Abstract
MSH2 is a key DNA mismatch repair protein, which plays an important role in genomic stability. In addition to its DNA repair function, MSH2 serves as a sensor for DNA base analogs-provoked DNA replication errors and binds to various DNA damage-induced adducts to trigger cell cycle arrest or apoptosis. Loss or depletion of MSH2 from cells renders resistance to certain DNA-damaging agents. Therefore, the level of MSH2 determines DNA damage response. Previous studies showed that the level of MSH2 protein is modulated by the ubiquitin-proteasome pathway, and histone deacetylase 6 (HDAC6) serves as an ubiquitin E3 ligase. However, the deubiquitinating enzymes, which regulate MSH2 remain unknown. Here we report that ubiquitin-specific peptidase 10 (USP10) interacts with and stabilizes MSH2. USP10 deubiquitinates MSH2 in vitro and in vivo. Moreover, the protein level of MSH2 is positively correlated with the USP10 protein level in a panel of lung cancer cell lines. Knockdown of USP10 in lung cancer cells exhibits increased cell survival and decreased apoptosis upon the treatment of DNA-methylating agent N-methyl-N′-nitro-N-nitrosoguanidine (MNNG) and antimetabolite 6-thioguanine (6-TG). The above phenotypes can be rescued by ectopic expression of MSH2. In addition, knockdown of MSH2 decreases the cellular mismatch repair activity. Overall, our results suggest a novel USP10-MSH2 pathway regulating DNA damage response and DNA mismatch repair.
Keywords: deubiquitylation (deubiquitination), DNA damage, DNA damage response, DNA repair, ubiquitin
Introduction
Mismatch repair (MMR)3 is a mutation avoidance mechanism that corrects DNA replication errors, and controls homologous recombination (HR) by aborting strand exchange between divergent DNA sequences (1). The MMR activity begins with mismatch recognition, which is carried out either by MutSα (MSH2-MSH6) or by MutSβ (MSH2-MSH3). MutSα recognizes DNA single base pair mismatches and small deletions/insertions, whereas MutSβ recognizes large deletions/insertions (2, 3). MutSα or MutSβ then recruits MutLα (MLH1-PMS2), proliferation cellular nuclear antigen (PCNA), and replication protein A (RPA) to form a complex leading to the recruitment of exonuclease 1 (EXO1) to the strand break. EXO1 then cut the nascent DNA from the nick forward and beyond the mismatch to generate a single strand gap, which is filled by polymerases δ using the parental DNA strand as a template. The repair is accomplished by filling the nick using DNA ligase I. Deletion or mutation of key DNA mismatch repair proteins, such as MSH2 and MLH1, can cause genomic instability (4).
Ιn addition to recognizing DNA mismatches MutSα can recognize certain DNA-damaging agents, such as 6-thioguanine (6-TG)-, N-methyl-N′-nitro-N-nitrosoguanidine (MNNG)-, and cisplatin-induced DNA adducts to trigger apoptosis (5). Thus, the level of MutSα controls cellular sensitivity to DNA damage. It has been reported that the level of MSH2 can be modulated by multiple means. First, when cells are treated with ultraviolet (UV) or phorbol ester (TPA), the mRNA level of MSH2 will be increased (6, 7). Second, MSH2 protein is more stable as a heterodimer with MSH6 than it exists as a monomer (8). Third, protein kinase C (PKC) phosphorylates MutSα and protects it from proteasome-dependent degradation (9). Fourth, histone deacetylase 6 (HDAC6) was recently identified as an E3 ligase of MSH2 to promote its degradation (8). However, the deubiquitinating enzyme (DUB), which counteracts HDAC6 to stabilize MSH2, has remained unknown.
Currently about 100 DUBs have been identified in the human genome and are classified into five families based on their sequence similarity and mechanism of action (10–14). They are 1) the ubiquitin C-terminal hydrolases (UCHs), 2) the ubiquitin-specific proteases/ubiquitin-specific processing proteases (USPs/UBPs), 3) the ovarian tumor proteases (OTUs), 4) the Josephin or Machado-Joseph disease protein domain proteases (MJDs), and 5) the Jab1/MPN domain-associated metalloisopeptidase (JAMM) domain proteins. The first four families are cysteine peptidases, while the last one comprises of zinc metalloisopeptidases.
