Abstract
Rab GTPases are critical regulators of membrane trafficking. The canonical view is that Rabs are soluble in their inactive GDP-bound form, and only upon activation and conversion to their GTP-bound state are they anchored to membranes through membrane insertion of a C-terminal prenyl group. Here we demonstrate that C-terminal prenylation is not required for Rab13 to associate with and traffic on vesicles. Instead, inactive Rab13 appears to associate with vesicles via protein-protein interactions. Only following activation does Rab13 associate with the plasma membrane, presumably with insertion of the C-terminal prenyl group into the membrane.
Keywords: endosome, guanine nucleotide exchange factor (GEF), protein isoprenylation, Rab, vesicles, DENN domain, DENND2B, GDI, GDP dissociation inhibitor, TI-VAMP
Introduction
Rab GTPases control all aspects of membrane trafficking, ranging from vesicle formation and transport to tethering and fusion with the target membrane (1). The understanding of Rab regulation is thus critical for a full understanding of membrane trafficking. Furthermore, disturbances in Rab function are observed in several diseases. Notably, alterations in the levels or activity of Rab5, Rab35, Rab8, and Rab13 have been implicated in phenotypes associated with cancer (2–5).
As for all small GTPases, Rabs cycle between an active GTP-bound form and an inactive GDP-bound form. They are activated by guanine nucleotide exchange factors (GEFs)3 that facilitate the exchange of GDP for GTP, and they are inactivated by GTPase-activating proteins, which enhance Rab-mediated hydrolysis of GTP to GDP. Furthermore, Rabs are posttranslationally modified by prenylation; that is, the covalent attachment of either a Cys-15 (farnesyl) or Cys-20 (geranylgeranyl) isoprenoid to cysteine residues at the carboxyl terminus (6). When the Rab is active, it associates with membranes with insertion of the prenyl group into the lipid bilayer, whereas effector molecules bind to the switch region (7). This allows the Rab to coordinate the function of the effector protein to the membrane compartment where it is anchored. When the Rab is inactive, it loses affinity for the effector molecules and is extracted from membrane by GDP dissociation inhibitors (GDIs) (8, 9). GDIs bind the hydrophobic prenyl group to prevent its reassociation with the membrane and keep the Rab soluble in the cytosol. Consistently, deletion or mutation of the C-terminal cysteine residues results in complete cytosolic localization of both Rab4 and Rab5 (10).
Rabs are thought to associate with membranes only upon activation (11). However, it seems that at least some Rabs can associate with the membrane in their inactive GDP-bound form. For example, inactive Rab35 exists on the plasma membrane, whereas inactive Rab11 can be found on cytoplasmic vesicles (12, 13). Here we show that inactive Rab13, in the absence of prenylation, traffics on vesicles derived from recycling and late endosomal compartments. It appears that Rab13 associates with these vesicles as part of a protein complex.
Experimental Procedures
Antibodies, DNA Constructs, and shRNA
Mouse monoclonal antibodies used were as follows: Rab8 and flotillin (BD Biosciences), Rab9 (Abcam; Cambridge, MA), Na+/K+ ATPase (Upstate Biotechnology, Lake Placid, NY), and actin (Chemicon). Rabbit polyclonal antibodies used were as follows: Rab5 (Abcam) and tubulin (Cell Signaling Technology). The chicken polyclonal antibody against Rab13 was from Abcam. The rat monoclonal Hsc70 (1B5) antibody was obtained from Enzo Life Sciences. Homemade rabbit polyclonal antibodies against Rab35 and RME-8 were described previously (12, 14). mCherry (mCh)-tagged Rab13 (human amino acids 1–203) WT, T22N, and Q67L were described previously (5). The Rab13 C-terminal deletion construct (human, amino acids 1–199) and hypervariable domain deletion constructs (human, amino acids 1–175) were PCR-amplified, digested with EcoRI and BamHI, and ligated into the mCherry-C2 vector. Cellubrevin and TI-VAMP in pEGFP-C3 were described previously and were a gift from Dr. T. Galli (15, 16). Rab7 and Rab9 (canine) in pEGFP-C1 were a gift from F. Meunier (University of Queensland, Brisbane, Australia). Rab5 in pEGFP-C2 was described previously (12). Stable Rab13 knockdown in HEK-293T cells was achieved using a lentivirus to drive the expression of Rab13 shRNA1 (TRCN0000381528), shRNA2 (TRCN0000382005), shRNA3 (TRCN0000343510), or a control vector of pLKO-TRC005 obtained from the Mission@TRC genome-wide shRNA collection (Sigma) as described previously (5). The primers used to generate the constructs and shRNAs can be found in supplemental Table S1. All constructs were verified by sequence analysis.
Subcellular Fractionation
Various rat tissues or cultured cells were homogenized in HEPES buffer (20 mm HEPES (pH 7.4) containing protease inhibitors (0.83 mm benzamidine, 0.23 mm phenylmethylsulfonyl fluoride, 0.5 μg/ml aprotinin, and 0.5 μg/ml leupeptin)) and centrifuged at 800 × g for 10 min at 4 °C to remove cell debris. The supernatant was centrifuged at 200,000 × g for 30 min, yielding the supernatant (S1) and pellet (P1) fractions. P1 was subsequently resuspended in HEPES buffer with or without NaCl, KCl, EDTA, or detergents (1% Triton X-100, 1% Nonidet P-40, 25 μg/ml digitonin or 1% SDS, and 0.5% deoxycholate) or in 50 mm NaCO3 at pH 11.0. Following incubation for 15 min, the samples were centrifuged at 200,000 × g for 30 min, yielding the S2 and P2 fractions. Equal protein aliquots of the fractions were analyzed by SDS-PAGE and Western blotting. The blots were analyzed using ImageJ 1.43m (National Institutes of Health).
Rab GDI Extraction
HEK-293T cells were transfected with His-myc-Rab GDI. At 18 h post-transfection, cells were lysed in binding buffer (50 mm HEPES (pH 8.0), 300 mm NaCl, 20 mm imidazole, 1% Triton X-100, and protease inhibitors), centrifuged at 200,000 × g for 15 min at 4 °C, and incubated with nickel-nitrilotriacetic acid-agarose beads (Qiagen) with rocking for 2 h at 4 °C. The beads were washed once in binding buffer and twice in wash buffer (50 mm HEPES (pH 6.5), 300 mm NaCl, 20 mm imidazole, and protease inhibitors) and eluted from beads in elution buffer (50 mm HEPES (pH 6.5), 300 mm NaCl, 200 mm NaCl, and protease inhibitors). Purified His-myc-Rab GDI was concentrated, and the buffer was exchanged into HEPES buffer using an Amicon® Ultra 10K centrifugal filter (Millipore). For GDI extraction, HEK-293T cells were treated as described above, and P1 was resuspended in HEPES buffer containing increasing concentrations of purified His-myc-Rab GDI, incubated for 30 min at 37 °C, and centrifuged at 200,000 × g for 30 min at 4 °C, yielding the S2 and P2 fractions. Equal protein aliquots of the fractions were analyzed by SDS-PAGE and Western blotting.
Discontinuous Sucrose Gradient
Sucrose gradients were performed as described in Ref. 17. In brief, HEK-293T cells were washed twice in PBS and lysed in HNEX buffer (20 mm HEPES, 150 mm NaCl, 5 mm EDTA, 10% Triton X-100, and protease inhibitors), sonicated, incubated on ice for 10 min, and spun at 800 × g for 10 min at 4 °C to remove cell debris. Sucrose solutions were prepared in HNE buffer (20 mm HEPES, 150 mm NaCl, and 5 mm EDTA). The cell extract was adjusted to 40% sucrose. 3 ml of 5% sucrose solution was underlaid with 6 ml of 30% sucrose solution followed by 4 ml of cell extract. Samples were centrifuged at 230,000 × g for 16 h at 4 °C, and 1-ml fractions were collected and analyzed by SDS-PAGE and Western blotting.
