Abstract
In a microarray study, we found that hepatic miR-291b-3p was significantly increased in leptin-receptor-deficient type 2 mice (db/db), a mouse model of diabetes. The function of miR-291b-3p is unknown. The potential role of miR-291b-3p in regulating hepatic lipid metabolism was explored in this study. High-fat diet (HFD)- and chow-fed mice were injected with an adenovirus expressing a miR-291b-3p inhibitor and a miR-291b-3p mimic through the tail vein. Hepatic lipids and lipogenic gene expression were analyzed. Additionally, gain- and loss-of-function studies were performed in vitro to identify direct targets of miR-291b-3p. MiR-291b-3p expression and the protein levels of sterol regulatory element-binding protein 1 (SREBP1) and fatty acid synthase (FAS) were increased in the steatotic liver of db/db mice and HFD-fed mice versus their respective controls. Inhibition of hepatic miR-291b-3p expression prevented increases in hepatic lipogenesis and steatosis in HFD-fed mice. The opposite was observed when miR-291b-3p was overexpressed in the livers of chow-fed C57BL/6J wild-type mice. In vitro studies revealed that silencing of miR-291b-3p in NCTC1469 hepatic cells ameliorated oleic acid/palmitic acid mixture-induced elevation of cellular triglycerides. Importantly, we identified AMP-activated protein kinase (AMPK)-α1 as a direct target of miR-291b-3p. Using metformin, an activator of AMPK, we showed that AMPK activation-induced inhibition of hepatic lipid accumulation was accompanied by reduced expression of miR-291b-3p in the liver. Liver miR-291b-3p promoted hepatic lipogenesis and lipid accumulation in mice. AMPKα1 is a direct target of miR-291b-3p. In conclusion, our findings indicate that miR-291b-3p promotes hepatic lipogenesis by suppressing AMPKα1 expression and activity, indicating the therapeutic potential of miR-291b-3p inhibitors in fatty liver disease.
Keywords: AMP-activated kinase (AMPK), fatty acid, fatty acid synthase (FAS), lipid metabolism, microRNA (miRNA), AMPKα liver disease, lipid accumulation liver disease, nonalcoholic fatty liver disease
Introduction
Nonalcoholic fatty liver disease (NAFLD)4 is characterized by excessive triglyceride accumulation in the liver. It is the most prevalent chronic liver disease worldwide and is strongly associated with various metabolic disorders, such as obesity, type 2 diabetes, and cardiovascular disease (1–3). The disease may progress to nonalcoholic steatohepatitis (NASH), hepatic fibrosis/cirrhosis, and hepatocellular carcinoma due to the co-existence of other detrimental factors (4, 5). Although the molecular mechanisms for NAFLD development and progression are not completely understood, increased synthesis of triglycerides is considered an important factor in driving hepatic lipid accumulation.
Mammalian AMP-activated protein kinase (AMPK) is a key enzyme in energy sensing and fatty acid biosynthesis (6–8). It is a heterotrimer of the α, β, and γ subunits (9–11). The α1 subunit has a catalytic domain that is activated and phosphorylated by an upstream kinase, which is highly expressed in liver (12). AMPK is known to inhibit fatty acid synthesis by phosphorylating serine 79 of acetyl-CoA carboxylase 1 (ACC1), a rate-limiting enzyme in de novo lipogenesis. A recent study showed that hepatic activation of AMPK by a synthetic polyphenol protects against hepatic steatosis, hyperlipidemia and atherosclerosis in low-density lipoprotein (LDL) receptor (LDLR)-deficient mice, in part through suppression of sterol regulatory element-binding protein-1 (SREBP-1)- and SREBP-2-dependent lipogenesis (13). While activation of SREBP2 primarily promotes cholesterol synthesis, SREBP1c activation stimulates fatty acid and triglyceride synthesis by up-regulating the transcription of lipogenic genes, such as fatty acid synthase (FAS) (14).
MicroRNAs (miRNAs or miRs) are endogenous non-coding single-stranded RNAs of ∼22 nucleotides that post-transcriptionally regulate gene expression (15–17). Recently, miR-122, miR-33, miR-30c, miR-24, and miR-130 have been identified as important regulators of lipid metabolism (18–22). In a previous microarray screening, we found that miR-291b-3p was increased in the steatotic liver of db/db mice (23). MiR-291b-3p is in the murine miR-290 cluster, and previous studies have suggested a role for this miRNA in the development of embryonic stem cells (24–26). No studies have linked miR-291b-3p to hepatic lipid accumulation. Considering its substantial increase in steatotic livers (23), in the present study, we examined whether miR-291b-3p regulates hepatic lipid metabolism. We found that miR-291b-3p promotes hepatic lipid accumulation by stimulating hepatic lipogenesis. Importantly, we identified AMPKα1 as a direct target of miR-291b-3p, which suggests that miR-291b-3p may promote hepatic lipogenesis by inhibiting AMPKα1 expression and activity.
Experimental Procedures
Animals and Treatments
db/db mice and wild-type (WT) C57BL/6J mice were purchased from the Peking University Health Science Center. These mouse lines were originally obtained from The Jackson Laboratory. 8–10-week-old male db/db mice (n = 5) and age-matched male wild-type (WT) mice (n = 5) were fed a standard chow diet for 4 weeks. In another experiment, C57BL/6J mice were fed a standard chow diet or a high-fat diet (HFD, D12451, 45% kcal from fat, Research Diet for 10 weeks starting at ∼5 weeks of age in a temperature- (20–24 °C) and humidity-controlled (45–55%) environment. A 12 h/12 h light/dark cycle was maintained in the animal housing rooms. HFD-fed mice were injected intravenously through the tail vein with an adenovirus expressing a miR-291b-3p inhibitor (Ad-291i) or negative control (NC) adenoviral vector (Ad-Con). In addition, 6–8-week-old male C57BL/6J mice fed a standard chow diet were injected intravenously through the tail vein with the adenovirus expressing a miR-291b-3p mimic or negative control adenovirus vector. On day 7 after adenovirus injection, the mice were anesthetized, and blood was collected via cardiac puncture. Livers were harvested, snap-frozen in liquid nitrogen, and stored at −80 °C for further analysis (19). Additionally, eight male C57BL/6J mice were intragastrically treated with metformin or vehicle at 3 mg/kg/day for 8 weeks in the presence of HFD after 10-week HFD feeding. All mouse procedures were approved by the Animal Ethics Committee at the Beijing Institute of Geriatrics.
Adenoviral Vector Construction
Recombinant adenoviruses expressing a miR-291b-3p mimic (Ad-miR-291b-3p-m) or miR-291b-3p inhibitor (Ad-miR-291b-3p-i) or a negative control (NC) adenovirus vector containing GFP (Ad-Con) were purchased from Shanghai GeneChem Co., Ltd.
Cell Culture
The NCTC1469 murine liver cell line (American Type Culture Collection) was cultured in low-glucose Dulbecco's modified Eagle's medium (L-DMEM) (Invitrogen) supplemented with 10% horse serum (HyClone), 100 units/ml penicillin (Invitrogen), and 0.1 mg/ml streptomycin (HyClone) at 37 °C with humidified air and 5% CO2.
RNA Isolation and Real-time PCR
Total RNA was extracted from liver tissue using TRIzol reagent (Invitrogen). 1 μg of total RNA was transcribed to cDNA using the M-MuLV transcriptase (NEB). Stem-loop reverse transcription-polymerase chain reaction (RT-PCR) was conducted with the samples to detect and quantify mature miRNAs using a stem-loop antisense primer mix and M-MuLV transcriptase (NEB).