USP10 belongs to the USP family and is identified as a new regulator of p53 in DNA damage response and tumor development (15–17). Yuan et al. found that phosphorylation of USP10 on Thr-42 and Ser-337 by ATM leads to USP10 translocation to the nucleus to stabilize p53 (15). The other USP10-interacting proteins include Ras GTPase-activating protein-binding protein 1 (G3BP1), cystic fibrosis transmembrane conductance regulator (CFTR), and a histone H2A variant, H2A.Z (18–21). Among these USP10 partners, G3BP1 binding to USP10 may block USP10's enzyme activity. CFTR and H2A.Z are substrates of USP10. USP10 deubiquitinates CFTR to regulate CFTR's post-endocytic sorting, while USP10 deubiquitinates H2A.Z to transcriptionally activate androgen receptor (AR)-regulated PSA and KLK3 genes (21).
Here, we report MSH2 as a new USP10-interacting protein and a new USP10 substrate. We also reveal a novel USP10-MSH2 pathway regulating MSH2 homeostasis, DNA damage response, and DNA MMR.
Experimental Procedures
Cell Culture and Transfection
All cell lines were grown in Dulbecco's modified Eagle's Medium (DMEM) with 10% fetal bovine serum, penicillin (100 U/ml), and streptomycin (100 g/ml), except H1299, which was grown in RPMI 1640 medium. Cells were incubated at 37 °C with 5% CO2. The plasmids were transiently or stably transfected into cells, except MEFs, using Lipofectamine 2000 (Invitrogen). The plasmids were transfected into MEFs by electroporation with a Nucleofector™ device (Lonza).
Plasmids, Antibodies, and Chemicals
The expression plasmids of F-USP10 and His-USP10 were described in Yuan et al. (15). F-USP10 (C424A) was generated using F-USP10 as a template. The F-HA-USP10 plasmid was purchased from Addgene (#22543). The HA-USP10 plasmid was constructed by inserting the USP10 full-length cDNA into the pCMV-HA-N vector (Clontech) between the XhoI and NotI sites. The full-length USP10 cDNA was the PCR product using F-USP10 as the template. Fragments of USP10-(1–600) and USP10-(601–798) were cloned into a pCMV-3XFlag vector (Sigma) to generate F-USP10-(1–600) and F-USP10-(601–798), respectively. The HA-MSH2 construct was described in Zhang et al. (8). MSH2 cDNA was cloned into the pcDNA4v-His vector (Addgene) to generate His-MSH2. MSH2 full-length and MSH2 fragments were cloned into the pGEX vector to generate GST-MSH2, GST-MSH2-(1–378), GST-MSH2-(200–700), and GST-MSH2-(624–934). Human USP10 shRNAs (RHS4533-NM_005153) in the pGIPZ lentiviral vector were purchased from Open Biosystems. The shUSP10–1 (TRCN0000007430) targets USP10 sequence: GCCTCTCTTTAGTGGCTCTTT; the shUSP10–2 (TRCN0000007431) targets sequence: CCTATGTGGAAACTAAGTA.
The anti-USP10 antibody (ab72486) was purchased from Abcam. The anti-MSH2 antibody was purchased from Calbiochem. The anti-HA antibody was purchased from Covance. The anti-Flag M2 antibody and agarose beads, the anti-β-actin antibody, MG132, cycloheximide, 6-TG, imidazole, urea, guanidine-HCL, ATP, and MTT were purchased from Sigma. MNNG was purchased from Pfaltz &Bauer, Inc. Ni-NTA resin was purchased from Clontech. Rabbit reticulocyte lysate was purchased from Promega. HA-UB was purchased from Boston Biochem.
MNNG and 6-TG Treatment
MNNG was diluted in water immediately before use. Cells were treated with MNNG in FBS-free medium. Cells were then washed with medium and incubated in fresh medium at 37 °C for different time periods as indicated in the figures. 6-TG was first dissolved in DMSO to make a stock solution and then dissolved in medium directly before use.
Immunoprecipitation and Immunoblotting
For immunoprecipitations, cells were lysed in the LS buffer (PBS, pH 7.5, 10% glycerol, 0.1% Nonidet P-40, protease inhibitor mixture). Lysates were incubated with protein A- or protein G-agarose for 2 h for pre-clearing prior to incubation with the indicated primary antibodies for 12 h at 4 °C. Immunocomplexes were collected, washed four times in TBST buffer (0.1% Tween-20 in TBS), and resolved by SDS-PAGE. For immunoblotting, samples were transferred to nitrocellulose membranes then probed with the indicated antibodies. Bound antibodies were detected using a Chemiluminescent Detection Kit (Pierce).
Establishment of USP10-knockdown Stable Clones
For Figs. 2B and 3B, the pGIPZ vector containing shUSP10–2 was transfected into A549 and H1299. One day after transfection, cells were split into 3 dishes. After 24 h, 1 μg/ml puromycin was added into the medium to select positive cells. Ten days later, stable cell lines were subcloned to 60-mm dishes, 0.5 μg/ml puromycin was added to the medium to maintain the stable clones in the subsequent culture.
