Abstract
Mouse CD1d is a nonclassical MHC molecule able to present lipids and glycolipids to a specialized subset of T cells known as natural killer T cells. The antigens presented by CD1d have been shown to cover a broad range of chemical structures and to follow precise rules determining the potency of the antigen in the context of T cell activation. Together with lipids, initial reports suggested that CD1d can also bind and present hydrophobic peptides with (F/W)XX(I/L/M)XXW. However, the exact location of peptide binding and the molecular basis for the required motif are currently unknown. Here we present the crystal structure of the first peptide identified to bind CD1d, p99, and show that it binds in the antigen-binding groove of CD1d in a manner compatible with its presentation to T cell receptors. Interestingly, the peptide adopts an α-helical conformation, which orients the motif residues toward its deep binding groove, therefore explaining the molecular requirements for peptide binding. Moreover, we demonstrate that a lipopeptide version of the same peptide is able to bind CD1d in a similar conformation, identifying another class of molecules binding this antigen-presenting molecule.
Keywords: antigen presentation, crystal structure, glycolipid, peptides, T cell receptor (TCR), CD1d, MHC Class I-like
Introduction
The presentation of self- and non-self-peptides by the MHC class of proteins to T cells represents a critical step in the adaptive immune response. Peptides are bound in a shallow antigen-binding groove generated by two antiparallel α-helices sitting on top of a β-sheet. MHC class I peptides are bound in an extended conformation with both N and C termini bound inside a closed groove, limiting their size to 8–10 residues (on average 9). MHC class II peptides are also bound in an extended conformation but, because of the open nature of the groove, tend to be longer (14–20 residues) (1). Binding of longer peptides has also been reported for MHC class I, and in those cases, the peptides either bulged out of the groove (2–5), bound in a zig-zag fashion (6), or induced the opening of the F pocket to protrude with their C termini into the solvent (7). In both MHC class I and II, peptides are bound with specific anchor residues inside allele-specific pockets (denominated A to F), thereby exposing alternating amino acids for T cell receptor recognition (5, 6). Allelic variation and polymorphism within both MHC classes greatly increases the peptide repertoire that can be presented to T cells. In contrast, the nonclassical MHC-like molecule CD1d is nonpolymorphic and presents lipid and glycolipid antigens, rather than peptides, to a specialized subset of T cells termed natural killer T (NKT)2 cells (1, 8, 9). This innate-like T cells can be divided in two main subsets, types I and II, based on their antigen specificity, with each type showing distinct and sometimes opposing immunomodulatory properties (8). Unlike MHC molecules, CD1d molecules are characterized by a deep, apolar groove divided in two pockets called A′ and F′ because of their similarity to the A and F pockets found in MHC-I molecules (10). Polar moieties of the antigens, such as sugar and phosphate-containing moieties, are generally exposed for recognition by a T cell receptor (TCR), whereas the hydrophobic portions of the antigens are bound inside the groove. Prior to the discovery of the prototypical CD1d-presented antigen α-GalCer (11), peptides were also reported to bind CD1d (12). However, these peptides were relatively long and hydrophobic, compared with classical MHC-presented peptides, and unlikely to bind in the similar mode as observed for MHC molecules. In particular, attempts to define the molecular requirements of peptide binding to CD1d by testing random peptides with a phage display approach identified p99, a 22-mer characterized by a hydrophobic binding motif of the nature (F/W)XX(I/L/M)XXW (12). Interestingly, the motif itself was not sufficient for CD1d binding, because truncation of the peptide to versions containing only the minimal motif showed reduced or abolished binding affinity for the protein. Later, a peptide from chicken ovalbumin carrying the same motif (p18) was also shown to bind to CD1d and elicit an immune response (13, 14). However, even though peptide CD1d-reactive T cells have been reported in these studies, their frequency and role in the context of the immune response remains controversial. Interestingly, other peptides carrying the same motif can be identified, including a portion of the leader sequence on mouse CD1d (15) or several viral peptides (16), but it is currently unclear whether these peptides bind CD1d. An interesting viral peptide that contains the CD1d binding motif is the HIV-1 MPER (membrane-proximal external region) peptide of GP41 (LELDKWASLWNWFDITNWLWYIK). Because HIV-1 down-regulates CD1d cell surface expression via Nef and Vpu (17, 18), it is tempting to speculate that CD1d could either present a viral antigen, such as a peptide or lipopeptide, or a glycolipid self-antigen that is generated upon infection to alert the immune system. However, so far, no HIV-derived antigen for CD1d-restricted T cells has been identified. More recently, a peptide derived from collagen, mCII707–721,was reported to be presented to T cells by mouse CD1d (19). Although the structural and biochemical details of the binding of lipid antigens to CD1d have been extensively characterized (8, 9), the molecular details of the binding of peptides to CD1d molecules have not yet been understood. Here we describe the crystal structure of the co-complex between the p99 peptide and mouse CD1d, highlighting an unusual mode of peptide binding by a nonclassical MHC molecule. Modifications of the p99 moiety generating lipopeptides were also shown to bind CD1d, suggesting new potential modifications of this CD1d-binding peptide able to modulate binding affinity for CD1d.
