Abstract
In mammals, circadian rhythms in physiological function are generated by a molecular oscillator driven by transcriptional-translational feedback loop consisting of negative and positive regulators. Disruption of this circadian clock machinery is thought to increase the risk of cancer development, but the potential contributions of each component of circadian clock to oncogenesis have been little explored. Here we reported that negative and positive transcriptional regulators of circadian feedback loop had different roles in oncogene-induced neoplastic transformation. Mouse embryonic fibroblasts prepared from animals deficient in negative circadian clock regulators, Period2 (Per2) or Cryptochrome1/2 (Cry1/2), were prone to transformation induced by co-expression of H-rasV12 and SV40 large T antigen (SV40LT). In contrast, mouse embryonic fibroblasts prepared from mice deficient in positive circadian clock regulators, Bmal1 or Clock, showed resistance to oncogene-induced transformation. In Per2 mutant and Cry1/2-null cells, the introduction of oncogenes induced expression of ATF4, a potent repressor of cell senescence-associated proteins p16INK4a and p19ARF. Elevated levels of ATF4 were sufficient to suppress expression of these proteins and drive oncogenic transformation. Conversely, in Bmal1-null and Clock mutant cells, the expression of ATF4 was not induced by oncogene introduction, which allowed constitutive expression of p16INK4a and p19ARF triggering cellular senescence. Although genetic ablation of either negative or positive transcriptional regulators of the circadian clock leads to disrupted rhythms in physiological functions, our findings define their different contributions to neoplastic cellular transformation.
Keywords: cellular senescence, circadian rhythm, clock gene, oncogene, tumor cell biology
Introduction
The circadian clock is a timekeeping system that allows organisms to adapt their physiological and behavioral functions to anticipatory changes in their environment. In mammals, the circadian clock system is hierarchically organized, consisting of a light-responsive central clock in the suprachiasmatic nuclei of the anterior hypothalamus and subsidiary clocks in other brain regions and peripheral tissues (1). The suprachiasmatic nuclei entrains and synchronizes subsidiary clocks with the environmental light-dark cycle, whereas peripheral clocks regulate tissue-specific functions in an anticipatory manner.
Central and peripheral clocks are both governed by inter-connected transcriptional and translational feedback loops (2). The gene products of Bmal1 (also known as Arntl) and Clock form a heterodimer that activates the transcription of Period (Per) and Cryptochrome (Cry) genes. Once PER and CRY proteins have reached a critical concentration, they attenuate BMAL1/CLOCK-mediated transactivation. The alternating activation and suppression of the BMAL1/CLOCK-driven positive loop and PER/CRY-controlled negative loop result in a circadian oscillation in the molecular clock and also regulate 24-h variations in output physiology through the periodic activation/repression of clock-controlled genes (3, 4).
Because the expression of up to 10% of genes has been suggested to be under the control of the circadian clock (5), it should not come as a surprise that disruptions in the circadian clock system lead to the onset of various diseases. In fact, several epidemiological analyses and laboratory animal studies also revealed a relationship between disruptions in circadian rhythms and cancer development. For example, human night shift workers are at an increased risk of developing breast, prostate, colon, and endometrial cancers as well as hepatocellular carcinoma and non-Hodgkin lymphoma (6–7). These epidemiological findings are also supported by animal studies in which repetitive changes in the light-dark cycle facilitate the growth of implanted tumors (8, 9). Furthermore, spontaneous as well as radiation-induced tumor development is enhanced in circadian gene-defective animals whose behavioral and physiological rhythms are abnormal phenotypes (10, 11).
Although genetic ablation of either negative or positive transcriptional regulators of the circadian feedback loop leads to disruption of the rhythms in physiological functions (1, 2), the potential contributions of each circadian clock regulator to oncogenesis have been little explored. In this study we found that negative and positive components of circadian feedback loop played different roles during oncogene-induced neoplastic transformation. Mouse embryonic fibroblasts (MEFs)3 prepared from Per2 mutant (Per2m/m) or Cry1/2-null (Cry1/2−/−) mice were prone to oncogenic transformation induced by coexpression of H-rasV12 and SV40 large T antigen (SV40LT); however, these oncogene induced expression of cellular senescence-associated proteins in Bmal1-null (Bmal1−/−) or Clock mutant (Clk/Clk) cells, resulting in failure of their neoplastic transformation. Because the expression of ATF4, a potent repressor of cell senescence-associated proteins, was altered in oncogene-introduced clock gene-defective cells, we further focused on this gene to investigate the roles of negative and positive components of the circadian clock in the neoplastic transformation of mouse fibroblasts.
Experimental Procedures
Treatments of Animals and Cells
Per2m/m, Bmal1−/−, and Clock mutant (Clk/Clk) mice with an ICR background and wild type mice of the same strain were housed in a temperature-controlled (24 ± 1 °C) room under a 12-h light:12-h dark cycle. MEFs were prepared by standard techniques (12) from littermate embryos of Per2m/m, Bmal1−/−, Clk/Clk, or wild-type mice, and cells were maintained in Dulbecco's modified Eagle's medium (DMEM, Sigma) supplemented with 10% fetal bovine serum (FBS). Cry-null MEFs, defective in both Cry1 and Cry2, were kindly provided by Dr. Ueda (13). The preparation of MEFs from each genotype was conducted at least three times. Cells were expanded for three or four passages before the experiments. To achieve oncogenic transformation, MEFs were infected with 1 × 106 colony-forming units/ml retroviral vectors expressing H-rasV12 and SV40LT (14). Plasmid vectors (pcDNA3.1; Invitrogen) expressing Per2, Bmal1, or Atf4 were also transfected into oncogene-introduced MEFs. Transgene-expressing cells were selected with G418 (Wako, Osaka, Japan), and individual colonies were expanded and maintained in media containing 4 μg/ml G418.
Male NOD-SCID mice were purchased from Charles River. They were inoculated with oncogenic-transformed MEFs (5 × 106 cells/mouse). Tumor volumes were measured by caliper, and values were obtained by multiplying the square of the smallest diameter with the largest diameter. Animals were cared for in accordance with the guidelines established by the Animal Care and Use Committee of Kyushu University (Fukuoka, Japan).
Measurement of Locomotor Activity Rhythm
Mice were housed individually in breeding cages with food and water ad libitum. The cages were placed into an infrared ray area sensor, and locomotor activity was measured every 10 min. Locomotor activity was recorded under the light and dark cycles for 10 days and then continuously recorded under the constant dark condition for 10 days.
Anchorage-independent Cell Growth Assay
To evaluate the oncogenic transformation of cells, the ability of cells to grow in an anchorage-independent manner was assessed using the Quantitative 3D Cell Culture Colony Assay Kit (Nihon-ika Co. Ltd., Osaka, Japan). Cells were seeded in 10% FBS containing DMEM soft agar at a density of 1 × 104 cells. The viability of cells in agar was determined by tetrazolium chloride on day 7 after seeding.
