, Hep-G2, and HEK-293) were incubated in the presence or absence of 100 mm trehalose for 24 h. Cells were lysed, and proteins were detected by Western immunoblotting. H, H4 cells were incubated in the presence or absence of 100 mm trehalose (Tre) for 2 h. After addition of cycloheximide (CHX; 20 μg/ml), cells were further incubated for the indicated time periods. Cellular membranes were isolated, and APP-FL and APP-CTFs detected by Western immunoblotting. Error bars represent S.D. *, p < 0.05; ***, p < 0.001. a.u., arbitrary units; sAPP, soluble APP; Ctr, control.
To test whether trehalose exerts similar effects in other cell types, we also used human neuroblastoma SH-SY5Y, liver Hep-G2, and embryonic kidney 293 cells. Trehalose induced accumulation of APP-FL and particularly of APP-CTFs in all tested cell lines (Fig. 1G), very similar to the effects observed in human H4 cells.
We next wanted to test whether the trehalose-dependent increase in APP-FL and APP-CTFs is due to altered degradation. To inhibit de novo protein biosynthesis, H4 cells were incubated with cycloheximide and chased in the absence or presence of trehalose. In the absence of trehalose, levels of APP-FL and its CTFs declined within 2 h after block of de novo protein synthesis (Fig. 1F). In the presence of trehalose, levels of APP-FL also declined. Notably, levels of APP-CTFs were stabilized and even increased after 1 and 2 h of cycloheximide treatment in the presence of trehalose (Fig. 1H). The increase in APP-CTFs after block of protein biosynthesis could be explained by the ongoing processing of APP-FL by α- and β-secretases. These data indicate that trehalose inhibits the degradation of APP-CTFs without inhibition of secretory processing of APP-FL.
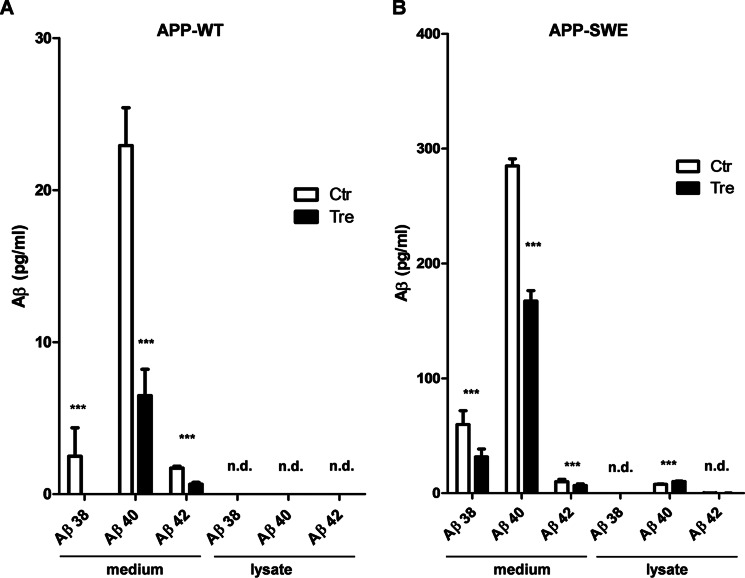
Levels of Aβ derived from endogenous APP in the different cell lines were below the detection limit (not shown). Thus, we used previously described fibroblasts that express either APP-WT or the familial Alzheimer disease-associated Swedish mutant variant of APP (APP-SW) (30). As expected, levels of Aβ in conditioned media from cells expressing the APP-SW variant were much higher than that in media from APP-WT-expressing cells (Fig. 2, A and B). Incubation of cells with trehalose reduced the secretion of the different length variants Aβ38, Aβ40, and Aβ42 from both APP-WT- and APP-SW-expressing cells (Fig. 2, A and B). Levels of cell-associated or intracellular Aβ were very low. Only Aβ40 could be detected in lysates of APP-SW-expressing cells. Here, levels were slightly increased upon cell treatment with trehalose (Fig. 2B). These data demonstrate that trehalose decreases the secretion of Aβ and impairs the degradation of APP-CTFs.
FIGURE 2.
Trehalose decreases the secretion of Aβ. Mouse embryonic fibroblast stably expressing APP-WT (A) or APP-SW (B) were incubated for 24 h in the absence (Ctr, control) or presence of trehalose (Tre). Levels of extracellular (medium) and intracellular (lysate) Aβ variants were determined by electrochemiluminescence. n.d., not detectable. Values represent means of three biological replicates. Error bars represent S.D. ***, p < 0.001.
Trehalose Does Not Inhibit γ-Secretase Activity or Stimulate mTOR-dependent Autophagy
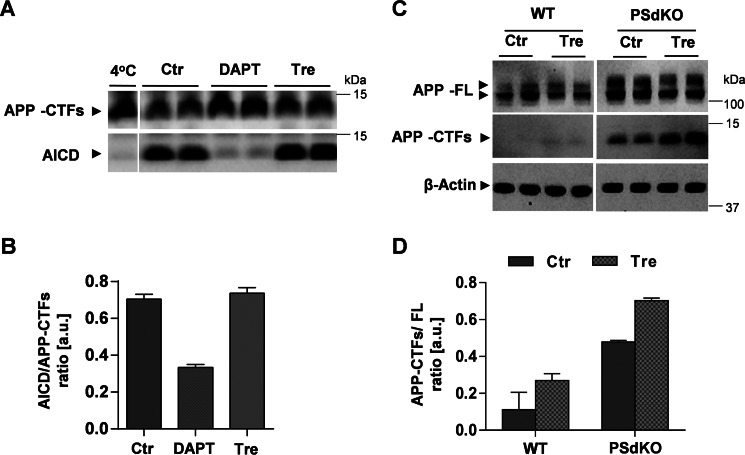
γ-Secretase is critically involved in the cleavage of APP-CTFs and Aβ generation. Thus, we next tested the potential effect of trehalose on γ-secretase activity. Purified membranes of H4 cells were incubated in the presence or absence of trehalose, and γ-secretase activity was monitored by the detection of the APP intracellular domain (AICD) released into the supernatant. In control samples, AICD was readily detected, indicating efficient cleavage of APP-CFTs by γ-secretase (Fig. 3, A and B). As expected, pharmacological inhibition of γ-secretase by the specific inhibitor N-[N-(3,5-difluorophenacetyl)-l-alanyl]-S-phenylglycine t-butyl ester efficiently reduced the generation of AICD (Fig. 3, A and B). However, trehalose did not decrease the formation of AICD (Fig. 3, A and B), demonstrating that trehalose does not inhibit γ-secretase activity. To further test whether trehalose induced accumulation of APP-CTFs independently of γ-secretase inhibition, we next used fibroblasts from WT and PSdKO mice. PSdKO cells lack both catalytically active variants of presenilins and thus have no γ-secretase activity. Accordingly, basal levels of APP-CTFs were higher in PSdKO than in WT cells (Fig. 3, C and D). Interestingly, trehalose further increased APP-CTF levels in PSdKO cells (Fig. 3, C and D), indicating that the observed accumulation of APP-CTFs upon cell treatment with trehalose is not caused by inhibition of γ-secretase activity. The combined data demonstrate that trehalose decreases the degradation of APP-CTFs without inhibiting γ-secretase.