Experimental Procedures
mCD1d Expression and Purification
The mouse CD1d extracellular portion (residues 1–278) was expressed recombinantly in Sf9 insect cells and purified as previously described (20).
Co-crystallization of the mCD1d-Peptide Complexes
The p99 peptide (in its biotinylated form, biotin-YEHDFHHIREWGNHWKNFLAVM) and the p99p peptide (YEHDFHHIREK{palm}GNHWKNFLAVM) were synthetized by Genscript at >95% purity and dissolved in DMSO at 5 mg/ml for further experiments. Purified mCD1d/β2m heterodimers at 5–10 mg/ml were incubated with a 6 molar excess of the peptide (DMSO concentration below 1%) for 16 h before removal of the excess peptide and concentration of the sample with a Millipore Amicon concentrator (molecular mass cutoff, 5 kDa). The CD1d-p99 crystals were grown in 0.2 m ammonium tartrate and 20% PEG 4000 at 4 °C using a sample at a concentration of 10 mg/ml. The CD1d-p99p crystals were grown in 0.2 m ammonium citrate and 20% PEG 4000 at 4 °C.
Structure Determination and Refinement
Diffraction data were processed with the iMosflm (21) and CCP4 software suites (22). Both the CD1d-p99 and CD1d-p99p crystals (P212121) contain one complex per asymmetric unit and diffracted to a resolution of 1.8Å. The structure was determined by molecular replacement using MOLREP (23) using the CD1d-β2m structure from the CD1d-OCH complex (Protein Data Bank code 3G08), with the ligand removed. Clear density was present above the CD1d binding groove, allowing the building of a peptide model covering residues 4–21 of p99 with iterative cycles of model building in Coot and refinement in REFMAC5 (24, 25). The final refinement statistics for both structures are presented in Table 1.
TABLE 1.
Data collection and refinement statistics
| mCD1d-p99 | mCD1d-p99p | |
|---|---|---|
| Data collection statistics | ||
| Space group | P212121 | P212121 |
| Cell dimension | ||
| a, b, c (Å) | 42.17, 107.57, 110.61 | 42.19, 107.85, 110.34 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 |
| Resolution range (Å) (outer shell) | 55–1.80 (1.9–1.8) | 49.1–1.8 (1.84–1.8) |
| No. of reflections | 47,272 | 46,178 |
| Rmerge (%) | 8.9 (83.8) | 5.6 (56.2) |
| Multiplicity | 6.1 (6.1) | 3.5 (3.2) |
| Average I/σI | 11.1 (2.4) | 10.9 (1.6) |
| Completeness (%) | 99.5 (99.6) | 97.4 (96.7) |
| Refinement statistics | ||
| R/Rfree | 0.205/0.234 | 0.219/0.244 |
| Ramachandran plot (%) | ||
| Favored | 98.4 | 98.4 |
| Allowed | 100.0 | 100.0 |
| Rmsa deviations | ||
| Bonds (Å) | 0.007 | 0.008 |
| Angles (°) | 1.267 | 1.334 |
a Rms, root mean square.
Competition Binding Assay
Recombinant CD1d was loaded with 6× molar excess of disulfatide (SLF2; Avanti Polar Lipids) at 37 °C for 1.5 h in 100 mm Tris, pH 7.0, 100 mm NaCl. After lipid loading, excess lipid was removed by ultrafiltration using Amicon filter cartridges (molecular mass weight cutoff, 30 kDa). CD1d-SLF2 complexes were then incubated overnight at room temperature with 10× molar excess of peptides either in the absence of any detergent or in the presence of 0.06 mm Tween 20 or 0.01 mm tyloxapol. Successful loading of SLF2, as well as its replacement by peptides was visualized using native isoelectric focusing gel electrophoresis on pH 3–9 gradient gels using the PHAST system (GE Healthcare).