Construction of Vectors
Retroviral plasmid vectors expressing H-rasV12 (Addgene plasmid#9051) and SV40LT (Addgene plasmid#13970) were transfected into G3T-hi packaging cells. All of the infected cells were cultured in medium containing the appropriate antibiotics. The mouse Atf4 promoter region spanning bp −346 to +170 (these numbers represent the distance in base pairs from the putative transcription start site, +1) was amplified by PCR, and the product was ligated into the pGL3-Basic luciferase reporter vector (Promega). The E-box of the Atf4 promoter was mutated at −83 to −78 (CACGTG to GAGTCT) using a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). Expression vectors for mouse Per2, Cry1, Bmal1, Clock, and ATF4 were also constructed using complementary DNAs (cDNAs) obtained by RT-PCR derived from mouse hepatic RNA. Coding regions were ligated into the pcDNA3.1 vectors (Invitrogen).
Histochemical Analysis
Cells were fixed with 4% paraformaldehyde/PBS for 10 min. Senescence-associated β-galactosidase (β-gal)-positive cells were detected using a senescence detection kit (BioVision, Inc. Palo Alto, CA). The percentage of β-gal-positive cells was determined by counting the number of positive cells in 3 randomly chosen fields at ×400 magnification.
Small Interfering RNA
Oncogene-transformed cells were transfected with siRNA against ATF4 (Santa Cruz Biotechnology, Inc. Dallas, TX). This siRNA is designed to prevent the expression of mouse ATF4 (15). Oncogene-introduced cells were transfected with the same amount of scrambled siRNA as the control and were then used in experiments 48 h after siRNA transfection.
RT-PCR Analysis
To quantify the mRNA levels of cellular senescence factors, cDNA was synthesized by reverse transcription using the ReverTra Ace quantitative real-time PCR kit (Toyobo, Osaka, Japan). Diluted cDNA samples were analyzed by PCR using the THUNDERBIRDSYBR qPCR Mix (Toyobo) and 7500 RT-PCR system (Applied Biosystems, Framingham, MA). To confirm the expression of H-rasV12 and SV40LT, diluted cDNA samples were also subjected to PCR as described above, and PCR products were run on agarose gel after staining with ethidium bromide. PCR primer sequences are described in Table 1.
TABLE 1.
Primer sets for PCR analysis
| Gene | Primers |
|---|---|
| H-rasV12 | Forward, 5′-GACGGAATATAAGCTGGTGGT-3′ |
| Reverse, 5′-GTCCTTCACCCGTTTGATCTG-3′ | |
| SV40LT | Forward, 5′-AAAGCTGCACTGCTATACAA-3′ |
| Reverse, 5′-AATTGTAGGCTATCAACCCG-3′ | |
| Mouse p16Ink4a | Forward, 5′-GAACTCTTTCGGTCGTACCC-3′ |
| Reverse, 5′-CAGTTCGAATCTGCACCGTAG-3′ | |
| Mouse p19Arf | Forward, 5′-CATGTTGTTGAGGCTAGAGAGG-3′ |
| Reverse, 5′-TGAGCAGAAGAGCTGCTACG-3′ | |
| Mouse p21 | Forward, 5′-TCTAGGGTGGGTCCTTGGTG-3′ |
| Reverse, 5′-CAGCCATTGCTCAGTGTCCT-3′ | |
| Mouse Bax | Forward, 5′-CAGGAGCGTCCACCAAGAA-3′ |
| Reverse, 5′-AGTAGAAGAGGGCAACCACG-3′ | |
| Mouse Atf4 | Forward, 5′-CGAATGGATGACCTGGAAAC-3′ |
| Reverse, 5′-GGCTGCAAGAATGTAAAGGG-3′ | |
| Mouse Per2 | Forward, 5′-CACCCTGAAAAGAAAGTGCGA-3′ |
| Reverse, 5′-CAACGCCAAGGAGCTCAAGT-3′ | |
| Mouse Cry1 | Forward, 5′-AAGTCATCGTGCGCATTTCA-3′ |
| Reverse, 5′-TCATCATGGTCGTCGGACAGA-3′ | |
| Mouse Cry2 | Forward, 5′-GGATAAGCACTTGGAACGGAA-3′ |
| Reverse, 5′-ACAAGTCCCACAGGCGGT-3′ | |
| Mouse Bmal1 | Forward, 5′-CCGATGACGAACTGAAACACCT-3′ |
| Reverse, 5′-TGCAGTGTCCGAGGAAGATAGC-3′ | |
| Mouse Clock | Forward, 5′-TCTCTTCCAAACCAGACGCC-3′ |
| Reverse, 5′-TGCGGCATACTGGATGGAAT-3′ | |
| Mouse β-actin | Forward, 5′-CACACCTTCTACAATGAGCTGC-3′ |
| Reverse, 5′-CATGATCTGGGTCATCTTTTCA-3′ |
Western Blotting
Nuclear or cytoplasmic proteins prepared from cells were separated on SDS-polyacrylamide gels and transferred to polyvinylidene difluoride membranes. The membranes were reacted with antibodies against activating transcription factor 4 (ATF4; sc-200, Santa Cruz Biotechnology), p16INK4a (sc-1207), p19ARF (sc-22784), retinoblastoma protein (pRB; sc-69791), p53 (sc-6243), PER2 (sc-25368), BMAL1 (sc-48790) ACTIN (sc-1616), or anti-phospho-pRB-Thr826 (44–576; BIOSOURCE International, Camarillo, CA). Specific antigen-antibody complexes were visualized using horseradish peroxidase-conjugated secondary antibodies and Chemi-Lumi One (Nacalai Tesque) or ImmunoStar reagent (Wako Chemical Co. Ltd., Tokyo, Japan).
Immunoprecipitation
Oncogene-introduced wild type, Per2m/m, and Bmal1−/− cells were lysed in 25 mmol/liter Tris-HCl, pH 8.0, 137 mmol/liter NaCl, 2.7 mmol/liter KCl, and 1% Triton X-100 supplemented with protease inhibitor mixtures and were then subjected to immunoprecipitation with anti-SV40LT antibodies (sc-58665, Santa Cruz Biotechnology). The amounts of p53, pRB, and SV40LT in cell lysates, supernatants, and immune complexes were detected by Western blotting.
Luciferase Reporter Assay
MEFs prepared from the wild type were seeded at a density of 1×105 cells/well on 24-well culture plates. Cells were transfected 18 h later with 100 ng/well of reporter vectors and 1–2 μg/well (total) of expression vectors. The pRL-TK vector (0.5 ng/well; Promega) was also co-transfected as an internal control reporter. Cells were then harvested, and cell lysates were analyzed using a dual-luciferase reporter assay system (Promega). The ratio of firefly (expressed from the reporter construct) to Renilla (expressed from pRL-TK) luciferase activities in each sample served as a measure of normalized luciferase activity.
Statistical and Data Analyses
To assess the endogenous period of each animal, locomotor counts were analyzed using ClockLab software (Actimetrics, Evanston, IL). The values presented are expressed as the means ± S.E. The significance of differences among groups was analyzed by analysis of variance followed by Tukey-Kramer's post hoc tests. Equal variances were not formally tested. p < 0.05 was considered significant.
