Abstract
N-Glycosylation is a post-translational modification common to all three domains of life. In many archaea, the oligosacharyltransferase (AglB)-dependent N-glycosylation of flagellins is required for flagella assembly. However, whether N-glycosylation is required for the assembly and/or function of the structurally related archaeal type IV pili is unknown. Here, we show that of six Haloferax volcanii adhesion pilins, PilA1 and PilA2, the most abundant pilins in pili of wild-type and ΔaglB strains, are modified under planktonic conditions in an AglB-dependent manner by the same pentasaccharide detected on H. volcanii flagellins. However, unlike wild-type cells, which have surfaces decorated with discrete pili and form a dispersed layer of cells on a plastic surface, ΔaglB cells have thick pili bundles and form microcolonies. Moreover, expressing PilA1, PilA2, or PilA6 in ΔpilA[1–6]ΔaglB stimulates microcolony formation compared with their expression in ΔpilA[1–6]. Conversely, expressing PilA3 or PilA4 in ΔpilA[1–6] cells results in strong surface adhesion, but not microcolony formation, and neither pilin stimulates surface adhesion in ΔpilA[1–6]ΔaglB cells. Although PilA4 assembles into pili in the ΔpilA[1–6]ΔaglB cells, these pili are, unlike wild-type pili, curled, perhaps rendering them non-functional. To our knowledge, this is the first demonstration of a differential effect of glycosylation on pilus assembly and function of paralogous pilins. The growth of wild-type cells in low salt media, a condition that decreases AglB glycosylation, also stimulates microcolony formation and inhibits motility, supporting our hypothesis that N-glycosylation plays an important role in regulating the transition between planktonic to sessile cell states as a response to stress.
Keywords: adhesion, archaea, biofilm, Haloferax volcanii, N-linked glycosylation, type IV pili, flagella, flagellin, pili, pilin
Introduction
In all three domains of life, N-glycosylation is a post-translational modification that is critical for the functions of many surface-bound and secreted proteins (1–3). N-Glycans are first assembled on a phosphorylated polyprenol lipid, followed by transfer of the assembled polysaccharide, by an oligosaccharyltransferase, to the target protein at an asparagine located in the consensus sequence NX(S/T) (4, 5). In eukaryotes, where protein N-glycosylation is common, the modification plays an important role in supporting the stability and folding of many secreted and membrane proteins (5). Although long thought to be limited to a small subset of bacteria, recent genomic studies have shown that a large percentage of bacteria possess homologs of N-glycosylation pathway components (2). Moreover, in vivo studies have confirmed that these homologs are indeed components of a bacterial N-glycosylation pathway, although the roles played by this post-translational modification in most bacteria remain poorly understood (6).
In archaea, pathways that build and transfer sugars to individual proteins are known as the archaeal glycosylation (Agl) pathways (7–9). N-Glycosylation is prevalent throughout the archaea and appears to be essential in some species, such as Sulfolobus acidocaldarius, where attempted deletions of the gene encoding the oligosaccharyltransferase, AglB, have been unsuccessful (1, 11). Conversely, in two methanogens, Methanococcus voltae and Methanococcus maripaludis, as well as in the haloarchaeon Haloferax volcanii, AglB is not essential under laboratory conditions, allowing the specific effects of this pathway to be studied in these genetically and biochemically tractable euryarchaea (8–10, 12). In M. voltae, the flagellins and a potential S-layer glycoprotein bear 779-Da trisaccharides added by AglB (13). M. maripaludis flagellins, which are modified with a tetrasaccharide, have been more extensively studied, and flagella assembly requires glycosylation, because an aglB deletion mutant is not flagellated (9, 10).
Subunits of archaeal type IV pili, structurally related surface filaments to archaeal flagella, also known as archaella (14, 15), are also glycosylated. These filaments are modified by a pentasaccharide that consists of the tetrasaccharide added to the M. maripaludis flagellins plus an extra hexose (16). Although cells lacking aglB have flagellins and pilins that lack AglB-dependent N-glycosylation, the surfaces of these cells exhibit ample piliation (9). Although the composition and functionality of the pili on this ΔaglB strain remain to be determined, the fact that these mutant cells lack flagella under the tested laboratory conditions but retain pili is intriguing, considering that pili and flagella play opposing functional roles in the context of biofilm formation. The pili are required for adhesion and microcolony formation, which are early steps in biofilm formation, although flagella are required for swimming motility, which is essential for biofilm dispersal (15).
In H. volcanii, AglB-dependent glycosylation of the S-layer as well as the flagellin FlgA1 results in proteins that are modified with a pentasaccharide. A tetrasaccharide, composed of a hexose (glucose), two hexuronic acids (glucuronic acid and galacturonic acid), and a methylated hexuronic acid (methyl-O-4-glucuronic acid), is first transferred to the protein, followed by the addition of the terminal mannose (12, 17, 18). Wild-type motility requires the presence of the entire pentasaccharide, and flagellum assembly requires AglB-dependent glycosylation of FlgA1. However, similar to M. maripaludis, an H. volcanii ΔaglB strain has remaining surface filaments that are not flagella (19). H. volcanii has six type IV adhesion pilins, PilA1–6, that contain a highly conserved hydrophobic domain required for pilus assembly, adhesion, and motility regulation (20, 21). Although each of these pilins can be assembled into a functional pilus in a ΔpilA[1–6] mutant expressing a single pilin in trans, the adhesion phenotypes differ (20). Here, we demonstrate that the surface filaments of a ΔaglB strain are indeed type IV pili, composed of at least a subset of the PilA pilins, despite the fact that five of the six pilins are glycosylated in an AglB-dependent manner in H. volcanii. Unlike the wild-type, the pili of the ΔaglB strain forms thick bundles containing multiple filaments, and the lack of AglB-dependent glycosylation in this strain promotes microcolony formation. Although this post-translational modification is crucial to the functions of PilA3 and PilA4, but not to the assembly of pili containing PilA4 subunits, other PilA pilins are functional in the ΔaglB strain. Our results also suggest that N-glycosylation plays a role in regulating the response to low salt conditions where wild-type H. volcanii is non-motile and forms microcolonies similar to ΔaglB under standard salt conditions used in the laboratory.
Experimental Procedures
Strains and Growth Conditions
The plasmids and strains used in this study are listed in Table 1. H. volcanii H53 and its derivatives were grown at 45 °C in liquid or on solid agar semi-defined casamino acid (CA)6 medium, supplemented with tryptophan and uracil (50 μg ml−1 final concentration), or complex Modified Growth Medium.7 For low salt medium, the total salt concentration of CA medium was reduced from 18% (∼2.5 m) to 12% (∼1.66 m). Strains transformed with pTA963 were grown on CA medium supplemented with tryptophan (50 μg ml−1 final concentration) (31). For selection of the deletion mutants (see below), 5-fluorootic acid was added at a final concentration of 150 μg ml−1 in CA medium, and uracil was added to a final concentration of 10 μg ml−1. Strain H53 and the deletion mutants transformed with pTA963 or its derivatives were grown in CA medium supplemented with tryptophan at a final concentration of 50 μg ml−1. Escherichia coli strains were grown at 37 °C in NZCYM medium supplemented with ampicillin (200 μg ml−1) (31).
TABLE 1.