Immunofluorescence and Live Cell Imaging
For live-cell imaging, MCF10A cells were plated on poly-l-lysine-coated, 35-mm, no. 1.5 glass-bottom dishes (MatTek Corp.). Cells were transfected using JetPrime reagent and incubated for 14 h. PC12 cells were differentiated in reduced serum medium containing 50 ng/ml NGF for 36 h, transfected using JetPrime reagent, and incubated for 14 h. For internalization of transferrin, transfected cells were serum-starved for 1 h and incubated in serum-free medium containing 25 μg/ml transferrin-Alexa Fluor 647 for 30 min. Cells were washed and imaged in complete medium. Live-cell imaging was performed using an AxioObserver Z1 (Zeiss) microscope equipped with a Plan-Aprochromat ×40 oil objective (numerical aperture, 1.4), a Definite Focus system, and an AxioCam MR3 camera (Zeiss). Cells were kept at 37 °C in 5% CO2 using Incubation System S (Pecon, Germany). GFP and mCherry were imaged using 470- and 591-nm LEDs respectively, from a Colibri.2 illumination source (Zeiss). Acquisition and analyses were performed using ZEN 11.0 software (Zeiss), whereas movies were made using ImageJ 1.43m. For co-localization analysis, MCF10A cells were plated on poly-l-lysine-coated coverglass, transfected using JetPrime reagent, and incubated for 14 h. Cells were subsequently washed in PBS at 37 °C and fixed for 10 min in 3% paraformaldehyde at 37 °C. Cells were mounted using Dako mounting medium (Agilent Technologies), and imaging was performed using a 710 laser-scanning confocal microscope equipped with a Plan-Aprochromat ×63 oil objective (numerical aperture, 1.4) (Zeiss). Acquisition was performed using ZEN 11.0 software (Zeiss), and the percentage of co-localization was determined using the co-localization analysis plugin on ImageJ.
Statistical Analysis
Statistics were analyzed using SPSS version 17. Mean ± S.D. was used to determine significant differences between pairs. Comparisons of groups were performed using Student's t test or one-way analysis of variance and Dunnett post-test for multiple comparisons (*, p < 0.05; **, p < 0.01; ***, p < 0.001).
Results and Discussion
Inactive Rab13 Traffics on Vesicles
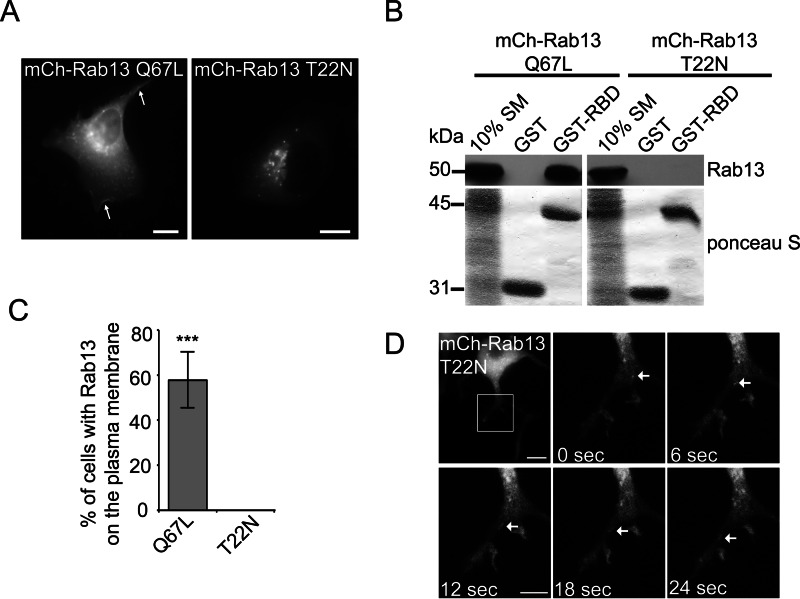
It is generally assumed that Rabs are soluble in their inactive GDP-bound form and that only upon activation and conversion to the GTP-bound form do they associate with membranes. However, as we have described previously (5), both constitutively active (Q67L) and constitutively inactive (T22N) mutant forms of Rab13 are present on vesicles in MCF10A epithelial cells (Fig. 1A). In fact, Rab13 T22N traffics on dynamic vesicles into and out of the perinuclear region, similar to Rab13 Q67L (5). The mutant Rabs in these experiments are commonly used to study GTPase function. The active Q67L mutation abolishes GTP hydrolysis, rendering the Rab GTP-locked, whereas the inactive T22N mutation disrupts Mg2+ binding, thereby reducing the affinity for GTP by 100-fold without affecting the affinity for GDP, resulting in a GDP-locked Rab (18, 19). However, in some cases, dominant-negative mutations may not completely inactivate the protein but, rather, slow its activation. This is the case in Drosophila, where Rab7 T22N overexpression can partially rescue Rab7-null mutants, showing that Rab7 T22N retains some of its wild-type function (20). Therefore, we sought to examine whether the mCh-Rab13 T22N mutant used here retains wild-type function by performing pulldown assays using the Rab-binding domain of the Rab13 effector MICAL-L1 (21). Although mCh-Rab13 Q67L binds strongly to the effector domain, mCh-Rab13 T22N is unable to bind (Fig. 1B). This is consistent with previous studies showing that expression of Rab13 T22N in functional assays acts in a dominant-negative fashion (22). Thus, Rab13 T22N is indeed in its inactive form as it traffics on vesicles.
FIGURE 1.
Inactive Rab13 traffics on vesicles. A, MCF10A cells expressing mCh-Rab13 Q67L or mCh-Rab13 T22N were imaged live. The arrows illustrate localization on the plasma membrane. Scale bars = 10 μm. B, the GST-Rab-binding domain (RBD) or GST alone, bound to glutathione-Sepharose, was incubated with HEK-293T cell lysates expressing mCh-Rab13 Q67L or T22N. Ponceau S staining revealed the level of GST-Rab-binding domain, whereas specifically bound Rab13 was detected by blot. Starting material (SM) equals 10% of the lysate used per condition. C, the percentage of transfected cells with mCh-Rab13 constructs on the plasma membrane. Data are mean ± S.D., measuring a minimum of 20 cells/experiment from a minimum of three independent experiments (Student's t test; ***, p < 0.001). D, PC12 cells differentiated for 24 h by addition of 50 ng/ml NGF and expressing mCh-Rab13 T22N were imaged live. The boxed region is magnified. The arrows follow a vesicle trafficking along a neurite. Scale bar = 10 μm (top panel) and 5 μm (magnified panel).
Interestingly, although both Rab13 mutants are found on cytoplasmic vesicles, only mCh-Rab13-Q67L is found on the plasma membrane (Fig. 1, A and C). Previously, we determined that Rab13 is activated specifically at the plasma membrane, where its GEFs DENND1C and DENND2B are localized (5, 23). This suggests that activation by its GEF is required for association of Rab13 with the plasma membrane. This is consistent with the notion that GEFs play an important role in targeting Rabs to specific membranes (24). For example, knockdown of the GEF for Rab32, BLOC-3, prevents the targeting of Rab32 to ring-like membrane structures in melanosomes (25). Interestingly, Rab32 could still be found on small puncta following BLOC-3 knockdown. Thus, BLOC-3 functions to target Rab32 to specific membrane compartments. The same appears to hold true for Rab13 because only the active form is found on the plasma membrane.
Rab13 has been predominantly studied in epithelial cells because Rab13 activity regulates tight junction assembly and stimulates cell invasion and migration (5, 26–28). However, Rab13 is also required for NGF-induced neurite outgrowth in PC12 cells and dorsal root ganglion neurons (22, 29). We thus sought to determine whether trafficking of inactive Rab13 on vesicles is a characteristic of cell types other than MCF10A. mCh-Rab13 T22N traffics on vesicles in neurites of differentiated PC12 cells (Fig. 1D). Therefore, the targeting and trafficking of inactive Rab13 on vesicles is likely to be characteristic of multiple cells types.
