Abstract
Translation initiation is a focal point of translational control and requires the binding of eIF4E to the 5′ cap of mRNA. Under conditions of extreme oxygen depletion (hypoxia), human cells repress eIF4E and switch to an alternative cap-dependent translation mediated by a homolog of eIF4E, eIF4E2. This homolog forms a complex with the oxygen-regulated hypoxia-inducible factor 2α and can escape translation repression. This complex mediates cap-dependent translation under cell culture conditions of 1% oxygen (to mimic tumor microenvironments), whereas eIF4E mediates cap-dependent translation at 21% oxygen (ambient air). However, emerging evidence suggests that culturing cells in ambient air, or “normoxia,” is far from physiological or “normal.” In fact, oxygen in human tissues ranges from 1–11% or “physioxia.” Here we show that two distinct modes of cap-dependent translation initiation are active during physioxia and act on separate pools of mRNAs. The oxygen-dependent activities of eIF4E and eIF4E2 are elucidated by observing their polysome association and the status of mammalian target of rapamycin complex 1 (eIF4E-dependent) or hypoxia-inducible factor 2α expression (eIF4E2-dependent). We have identified oxygen conditions where eIF4E is the dominant cap-binding protein (21% normoxia or standard cell culture conditions), where eIF4E2 is the dominant cap-binding protein (1% hypoxia or ischemic diseases and cancerous tumors), and where both cap-binding proteins act simultaneously to initiate the translation of distinct mRNAs (1–11% physioxia or during development and stem cell differentiation). These data suggest that the physioxic proteome is generated by initiating translation of mRNAs via two distinct but complementary cap-binding proteins.
Keywords: eIF4E, hypoxia, mammalian target of rapamycin (mTOR), mRNA, physiology, translation, translation control, oxygen
Introduction
In the laboratory, we take measures to control the cellular environment to better reflect what cells experience in nature. In human cell culture, cells are grown at 37 °C, and ambient air is supplemented with 5% CO2 to mimic body temperature and pH, respectively. Oxygen is a surprisingly neglected parameter. Because we breathe air (21% oxygen), cells are routinely cultured in the same air. However, emerging evidence suggests that culturing cells in ambient air, or “normoxia,” is far from physiological. In fact, oxygen in arterial blood is only about 12% (1). The oxygen supply to tissues varies from 2–6% in the brain (2), 3–12% in the lungs (3), 3.5–6% in the intestine (4), 4% in the liver (5), 7–12% in the kidney (6), 4% in muscle (7), and 6–7% in bone marrow (8). A single cell may have as little as 1–2.5% available oxygen (9). Low oxygen has been linked to proper fetal development, and it is not surprising that umbilical cord blood contains only 2.5–4% oxygen (10). Therefore, human cells in their tissue of origin are exposed to oxygen levels, termed physioxia, that are much closer to what is defined in the literature as hypoxia (1% oxygen) rather than normoxia. This begs one to consider whether experiments performed in so-called normoxia might be misleading or that physiological phenotypes could be masked.
Considering the contributions from oxygen in cell culture is not a new idea, but it has not been widely adopted because cells cultured under atmospheric conditions have historically grown quite well. It was first noted that culturing cells in low oxygen increased plating efficiency (11). Culturing human fibroblasts at 10% oxygen dramatically increased their life span by 25% relative to cells grown at 21% oxygen (12). In the last 15 years, there have been examples that demonstrate the many benefits of culturing different cell types in low oxygen. For example, mouse embryonic fibroblasts cultured in 3% oxygen avoided the senescence that occurs after 28 days of growth, grew faster, showed less DNA damage, and had fewer stress responses relative to cells cultured in 21% oxygen (13). Immune cells at low oxygen behaved as though they were in a healthy body, but the same cells cultured at atmospheric oxygen sent signals as though they were fighting off an infection (14). Stem cells are routinely cultured in low oxygen to maintain their normal stem cell characteristics and to keep them from differentiating. Recently, it was shown that human embryonic stem cells must be cultured in 5% oxygen to retain their pluripotency and reduce chromosomal aberrations (15). These findings have led in vitro fertilization clinics to culture human embryos at 3–5% oxygen to ensure proper development.
Human cells appear “healthier” when cultured in the physioxic range, but little is known about what is happening behind these phenotypic observations at the molecular level. Here we investigate the physioxic activity of a fundamental mechanism of gene expression: cap-dependent translation initiation. Cap-dependent translation is the most common pathway to initiate protein synthesis in eukaryotic cells, and its initiation is a principal point of regulation (16). The first steps require the binding of eIF4E to the 7-methylguanosine (m7-GTP)3 5′ cap of mRNAs, but the experiments that contributed to this model were performed in normoxia (16, 17). More recently, studies have shown that translation initiation under hypoxic conditions can be quite different in normoxia. For example, cap-independent mechanisms of initiation, such as internal ribosome entry site-mediated translation, compensate for the stress-mediated repression of eIF4E by the mammalian target of rapamycin complex 1 (mTORC1) (18). Moreover, we have previously characterized an eIF4E homolog, eIF4E2, as a hypoxia-activated cap-binding protein responsible for the selective recruitment of hundreds of mRNAs for translation (19). eIF4E2 is an inhibitor of translation in normoxia (20) but gains the ability to initiate translation via the 5′ cap of hundreds of mRNAs with 3′ UTR RNA hypoxia response elements (19). This change in activity requires the hypoxia-stabilized hypoxia-inducible factor (HIF) 2α, not HIF-1α, and perhaps other hypoxia-induced factors (19). There is increasing evidence that the HIFα subunits are not redundant homologs but have specialized roles in the molecular response to low oxygen. HIF-2α accumulates in the cytoplasm of cells (21) in all tissues under chronic hypoxia (>24 h) (22) and can be stabilized within the physioxic range (5% oxygen) (23). Conversely, HIF-1α accumulates primarily in the nucleus under acute hypoxia (<6 h) and is stabilized at ≤1% oxygen. These observations led us to hypothesize that eIF4E2-mediated translation initiation is active under physiological conditions and may contribute to the proteomes of human tissues.
In this report, we show that two distinct modes of cap-dependent translation initiation are active during physioxia and act on separate mRNAs. The oxygen-dependent activities of eIF4E and eIF4E2 were determined by their polysome association and the status of mTORC1 (eIF4E-dependent) or HIF-2α expression (eIF4E2-dependent). We have identified conditions where eIF4E is the dominant cap-binding protein (21% normoxia or standard cell culture conditions), where eIF4E2 is the dominant cap-binding protein (1% hypoxia or ischemic diseases such as cancer), and where both cap-binding proteins act simultaneously to initiate the translation of distinct mRNAs (1–12% physioxia or during development and stem cell differentiation). These data suggest that the physioxic proteome is generated by initiating mRNA translation via two distinct but complementary cap-binding proteins. This new layer of translational regulation likely occurs in situ, during development, and during tumor progression as a way to selectively express different classes of mRNAs through the 5′ cap.