Results
The p99 Peptide Binds CD1d Above the Antigen-binding Groove
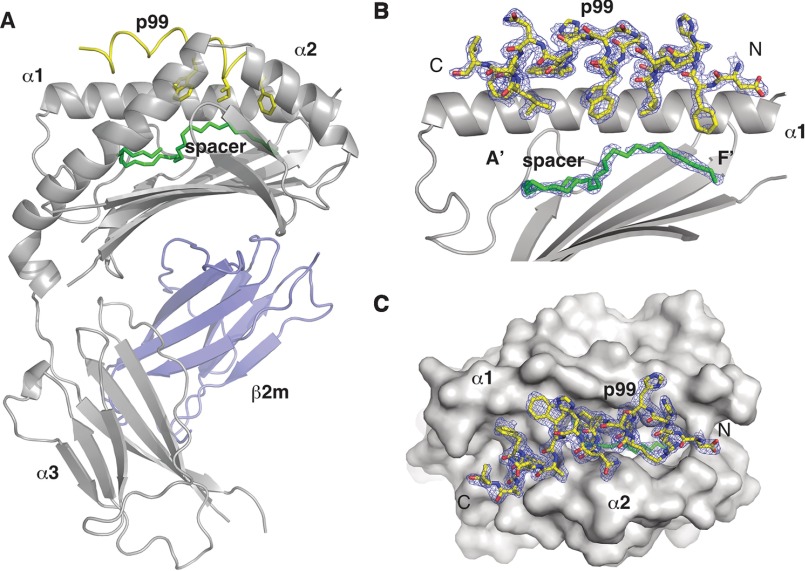
The crystal structure of the mCD1d-p99 complex, solved at 1.8Å of resolution, revealed that the peptide binds between the α1 and α2 helices of the antigen binding groove, straddling the whole length of the groove, its N terminus sitting above the F′ pocket and its C terminus at the opposite extremity, above the A′ pocket (Fig. 1A, residues are numbered −3 to 19, with the first residue of the motif as residue 1). The peptide binding orientation is reversed compared with MHC I, where the N terminus binds at the A pocket and the C terminus in the F pocket. The strong positive electron density after the molecular replacement step clearly supported this location of the peptide with well defined electron density observed for all but the first three residues and the C-terminal residue of the peptide, allowing for the unambiguous determination of the peptide orientation and the placing of almost all side chains (Fig. 1, B and C). Unexpectedly, the p99 peptide adopts a right-handed α-helical conformation in the complex. In particular two α-helical segments, corresponding to residues 0–10 and 11–17 in the structure, can be distinguished. The first segment contains the FXXIXXW motif and is situated at the entrance of the deep A′ and F′ binding pockets. The second segment contains the residues WKNFLAVM and sits above the surface of the protein, correspondence to the A′ pocket (Fig. 1, B and C). Additional electron density suggesting the presence of a spacer lipid was also present at the bottom of the A′ and F′ pockets not yet occupied by the peptide. The nature of this spacer is currently unclear, and a C32 aliphatic chain was modeled into the density.
FIGURE 1.
Crystal structure of the CD1d-p99 complex. A, overall view of the mCD1d-p99 complex with the CD1d chain in gray, β2m in blue, the p99 peptide in yellow, and the spacer in green. The p99 peptide binds in the antigen-binding groove, between the α1 and α2 helices and above the spacer filling the remaining of the cavity. B, side view of the antigen-binding groove with the α2 helix removed for clarity. 2 Fo − Fc density at 1.0 σ is shown for the peptide and spacer molecule. C, top view of the CD1d-p99 complex with the CD1d molecule shown as a surface and the 2Fo − Fc density at 1.0σ for the peptide.
The p99 Residue Orientation Explains the Requirements for Binding
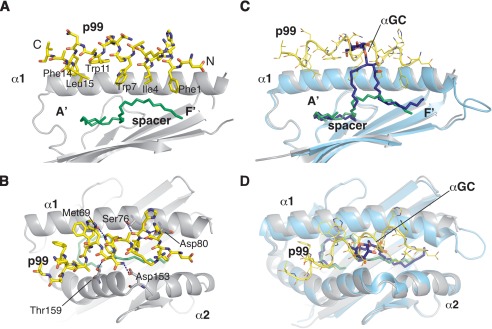
Although unexpected, the α-helical conformation adopted by the p99 peptide provides a rational framework to understand the molecular requirements for the peptide-binding motif. Because of their position along the helix, all three key residues (Phe1, Ile4, and Trp7) are oriented in the same direction and point toward the CD1d binding groove, forming an hydrophobic surface that mimics the acyl chains of the lipids antigens commonly bound by CD1d (Fig. 2A). Together, these three residues fill the upper portion of the F′ pocket, which is generally open to the exterior, and contact the spacer molecule underneath. On the other side, the second segment of the peptide binds above the A′ roof and does not contact directly the spacer molecule. In particular, Trp11, Phe14, and Leu15 form a second hydrophobic interface, anchoring the peptide between the α1 and α2 helices. Mutational studies done on the p99 peptide revealed that the motif alone is not sufficient to promote binding to CD1d (12), possibly as a result of the requirement for the second “minor” binding motif or, alternatively, because of a requirement for sufficient peptide length to stabilize the secondary structure. Contacts between the peptide and CD1d are mainly of a hydrophobic nature, and only a handful of residues on both α1 and α2 helices engage in polar contacts with the peptide (Fig. 2B). Residues Met69, Ser76, and Asp80 on the α1 helix and Asp153 and Thr159 on the α2 helix form H bond interactions with the core motif of the peptide. Interestingly, only Met69 and Asp153 make H bonds with side chains of the peptide, Trp7 and Gln8 respectively, consistent with the finding that most of the nonanchoring positions are not restricted to a specific moiety (12).