Inactive Rab13 Traffics on Vesicles Derived from Multiple Endosomal Compartments
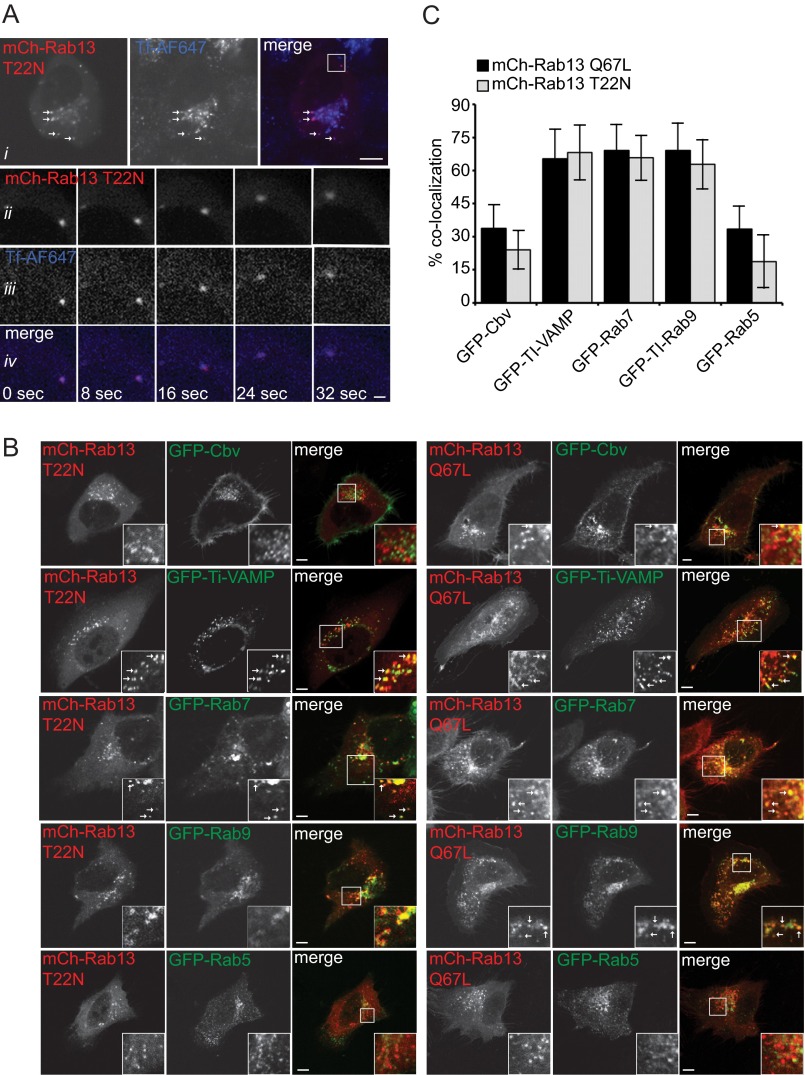
Rab13 controls the delivery of cargo that traffics through recycling endosomes to the plasma membrane (30). Consistently, Rab13 is localized to the trans-Golgi network, recycling endosomes and the plasma membrane (5, 30). To confirm that the vesicles carrying inactive Rab13 are functional, we tested for co-trafficking with internalized transferrin, a major recycling cargo that passes through recycling endosomes and has been shown previously to co-localize with wild-type Rab13 (30, 31). mCh-Rab13 T22N co-localizes with internalized transferrin (Fig. 2A, i) and traffics on vesicles that contain the internalized ligand (Fig 2A, ii–iv). Thus, inactive Rab13 is present on functional vesicles, likely derived from recycling endosomes.
FIGURE 2.
Rab13 is found on vesicles with multiple endosomal origins. A, MCF10A cells expressing mCh-Rab13 T22N and internalized Alexa Fluor (AF) 647-labeled transferrin were imaged live (i). The arrows illustrate co-localization. The boxed region from i is magnified below over a time-lapse imaging series (ii–iv). Scale bars = 10 μm (top panel) and 1 μm (magnified panels). B, MCF10A cells expressing mCh-Rab13 T22N or mCh-Rab13 Q67L co-expressed with GFP-cellubrevin (Cbv), GFP-TI-VAMP, GFP-Rab7, GFP-Rab9, or GFP-Rab5 were fixed and imaged. The boxed region in each image is magnified in the insets. The arrows illustrate co-localization. Scale bars = 10 μm. C, quantification of percent co-localization as determined in experiments such as those in B. Data are mean ± S.D., measuring a minimum of 8 cells/experiment from a minimum of two independent experiments.
Because several trafficking routes exist to recycle cargo back to the plasma membrane, we sought to further define the identity of vesicles carrying Rab13. Exocytosis of transferrin-containing vesicles is partially regulated by the tetanus toxin-sensitive v-SNARE cellubrevin (16). However, there was limited co-localization of Rab13 (inactive or active) with cellubrevin (Fig. 2, B and C). Instead, we discovered that a large fraction of both mutants of Rab13 co-localize with tetanus-insensitive TI-VAMP (VAMP7) (Fig. 2, B and C). This result was particularly interesting given that TI-VAMP regulates many of the same physiological functions as Rab13, including plasma membrane delivery of GLUT4, neurite outgrowth, and cell migration (32). Although TI-VAMP can be found at the plasma membrane and trans-Golgi network, where Rab13 has been localized, TI-VAMP is predominantly found on late endosomal compartments (32). Intriguingly, wild-type Rab13 also localizes in part to Rab7-positive late endosomes (21). In fact, we found that both active and inactive Rab13 constructs co-localize with the late endosomal markers Rab7 and Rab9 with much less co-localization than with the early endosomal marker Rab5 (Fig. 2, B and C). Together, our data suggest that both active and inactive Rab13 traffic on two distinct populations of vesicles: those carrying transferrin receptor from recycling endosomes and TI-VAMP/Rab7-positive vesicles derived from late endosomes. However, it remains unknown how inactive Rab13 associates with these vesicles.
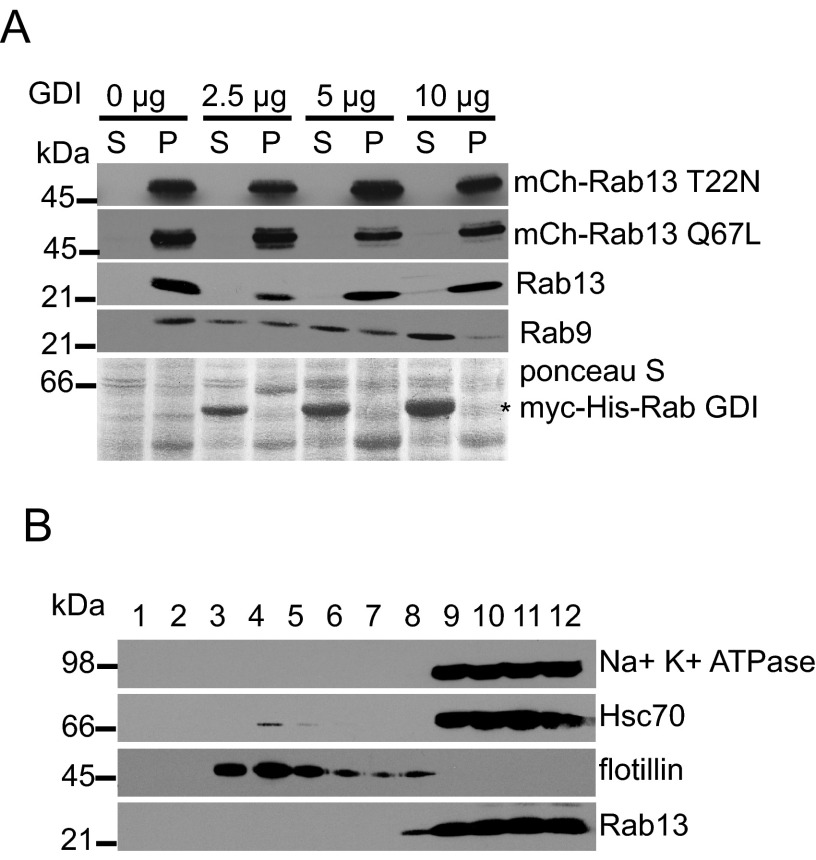
Inactive Rab13 Resists Membrane Extraction by Rab GDI
We next sought to characterize the association of inactive Rab13 on endosome-derived vesicles. Classically, following inactivation, Rabs are solubilized from the membrane by GDI. GDI binds to prenylated Rabs in their inactive GDP-bound form, extracts them from the membrane, and holds them soluble in the cytosol until reactivation (33). However, endogenous Rab13 resists extraction from membranes by GDI (34). Here we show that, in addition to endogenous Rab13, both mCh-Rab13 Q67L and mCh-Rab13 T22N resist extraction from membranes by purified GDI, whereas Rab9 shows a clear dose-dependent extraction (Fig. 3A). Although phosphodiesterase-δ can partially solubilize Rab13 from the particulate fraction (34), GDIs bind Rabs with little to no specificity (35). We therefore reasoned that some mechanism must be in place to prevent the solubilization of Rab13 by GDI. One possibility is that Rab13 traffics on lipid rafts. Lipid rafts are stable microdomains within membranes that are enriched in cholesterol and sphingolipids (36). Elevated levels of cholesterol on these membrane domains reduce the ability of GDI to extract certain Rabs from membranes (37, 38). Because lipid raft markers cycle between the trans-Golgi network and the plasma membrane, where Rab13 also functions, we tested whether Rab13 was present on lipid rafts (39). Using sucrose gradients, we found that Rab13 from HEK-293T cell lysates does not co-fractionate (float) with detergent-resistant membranes, revealed by the raft marker flotillin (Fig. 3B). Although we cannot exclude that Rab13 associates with lipid rafts in other cell types, it does not in HEK-293T cells, and therefore another mechanism must exist to mediate the resistance to GDI extraction.