Experimental Procedures
Cell Culture and Cell Lines
HCT116 colorectal carcinoma (CCL-247) cells, U87MG glioblastoma (HTB-14) cells, human renal proximal tubular epithelial cells (PCS-400-010), primary dermal fibroblasts (PCS-201-012), and primary bronchial/tracheal epithelial cells (PCS-300-01) were used within 6 months of being obtained from the American Type Culture Collection and maintained as suggested. Hypoxia was induced by incubating at a specified percentage of oxygen in a constant 5% CO2 environment for 24 h in a Whitley HypOxystation H35.
Western Blotting Analysis
Standard Western blotting protocols were used. Primary antibodies were as follows: anti-EGFR (Ab-12, LabVision), anti-PDGFRA (D1E1E, Cell Signaling Technology), anti-GAPDH (D16H11, Cell Signaling Technology), anti-HIF-2α (NB100-122, Novus), anti-HIF-1α (NB100-123, Novus), anti-rpL5 (AB137617, Abcam), anti-4EBP1-P-Thr37/46 (236B4, Cell Signaling Technology), anti-4EBP1-Ser(P)65 (Ser65, Cell Signaling Technology), anti-rpS6-Ser(P)235/236 (Ser235/236, Cell Signaling Technology), anti-4EBP1 (53H11, Cell Signaling Technology), anti-rpS6 (5G10, Cell Signaling Technology), anti-eIF4E (C46H6, Cell Signaling Technology), and anti-eIF4E2 (N1C3, Genetex). Band intensities were quantified using ImageJ software.
Polysomal Analysis
Cells were isolated from four 15-cm culture dishes at 60–80% confluence. For isolation of intact polysomes, 0.1 mg/ml of cycloheximide was added to cells for 5 min at 37 °C before harvesting. Polysome lysates were prepared in RNA lysis buffer (15 mm Tris-HCl (pH 7.4), 15 mm MgCl2, 0.3 M NaCl, 1% Triton X-100, 0.1 mg/ml cycloheximide, and 100 units/ml RNasein) and loaded onto gradients based on equal total RNA. Sucrose gradients (7–47%) were centrifuged at 39,000 rpm with a SW-41-Ti Rotor (Beckman Coulter, Fullerton, CA) for 90 min at 4 °C. Gradients were then collected into nine equal fractions while the absorbance at 254 nm was continuously monitored with a Brandel BR-188 density gradient fractionation system. The baseline absorbance (blank RNA lysis buffer loaded onto a 7–47% sucrose density gradient) was calculated by Peakchart software and subtracted from the absorbance reading of each sample. For RNA isolation, each fraction was digested with proteinase K, and total RNA was isolated by phenol-chloroform extraction and ethanol precipitation. Equal volumes of total RNA were then used for RT-PCR analysis of select mRNAs. Semiquantitative analysis of the polysome gradients was performed by measurement of band intensities, less the background readings for equivalent areas, using ImageJ software and represented as percent mRNA in each fraction relative to the total signal from all fractions. For protein analysis, proteins from each fraction were concentrated by trichloroacetic acid precipitation. 1:4 volume of trichloroacetic acid was added to each fraction and incubated for 10 min at 4 °C. Samples were centrifuged at 12,000 × g for 5 min, and pellets were washed twice with 200 μl of cold acetone with centrifugation at 12,000 × g for 5 min in between. Pellets were dried at 95 °C to evaporate the acetone. Pellets were resuspended in 2× SDS-PAGE sample buffer and boiled before performing the Western blotting analysis. 250 nm Torin 1 (Tocris) was added to cells in the final 2 h of 5% oxygen treatment to inhibit mTORC1. Validated siRNAs (Dharmacon) targeting HIF-2α (19) were used in cells exposed to 5% oxygen. Lipofectamine 2000 (Life Technologies) was used to transfect siRNAs 24 h prior to 5% oxygen exposure according to the directions of the manufacturer. Protein integrity in each fraction was verified by blotting for ribosomal protein L5. All siRNAs were purchased from GE Dharmacon and validated previously (19, 24). Polysome-to-monosome (P/M) ratios were calculated by measuring the area under the 254 nm absorbance curve using ImageJ. Polysomes and monosomes are separated by dotted lines in the figures.
RNA Isolation, RT-PCR, and Real-time PCR
RT-PCR was performed using the two-step high-capacity cDNA reverse transcription kit (Applied Biosystems) followed by standard PCR conditions. Quantitative PCR reactions were performed using iQ SYBR Green SuperMix (Bio-Rad). Transcript levels were normalized to GAPDH and RPLP0. The relative -fold change in expression was calculated using the ΔΔCT method. Primer sequences (5′ → 3′) were as follows: EGFR, GGA GAA CTG CCA GAA ACT GAC (forward) and GGG GTT CAC ATC CAT CTG (reverse); PDGFRA, GCA GAC AGG GCT TTA ATG GG (forward) and GCC TTT GCC TTT CAC TTC T (reverse); EEF2, TTC AAG TCA TTC TCC GAG A (forward) and AGA CAC GCT TCA CTG ATA (reverse); HSP90ab1, TGT CCC TCA TCA TCS ATA CC (forward) and TCT TTA CCA CTG TCC AAC TT (reverse); GAPDH, GTC AAG GCT GAG AAC GGG A (forward) and CAA ATG AGC CCC AGC CTT C (reverse); and RPLP0, AAC ATC TCC CCC TTC TCC (forward) and CCA GGA AGC GAG AAT GC (reverse).
m7-GTP Cap-binding Assays
Cells on two 100-mm plates were washed with PBS and lysed in 1 ml of lysis buffer (20 mm Tris-HCl, 100 mm NaCl, 25 mm MgCl2, 0.5% Nonidet P-40, and standard protease and phosphatase inhibitors). Extracts were clarified by centrifugation at 10,000 × g for 10 min at 4 °C. Supernatants were precleared with 30 μl of blank agarose beads (Jena Bioscience) for 10 min at 4 °C. Beads were removed by centrifugation at 500 × g for 30 s, and supernatants were incubated with 50 μl m7-GTP-agarose beads (Jena Bioscience) for 1 h at 4 °C. Pelleted beads were washed four times with 0.5 ml of lysis buffer and resuspended in 0.6 ml of lysis buffer and 1 mm GTP for 1 h at 4 °C. Following four final washes with lysis buffer, the beads were resuspended in sample buffer and boiled for 1 min. Concentrated GTP wash, m7-GTP-bound proteins, as well as 5% input taken just before m7-GTP beads were added were run on an SDS-PAGE.
Statistical Analysis
Results are expressed as mean ± S.E. of at least three independent experiments. Experimental samples were compared with controls by unpaired two-tailed Student's t test. p < 0.05 was considered statistically significant.
Results
The Positive Regulators of eIF4E- and eIF4E2-mediated Translation Are Both Active during Physioxia
There is limited knowledge about how physioxic cells initiate translation, including the involvement of the eIF4E and eIF4E2 cap-binding proteins and their activators mTORC1 and HIF-2α, respectively. We cultured a panel of five human cell lines of different tissue of origin and cell type for 24 h at eight different oxygen concentrations: three primary cell lines (human renal proximal tubular epithelial cells (HRPTEC), dermal fibroblasts, and bronchial/tracheal epithelial cells) and two cancer cell lines (U87MG glioblastoma and HCT116 colorectal carcinoma). We used a hypoxia work station to perform cell culture at oxygen concentrations spanning normoxia, physioxia, and hypoxia (21%, 15%, 12%, 8%, 5%, 3%, 1%, and 0.1% oxygen). Even in a hypoxia system, 24 h are required to ensure that the dissolved oxygen in the culture medium has equilibrated with the hypoxic air (25).