Results
Different Oncogenic Phenotypes of Per2m/m and Bmal1−/− Cells
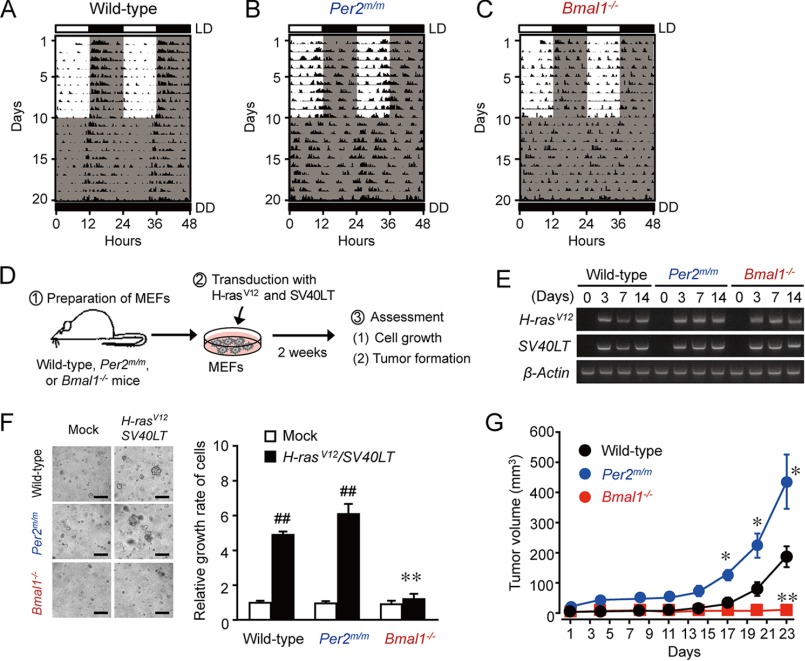
Spontaneous locomotor activity, which is an accurate measure of circadian activity, was monitored in wild-type, Per2m/m, and Bmal1−/− mice. Animals were initially maintained on a 12-h light/dark cycle (LD) for 10 days and were subsequently maintained under constant darkness (DD) (Fig. 1). The locomotor activity of wild-type mice increased during the dark phase, so that they entrained to the scheduled dark lighting cycle (Fig. 1A). Under constant darkness condition, wild-type mice also exhibited robust circadian rhythms of locomotor activity, and their free-running periods were 23.67 ± 0.34 h (n = 4, mean ± S.E.). In contrast, neither Per2m/m nor Bmal1−/− mice failed to show entrained rhythms of locomotor activity even under LD cycle condition (Fig. 1, B and C). Furthermore, a spectral analysis of locomotor activity did not detect periodicity in the circadian range for Per2m/m or Bmal1−/− mice, suggesting that the apparent circadian rhythms are similarly disrupted even though the negative or positive components of the circadian clock are deleted.
FIGURE 1.
Different oncogenic phenotypes of Per2m/m and Bmal1−/− cells after introduction of H-rasV12 and SV40LT. A–C, representative locomotor activity records of wild-type (A), Per2m/m (B), or Bmal1−/− (C) mice. Light regime: 10 days in a 12-h light and 12-h dark cycle followed by 10 days in a constant dark cycle. D, schematic experimental procedure for evaluating the oncogenesis of clock gene-deficient cells. MEFs were prepared from wild-type, Per2m/m, or Bmal1−/− mice. Cells were infected with retroviral vectors expressing H-rasV12 and SV40LT. E, the time course of mRNA expressions of H-rasV12 and SV40LT in wild-type, Per2m/m, or Bmal1−/− cells after infection with retrovirus vectors expressing oncogenes. F, anchorage-independent growth of wild-type, Per2m/m, and Bmal1−/− cells after the concomitant introduction of H-rasV12 and SV40LT. Cells infected with oncogenes were subjected to a soft-agar colony assay, and their colony formation and viability were assessed 14 days after seeding. Control cells were infected with mock vectors. The left panels show representative microscopic photographs of colony formation in each type of cells. The scale bars indicate 100 μm. The right panel shows viability of cells. Values are shown as means ± S.E. (n = 3–6). Mean values of mock-transfected wild-type cells were set at 1.0. ##, p < 0.01 significantly different from the mock-transfected group. **, p < 0.01 significantly different from oncogene-transfected wild-type and Per2m/m cells. G, tumor formation by oncogene-introduced wild-type, Per2m/m, or Bmal1−/− cells. Equal numbers of cells (5 × 106 cells) were implanted into the dorsal air sacs of NOD/SCID mice. Values are the means ± S.E. (n = 4). **, p < 0.01; *, p < 0.05 significantly different from the other groups at the corresponding time points.
To compare the roles of Per2m/m and Bmal1−/− in oncogenic transformation, we prepared MEFs from wild-type, Per2m/m, and Bmal1−/− mice and infected cells concomitantly with retrovirus vectors expressing H-rasV12 and SV40LT (Fig. 1D). The expression of mRNAs for these oncogenes was detected on day 3 after infection, and they were equally expressed in all types of cells (Fig. 1E). The concomitant introduction of H-rasV12 and SV40LT significantly enhanced the anchorage-independent growth of wild-type and Per2m/m cells (p < 0.01, respectively, Fig. 1F), whereas the introduction of these oncogenes failed to enhance the growth of Bmal1−/− cells. These findings suggested that the loss of Bmal1 resulted in the phenotype resisting oncogene-induced malignant transformation.
We also tested the tumorigenicity of oncogene-introduced cells using an implant model in mice. Equal numbers of wild-type, Per2m/m, or Bmal1−/− cells infected with the oncogenes (H-rasV12 and SV40LT) were inoculated into the flanks of NOD-SCID mice. Although mice inoculated with wild-type and Per2m/m cells infected with oncogenes showed the significant growth of tumor masses (Fig. 1G), the average tumor volume in mice inoculated with Per2m/m cells was significantly larger than that in mice inoculated with wild-type cells (p < 0.05 on days 17, 20, and 23, respectively). In contrast, mice inoculated with oncogene-introduced Bmal1−/− cells showed no palpable tumor masses throughout the experimental period. Taken together, these results indicated that a deficiency in PER2 tended to enhance tumorigenicity, whereas the loss of BMAL1 resulted in the phenotype resisting oncogene-induced malignant transformation.
Oncogene Introduction Induces Cellular Senescence of Bmal1−/− Cells
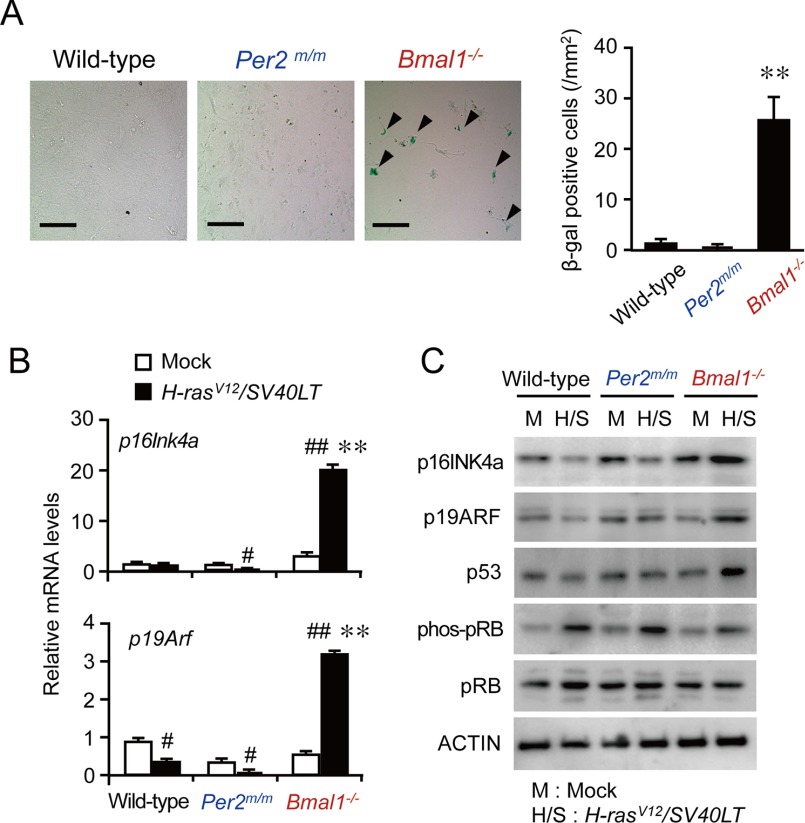
During oncogenic transformation, growth signals that override the normal cell-cycle mechanism are activated in cancer cells, resulting in abnormal proliferation. Failure of cells to override this mechanism often causes reversible cell-cycle arrest, apoptotic cell death, or cellular senescence (16). In contrast to reversible cell-cycle arrest, cellular senescence is defined by the irreversible loss of proliferative potential, acquisition of a characteristic morphology, and expression of specific biomarkers, such as β-gal (17, 18). On day 14 after the infection with retrovirus vectors expressing H-rasV12 and SV40LT, the number of β-gal-positive Bmal1−/− cells was significantly higher than those among wild-type and Per2m/m cells (p < 0.01, Fig. 2A).