FIGURE 3.
Trehalose impairs degradation of APP CTFs without inhibition of γ-secretase activity. A, γ-secretase in vitro assay. Isolated membranes of H4 cells were incubated in citrate buffer supplemented with either 10 μm N-[N-(3,5-difluorophenacetyl)-l-alanyl]-S-phenylglycine t-butyl ester (DAPT) or 100 mm trehalose (Tre) at 37 °C for 2 h and then centrifuged to separate pellets and supernatants. Proteins of the pellet and supernatant fractions were separated by SDS-PAGE. Subsequently, APP-CTFs and AICD were detected by immunoblotting in pellet and supernatant fractions, respectively. B, quantification of the AICD/APP-CTF ratio from the blot (A) (n = 2). C, immunoblotting of APP in lysates of WT and PSdKO MEFs after incubation in the absence or presence of 100 mm trehalose for 24 h. D, quantification of the APP-CTF/APP-FL ratio from C. Values represent means of duplicate experiments. Error bars represent S.D. a.u., arbitrary units; Ctr, control.
We next analyzed the effects of trehalose on LC3, which is widely used as a marker for autophagosomes. Upon induction of autophagy, LC3 is converted from a cytosolic form (LC3-I) to a phosphatidylethanolamine-conjugated form (LC3-II) and thereby recruited to membranes of autophagosomes (31). Consistent with previous findings (21, 22), trehalose induced a strong increase of LC3-II levels over time (Fig. 4A), resulting in elevated LC3-II/LC3-I ratios (Fig. 4B). However, this result would indicate that trehalose either increases autophagosome formation and/or decreases autophagic flux. To further verify the effects of trehalose on autophagy, we assessed p62 levels by immunoblotting. p62 binds directly to LC3 and is also degraded during autophagy (32). Consistent with increased LC3-II, we detected a significant increase of p62 upon treatment with trehalose (Fig. 4, A and C). Thus, the accumulation of p62 supports a decreased clearance capacity of trehalose-treated cells. Interestingly, immunocytochemistry revealed that accumulated LC3 partially co-localized with APP-positive vesicles upon trehalose treatment, whereas both proteins were segregated in control cells (Fig. 4, D–F).
FIGURE 4.
Trehalose increases levels of autophagy markers. A, Western immunoblotting analyses of the autophagosomal marker proteins LC3 and p62 in H4 cells treated with 100 mm trehalose (Tre) for indicated time periods. B and C, quantification of the LC3-II/LC3-I ratio (B) and p62 (C) was performed by ECL imaging. Values represent means ± S.D. of triplicates. D, immunocytochemical analyses of LC3 and APP in control and trehalose-treated cells. Cells were co-stained with anti-LC3 and anti-APP C-terminus-specific primary antibodies and Alexa Fluor 546- and Alexa Fluor 488-coupled secondary antibodies, respectively. Scale bar, 10 μm. E, the co-localization of APP and LC3 in control and trehalose-treated cells was estimated through the Pearson's correlation coefficient by using GraphPad Prism software. F, quantification of LC3/APP puncta in control and trehalose-treated cells (n = 20). Error bars represent S.D. ***, p < 0.001. a.u., arbitrary units; Ctr, control.
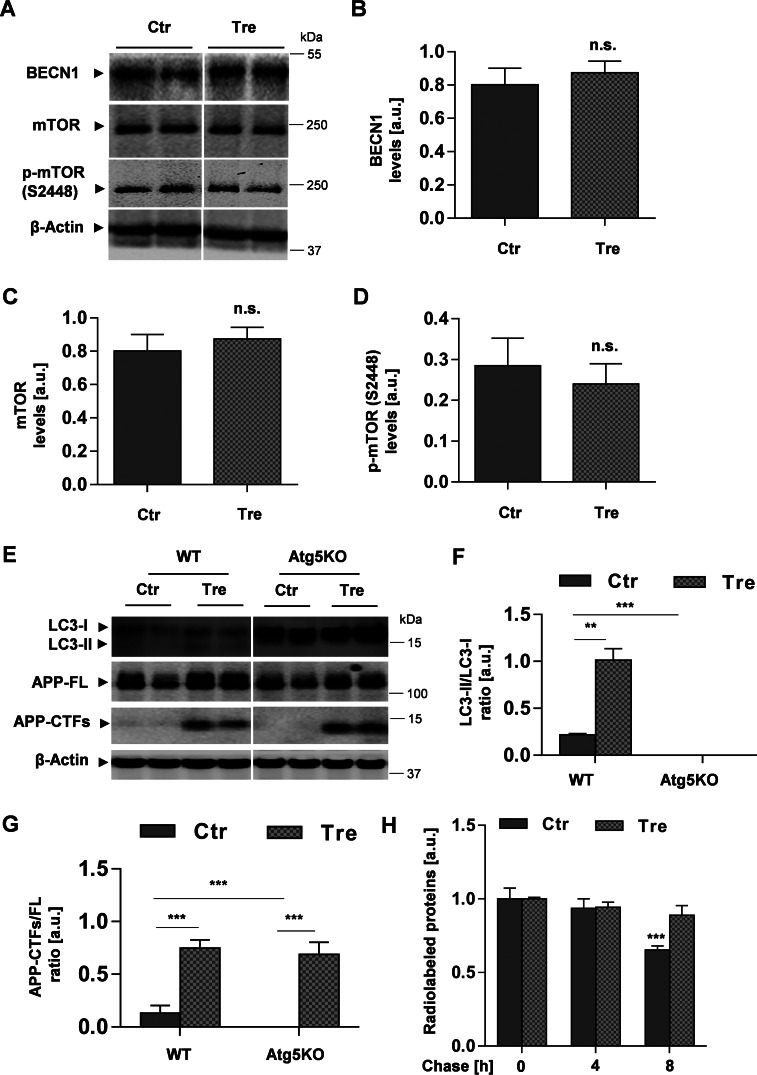
Although trehalose is commonly used to modulate autophagic activity, very little is known regarding how it regulates the autophagic machinery. We next analyzed proteins involved in the induction of autophagy, including the mTOR found in mTOR complexes and beclin-1 (BECN1), a major regulator protein of the class III phosphatidylinositol 3-kinase (PI3K) (8, 33). Western blotting analyses revealed no significant changes in the levels of BECN1 (Fig. 5, A and B), total mTOR, and phosphorylated mTOR (Ser-2448) (Fig. 5, A, C, and D) upon cell treatment with trehalose, suggesting that trehalose did not affect these proteins involved in the induction of autophagy. The results are consistent with previous findings showing that effects of trehalose are independent of mTOR activity (21).
FIGURE 5.