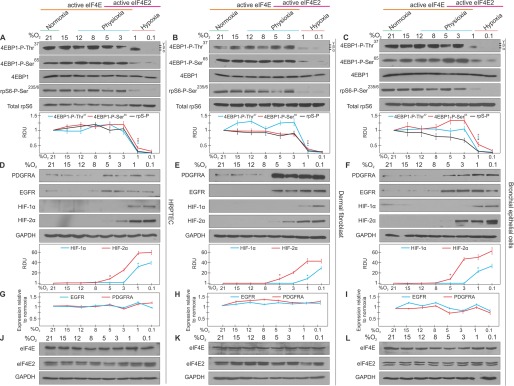
We measured the activity of eIF4E and eIF4E2 by determining the oxygen levels required to turn them “on” and “off.” These proteins have on/off switches in mTORC1 for eIF4E and HIF-2α for eIF4E2. We first monitored the activity of mTORC1 through the phosphorylation status of two downstream targets: 4E-binding protein 1 (4EBP1) and ribosomal protein S6 (rpS6). In all three primary cell lines, mTORC1 activity significantly decreased between 1–3% oxygen, as evidenced by 4EBP1 and rpS6 loss of phosphorylation relative to total 4EBP1 and rpS6 (Fig. 1, A–C). The activity of eIF4E2 was monitored by observing the oxygen-dependent stabilization of its activator, HIF-2α, and the accumulation of protein from two of its bona fide mRNA targets, epidermal growth factor receptor (EGFR) and platelet-derived growth factor receptor α (PDGFRA) (19). HIF-2α displayed significant stabilization between 5–8% oxygen, which coincided with the accumulation of EGFR and PDGFRA, suggesting that eIF4E2-dependent translation is active during physioxia (Fig. 1, D–F). Significant HIF-1α stabilization was only observed during hypoxia (≤1% oxygen), suggesting that HIF-2α has a more prominent role during physioxic gene expression. The increase in EGFR and PDGFRA protein appeared to be posttranscriptional because their mRNA levels did not change more than 1.5-fold across the oxygen spectrum (Fig. 1, G–I). eIF4E and eIF4E2 protein levels displayed little variability across the oxygen gradient (Fig. 1, J–L), consistent with their posttranslational regulation via the 4EBPs and HIF-2α, respectively. These data support the intriguing possibility that there exists a window within physioxia (1–8% oxygen) where both eIF4E and eIF4E2 are initiating translation in human cells.
FIGURE 1.
The positive regulators of eIF4E- and eIF4E2-mediated translation are both active during physioxia in human primary cells. A–C, HRPTEC, dermal fibroblasts, and bronchial/tracheal epithelial cells were exposed to [oxygen] spanning normoxia, physioxia, and hypoxia for 24 h. The activity of eIF4E was measured by observing the phosphorylation of mTORC1 targets. The antibody against 4EBP1-Thr(P)37 recognizes the α, β, and γ phosphates. Total 4EBP1 and rpS6 were used to indicate unchanging total levels and as loading controls. RDU, relative density unit. D--F, the activity of eIF4E2 was measured by detecting the stabilization of its hypoxic activator, HIF-2α, and proteins from two of its mRNA targets, EGFR and PDGFRA. GAPDH was used as a loading control. Western blots from at least three independent experiments were quantified by ImageJ and expressed as relative density units relative to 21% oxygen. *, p < 0.05 when comparing a data point to the preceding oxygen concentration. G–I, quantitative RT-PCR of EGFR and PDGFRA mRNA levels in a gradient of oxygen availability relative to normoxia. GAPDH and RPLP0 were used as endogenous controls. Data are mean ± S.E. of three independent experiments. J–L, oxygen-dependent eIF4E and eIF4E2 protein levels. GAPDH was used as a loading control. Results are representative of three independent experiments.
A similar response was observed in colorectal carcinoma and glioblastoma cancer cells, except that 4EBP1 and rpS6 lost significant phosphorylation at a higher oxygen availability (between 5–8% instead of 1–3% oxygen) (Fig. 2, A and B). As in primary cells, HIF-2α was significantly stabilized between 5–8% oxygen, but traces could be observed by Western blotting as high as 12% oxygen in HCT116 cells, which may have led to the accumulation of EGFR and PDGFRA at the same oxygen concentration (Fig. 2, C and D). This was further examined in Fig. 3. Consistent with our observations in primary cells, EGFR and PDGFRA mRNA levels did not change more than 1.5-fold throughout the oxygen spectrum and neither did eIF4E and eIF4E2 protein levels (Fig. 2, E–H). Therefore, these two cancer cell lines differ in their cap-binding protein usage relative to primary cells, whereby eIF4E is repressed at a higher oxygen availability.
FIGURE 2.
The positive regulators of eIF4E- and eIF4E2-mediated translation are both active during physioxia in human cancer cells. HCT116 colorectal carcinoma and U87MG glioblastoma cells were exposed to [O2] spanning normoxia, physioxia, and hypoxia for 24 h. A and B, the activity of eIF4E was measured by observing the phosphorylation of mTORC1 targets. The antibody against 4EBP1-Thr(P)37 recognizes the α, β, and γ phosphates. Total 4EBP1 and rpS6 were used to indicate unchanging total levels and as loading controls. RDU, relative density unit. C and D, the activity of eIF4E2 was measured by detecting the stabilization of its hypoxic activator, HIF-2α, and proteins from two of its mRNA targets, EGFR and PDGFRA. GAPDH was used as a loading control. Western blots from at least three independent experiments were quantified by ImageJ and expressed as relative density units relative to 21% oxygen. *, p < 0.05 when comparing a data point to the preceding oxygen concentration. E and F, quantitative RT-PCR of EGFR and PDGFRA mRNA levels at various oxygen concentrations relative to normoxia. GAPDH and RPLP0 were used as endogenous controls. Data are mean ± S.E. of three independent experiments. G and H, oxygen-dependent eIF4E and eIF4E2 protein levels. GAPDH was used as a loading control. Results are representative of three independent experiments.
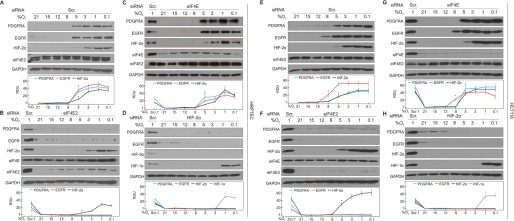
FIGURE 3.