SYBR Green I was used for real-time PCR according to the manufacturer's instructions (TaKaRa) with the Bio-Rad iQ5 system (Bio-Rad). The relative expression level of a miRNA or mRNA was normalized to an internal invariant control, U6 small nucleolar RNA or GAPDH. Each reaction was performed in triplicate, and analysis was performed by the 2−▵▵CT method.
Nucleotide primers (5′-3′) used for reverse transcription were GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACACAAAC for miR-291b-3p and GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAAATATG for U6.
The primers (5′-3′) used for real-time PCR were miR-291b-3p forward: GCAAAGTGCATCCATTTTGTTTGT;U6 forward: GCGCGTCGTGAAGCGTTC; universal reverse primer: GTGCAGGGTCCGAGGT. IL6 forward: 5′-CAATATGAATGTTGGGACACT-3′; IL6 Reverse: ACATTCCAAGAAACCATCTG); TNFα forward: 5′-GCTCAGAGCTTTCAACAACT-3′; TNFα reverse: 5′-AAGGTCTGAAGGTAGGAAGG-3′); GAPDH forward: 5′-GTCGGTGTGAACGGATTTG-3′; GAPDH reverse: 5′-AAGATGGTGATGGGCTTCC-3′.
Biochemical Analysis
The serum biochemical profiles, such as aspartate aminotransferase (AST) and alanine aminotrans (ALT), were detected with a Biochem-Immunoautoanalyser (Brea, CA).
Western Blotting Analysis
Cell lysates (15–30 μg of proteins) were separated by 10% SDS-PAGE, transferred to PVDF membranes (Millipore), blocked with 8% nonfat dry milk, and then incubated with a primary antibody at 4 °C overnight. The blot was incubated with HRP-conjugated anti-IgG, followed by detection with ECL (Millipore). Antibodies against ACC1 (ab45174), phosphorylated ACC1 (ab68191), FAS (ab128870), GAPDH (ab181602) and AMPKα1 (ab32047) were purchased from Abcam. Antibodies against β-actin (#4970), AMPK (#5831), and phosphorylated AMPK (Thr172) (#2531) were obtained from Cell Signaling Technology. An antibody to SREBP1 was purchased from Santa Cruz Biotechnology (sc-366).
Triglyceride Measurement
The content of tissue triglycerides was measured as described previously (18–20) using a triglyceride enzymatic assay kit (ShenSuoYouFu Medical Diagnostic Products Co., Ltd., Shanghai, China).
Histological Analysis of Tissues
Frozen sections of liver specimens were fixed in paraformaldehyde for Oil Red O or hematoxylin and eosin (H&E) staining. In brief, liver tissue was fixed in 4% paraformaldehyde buffer for 1 h at 37 °C. Then, they were embedded in optimal cutting temperature (OCT) solution on dry ice and cut into 5 μm sections. To further confirm lipid droplet accumulation, they were incubated with pre-warmed Oil Red O (Solarbio, Beijing, China) for 30 min at 37 °C and washed with 60% 1,2-propanediol. Then, the droplets stained with Oil Red O were visualized under a microscope (CK Microscope, Olympus, Tokyo, Japan) (27). For H&E staining, the slides were first incubated with hematoxylin for 5 min and then washed with 1% ethanol hydrochloride for 3 s. After washing with water, the slides were stained with eosin for 3 min and dehydrated with an alcohol gradient. The vacuoles were considered the lipid droplets (28).
Luciferase Reporter Assay
The 3′-UTR sequence of AMPKα1 predicted to interact with miR-291b-3p was PCR-cloned from the genomic DNA of the NCTC1469 cell line and then inserted into the XbaI and SacI sites of the pmir-GLO control luciferase reporter vector (Promega, Madison, WI). HEK293 cells were co-transfected with the internal control vector pmir-GLO (Promega), pmir-GLO-AMPKα1 3′UTR and a miR-291b-3p mimic (or negative control) using Effectene (Qiagen) for 48 h. Luciferase reporter assays were performed using the Dual-Glo luciferase assay system according to standard protocols (Promega).
Cell Transfections
miR mimics and inhibitors or the negative control (NC) as well as AMPKα1 siRNA or a nonspecific siRNA (NC) were purchased from GenePharma (Shanghai). Transfection of miRs or siRNA was performed with HiPerFect transfection reagent (Qiagen) as previously described (23).
Oleic Acid/Palmitic Acid (O/P) Treatment
0.25 M Oleic acid (Sigma) and palmitic acid (Sigma) were dissolved in 100% ethyl alcohol. Before using, 0.25 m oleic acid, palmitic acid stock, and 5% BSA were incubated in 60 °C water bath for 5–10 min. Then, 640 μl or 320 μl of oleic acid and palmitic acid stock were dropwise added into 20 ml of 5% BSA to make 8 mm and 4 mm oleic acid and palmitic acid, respectively. Before experiments, 8 mm and 4 mm oleic acid and palmitic acid were incubated in a 60 °C water bath for 5–10 min. Then, equal volumes of oleic acid and palmitic acid were mixed together. And a mixture of oleic acid/palmitic acid was added at a total concentration of 300 μm (2:1, m/m).
Statistics
The data are represented as the means ± S.E. The two-tailed unpaired Student's t-tests were used for comparisons of two groups. The ANOVA multiple comparison test (SPSS 13.0) followed by Turkey post hoc test were used for comparisons of two more groups. p < 0.05 was considered to be statistically significant.
Results
MiR-291b-3p Expression Is Up-regulated in the Steatotic Liver of db/db Mice and HFD-fed Mice
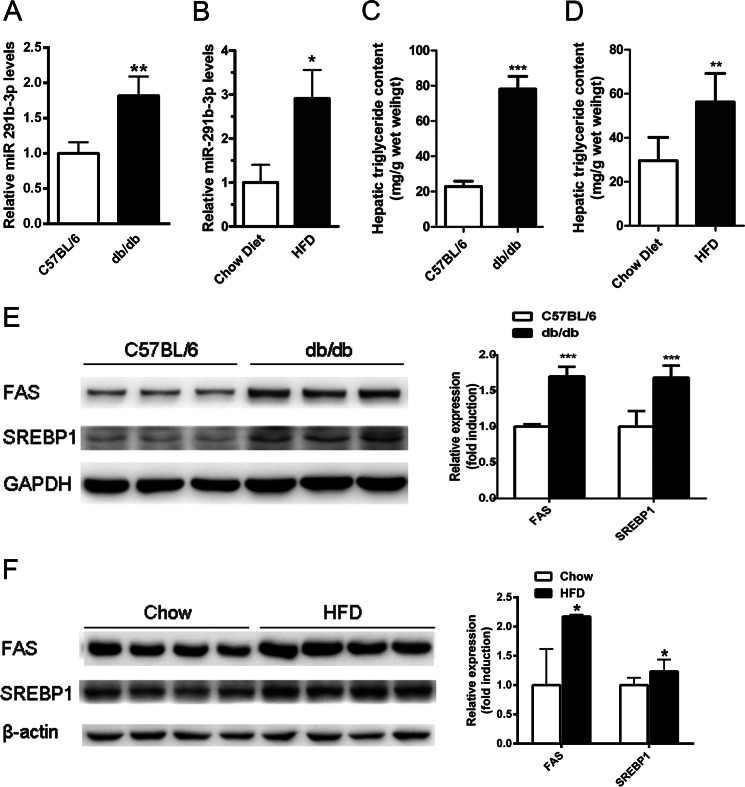
To identify the potential role of miR-291b-3p in lipid metabolism, we first examined its expression in the steatotic liver of db/db mice and HFD-fed mice by real-time PCR. Compared with the respective control mice, db/db mice and HFD-fed mice displayed a significant increase in hepatic miR-291b-3p (Fig. 1, A and B). As expected, db/db and HFD-fed mice accumulated excessive amounts of lipids (Fig. 1, C and D) and exhibited elevated levels of lipogenic proteins, including SREBP-1 and FAS, in liver relative to their respective controls (Fig. 1, E and F).