FIGURE 2.
Induction of cellular senescence in oncogene-introduced Bmal1−/− cells. A, β-gal staining of wild-type, Per2m/m, or Bmal1−/− cells infected with H-rasV12 and SV40LT. Arrows in the microscopic photograph indicate β-gal-positive cells. The scale bars indicate 50 μm. Values are the means ± S.E. (n = 3). **, p < 0.01 significantly different from other oncogene-introduced cells. B, the mRNA levels of p16Ink4a and p19Arf in wild-type, Per2m/m, or Bmal1−/− cells infected with oncogenes. Values are the means ± S.E. (n = 3). ##, p < 0.01 significantly different from mock-transfected group. **, p < 0.01 significantly different from other oncogene-introduced cells. C, the protein abundance of p16INK4a, p19ARF, p53, phosphorylated pRB (phos-pRB), and pRB in wild-type, Per2m/m, or Bmal1−/− cells infected with oncogenes.
The tumor suppressor gene Cdkn2a acts as a canonical inducer of cellular senescence (18, 19). Cdkn2a generates the different transcript variants, p16Ink4a and p19Arf (also known as p14ARF in humans), by using different first exons and alternate polyadenylation sites (20, 21). The p16Ink4a variants encode structurally related protein isoforms that inhibit CDK4 kinase (19). The CDK4 inhibitor prevents the phosphorylation of pRB by disrupting the activity of the CDK4-cyclin D complex. On the other hand, p19ARF stabilizes p53 by sequestering MDM2 (22). The mRNA levels of p16Ink4a and p19Arf were significantly increased in oncogene-introduced Bmal1−/− cells (p < 0.01, respectively; Fig. 2B), whereas the mRNA levels of both transcript variants of Cdkn2a in wild-type and Per2m/m cells were slightly but significantly decreased by the introduction of oncogenes. Consistent with these results, marked increases in the protein levels of p16INK4a and p19ARF were also detected in oncogene-introduced Bmal1−/− cells (Fig. 2C), whereas those protein levels were decreased in oncogene-introduced wild-type and Per2m/m cells. The induction of cellular senescence factors in oncogene-introduced Bmal1−/− cells was accompanied by the suppressed phosphorylation of pRB as well as increases in the protein levels of p53 (Fig. 2C). These results suggested that oncogene-introduced Bmal1−/− cells failed to override the normal cell-cycle mechanism, resulting in cellular senescence.
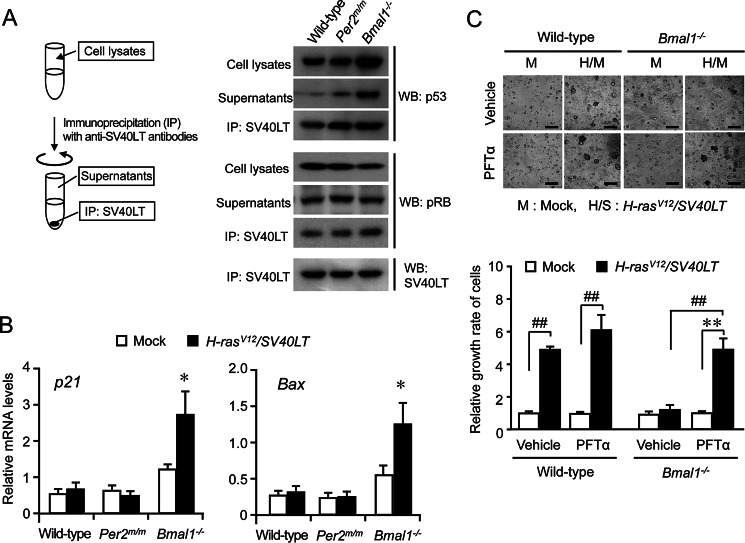
SV40LT-transduced cells are considered to be insensitive to growth arrest by p16INK4a and p19ARF, because SV40LT inactivates p53 by protein-protein interaction (23). The results of an immunoprecipitation analysis revealed that the most amounts of p53 protein in oncogene-introduced wild-type and Per2m/m cells were precipitated together with SV40LT (Fig. 3A). On the other hand, large amounts of p53 protein in Bmal1−/− cells was unable to be precipitated together with SV40LT, suggesting that extensive expression of p53 in oncogene-introduced Bmal1−/− cells overrides the binding capacity of SV40LT. In fact, the expression of the p53 target genes, p21 and Bax, was increased in oncogene-introduced Bmal1−/− cells (Fig. 3B).
FIGURE 3.
The overexpression of p53 in oncogene-introduced Bmal1−/− cells overrides the binding capacity of SV40LT. A, the oncogene-introduced wild-type, Per2m/m, or Bmal1−/− cells were lysed and were then subjected to immunoprecipitation with anti-SV40LT antibodies. The amounts of p53, pRb, and SV40LT in cell lysates, supernatants, and immune complexes were detected by Western blotting (WB). B, the mRNA levels of p53 target genes, p21 and Bax, in wild-type, Per2m/m, or Bmal1−/− cells infected with oncogenes. Values are the means ± S.E. (n = 3). *, p < 0.05 significantly different from other oncogene-introduced cells. C, influence of p53 inhibitor Pifithrin-α (PFTα) on the anchorage-independent growth of wild-type and Bmal1−/− cells after the concomitant introduction of H-rasV12 and SV40LT. Cells infected with oncogenes were subjected to a soft agar colony assay in the presence or absence of 30 μm pifithrin-α. The colony formation of cells and their viability were assessed 14 days after seeding. Control cells were infected with mock vectors. The upper panels show representative microscopic photographs of colony formation in each type of cells. The scale bars indicate 100 μm. The lower panel shows viability of cells. Values are shown as the means ± S.E. (n = 4). Mean values of mock-transfected wild-type cells were set at 1.0. ##, p < 0.01; **, p < 0.01 significantly different between the two groups.