EGFR and PDGFRA protein accumulate during physioxia in an eIF4E2- and HIF-2α-dependent manner. HRPTEC and HCT116 colorectal carcinoma cells were exposed to eight different oxygen conditions spanning normoxia, physioxia, and hypoxia for 24 h. A–G, Western blotting of EGFR, PDGFRA, eIF4E, eIF4E2, and HIF-2α protein levels in (A and E) cells treated with non-targeting control siRNA, (B and F) eIF4E2-depleted cells, and (C and G) eIF4E-depleted cells. Scr, scrambled; RDU, relative density unit. D and H, the activator of eIF4E2, HIF-2α, was depleted via siRNA, and EGFR, PDGFRA, HIF-2α, and HIF-1α protein levels were measured. GAPDH was used as a loading control. Cells transiently expressing a non-targeting scrambled siRNA and exposed to 1% oxygen (Scr. 1) were used as a positive control in many blots that displayed little to no signal in the other lanes for some markers. Western blots from at least three independent experiments were quantified by ImageJ and expressed as relative density units relative to 21% oxygen. Data are mean ± S.E. of at least three independent experiments. *, p < 0.05 when comparing a data point to the preceding oxygen concentration. Results are representative of three independent experiments.
EGFR and PDGFRA Protein Accumulate during Physioxia in an eIF4E2- and HIF-2α-dependent Manner
The accumulation of EGFR and PDGFRA during physioxia is synchronous with the accumulation of HIF-2α, suggesting that eIF4E2 contributes to the physioxic proteome. We continued this study with HCT116 cells and one primary cell line (HRPTEC) because they responded similarly to the other cancer cell line and primary cell lines, respectively. We selectively impaired eIF4E- or eIF4E2-dependent translation initiation to monitor the effect on physioxic EGFR and PDGFRA expression. eIF4E2, eIF4E, or HIF-2α was depleted independently via commercially available siRNA. Control cells expressing non-targeting siRNA displayed significant EGFR and PDGFRA protein accumulation between 5–8% oxygen for HRPTEC and between 5–12% for HCT116 cells, as observed in Figs. 1 and 2, respectively (Fig. 3, A and E). eIF4E2 depletion reduced EGFR and PDGFRA accumulation from 0.1–5% oxygen in HRPTEC, and 0.1–8% in HCT116 for EGFR, even in the presence of HIF-2α (Fig. 3, B and F). eIF4E-depleted cells were not impaired in their physioxic or hypoxic accumulation of EGFR and PDGFRA in both cell lines but displayed reduced expression in the 8–21% oxygen range (Fig. 3, C and G), where eIF4E is the dominant cap-binding protein (Figs. 1 and 2). HIF-2α depletion prevented EGFR and PDGFRA accumulation from 0.1–5% oxygen in HRPTEC and 0.1–8% in HCT116 cells for EGFR (Fig. 3, D and H). However, HIF-2α depletion had little effect on the EGFR and PDGFRA levels between 8–21% oxygen. This highlights the dependence of eIF4E2-mediated translation on the hypoxic stabilization of HIF-2α. These data demonstrate that the physioxic expression of EGFR and PDGFRA relies on eIF4E2-mediated translation, whereas their normoxic expression is impaired by eIF4E depletion.
eIF4E and eIF4E2 Activities Overlap during Physioxia
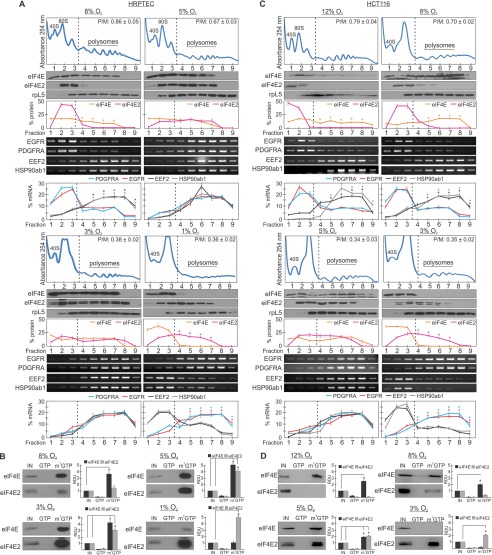
The activity of translation factors can be observed by their association with mRNA bound to actively translating ribosomes called polysomes. Translation initiation factors, including eIF4E and eIF4E2, remain bound to polysomes to aid in ribosome recycling (26). The data from Fig. 1 suggest that eIF4E is the dominant cap-binding protein at ≥8% oxygen, that eIF4E2 is the dominant cap-binding protein at ≤1% oxygen, and that both cap-binding proteins participate in translation initiation between 1–8% oxygen. To determine the activity of eIF4E and eIF4E2, we measured their polysome association in HRPTEC exposed to 8%, 5%, 3%, and 1% oxygen. The continuous absorbance reading at 254 nm measures RNA during fraction collection to identify polysome-containing fractions (typically fractions ≥4). We observed that eIF4E was significantly associated with polysome fractions at 8% oxygen relative to eIF4E2, which was in the untranslated pool (Fig. 4A). At 5% and 3% oxygen, both eIF4E and eIF4E2 were observed in polysome fractions. Finally, eIF4E2 was significantly associated with polysome fractions at 1% oxygen relative to eIF4E. Polysome disassembly via puromycin treatment removes eIF4E and eIF4E2 from polysome-containing fractions, demonstrating that they are polysome-bound (19). Therefore, there appears to be a switch in cap-binding protein usage within the physioxic range and three overall “zones”: hypoxia (≤1% oxygen, eIF4E2 only), low- to mid-physioxia (1–8% oxygen, both eIF4E and eIF4E2), and upper-range physioxia/normoxia (8–21% oxygen, eIF4E only). The P/M ratio decreased with oxygen availability up to 1–3% oxygen for HRPTEC and 3–5% oxygen for HCT116 cells (Fig. 4, A and C). Even as eIF4E2 became the dominant cap-binding protein at 1% oxygen in HRPTEC and 3% oxygen in HCT116 cells, the P/M ratio remained unchanged, suggesting that eIF4E2 can sustain overall translation initiation rates at low oxygen. These data suggest that cells exposed to physiological oxygen utilize two cap-binding proteins simultaneously to initiate translation.
FIGURE 4.
eIF4E and eIF4E2 activities overlap during physioxia. A and C, polysomal distribution of eIF4E and two of its mRNA targets (EEF2 and HSP90ab1) and eIF4E2 protein and two of its mRNA targets (EGFR and PDGFRA) in (A) HRPTEC exposed to 8%, 5%, 3%, or 1% O2 or (C) HCT116 cells exposed to 12%, 8%, 5%, or 3% O2 for 24 h. The polysomal association of proteins eIF4E and eIF4E2 and mRNAs EGFR, PDGFRA, EEF2, and HSP90ab1 were observed by Western blotting or RT-PCR, respectively. Ribosomal protein L5 (rpL5) was used as a marker of protein integrity in each fraction. The percentage of mRNA or protein in each fraction relative to the total was quantified by ImageJ and plotted. Representative Western blots and RT-PCR gels are shown. *, p < 0.05 was considered a significant change between eIF4E and eIF4E2, or their mRNA targets, within polysome fractions. P/M ratios are displayed in the top right corner of each representative polysome profile and were quantified by measuring the area under the curve by ImageJ. The separation used between polysomes and monosomes is indicated by a dotted line. B and D, capture assays using m7-GTP beads in (B) HRPTEC lysates exposed to 8%, 5%, 3%, or 1% O2 or (D) HCT116 cells exposed to 12%, 8%, 5%, or 3% O2. GTP, GTP wash to measure specificity for m7-GTP; m7-GTP, proteins bound to m7-GTP beads after the GTP wash. Western blots from at least three independent experiments were quantified by ImageJ and expressed as relative density units (RDU) relative to 10% input (IN) of whole cell lysate. *, p < 0.05 when comparing the enrichment of eIF4E or eIF4E2 in the cap-bound fraction (m7GTP) to the input. Data are mean ± S.E. of three independent experiments. Results are representative of three independent experiments.