FIGURE 1.
MiR-291b-3p expression is increased in the steatotic liver of db/db mice and HFD-fed mice. A and B, hepatic levels of miR-291b-3p were determined by real-time PCR in chow-fed 8–10-week-old male db/db mice (A) and male C57BL/6J mice fed a HFD for 10 weeks starting at ∼5 weeks of age (B). C and D, hepatic contents of triglycerides in db/db mice (C) and HFD-fed mice (D). E and F, Western blots of hepatic protein levels of lipogenic genes in db/db mice (E) and HFD-fed mice (F). **, p < 0.01; ***, p < 0.001. n = 5 in each group.
Inhibition of miR-291b-3p Expression in the Liver Prevents Hepatic Lipid Accumulation in HFD-fed Mice
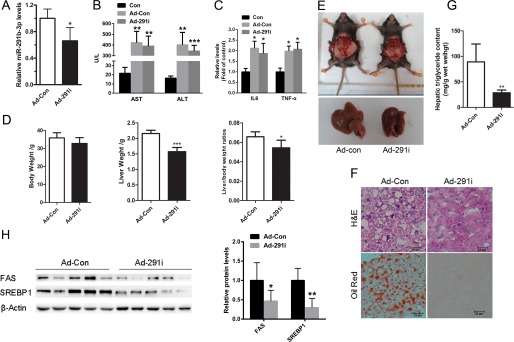
To determine if miR-219b-3p is involved in the hepatic lipid accumulation, we inhibited miR-219b-3p expression in the liver of HFD-fed mice with a recombinant adenovirus expressing a miR-291b-3p inhibitor (Ad-291i). Seven days after tail vein injection of the Ad-291i adenoviruses, hepatic levels of miR-291b-3p were reduced by ∼40% (Fig. 2A). Of note, biochemical analysis revealed that the liver enzymes including AST and ALT (Fig. 2B), and hepatic cytokines such as IL6 and TNF α (Fig. 2C) were elevated in mice treated with the adenovirus vectors compared with in control mice injected with vehicle (no virus), indicating that adenovirus vectors caused inflammatory processes in the liver. Therefore, to exclude the effects of adenovirus vectors on inflammation, adenovirus vectors containing GFP were used as control (Ad-Con) to compare with adenovirus vectors expressing miR-291b-3p inhibitor (Ad-291i). More importantly, this 40% reduction in hepatic miR-219b-3p expression was associated with a significant reduction in liver weight and liver weight-to-body weight ratios (Fig. 2, D and E). H&E and Oil Red O staining revealed a significant decrease in hepatic lipid deposition (Fig. 2F) in these animals. Importantly, the hepatic triglyceride content was significantly reduced (Fig. 2G), which was associated with reduced protein levels of SREBP-1 and FAS in the liver (Fig. 2H). These results suggest that inhibition of miR-291b-3p expression in the liver may prevent hepatic triglyceride accumulation in HFD-fed mice by suppressing lipogenesis.
FIGURE 2.
Inhibition of miR-291b-3p expression in the liver prevents hepatic steatosis in HFD-fed mice. 6–8-week-old male C57BL/6J mice fed a HFD for 10 weeks were injected with Ad-291i or Ad-Con through the tail vein and sacrificed 7 days post injection. A, hepatic levels of miR-291b-3p. B, levels of AST and ALT were analyzed in mice treated with the adenoviral vectors. C, quantification of IL6 and TNFα expression in the livers. D and E, body weight, liver weight and liver weight-to-body weight ratio. F, H&E and Oil Red O staining of liver frozen sections. G, hepatic triglyceride content. H, Western blots of hepatic SREBP1 and FAS. **, p < 0.01; ***, p < 0.001. n = 6.
Hepatic Overexpression of miR-291b-3p Induces Lipid Accumulation in the Liver
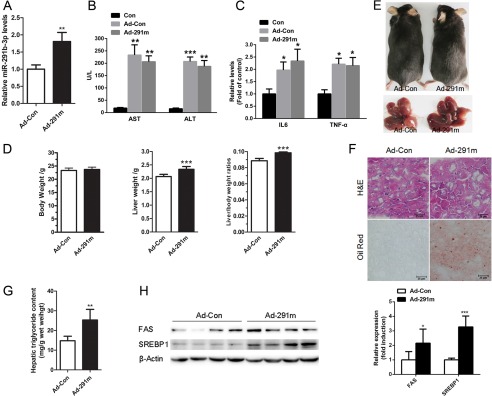
To determine if miR-219b-3p directly causes hepatic steatosis, we overexpressed it in the liver of C57BL/6J mice fed a standard chow diet via the recombinant adenovirus expressing a miR-291b-3p mimic (Ad-291m). Seven days after adenoviral injection, miR-291b-3p was elevated by ∼80% in the liver (Fig. 3A). The levels of AST and ALT were also found to be enhanced in mice treated with the adenovirus vectors (Fig. 3B), accompanied by elevated hepatic cytokine production, such as IL6 and TNFα, suggesting that injection of adenovirus vectors led to hepatic inflammation in the liver of C57BL/6J mice (Fig. 3C). To exclude the effects of adenovirus vectors on inflammation, adenovirus vectors containing GFP were used as control (Ad-Con) to compare with adenovirus vectors expressing miR-291b-3p mimic (Ad-291m). The injection of Ad-291m did not alter body weight but did increase liver weight and the liver weight-to-body weight ratio compared with Ad-Con (Fig. 3, D and E). H&E and Oil Red O staining showed that the mice injected with Ad-291m accumulated lipids in the liver (Fig. 3F), which was consistent with a significant increase in hepatic triglycerides (Fig. 3G). These changes in hepatic lipids were associated with increased hepatic levels of the SREBP-1 and FAS proteins (Fig. 3H). These findings indicate that hepatic overexpression of miR-291b-3p may cause lipid accumulation in the liver by up-regulating lipogenic gene expression.
FIGURE 3.
Hepatic overexpression of miR-291b-3p induces lipid accumulation in the liver. 6–8-week-old male C57BL/6J mice on a chow diet were injected with the recombinant adenovirus expressing a miR-291b-3p mimic (Ad-291m) or Ad-Con and sacrificed 7 days after injection. A, hepatic levels of miR-291b-3p. B, biochemical analysis of AST and ALT in mice treated with the adenovirus vectors. C, quantification of IL6 and TNFα expression in the livers. D and E, body weight, liver weight, and liver weight-to-body weight ratio. F, H&E and Oil Red O staining of liver frozen sections. G, hepatic triglyceride content. H, Western blots of hepatic SREBP1 and FAS protein levels. **, p < 0.01; ***, p < 0.001. n = 6.