FIGURE 2.
Binding mode of the p99 peptide to CD1d. A, side view of the antigen-binding groove with the α2 helix removed for clarity. The residues composing the CD1d-binding motif are labeled, highlighting how they are all oriented toward the cavity of the A′ and F′ pockets. B, top view of the antigen-binding groove with highlighted the polar contacts between CD1d and the p99 peptide. C, superposition of the CD1d-p99 and CD1d-α-GalCer structure. In the side view, α2 helix was removed for clarity. Note how the peptide and α-GalCer overlap and protrude to similar extents from the antigen-binding groove. D, top view of the antigen-binding groove with the superposition of the CD1d-p99 and CD1d-α-GalCer structures.
p99 Binding Does Not Require Major CD1d Structural Rearrangements and Is Compatible with Antigen Presentation
The binding site of the p99 binding site overlaps significantly with the binding site of the well characterized lipid and glycolipid antigens presented by CD1d, including the protypical antigen α-GalCer (Fig. 2C). We therefore checked whether the binding of the p99 peptide resulted in major rearrangement in the α1α2 domain compared with previously reported CD1d-lipid complexes. Superposition of the CD1d-p99 complex with the mCD1d-αGC structure (Protein Data Bank code 1Z5L) showed no gross changes in the conformation of the antigen-binding region (Fig. 2, C and D). We observed a minor shift (4.3 Å between Cα atoms of Met87) at the C terminus of the α1 helix. However, this region is followed by a rather disordered loop, possibly as a result of different packing contacts between the molecules in the two crystal structures. We next asked whether the binding of p99 would allow for binding of NKT cell TCRs to CD1d. Superposition of the CD1d-p99 structure on the mCD1d-α-GalCer-type I-NKT TCR structure showed the presence of several important steric clashes between the peptide and the α chain of the TCR, suggesting that the p99 cannot be recognized by this TCR (supplemental Fig. S1). This is consistent with reports suggesting that this peptide could elicit only CD8+ T cell responses in mouse and our own findings that CD1d-presented p99 cannot activate iNKT cell hybridomas (12) (data not shown).
Structure of Lipopeptide Bound to CD1d Suggests Novel Peptide-like Ligands
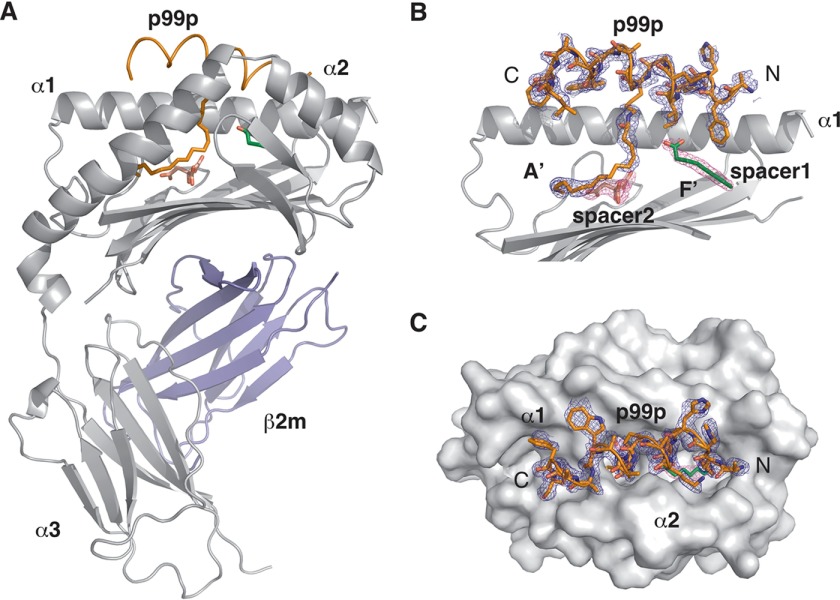
To explore further the molecular determinants of peptide binding to CD1d, we modified the p99 peptide at the anchor residue Trp7 by mutating the residue to Lys and covalently attaching to it a palmitate moiety. The lipopeptide generated (p99p) was crystallized in complex with CD1d at a resolution of 1.8 Å. p99p binds in a similar orientation and conformation to p99, adopting an α-helical conformation and sitting above the antigen binding groove (Fig. 3). The palmitate chain binds in the antigen binding groove, and it is best modeled as occupying the A′ pocket. Interestingly, binding of p99p to CD1d recruited a different set of spacer molecules in the A′ and F′ pocket compared with p99. In this case, a short linear molecule (modeled as caprylic acid, C8) and a citrate molecule probably obtained from the crystallization mother liquor are filling the remaining cavities in the two pockets of the groove. The C8 spacer lipid found in the F′ pocket has previously been reported to be recruited when short chain glycolipids, such as OCH, do not fully occupy the F′ pocket and appears to be a general feature required for the stabilization of the F′ pocket of CD1d (26).