The transforming activity of SV40LT has also been attributed to its perturbation of pRb (24). Although large amounts of pRb were precipitated together with SV40LT, the protein was also detected in the supernatant fractions of all cell types (Fig. 3A). These results indicated that the growth inhibitory function of pRb still remained in oncogene-introduced wild-type, Per2m/m, and Bmal1−/− cells. The retroviral transfer of SV40LT into these cells appeared to partially suppress pRb function.
In an attempt to determine whether accumulation of p53 in oncogene-introduced Bmal1−/− cells contributes to the induction of cellular senescence, oncogene-introduced wild-type and Bmal1−/− cells were incubated in the presence and absence of the p53 inhibitor pifithrin-α. The treatment with 30 μm pifithrin-α had a negligible effect on the anchorage-independent growth of oncogene-introduced wild-type cells but significantly enhanced the growth of oncogene-introduced Bmal1−/− cells (p < 0.01, Fig. 3C). Extensive expression of p53 in oncogene-introduced Bmal1−/− cells appeared to override the binding capacity of SV40LT and allowed the induction of cellular senescence.
ATF4 Determines the Oncogenic Phenotypes of Per2m/m and Bmal1−/− Cells
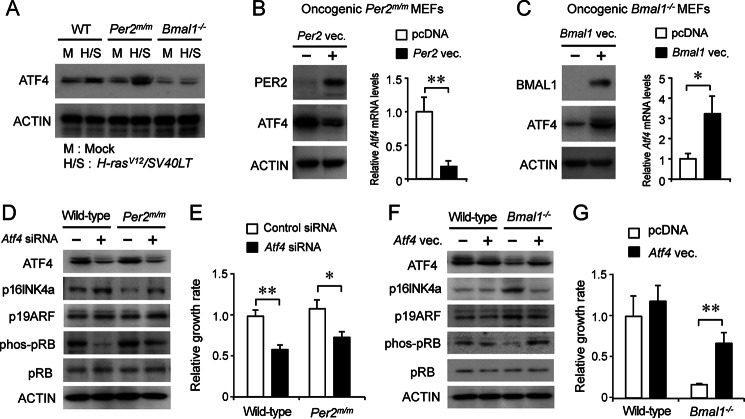
A member of the cAMP-binding protein family, ATF4, promotes oncogene-induced transformation by suppressing the expression of p16INK4a and p19ARF (15). The protein levels of ATF4 were substantially increased in oncogene-introduced wild-type and Per2m/m cells but not in Bmal1−/− cells (Fig. 4A). The excessive expression of ATF4 in oncogene-introduced Per2m/m cells was repressed by the transfection of functional “native” Per2-expressing vectors (Fig. 4B), whereas the transfection of oncogene-introduced Bmal1−/− cells with Bmal1-expressing vectors caused an increase in ATF4 levels (Fig. 4C). The modulation of ATF4 protein levels by PER2 and BMAL1 occurred at the transcriptional level because the mRNA levels of Atf4 were also down- and up-regulated by PER2 and BMAL1, respectively (Fig. 4, B and C).
FIGURE 4.
Role of ATF4 in the oncogene-induced transformation of Per2 or Bmal1-defective cells. A, the protein levels of ATF4 in wild-type, Per2m/m, or Bmal1−/− cells infected with H-rasV12 and SV40LT. B, the overexpression of PER2 in oncogene-introduced Per2m/m cells suppresses the expression of ATF4. C, the overexpression of BMAL1 in oncogene-introduced Bmal1−/− cells enhances the expression of ATF4. For panels B and C, the mean value of empty vectors (pcDNA)-transfected cells were set at 1.0. D, influence of siRNA-induced down-regulation of Atf4 on the protein abundance of p16INK4a, p19ARF, p53, phosphorylated pRB (phos-pRB), and pRB in wild-type or Per2m/m cells. E, down-regulation of ATF4 in oncogene-introduced wild-type or Per2m/m cells by siRNA attenuated anchorage-independent growth ability. F, the protein abundance of p16INK4a, p19ARF, p53, phosphorylated pRB (phos-pRB), and pRB in oncogenic wild-type or Bmal1−/− cells transfected with Atf4-expressing vectors. G, overexpression of ATF4 in oncogene-introduced Bmal1−/− cells enhanced the anchorage-independent growth ability. For panels B, C, E, and G, values are shown as the mean ± S.E. (n = 4). **, p < 0.01, *, p < 0.05 significantly different between the two groups.
The transfection of oncogenic wild-type and Per2m/m cells with siRNA against Atf4 decreased ATF4 protein levels as well as the phosphorylation of pRB (Fig. 4D). The down-regulation of ATF4 in both types of cells increased the protein levels of p16INK4a and p19ARF, but the effect was greater on p16INK4a than on p19ARF (Fig. 4D). Furthermore, anchorage-independent growth of oncogene-introduced wild-type and Per2m/m cells was significantly suppressed by down-regulation of ATF4 (p < 0.01 for wild-type cells; p < 0.05 for Per2m/m cells, Fig. 4E). On the other hand, the transfection of oncogene-introduced Bmal1−/− cells with ATF4-expressing vectors decreased the protein levels of p16INK4a and p19ARF (Fig. 4F), whereas these protein levels in oncogene-introduced wild-type cells were not markedly affected by the transfection of ATF4-expressing vectors. The overexpression of ATF4 in Bmal1−/− cells also significantly enhanced their anchorage-independent growth ability (p < 0.01, Fig. 4G). These results suggested that the tumorigenicity of clock gene-defective cells was dependent on the responsiveness of ATF4 to the introduction of oncogenes.
Different Oncogenic Phenotypes of Cry1/2−/− and Clk/Clk Cells
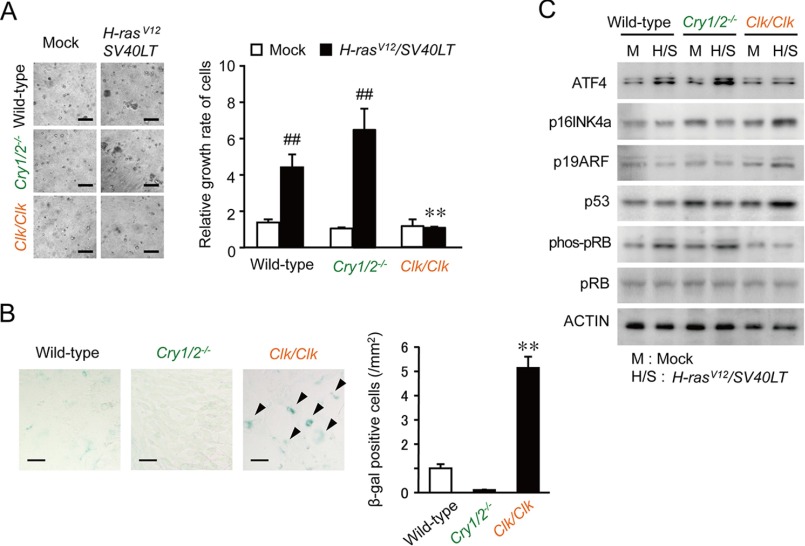
Next, we also investigated the tumorigenicity of other clock gene-defective cells. CRY proteins act as negative transcriptional regulators of circadian feedback loops. Cry-null mice, defective in both Cry1 and Cry2 (Cry1/2−/−), accordingly show arrhythmic behavior, physiology, and metabolism (25). In addition, the CLOCK protein heterodimerizes with BMAL1 and acts as a positive transcriptional regulator in the circadian clock machinery. Clk/Clk mice also exhibit abnormal rhythms in physiology and behavior (26, 27). The concomitant introduction of H-rasV12 and SV40LT significantly enhanced the anchorage-independent growth of Cry1/2−/− cells (p < 0.01) but did not promote the growth of Clk/Clk cells (Fig. 5A). In contrast, on day 14 after the infection with retrovirus vectors expressing oncogenes, the number of β-gal-positive Clk/Clk cells was significantly higher than those among wild-type and Cry1/2−/− cells (p < 0.01 respectively; Fig. 5B). The retroviral transfer of H-rasV12 and SV40LT into Clk/Clk cells had a negligible effect on ATF4 protein levels, whereas oncogene introduction enhanced the expression of p16INK4a, p19AFR, and p53 (Fig. 5C). These results also supported negative and positive regulators of circadian clock playing different roles in neoplastic transformation induced by the introduction of H-rasV12 and SV40LT.