Suppression of miR-291b-3p Expression Ameliorates Oleic Acid/Palmitic Acid-induced Lipid Accumulation in NCTC1469 Cells
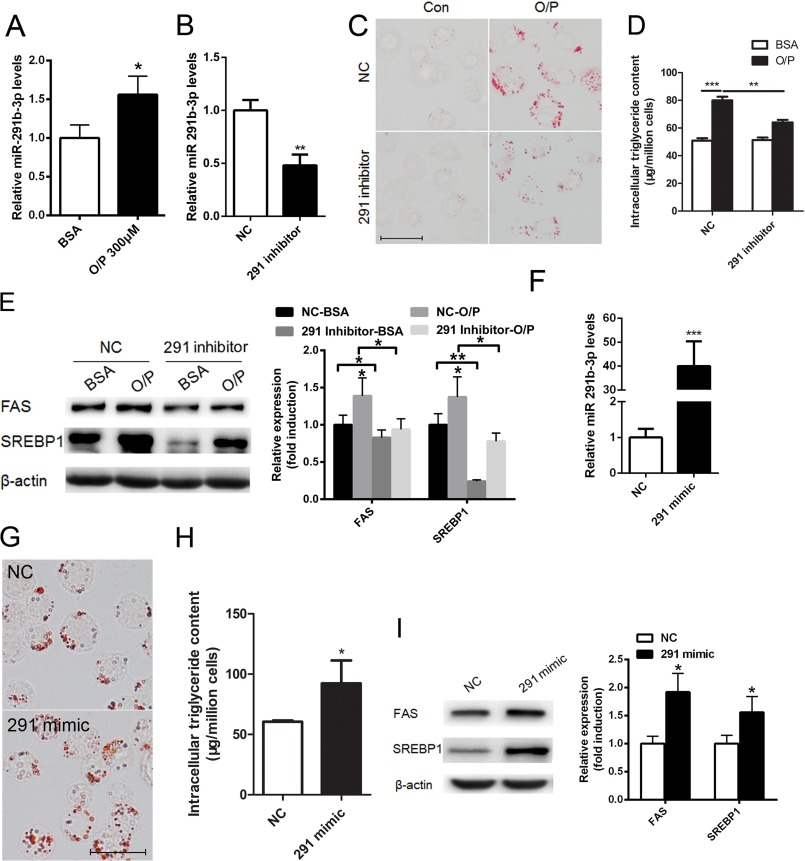
To determine whether our in vivo findings occur in a cell-autonomous manner, we employed the NCTC1469 mouse liver cell line as a model. We treated NCTC1469 cells with a mixture of oleic acid/palmitic acid (2:1, m/m) for 24 h, as previously described (29). The expression of miR-291b-3p was significantly up-regulated in NCTC1469 cells treated with 300 μm of the oleic acid/palmitic acid mixture (Fig. 4A). When a miR-291b-3p inhibitor was added for 48 h, miR-291b-3p expression was suppressed in NCTC1469 cells (Fig. 4B), accompanied by reduced lipid accumulation (Fig. 4C) and cellular triglyceride content (Fig. 4D) in the presence of oleic acid/palmitic acid. Moreover, suppression of miR-291b-3p reversed the oleic acid/palmitic acid-induced elevation of protein and mRNA expression of SREBP-1 and FAS (Fig. 4E).
FIGURE 4.
Suppression of miR-291b-3p expression ameliorates oleic acid/palmitic acid mixture-induced lipid accumulation in NCTC1469 cells. A–E, NCTC1469 cells were treated with the mixture of oleic acid/palmitic acid (2:1, m (mol/liter)/m (mol/liter)] for 24 h in DMEM containing 10% horse serum, followed by transfection with a miR-291b-3p inhibitor or NC for 48 h in the presence of oleic acid/palmitic acid. A and B, real-time PCR analysis of miR-291b-3p expression. C, oil Red O staining. D, intracellular triglyceride content. E, Western blotting analysis of SREBP1 and FAS. F–I, NCTC1469 cells were grown in normal medium without oleic acid/palmitic acid and were transfected with the miR-291b-3p mimic or NC for 48 h. F, miR-291b-3p expression measured by real-time PCR. G, lipid accumulation revealed by Oil red O staining. H, cellular triglyceride content. I, levels of SREBP-1 and FAS expression. n = 3 independent experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
To determine whether miR-291b-3p is capable of inducing triglyceride deposition, miR-291b-3p was overexpressed in NCTC1469 cells by transfection of a miR-291b-3p mimic (Fig. 4F). As revealed by Oil Red O staining, the miR-291b-3p mimic directly induced lipid accumulation in NCTC1469 cells (Fig. 4G). Similarly, it increased the cellular triglyceride content (Fig. 4H). This increase in cellular triglycerides was associated with increased expression of SREBP-1 and FAS (Fig. 4I).
MiR-291b-3p Mediates Lipid Accumulation by Directly Targeting AMPKα1
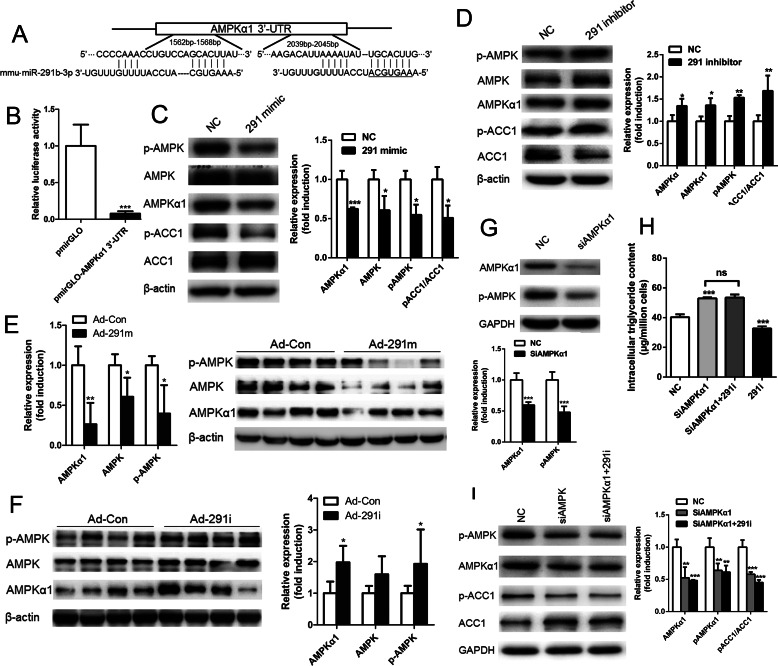
To identify the direct effectors of miR-291b-3p-mediated lipid accumulation, we employed a bioinformatics approach. Based on the analysis of predicted target genes and sequences, murine miR-291b-3p was aligned with the 3′-UTR of AMPKα1 mRNA (Fig. 5A), indicating that AMPKα1, an important catalytic subunit of AMPK, may be a potential target gene of miR-291b-3p. To determine if AMPKα1 is a true target of miR-291b-3p, we generated a Renilla luciferase reporter plasmid containing the 3′-UTR of mouse AMPKα1 with miR-291b-3p binding sites. A reporter assay performed in HEK 293T cells showed that overexpression of miR-291b-3p significantly reduced the luciferase activity in HEK 293T cells transfected with the luciferase reporter plasmid (Fig. 5B). To further determine whether miR-291b-3p can target endogenous AMPKα1, NCTC1469 cells were transfected with a miR-291b-3p mimic or miR-291b-3p inhibitor. As shown in Fig. 5C, overexpression of miR-291b-3p in NCTC1469 cells led to a significant reduction in levels of phosphorylated AMPK and total AMPKα1 protein. Importantly, the level of phosphorylated ACC1 (p-ACC1), a target of p-AMPK, was substantially decreased in NCTC1469 cells transfected with the miR-291b-3p mimic. The opposite was observed when miR-291b-3p was silenced by a miR-291b-3p inhibitor (Fig. 5D). To determine if these in vitro findings are valid in vivo, we overexpressed miR-291b-3p in the liver of C57BL/6J mice by Ad-291m injections. Adenovirus-mediated overexpression of miR-291b-3p substantially reduced the levels of AMPKα1, AMPK, and p-AMPK in the mouse liver (Fig. 5E). The opposite was observed when miR-291b-3p was inhibited in the liver of HFD mice (Fig. 5F). These observations establish AMPKα1 as a direct target of miR-291b-3p.