FIGURE 3.
Crystal structure of the CD1d-p99p complex. A, overall view of the mCD1d-p99p complex with the CD1d chain in gray, β2m in blue, and the p99p lipopeptide in orange. The caprylic spacer is shown in green, and the citrate spacer is in pink. B, side view of the antigen-binding groove with the α2 helix removed for clarity. 2 Fo − Fc density at 0.8 σ is shown for the lipopeptide and spacer molecules. C, top view of the CD1d-p99p complex with the CD1d molecule shown as a surface and the 2 Fo − Fc density at 0.8 σ for the lipopeptide.
Peptides and Lipopeptides Can Compete with Short Chain Lipids for CD1d Binding
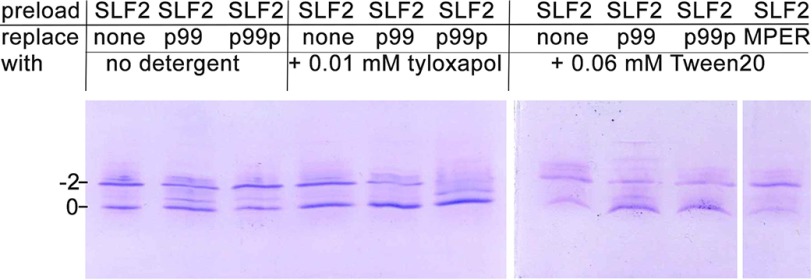
We next assessed whether either p99 or p99p could compete with lipids for CD1d binding. We first preloaded CD1d with a charged lipid, which upon CD1d binding results in a gel shift of CD1d using isoelectric focusing gel electrophoresis (27, 28). We first tested the glycosphingolipid sulfatide and the diacylglycerolipid phosphatidyl inositol. However, because sulfatide fully fills the CD1d binding groove (C24:1 and C18:0 alkyl chains) and binds very intimately to CD1d (29), no competition by p99 or p99p could be observed. Similarly, competition with phosphatidyl inositol (C16/C18 alkyl chains) was poor (not shown). We then chose a lipid that does not fully fill the CD1d binding groove, similar to p99 or p99p, which both only insert slightly, or only with one alkyl chain into the binding groove. We used C12-SLF2, because we expected it to bind CD1d less tightly, allowing for the presence of the A′ pocket spacer lipid that is also seen in the p99 and p99p structure. Binding of SLF2 resulted in shift of ∼60% of all CD1d molecules to the −2 charge position (Fig. 4). This indicates that the competition of SLF2 with the endogenously loaded lipid (30) is not complete. However, incubation of CD1d-SLF2 complexes with p99 resulted in a slight gel shift of CD1d back to the 0 charge position. p99p did not compete for SLF2 in this assay, likely because it is less soluble in the absence of any detergent. Because it was recently shown that the nature of the detergent can influence lipid loading efficiencies (31), we performed the p99 and p99p competition assay in the presence of 0.01 mm tyloxapol or 0.06 mm Tween 20. Although p99 and p99p compete equally well with SLF2 in the presence of Tween 20, p99p competes more efficiently than p99 in the presence of tyloxapol. We noticed a full migration back to the starting position on the gel (Fig. 4). We also tested binding of the HIV-1 MPER peptide, which contains the CD1d binding motif, in this assay. However, we failed to see any competition using MPER, suggesting that it does not bind CD1d. We generated a MPER-CD1d model and noticed that this peptide has two bulky side chains around the CD1d binding motif that likely leads to steric clashes with CD1d and explains the lack of CD1d binding (supplemental Fig. S2). In summary, our data suggest that the lipopeptide p99p can compete better with lipids than the peptide 99 itself, because its single alkyl chain assists in the competition. However, solubility of p99p in aqueous buffers is likely reduced compared with p99, necessitating the presence of detergent in this assay. Therefore, p99p should be able to compete well with lysolipids, an emerging family of NKT cell antigens that contain only one fatty acid (32). However, because we did not see any gel shift upon binding of lyso-phosphatidyl inositol and lyso-phosphatidyl glycerol in our isoelectric focusing assay (not shown), likely because of their transient interaction with CD1d, we could not test competition by p99 or p99p directly.