FIGURE 3.
Rab13 resists membrane extraction by GDI. A, HEK-293T cell homogenates were spun for 30 min at 200,000 × g, and the resulting pellet was resuspended in HEPES buffer and incubated without or with increasing concentrations of purified myc-His-GDI as indicated. After 30 min of incubation at 37 °C, samples were spun for 30 min at 200,000 × g, and the resulting supernatant (S) and pellet (P) fractions were analyzed by Western blotting using the indicated antibodies. The Ponceau S-stained transfer revealed the presence of the purified GDI. B, HEK-293T cells were lysed in buffer containing 1% Triton X-100. Cell lysates were placed at the bottom of a discontinuous sucrose gradient and centrifuged at 230,000 × g for 16 h at 4 °C. Twelve fractions of 1 ml each were collected with fraction 1 from the top and fraction 12 from the bottom of the gradient. The fractions were analyzed by Western blotting with the indicated antibodies.
C-terminal Prenylation Is Not Required for the Targeting of Inactive Rab13 to Vesicles
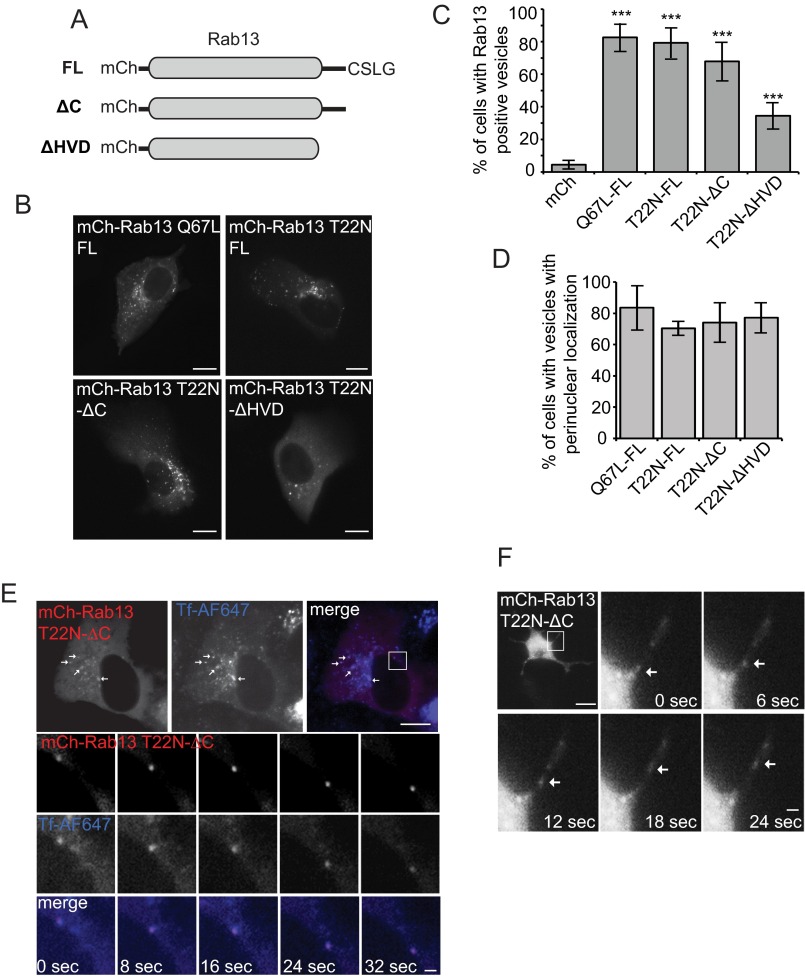
Because GDIs bind to the hydrophobic prenyl group on Rabs, and Rab13 resists extraction by GDI, we tested whether Rab13 requires C-terminal prenylation to interact with cytoplasmic vesicles. Rab13 is geranylgeranylated on the cysteine residue of its CAAX motif found at the extreme C terminus (40). Therefore, we deleted the last four amino acids of the protein containing the prenylation site (Rab13ΔC) (Fig. 4A). Interestingly, inactive mCh-Rab13 T22N-ΔC remains associated with vesicles (Fig. 4, B and C). These vesicles retain the proper perinuclear localization and have no obvious trafficking defects (Fig. 4, B and D, and supplemental Movie 1). Again, we verified that these vesicles were functional as mCh-Rab13 T22N-ΔC co-traffics with internalized transferrin (Fig. 4E). Thus, inactive Rab13 is present on vesicles carrying recycling cargo even in the absence of prenylation. Furthermore, we found that mCh-Rab13 T22N-ΔC traffics on vesicles in neurites of differentiated PC12 cells, showing that this is a general feature of Rab13 and not cell type-specific (Fig. 4F). These data indicate that inactive Rab13 is recruited to vesicles in a prenylation-independent manner.
FIGURE 4.
Inactive Rab13 traffics on vesicles following deletion of the C-terminal prenylation motif. A, schematic of the constructs used in the figure. FL, full-length; ΔC, C-terminal deletion of the last four amino acids CSLG (Cys-Ser-Leu-Gly); ΔHVD, hypervariable domain deletion. B, MCF10A cells expressing mCh-Rab13 Q67L FL, T22N FL, or T22N deletion constructs, as indicated, were imaged live. Scale bars = 10 μm. C, the percentage of transfected cells with mCh-Rab13 constructs on vesicles. Data are mean ± S.D., measuring a minimum of 20 cells/experiment from a minimum of three independent experiments. ***, p < 0.001. D, percentage of cells from experiments as in B with mCh-Rab13 on vesicles that exhibit perinuclear localization. Data are mean ± S.D., measuring a minimum of 20 cells/experiment from a minimum of three independent experiments. E, MCF10A cells expressing mCh-Rab13 T22N ΔC and internalized Alexa Fluor (AF) 647-labeled transferrin were imaged live. The arrows illustrate co-localization. The boxed region is magnified below over a time-lapse imaging series. Scale bars = 10 μm (top panel) and 1 μm (magnified panels). F, PC12 cells differentiated for 24 h with 50 ng/ml NGF and expressing mCh-Rab13 T22N-ΔC were imaged live. The boxed region is magnified. The arrows follow vesicle trafficking along a neurite. Scale bars = 10 μm (top panel) and 2.5 μm (magnified panel).