Plasmids and strains
| Strain or plasmid | Relevant characteristic(s) | Ref. or source |
|---|---|---|
| pTA131 | Ampr; pyrE2 under a ferredoxin promoter | 23 |
| pTA963 | Ampr, pyrE2 and hdrB markers, inducible ptna promoter | 24 |
| pRE3 | pTA963 containing pilA2His | 20 |
| pRE32 | pTA963 containing pilA3His | 20 |
| pRE33 | pTA963 containing pilA4His | 20 |
| pRE34 | pTA963 containing pilA5His | 20 |
| pRE35 | pTA963 containing pilA6His | 20 |
| pMT24 | pTA963 containing pilA1His | 20 |
| pFH27 | pTA131 containing chromosomal aglB flanking regions | This study |
| E. coli | ||
| DH5α | F− 80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA hsdR17(rK− mK−) supE44 thi-1 gyrA relA1 | Invitrogen |
| DL739 | MC4100 recA dam-13::Tn9 | 25 |
| H. volcanii | ||
| H53 | ΔpyrE2 Δtrp | 23 |
| RE13 | H53 containing pMT24 | 20 |
| RE15 | ΔaglB containing pMT24 | This study |
| RE19 | H53 containing pRE3 | 20 |
| RE21 | ΔaglB containing pRE3 | This study |
| RE30 | RE43 containing pTA963 | 20 |
| RE37 | H53 containing pRE34 | 20 |
| RE40 | H53 containing pRE32 | 20 |
| RE41 | H53 containing pRE33 | 20 |
| RE43 | H53 ΔpilA1 ΔpilA2 ΔpilA3 ΔpilA4 ΔpilA5 ΔpilA6 | 20 |
| RE44 | RE43 containing pRE34 | 20 |
| RE45 | RE43 containing pMT24 | 20 |
| RE48 | H53 containing pRE35 | 20 |
| RE50 | RE43 containing pRE35 | 20 |
| RE51 | RE43 containing pRE32 | 20 |
| RE52 | RE43 containing pRE33 | 20 |
| RE53 | RE43 containing pRE3 | 20 |
| RE130 | H53 ΔpilA[1–6]ΔaglB | This study |
| RE137 | ΔaglB containing pRE35 | This study |
| RE138 | ΔaglB containing pRE34 | This study |
| RE139 | ΔaglB containing pRE33 | This study |
| RE140 | ΔaglB containing pRE32 | This study |
| RE141 | ΔaglB containing pTA963 | This study |
| RE142 | RE130 containing pTA963 | This study |
| RE143 | RE130 containing pMT24 | This study |
| RE144 | RE130 containing pRE3 | This study |
| RE145 | RE130 containing pRE32 | This study |
| RE146 | RE130 containing pRE33 | This study |
| RE147 | RE130 containing pRE34 | This study |
| RE148 | RE130 containing pRE35 | This study |
| MT13 | H53 containing pTA963 | 19 |
| ΔaglB | H53 ΔaglB | 7 |
| ΔaglD | H53 ΔaglD | 7 |
| ΔaglE | H53 ΔaglE | 26 |
| ΔaglF | H53 ΔaglF | 27 |
| ΔaglG | H53 ΔaglG | 27 |
| ΔaglI | H53 ΔaglI | 27 |
| ΔaglJ | H53 ΔaglJ | 28 |
| ΔaglM | H53 ΔaglM | 29 |
| ΔaglP | H53 ΔaglP | 30 |
Generation of Chromosomal Deletions
Chromosomal deletions of the PilA pilins and aglB in the ΔpilA[1–6] strain were generated by using a homologous recombination (pop-in pop-out) method previously described (32). The agl mutant strains were received from Eichler and co-workers (1 and references therein). Plasmid constructs were generated using overlap PCR (33) with modifications (34). To confirm the chromosomal replacement event occurred at the proper location on the chromosome, genomic DNA isolated from colonies derived using these techniques was screened by PCR using primers lying outside the gene of interest (primers used are listed in Table 2). The identities of the PCR products were verified by sequencing.
TABLE 2.
Primers used for PCR amplification
| Primer name | Sequence (5′–3′)a | Target sequences |
|---|---|---|
| FWaglBXhoI | ATAAAACTCGAGCAGACGGGTGCTTCG | 700 bp downstream of aglB start codon, extension toward gene |
| RVaglBXbaI | ATCTTATCTAGAGACTCCCGTATCGGG | 700 bp upstream of aglB start codon, extension toward gene |
| FWOverlapAglB | CGGTTGTTGGTCACACACGAGCCGAGACGG | 1 bp upstream aglB start codon, extends away from gene (fused with aglB stop codon) |
| RVOverlapAglB | CCGTCTCGGCTCGTGTGTGACCAACAACCG | aglB stop codon extends away from gene (fused with 1 bp downstream of aglB start codon, extends away from gene) |
a Restriction endonuclease sites are underlined.
Surface Adhesion Assay
H. volcanii surface adhesion was assayed using a modified air-liquid interface assay (35) as described previously (20). Briefly, 3 ml of culture in CA medium supplemented with tryptophan or tryptophan and uracil as necessary in the absence of pTA963, at an absorbance at 600 nm (A600) of ∼0.3, Fisher Scientific was incubated in each well of a covered 12-well plate. Plastic coverslips (22 × 22 mm; 0.19–0.25 mm thick) were inserted into each well and incubated overnight at 45 °C without shaking. Upon acetic acid fixing, coverslips were stained in 0.1% w/v crystal violet solution for 10 min. The coverslips were then washed with distilled water, air-dried, and examined using light microscopy.
Isolation and Purification of Surface Filaments
The isolation of H. volcanii flagella or type IV pili supernatant fractions was performed by cesium chloride (CsCl) gradient purification (36), with modifications (37). Briefly, to select for motile cells, colonies from a solid-agar plate were stab-inoculated into motility plates and incubated for 3 days at 45 °C; subsequently, cells from the outer motility ring were inoculated into 5 ml of CA liquid medium and grown to late log-phase. Two liters of CA medium were inoculated with a 5-ml culture each, and the cultures were harvested at an A600 of ∼0.3 by centrifugation at 8,700 rpm (JA-10 rotor; Beckman) for 30 min at 4 °C. The supernatant was centrifuged again (8,700 rpm for 30 min) and incubated at 4 °C with 4% w/v polyethylene glycol 6000 for 1 h. The polyethylene glycol-precipitated proteins were then centrifuged at 16,000 rpm (JLA-16.250 rotor; Beckman) for 50 min at 4 °C. Cell pellets were resuspended in 10 ml of CsCl dissolved in a 3 m NaCl saline solution to a final density of 1.37 g cm−1. The surface filaments were purified by CsCl density gradient centrifugation (overnight centrifugation at 50,000 rpm) (VTI-65.1 rotor; Beckman). Fractions were collected in 1-ml samples on ice.
LC-MS/MS Analysis of Surface Filaments
Proteins from fractions 4 and 5 from CsCl density gradients were digested with Glu-C or trypsin (sequencing grade, Promega) according to the filter-aided sample preparation method (38) with slight changes. The complete fraction was loaded onto a centrifugal filter unit with a 30-kDa molecular mass cutoff (Amicon ultra centrifugal filters, 0.5 ml, Millipore). Centrifugation at 14,000 × g for 15 min was performed between each step. The filter unit was washed with 200 μl of UA buffer (6 m urea and 2 m thiourea in 10 mm HEPES (pH 6.5)), followed by incubation for 20 min with 50 mm iodoacetamide in UA in the dark for carbamidomethylation of thiol groups. After washing twice with UA, samples were washed five times with 300 μl of either 10 mm phosphate buffer (pH 8.0) for Glu-C or 50 mm ammonium bicarbonate for trypsin digests, respectively. Digestion with 1 μg of either Glu-C or trypsin in 50 μl of the respective buffer was performed overnight at 37 °C. Peptides were eluted two times with 50 μl of H2O, dried in a vacuum centrifuge, and reconstituted in 6 μl of 2% (v/v) acetonitrile, 0.1% (v/v) formic acid in ultrapure water.
Chromatographic separation was performed on an Ultimate 3000 nanoflow HPLC system (Dionex) directly coupled via a nanospray source to a Q ExactiveTM Plus (Thermo Scientific) mass spectrometer. The mobile phases were composed of 0.1% (v/v) formic acid in ultrapure water (A) and 80% acetonitrile, 0.08% formic acid in ultrapure water (B). The sample (1 μl of reconstituted peptides) was desalted on a trap column (C18 PepMapTM 5 mm × 300 μm, 5-μm particle size, 100-Å pore size) for 5 min at a flow rate of 10 μl/min using 0.05% (v/v) TFA in ultrapure water followed by separation on an Acclaim PepMapTM RSLC C18 capillary column (75 μm × 15 cm, 2-μm particle size, 100-Å pore size). The following gradient was applied using a flow rate of 0.3 μl/min: 2.5% buffer B (5 min); 2.5–7.5% buffer B (4 min); 7.5–40% buffer B (26 min); 40–99% buffer B (1 min); and 99% buffer B (10 min).
The mass spectrometer was operated in positive ion mode using two different methods. In the first method, in-source collision-induced dissociation (IS-CID) was performed similarly as described by Mathieu-Rivet et al. (39) by applying an IS voltage of 80 V. Some samples were reanalyzed using 60, 70, and 90 V. MS1 scans were obtained from 400 to 2,000 m/z at a resolution of 70,000 full width at half-maximum, an AGC target of 106, and a maximum ion injection time of 100 ms. IS-CID of N-glycopeptides leads to the fragmentation of glycosidic bonds, which results in a series of neutral losses on MS1 level. The “Mass Tags” option (accuracy of 5 ppm) was enabled to select ion pairs differing by the mass of a monosaccharide (hexose, 162.0528 Da; hexuronic acid, 176.0321 Da; charges 1–4) for further fragmentation by higher energy collisional dissociation (HCD). The six most intense ion pairs were fragmented by HCD with 30% normalized collision energy. Unassigned charge states and charge states of >4 were rejected. MS2 spectra were then acquired at a resolution of 17,500 full width at half-maximum with a fixed first m/z 150, an AGC target of 5 × 105, and a maximum ion injection time of 120 ms. Fragmented ions were dynamically excluded for 5 s with a tolerance of 5 ppm.