FIGURE 4.
p99 and p99p can compete with disulfatide for binding to CD1d. Isoelectric focusing gel showing mouse CD1d samples loaded with SLF2 and incubated with peptides and surfactants. p99 and p99p are able to compete with disulfatide antigens, as shown by the shift in the ratio between the two bands observed toward the lower band. No competition was observed with the MPER peptide with disulfatide preloaded CD1d.
Discussion
We present here the structure of the p99 α-helical peptide bound to CD1d, the first case in which an α-helical peptide binds the antigen-binding groove of an MHC fold. The structure revealed that the secondary structure of the peptide is critical for the orientation of hydrophobic side chains within the antigen-binding groove of the protein, therefore providing a rationale for the previously determined CD1d-binding motif ((F/W)XX(I/L/M)XXW) found in the peptide.
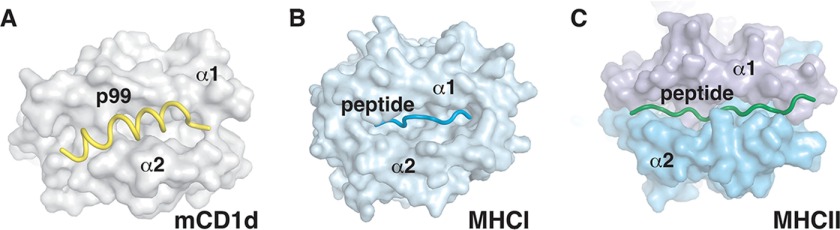
It was initially proposed that MHC molecules could also present α-helical peptides of amphiphatic nature (33, 34), but the determination of the first MHC peptide co-crystal structures revealed that peptides bound the proteins in an extended conformation, with MHC-I-specific peptides binding within the groove, whereas MHC-II-specific peptides bind with their extremities exposed outside of the binding groove (Fig. 5). The CD1d-p99 structure highlighted here shows that CD1d can, with minimal structural rearrangements, accommodate a third helix between the α1 and α2 helices, therefore creating a composite surface that could be involved in further protein-protein interactions, possibly involving TCRs or NK cell receptors. Interestingly, the reverse N- to C-terminal orientation of the peptide, compared with MHC-I-peptide complexes, is possibly the result of the lack of N- or C-terminal anchors, because reverse peptide binding orientations have been previously reported for MHC-II complexes (35). Moreover, we show that it is possible to modify the p99 backbone to generate lipopeptides able to bind mouse CD1d. Further modifications of this backbone, for example by extending the aliphatic portion to completely fill the A′ pocket now only partly occupied by the palmitate residue, could result in novel ligands for CD1d with increased stability and affinity. Consistent with the structural information presented here, p99 and p99p are able to compete with short chain glycolipids for binding within the antigen binding groove (Fig. 4).
FIGURE 5.
Peptide binding to MHC and CD1d molecules. A, top view of the CD1d-p99 complex with the peptide in yellow. B, top view of a MHCI-peptide complex (Protein Data Bank code 1FZK) with the peptide fully contained with the antigen-binding groove. C, top view of a MHCII-peptide complex (Protein Data Bank code 1ES0) with the peptide straddling the whole antigen-binding groove.
Several other peptides have been identified that carry a similar motif to p99, including a peptide derived from chicken albumin, p18, that was reported to induce a CD1d-dependent immune response (13) and several viral peptides including the HIV MPER peptide. However, we were unable to clearly demonstrate mouse or human CD1d binding of p18 (not shown) or the MPER peptide (Fig. 4), possibly because of further steric clashes between peptide side chains and the CD1d binding groove (supplemental Fig. S2). Also, attempts at crystallizing the complexes of p18 and MPER with CD1d have so far proven unsuccessful. It is therefore possible that such peptides have a reduced binding affinity for CD1d compared with p99, because the latter was the result of several rounds of optimization by phage display technique against mouse CD1d expressed in insect cells, the same source of the material used for crystallization. Accordingly, the p99 peptide is not expected to bind to human CD1d in the same conformation observed for mouse CD1d because of several sterical clashes (supplemental Fig. S3). Further functional, biochemical, and structural studies, possibly involved improved versions of the (lipo)peptides studied here, would therefore be required to advance our understanding of the nature of CD1d-peptides interactions.