FIGURE 5.
MiR-291b-3p mediates cellular lipid accumulation by directly targeting AMPKα1. A, bioinformatics-predicted binding sites of miR-291b-3p in the 3′-UTR of AMPKα1. B, luciferase activity in HEK 293T cells co-transfected with a miR-291b-3p mimic and pmir-GLO (Promega) or pmir-GLO-AMPKα1 3′UTR. C and D, levels of AMPKα1, total AMPK, AMPK phosphorylated at threonine 172 (p-AMPK), total ACC1 and p-ACC1 in NCTC1469 cells transfected with a miR-291b-3p mimic (C) or miR-291b-3p inhibitor (D). E and F, levels of AMPKα1, total AMPK and p-AMPK in the livers of C57BL/6J mice treated with Ad-291m (E) or HFD-fed mice treated with Ad-291i (F). G, expression and phosphorylation of AMPKα1 in NCTC1469 cells treated with an AMPKα1 siRNA. H and I, intracellular triglyceride content (H) and protein levels of p-ACC1 and total ACC1 (I) in NCTC1469 cells treated with a miR-291b-3p inhibitor and the AMPKα1 siRNA. n = 3 independent experiments. *, p < 0.05 (versus NC); **, p < 0.01(versus NC); ***, p < 0.001 (versus NC).
To determine whether AMPKα1 mediates miR-291b-3p-induced lipid accumulation, AMPKα1 expression was silenced by AMPKα1 siRNA. Western blotting analysis indicated that expression levels of AMPKα1 and p-AMPK were suppressed in NCTC1469 cells transfected with AMPKα1 siRNA (Fig. 5G). Interestingly, silencing of AMPKα1 reversed miR-291b-3p inhibitor-induced decreases in cellular triglyceride content (Fig. 5H) and reduced the levels of p-ACC1 and ACC1 (Fig. 5I). Taken together, these findings indicate that miR-291b-3p may mediate lipid accumulation by inhibiting AMPKα1 expression.
Reducing miR-291b-3p Expression Is Associated with Metformin-reversed Hepatic Lipid Accumulation in HFD-fed Mice
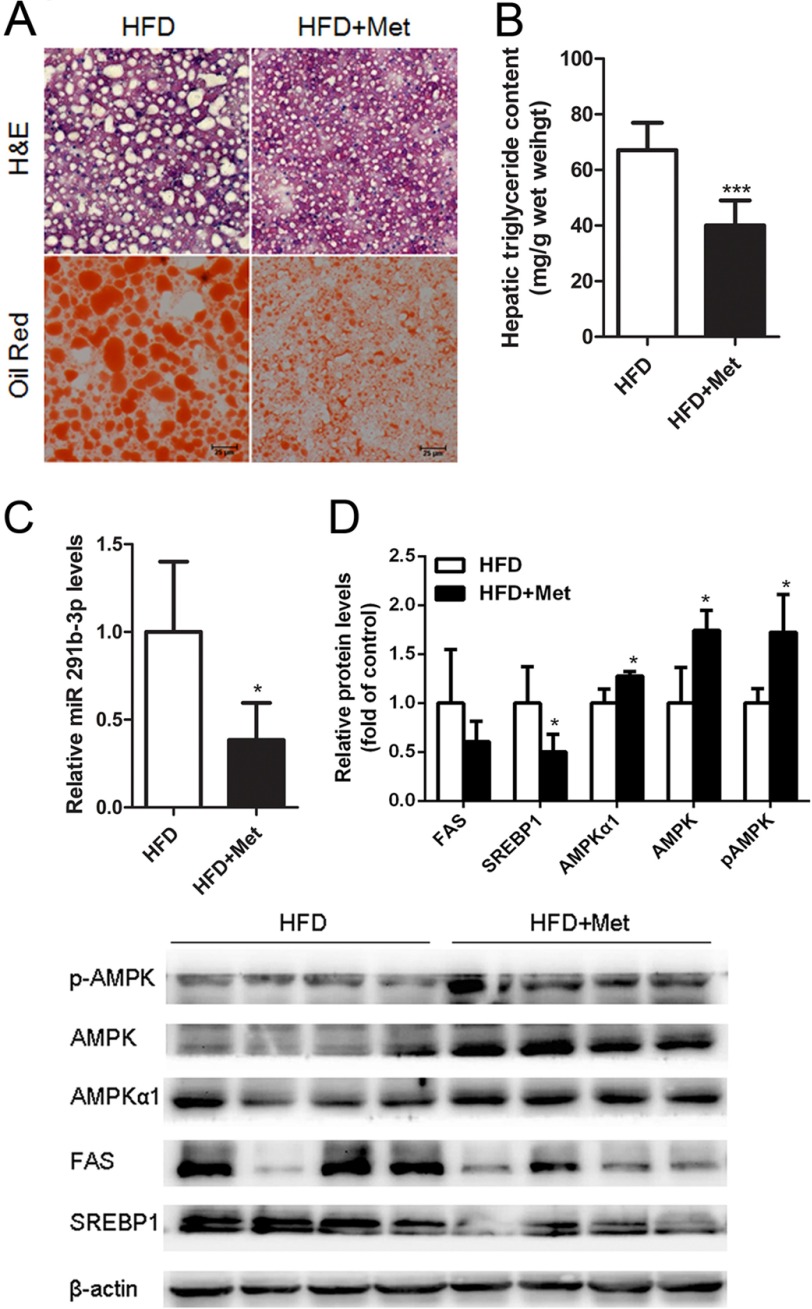
Metformin is an activator of AMPK. HFD-fed mice treated with metformin displayed reduced lipid accumulation (Fig. 6, A and B). To examine the potential relationship between miR-291b-3p and metformin, we measured miR-291b-3p expression levels in the livers of HFD-fed mice treated with metformin for 8 weeks and found that metformin significantly reduced miR-291b-3p expression (Fig. 6C). As expected, metformin increased hepatic levels of AMPKα1, AMPK, and p-AMPK and decreased hepatic expression of SREBP-1, FAS, and p-ACC1 in the HFD-fed mice (Fig. 6D). These data suggest that metformin may activate AMPK via a novel miR-291b-3p-dependent mechanism in liver.
FIGURE 6.
Metformin-induced reduction of hepatic lipids and lipogenic gene expression is associated with suppression of miR-291b-3p expression in the liver. Eight male C57BL/6J mice were intragastrically treated with metformin or vehicle at 3 mg/kg/day for 8 weeks in the presence of HFD after 10-week-HFD feeding and then sacrificed for sample collection. A, H&E and Oil Red O staining of frozen liver sections. B, hepatic triglyceride content. C, hepatic levels of miR-291b-3p. D, Western blotting analysis of AMPKα1, AMPK, p-AMPK, SREBP1, and FAS. *, p < 0.05; ***, p < 0.001. n = 4.
Discussion
In this study, we identified miR-291b-3p as a novel mediator of hepatic lipid accumulation. We provided evidence supporting a potential role for miR-291b-3p in driving hepatic lipogenesis associated with overnutrition or obesity. In addition, we established AMPKα1 as a direct target of miR-291b-3p and showed for the first time that metformin, a well-established activator of AMPK, suppresses hepatic miR-291b-3p expression in HFD-fed mice. This finding suggests that the activation of AMPK by metformin may at least partly be due to suppression of miR-291b-3p. Clearly, further studies are needed to address this novel question.