In the second method, no IS-CID was applied. MS1 scans were obtained with the same parameters from 375 to 3,000 m/z. The 12 most intense ions were selected for HCD fragmentation with 27% normalized collision energy. Unassigned charge states as well as charge state 1 and >6 were rejected. MS2 spectra were acquired with the same parameters as before. Dynamic exclusion was enabled for 15 s. Some samples were reanalyzed using an inclusion list with the mass of intact glycopeptides as identified in the IS-CID experiments.
Peptides were identified using the search engine X! Tandem Sledgehammer (40) incorporated into Proteomatic (41). Obtained MS2 spectra were matched against a target-decoy database consisting of the UniProt H. volcanii proteome (proteome ID UP000008243, download date April 21, 2015) merged with sequences from the Common Repository of Adventitious Proteins (January 30, 2015) resulting in a total database of 4,984 proteins. Reverse protein sequences were used as decoy sequences. Search parameters included a mass accuracy of 5 ppm for MS1 precursor ions, 20 ppm for MS2 product ions, a maximum number of missed cleavages of three, carbamidomethylation of cysteine (static modification), and oxidation of methionine (variable). The mass of N-glycans ranging from mono- to pentasaccharide (162.0528, 338.0849, 514.117, 704.1647, and 866.2176 Da) was added as a variable modification of asparagine in separate identification runs. A statistical evaluation of peptide spectral matches was carried out using Quality (version 2.02) (42) with a q-value threshold of 0.05. In addition, all peptide spectral matches were filtered by a mass accuracy of 5 ppm.
If IS-CID was applied, MS2 spectra were used for the identification of peptide sequences, and the corresponding MS1 spectra of N-glycopeptides were manually annotated to identify the glycan composition. If intact N-glycopeptides were only fragmented by HCD, the identification of peptide sequences and glycan composition was based on MS2 peptide spectral matches. Peptide spectral counts were only analyzed if no IS-CID was applied, and no inclusion list was used. Peptide spectral counts from analyzing fractions 4 and 5 of CsCl gradients were summed up, as well as, on the protein level, spectral counts of glycopeptides with varying N-glycan length.
Protein Extraction, LDS-PAGE, and Western Blotting
Proteins from cell pellets of H. volcanii strains expressing His-tagged constructs were separated by lithium dodecyl sulfate (LDS) gel electrophoresis and analyzed by Western blot using anti-His antibodies (19). Liquid cultures were grown to early log phase (A600 ∼0.3). Cells were collected by centrifugation at 4,300 × g for 10 min at 4 °C. Cell pellets were resuspended and lysed in 1% v/v NuPAGE LDS supplemented with 50 mm dithiothreitol. The electrophoresis of protein samples was performed with 12% v/v BisTris NuPAGE gels under denaturing conditions using MOPS buffer (pH 7.7). Proteins were transferred from the gel onto a polyvinylidene difluoride membrane using a Transblot-SD semidry transfer cell (Bio-Rad) at 15 V for 30 min. Western blots of whole-cell lysates of strains expressing C-terminally His-tagged constructs with a three-amino acid linker sequence were probed with an anti-His antibody (Qiagen) at a dilution of 1:1,000, followed by a secondary anti-mouse antibody (Amersham Biosciences) at a dilution of 1:10,000. Antibody-labeled protein bands were identified using the Amersham Biosciences ECL Plus Western blotting detection system.
Transmission Electron Microscopy (TEM)
H. volcanii whole cells were prepared as described (37). To allow for observation of assembled PilA pili in ΔpilA[1–6]ΔaglB strains, 750 μl of liquid culture was incubated overnight in 12-well plates. Cell cultures from the 12-well plates were fixed in CA medium with 2% v/v glutaraldehyde and 1% v/v paraformaldehyde for 1 h. Ten microliters of the fixed culture was transferred onto glow-discharged copper grids coated with carbon-Formvar for 10 min. The grids were rinsed two times in double distilled H2O and negatively stained using 1% w/v uranyl acetate. Grids were then analyzed using a JEOL JEM-1010 operated at 80 keV with a side mount 1,000 AMT video rate camera. ImageJ software was used to measure the diameter of 10 ΔaglB bundles.
Results
Non-glycosylated Pilins Assemble into Pili and Form Pilus Bundles
In H. volcanii, the difference between pili and flagella cannot be easily discerned through electron microscopy, each being about 8–12 nm in diameter and varying substantially in length (21, 34). TEM of ΔaglB cell-associated filaments and CsCl-purified filaments revealed that their diameters are similar to those of type IV pili or flagella (8–12 nm) (19). Unlike wild-type or ΔflgA[1–2] strains, where cells have 1–12 individual filaments dispersed about the surface of the cell, the filaments of the ΔaglB strain are found in thick bundles that often appear at the poles of the rod-shaped cells (Fig. 1). As the ΔaglB strain is non-motile, these filaments were likely type IV pili.
FIGURE 1.
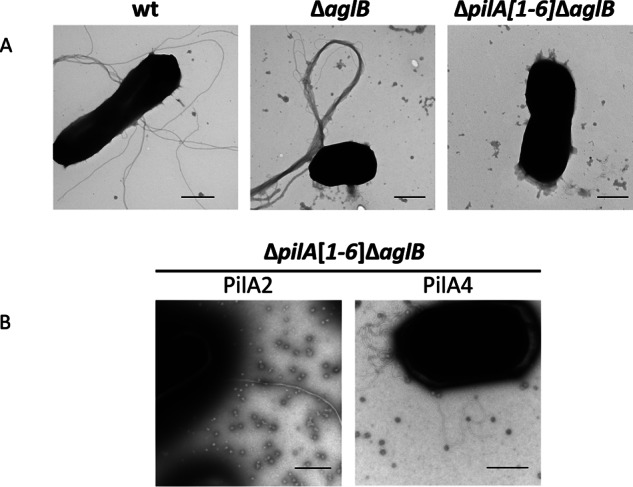
Glycosylation differentially affects pilus structures and interactions. A, TEM of whole cells of wild-type (wt), ΔaglB, and ΔpilA[1–6]ΔaglB strains transformed with pTA963; B, pTA963 expressing PilA2 or PilA4. WT cells, which have both pili and flagella on the surface (as determined by MS) (Fig. 2) (37), form long single filaments, although the pili on ΔaglB cells (as determined by MS (Fig. 2)) form long thick bundles. The ΔpilA[1–6]ΔaglB strain lacks pili and flagella. Although PilA2 forms long bundles of pili when expressed in the ΔpilA[1–6]ΔaglB strain, PilA4 has individual pili that unlike WT are curly when expressed in this background. Bars, 500 nm (n ≥2).
Indeed, mass spectrometry (MS) of CsCl gradient-purified trypsin-digested surface filaments revealed that FlgA1 and FlgA2 flagellin peptides, which are readily detectable in wild-type cells, were barely detectable in the ΔaglB strain (Table 3) (19). Consistent with these filaments being type IV pili, we identified PilA pilin peptides in both the wild-type and ΔaglB strains. In both strains, the majority of peptide spectral counts, which are an indicator for protein abundance (43), were derived from PilA1 and PilA2 (Table 3). PilA3, PilA5, and PilA6 peptide spectral counts suggest that these pilins are also incorporated into wild-type pilus structures; however, they are not present or are only barely detectable in the ΔaglB strain (Table 3). PilA4 was not detected in the wild-type and only two peptide spectral counts could be detected for this pilin in the ΔaglB strain. It is possible that the low peptide spectral counts are the result of cell-associated pilins due to cell lysis. To determine whether AglB-dependent N-glycosylation is present on the PilA pili, we carried out mass spectrometry of CsCl-purified filaments from wild-type and the ΔaglB strain.
TABLE 3.
Mass spectrometric analysis of the wild-type and ΔaglB strain indicates differences in the abundance of pilins
CsCl gradient fractions 4 and 5 from WT and the ΔaglB mutant were analyzed by MS applying HCD fragmentation. The number of spectra leading to the identification of the corresponding protein are given for the different pilins and flagellins as well as their UniProt ID. Peptide spectral counts from fraction 4 and 5 were combined (n = 1).
Additional N-Glycosites on FlgA1 and FlgA2
Previous MS analyses of CsCl gradient-purified supernatant of wild-type cells identified N-glycosylation of H. volcanii flagellin FlgA1 with a pentasaccharide composed of a hexose, two hexuronic acids, a methylated hexuronic acid, and a mannose at three of six predicted glycosylation sites in an AglB-dependent manner. Glycosylation at each of these sites is required for swimming motility (19).