We next investigated how the physioxic switch in cap-binding protein usage might affect the translation of specific mRNAs. eIF4E prefers mostly terminal oligopyrimidine tract mRNAs that have specific nucleotide motifs surrounding the transcriptional start site, such as eukaryotic elongation factor 2 (EEF2) and heat shock protein 90ab1 (HSP90ab1) (27), whereas eIF4E2 recognizes mRNAs based on structural and sequence motifs within 3′ UTR RNA hypoxia response elements such as in EGFR and PDGFRA (19). We monitored the polysome association of eIF4E and eIF4E2 mRNA targets under oxygen conditions where only one or the other is active (8% and 1% oxygen) or where both are active (5 and 3% oxygen). At 8% oxygen, where only eIF4E is active, EEF2 and HSP90ab1 mRNAs were significantly enriched in polysome fractions relative to the eIF4E2 mRNA targets EGFR and PDGFRA, which were concentrated in the untranslated pool (Fig. 4A). Conversely, at 1% oxygen, EGFR and PDGFRA were significantly enriched in polysome fractions relative to EEF2 and HSP90ab1, which were in the untranslated pool. At 3% and 5% oxygen, conditions where eIF4E and eIF4E2 were both associated with polysomes, all four mRNA targets were enriched in polysomes (Fig. 4A).
In vitro, eIF4E has a 100-fold higher affinity for the 5′ cap of mRNA than eIF4E2 because of two amino acid differences in the cap-binding pocket (28). However, in vivo experiments have shown that their cap-binding affinities can be altered by changes in oxygen availability. eIF4E binds m7-GTP cap beads strongly in normoxia but poorly in hypoxia and vice versa for eIF4E2 (19). As another measure of their physioxic activity, we investigated whether the cap-binding affinities of eIF4E and eIF4E2 changed according to their polysome association. In HRPTEC exposed to 8% oxygen for 24 h, eIF4E was significantly enriched in the m7-GTP-bound fraction (Fig. 4B). Conversely, eIF4E2 weakly associated with m7-GTP beads. In HRPTEC exposed to 1% oxygen, eIF4E2 was significantly enriched in the m7-GTP-bound fraction, whereas the opposite was observed for eIF4E (Fig. 4B). Under conditions of 3% and 5% oxygen where both cap-binding proteins were active, eIF4E and eIF4E2 were significantly bound to m7-GTP. These data are consistent with the oxygenation required for eIF4E and eIF4E2 polysome association (Fig. 4A) and their activation (Fig. 1A). Our data suggest that eIF4E is impaired in its cap-binding ability in hypoxia (Fig. 4B).
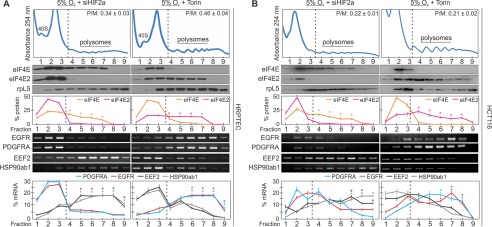
We next demonstrated that, when both eIF4E and eIF4E2 are active, either could be selectively removed from polysome fractions by inhibiting or knocking down their activator. For eIF4E2, knocking down its hypoxic activator, HIF-2α, significantly depleted it from polysomes relative to eIF4E and shifted it into monosomes (Fig. 5, A and B) compared with untreated cells exposed to 5% oxygen (Fig. 4, A and C). Conversely, treating cells exposed to 5% oxygen and the mTORC1 inhibitor Torin 1 for 2 h prior to polysome isolation significantly depleted eIF4E from polysomes relative to eIF4E2. Furthermore, the mRNA targets were significantly depleted from polysomes along with their respective cap-binding protein (Fig. 5, A and B). The decrease in P/M ratio observed when either cap-binding protein was repressed (Fig. 5, A and B) relative to the 5% oxygen control (Fig. 4C) gives some indication of their individual contributions to translation initiation at this oxygen concentration. The P/M ratio at 5% oxygen in HRPTEC decreased from 0.67 ± 0.03 (Fig. 4A) to 0.34 ± 0.03 when eIF4E2 is repressed and 0.46 ± 0.04 when eIF4E is repressed (Fig. 5A). The P/M ratio in HCT116 cells at 5% oxygen decreased from 0.34 ± 0.03 (Fig. 4C) to 0.22 ± 0.01 when eIF4E2 was repressed and 0.21 ± 0.02 when eIF4E was repressed (Fig. 5B).
FIGURE 5.
eIF4E and eIF4E2 can be selectively disabled during physioxia. HRPTEC (A) or HCT116 (B) cells exposed to 5% oxygen and 250 nm Torin 1 to selectively inhibit eIF4E-dependent translation, or treated with siHIF-2α to selectively inhibit eIF4E2-dependent translation. The polysomal association of proteins eIF4E and eIF4E2 and mRNAs EGFR, PDGFRA, EEF2, and HSP90ab1 were observed by Western blotting or RT-PCR, respectively. Ribosomal protein L5 (rpL5) used as a marker of protein integrity. The percentage of mRNA or protein in each fraction relative to the total was quantified by ImageJ and plotted. Representative Western blots and RT-PCR gels are shown. *, p < 0.05 was considered a significant change between eIF4E and eIF4E2 or their mRNA targets within polysome fractions. P/M ratios are displayed in the top right corner of each representative polysome profile and were quantified by measuring the area under the curve by ImageJ. The separation used between polysomes and monosomes is indicated by a dotted line. Data are mean ± S.E. of three independent experiments. Results are representative of three independent experiments.
HCT116 cells displayed similar eIF4E and eIF4E2 activities but in a slightly shifted oxygen range (Fig. 4, C and D). eIF4E2 remained partially associated with polysomes as high as 8% oxygen, and although PDGFRA and EGFR mRNAs were not significantly enriched in polysomes, moderate levels of EGFR protein could be detected (Figs. 2C and 3, E and G). Differences in PDGFRA and EGFR mRNA 3′ UTR RNA hypoxia response element secondary structure could play a role in their sensitivity to eIF4E2. These data are consistent with previous studies describing several mRNA classes that differ in their mode of mRNA recruitment onto 40S ribosomes (29, 30). Here we focused on the classes belonging to the major mode of mRNA recruitment, cap-dependent: class I (eIF4E-dependent) and class III (mRNAs controlled by variants of eIF4E). Our findings reveal a new layer of complexity for the regulation of cap-dependent translation initiation in cells exposed to physiological oxygen.