MiRNAs, small RNAs with ∼22 nucleotides that repress gene expression though incomplete base pairing, have been found to be involved in lipid metabolism. For example, as a predominant liver miRNA, miR-122 has been shown to have essential metabolic, anti-inflammatory, and anti-tumorigenic roles (30). MiR-33a/b was also reported to control lipid/cholesterol metabolism, mainly by repressing human SREBF genes (31). In addition, miR-181a transgenic mice demonstrated reduced lipid accumulation, mainly by targeting isocitrate dehydrogenase 1 (IDH1), a novel metabolic enzyme in the TCA cycle (32). In this study, using miRNA analysis of the livers of db/db mice and HFD-fed mice and NCTC1469 cells treated with an oleic acid/palmitic acid mixture, we examined the role of miR-291b-3p which belongs to the murine miR-290 cluster, in lipid metabolism. The miR-290 cluster is a 2.2-kb region on chromosome 7, and miRNAs of the miR-290 cluster are the most abundant miRNAs in murine embryonic stem cells (ESCs). Among 14 mature miRNAs transcribed from the cluster, miR-290–3p, miR-291a-3p, miR-291b-3p, miR-292–3p, miR-294, and miR-295 exhibit the same seed sequence (AAGUGC), while the other miRNAs (miR-290–5p, miR-291a-5p, miR-291b-5p, miR-292–5p, miR-293, miR-293*, miR-294*, and miR-295*) contain different seed sequences (33). Knock-out of this cluster led to partial embryonic lethality and germ cell defects, suggesting a key role in early embryonic development (34). Recent studies have indicated that the miR-290 cluster is involved in different cellular pathways by widely repressing various target genes (35–37). For example, p65, which is involved in modulating stem cell pluripotency, was identified as a new target for the miR-290 cluster (38). The miR-290 cluster has also been reported to stimulate glycolysis by regulating the glycolytic enzymes PKM2 and LDHA (39). To date, the study of the miR-290–295 cluster has been limited to early embryos and stem cells. In the present study, our data indicate that miR-291b-3p contributes to the accumulation of triglycerides in hepatocytes. More importantly, knockdown of miR-291b-3p has a therapeutic effect on lipid accumulation in HFD-fed mice, suggesting the potential application of miRNA therapy for NAFLD.
MiR-291b-3p is not the only miRNA that regulates obesity-associated NAFLD. In the present study, we identified AMPKα1 as a direct target of miR-291b-3p. As a sensor of cellular energy and nutritional status (40), AMPK activation can directly inhibit the transcriptional activity of the SREBP family. Members of the SREBP family are key regulators of fatty acid, triglyceride, phospholipid, and cholesterol biosynthesis and uptake (41, 42). There are 2 members of the mammalian SREBP family: SREBP1 (including 2 isoforms, SREBP-1c and SREBP-1a) and SREBP2. Importantly, SREBP1 mainly activates fatty acid and triglyceride biosynthesis. In comparison, SREBP2 mainly participates in cholesterol biosynthesis by regulating 3-hydroxy-3-methylglutaryl-coenzyme A reductase (HMGCR) expression and uptake (43). In rat hepatoma McA-RH7777 cells, AMPK was reported to inhibit liver X receptor (LXR)-dependent SREBP-1c mRNA expression, mainly by suppressing endogenous LXR ligand production (42, 44). Recently, AMPK was shown to phosphorylate and inhibit SREBP activity, preventing hepatic steatosis (13). SREBP-1c is a transcriptional activator of lipogenic genes, such as FAS (7); thus, its inhibition in liver is expected to suppress hepatic de novo lipogenesis. Additionally, AMPK activation may inhibit fatty acid synthesis by phosphorylating ACC, the rate-limiting enzyme in the fatty acid biosynthetic pathway (45). Thus, it is tempting to speculate that miR-291b-3p may promote hepatic steatosis in part by increasing de novo lipogenesis via suppressing AMPKα1 expression.
Metformin derived from biguanide is the most commonly prescribed medication for type 2 diabetes. Through activation of AMPK, metformin reduces hepatic glucose release and enhances glucose uptake in skeletal muscle (46). More recently, metformin was shown to be a therapeutic agent in a variety of diseases, including NAFLD and cancers (47). However, the mechanism underlying the enhanced AMPK activation by metformin has not been elucidated. Recent studies have found that multiple miRNAs are modulated by metformin in various diseases (48, 49). For example, metformin was shown to increase the expression of let-7a, let-7b, miR-26a, miR-101, miR-200b, and miR-200c in a dose-dependent manner in pancreatic cancer cells (47). In colon cancer, metformin treatment reduced the relative level of miR-21 and miR-145 (48). In this study, gastric administration of metformin increased AMPK activity and resulted in significantly impaired lipid deposition in the livers of HFD-fed mice. Most importantly, we found that metformin could down-regulate the expression of miR-291b-3p in the liver of HFD-fed mice, suggesting a potential role for miR-291b-3p in the metformin-mediated hepatic phenotypic shift away from lipid accretion via AMPK activation. However, we need to further investigate the effects of miR-291b-3p overexpression on metformin and the mechanisms by which metformin down-regulates miR-291b-3p expression in the liver of HFD-fed mice.
In addition, we observed that the mice injected with Ad-291m and Ad-291i exhibited increased serum levels of hepatic enzymes (AST and ALT) and elevated content of cytokines (TNFα and IL6) in liver compared with in control mice injected with vehicle (no virus). Consistently, Zaiss et al. (50) reported that in human HeLa cells and murine renal epithelium-derived cells (REC cells) the adenovirus vector AdlacZ induced the expression of multiple inflammatory chemokines including RANTES, interferon-inducible protein 10 (IP-10), IL-8, MIP-1and MIP-2 in a dose-dependent manner. Moreover, injection of adenovirus vector in mice resulted in increased chemokine and cytokine expression, and elevated serum AST and ALT levels. These findings indicated that adenovirus vectors significantly induced the vector-specific immune responses, which limited the potential therapeutic use of adenovirus vectors for gene therapy.
In conclusion, miR-291b-3p is critically involved in the regulation of obesity-associated NAFLD and is an inhibitor of AMPKα1. MiR-291b-3p inhibitors may have therapeutic effects on fatty liver disease. The beneficial effects of metformin in NAFLD prevention and treatment may be partly through down-regulation of hepatic miR-291b-3p expression. Detailed future studies on the molecular mechanisms by which miR-291b-3p promotes hepatic lipid accumulation should be carried out.
Author Contributions
X. M. and J. G. conducted most of the experiments, analyzed the results and wrote most of the paper. W. F., L. D., and M. L. conducted experiments on the adenovirus injection of the animal model. Y. M. and S. Z. constructed the metformin-treated HFD-fed mice model. X. H. and W. T. conducted the triglyceride content analysis. L. Y. and J. L. conceived the idea for the project.
This work was supported by 973 program grants from the National Basic Research Program of China (2012CB517502, 2014CB910503, to J. L.), the National Natural Science Foundation of China (81270887, 81270495, 81170381, to J. L.), the Specific Fund of Clinical Medical Research of the Chinese Medical Association (13060970482, to M. S.), and the NIDDK, National Institutes of Health (R01DK085176, to L. Y.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- NAFLD
- nonalcoholic fatty liver disease
- AMPK
- AMP-activated kinase
- FAS
- fatty acid synthase
- SREBP
- sterol regulatory element-binding protein
- HFD
- high-fat diet
- LDL
- low-density lipoprotein
- miRNA
- microRNA.