Effects of trehalose are independent of mTOR and Atg5. A, Western immunoblot of BECN1 and mTOR from lysates of H4 cells after cell incubation with or without trehalose (Tre) for 24 h. B–D, quantification of BECN1 (B), mTOR (C), and phosphorylated mTOR (p-mTOR) (Ser-2448) (D) by ECL imaging. E, Western immunoblot of LC3 and APP from lysates of MEF-WT and MEF-Atg5KO. F and G, quantification of the ratios LC3-II/LC3-I (F) and APP-CTFs/FL (G). Values represent means of triplicate experiments. H, trehalose reduces the degradation of long lived proteins. H4 cells were pulse-labeled with [35S]methionine/cysteine and then chased in the presence or absence of 100 mm trehalose for the indicated time periods as described under “Experimental Procedures.” Values represent means of triplicate experiments; p < 0.001. Error bars represent S.D. **, p < 0.01; ***, p < 0.001; n.s., not significant. a.u., arbitrary units; Ctr, control.
To further analyze the role of trehalose in the autophagic process and the metabolism of APP, we used Atg5KO cells. Atg5 plays a role at early steps of autophagosome formation and is required for the lipidation of LC3 (31). Accordingly, cells deficient for Atg5 are defective in the formation of autophagosomes (34). Fibroblasts of WT and Atg5-KO mice were treated with trehalose, and LC3 and APP were analyzed by Western immunoblotting. In WT cells, trehalose induced an increase in LC3-II, resulting in an increased LC3-II/LC3-I ratio (Fig. 5, E and F). In contrast, LC3-II was not detected in Atg5 KO cells. Due to defective conversion of LC3-I to LC3-II, LC3-I levels were strongly elevated in Atg5-KO cells (Fig. 5, E and F). Interestingly, the treatment of Atg5KO cells with trehalose still led to accumulation of APP-FL and particularly of its CTFs (Fig. 5, E and G). The response of Atg5KO cells was very similar to that of WT cells, indicating that the effects of trehalose on APP metabolism are independent of Atg5-dependent induction of autophagy. Together with the unchanged levels of BECN1 and mTOR phosphorylation (Ser-2448) in trehalose-treated cells, increases in LC3-II, APP-CTFs, and p62 strongly suggest alterations of vesicular transport and/or autophagic flux upon cell treatment with trehalose. Trehalose also decreased the degradation of long lived proteins (Fig. 5H), further indicating an impairment of autophagic flux.
Inhibition of APP-CTF Degradation Is Selective for the Disaccharide Trehalose
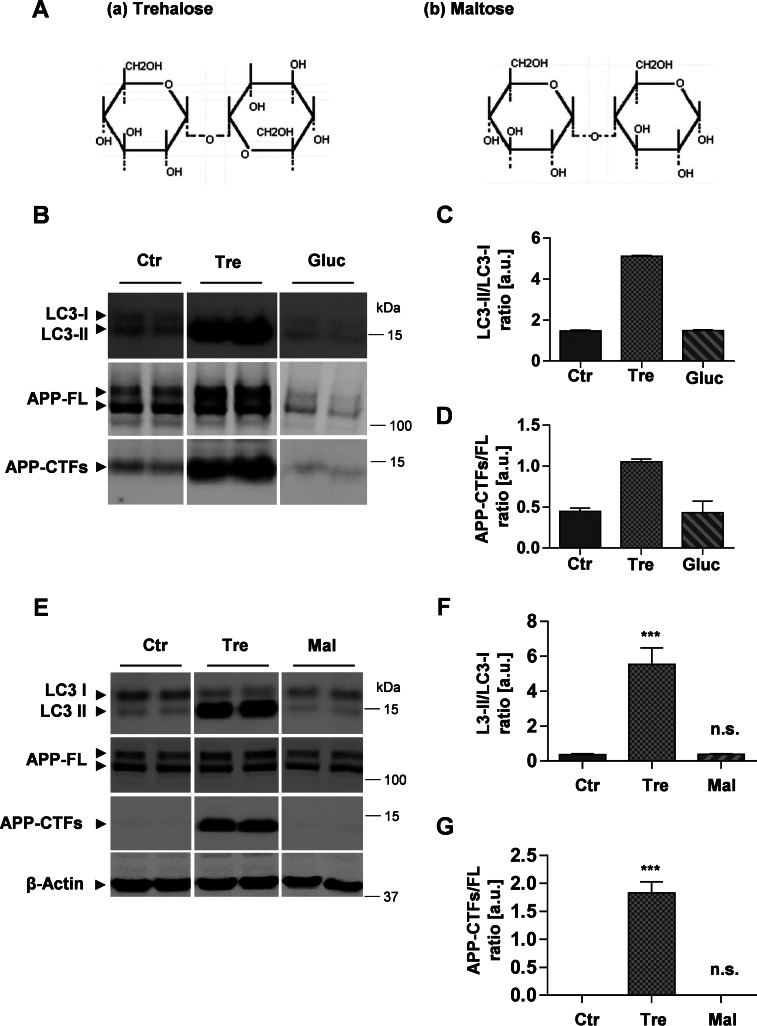
The intracellular pathways by which trehalose is metabolized and regulates cell signaling are still unknown. Because trehalose is a disaccharide composed of two d-glucose residues, we tested whether supplementation of glucose induces similar effects on the metabolism of APP and LC3. H4 cells were incubated with trehalose or additional glucose in the culture media. As observed before, trehalose treatment led to strong accumulation of LC3-II, APP-FL, and APP-CTFs. In contrast, glucose treatment did not change the ratio of LC3-II/LC3-I (Fig. 6, B and C). Similarly, glucose also did not alter the ratio of APP-CTFs/APP-FL (Fig. 6, B and D). Together, the data indicate that the effects of trehalose on autophagy-lysosomal function did not involve increased generation of glucose.
FIGURE 6.
Selective effect of trehalose on the degradation of LC3 and APP-CTFs. A, chemical structures of trehalose, characterized by an α,α-1,1 glycosidic linkage between two glucose (Gluc) residues (a), and maltose, characterized by an α,α-1,4 linkage between two glucose residues (b) (54). B, Western immunoblot of LC3 and APP from H4 cellular lysates after treatment in the absence or presence of 100 mm trehalose (Tre) or 150 mm glucose (Gluc) for 24 h. C and D, quantification of the ratios LC3-II/LC3-I (C) and APP-CTFs/APP-FL (D). E, Western immunoblot of LC3 and APP from H4 cellular lysates after cell incubation in the absence or presence of 100 mm trehalose or 100 mm maltose for 24 h. F and G, quantification of LC3-II/LC3-I (F) and APP-CTF/APP-FL (G) ratios. Values represent means of triplicate experiments. Error bars represent S.D. ***, p < 0.001; n.s., not significant. a.u., arbitrary units; Ctr, control.
Maltose is a disaccharide similar to trehalose that is also composed of two glucose molecules. The two disaccharides only differ in the type of glycosidic linkage between the two glucose residues. Although the glucose residues in maltose are coupled by an α,α-1,4 linkage, they are coupled by an α,α-1,1 glycosidic linkage in trehalose (35) (Fig. 6A). Therefore, we tested whether trehalose-induced effects could be mimicked by the disaccharide maltose. However, in contrast to trehalose, maltose did not affect the levels of LC3 and APP (Fig. 6, E–G). These findings indicate that the inhibition of LC3 and APP degradation is specific for the disaccharide trehalose.