Several studies have indicated that the hypervariable domain (HVD) of Rabs contributes to membrane targeting (41, 42). The HVD is an unstructured region directly adjacent to the prenylation motif and is the most divergent region of Rabs. The HVD is required for membrane targeting of several Rabs, including Rab7 and Rab35, but is not required for membrane targeting of Rab1 or Rab5 (42, 43). Deletion of both the HVD and C terminus of inactive Rab13 (Fig. 4A) reduced the number of cells containing Rab13-positive vesicles but did not lead to a complete redistribution to the cytosol (Fig. 4, B and C, and supplemental Movie 2). Thus, inactive Rab13 can be recruited to vesicles in the absence of its HVD and prenylation.
Endogenous Rab13 Associates with Membranes through Protein-Protein Interactions
The retention of inactive Rab13 on vesicles despite deletion of the HVD and the C-terminal prenylation site suggests that interactions within the switch regions are important for vesicle association. Proteins that interact preferentially with the GDP-bound form of Rab13 could mediate this vesicle association. Although GEFs and GDIs are thought to be the predominant binding partners for GDP-bound Rabs, other interacting proteins have been described. For instance, the GRAM domain of myotubularin-related protein 6 (MTMR6) interacts preferentially with GDP-bound Rab1b (44). Furthermore, protrudin interacts preferentially with GDP-bound Rab11 (45). In fact, GDP-bound Rab11 binds protrudin and KIF5 to facilitate plus end microtubule transport, whereas GTP-bound Rab11 that binds FIP3 and dynein to facilitate minus end microtubule transport, suggesting that GTP/GDP cycling of Rabs is not as simple as an on/off switch (45, 46).
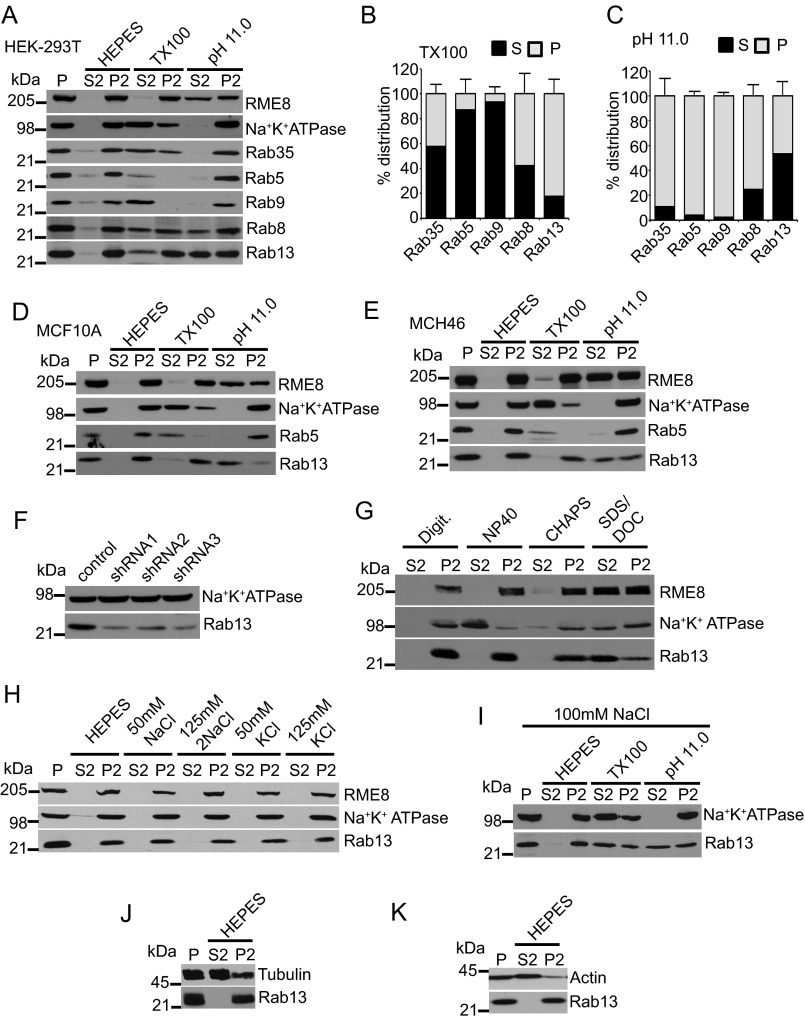
Thus, we speculate that inactive Rab13 associates with vesicle membranes through interactions with proteins. We therefore used subcellular fractionation and classic membrane extraction protocols to examine the association of endogenous Rab13 with membranes. Ultraspeed centrifugation was employed to separate HEK-293 cell lysates into particulate (P) and cytosolic (S) fractions. The P fraction was resuspended in various buffers, and a second high-speed centrifugation yielded S2 and P2 fractions. We first used Triton X-100 in the absence of salt, which will solubilize integral membrane proteins such as the Na+K+ATPase as well as proteins anchored to membranes through insertion of prenyl groups, such as Rab5 and Rab9 (Fig. 5, A and B) (14). Consistent with our transfection studies, endogenous Rab13 largely resisted extraction under this condition (Fig. 5, A and B). We next utilized carbonate buffer at pH 11.0, which disrupts most protein-protein complexes and solubilizes large scaffolding proteins that are anchored to membranes through protein-protein interactions, such as RME-8, but leaves membranes intact and will thus not alter the pelleting of prenylated proteins such as Rab9 or integral membrane proteins such as Na+K+ATPase (Fig. 5, A and C) (14). This condition leads to the extraction of a significant percentage of Rab13, whereas Rab35, Rab5, and Rab9 remain entirely in the P2 fraction (Fig. 5, A and C). In the epithelial cell lines MCF10A and MCH46, we also found that the majority of Rab13 extracts at pH 11.0 and not in Triton X-100, demonstrating that the observed results are a general characteristic of Rab13 and are not cell type-specific (Fig. 5, D and E). To confirm that the Rab13 antibody is indeed targeting endogenous Rab13, we performed knockdown of Rab13 using three different shRNAs and observed a large reduction of the immunoreactive band (Fig. 5F). Furthermore, Rab13 resists extraction in multiple non-ionic detergents in the absence of salt but is extracted in a mixture of the anionic/denaturing detergents sodium deoxycholate and sodium dodecyl sulfate, similar to the extrinsic membrane protein RME-8 (Fig. 5G) (14). Rab13 also resists extraction with physiological concentrations of salt and only partially extracts in 100 mm NaCl containing Triton X-100 (Fig. 5, H and I). Thus, Rab13 behaves as a peripheral membrane protein in cell lines.
FIGURE 5.
Rab13 associates with membranes via protein-protein interactions in cells. A, HEK-293T cell homogenates in HEPES buffer were spun for 30 min at 200,000 × g, and equal protein aliquots of the resulting pellet (P) were resuspended in ice-cold HEPES buffer with or without 1% Triton X-100 (TX100) or NaCO3 at pH 11. 0. After 15 min of incubation, samples were spun for 30 min at 200,000 × g, and the resulting supernatant (S2) and pellet (P2) fractions were analyzed by Western blotting using the indicated antibodies. B and C, quantification of the distribution of Rab proteins resuspended in Triton X-100 (B) and at pH 11.0 (C). Data are mean ± S.D. (n = 7 for Rab13, n = 5 for Rab5, n = 4 for Rab8 and Rab35, and n = 3 for Rab9). D--E, lysates of MCF10A cells (D) and MCH46 cells (E) were processed and analyzed as in A. F, HEK-293T cells were stably transduced with a lentivirus driving the expression of control shRNA or three different shRNAs targeting Rab13, and the indicated proteins were detected by blotting. G, HEK-293T cell homogenates in HEPES buffer were spun for 30 min at 200,000 × g, and equal protein aliquots of the resulting pellet were resuspended in ice-cold HEPES buffer with digitonin (Digit.), Nonidet P-40, CHAPS, or SDS/deoxycholate (DOC). After 15 min of incubation, samples were spun for 30 min at 200,000 × g, and the resulting supernatant and pellet fractions were analyzed by Western blotting using the indicated antibodies. H, HEK-293T cell homogenates in HEPES buffer were spun for 30 min at 200,000 × g, and equal protein aliquots of the resulting pellet were resuspended in ice-cold HEPES buffer with or without NaCl or KCl at the indicated concentrations. After 15 min of incubation, the samples were spun for 30 min at 200,000 × g, and the resulting supernatant and pellets were analyzed by Western blotting using the indicated antibodies. I, HEK-293T cell homogenates in HEPES buffer were spun for 30 min at 200,000 × g, and equal protein aliquots of the resulting pellet were resuspended in ice-cold HEPES buffer containing 100 mm NaCl with or without 1% Triton X-100 or NaCO3 at pH 11.0. After 15 min of incubation, samples were spun for 30 min at 200,000 × g, and the resulting supernatant and pellet fractions were analyzed by Western blotting using the indicated antibodies. J and K, HEK-293T cell homogenates in HEPES buffer were spun for 30 min at 200,000 × g, and equal protein aliquots of the resulting pellet were resuspended in ice-cold HEPES buffer. After 15 min of incubation, samples were spun for 30 min at 200,000 × g, and the resulting supernatant and pellets were analyzed by Western blotting using the indicated antibodies.