Author Contributions
E. G. prepared and crystallized the CD1d-peptide complexes and determined the crystal structures. J. W. assisted with protein production and purification and performed competition binding experiments. E. G. and D. M. Z. conceived the study and wrote the paper.
Supplementary Material
Acknowledgment
We acknowledge Ian A. Wilson (TSRI) for discussions regarding the HIV-1 MPER peptide of GP41.
This work was supported by National Institutes of Health Grant AI107318 (to D. M. Z.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Figs. S1–S3.
The atomic coordinates and structure factors (codes 5FKP and 5EFI) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- NKT
- natural killer T
- TCR
- T cell receptor
- α-GalCer
- α-galactosylceramide
- SLF2
- disulfatide.
References
- 1. Adams E. J., and Luoma A. M. (2013) The adaptable major histocompatibility complex (MHC) fold: structure and function of nonclassical and MHC class I-like molecules. Annu. Rev. Immunol. 31, 529–561 [DOI] [PubMed] [Google Scholar]
- 2. Guo H. C., Jardetzky T. S., Garrett T. P., Lane W. S., Strominger J. L., and Wiley D. C. (1992) Different length peptides bind to HLA-Aw68 similarly at their ends but bulge out in the middle. Nature 360, 364–366 [DOI] [PubMed] [Google Scholar]
- 3. Madden D. R. (1995) The three-dimensional structure of peptide-MHC complexes. Annu. Rev. Immunol. 13, 587–622 [DOI] [PubMed] [Google Scholar]
- 4. Zhang W., Young A. C., Imarai M., Nathenson S. G., and Sacchettini J. C. (1992) Crystal structure of the major histocompatibility complex class I H-2Kb molecule containing a single viral peptide: implications for peptide binding and T-cell receptor recognition. Proc. Natl. Acad. Sci. U.S.A. 89, 8403–8407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rossjohn J., Gras S., Miles J. J., Turner S. J., Godfrey D. I., and McCluskey J. (2015) T cell antigen receptor recognition of antigen-presenting molecules. Annu. Rev. Immunol. 33, 169–200 [DOI] [PubMed] [Google Scholar]
- 6. Rudolph M. G., Stanfield R. L., and Wilson I. A. (2006) How TCRs bind MHCs, peptides, and coreceptors. Annu. Rev. Immunol. 24, 419–466 [DOI] [PubMed] [Google Scholar]
- 7. McMurtrey C., Trolle T., Sansom T., Remesh S. G., Kaever T., Bardet W., Jackson K., McLeod R., Sette A., Nielsen M., Zajonc D. M., Blader I. J., Peters B., and Hildebrand W. (2016) Toxoplasma gondii peptide ligands open the gate of the HLA class I binding groove. eLife 5, e12556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Girardi E., and Zajonc D. M. (2012) Molecular basis of lipid antigen presentation by CD1d and recognition by natural killer T cells. Immunol. Rev. 250, 167–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rossjohn J., Pellicci D. G., Patel O., Gapin L., and Godfrey D. I. (2012) Recognition of CD1d-restricted antigens by natural killer T cells. Nat. Rev. Immunol. 12, 845–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zajonc D. M., and Wilson I. A. (2007) Architecture of CD1 proteins. Curr. Top. Microbiol. Immunol. 314, 27–50 [DOI] [PubMed] [Google Scholar]
- 11. Kawano T., Cui J., Koezuka Y., Toura I., Kaneko Y., Motoki K., Ueno H., Nakagawa R., Sato H., Kondo E., Koseki H., and Taniguchi M. (1997) CD1d-restricted and TCR-mediated activation of vα14 NKT cells by glycosylceramides. Science 278, 1626–1629 [DOI] [PubMed] [Google Scholar]
- 12. Castaño A. R., Tangri S., Miller J. E., Holcombe H. R., Jackson M. R., Huse W. D., Kronenberg M., and Peterson P. A. (1995) Peptide binding and presentation by mouse CD1. Science 269, 223–226 [DOI] [PubMed] [Google Scholar]
- 13. Lee D. J., Abeyratne A., Carson D. A., and Corr M. (1998) Induction of an antigen-specific, CD1-restricted cytotoxic T lymphocyte response in vivo. J. Exp. Med. 187, 433–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tangri S., Brossay L., Burdin N., Lee D. J., Corr M., and Kronenberg M. (1998) Presentation of peptide antigens by mouse CD1 requires endosomal localization and protein antigen processing. Proc. Natl. Acad. Sci. U.S.A. 95, 14314–14319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bendelac A., Rivera M. N., Park S. H., and Roark J. H. (1997) Mouse CD1-specific NK1 T cells: development, specificity, and function. Annu. Rev. Immunol. 15, 535–562 [DOI] [PubMed] [Google Scholar]
- 16. Brossay L., Burdin N., Tangri S., and Kronenberg M. (1998) Antigen-presenting function of mouse CD1: one molecule with two different kinds of antigenic ligands. Immunol. Rev. 163, 139–150 [DOI] [PubMed] [Google Scholar]