Trehalose Alters the Subcellular Localization and Degradation of APP-CTFs
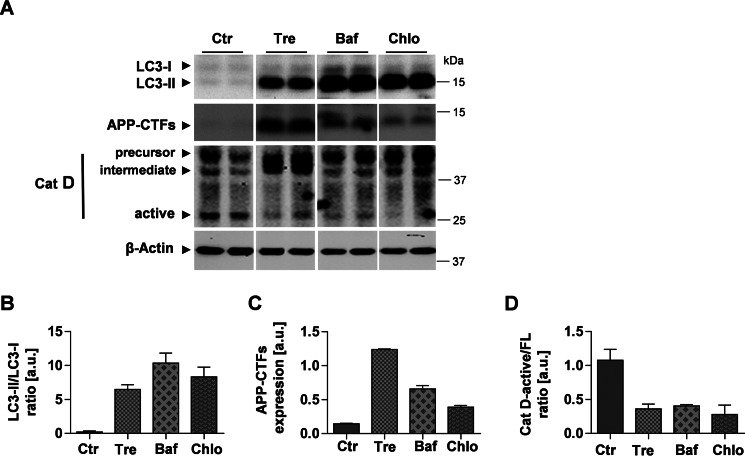
The present data strongly suggest an inhibitory effect of trehalose on APP metabolism in endolysosomal compartments. To test whether direct inhibition of lysosomal degradation could mimic the effects of trehalose on LC3 and APP, H4 cells were treated with trehalose or different lysosomal inhibitors including bafilomycin A1 and chloroquine. Bafilomycin A1 is a specific inhibitor of the vacuolar-type H+-ATPase, which inhibits acidification and therefore protein degradation in lysosomes (36). Chloroquine is a lysosomotropic compound that also elevates/neutralizes the intraluminal pH of endolysosomal compartments (37). Both bafilomycin A1 and chloroquine led to accumulation of LC3-II and APP-CTFs (Fig. 7, A–C). However, the effects of bafilomycin A1 and chloroquine differed quantitatively from that of trehalose. Although LC3-II showed strongest accumulation upon direct inhibition of lysosomal activity with bafilomycin A1 or chloroquine, APP-CTF showed strongest accumulation upon cell treatment with trehalose (Fig. 7, A–C).
FIGURE 7.
Impairment of cathepsin D processing upon trehalose treatment or inhibition of lysosomal acidification. A, H4 cells were incubated in the absence or presence of 100 mm trehalose (Tre) for 24 h, 50 nm bafilomycin A1 (Baf) for 12 h, or 50 μm chloroquine (Chlo) for 12 h. Western immunoblotting detection of LC3, APP, and Cat D is shown. B and D, quantification of LC3 (B), APP (C), and Cat D (D). Values represent means of duplicate experiments. Error bars represent S.D. a.u., arbitrary units; Ctr, control.
Cat D is an abundant aspartic endopeptidase in lysosomes (38, 39). The precursor form of Cat D is proteolytically processed to an intermediate during its transport from the Golgi to acidic endolysosomal compartments. In the lysosomes, the intermediate Cat D is further processed to generate the active fragment as the C-terminal heavy chain (40, 41). Thus, we assessed the effect of trehalose on the proteolytic processing of Cat D. Interestingly, cell treatment with trehalose strongly decreased the processing of Cat D as indicated by an elevated level of the Cat D intermediate form with concurrent decrease of the catalytically active fragment of Cat D (Fig. 7A). Accordingly, ratios of fully processed active Cat D to precursor and intermediate forms were strongly reduced upon cell incubation with trehalose (Fig. 7D). Consistent with previous reports showing that Cat D processing is affected by lysosomal pH (40), bafilomycin A1 and chloroquine also decreased the levels of the active forms of Cat D (Fig. 7, A and D).
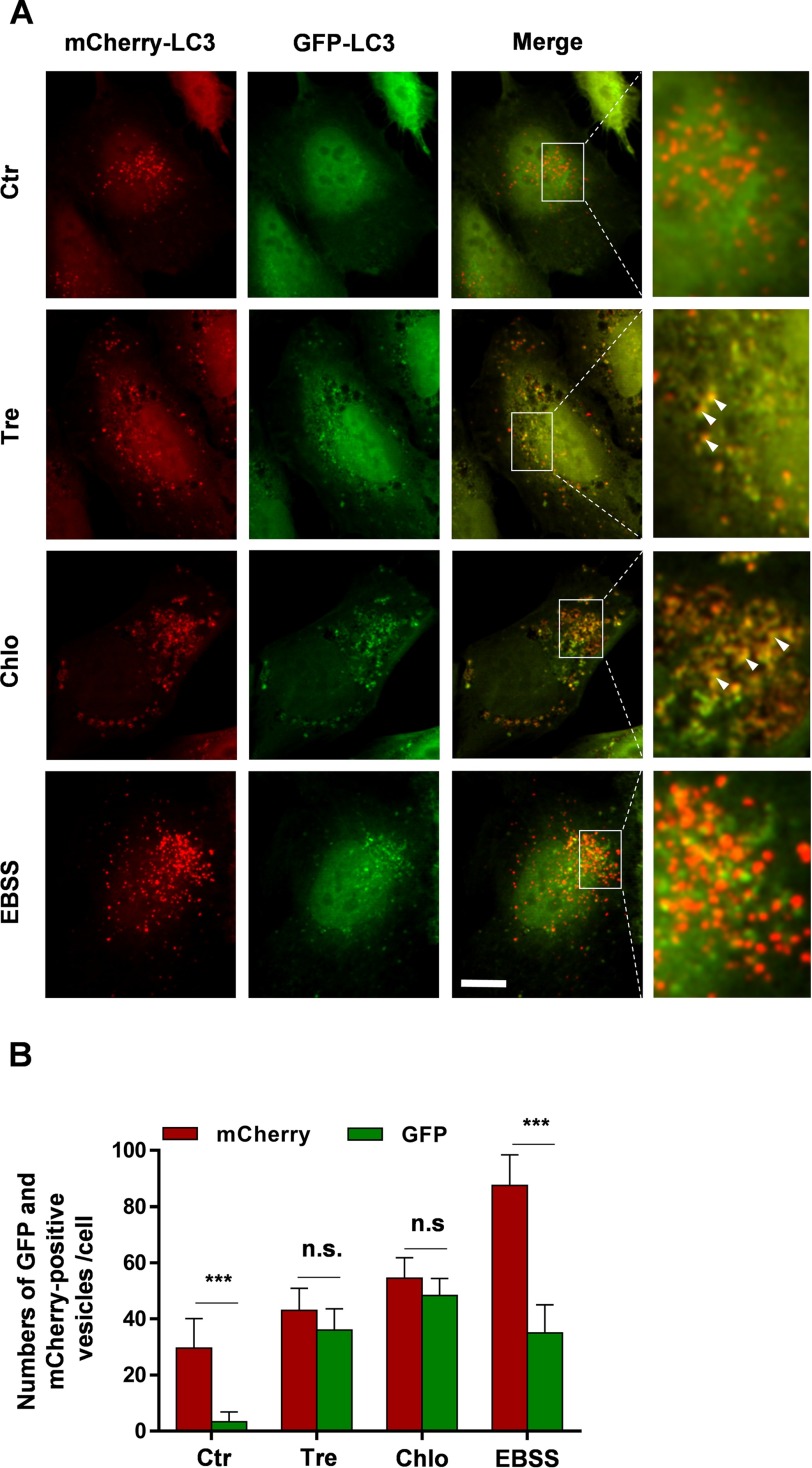
GFP-LC3 is being widely used as a fluorescent marker of autophagosomes. However, GFP is acid-sensitive, and thus its fluorescence decreases in acidic compartments, i.e. when GFP-LC3-containing autophagosomes fuse with endosomes and lysosomes (26). In contrast, the monomeric red fluorescent protein mCherry is acid-stable. Therefore, a tandem fusion of the acid-insensitive red fluorescent protein (mCherry) and the acid-sensitive GFP at the N terminus of LC3 is a useful tool to assess fusion of LC3-positive autophagic vesicles with lysosomes by microscopy (37). H4 cells stably expressing mCherry-GFP-LC3 were cultured in the absence or presence of trehalose. As additional controls, cells were also incubated with chloroquine or starved in EBSS medium. Signals for mCherry and GFP were analyzed by fluorescence microscopy. Control cells showed predominantly