Some Rabs may pellet because of interactions with the cytoskeleton. For example, Rab24 distributes to the particulate fraction because of interactions with microtubules (47). Because Rab13 interacts with actin-binding proteins (5), we wondered whether the extraction behavior of Rab13 in our biochemical experiments could be attributed to cytoskeletal associations. Resuspension of the particulate fraction in ice-cold HEPEs leads to spontaneous disassembly of the cytoskeleton and redistribution of the majority of both actin and tubulin into the supernatant following a second high-speed centrifugation (Fig. 5, J and K). However, Rab13 remains entirely in the pellet under these conditions (Fig. 5, J and K). Because Rab13 does not solubilize along with the cytoskeleton, this suggests that protein-protein interactions other than those with the cytoskeleton are responsible for the extraction behavior of Rab13.
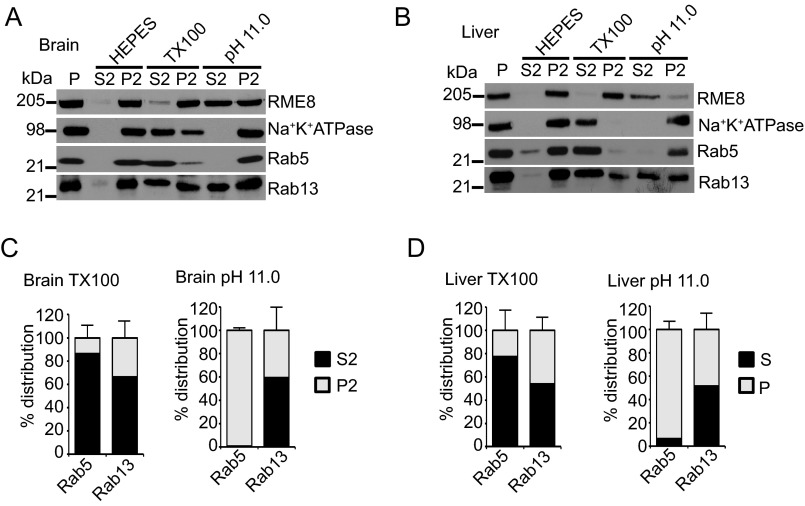
Next, we wondered whether this phenotype was specific to cultured cells or could also be observed with endogenous Rab13 from tissue. In fact, Rab13 extracts from the particulate fraction at pH 11.0 and only partially extracts in Triton X-100 in both rat brain (Fig. 6, A and C) and liver (Fig. 6, B and D). Consistent with our cell line data, Rab5 extracted only with Triton X-100 and not at pH 11.0 (Fig 6, A–D). Thus, our data suggest that a pool Rab13 is likely stabilized on membranes through protein-protein interactions and not through insertion of a hydrophobic prenyl group.
FIGURE 6.
Rab13 associates with membranes via protein-protein interactions in tissue. A and B, equal protein aliquots of rat brain (A) and rat liver (B) homogenate in HEPES buffer were spun for 30 min at 200,000 × g, and the resulting pellet fraction (P) was resuspended in ice-cold HEPES buffer with or without 1% Triton X-100 (TX100) or NaCO3 at pH 11.0. After 15 min of incubation, the samples were spun for 30 min at 200,000 × g, and the resulting supernatant (S2) and pellets (P2) were analyzed by Western blotting using the indicated antibodies. C, quantification of the distribution of Rab proteins from extraction experiments as in A. Data are mean ± S.D. (n = 3 for Rab13 and Rab5). D, quantification of the distribution of Rab proteins from extraction experiments as in B. Data are mean ± S.D. (n = 5 for Rab5 and n = 3 Rab13).
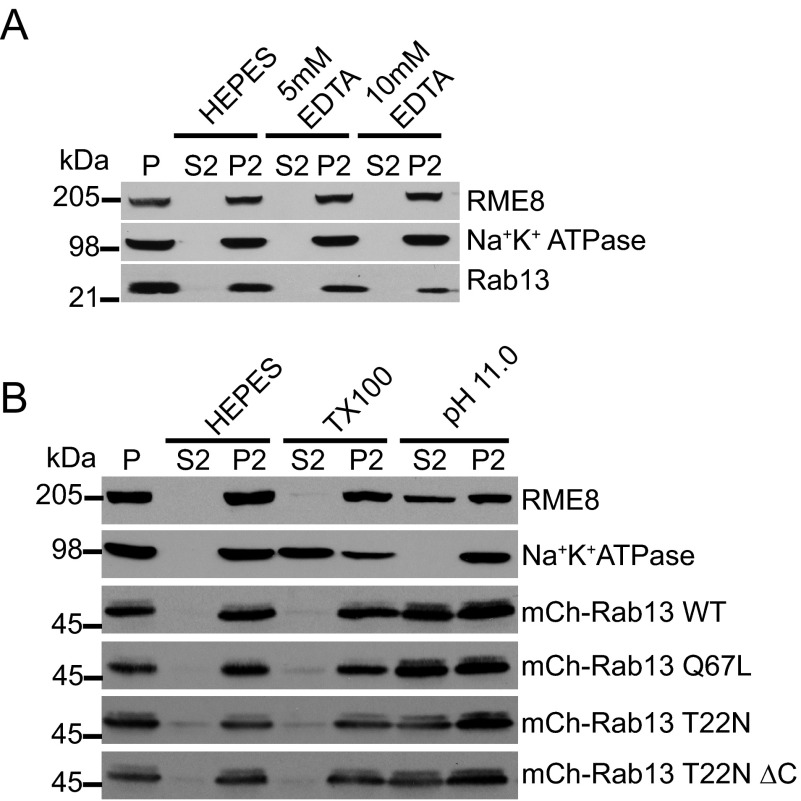
We also wondered whether the nucleotide status of Rab13 affects its ability to associate with protein complexes. We repeated our ultraspeed centrifugation of HEK-293 cell lysates and found that Rab13 resists extraction with the addition of EDTA to render the protein nucleotide free (Fig. 7A). Furthermore, we found that mCh-Rab13 WT, mCh-Rab13 Q67L, mCh-Rab13 T22N, and mCh-Rab13 T22N-ΔC all resist extraction with Triton X-100 and largely extract at pH 11.0 (Fig. 7A). Therefore, Rab13 associates with membranes likely through protein-protein interactions independent of its nucleotide status or prenylation.
FIGURE 7.
Rab13 associates in a protein complex independent of nucleotide status. A, HEK-293T cell homogenates in HEPES buffer were spun for 30 min at 200,000 × g, and equal protein aliquots of the resulting pellet (P) were resuspended in ice-cold HEPES buffer with or without EDTA at the indicated concentrations. After 15 min of incubation, the samples were spun for 30 min at 200,000 × g, and the resulting supernatant (S2) and pellets (P2) were analyzed by Western blotting using the indicated antibodies. B, HEK-293T cells were left untransfected or were transfected with various mCh-Rab13 constructs as indicated. Cell homogenates in HEPES buffer were spun for 30 min at 200,000 × g, and equal protein aliquots of the resulting pellet were resuspended in ice-cold HEPES buffer with or without 1% Triton X-100 (TX100) or NaCO3 at pH 11.0. After 15 min of incubation, the samples were spun for 30 min at 200,000 × g, and the resulting supernatant (S2) and pellets (P2) were analyzed by Western blotting using the indicated antibodies for the first and second panels and an antibody against Rab13 to detect the various Rab13 constructs.