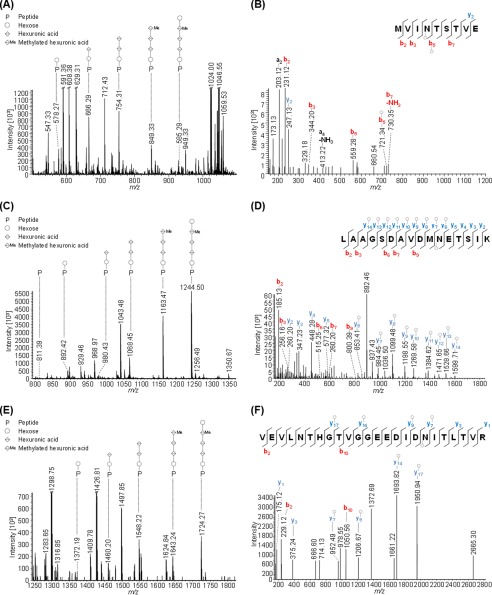
Using a more sensitive mass spectrometer, combined with IS-CID and HCD, has now allowed us to confirm an additional predicted N-glycosylation site for FlgA1. The peptide MVIN@TSTVE, where @ corresponds to the N-glycosite, is glycosylated with the same pentasaccharide identified at the other three glycosites (Fig. 2, A and B). Moreover, this work identified the same pentasaccharide N-glycan on the peptides VEVLNTHGTVGGEEDIDN@ITLTVR and LAAGSDAVDMN@ETSIK of FlgA2 that had been predicted to be glycosylated (Fig. 2, C–F) (19).
FIGURE 2.
Mass spectrometric identification of novel N-glycopeptides from FlgA1 and FlgA2. Peptides of GluC or trypsin-digested proteins from CsCl density gradients obtained from WT cells were analyzed by MS applying IS-CID (80 V). The N-glycan composition could be analyzed by a series of neutral losses in the MS1 spectra (A, C, and E) and resulted in the identification of a pentasaccharide composed of hexose (○), hexuronic acid (♢), and a methylated hexuronic acid (♢Me). The corresponding N-glycopeptide sequence was analyzed by further fragmentation (HCD) of a precursor ion corresponding to the peptide (P) + hexose and led to the identification of MVIN@TSTVE from FlgA1 (B, precursor m/z 578.27, charge 2) as well as LAAGSDAVDMN@ETSIK (D, precursor m/z 892.42, charge 2) and VEVLNTHGTVGGEEDIDN@ITLTVR (F, precursor m/z 1372.19, charge 2). @ corresponds to the N-glycosite. The observed b-ions (red) and y-ions (blue) are indicated in the spectrum as well as the fragmentation scheme, and the circle indicates the presence of a hexose attached to the peptide or fragment ion. Some peaks (slanted parallel bars) are not shown with their full intensity for better visualization (n = 1).
PilA2 Is N-Glycosylated with the Same Pentasaccharide Detected on the Flagellins
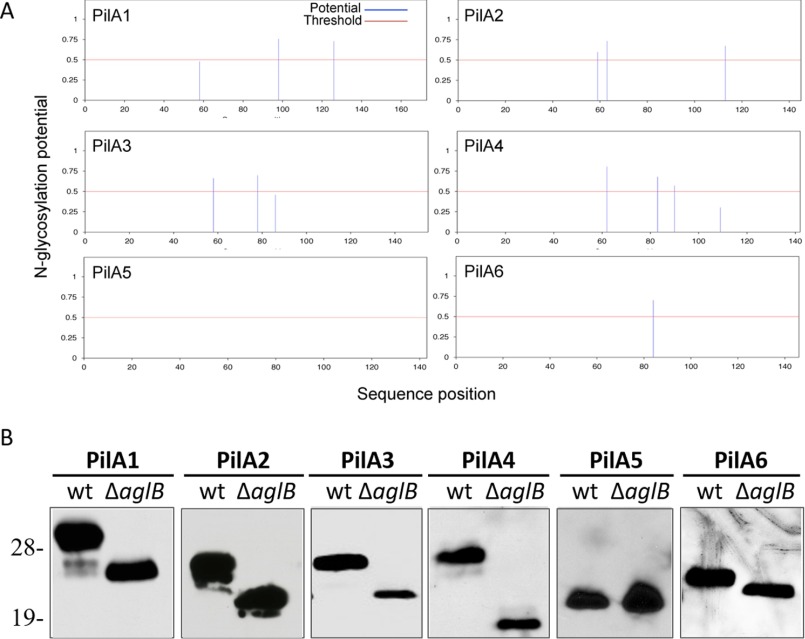
NetNGlyc 1.0, software used to predict N-glycosylation sites, also predicts that PilA1–4 and PilA6 are N-glycosylated (Fig. 3A). To determine whether the PilA pilins may be modified by AglB-dependent N-glycosylation, we transformed a plasmid expressing a single His-tagged pilin into wild-type and ΔaglB strains, and we determined the migration of these pilins using LDS-PAGE and Western blotting. Although PilA1–2 and PilA4–6 are expressed at similar levels in wild-type and ΔaglB, PilA3 expression is decreased in the mutant strain, correlating with the lack of PilA3 peptides detected in the ΔaglB strain using MS. Consistent with NetNGlyc predictions, Western blot analysis showed that all His-tagged pilins expressed in the strain lacking AglB migrate faster than those expressed in the wild type, except for PilA5, which lacks predicted N-glycosylation sites (Fig. 3B). This faster migration of these pilins, reminiscent of what was previously shown for flagellins expressed in a ΔaglB strain (19), is AglB-dependent, suggesting that the reduced molecular mass of the pilins is due to the lack of N-glycosylation.
FIGURE 3.
Only PilA5 lacks AglB-dependent N-glycosylation. A, PilA1–4 and PilA6 each have at least one N-glycosylation site predicted by NetNGlyc 1.0. B, Western blot analysis using anti-His antibodies was performed on protein extracts from cell lysates separated by LDS-PAGE of WT or ΔaglB strains expressing one of the six His-tagged PilA pilins in trans. Molecular mass standard indicated on the left (in kDa) (n ≥ 3).
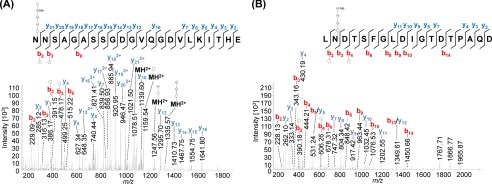
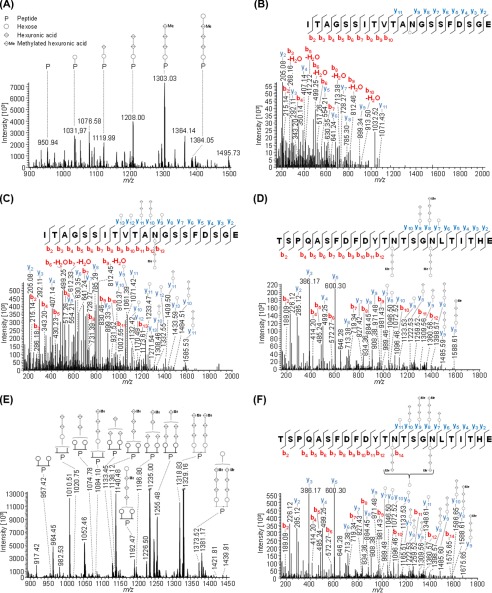
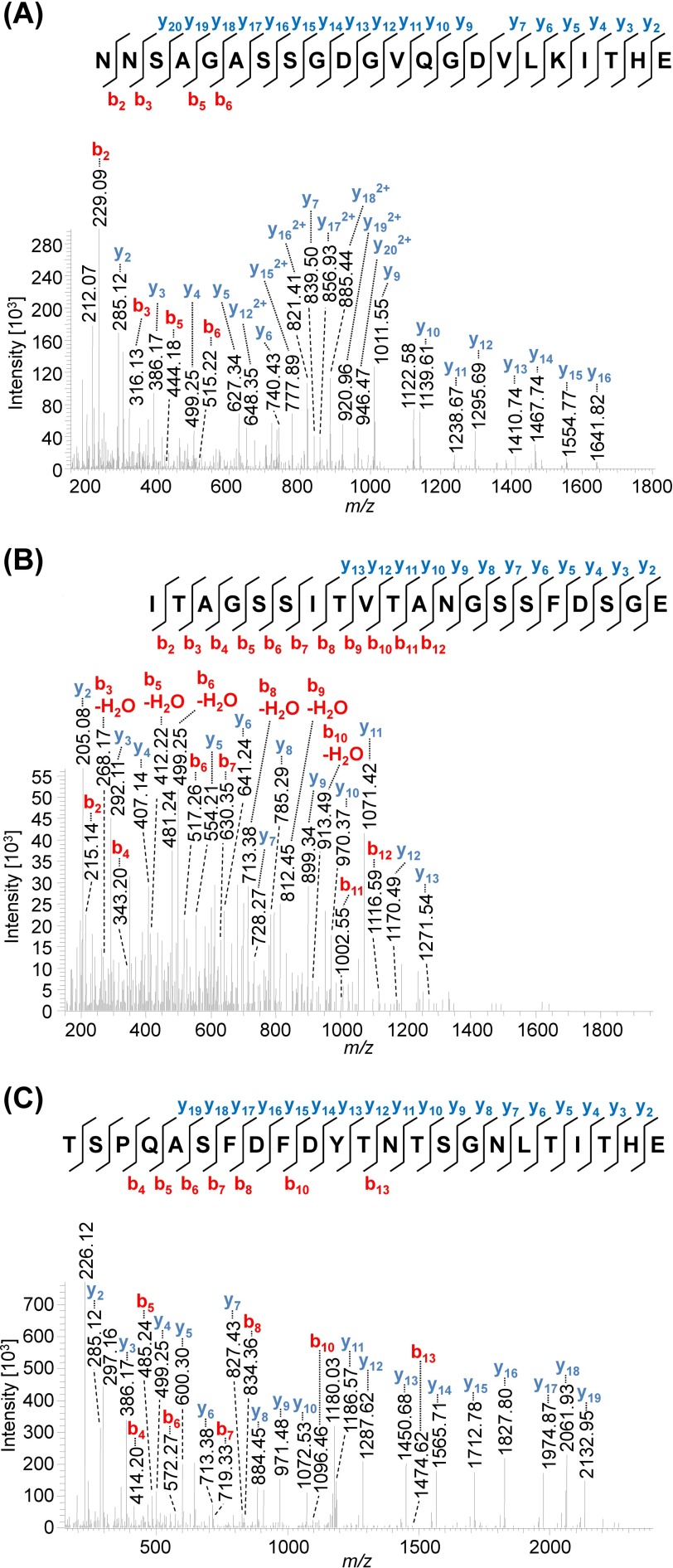
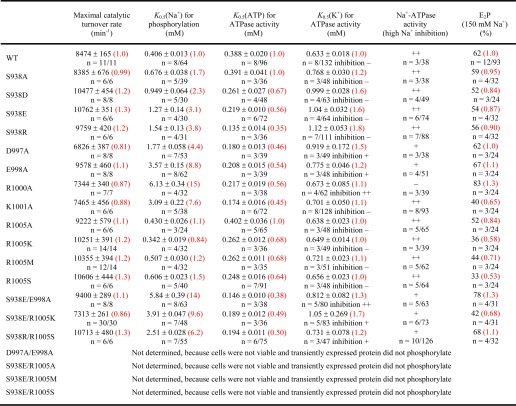
Indeed, MS data presented here identified both PilA1 (Fig. 4) and PilA2 (Fig. 5) peptides that were glycosylated at each of the two and three predicted sites, respectively. The same pentasaccharide found on the flagellins was identified on the PilA1 peptide, N@NSAGASSGDGVQGDVLKITHE (Fig. 4A), and on two PilA2 peptides, ITAGSSITVTAN@GSSFDSGE (Fig. 5, A–C) and TSPQASFDFDYTN@TSGN@LTITHE (Fig. 5D). Consistent with these peptides containing AglB-dependent glycosylation, only non-glycosylated peptides from PilA1 and PilA2 could be identified in the ΔaglB mutant (Fig. 6).
FIGURE 4.
Mass spectrometric identification of N-glycosylated peptides from PilA1. Peptides of GluC-digested proteins from CsCl density gradients obtained from WT cells were analyzed by MS. HCD fragmentation of intact glycopeptides allowed the identification of the peptide sequence by analyzing the resulting fragment ions, although the mass difference between the peptide backbone and the precursor ion led to the identification of the glycan composition. MS2 spectra corresponding to the fragmentation of N@NSAGASSGDGVQGDVLKITHE (A, precursor m/z 1008.74, charge 3) and LN@DTSFGLDIGTDTPAQD (B, precursor m/z 1292.51, charge 2) from PilA1 show the modification with a penta- and tetrasaccharide, respectively, at the marked glycosylation sites. An overview about the observed N-glycans with varying lengths is shown in Table 4. The observed b-ions (red) and y-ions (blue) are indicated in the spectra as well as the fragmentation scheme, including glycan fragments composed of hexose (○), hexuronic acid (♢), and methylated hexuronic acid (♢Me) that were attached to the peptide or fragment ions (n = 2).