References
- 1. Younossi Z. M., Stepanova M., Afendy M., Fang Y., Younossi Y., Mir H., and Srishord M. (2011) Changes in the prevalence of the most common causes of chronic liver diseases in the United States from 1988 to 2008. Clin. Gastroenterol. Hepatol. 9, 524–530 [DOI] [PubMed] [Google Scholar]
- 2. Ajmal M. R., Yaccha M., Malik M. A., Rabbani M. U., Ahmad I., Isalm N., and Abdali N. (2014) Prevalence of nonalcoholic fatty liver disease (NAFLD) in patients of cardiovascular diseases and its association with hs-CRP and TNF-alpha. Indian Heart J. 66, 574–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Targher G., Bertolini L., Poli F., Rodella S., Scala L., Tessari R., Zenari L., and Falezza G. (2005) Nonalcoholic fatty liver disease and risk of future cardiovascular events among type 2 diabetic patients. Diabetes 54, 3541–3546 [DOI] [PubMed] [Google Scholar]
- 4. Adams L. A., Lymp J. F., St Sauver J., Sanderson S. O., Lindor K. D., Feldstein A., and Angulo P. (2005) The natural history of nonalcoholic fatty liver disease: a population-based cohort study. Gastroenterology 129, 113–121 [DOI] [PubMed] [Google Scholar]
- 5. Harrison S. A., Torgerson S., and Hayashi P. H. (2003) The natural history of nonalcoholic fatty liver disease: a clinical histopathological study. Am. J. Gastroenterol. 98, 2042–2047 [DOI] [PubMed] [Google Scholar]
- 6. Foretz M., Carling D., Guichard C., Ferré P., and Foufelle F. (1998) AMP-activated protein kinase inhibits the glucose-activated expression of fatty acid synthase gene in rat hepatocytes. J. Biol. Chem. 273, 14767–14771 [DOI] [PubMed] [Google Scholar]
- 7. An Z., Wang H., Song P., Zhang M., Geng X., and Zou M. H. (2007) Nicotine-induced activation of AMP-activated protein kinase inhibits fatty acid synthase in 3T3L1 adipocytes: a role for oxidant stress. J. Biol. Chem. 282, 26793–26801 [DOI] [PubMed] [Google Scholar]
- 8. Zhou W., Han W. F., Landree L. E., Thupari J. N., Pinn M. L., Bililign T., Kim E. K., Vadlamudi A., Medghalchi S. M., El Meskini R., Ronnett G. V., Townsend C. A., and Kuhajda F. P. (2007) Fatty acid synthase inhibition activates AMP-activated protein kinase in SKOV3 human ovarian cancer cells. Cancer Res. 67, 2964–2971 [DOI] [PubMed] [Google Scholar]
- 9. Stapleton D., Mitchelhill K. I., Gao G., Widmer J., Michell B. J., Teh T., House C. M., Fernandez C. S., Cox T., Witters L. A., and Kemp B. E. (1996) Mammalian AMP-activated protein kinase subfamily. J. Biol. Chem. 271, 611–614 [DOI] [PubMed] [Google Scholar]
- 10. Chen Z., Heierhorst J., Mann R. J., Mitchelhill K. I., Michell B. J., Witters L. A., Lynch G. S., Kemp B. E., and Stapleton D. (1999) Expression of the AMP-activated protein kinase β1 and β2 subunits in skeletal muscle. FEBS Lett. 460, 343–348 [DOI] [PubMed] [Google Scholar]
- 11. Cheung P. C., Salt I. P., Davies S. P., Hardie D. G., and Carling D. (2000) Characterization of AMP-activated protein kinase γ-subunit isoforms and their role in AMP binding. Biochem. J. 346, 659–669 [PMC free article] [PubMed] [Google Scholar]
- 12. Tangeman L., Wyatt C. N., and Brown T. L. (2012) Knockdown of AMP-activated protein kinase α1 and α2 catalytic subunits. J. RNAi. Gene. Silencing 8, 470–478 [PMC free article] [PubMed] [Google Scholar]
- 13. Li Y., Xu S., Mihaylova M. M., Zheng B., Hou X., Jiang B., Park O., Luo Z., Lefai E., Shyy J. Y., Gao B., Wierzbicki M., Verbeuren T. J., Shaw R. J., Cohen R. A., and Zang M. (2011) AMPK phosphorylates and inhibits SREBP activity to attenuate hepatic steatosis and atherosclerosis in diet-induced insulin-resistant mice. Cell Metab. 13, 376–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Horton J. D., Goldstein J. L., and Brown M. S. (2002) SREBPs: transcriptional mediators of lipid homeostasis. Cold Spring Harb. Symp. Quant. Biol. 67, 491–498 [DOI] [PubMed] [Google Scholar]
- 15. Kong Y., and Han J. H. (2005) MicroRNA: biological and computational perspective. Genomics, Proteomics Bioinformatics 3, 62–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lee R. C., Feinbaum R. L., and Ambros V. (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854 [DOI] [PubMed] [Google Scholar]
- 17. Bartel D. P. (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 [DOI] [PubMed] [Google Scholar]
- 18. Xiao F., Yu J., Liu B., Guo Y., Li K., Deng J., Zhang J., Wang C., Chen S., Du Y., Lu Y., Xiao Y., Zhang Z., and Guo F. (2014) A novel function of microRNA 130a-3p in hepatic insulin sensitivity and liver steatosis. Diabetes 63, 2631–2642 [DOI] [PubMed] [Google Scholar]
- 19. Ng R., Wu H., Xiao H., Chen X., Willenbring H., Steer C. J., and Song G. (2014) Inhibition of microRNA-24 expression in liver prevents hepatic lipid accumulation and hyperlipidemia. Hepatology 60, 554–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Soh J., Iqbal J., Queiroz J., Fernandez-Hernando C., and Hussain M. M. (2013) MicroRNA-30c reduces hyperlipidemia and atherosclerosis in mice by decreasing lipid synthesis and lipoprotein secretion. Nat. Med. 19, 892–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ramírez C. M., Goedeke L., Rotllan N., Yoon J. H., Cirera-Salinas D., Mattison J. A., Suárez Y., de Cabo R., Gorospe M., and Fernández-Hernando C. (2013) MicroRNA 33 regulates glucose metabolism. Mol. Cell. Biol. 33, 2891–2902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wen J., and Friedman J. R. (2012) miR-122 regulates hepatic lipid metabolism and tumor suppression. J. Clin. Invest. 122, 2773–2776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Guo J., Li M., Meng X., Sui J., Dou L., Tang W., Huang X., Man Y., Wang S., and Li J. (2014) MiR-291b-3p induces apoptosis in liver cell line NCTC1469 by reducing the level of RNA-binding protein HuR. Cell. Physiol. Biochem. 33, 810–822 [DOI] [PubMed] [Google Scholar]
- 24. Kaspi H., Chapnik E., Levy M., Beck G., Hornstein E., and Soen Y. (2013) Brief report: miR-290–295 regulate embryonic stem cell differentiation propensities by repressing Pax6. Stem cells 31, 2266–2272 [DOI] [PubMed] [Google Scholar]
- 25. Lichner Z., Páll E., Kerekes A., Pállinger E., Maraghechi P., Bosze Z., and Gócza E. (2011) The miR-290–295 cluster promotes pluripotency maintenance by regulating cell cycle phase distribution in mouse embryonic stem cells. Differentiation 81, 11–24 [DOI] [PubMed] [Google Scholar]
- 26. Zovoilis A., Smorag L., Pantazi A., and Engel W. (2009) Members of the miR-290 cluster modulate in vitro differentiation of mouse embryonic stem cells. Differentiation. 78, 69–78 [DOI] [PubMed] [Google Scholar]