Conclusion
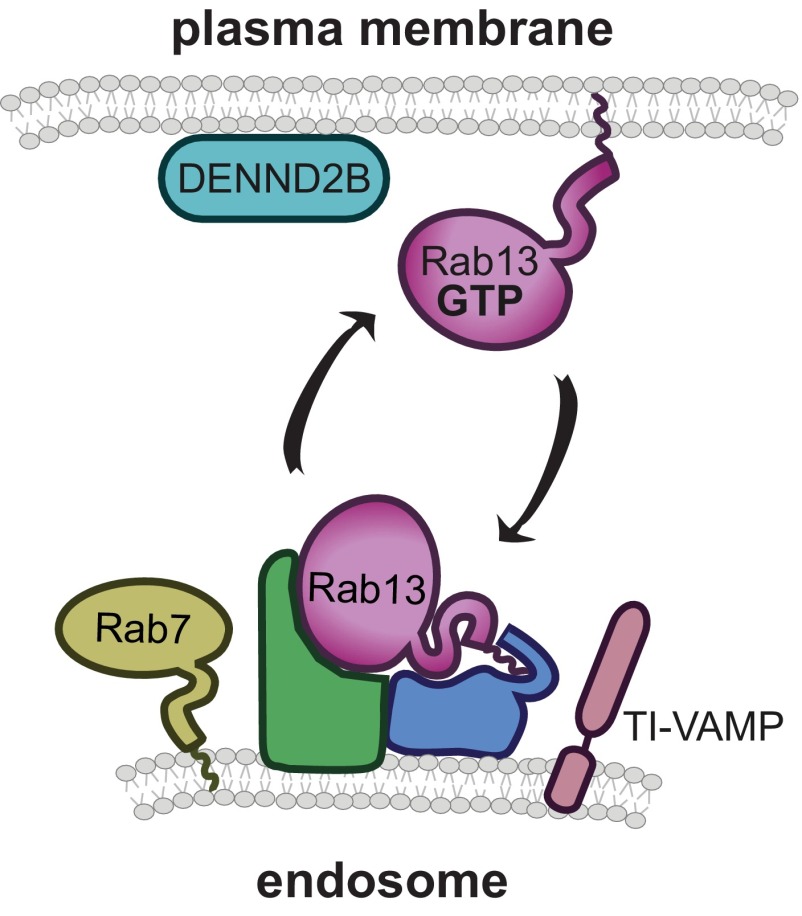
Here we show that Rab13 traffics on vesicles derived from both recycling and late endosomal compartments in both its inactive and active form, whereas only active Rab13 is found on the plasma membrane. We discovered that inactive Rab13 does not require C-terminal prenylation to traffic on these vesicles. Instead, Rab13 exists on vesicles likely as part of a large protein complex (Fig. 8). It will be interesting in the future to investigate whether other Rabs can associate with membranes independent of prenylation. With roughly 70 mammalian Rabs, the largest family of small GTPases, this could represent a conserved mechanism used by a subset of Rabs to regulate membrane trafficking.
FIGURE 8.
Model of Rab13 trafficking. Rab13 is targeted to and traffics on vesicles derived from late endosomes (depicted here) or recycling endosomes as part of a protein complex. When Rab13-positive vesicles reach the plasma membrane, Rab13 is activated locally by its GEF DENND2B, allowing active Rab13 to anchor to the plasma membrane via its C-terminal hydrophobic prenyl group.
Author Contributions
M. S. I. conceived and performed experiments and wrote the manuscript. M. G. performed experiments. P. S. M. conceived the experiments and wrote the manuscript.
Supplementary Material
Acknowledgments
We thank Jacynthe Philie for technical assistance and Dr. Martin Schmeing, Dr. Andrea L. Marat, and all members of the McPherson laboratory for helpful discussions regarding the project.
This work was supported by Canadian Institutes of Health Research Grant MOP-62684 (to P. S. M.). The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Table S1 and Movies S1 and S2.
- GEF
- guanine nucleotide exchange factor
- GDI
- GDP dissociation inhibitor
- mCh
- mCherry
- HVD
- hypervariable domain
- TI-VAMP
- tetanus-insensitive vesicle-associated membrane protein.
References
- 1. Stenmark H. (2009) Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 10, 513–525 [DOI] [PubMed] [Google Scholar]
- 2. Yang P. S., Yin P. H., Tseng L. M., Yang C. H., Hsu C. Y., Lee M. Y., Horng C. F., and Chi C. W. (2011) Rab5A is associated with axillary lymph node metastasis in breast cancer patients. Cancer Sci. 102, 2172–2178 [DOI] [PubMed] [Google Scholar]
- 3. Allaire P. D., Seyed Sadr M., Chaineau M., Seyed Sadr E., Konefal S., Fotouhi M., Maret D., Ritter B., Del Maestro R. F., and McPherson P. S. (2013) Interplay between Rab35 and Arf6 controls cargo recycling to coordinate cell adhesion and migration. J. Cell Sci. 126, 722–731 [DOI] [PubMed] [Google Scholar]
- 4. Bravo-Cordero J. J., Marrero-Diaz R., Megías D., Genís L., García-Grande A., García M. A., Arroyo A. G., and Montoya M. C. (2007) MT1-MMP proinvasive activity is regulated by a novel Rab8-dependent exocytic pathway. EMBO J. 26, 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ioannou M. S., Bell E. S., Girard M., Chaineau M., Hamlin J. N., Daubaras M., Monast A., Park M., Hodgson L., and McPherson P. S. (2015) DENND2B activates Rab13 at the leading edge of migrating cells and promotes metastatic behavior. J. Cell Biol. 208, 629–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pereira-Leal J. B., Hume A. N., and Seabra M. C. (2001) Prenylation of Rab GTPases: molecular mechanisms and involvement in genetic disease. FEBS Lett. 498, 197–200 [DOI] [PubMed] [Google Scholar]
- 7. Stenmark H., and Olkkonen V. M. (2001) The Rab GTPase family. Genome Biol. 2, REVIEWS3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Soldati T., Riederer M. A., and Pfeffer S. R. (1993) Rab GDI: a solubilizing and recycling factor for rab9 protein. Mol. Biol. Cell 4, 425–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pfeffer S. R., Dirac-Svejstrup A. B., and Soldati T. (1995) Rab GDP dissociation inhibitor: putting rab GTPases in the right place. J. Biol. Chem. 270, 17057–17059 [DOI] [PubMed] [Google Scholar]
- 10. Gomes A. Q., Ali B. R., Ramalho J. S., Godfrey R. F., Barral D. C., Hume A. N., and Seabra M. C. (2003) Membrane targeting of Rab GTPases is influenced by the prenylation motif. Mol. Biol. Cell 14, 1882–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Goody R. S., Rak A., and Alexandrov K. (2005) The structural and mechanistic basis for recycling of Rab proteins between membrane compartments. Cell. Mol. Life Sci. 62, 1657–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Allaire P. D., Marat A. L., Dall'Armi C., Di Paolo G., McPherson P. S., and Ritter B. (2010) The Connecdenn DENN domain: a GEF for Rab35 mediating cargo-specific exit from early endosomes. Mol. Cell 37, 370–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Savina A., Vidal M., and Colombo M. I. (2002) The exosome pathway in K562 cells is regulated by Rab11. J. Cell Sci. 115, 2505–2515 [DOI] [PubMed] [Google Scholar]
- 14. Girard M., Poupon V., Blondeau F., and McPherson P. S. (2005) The DnaJ-domain protein RME-8 functions in endosomal trafficking. J. Biol. Chem. 280, 40135–40143 [DOI] [PubMed] [Google Scholar]
- 15. Martinez-Arca S., Alberts P., Zahraoui A., Louvard D., and Galli T. (2000) Role of tetanus neurotoxin insensitive vesicle-associated membrane protein (TI-VAMP) in vesicular transport mediating neurite outgrowth. J. Cell Biol. 149, 889–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Galli T., Chilcote T., Mundigl O., Binz T., Niemann H., and De Camilli P. (1994) Tetanus toxin-mediated cleavage of cellubrevin impairs exocytosis of transferrin receptor-containing vesicles in CHO cells. J. Cell Biol. 125, 1015–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kumari R., and Francesconi A. (2011) Identification of GPCR localization in detergent resistant membranes. Methods Mol. Biol. 746, 411–423 [DOI] [PubMed] [Google Scholar]