FIGURE 5.
Mass spectrometric identification of PilA2 N-glycosylation. Peptides of GluC-digested proteins from CsCl density gradients obtained from WT cells were analyzed by MS. IS-CID was applied for A and B (80 V) and E (60 V); therefore, the N-glycan composition could be analyzed by a series of neutral losses from the N-glycosylated peptide ion (P) in the MS1 spectrum (A and E) and is depicted with symbols: hexose (○), hexuronic acid (♢), methylated hexuronic acid (♢Me). A pentasaccharide glycan could be identified (A), and the corresponding N-glycopeptide sequence was analyzed by further HCD fragmentation of m/z 1031.97 (charge 2) corresponding to the peptide + hexose leading to the identification of ITAGSSITVTAN@GSSFDSGE (B) from PilA2. The pentasaccharide N-glycosylation could be confirmed by HCD fragmentation of intact glycopeptides without IS-CID for PilA2 peptides ITAGSSITVTAN@GSSFDSGE (C, precursor m/z 1384.55, charge 2) and TSPQASFDFDYTN@TSGN@LTITHE (D, precursor m/z 1427.86, charge 3), which contains two glycosites. The two glycosites from TSPQASFDFDYTN@TSGN@LTITHE can harbor N-glycans of different length as observed in the MS1 spectrum (E) after applying IS-CID. The presence of a tetrasaccharide, lacking the terminal hexose, and a pentasaccharide on the same peptide was confirmed without IS-CID by HCD fragmentation of the triply charged precursor m/z 1075.44 (F). It could not be specified which of the two glycosites was modified by the tetra- and pentasaccharide, respectively. The observed b-ions (red) and y-ions (blue) are indicated in the spectra as well as the fragmentation schemes, including attached glycan fragments. Some peaks (slanted parallel bars) are not shown with their full intensity for better visualization (n = 2).
FIGURE 6.
Mass spectrometric identification of non-glycosylated peptides from PilA1 and PilA2 in the ΔaglB strain. Peptides of GluC-digested proteins from CsCl density gradients obtained from cells from the ΔaglB strain were analyzed by MS using HCD fragmentation. Consistent with the peptides containing AglB-dependent glycosylation, only the non-glycosylated peptides NNSAGASSGDGVQGDVLKITHE (A, precursor m/z 719.68, charge 3) from PilA1 as well as ITAGSSITVTANGSSFDSGE (B, precursor m/z 950.94, charge 2) and TSPQASFDFDYTNTSGNLTITHE (C, precursor m/z 1427.86, charge 3) from PilA2 were identified. The observed b-ions (red) and y-ions (blue) are indicated in the presented MS2 spectra as well as the fragmentation scheme (n = 2).
In addition to the pentasaccharide modification, we found that a tetrasaccharide can alternatively decorate both of the two N-glycosites found on each of the PilA1 and PilA2 peptides (Table 4). Furthermore, both the pentasaccharide and the tetrasaccharide can be present on the same protein, which was observed for the TSPQASFDFDYTN@TSGN@LTITHE peptide from PilA2, which harbors both one tetra- and one pentasaccharide on its two glycosites (Fig. 5, E and F). Corresponding peptide spectral counts of shortened N-glycans ranging from one to three carbohydrate residues suggest a higher abundance of the tetra- and pentasaccharide modification compared with the shortened N-glycans, which were only found with strongly decreased peptide spectral counts in PilA2 (Table 4). To our knowledge, chemical partial degradation of the N-glycan during sample preparation under the applied conditions is not known to occur, as a strong acid would be required (44). It is more likely to represent a variable length of native N-glycans in H. volcanii because the addition of shortened N-glycans has been previously observed for FlgA1 and FlgA2 in different agl knock-out strains (19). Moreover, a tetrasaccharide with the same mass has also recently been reported for the S-layer glycoprotein (18). However, enzymatic degradation cannot be excluded, because molecular insights into the N-glycan degradation pathway are missing for H. volcanii.
TABLE 4.
Peptide spectral counts for identified N-glycopeptides with varying N-glycan lengths
CsCl gradient fractions 4 and 5 from the WT were analyzed by MS applying HCD fragmentation. N-Glycopeptides with N-glycans ranging from one to five carbohydrate residues (depicted as symbols: hexose (○), hexuronic acid (♢), methylated hexuronic acid (♢Me), as well as non-glycosylated peptides (P)) were identified in the given number of spectra. Peptide spectral counts from fractions 4 and 5 were combined. TSPQASFDFDYTN@TSGN@LTITHE from PilA2 was not included as its two N-glycosites can harbor different glycans (see Fig. 5) (n = 1).