- 17. Cho S., Knox K. S., Kohli L. M., He J. J., Exley M. A., Wilson S. B., and Brutkiewicz R. R. (2005) Impaired cell surface expression of human CD1d by the formation of an HIV-1 Nef/CD1d complex. Virology 337, 242–252 [DOI] [PubMed] [Google Scholar]
- 18. Moll M., Andersson S. K., Smed-Sörensen A., and Sandberg J. K. (2010) Inhibition of lipid antigen presentation in dendritic cells by HIV-1 Vpu interference with CD1d recycling from endosomal compartments. Blood 116, 1876–1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu Y., Teige A., Mondoc E., Ibrahim S., Holmdahl R., and Issazadeh-Navikas S. (2011) Endogenous collagen peptide activation of CD1d-restricted NKT cells ameliorates tissue-specific inflammation in mice. J. Clin. Invest. 121, 249–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zajonc D. M., Cantu C. 3rd, Mattner J., Zhou D., Savage P. B., Bendelac A., Wilson I. A., and Teyton L. (2005) Structure and function of a potent agonist for the semi-invariant natural killer T cell receptor. Nat. Immunol. 6, 810–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Battye T. G., Kontogiannis L., Johnson O., Powell H. R., and Leslie A. G. (2011) iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr. 67, 271–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., et al. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vagin A., and Teplyakov A. (2010) Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 66, 22–25 [DOI] [PubMed] [Google Scholar]
- 24. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Murshudov G. N., Skubák P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., and Vagin A. A. (2011) REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sullivan B. A., Nagarajan N. A., Wingender G., Wang J., Scott I., Tsuji M., Franck R. W., Porcelli S. A., Zajonc D. M., and Kronenberg M. (2010) Mechanisms for glycolipid antigen-driven cytokine polarization by Vα14i NKT cells. J. Immunol. 184, 141–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhou D., Cantu C. 3rd, Sagiv Y., Schrantz N., Kulkarni A. B., Qi X., Mahuran D. J., Morales C. R., Grabowski G. A., Benlagha K., Savage P., Bendelac A., and Teyton L. (2004) Editing of CD1d-bound lipid antigens by endosomal lipid transfer proteins. Science 303, 523–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang J., Guillaume J., Pauwels N., Van Calenbergh S., Van Rhijn I., and Zajonc D. M. (2012) Crystal structures of bovine CD1d reveal altered αGalCer presentation and a restricted A′ pocket unable to bind long-chain glycolipids. PLoS One 7, e47989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zajonc D. M., Maricic I., Wu D., Halder R., Roy K., Wong C.-H., Kumar V., and Wilson I. A. (2005) Structural basis for CD1d presentation of a sulfatide derived from myelin and its implications for autoimmunity. J. Exp. Med. 202, 1517–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cox D., Fox L., Tian R., Bardet W., Skaley M., Mojsilovic D., Gumperz J., and Hildebrand W. (2009) Determination of cellular lipids bound to human CD1d molecules. PLoS One 4, e5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Paletta D., Fichtner A. S., Starick L., Porcelli S. A., Savage P. B., and Herrmann T. (2015) Species specific differences of CD1d oligomer loading in vitro. PLoS One 10, e0143449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fox L. M., Cox D. G., Lockridge J. L., Wang X., Chen X., Scharf L., Trott D. L., Ndonye R. M., Veerapen N., Besra G. S., Howell A. R., Cook M. E., Adams E. J., Hildebrand W. H., and Gumperz J. E. (2009) Recognition of lyso-phospholipids by human natural killer T lymphocytes. PLoS Biol. 7, e1000228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. DeLisi C., and Berzofsky J. A. (1985) T-cell antigenic sites tend to be amphipathic structures. Proc. Natl. Acad. Sci. U.S.A. 82, 7048–7052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cease K. B., Berkower I., York-Jolley J., and Berzofsky J. A. (1986) T cell clones specific for an amphipathic α-helical region of sperm whale myoglobin show differing fine specificities for synthetic peptides. A multiview/single structure interpretation of immunodominance. J. Exp. Med. 164, 1779–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Günther S., Schlundt A., Sticht J., Roske Y., Heinemann U., Wiesmüller K.-H., Jung G., Falk K., Rötzschke O., and Freund C. (2010) Bidirectional binding of invariant chain peptides to an MHC class II molecule. Proc. Natl. Acad. Sci. U.S.A. 107, 22219–22224 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.