FIGURE 5.
Different responses of Cry1/2- or Clock-defective cells to oncogene-induced transformation. A, anchorage-independent growth of wild-type, Cry1/2−/−, or Clk/Clk cells after the concomitant introduction of H-rasV12 and SV40LT. Cells infected with oncogenes were subjected to a soft-agar colony assay, and their colony formation and viability were assessed 14 days after seeding. Control cells were infected with mock vectors. The left panels show representative microscopic photographs of colony formation in each type of cells. The scale bars indicate 100 μm. The right panel shows viability of cells. Values are shown as the means ± S.E. (n = 4). Mean values of mock-transfected wild-type cells were set at 1.0. ##, p < 0.01 significantly different from the mock-transfected group. **, p < 0.01 significantly different from oncogene-transfected wild-type and Cry1/2−/− cells. B, β-gal staining of wild-type, Cry1/2−/−, or Clk/Clk cells infected with oncogenes. Arrows in the microscopic photograph indicate β-gal-positive cells. The scale bars indicate 50 μm. Values are the means ± S.E. (n = 3). **, p < 0.01 significantly different from other oncogene-introduced cells. C, the protein abundance of p16INK4a, p19ARF, p53, phosphorylated pRB (phos-pRB), and pRB in wild-type, Cry1/2−/−, or Clk/Clk cells infected with oncogenes.
Transcriptional Regulation of Atf4 by Products of Clock Genes
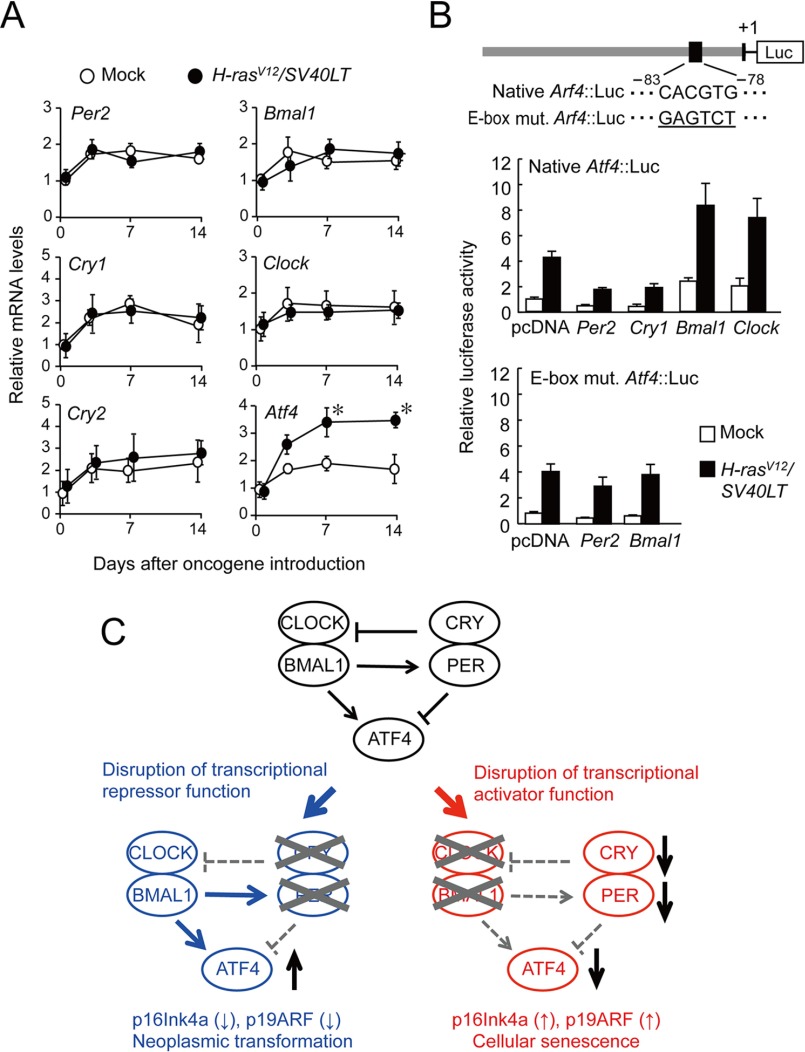
In the final set of experiments, we investigated the mechanisms by which circadian clock genes modulate ATF4 expression during oncogenic transformation. The mRNAs for Per2, Cry1, Cry2, Bmal1, and Clock were detected in normal wild-type cells (day 0). Their mRNA levels were elevated after the concomitant introduction of H-rasV12 and SV40LT; however, the time courses of mRNA elevations in oncogene-introduced cells were similar to those observed in cells that were infected with mock vectors (Fig. 6A). In contrast, the introduction of H-rasV12 and SV40LT into wild-type cells significantly increased the mRNA levels of Atf4 (p < 0.05).
FIGURE 6.
Transcriptional regulation of Atf4 gene by clock gene products. A, the time course of mRNA expressions of Per2, Cry1, Cry2, Bmal1, Clock, and Atf4 in wild-type cells after infection with retrovirus vectors expressing H-rasV12 and SV40LT. Mean values of mock-transfected cells on day 0 are set at 1.0. Values are shown as the mean ± S.E. (n = 3). *, p < 0.05 as compared with mock-transfected group at corresponding time points. B, luciferase reporter assay of the mouse Atf4 promoter in oncogene introduced wild-type cells. The upper panel shows a schematic representation of the mouse Atf4 promoter. Numbers below the boxes are nucleotide residues positioned relative to transcription start site (+1). Underlined nucleotide residues indicate a mutated sequence of E-box. Wild-type MEFs and oncogene-introduced wild-type MEFs were transfected with 0.1 μg of each native Atf4::Luc or E-box mutated Atf4::Luc (E-box mut. Atf4::Luc). Cells transfected with reporter constructs were also co-transfected with expressing constructs for Per2, Cry1, Bmal1, or Clock. Values are expressed as relative ratio to the Atf4::Luc activity of wild-type MEFs in the absence of expressing vectors (set at 1.0). All values are shown as the means ± S.E. (n = 4). C, possible mechanisms for the promotion of oncogenicity by disruptions in the molecular circadian clock. When the circadian clock is disrupted due a dysfunction in the transcriptional repressor PER or CRY, ATF4 is markedly expressed by oncogenic stimuli, and cells easily undergo malignant transformation. In contrast, when the circadian clock is disrupted due to a dysfunction in the transcriptional activator CLOCK or BMAL1, ATF4 is hardly expressed by oncogenic stimuli, and p16INK4a and p19ARF induce cellular senescence.