Only a Subset of Pilins Require AglB-dependent Glycosylation to Mediate Surface Adhesion
Incubation of a glass coverslip in a wild-type H. volcanii culture results in evenly dispersed attachment of cells at the air-liquid interface. The ΔpilA[1–6] strain, which lacks all adhesive pili expressed under the assay conditions, displays varied adhesion phenotypes when complemented with individual PilA pilins in trans. These phenotypes include microcolony formation when PilA5 or PilA6 are individually expressed (Fig. 7) (20). Interestingly, microcolonies are also formed by the ΔaglB strain (Fig. 7).
FIGURE 7.
PilA3 and PilA4 require AglB-dependent N-glycosylation for function. Surface adhesion assays of WT, ΔpilA[1–6], and ΔpilA[1–6]ΔaglB strains transformed with pTA963 or pTA963 encoding one of the six His-tagged PilA pilins. Adhesion to plastic coverslips was tested using a modified air-liquid interface assay (35). Coverslips were placed in individual wells of 12-well plates, each containing 3 ml of a mid-log phase liquid CA culture. After overnight incubation, cells were fixed with 2% acetic acid, stained with 0.1% crystal violet, and observed by light microscopy (×400 magnification) (n = 5).
To determine the role that glycosylation plays in adhesion of individual pilins, we introduced the aglB deletion into the ΔpilA[1–6] strain. Like the ΔpilA[1–6] strain, ΔpilA[1–6]ΔaglB lacks surface filaments and does not adhere (Fig. 1) (20). We then transformed this strain with a plasmid expressing one of the six PilA pilins. When expressed in trans, PilA5 similarly affects the adhesion phenotype of both the ΔpilA[1–6] and the ΔpilA[1–6]ΔaglB strains, consistent with PilA5 not being glycosylated in an AglB-dependent manner. Conversely, although ΔpilA[1–6]ΔaglB cells expressing either PilA1, PilA2, or PilA6 are still adherent, they form more microcolonies compared with ΔpilA[1–6] cells that express one of these three pilins (Fig. 7). Finally, adhesion is markedly reduced when PilA3 or PilA4 are expressed in ΔpilA[1–6]ΔaglB compared with the effect of the expression of these pilins in the ΔpilA[1–6] strain, suggesting that N-glycosylation plays a major role in the assembly and/or function of pili containing these pilins.
AglB-dependent N-Glycosylation Is Not Required for PilA4 Assembly
To determine whether lack of adhesion by PilA4 in the ΔaglB mutant is due to the inability to assemble pili or rather to the assembly of non-functional pili, we observed ΔpilA[1–6]ΔaglB cells that express PilA4 using TEM. Piliated cells expressed multiple pili on the surfaces of individual cells. These pili were not bundled but appeared to have a curlier structure than pili observed on wild-type cells (Fig. 1). This curly phenotype appears specific to the effect that the lack of AglB-dependent N-glycosylation has on PilA4 because prior TEM studies of PilA4 in the ΔpilA[1–6] strain revealed that these cells have short individual pili (20). No PilA3 pili are observed on the surface of ΔpilA[1–6]ΔaglB when PilA3 is expressed in trans. This may be due to the low levels of PilA3 expression in strains lacking aglB as seen by Western blotting. TEM of ΔpilA[1–6]ΔaglB cells expressing PilA2 revealed long bundles of filaments having a more rigid appearance, similar to those seen on cells of a ΔaglB strain (Fig. 1).
Flagellin and Pilin Function Is Dependent on the Presence of Different N-Glycans
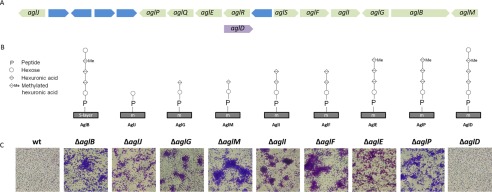
Data presented above strongly suggest that flagellins and pilins are glycosylated using the same AglB glycosylation pathway. Although three or four sugars, where the fourth sugar is not methylated, are sufficient for moderate swimming motility, modification of the flagellins by a tetrasaccharide, where the fourth sugar is methylated, results in a severe motility defect (Fig. 8, A and B) (19). To determine the importance of each sugar within the oligosaccharide for adhesion, we tested individual agl mutants within the AglB pathway. The knock-out of specific agl biosynthesis components involved in assembling the tetrasaccharide results in microcolony formation, similar to ΔaglB (Fig. 8). However, the strain lacking AglD, which is a dolichol-phosphate-mannose synthase required for transfer of the final mannose (12, 17), produces an evenly dispersed attachment layer, similar to wild-type and the complemented ΔaglB strain.
FIGURE 8.
An aglD knock-out does not cause microcolony formation. A, H. volcanii agl genomic region contains all known agl genes encoding components required for AglB-dependent glycosylation, except for aglD. Hypothetical genes are blue. B, schematic of the successive addition by individual Agl proteins in the AglB pathway of oligosaccharides and the methyl modification required to assemble the pentasaccharide decorating the S-layer glycoprotein. The final mannose is assembled separately to a dolichol phosphate carrier by AglD and then transferred to the tetrasaccharide (1). C, surface adhesion assays, as described in Fig. 7, of individual agl mutants (×400 magnification) (n = 3).
H. volcanii Is Non-motile and Forms Microcolonies at Low Salt
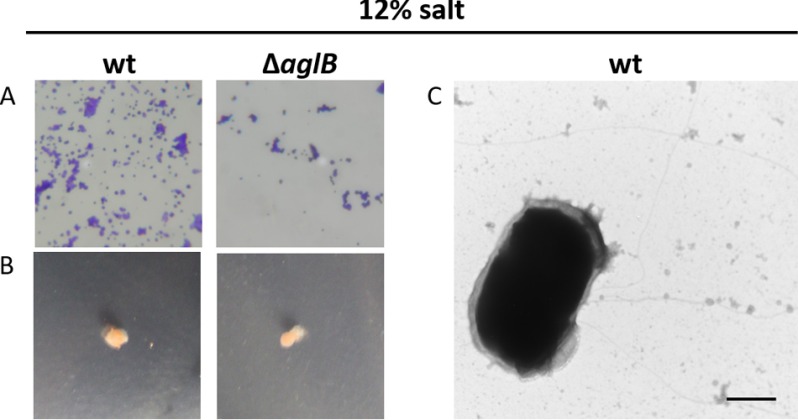
Results presented above suggest that N-glycosylation may regulate the transition from planktonic cell to biofilm formation under stress conditions. In fact, previous studies have shown AglB-dependent N-glycosylation of the S-layer decreases under low salt conditions (45, 46). Consistent with a lack of AglB-dependent flagellin and pilin N-glycosylation resulting in non-motile microcolony-forming cells, we demonstrated that under low salt conditions both the ΔaglB and the wild-type H. volcanii strains were indeed non-motile and formed microcolonies on coverslips (Fig. 9, A and B). When grown in low salt conditions, wild-type cells do not form bundles of pili, as determined by TEM (Fig. 9C). As reported previously, the S-layer glycoprotein still displays some AglB-dependent glycosylation in cells grown under low salt conditions (46). Assays on these cells, including the TEM, were performed using planktonic cells. Conversely, the air-liquid interface assay is used to examine cells during the early stages of biofilm formation, where AglB-dependent N-glycosylation may be further reduced or completely lacking.
FIGURE 9.
ΔaglB as well as wild-type H. volcanii are non-motile and form microcolonies under low salt conditions. Motility assays (n = 10) (A) and adhesion assays (×400 magnification) (n = 5) (B) of H. volcanii WT and ΔaglB in CA medium with 12% salt. Although, under standard salt conditions, WT cells form a visible halo after 2 days (34), in low salt, no halo had formed after 10 days of incubation at 45 °C. Surface adhesion assays were performed as described in Fig. 7 using low salt medium for culturing and incubation. TEM of WT cells (n = 2); bar, 500 nm (C).
Discussion
We have identified H. volcanii type IV pilins and flagellins decorated with an identical pentasaccharide, synthesized using the same AglB pathway. Although the presence of at least part of this pentasaccharide is required for motility, cells lacking AglB can still adhere to abiotic surfaces, and unlike wild-type cells, these cells form microcolonies at a high rate.