mCherry-positive puncta but very few GFP-positive puncta (Fig. 8A). Quantification revealed that the number of mCherry-positive puncta was approximately 10 times higher than that of GFP-positive puncta (Fig. 8B). This is consistent with efficient quenching of the GFP fluorescence in acidic autolysosomes, whereas the mCherry fluorescence is not affected in these compartments. Interestingly, cell treatment with trehalose not only increased the number of GFP-positive puncta as compared with control cells but also led to a large overlap with mCherry-positive structures (Fig. 8, A and B), indicating inefficient quenching of the GFP signal. Cell incubation with chloroquine had very similar effects as trehalose (Fig. 8, A and B). Cell incubation with EBSS increased both mCherry- and GFP-positive puncta compared with control cells (Fig. 8, A and B). However, the number of mCherry-positive puncta was approximately 3 times higher than the number of GFP-LC3-positive puncta (Fig. 8B). As the incubation with EBSS enhances the induction of autophagy, LC3-I is efficiently converted to LC3-II and associates with autophagosomes. However, the efficient acidification of lysosomes and autolysosomes could lead to quenching of the GFP signal, thereby explaining the higher number of mCherry-positive vesicles as compared with that of GFP-positive puncta. The combined data strongly indicate that trehalose impairs the delivery of LC3 to and its degradation in lysosomal compartments.
FIGURE 8.
Altered trafficking of LC3 upon cell treatment with trehalose. A, H4 cells stably expressing mCherry-GFP-LC3 were treated with 100 mm trehalose (Tre) for 24 h, 50 μm chloroquine (Chlo) for 12 h, or starved in EBSS medium for 3 h. Images are representative maximum intensity layers of ApoTome optical sections. White arrowheads indicate mCherry- and GFP-positive vesicles. Scale bar, 10 μm. B, mCherry- and GFP-positive puncta were quantified as described under “Experimental Procedures.” Error bars represent S.D. ***, p < 0.001; n.s., not significant. Ctr, control.
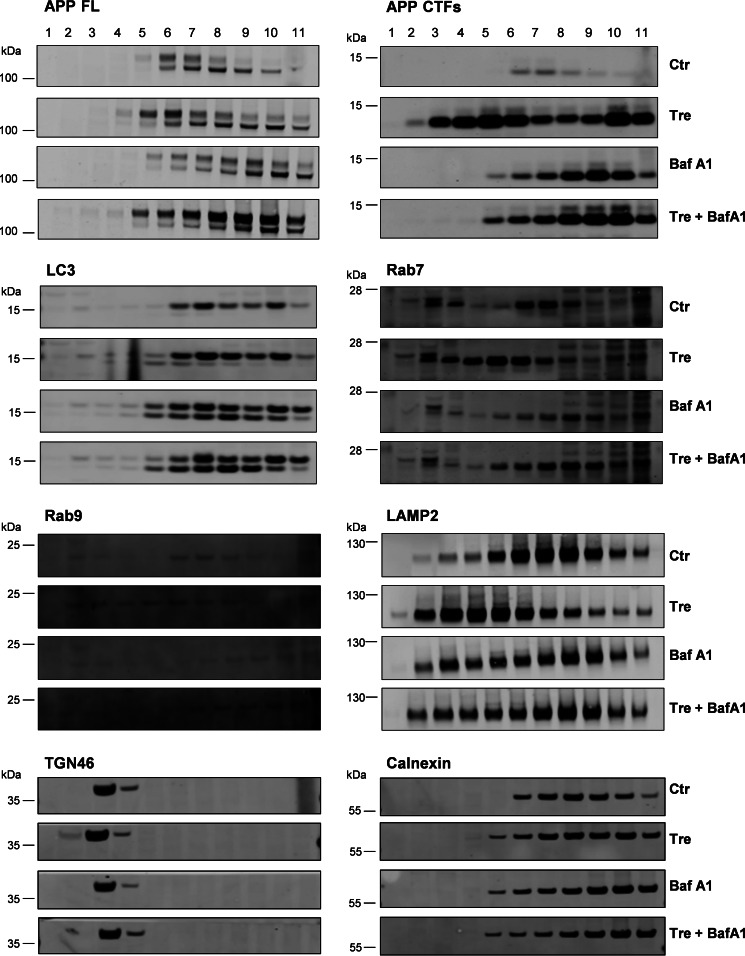
To further test alterations in the subcellular distribution of APP-CTFs upon cell treatment with trehalose, we performed fractionation of vesicular compartments. In control cells, mature full-length APP and CTFs were detected in LC3-, Rab7-, Rab9-, and LAMP2-positive fractions (Fig. 9). The treatment with trehalose resulted in redistribution of APP-CTFs, Rab7, Rab9, and LAMP2. Cell incubation with bafilomycin A1 also led to accumulation of APP-CTFs in fractions that clearly segregated from APP-CTF-positive fractions from trehalose-treated cells (Fig. 9). The combined treatment of cells with trehalose and Baf A1 resulted in an APP-CTF distribution similar to that from cells treated with Baf A1 alone. Fractionation of marker proteins for the trans-Golgi network (TGN46) and endoplasmic reticulum (calnexin) was not affected by the treatment with trehalose or Baf A1. These data suggest that cell treatment with trehalose affects the subcellular distribution and/or trafficking of APP in late endosomal and lysosomal compartments.
FIGURE 9.
Trehalose alters subcellular localization of APP-CTFs. H4 cells were incubated in medium only (Ctr), with trehalose (Tre) or Baf A1 separately, or with a combination of trehalose and bafilomycin A1 (Tre + BafA1) for 24 h and subjected to subcellular fractionation by density gradient centrifugation (see “Experimental Procedures”). The indicated proteins were detected by Western immunoblotting. Results are representative of two independent experiments.