- 27. Lee K. H., Song J. L., Park E. S., Ju J., Kim H. Y., and Park K. Y. (2015) Anti-obesity effects of starter fermented Kimchi on 3T3-L1 adipocytes. Prev. Nutr. Food. Sci. 20, 298–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kleiner D. E., Brunt E. M., Van Natta M., Behling C., Contos M. J., Cummings O. W., Ferrell L. D., Liu Y. C., Torbenson M. S., Unalp-Arida A., Yeh M., McCullough A. J., Sanyal A. J., and Nonalcoholic Steatohepatitis Clinical Research, N. (2005) Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology 41, 1313–1321 [DOI] [PubMed] [Google Scholar]
- 29. Ricchi M., Odoardi M. R., Carulli L., Anzivino C., Ballestri S., Pinetti A., Fantoni L. I., Marra F., Bertolotti M., Banni S., Lonardo A., Carulli N., and Loria P. (2009) Differential effect of oleic and palmitic acid on lipid accumulation and apoptosis in cultured hepatocytes. J. Gastroenterol. Hepatol. 24, 830–840 [DOI] [PubMed] [Google Scholar]
- 30. Hsu S. H., Wang B., Kota J., Yu J., Costinean S., Kutay H., Yu L., Bai S., La Perle K., Chivukula R. R., Mao H., Wei M., Clark K. R., Mendell J. R., Caligiuri M. A., Jacob S. T., Mendell J. T., and Ghoshal K. (2012) Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver. J. Clin. Invest. 122, 2871–2883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rottiers V., Najafi-Shoushtari S. H., Kristo F., Gurumurthy S., Zhong L., Li Y., Cohen D. E., Gerszten R. E., Bardeesy N., Mostoslavsky R., and Näär A. M. (2011) MicroRNAs in metabolism and metabolic diseases. Cold Spring Harb. Symp. Quant. Biol. 76, 225–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chu B., Wu T., Miao L., Mei Y., and Wu M. (2015) MiR-181a regulates lipid metabolism via IDH1. Sci. Rep. 5, 8801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Landgraf P., Rusu M., Sheridan R., Sewer A., Iovino N., Aravin A., Pfeffer S., Rice A., Kamphorst A. O., Landthaler M., Lin C., Socci N. D., Hermida L., Fulci V., Chiaretti S., Foà R., Schliwka J., Fuchs U., Novosel A., Müller R. U., Schermer B., Bissels U., Inman J., Phan Q., Chien M., Weir D. B., Choksi R., De Vita G., Frezzetti D., Trompeter H. I., Hornung V., Teng G., Hartmann G., Palkovits M., Di Lauro R., Wernet P., Macino G., Rogler C. E., Nagle J. W., Ju J., Papavasiliou F. N., Benzing T., Lichter P., Tam W., Brownstein M. J., Bosio A., Borkhardt A., Russo J. J., Sander C., Zavolan M., and Tuschl T. (2007) A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129, 1401–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Medeiros L. A., Dennis L. M., Gill M. E., Houbaviy H., Markoulaki S., Fu D., White A. C., Kirak O., Sharp P. A., Page D. C., and Jaenisch R. (2011) Mir-290–295 deficiency in mice results in partially penetrant embryonic lethality and germ cell defects. Proc. Natl. Acad. Sci. U.S.A. 108, 14163–14168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sinkkonen L., Hugenschmidt T., Berninger P., Gaidatzis D., Mohn F., Artus-Revel C. G., Zavolan M., Svoboda P., and Filipowicz W. (2008) MicroRNAs control de novo DNA methylation through regulation of transcriptional repressors in mouse embryonic stem cells. Nat. Struct. Mol. Biol. 15, 259–267 [DOI] [PubMed] [Google Scholar]
- 36. Wang Y., Baskerville S., Shenoy A., Babiarz J. E., Baehner L., and Blelloch R. (2008) Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat. Genet. 40, 1478–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zheng G. X., Ravi A., Calabrese J. M., Medeiros L. A., Kirak O., Dennis L. M., Jaenisch R., Burge C. B., and Sharp P. A. (2011) A latent pro-survival function for the mir-290–295 cluster in mouse embryonic stem cells. PLoS Genet. 7, e1002054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lüningschröor P., Stöcker B., Kaltschmidt B., and Kaltschmidt C. (2012) miR-290 cluster modulates pluripotency by repressing canonical NF-κB signaling. Stem cells. 30, 655–664 [DOI] [PubMed] [Google Scholar]
- 39. Cao Y., Guo W. T., Tian S., He X., Wang X. W., Liu X., Gu K. L., Ma X., Huang D., Hu L., Cai Y., Zhang H., Wang Y., and Gao P. (2015) miR-290/371-Mbd2-Myc circuit regulates glycolytic metabolism to promote pluripotency. EMBO J. 34, 609–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hu M., Wang F., Li X., Rogers C. Q., Liang X., Finck B. N., Mitra M. S., Zhang R., Mitchell D. A., and You M. (2012) Regulation of hepatic lipin-1 by ethanol: role of AMP-activated protein kinase/sterol regulatory element-binding protein 1 signaling in mice. Hepatology 55, 437–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shimano H., Horton J. D., Hammer R. E., Shimomura I., Brown M. S., and Goldstein J. L. (1996) Overproduction of cholesterol and fatty acids causes massive liver enlargement in transgenic mice expressing truncated SREBP-1a. J. Clin. Invest. 98, 1575–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yang J., Craddock L., Hong S., and Liu Z. M. (2009) AMP-activated protein kinase suppresses LXR-dependent sterol regulatory element-binding protein-1c transcription in rat hepatoma McA-RH7777 cells. J. Cell. Biochem. 106, 414–426 [DOI] [PubMed] [Google Scholar]
- 43. Horton J. D., Goldstein J. L., and Brown M. S. (2002) SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest. 109, 1125–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cho K., Kim S. J., Park S. H., Kim S., and Park T. (2009) Protective effect of Codonopsis lanceolata root extract against alcoholic fatty liver in the rat. J. Med. Food. 12, 1293–1301 [DOI] [PubMed] [Google Scholar]
- 45. Ha J., Daniel S., Broyles S. S., and Kim K. H. (1994) Critical phosphorylation sites for acetyl-CoA carboxylase activity. J. Biol. Chem. 269, 22162–22168 [PubMed] [Google Scholar]
- 46. Aljada A., and Mousa S. A. (2012) Metformin and neoplasia: implications and indications. Pharmacol. Therap. 133, 108–115 [DOI] [PubMed] [Google Scholar]
- 47. Ford R. J., Fullerton M. D., Pinkosky S. L., Day E. A., Scott J. W., Oakhill J. S., Bujak A. L., Smith B. K., Crane J. D., Blümer R. M., Marcinko K., Kemp B. E., Gerstein H. C., and Steinberg G. R. (2015) Metformin and salicylate synergistically activate liver AMPK, inhibit lipogenesis and improve insulin sensitivity. Biochem. J. 468, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nangia-Makker P., Yu Y., Vasudevan A., Farhana L., Rajendra S. G., Levi E., and Majumdar A. P. (2014) Metformin: a potential therapeutic agent for recurrent colon cancer. PLoS ONE 9, e84369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bao B., Wang Z., Ali S., Ahmad A., Azmi A. S., Sarkar S. H., Banerjee S., Kong D., Li Y., Thakur S., and Sarkar F. H. (2012) Metformin inhibits cell proliferation, migration and invasion by attenuating CSC function mediated by deregulating miRNAs in pancreatic cancer cells. Cancer Prev. Res. 5, 355–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zaiss A. K., Liu Q., Bowen G. P., Wong N. C., Bartlett J. S., and Muruve D. A. (2002) Differential activation of innate immune responses by adenovirus and adeno-associated virus vectors. J. Virol. 76, 4580–4590 [DOI] [PMC free article] [PubMed] [Google Scholar]