The products of Bmal1 and Clock have been shown to positively regulate the expression of their target genes through an E-box (CACGTG) element (1, 2). BMAL1/CLOCK-mediated transactivation is repressed by PERs or CRYs. An E-box motif has been detected upstream of the Atf4 gene in mice and all mammals examined, including rats, chimpanzees, and humans (28, 29). The activity of the luciferase reporter of the mouse Atf4 promoter region containing the E-box (native Atf4::Luc) was enhanced when it was transfected into oncogene-transformed wild-type cells (Fig. 6B). The enhanced reporter activity of native Atf4::Luc was repressed by the co-transfection with Per2- or Cry1-expressing vectors. In contrast, the co-transfection with Bmal1- or Clock-expressing vectors further enhanced the reporter activity of native Atf4::Luc in oncogene-transformed wild-type cells. Although the reporter activity of E-box-mutated Atf4 luciferase constructs (E-box mut. Atf4::Luc) was also enhanced in oncogene-transformed wild-type cells (Fig. 6B), enhanced reporter activity was not further modulated by the co-transfection with Per2- and Bmal1-expressing vectors. These results indicated that the oncogene-induced transformation of cells resulted in elevations in ATF4 levels without affecting clock gene expression, whereas components of the circadian clock appeared to influence the expression of Atf4 in oncogenic cells through E-box elements.
Discussion
The results of the present study suggest the opposite roles for the negative and positive components of the circadian clock in oncogene-induced transformation. As shown in Fig. 6C, when the circadian rhythms are disrupted due to a dysfunction in the transcriptional repressor PER or CRY, the expression of ATF4 is facilitated by oncogenic stimuli, and cells override normal cell cycle mechanism, and easily undergo malignant transformation. By contrast, when the circadian rhythms are disrupted due to a dysfunction in the transcriptional activator BMAL1 or CLOCK, ATF4 is unresponsive to oncogenic stimuli, and elevations in the expression of p16INK4a and p19ARF causes cellular senescence. Consequently, oncogene-introduced Bmal1- or Clock-defective cells fail to override the normal cell-cycle mechanism.
ATF4 is ubiquitously expressed throughout the body and is induced in response to various stress signals, including anoxia, hypoxia, endoplasmic reticulum stress, amino acid deprivation, and oxidative stress (29). The stress-induced expression of ATF4 causes adaptive responses in cells through regulating the expression of target genes involved in amino acid synthesis, differentiation, metastasis, angiogenesis, and drug resistance (30). The expression of ATF4 is also under the control of the circadian clock (28). BMAL1:CLOCK heterodimers activate the transcription of Atf4 through E-box enhancer elements, but this transactivation is repressed by PER and CRY. The reporter activity of native Atf4::Luc in oncogene-introduced cells was further enhanced by Bmal1- and Clock-expressing vectors. Therefore, the expression of ATF4 appeared to be diminished in Bmal1- or Clock-defective cells even when those cells were introduced with oncogenes. On the other hand, in the absence of negative circadian regulator PER and CRY, ATF4 seemed to be markedly expressed in response to oncogenic stimuli. Excessive expression of ATF4 is often observed in malignant tumors in humans and rodents (30). The highly expressed ATF4 is thought to facilitate tumor progression, because transcription of genes involved in tumor cell proliferation is modulated by ATF4 (31).
We previously reported that ATF4 promotes the oncogene-induced neoplastic transformation of MEFs by suppressing the expression of p16INK4a and p19ARF (15). The introduction of H-rasV12 and SV40LT into wild-type MEFs induces the expression of the Atf4 gene along with that of p16INK4a and p19ARF. Elevations in ATF4 protein levels suppress the expression of these cellular senescence factors and drive oncogenic transformation. Therefore, elevated levels of ATF4 in oncogene-introduced wild-type and Per2m/m cells may be sufficient to suppress the expression of p16INK4a and p19ARF. Conversely, low levels of ATF4 in Bmal1−/− cells appear to allow the constitutive expression of these cellular senescence factors.
In this study we used retrovirus vectors expressing SV40LT as a transforming agent together with H-rasV12. Because SV40LT inactivates p53 by protein-protein interactions and also perturbs the function of pRb (23, 24), SV40LT-transduced cells are considered to be insensitive to growth arrest by p16INK4a and p19ARF. Inactivation of the growth-suppressive properties of p53 and pRb was previously shown to be essential for the immortalization of MEFs by SV40LT. The concomitant introduction of H-rasV12 and SV40LT induced elevations in p53 protein levels in Bmal1−/− cells. p53 protein levels were markedly larger in oncogene-introduced Bmal1−/− cells than in wild-type and Per2m/m cells. Significant amounts of p53 protein in Bmal1−/− cells was unable to be precipitated together with SV40LT. In addition, there were no significant mutations in the p53 gene or p16Ink4a gene among wild-type, Per2m/m, and Bmal1−/− cells (supplemental Fig. S1). Extensive expression of p53 in oncogene-introduced Bmal1−/− cells overrode the binding capacity of SV40LT. Therefore, untrapped/free p53 protein may allow the induction of cellular senescence. On the other hand, in wild-type, Per2m/m, and Bmal1−/− cells, a large amount of pRb was unable to be precipitated together with SV40LT. The growth inhibitory function of pRb still remained in all cell types after the infection of oncogenes. The function of the remained pRb may be modulated by p16INK4a.
Although genetic ablation of either negative or positive components of the circadian clock leads to disrupted rhythm in physiological functions, our findings define the different contributions of each component of the circadian clock to neoplastic transformation. The fundamental mechanism of the mammalian circadian clock is conserved beyond the species (32). Similar mechanism may function in human cells during oncogenic transformation associated with the disruption of circadian clock machinery.
Author Contributions
C. K., S. K., and S. O. designed the study and wrote the paper. C. K. and S. K. performed and analyzed the experiments shown in Figs. 1–6. S. Shiromizu, N. Matsunaga, S. Shimba S., and S. Shibata provided technical assistance and contributed to the preparation of the figures. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
Cry-null MEFs were kindly provided by Dr. Ueda (The University of Tokyo). We thank the Research Support Center, Graduate School of Medical Sciences, Kyushu University for technical support.
This work was supported in part of Japan Society for the Promotion of Science (JSPS) KAKENHI Grants FCG6670317 (to S. K.) and 25253038 (to S. O.) and the Mandom International Research Grants on Alternative to Animal Experiments. The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Fig. S1.
- MEF
- mouse embryonic fibroblast
- ATF4
- activating transcription factor 4.