Discussion
The present data demonstrate that trehalose decreases clearance of APP-CTFs and the secretion of Aβ. Cell biological experiments showed redistribution of APP-CTFs and other endosomal and lysosomal proteins upon trehalose treatment. Trehalose also led to accumulation of LC3-II and decreased the processing of Cat D. These data are consistent with impaired vesicular transport and fusion of endosomal and lysosomal compartments. The cleavage of APP by α- and β-secretases can occur at distinct cellular locations. Although α-secretase could predominantly cleave APP in secretory vesicles and at the cell surface, β-secretase has an acidic pH optimum and shows preferential cleavage in acidic compartments, including late endosomes and lysosomes. The C-terminal fragments of APP generated by α- and β-secretase cleavage represent substrates for γ-secretase that resides predominantly in endolysosomal compartments or at the cell surface (42–46). Although trehalose led to strong accumulation of APP-CTFs, we observed a rather decreased secretion of Aβ. Because trehalose did not inhibit γ-secretase, the findings suggest that trehalose rather decreases the delivery of the APP-CTFs to vesicular compartments containing γ-secretase activity. We also observed decreased proteolytic activation of cathepsin D that also occurs during endosomal transport and delivery to lysosomes upon cell treatment with trehalose. Subcellular fractionation indeed showed redistribution of APP-CTFs together with marker proteins of late endosomal (Rab7 and Rab9) and lysosomal (LAMP2) compartments. The molecular mechanisms underlying these effects remain to be determined in more detail. However, the effects of trehalose on subcellular distribution differ from those induced by bafilomycin A1, suggesting that trehalose does not or at least does not only alter lysosomal pH regulation.
Additional experiments using a tandem fusion mCherry-GFP-tagged LC3 also demonstrated significant changes in the transport of LC3 to lysosomes upon trehalose treatment. Although control cells showed predominantly mCherry puncta due to quenching of GFP in efficiently acidified autolysosomes, trehalose treatment increased the co-localization of mCherry and GFP. These data indicate inefficient delivery of LC3 to lysosomes.
Trehalose is a disaccharide composed two d-glucose molecules linked by an α,α-1,1 glycosidic linkage (35). Therefore, we tested whether trehalose-regulated effects might be caused by potential metabolites of trehalose or could be mimicked by other disaccharides. However, neither addition of glucose itself nor addition of the disaccharide maltose had overt effects on APP and LC3 levels, indicating that the observed effects are specific for trehalose and not due to increased supply with glucose. Our data also indicate that the effects of trehalose are independent of mTOR, BECN1, and Atg5. The data thus confirm and extend that trehalose-induced effects do not involve changes in mTOR complex 1 activity (21).
The present findings are surprising because trehalose has been shown to decrease cellular levels of the neurodegenerative disease-associated proteins huntingtin, α-synuclein, and phosphorylated tau in cellular and animal models (22–24), suggesting that trehalose promoted autophagy and lysosomal clearance of these proteins. A possible explanation for this contradiction might emerge from a recent study demonstrating the involvement of unconventional secretion of aggregated α-synuclein via exophagy in cases of impaired autophagosome-lysosome fusion or lysosomal function. Importantly, trehalose was shown to enhance α-synuclein secretion in PC12 catecholaminergic nerve cells (47). In addition, increased secretion of tau was also observed upon starvation and lysosomal inhibition in primary neurons, likely caused by release of autophagosomal contents into extracellular fluids (48). In our experiments, cellular levels of endogenously expressed tau and α-synuclein were not affected by trehalose (data not shown). Both proteins were not detected in conditioned media. However, we observed elevated levels of an overexpressed reporter protein for poly(Gln)-containing aggregates upon cell treatment with trehalose (data not shown), suggesting that trehalose might increase the release of protein aggregates. Secretion of pathogenic poly(Gln) protein from cultured cells has been reported previously (49).
Decreased levels of neurodegeneration-associated protein aggregates upon trehalose treatment might also result from up-regulation of the ubiquitin-proteasome system. In contrast to APP and its CTFs, tau, huntingtin, and α-synuclein are cytosolic proteins. It has been shown that inhibition of the proteasome by epoxomicin increased the levels of α-synuclein and phosphorylated tau and correlated with enhanced cell death (22). Thus, although autophagy is considered as the major pathway to degrade protein aggregates, a contribution of proteasomal degradation to the reduction of aggregate-prone proteins could not be excluded. APP as an integral membrane protein is mainly targeted to endosomal-lysosomal compartments via endocytosis (42) and eventually degraded by lysosomal hydrolases (17–19, 44, 50, 51).
Decreased formation of cytosolic aggregates and lower toxicity were also observed with a mouse model for Huntington disease upon treatment with trehalose (21). However, this effect was at least partially attributed to the function of trehalose as a chemical chaperone that could bind to expanded poly(Gln) stretches and thereby stabilize the partially unfolded protein (52, 53).
Trehalose appears to exert complex functions in cellular protein homeostasis. The effects of trehalose on vesicular trafficking and protein metabolism shown in this study should be considered not only for the interpretation of experimental results obtained with this disaccharide but also for its application in future therapeutic approaches.
Author Contributions
N. T. T. and I. K. contributed equally to this study, performed and designed the experiments, and analyzed data. I. Y. T. also designed and analyzed the experiments. J. W. conceived the study, designed experiments, interpreted data, and wrote the manuscript. All authors read and edited the manuscript.
Acknowledgments
We thank Drs. S. Höning and J. Höhfeld for providing cathepsin D antibodies and mCherry-GFP-LC3 cDNA, respectively. We are also grateful to Dr. B. de Strooper and the RIKEN Cell Bank for providing PS1/2- and Atg5-deficient cells, respectively.
This work was supported by Deutsche Forschungsgemeinschaft Grants SFB645 and KFO177 (to J. W.). The authors declare that they have no conflicts of interest with the contents of this article.
- Aβ
- amyloid β-peptide
- APP
- amyloid precursor protein
- AICD
- APP intracellular domain
- Atg
- autophagy-related gene
- Baf A1
- bafilomycin A1
- BECN1
- beclin-1
- CTF
- C-terminal fragment
- EBSS
- Earle's balanced salt solution
- FL
- full length
- Cat D
- cathepsin D
- LC3
- microtubule-associated protein 1 light chain 3
- LAMP2
- lysosome-associated membrane protein 2
- mTOR
- mechanistic target of rapamycin
- PS
- presenilin
- MEF
- mouse embryonic fibroblast
- dKO
- double knock-out
- APP-SW
- Swedish mutant variant of APP.