Despite each PilA pilin mediating adhesion, the effect that AglB-dependent N-glycosylation has on the individual H. volcanii type IV pilins varies. Although AglB-dependent N-glycosylation is required for PilA3 and PilA4 to complement the adhesion defect of a ΔpilA[1–6] strain, the other four adhesion pilins do not require AglB-dependent N-glycosylation to rescue this defect. Surprisingly, PilA4 still assembles into pili, albeit curlier than wild-type pili. When either PilA1 or PilA2, which appear to be the most abundant pilins in planktonic cells, is expressed in trans in a ΔpilA[1–6]ΔaglB mutant strain, microcolony formation increases. One function of AglB-dependent pilin N-glycosylation may be to inhibit cell-cell adherence in planktonic cultures. This hypothesis is also consistent with the observation that the H. volcanii pili form large bundles in a ΔaglB strain.
Previous studies have shown that the AglB-dependent glycosylation of the S-layer glycoprotein is reduced under low salt conditions, suggesting that AglB is down-regulated under stress (45, 46). In sessile cells, the down-regulation of AglB-dependent N-glycosylation might also promote the formation of bundles of type IV pili. These bundles may contain not only other pili on the same cell, but also pili on other cells, bringing cells into close contact and facilitating microcolony formation. This observation is reminiscent of the bundle formation of Neisseria gonorrhoeae pili when a phosphoglycerol post-translational modification is lacking (22). Our observation that wild-type cells form microcolonies at low salt supports this hypothesis.
Mass spectrometric analyses failed to identify a significant number of PilA3 and PilA4 peptides in planktonic cells lacking AglB. Moreover, when either of these pilins are glycosylated in a ΔpilA1–6 mutant strain, cells adhere as a strong even layer to abiotic surfaces, where no microcolonies are observed. Perhaps PilA3 and PilA4 are highly expressed when conditions favor cell surface attachment. Once a cell is attached to the surface, AglB-dependent N-glycosylation is repressed, promoting cell-cell interactions rather than surface attachment. This repression of N-glycosylation would also prevent flagella assembly (19), allowing the cells to more readily establish a biofilm. Under stress conditions where AglB-dependent N-glycosylation may be down-regulated, some of the pilin genes, including pilA3 and pilA4, and flagellin genes might also be down-regulated. In contrast, expressing PilA5, which is not predicted to be N-glycosylated, in trans in a ΔpilA[1–6] strain promotes a high rate of microcolony formation, similar to that observed for a ΔaglB strain. Additionally, expressing PilA6 in a ΔpilA[1–6]ΔaglB strain increases microcolony formation compared with the ΔpilA[1–6] strain. It may be that the expression of these pilins increases under sessile conditions.
As conditions become conducive for growth outside of the biofilm, expression of the components of the AglB-dependent N-glycosylation pathway would facilitate the assembly of flagella and allow cells in the biofilm to disperse. Using this post-translational modification as a switch offers a rapid and reversible mechanism for regulating the transition of H. volcanii cells between a sessile state and a planktonic one. As AglD is required for the flagella-dependent motility (19), but not for microcolony formation, it would be interesting if the terminal saccharide was not present on all of the pili. As shown in this study, PilA1 and PilA2 are N-glycosylated with a tetrasaccharide or pentasaccharide more abundantly than other subsets of the pentasaccharide. The high peptide spectral count for tetrasaccharides attached to pilin peptides indicates that it is possible that the tetrasaccharide, which is inhibitory to bundle formation, can decorate H. volcanii pilins in nature. The tetrasaccharide may facilitate planktonic cell transitions to a sessile state as a precursor to forming a biofilm and suppressing motility, although microcolony formation is not yet promoted. Thus, cells would not have to fully commit to forming a mature biofilm until AglB-dependent glycosylation was significantly decreased.
To test this hypothesis, mass spectrometric analyses on cells during the early stages of biofilm formation and in the mature biofilm must be performed to determine which pilins are expressed and the extent to which these pilins are N-glycosylated during these stages. An important consideration, based on recent studies showing S-layer N-glycosylation by an alternative pathway under low salt conditions (45, 46), is that the pili may be modified by an oligosaccharide moiety other than the pentasaccharide under low salt conditions, where increased osmotic stress may promote biofilm formation. Future studies on the regulation of N-glycosylation may result in the discovery of novel mechanisms for regulating biofilm formation and dispersal, and because N-glycosylation is common throughout all three domains of life, this mechanism may be broadly conserved.
Author Contributions
M. H. and S. S. designed and interpreted mass spectrometry experiments. R. N. E. and M. P. conceived of the idea for the project and designed and interpreted results of all other experiments. R. X. performed the initial adhesion assays of the agl mutant strains. R. N. E. developed the strains and performed TEM, adhesion assays, as well as Western blots and wrote the manuscript together with M. P. and S. S.
Acknowledgments
We thank Dewight Williams for advice on microscopy and the Pohlschroder laboratory for helpful discussions.
The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
M. Dyall-Smith, personal communication.
- CA
- casamino acid
- TEM
- transmission electron microscopy
- HCD
- higher energy collisional dissociation
- IS-CID
- in-source collision-induced dissociation
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
References
- 1. Jarrell K. F., Ding Y., Meyer B. H., Albers S. V., Kaminski L., and Eichler J. (2014) N-Linked glycosylation in archaea: a structural, functional, and genetic analysis. Microbiol. Mol. Biol. Rev. 78, 304–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nothaft H., and Szymanski C. M. (2010) Protein glycosylation in bacteria: sweeter than ever. Nat. Rev. Microbiol. 8, 765–778 [DOI] [PubMed] [Google Scholar]
- 3. Weerapana E., and Imperiali B. (2006) Asparagine-linked protein glycosylation: from eukaryotic to prokaryotic systems. Glycobiology 16, 91R–101R [DOI] [PubMed] [Google Scholar]
- 4. Marshall R. D. (1974) The nature and metabolism of the carbohydrate-peptide linkages of glycoproteins. Biochem. Soc. Symp. 40, 17–26 [PubMed] [Google Scholar]
- 5. Breitling J., and Aebi M. (2013) N-Linked protein glycosylation in the endoplasmic reticulum. Cold Spring Harb. Perspect. Biol. 5, a013359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nothaft H., and Szymanski C. M. (2013) Bacterial protein N-glycosylation: new perspectives and applications. J. Biol. Chem. 288, 6912–6920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Abu-Qarn M., and Eichler J. (2006) Protein N-glycosylation in archaea: defining Haloferax volcanii genes involved in S-layer glycoprotein glycosylation. Mol. Microbiol. 61, 511–525 [DOI] [PubMed] [Google Scholar]
- 8. Chaban B., Voisin S., Kelly J., Logan S. M., and Jarrell K. F. (2006) Identification of genes involved in the biosynthesis and attachment of Methanococcus voltae N-linked glycans: insight into N-linked glycosylation pathways in archaea. Mol. Microbiol. 61, 259–268 [DOI] [PubMed] [Google Scholar]
- 9. VanDyke D. J., Wu J., Logan S. M., Kelly J. F., Mizuno S., Aizawa S., and Jarrell K. F. (2009) Identification of genes involved in the assembly and attachment of a novel flagellin N-linked tetrasaccharide important for motility in the archaeon Methanococcus maripaludis. Mol. Microbiol. 72, 633–644 [DOI] [PubMed] [Google Scholar]
- 10. Kelly J., Logan S. M., Jarrell K. F., VanDyke D. J., and Vinogradov E. (2009) A novel N-linked flagellar glycan from Methanococcus maripaludis. Carbohydr. Res. 344, 648–653 [DOI] [PubMed] [Google Scholar]
- 11. Meyer B. H., and Albers S. V. (2014) AglB, catalyzing the oligosaccharyl transferase step of the archaeal N-glycosylation process, is essential in the thermoacidophilic crenarchaeon Sulfolobus acidocaldarius. MicrobiologyOpen 3, 531–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Abu-Qarn M., Yurist-Doutsch S., Giordano A., Trauner A., Morris H. R., Hitchen P., Medalia O., Dell A., and Eichler J. (2007) Haloferax volcanii AglB and AglD are involved in N-glycosylation of the S-layer glycoprotein and proper assembly of the surface layer. J. Mol. Biol. 374, 1224–1236 [DOI] [PubMed] [Google Scholar]