The Switch between eIF4E and eIF4E2 Is Dynamic
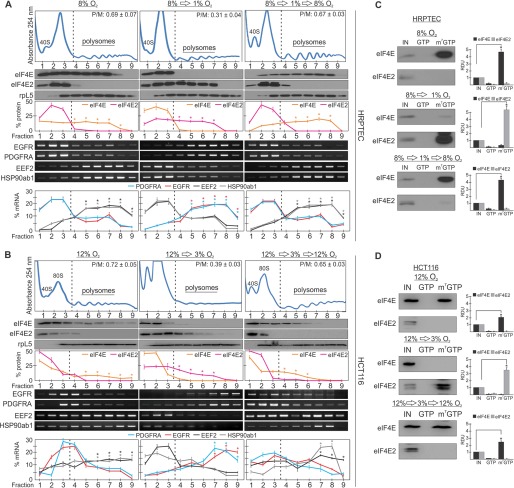
We next asked whether cells could dynamically switch between eIF4E as the major cap-binding protein to eIF4E2 and then back to eIF4E again. The reversible phosphorylation of 4EBPs regulates the translation of eIF4E transcripts, but no study has monitored the on/off switching between eIF4E- or eIF4E2-dependent translation upon reoxygenation. We exposed HRPTEC to oxygen conditions of 8% for 24 h, 8% to 1% for an additional 24 h, or 8% to 1% to 8% oxygen for a total of 72 h and monitored the polysome association of eIF4E, eIF4E2, and their mRNA targets. eIF4E and two of its mRNA targets were significantly enriched in 8% oxygen polysome fractions relative to eIF4E2 and its mRNA targets (Fig. 6A). eIF4E then shifted to monosomes in 1% oxygen and back into polysomes in 8% oxygen (Fig. 6A). Conversely, eIF4E2 and two of its mRNA targets were significantly depleted from polysome fractions at 8% oxygen, shifted to polysomes at 1% oxygen, and then back into monosomes at 8% oxygen. The same was observed in HCT116 cells but in a shifted oxygen range of 12% oxygen to 3% oxygen to accommodate the eIF4E and eIF4E2 activities observed in Fig. 4 for these cells (Fig. 6B). In addition, the affinity of eIF4E and eIF4E2 for m7-GTP-linked beads was reversible in an oxygen-dependent manner in both cell lines (Fig. 6, C and D). We show that a primary human cell line and a cancer cell line can transiently turn on and off eIF4E and eIF4E2 translation initiation in response to broad oxygen fluctuations.
FIGURE 6.
The switch between eIF4E and eIF4E2 is dynamic. A, polysomal distribution of eIF4E and two of its mRNA targets (EEF2 and HSP90ab1), and eIF4E2 protein and two of its mRNA targets (EGFR and PDGFRA) in HRPTEC cultured in three separate conditions: 1) 8% oxygen for 24 h, 2) 8% oxygen for 24 h followed by 1% oxygen for another 24 h, or 3) 8% oxygen for 24 h, then 1% oxygen for an additional 24 h followed by another 24 h of 8% oxygen exposure. Ribosomal protein L5 (rpL5) was used as a marker of protein integrity. The percentage of mRNA or protein in each fraction relative to the total was quantified by ImageJ and plotted. Representative Western blots and RT-PCR gels are shown. *, p < 0.05 was considered a significant change between eIF4E and eIF4E2 or their mRNA targets within polysome fractions. P/M ratios are displayed in the top right corner of each representative polysome profile and were quantified by measuring the area under the curve by ImageJ. The separation used between polysomes and monosomes is indicated by a dotted line. B, the same was performed in HCT116 cells but in a range of 12% to 3% oxygen. C and D, capture assays using m7-GTP beads in (C) HRPTEC or (D) HCT116 lysates exposed to the above oxygen gradients. GTP, GTP wash to measure the specificity for m7-GTP; m7-GTP, proteins bound to m7-GTP beads after the GTP wash. Western blots from at least three independent experiments were quantified by ImageJ and expressed as relative density units (RDU) relative to 10% input (IN) of whole cell lysate. *, p < 0.05 was considered a significant change when comparing the enrichment of eIF4E or eIF4E2 in the cap-bound fraction (m7-GTP) to the input. Data are mean ± S.E. of three independent experiments. Results are representative of three independent experiments.
Discussion
In light of recent tissue oxygenation measurements, culturing cells in ambient air could be far from physiological with respect to oxygen (1). We demonstrate that cap-dependent translation initiation in physioxic cells is not performed solely by the canonical eIF4E, as previously assumed, but by a complementary duo of cap-binding proteins to express distinct genes. We identify three overall oxygen zones of cap-binding protein usage: Hypoxia (≤1% oxygen, eIF4E2 only), low- to mid-physioxia (1–8% oxygen, both eIF4E and eIF4E2), and upper-range physioxia/normoxia (8–21% oxygen, eIF4E only). This new layer of translational regulation likely occurs in situ, during development, and during tumor progression as a way to selectively express different classes of mRNAs through the 5′ cap. Here we focus on the classes belonging to the major mode of mRNA recruitment, cap-dependent: class I (eIF4E-dependent) and class III (mRNAs controlled by variants of eIF4E) (29, 30). eIF4E-dependent mRNAs tend to code for “housekeeping” proteins such as ribosomal proteins, translation factors, and proteins indispensable for cell growth. On the other hand, many eIF4E2-dependent mRNAs code for signaling molecules, growth factors, and growth factor receptors required in rate-limiting quantities to respond to environmental change (19). The third member of the eIF4E family, eIF4E3, was not included in this study because it has a restricted tissue distribution as opposed to the ubiquitous eIF4E and eIF4E2 (31). These two modes of cap-dependent mRNA recruitment could allow cells in a physiological setting to selectively control the translation of different classes of essential mRNAs. Some indication of the individual contribution of eIF4E and eIF4E2 to translation initiation during physioxia could be observed by monitoring the decrease in P/M ratio when each cap-binding protein was selectively repressed at 5% oxygen concentration (Fig. 5, A and B) relative to the control (Fig. 4, A and C). Further mechanistic insight into the low oxygen-dependent activation and dominance of eIF4E2 will be required to understand the relationship between these two modes of cap-dependent mRNA recruitment. An increase in eIF4E2 cap-binding affinity during reduced oxygen may create more competition for eIF4E. Some recent progress has been made by demonstrating that eIF4E2 requires HIF-2α for activation (19) and gains affinity for the 5′ cap with the addition of ISG15 (32), which has several hypoxia response elements in its promoter. It would also be valuable to investigate cap-independent modes of mRNA recruitment during physioxia, such as internal ribosome entry site-mediated translation, because this mechanism is active at low oxygen.