- 18. Farnsworth C. L., and Feig L. A. (1991) Dominant inhibitory mutations in the Mg2+-binding site of RasH prevent its activation by GTP. Mol. Cell. Biol. 11, 4822–4829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nuoffer C., Davidson H. W., Matteson J., Meinkoth J., and Balch W. E. (1994) A GDP-bound of rab1 inhibits protein export from the endoplasmic reticulum and transport between Golgi compartments. J. Cell Biol. 125, 225–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cherry S., Jin E. J., Ozel M. N., Lu Z., Agi E., Wang D., Jung W. H., Epstein D., Meinertzhagen I. A., Chan C. C., and Hiesinger P. R. (2013) Charcot-Marie-Tooth 2B mutations in rab7 cause dosage-dependent neurodegeneration due to partial loss of function. eLife 2, e01064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Abou-Zeid N., Pandjaitan R., Sengmanivong L., David V., Le Pavec G., Salamero J., and Zahraoui A. (2011) MICAL-like1 mediates epidermal growth factor receptor endocytosis. Mol. Biol. Cell 22, 3431–3441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sakane A., Honda K., and Sasaki T. (2010) Rab13 regulates neurite outgrowth in PC12 cells through its effector protein, JRAB/MICAL-L2. Mol. Cell. Biol. 30, 1077–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Marat A. L., Ioannou M. S., and McPherson P. S. (2012) Connecdenn 3/DENND1C binds actin linking Rab35 activation to the actin cytoskeleton. Mol. Biol. Cell 23, 163–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Blümer J., Rey J., Dehmelt L., Mazel T., Wu Y. W., Bastiaens P., Goody R. S., and Itzen A. (2013) RabGEFs are a major determinant for specific Rab membrane targeting. J. Cell Biol. 200, 287–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gerondopoulos A., Langemeyer L., Liang J. R., Linford A., and Barr F. A. (2012) BLOC-3 mutated in Hermansky-Pudlak syndrome is a Rab32/38 guanine nucleotide exchange factor. Curr. Biol. 22, 2135–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Marzesco A. M., Dunia I., Pandjaitan R., Recouvreur M., Dauzonne D., Benedetti E. L., Louvard D., and Zahraoui A. (2002) The small GTPase Rab13 regulates assembly of functional tight junctions in epithelial cells. Mol. Biol. Cell 13, 1819–1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Köhler K., Louvard D., and Zahraoui A. (2004) Rab13 regulates PKA signaling during tight junction assembly. J. Cell Biol. 165, 175–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wu C., Agrawal S., Vasanji A., Drazba J., Sarkaria S., Xie J., Welch C. M., Liu M., Anand-Apte B., and Horowitz A. (2011) Rab13-dependent trafficking of RhoA is required for directional migration and angiogenesis. J. Biol. Chem. 286, 23511–23520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Di Giovanni S., De Biase A., Yakovlev A., Finn T., Beers J., Hoffman E. P., and Faden A. I. (2005) In vivo and in vitro characterization of novel neuronal plasticity factors identified following spinal cord injury. J. Biol. Chem. 280, 2084–2091 [DOI] [PubMed] [Google Scholar]
- 30. Nokes R. L., Fields I. C., Collins R. N., and Fölsch H. (2008) Rab13 regulates membrane trafficking between TGN and recycling endosomes in polarized epithelial cells. J. Cell Biol. 182, 845–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yamashiro D. J., Tycko B., Fluss S. R., and Maxfield F. R. (1984) Segregation of transferrin to a mildly acidic (pH 6.5) para-Golgi compartment in the recycling pathway. Cell 37, 789–800 [DOI] [PubMed] [Google Scholar]
- 32. Chaineau M., Danglot L., and Galli T. (2009) Multiple roles of the vesicular-SNARE TI-VAMP in post-Golgi and endosomal trafficking. FEBS Lett. 583, 3817–3826 [DOI] [PubMed] [Google Scholar]
- 33. Pfeffer S., and Aivazian D. (2004) Targeting Rab GTPases to distinct membrane compartments. Nat. Rev. Mol. Cell Biol. 5, 886–896 [DOI] [PubMed] [Google Scholar]
- 34. Marzesco A. M., Galli T., Louvard D., and Zahraoui A. (1998) The rod cGMP phosphodiesterase δ subunit dissociates the small GTPase Rab13 from membranes. J. Biol. Chem. 273, 22340–22345 [DOI] [PubMed] [Google Scholar]
- 35. Ullrich O., Stenmark H., Alexandrov K., Huber L. A., Kaibuchi K., Sasaki T., Takai Y., and Zerial M. (1993) Rab GDP dissociation inhibitor as a general regulator for the membrane association of rab proteins. J. Biol. Chem. 268, 18143–18150 [PubMed] [Google Scholar]
- 36. Simons K., and Ikonen E. (1997) Functional rafts in cell membranes. Nature 387, 569–572 [DOI] [PubMed] [Google Scholar]
- 37. Lebrand C., Corti M., Goodson H., Cosson P., Cavalli V., Mayran N., Fauré J., and Gruenberg J. (2002) Late endosome motility depends on lipids via the small GTPase Rab7. EMBO J. 21, 1289–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ganley I. G., and Pfeffer S. R. (2006) Cholesterol accumulation sequesters Rab9 and disrupts late endosome function in NPC1-deficient cells. J. Biol. Chem. 281, 17890–17899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nichols B. J., Kenworthy A. K., Polishchuk R. S., Lodge R., Roberts T. H., Hirschberg K., Phair R. D., and Lippincott-Schwartz J. (2001) Rapid cycling of lipid raft markers between the cell surface and Golgi complex. J. Cell Biol. 153, 529–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Joberty G., Tavitian A., and Zahraoui A. (1993) Isoprenylation of Rab proteins possessing a C-terminal CaaX motif. FEBS Lett. 330, 323–328 [DOI] [PubMed] [Google Scholar]
- 41. Chavrier P., Gorvel J. P., Stelzer E., Simons K., Gruenberg J., and Zerial M. (1991) Hypervariable C-terminal domain of rab proteins acts as a targeting signal. Nature 353, 769–772 [DOI] [PubMed] [Google Scholar]
- 42. Li F., Yi L., Zhao L., Itzen A., Goody R. S., and Wu Y. W. (2014) The role of the hypervariable C-terminal domain in Rab GTPases membrane targeting. Proc. Natl. Acad. Sci. U. S. A. 111, 2572–2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ali B. R., Wasmeier C., Lamoreux L., Strom M., and Seabra M. C. (2004) Multiple regions contribute to membrane targeting of Rab GTPases. J. Cell Sci. 117, 6401–6412 [DOI] [PubMed] [Google Scholar]
- 44. Mochizuki Y., Ohashi R., Kawamura T., Iwanari H., Kodama T., Naito M., and Hamakubo T. (2013) Phosphatidylinositol 3-phosphatase myotubularin-related protein 6 (MTMR6) is regulated by small GTPase Rab1B in the early secretory and autophagic pathways. J. Biol. Chem. 288, 1009–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shirane M., and Nakayama K. I. (2006) Protrudin induces neurite formation by directional membrane trafficking. Science 314, 818–821 [DOI] [PubMed] [Google Scholar]
- 46. Welz T., Wellbourne-Wood J., and Kerkhoff E. (2014) Orchestration of cell surface proteins by Rab11. Trends Cell Biol. 24, 407–415 [DOI] [PubMed] [Google Scholar]
- 47. Militello R. D., Munafó D. B., Berón W., López L. A., Monier S., Goud B., and Colombo M. I. (2013) Rab24 is required for normal cell division. Traffic 14, 502–518 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.