- 13. Voisin S., Houliston R. S., Kelly J., Brisson J. R., Watson D., Bardy S. L., Jarrell K. F., and Logan S. M. (2005) Identification and characterization of the unique N-linked glycan common to the flagellins and S-layer glycoprotein of Methanococcus voltae. J. Biol. Chem. 280, 16586–16593 [DOI] [PubMed] [Google Scholar]
- 14. Jarrell K. F., and Albers S. V. (2012) The archaellum: an old motility structure with a new name. Trends Microbiol. 20, 307–312 [DOI] [PubMed] [Google Scholar]
- 15. Pohlschroder M., and Esquivel R. N. (2015) Archaeal type IV pili and their involvement in biofilm formation. Front. Microbiol. 6, 190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ng S. Y., Wu J., Nair D. B., Logan S. M., Robotham A., Tessier L., Kelly J. F., Uchida K., Aizawa S., and Jarrell K. F. (2011) Genetic and mass spectrometry analyses of the unusual type IV-like pili of the archaeon Methanococcus maripaludis. J. Bacteriol. 193, 804–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Guan Z., Naparstek S., Kaminski L., Konrad Z., and Eichler J. (2010) Distinct glycan-charged phosphodolichol carriers are required for the assembly of the pentasaccharide N-linked to the Haloferax volcanii S-layer glycoprotein. Mol. Microbiol. 78, 1294–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kandiba L., Lin C. W., Aebi M., Eichler J., and Guerardel Y. (2016) Structural characterization of the N-linked pentasaccharide decorating glycoproteins of the halophilic archaeon Haloferax volcanii. Glycobiology 10.1093/glycob/cww014 [DOI] [PubMed] [Google Scholar]
- 19. Tripepi M., You J., Temel S., Önder Ö., Brisson D., and Pohlschröder M. (2012) N-Glycosylation of Haloferax volcanii flagellins requires known Agl proteins and is essential for biosynthesis of stable flagella. J. Bacteriol. 194, 4876–4887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Esquivel R. N., Xu R., and Pohlschroder M. (2013) Novel archaeal adhesion pilins with a conserved N terminus. J. Bacteriol. 195, 3808–3818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Esquivel R. N., and Pohlschroder M. (2014) A conserved type IV pilin signal peptide H-domain is critical for the post-translational regulation of flagella-dependent motility. Mol. Microbiol. 93, 494–504 [DOI] [PubMed] [Google Scholar]
- 22. Chamot-Rooke J., Mikaty G., Malosse C., Soyer M., Dumont A., Gault J., Imhaus A. F., Martin P., Trellet M., Clary G., Chafey P., Camoin L., Nilges M., Nassif X., and Duménil G. (2011) Posttranslational modification of pili upon cell contact triggers N. meningitidis dissemination. Science 331, 778–782 [DOI] [PubMed] [Google Scholar]
- 23. Allers T., Ngo H. P., Mevarech M., and Lloyd R. G. (2004) Development of additional selectable markers for the halophilic archaeon Haloferax volcanii based on the leuB and trpA genes. Appl. Environ. Microbiol. 70, 943–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Allers T., Barak S., Liddell S., Wardell K., and Mevarech M. (2010) Improved strains and plasmid vectors for conditional overexpression of His-tagged proteins in Haloferax volcanii. Appl. Environ. Microbiol. 76, 1759–1769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Blyn L. B., Braaten B. A., and Low D. A. (1990) Regulation of pap pilin phase variation by a mechanism involving differential dam methylation states. EMBO J. 9, 4045–4054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Abu-Qarn M., Giordano A., Battaglia F., Trauner A., Hitchen P. G., Morris H. R., Dell A., and Eichler J. (2008) Identification of AglE, a second glycosyltransferase involved in N glycosylation of the Haloferax volcanii S-layer glycoprotein. J. Bacteriol. 190, 3140–3146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yurist-Doutsch S., Abu-Qarn M., Battaglia F., Morris H. R., Hitchen P. G., Dell A., and Eichler J. (2008) aglF, aglG and aglI, novel members of a gene island involved in the N-glycosylation of the Haloferax volcanii S-layer glycoprotein. Mol. Microbiol. 69, 1234–1245 [DOI] [PubMed] [Google Scholar]
- 28. Kaminski L., Abu-Qarn M., Guan Z., Naparstek S., Ventura V. V., Raetz C. R., Hitchen P. G., Dell A., and Eichler J. (2010) AglJ adds the first sugar of the N-linked pentasaccharide decorating the Haloferax volcanii S-layer glycoprotein. J. Bacteriol. 192, 5572–5579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yurist-Doutsch S., Magidovich H., Ventura V. V., Hitchen P. G., Dell A., and Eichler J. (2010) N-Glycosylation in archaea: on the coordinated actions of Haloferax volcanii AglF and AglM. Mol. Microbiol. 75, 1047–1058 [DOI] [PubMed] [Google Scholar]
- 30. Magidovich H., Yurist-Doutsch S., Konrad Z., Ventura V. V., Dell A., Hitchen P. G., and Eichler J. (2010) AglP is an S-adenosyl-l-methionine-dependent methyltransferase that participates in the N-glycosylation pathway of Haloferax volcanii. Mol. Microbiol. 76, 190–199 [DOI] [PubMed] [Google Scholar]
- 31. Blattner F. R., Williams B. G., Blechl A. E., Denniston-Thompson K., Faber H. E., Furlong L., Grunwald D. J., Kiefer D. O., Moore D. D., Schumm J. W., Sheldon E. L., and Smithies O. (1977) Charon phages: safer derivatives of bacteriophage λ for DNA cloning. Science 196, 161–169 [DOI] [PubMed] [Google Scholar]
- 32. Allers T., and Ngo H. P. (2003) Genetic analysis of homologous recombination in Archaea: Haloferax volcanii as a model organism. Biochem. Soc. Trans. 31, 706–710 [DOI] [PubMed] [Google Scholar]
- 33. Hammelmann M., and Soppa J. (2008) Optimized generation of vectors for the construction of Haloferax volcanii deletion mutants. J. Microbiol. Methods 75, 201–204 [DOI] [PubMed] [Google Scholar]
- 34. Tripepi M., Imam S., and Pohlschröder M. (2010) Haloferax volcanii flagella are required for motility but are not involved in PibD-dependent surface adhesion. J. Bacteriol. 192, 3093–3102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. O'Toole G. A., Pratt L. A., Watnick P. I., Newman D. K., Weaver V. B., and Kolter R. (1999) Genetic approaches to study of biofilms. Methods Enzymol. 310, 91–109 [DOI] [PubMed] [Google Scholar]
- 36. Fedorov O. V., Pyatibratov M. G., Kostyukova A. S., Osina N. K., and Tarasov V. Y. (1994) Protofilament as a structural element of flagella of haloalkalophilic archaebacteria. Can. J. Microbiol. 40, 45–53 [Google Scholar]
- 37. Tripepi M., Esquivel R. N., Wirth R., and Pohlschröder M. (2013) Haloferax volcanii cells lacking the flagellin FlgA2 are hypermotile. Microbiology 159, 2249–2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wiśniewski J. R., Zougman A., Nagaraj N., and Mann M. (2009) Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362 [DOI] [PubMed] [Google Scholar]
- 39. Mathieu-Rivet E., Scholz M., Arias C., Dardelle F., Schulze S., Le Mauff F., Teo G., Hochmal A. K., Blanco-Rivero A., Loutelier-Bourhis C., Kiefer-Meyer M. C., Fufezan C., Burel C., Lerouge P., Martinez F., Bardor M., and Hippler M. (2013) Exploring the N-glycosylation pathway in Chlamydomonas reinhardtii unravels novel complex structures. Mol. Cell. Proteomics 12, 3160–3183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Craig R., and Beavis R. C. (2004) TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20, 1466–1467 [DOI] [PubMed] [Google Scholar]
- 41. Specht M., Kuhlgert S., Fufezan C., and Hippler M. (2011) Proteomics to go: proteomatic enables the user-friendly creation of versatile MS/MS data evaluation workflows. Bioinformatics 27, 1183–1184 [DOI] [PubMed] [Google Scholar]
- 42. Käll L., Storey J. D., MacCoss M. J., and Noble W. S. (2008) Assigning significance to peptides identified by tandem mass spectrometry using decoy databases. J. Proteome Res. 7, 29–34 [DOI] [PubMed] [Google Scholar]
- 43. Wang G., Wu W. W., Zeng W., Chou C. L., and Shen R. F. (2006) Label-free protein quantification using LC-coupled ion trap or FT mass spectrometry: reproducibility, linearity, and application with complex proteomes. J. Proteome Res. 5, 1214–1223 [DOI] [PubMed] [Google Scholar]
- 44. Sojar H. T., and Bahl O. P. (1987) A chemical method for the deglycosylation of proteins. Arch. Biochem. Biophys. 259, 52–57 [DOI] [PubMed] [Google Scholar]
- 45. Guan Z., Naparstek S., Calo D., and Eichler J. (2012) Protein glycosylation as an adaptive response in archaea: growth at different salt concentrations leads to alterations in Haloferax volcanii S-layer glycoprotein N-glycosylation. Environ. Microbiol. 14, 743–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kaminski L., Guan Z., Yurist-Doutsch S., and Eichler J. (2013) Two distinct N-glycosylation pathways process the Haloferax volcanii S-layer glycoprotein upon changes in environmental salinity. MBio 4, e00716–13 [DOI] [PMC free article] [PubMed] [Google Scholar]