We observed differences in the regulation of eIF4E and eIF4E2 activities between primary cell lines and cancer cell lines. The two cancer cell lines in this study repressed eIF4E at a higher oxygen availability relative to primary cell lines. Essentially, the three oxygen zones of cap-binding usage are modified in the two cancer cell lines (Figs. 1–4): “eIF4E2 alone” is shifted up from ≤1% oxygen to ≤3% oxygen, “both eIF4E and eIF4E2 active” is restricted from 1–8% oxygen to 3–5% oxygen, and “eIF4E alone” is shifted down from ≥8% oxygen to ≥5% oxygen. eIF4E overexpression drives cancer cell growth and tumor progression but selectively kills hypoxic cancer cells (33). Therefore, repressing eIF4E sooner during tumor hypoxification (3% versus 1% oxygen) to rely more heavily on eIF4E2 could be advantageous to cancer cells. eIF4E is a strong competitor of eIF4E2 because of its 100-fold higher affinity for the 5′ cap of mRNA (31). HIF-2α and eIF4E2 mediate the expression of hundreds of genes required for hypoxic survival and tumor progression (24, 34). In support of this argument, traces of HIF-2α were detected as high as 12% oxygen in HCT116, which likely led to the accumulation of EGFR and PDGFRA at the same oxygen concentration (Fig. 2C). Proliferation is stimulated in many epithelial cancers because of EGFR overexpression. Therefore, an elevated EGFR expression in a broader oxygen range would affect a larger tumor volume. PDGFRA has also been linked to aggressive colorectal carcinomas (35). We did not observe noticeable differences in eIF4E protein levels across all five cell lines; however, 3-fold increases in eIF4E in a HIF1α-dependent manner in a hypoxic breast cancer cell line suggest that cancer-specific mutation profiles may have different effects on the regulation of cap-dependent translation (36).
The only human organ that has been reported to receive a mean percentage of oxygen greater than 8% where eIF4E is the dominant cap-binding protein is the kidney, at 9.5% oxygen (1). The primary cell lines used in this study differ in their cell type and tissue of origin (including kidney HRPTEC), but they responded similarly to oxygen (Fig. 1). The cellular response to oxygen appears to be dependent on the oxygenation of the tissue where a cell resides rather than inherent differences between cells. However, tissue oxygenation is likely more complex than what has been reported recently (1). Although some tissues, such as brain, have uniform energy requirements, the blood delivery and energy expenditure of other tissues, such as liver and bowel, vary largely depending on their functional state (1). Therefore, precise knowledge of the functional changes in oxygen delivery is mandatory to fully grasp how biochemical pathways may be affected. We show that the oxygen-dependent switch between eIF4E and eIF4E2 occurs within the physioxic range and is reversible. This observation is likely relevant in many organs based on changes in oxygen supply and consumption or during tissue injury and repair 1. Tumors most likely also experience these dynamic changes in cap-binding protein usage because their abnormal vasculature causes heterogeneous oxygenation. It has been noted over the past several decades that human cells in culture benefit from low oxygen by displaying longer life spans, higher growth rates, and more rapid proliferation (12–15). However, physioxic cell culture studies investigating the molecular mechanisms behind these phenotypes have been lacking. We demonstrate that a fundamental mechanism of gene expression, cap-dependent translation initiation, is regulated differently in physioxia than in cells cultured in ambient air. This study highlights the importance of oxygen as a cell culture parameter when making physiological inferences.
Author Contributions
S. T. conducted most of the experiments and analyzed the results. J. U. conceived the idea for the project, conducted some experiments, analyzed the data, and wrote the paper.
Acknowledgments
We thank Erin Specker for technical assistance. We also thank Cezar Khursigara for critical review of the manuscript.
This work was funded by Ontario Ministry of Research and Innovation and the Natural Sciences and Engineering Council of Canada Grant 04807 (to J. U.). The authors declare that they have no conflicts of interest with the contents of this article.
- m7-GTP
- 7-methylguanosine
- mTORC
- mammalian target of rapamycin complex
- HIF
- hypoxia-inducible factor
- P/M
- polysome-to-monosome ratio
- EGFR
- epidermal growth factor receptor
- PDGFRA
- platelet-derived growth factor receptor α
- HRPTEC
- human renal proximal tubular epithelial cell(s).
References
- 1. Carreau A., El Hafny-Rahbi B., Matejuk A., Grillon C., and Kieda C. (2011) Why is the partial oxygen pressure of human tissues a crucial parameter? Small molecules and hypoxia. J. Cell Mol. Med. 15, 1239–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dings J., Meixensberger J., Jäger A., and Roosen K. (1998) Clinical experience with 118 brain tissue oxygen partial pressure catheter probes. Neurosurgery 43, 1082–1095 [DOI] [PubMed] [Google Scholar]
- 3. Le Q. T., Chen E., Salim A., Cao H., Kong C. S., Whyte R., Donington J., Cannon W., Wakelee H., Tibshirani R., Mitchell J. D., Richardson D., O'Byrne K. J., Koong A. C., and Giaccia A. J. (2006) An evaluation of tumor oxygenation and gene expression in patients with early stage non-small cell lung cancers. Clin. Cancer Res. 12, 1507–1514 [DOI] [PubMed] [Google Scholar]
- 4. Müller M., Schindler E., Roth S., Schürholz A., Vollerthun M., and Hempelmann G. (2002) Effects of desflurane and isoflurane on intestinal tissue oxygen pressure during colorectal surgery. Anaesthesia 57, 110–115 [DOI] [PubMed] [Google Scholar]
- 5. Brooks A. J., Eastwood J., Beckingham I. J., and Girling K. J. (2004) Liver tissue partial pressure of oxygen and carbon dioxide during partial hepatectomy. Br. J. Anaesth. 92, 735–737 [DOI] [PubMed] [Google Scholar]
- 6. Müller M., Padberg W., Schindler E., Sticher J., Osmer C., Friemann S., and Hempelmann G. (1998) Renocortical tissue oxygen pressure measurements in patients undergoing living donor kidney transplantation. Anesth. Analg. 87, 474–476 [DOI] [PubMed] [Google Scholar]
- 7. Richardson R. S., Duteil S., Wary C., Wray D. W., Hoff J., and Carlier P. G. (2006) Human skeletal muscle intracellular oxygenation: the impact of ambient oxygen availability. J. Physiol. 571, 415–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Harrison J. S., Rameshwar P., Chang V., and Bandari P. (2002) Oxygen saturation in the bone marrow of healthy volunteers. Blood 99, 394. [DOI] [PubMed] [Google Scholar]
- 9. Gleadle J., and Ratcliffe P. (2001) Hypoxia, pp. 3–5, John Wiley & Sons, Ltd., Chichester, UK [Google Scholar]
- 10. Gluckman E., Broxmeyer H. A., Auerbach A. D., Friedman H. S., Douglas G. W., Devergie A., Esperou H., Thierry D., Socie G., and Lehn P. (1989) Hematopoietic reconstitution in a patient with Fanconi's anemia by means of umbilical-cord blood from an HLA-identical sibling. N. Engl. J. Med. 321, 1174–1178 [DOI] [PubMed] [Google Scholar]