References
- 1. Selkoe D. J. (2001) Alzheimer's disease: genes, proteins, and therapy. Physiol. Rev. 81, 741–766 [DOI] [PubMed] [Google Scholar]
- 2. O'Brien R. J., and Wong P. C. (2011) Amyloid precursor protein processing and Alzheimer's disease. Annu. Rev. Neurosci. 34, 185–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pasternak S. H., Bagshaw R. D., Guiral M., Zhang S., Ackerley C. A., Pak B. J., Callahan J. W., and Mahuran D. J. (2003) Presenilin-1, nicastrin, amyloid precursor protein, and γ-secretase activity are co-localized in the lysosomal membrane. J. Biol. Chem. 278, 26687–26694 [DOI] [PubMed] [Google Scholar]
- 4. Almeida C. G., Takahashi R. H., and Gouras G. K. (2006) β-Amyloid accumulation impairs multivesicular body sorting by inhibiting the ubiquitin-proteasome system. J. Neurosci. 26, 4277–4288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Torres M., Jimenez S., Sanchez-Varo R., Navarro V., Trujillo-Estrada L., Sanchez-Mejias E., Carmona I., Davila J. C., Vizuete M., Gutierrez A., and Vitorica J. (2012) Defective lysosomal proteolysis and axonal transport are early pathogenic events that worsen with age leading to increased APP metabolism and synaptic Aβ in transgenic APP/PS1 hippocampus. Mol. Neurodegener. 7, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mizushima N., Levine B., Cuervo A. M., and Klionsky D. J. (2008) Autophagy fights disease through cellular self-digestion. Nature 451, 1069–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Klionsky D. J. (2007) Autophagy: from phenomenology to molecular understanding in less than a decade. Nat. Rev. Mol. Cell Biol. 8, 931–937 [DOI] [PubMed] [Google Scholar]
- 8. Levine B., and Kroemer G. (2008) Autophagy in the pathogenesis of disease. Cell 132, 27–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kroemer G., Mariño G., and Levine B. (2010) Autophagy and the integrated stress response. Mol. Cell 40, 280–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Murrow L., and Debnath J. (2013) Autophagy as a stress-response and quality-control mechanism: implications for cell injury and human disease. Annu. Rev. Pathol. 8, 105–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hara T., Nakamura K., Matsui M., Yamamoto A., Nakahara Y., Suzuki-Migishima R., Yokoyama M., Mishima K., Saito I., Okano H., and Mizushima N. (2006) Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441, 885–889 [DOI] [PubMed] [Google Scholar]
- 12. Ebato C., Uchida T., Arakawa M., Komatsu M., Ueno T., Komiya K., Azuma K., Hirose T., Tanaka K., Kominami E., Kawamori R., Fujitani Y., and Watada H. (2008) Autophagy is important in islet homeostasis and compensatory increase of β cell mass in response to high-fat diet. Cell Metab. 8, 325–332 [DOI] [PubMed] [Google Scholar]
- 13. Komatsu M., Wang Q. J., Holstein G. R., Friedrich V. L. Jr., Iwata J., Kominami E., Chait B. T., Tanaka K., and Yue Z. (2007) Essential role for autophagy protein Atg7 in the maintenance of axonal homeostasis and the prevention of axonal degeneration. Proc. Natl. Acad. Sci. U.S.A. 104, 14489–14494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nakai A., Yamaguchi O., Takeda T., Higuchi Y., Hikoso S., Taniike M., Omiya S., Mizote I., Matsumura Y., Asahi M., Nishida K., Hori M., Mizushima N., and Otsu K. (2007) The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat. Med. 13, 619–624 [DOI] [PubMed] [Google Scholar]
- 15. Kundu M., and Thompson C. B. (2008) Autophagy: basic principles and relevance to disease. Annu. Rev. Pathol. 3, 427–455 [DOI] [PubMed] [Google Scholar]
- 16. Jaeger P. A., and Wyss-Coray T. (2009) All-you-can-eat: autophagy in neurodegeneration and neuroprotection. Mol. Neurodegener. 4, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tian Y., Chang J. C., Fan E. Y., Flajolet M., and Greengard P. (2013) Adaptor complex AP2/PICALM, through interaction with LC3, targets Alzheimer's APP-CTF for terminal degradation via autophagy. Proc. Natl. Acad. Sci. U.S.A. 110, 17071–17076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tamboli I. Y., Hampel H., Tien N. T., Tolksdorf K., Breiden B., Mathews P. M., Saftig P., Sandhoff K., and Walter J. (2011) Sphingolipid storage affects autophagic metabolism of the amyloid precursor protein and promotes Aβ generation. J. Neurosci. 31, 1837–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jaeger P. A., Pickford F., Sun C. H., Lucin K. M., Masliah E., and Wyss-Coray T. (2010) Regulation of amyloid precursor protein processing by the Beclin 1 complex. PLoS One 5, e11102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Richards A. B., Krakowka S., Dexter L. B., Schmid H., Wolterbeek A. P., Waalkens-Berendsen D. H., Shigoyuki A., and Kurimoto M. (2002) Trehalose: a review of properties, history of use and human tolerance, and results of multiple safety studies. Food Chem. Toxicol. 40, 871–898 [DOI] [PubMed] [Google Scholar]
- 21. Sarkar S., Davies J. E., Huang Z., Tunnacliffe A., and Rubinsztein D. C. (2007) Trehalose, a novel mTOR-independent autophagy enhancer, accelerates the clearance of mutant huntingtin and α-synuclein. J. Biol. Chem. 282, 5641–5652 [DOI] [PubMed] [Google Scholar]
- 22. Casarejos M. J., Solano R. M., Gómez A., Perucho J., de Yébenes J. G., and Mena M. A. (2011) The accumulation of neurotoxic proteins, induced by proteasome inhibition, is reverted by trehalose, an enhancer of autophagy, in human neuroblastoma cells. Neurochem. Int. 58, 512–520 [DOI] [PubMed] [Google Scholar]
- 23. Krüger U., Wang Y., Kumar S., and Mandelkow E. M. (2012) Autophagic degradation of tau in primary neurons and its enhancement by trehalose. Neurobiol. Aging 33, 2291–2305 [DOI] [PubMed] [Google Scholar]
- 24. Schaeffer V., Lavenir I., Ozcelik S., Tolnay M., Winkler D. T., and Goedert M. (2012) Stimulation of autophagy reduces neurodegeneration in a mouse model of human tauopathy. Brain 135, 2169–2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mathews P. M., Guerra C. B., Jiang Y., Grbovic O. M., Kao B. H., Schmidt S. D., Dinakar R., Mercken M., Hille-Rehfeld A., Rohrer J., Mehta P., Cataldo A. M., and Nixon R. A. (2002) Alzheimer's disease-related overexpression of the cation-dependent mannose 6-phosphate receptor increases Aβ secretion: role for altered lysosomal hydrolase distribution in β-amyloidogenesis. J. Biol. Chem. 277, 5299–5307 [DOI] [PubMed] [Google Scholar]
- 26. Pankiv S., Clausen T. H., Lamark T., Brech A., Bruun J. A., Outzen H., Øvervatn A., Bjørkøy G., and Johansen T. (2007) p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282, 24131–24145 [DOI] [PubMed] [Google Scholar]
- 27. Nyabi O., Pype S., Mercken M., Herreman A., Saftig P., Craessaerts K., Serneels L., Annaert W., and De Strooper B. (2002) No endogenous Aβ production in presenilin-deficient fibroblasts. Nat. Cell Biol. 4, E164. [DOI] [PubMed] [Google Scholar]