References
- 1. Dibner C., Schibler U., and Albrecht U. (2010) The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu. Rev. Physiol. 72, 517–549 [DOI] [PubMed] [Google Scholar]
- 2. Partch C. L., Green C. B., and Takahashi J. S. (2014) Molecular architecture of the mammalian circadian clock. Trends Cell Biol. 24, 90–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gachon F., Olela F. F., Schaad O., Descombes P., and Schibler U. (2006) The circadian PAR-domain basic leucine zipper transcription factors DBP, TEF, and HLF modulate basal and inducible xenobiotic detoxification. Cell Metab. 4, 25–36 [DOI] [PubMed] [Google Scholar]
- 4. Gréchez-Cassiau A., Rayet B., Guillaumond F., Teboul M., and Delaunay F. (2008) The circadian clock component BMAL1 is a critical regulator of p21WAF1/CIP1 expression and hepatocyte proliferation. J. Biol. Chem. 283, 4535–4542 [DOI] [PubMed] [Google Scholar]
- 5. Panda S., Antoch M. P., Miller B. H., Su A. I., Schook A. B., Straume M., Schultz P. G., Kay S. A., Takahashi J. S., and Hogenesch J. B. (2002) Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 109, 307–320 [DOI] [PubMed] [Google Scholar]
- 6. Masri S., Kinouchi K., and Sassone-Corsi P. (2015) Circadian clocks, epigenetics, and cancer. Curr. Opin. Oncol. 27, 50–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lahti T. A., Partonen T., Kyyrönen P., Kauppinen T., and Pukkala E. (2008) Night-time work predisposes to non-Hodgkin lymphoma. Int. J. Cancer 123, 2148–2151 [DOI] [PubMed] [Google Scholar]
- 8. Kloog I., Haim A., Stevens R. G., and Portnov B. A. (2009) Global co-distribution of light at night (LAN) and cancers of prostate, colon, and lung in men. Chronobiol. Int. 26, 108–125 [DOI] [PubMed] [Google Scholar]
- 9. Filipski E., Innominato P. F., Wu M., Li X. M., Iacobelli S., Xian L. J., and Lévi F. (2005) Effects of light and food schedules on liver and tumor molecular clocks in mice. J. Natl. Cancer Inst. 97, 507–517 [DOI] [PubMed] [Google Scholar]
- 10. Lee S., Donehower L. A., Herron A. J., Moore D. D., and Fu L. (2010) Disrupting circadian homeostasis of sympathetic signaling promotes tumor development in mice. PLoS ONE 5, e10995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fu L., Pelicano H., Liu J., Huang P., and Lee C. (2002) The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell 111, 41–50 [DOI] [PubMed] [Google Scholar]
- 12. Jozefczuk J., Drews K., and Adjaye J. (2012) Preparation of mouse embryonic fibroblast cells suitable for culturing human embryonic and induced pluripotent stem cells. J. Vis. Exp. 64, 3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ukai-Tadenuma M., Yamada R. G., Xu H., Ripperger J. A., Liu A. C., and Ueda H. R. (2011) Delay in feedback repression by cryptochrome 1 is required for circadian clock function. Cell 144, 268–281 [DOI] [PubMed] [Google Scholar]
- 14. Kakumoto K., Sasai K., Sukezane T., Oneyama C., Ishimaru S., Shibutani K., Mizushima H., Mekada E., Hanafusa H., and Akagi T. (2006) FRA1 is a determinant for the difference in RAS-induced transformation between human and rat fibroblasts. Proc. Natl. Acad. Sci. U.S.A. 103, 5490–5495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Horiguchi M., Koyanagi S., Okamoto A., Suzuki S. O., Matsunaga N., and Ohdo S. (2012) Stress-regulated transcription factor ATF4 promotes neoplastic transformation by suppressing expression of the INK4a/ARF cell senescence factors. Cancer Res. 72, 395–401 [DOI] [PubMed] [Google Scholar]
- 16. Hanahan D., and Weinberg R. A. (2011) Hallmarks of cancer: the next generation. Cell 144, 646–674 [DOI] [PubMed] [Google Scholar]
- 17. Collado M., Gil J., Efeyan A., Guerra C., Schuhmacher A. J., Barradas M., Benguría A., Zaballos A., Flores J. M., Barbacid M., Beach D., and Serrano M. (2005) Tumour biology: senescence in premalignant tumours. Nature 436, 642. [DOI] [PubMed] [Google Scholar]
- 18. Collado M., and Serrano M. (2010) Senescence in tumours: evidence from mice and humans. Nat. Rev. Cancer 10, 51–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kim W. Y., and Sharpless N. E. (2006) The regulation of INK4/ARF in cancer and aging. Cell 127, 265–275 [DOI] [PubMed] [Google Scholar]
- 20. Kamijo T., Zindy F., Roussel M. F., Quelle D. E., Downing J. R., Ashmun R. A., Grosveld G., and Sherr C. J. (1997) Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell 91, 649–659 [DOI] [PubMed] [Google Scholar]
- 21. Sharpless N. E. (2005) INK4a/ARF: a multifunctional tumor suppressor locus. Mutat. Res. 576, 22–38 [DOI] [PubMed] [Google Scholar]
- 22. Sherr C. J. (2006) Divorcing ARF and p53: an unsettled case. Nat. Rev. Cancer 6, 663–673 [DOI] [PubMed] [Google Scholar]
- 23. Zhu J. Y., Abate M., Rice P. W., and Cole C. N. (1991) The ability of simian virus 40 large T antigen to immortalize primary mouse embryo fibroblasts cosegregates with its ability to bind to p53. J. Virol. 65, 6872–6880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zalvide J., and DeCaprio J. A. (1995) Role of pRb-related proteins in simian virus 40 large-T-antigen-mediated transformation. Mol. Cell Biol. 15, 5800–5810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. van der Horst G. T., Muijtjens M., Kobayashi K., Takano R., Kanno S., Takao M., de Wit J., Verkerk A., Eker A. P., van Leenen D., Buijs R., Bootsma D., Hoeijmakers J. H., and Yasui A. (1999) Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature 398, 627–630 [DOI] [PubMed] [Google Scholar]
- 26. Vitaterna M. H., King D. P., Chang A. M., Kornhauser J. M., Lowrey P. L., McDonald J. D., Dove W. F., Pinto L. H., Turek F. W., and Takahashi J. S. (1994) Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science 264, 719–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Turek F. W., Joshu C., Kohsaka A., Lin E., Ivanova G., McDearmon E., Laposky A., Losee-Olson S., Easton A., Jensen D. R., Eckel R. H., Takahashi J. S., and Bass J. (2005) Obesity and metabolic syndrome in circadian Clock mutant mice. Science 308, 1043–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Koyanagi S., Hamdan A. M., Horiguchi M., Kusunose N., Okamoto A., Matsunaga N., and Ohdo S. (2011) cAMP-response element (CRE)-mediated transcription by activating transcription factor-4 (ATF4) is essential for circadian expression of the Period2 gene. J. Biol. Chem. 286, 32416–32423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ameri K., and Harris A. L. (2008) Activating transcription factor 4. Int. J. Biochem. Cell Biol. 40, 14–21 [DOI] [PubMed] [Google Scholar]
- 30. Ye J., Kumanova M., Hart L. S., Sloane K., Zhang H., De Panis D. N., Bobrovnikova-Marjon E., Diehl J. A., Ron D., and Koumenis C. (2010) The GCN2-ATF4 pathwayis critical for tumourcell survival and proliferation in response to nutrient deprivation. EMBO J. 29, 2082–2096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bi M., Naczki C., Koritzinsky M., Fels D., Blais J., Hu N., Harding H., Novoa I., Varia M., Raleigh J., Scheuner D., Kaufman R. J., Bell J., Ron D., Wouters B. G., and Koumenis C. (2005) ER stress-regulated translation increases tolerance to extreme hypoxia and promotes tumor growth. EMBO J. 24, 3470–3481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dibner C., and Schibler U. (2015) Circadian timing of metabolism in animal models and humans. J. Intern. Med. 277, 513–527 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.