- 11. Richter A., Sanford K. K., and Evans V. J. (1972) Influence of oxygen and culture media on plating efficiency of some mammalian tissue cells. J. Natl. Cancer Inst. 49, 1705–1712 [DOI] [PubMed] [Google Scholar]
- 12. Packer L., and Fuehr K. (1977) Low oxygen concentration extends the lifespan of cultured human diploid cells. Nature 267, 423–425 [DOI] [PubMed] [Google Scholar]
- 13. Parrinello S., Samper E., Krtolica A., Goldstein J., Melov S., and Campisi J. (2003) Oxygen sensitivity severely limits the replicative lifespan of murine fibroblasts. Nat. Cell Biol. 5, 741–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Atkuri K. R., Herzenberg L. A., Niemi A. K., Cowan T., and Herzenberg L. A. (2007) Importance of culturing primary lymphocytes at physiological oxygen levels. Proc. Natl. Acad. Sci. U.S.A. 104, 4547–4552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lengner C. J., Gimelbrant A. A., Erwin J. A., Cheng A. W., Guenther M. G., Welstead G. G., Alagappan R., Frampton G. M., Xu P., Muffat J., Santagata S., Powers D., Barrett C. B., Young R. A., Lee J. T., Jaenisch R., and Mitalipova M. (2010) Derivation of pre-X inactivation human embryonic stem cells under physiological oxygen concentrations. Cell 141, 872–883 [DOI] [PubMed] [Google Scholar]
- 16. Gebauer F., and Hentze M. W. (2004) Molecular mechanisms of translational control. Nat. Rev. Mol. Cell Biol. 5, 827–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sonenberg N., Rupprecht K. M., Hecht S. M., and Shatkin A. J. (1979) Eukaryotic mRNA cap binding protein: purification by affinity chromatography on Sepharose-coupled m7GDP. Proc. Natl. Acad. Sci. U.S.A. 76, 4345–4349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Holcik M., and Sonenberg N. (2005) Translational control in stress and apoptosis. Nat. Rev. Mol. Cell Biol. 6, 318–327 [DOI] [PubMed] [Google Scholar]
- 19. Uniacke J., Holterman C. E., Lachance G., Franovic A., Jacob M. D., Fabian M. R., Payette J., Holcik M., Pause A., and Lee S. (2012) An oxygen-regulated switch in the protein synthesis machinery. Nature 486, 126–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Morita M., Ler L. W., Fabian M. R., Siddiqui N., Mullin M., Henderson V. C., Alain T., Fonseca B. D., Karashchuk G., Bennett C. F., Kabuta T., Higashi S., Larsson O., Topisirovic I., Smith R. J., Gingras A. C., and Sonenberg N. (2012) A novel 4EHP-GIGYF2 translational repressor complex is essential for mammalian development. Mol. Cell Biol. 32, 3585–3593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Talks K. L., Turley H., Gatter K. C., Maxwell P. H., Pugh C. W., Ratcliffe P. J., and Harris A. L. (2000) The expression and distribution of the hypoxia-inducible factors HIF-1α and HIF-2α in normal human tissues, cancers, and tumor-associated macrophages. Am. J. Pathol. 157, 411–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wiesener M. S., Jürgensen J. S., Rosenberger C., Scholze C. K., Hörstrup J. H., Warnecke C., Mandriota S., Bechmann I., Frei U. A., Pugh C. W., Ratcliffe P. J., Bachmann S., Maxwell P. H., and Eckardt K. U. (2003) Widespread hypoxia-inducible expression of HIF-2alpha in distinct cell populations of different organs. FASEB J. 17, 271–273 [DOI] [PubMed] [Google Scholar]
- 23. Holmquist-Mengelbier L., Fredlund E., Löfstedt T., Noguera R., Navarro S., Nilsson H., Pietras A., Vallon-Christersson J., Borg A., Gradin K., Poellinger L., and Påhlman S. (2006) Recruitment of HIF-1α and HIF-2α to common target genes is differentially regulated in neuroblastoma: HIF-2α promotes an aggressive phenotype. Cancer Cell 10, 413–423 [DOI] [PubMed] [Google Scholar]
- 24. Uniacke J., Perera J. K., Lachance G., Francisco C. B., and Lee S. (2014) Cancer cells exploit eIF4E2-directed synthesis of hypoxia response proteins to drive tumor progression. Cancer Res. 74, 1379–1389 [DOI] [PubMed] [Google Scholar]
- 25. Newby D., Marks L., and Lyall F. (2005) Dissolved oxygen concentration in culture medium: assumptions and pitfalls. Placenta 26, 353–357 [DOI] [PubMed] [Google Scholar]
- 26. Gray N. K., Coller J. M., Dickson K. S., and Wickens M. (2000) Multiple portions of poly(A)-binding protein stimulate translation in vivo. EMBO J. 19, 4723–4733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Thoreen C. C., Chantranupong L., Keys H. R., Wang T., Gray N. S., and Sabatini D. M. (2012) A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 485, 109–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zuberek J., Kubacka D., Jablonowska A., Jemielity J., Stepinski J., Sonenberg N., and Darzynkiewicz E. (2007) Weak binding affinity of human 4EHP for mRNA cap analogs. RNA 13, 691–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shatsky I. N., Dmitriev S. E., Andreev D. E., and Terenin I. M. (2014) Transcriptome-wide studies uncover the diversity of modes of mRNA recruitment to eukaryotic ribosomes. Crit. Rev. Biochem. Mol. Biol. 49, 164–177 [DOI] [PubMed] [Google Scholar]
- 30. Ho J. J., Wang M., Audas T. E., Kwon D., Carlsson S. K., Timpano S., Evagelou S. L., Brothers S., Gonzalgo M. L., Krieger J. R., Chen S., Uniacke J., and Lee S. (2016) Systemic reprogramming of translation efficiencies on oxygen stimulus. Cell Rep. 14, 1293–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Joshi B., Cameron A., and Jagus R. (2004) Characterization of mammalian eIF4E-family members. Eur. J. Biochem. 271, 2189–2203 [DOI] [PubMed] [Google Scholar]
- 32. Okumura F., Zou W., and Zhang D. E. (2007) ISG15 modification of the eIF4E cognate 4EHP enhances cap structure-binding activity of 4EHP. Genes Dev. 21, 255–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rouschop K. M., Dubois L., Schaaf M. B., van den Beucken T., Lieuwes N., Keulers T. G., Savelkouls K. G., Bussink J., van der Kogel A. J., Koritzinsky M., and Wouters B. G. (2011) Deregulation of cap-dependent mRNA translation increases tumour radiosensitivity through reduction of the hypoxic fraction. Radiother. Oncol. 99, 385–391 [DOI] [PubMed] [Google Scholar]
- 34. Rankin E. B., Rha J., Unger T. L., Wu C. H., Shutt H. P., Johnson R. S., Simon M. C., Keith B., and Haase V. H. (2008) Hypoxia-inducible factor-2 regulates vascular tumorigenesis in mice. Oncogene 27, 5354–5358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Heinrich M. C., Corless C. L., Duensing A., McGreevey L., Chen C. J., Joseph N., Singer S., Griffith D. J., Haley A., Town A., Demetri G. D., Fletcher C. D., and Fletcher J. A. (2003) PDGFRA activating mutations in gastrointestinal stromal tumors. Science 299, 708–710 [DOI] [PubMed] [Google Scholar]
- 36. Yi T., Papadopoulos E., Hagner P. R., and Wagner G. (2013) Hypoxia-inducible factor-1α (HIF-1α) promotes cap-dependent translation of selective mRNAs through up-regulating initiation factor eIF4E1 in breast cancer cells under hypoxia conditions. J. Biol. Chem. 288, 18732–18742 [DOI] [PMC free article] [PubMed] [Google Scholar]