- 28. Tamboli I. Y., Prager K., Thal D. R., Thelen K. M., Dewachter I., Pietrzik C. U., St George-Hyslop P., Sisodia S. S., De Strooper B., Heneka M. T., Filippov M. A., Müller U., van Leuven F., Lütjohann D., and Walter J. (2008) Loss of γ-secretase function impairs endocytosis of lipoprotein particles and membrane cholesterol homeostasis. J. Neurosci. 28, 12097–12106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wunderlich P., Glebov K., Kemmerling N., Tien N. T., Neumann H., and Walter J. (2013) Sequential proteolytic processing of the triggering receptor expressed on myeloid cells-2 (TREM2) protein by ectodomain shedding and γ-secretase-dependent intramembranous cleavage. J. Biol. Chem. 288, 33027–33036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Karaca I., Tamboli I. Y., Glebov K., Richter J., Fell L. H., Grimm M. O., Haupenthal V. J., Hartmann T., Gräler M. H., van Echten-Deckert G., and Walter J. (2014) Deficiency of sphingosine-1-phosphate lyase impairs lysosomal metabolism of the amyloid precursor protein. J. Biol. Chem. 289, 16761–16772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Otomo C., Metlagel Z., Takaesu G., and Otomo T. (2013) Structure of the human ATG12∼ATG5 conjugate required for LC3 lipidation in autophagy. Nat. Struct. Mol. Biol. 20, 59–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bjørkøy G., Lamark T., Brech A., Outzen H., Perander M., Overvatn A., Stenmark H., and Johansen T. (2005) p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 171, 603–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Laplante M., and Sabatini D. M. (2009) mTOR signaling at a glance. J. Cell Sci. 122, 3589–3594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mizushima N., Yamamoto A., Hatano M., Kobayashi Y., Kabeya Y., Suzuki K., Tokuhisa T., Ohsumi Y., and Yoshimori T. (2001) Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J. Cell Biol. 152, 657–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Venables M. C., Brouns F., and Jeukendrup A. E. (2008) Oxidation of maltose and trehalose during prolonged moderate-intensity exercise. Med. Sci. Sports Exerc. 40, 1653–1659 [DOI] [PubMed] [Google Scholar]
- 36. Yoshimori T., Yamamoto A., Moriyama Y., Futai M., and Tashiro Y. (1991) Bafilomycin A1, a specific inhibitor of vacuolar-type H+-ATPase, inhibits acidification and protein degradation in lysosomes of cultured cells. J. Biol. Chem. 266, 17707–17712 [PubMed] [Google Scholar]
- 37. Klionsky D. J., Abdalla F. C., Abeliovich H., Abraham R. T., Acevedo-Arozena A., Adeli K., Agholme L., Agnello M., Agostinis P., Aguirre-Ghiso J. A., Ahn H. J., Ait-Mohamed O., Ait-Si-Ali S., Akematsu T., Akira S., et al. (2012) Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 8, 445–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Benes P., Vetvicka V., and Fusek M. (2008) Cathepsin D—many functions of one aspartic protease. Crit. Rev. Oncol. Hematol. 68, 12–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Metcalf P., and Fusek M. (1993) Two crystal structures for cathepsin D: the lysosomal targeting signal and active site. EMBO J. 12, 1293–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gieselmann V., Hasilik A., and von Figura K. (1985) Processing of human cathepsin D in lysosomes in vitro. J. Biol. Chem. 260, 3215–3220 [PubMed] [Google Scholar]
- 41. Richo G. R., and Conner G. E. (1994) Structural requirements of procathepsin D activation and maturation. J. Biol. Chem. 269, 14806–14812 [PubMed] [Google Scholar]
- 42. Walter J., Kaether C., Steiner H., and Haass C. (2001) The cell biology of Alzheimer's disease: uncovering the secrets of secretases. Curr. Opin. Neurobiol. 11, 585–590 [DOI] [PubMed] [Google Scholar]
- 43. Walter J., and van Echten-Deckert G. (2013) Cross-talk of membrane lipids and Alzheimer-related proteins. Mol. Neurodegener. 8, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Haass C., Koo E. H., Mellon A., Hung A. Y., and Selkoe D. J. (1992) Targeting of cell-surface β-amyloid precursor protein to lysosomes: alternative processing into amyloid-bearing fragments. Nature 357, 500–503 [DOI] [PubMed] [Google Scholar]
- 45. De Strooper B., Vassar R., and Golde T. (2010) The secretases: enzymes with therapeutic potential in Alzheimer disease. Nat. Rev. Neurol. 6, 99–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Haass C., Kaether C., Thinakaran G., and Sisodia S. (2012) Trafficking and proteolytic processing of APP. Cold Spring Harb. Perspect. Med. 2, a006270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ejlerskov P., Rasmussen I., Nielsen T. T., Bergström A. L., Tohyama Y., Jensen P. H., and Vilhardt F. (2013) Tubulin polymerization-promoting protein (TPPP/p25α) promotes unconventional secretion of α-synuclein through exophagy by impairing autophagosome-lysosome fusion. J. Biol. Chem. 288, 17313–17335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mohamed N. V., Plouffe V., Rémillard-Labrosse G., Planel E., and Leclerc N. (2014) Starvation and inhibition of lysosomal function increased tau secretion by primary cortical neurons. Sci. Rep. 4, 5715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Popiel H. A., Takeuchi T., Fujita H., Yamamoto K., Ito C., Yamane H., Muramatsu S., Toda T., Wada K., and Nagai Y. (2012) Hsp40 gene therapy exerts therapeutic effects on polyglutamine disease mice via a non-cell autonomous mechanism. PLoS One 7, e51069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yu W. H., Cuervo A. M., Kumar A., Peterhoff C. M., Schmidt S. D., Lee J. H., Mohan P. S., Mercken M., Farmery M. R., Tjernberg L. O., Jiang Y., Duff K., Uchiyama Y., Näslund J., Mathews P. M., et al. (2005) Macroautophagy—a novel β-amyloid peptide-generating pathway activated in Alzheimer's disease. J. Cell Biol. 171, 87–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yu W. H., Kumar A., Peterhoff C., Shapiro Kulnane L., Uchiyama Y., Lamb B. T., Cuervo A. M., and Nixon R. A. (2004) Autophagic vacuoles are enriched in amyloid precursor protein-secretase activities: implications for β-amyloid peptide over-production and localization in Alzheimer's disease. Int. J. Biochem. Cell Biol. 36, 2531–2540 [DOI] [PubMed] [Google Scholar]
- 52. Tanaka M., Machida Y., Niu S., Ikeda T., Jana N. R., Doi H., Kurosawa M., Nekooki M., and Nukina N. (2004) Trehalose alleviates polyglutamine-mediated pathology in a mouse model of Huntington disease. Nat. Med. 10, 148–154 [DOI] [PubMed] [Google Scholar]
- 53. Singer M. A., and Lindquist S. (1998) Multiple effects of trehalose on protein folding in vitro and in vivo. Mol. Cell 1, 639–648 [DOI] [PubMed] [Google Scholar]
- 54. Magazù S., Migliardo F., and Telling M. T. (2007) Study of the dynamical properties of water in disaccharide solutions. Eur. Biophys. J. 36, 163–171 [DOI] [PubMed] [Google Scholar]