FIGURE 2.
USP10 regulates the protein level and half-life of MSH2. A, USP10 up-regulates the protein level of MSH2 in 293T cells and MEFs. 293T cells and MEFs were transfected with indicated constructs and the Western blotting analyses were performed by indicated antibodies. B, knockdown of USP10 decreases the protein level of MSH2 in A549 and H1299 cells. A549 and H1299 cells were stably transfected with empty vector or vector containing USP10 shRNA (shUSP10–2). The cells were lysed, and the anti-MSH2, anti-USP10, and anti-β-actin Western blotting analyses were performed as indicated. C, knockdown of USP10 shortens MSH2 half-life in A549 cells. A549 cells transduced with lentiviruses containing empty vector or USP10 shRNA (shUSP10–2) were treated with 20 μg/ml cycloheximide (CHX) with the indicated times followed by Western blotting analyses with indicated antibodies. The MSH2 and USP10 bands were quantified by densitometry. The level of MSH2 and USP10 at time point 0 was designated as 1. The levels of USP10 and MSH2 at other times were quantified, relative to the time point zero, and were shown in fold changes below the panels of MSH2 and USP10. D, a graph of the mean band intensities from three independent experiments as measured by Image-Pro Plus 6.0 shows the approximate half-lives in the presence of CHX. The error bars represent the standard deviation. E, Spautin-1 decreases the level of MSH2 but not USP10. A549 cells were treated with 10 μm Spautin-1 at indicated times. Cells were lysed, and the anti-MSH2, anti-USP10, and anti-β-actin Western blotting analyses were performed. The MSH2 and USP10 bands were quantified as in C. F, expression of USP10 and MSH2 in a panel of lung cancer cell lines. The indicated cell lines, H23, H2122, A549, H1975, H292, H441, H157, H1650, and H661, were lysed, and the anti-USP10, anti-MSH2, and anti-β-actin Western blotting analyses were performed. G, MSH2 expression is positively correlated with the USP10 expression in a panel of lung cancer cells. The bands of USP10 and MSH2 in 2F were quantified by densitometry as 2C. The correlation analysis was performed by the SPSS software, and the results were shown as a scatter plot indicating the correlation of MSH2 and USP10 expression in these cell lines. Pearson correlation (0.85) and p value (0.004) were indicated.
FIGURE 3.
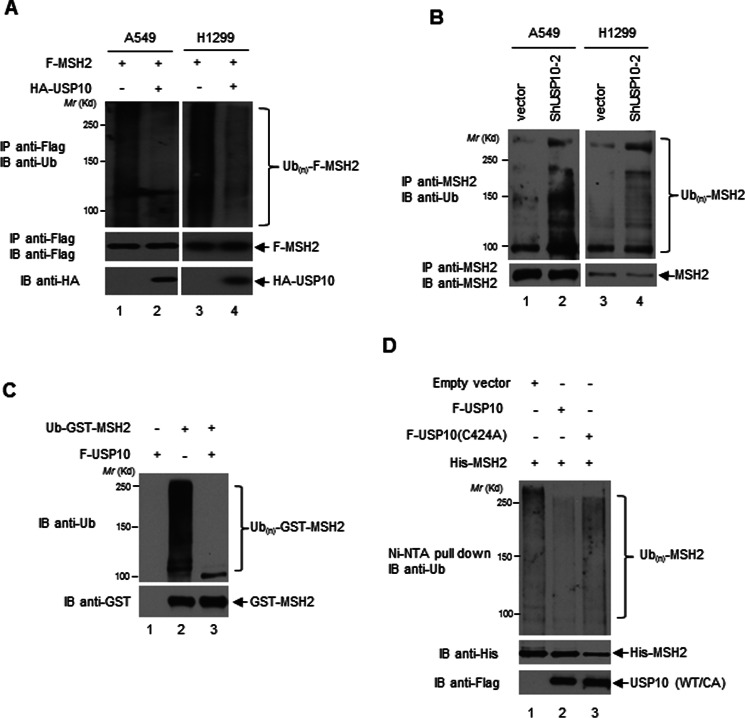
USP10 deubiquitinates MSH2. A, USP10 deubiquitinates MSH2 in vivo. A549 and H1299 cells were transfected with the indicated plasmids. Twelve hours prior to harvest, 10 μm MG132 was added to the cells. F-MSH2 was IP-ed by anti-Flag antibodies. The immunoprecipitates were then subjected to the anti-Ub Western blotting analysis (upper panels). The blot was stripped and reprobed with anti-Flag antibodies (middle panels). The expression of HA-USP10 was detected by the anti-HA Western blotting analysis (lower panels). B, knockdown of USP10 increases MSH2 ubiquitination in vivo. USP10 was stably transfected by vector control or shRNA against USP10 (shUSP10–2) in A549 and H1299. The cells were treated overnight with 25 μm MG132 prior to harvest. MSH2 was IP-ed by anti-MSH2 antibodies. The immunoprecipitates were then subjected to an anti-Ub Western blotting analysis (upper panels). Ten percent of the above IP-ed samples was subjected to the anti-MSH2 Western blotting analysis for determining IP efficiencies (lower panels). C, USP10 deubiquitinates ubiquitinated MSH2 in vitro. The Ub-GST-MSH2 protein was prepared as described in the “Experimental Procedures.” Ub-GST-MSH2 was incubated in the absence or presence of F-USP10 protein purified from 293T cells in the deubiquitination buffer (50 mm Tris-HCl, pH 8.0, 50 mm NaCl, 1 mm EDTA, 10 mm DTT, 5% glycerol) at 37 °C for 1 h. The reaction was resolved on SDS-PAGE followed by an anti-Ub Western analysis (upper panel). The amount of GST-MSH2 was examined by an anti-GST Western blotting analysis (lower panel). D, USP10-WT, but not USP10-(C424A) mutant, deubiquitinates MSH2 in vivo. 293T cells were transfected with indicated constructs. His-MSH2 was pulled down with Ni-NTA-agarose beads under denaturing conditions followed by anti-Ub Western blotting analysis (upper panel). The expression of His-MSH2 and USP10-WT and the USP10-CA mutant was examined by anti-His (middle panel) and anti-Flag (bottom panel) Western blotting analyses.
Establishment of the USP10-knockdown A549 Pool
Addgene's protocol was followed to produce lentiviral particles and infect A549 cells. Briefly, pGIPZ vector containing shUSP10–1 or shUSP10–2 was co-transfected with psPAX2 and pMD2.G into HEK-293T cells. Lentiviral particles in the HEK-293T cell medium were harvested 24 and 36 h after transfection. Then, the lentiviral particle solution was added into A549 cells. About 24 h after infection, puromycin was added to the medium at the concentration of 1 μg/ml for A549 cells. After a 2-week selection, Western blot analyses were used to determine the knockdown efficiency.
GST Pull-down Assay
GST fusion proteins were purified as previously described (22). For in vitro binding assays, glutathione Sepharose-bound GST-MSH2 proteins were incubated with cell lysates. After washing extensively with PBST (0.1% Tween 20 in PBS), the proteins bound to GST-MSH2 were resolved by SDS-PAGE and immunoblotted with indicated antibodies.
In Vitro Ubiquitination and Deubiquitination Assays
The in vitro ubiquitination assay is described in our previous report (23). Briefly, glutathione-Sepharose-bound GST-MSH2 was incubated with 5 mm ATP, 200 μm HA-Ub, and rabbit reticulocyte lysate (RRL) in ubiquitination buffer (50 mm Tris-HCl, pH 7.5, 100 mm NaCl, 2.5 mm MgCl2, and 1 mm DTT) for 2 h at 37 °C. Beads were washed with PBST 3 times followed by the deubiquitination buffer (50 mm Tris-HCl, pH 8.0, 50 mm NaCl, 1 mm EDTA, 10 mm DTT, 5% glycerol) once. Ubiquitinated GST-MSH2 as a substrate was incubated with purified F-USP10 from 293T cells in deubiquitination buffer at 37 °C for 1 h. The reaction was stopped by SDS-loading buffer followed by immunoblotting.
In Vivo Ubiquitination Assay
His-MSH2 was co-transfected with empty vector, F-USP10 or F-USP10 (C424A) into 293T cells. Thirty-six hours post-transfection, cells were harvested. Cell pellets were lysed in Buffer A (6 m guanidine-HCl, 0.1 m Na2HPO4/NaH2PO4, 0.01 m Tris-HCl, pH 8.0, 10 mm imidazole, 10 mm β-mecaptoethanol) and 30 μl of Ni-NTA agarose beads were added, and the mixture was rotated at room temperature for 12 h. The beads were sequentially washed with Buffer A, Buffer B (8 μm urea, 0.1 m Na2HPO4/NaH2PO4, 0.01 m Tris-HCl, pH 8.0, 10 mm β-mercaptoethanol), Buffer C (8 m urea, 0.1 m Na2HPO4/NaH2PO4, 0.01 m Tris-HCl, pH 6.3, 10 mm β-mercaptoethanol), and Buffer C plus 0.1% Tween-20, then resolved on SDS-PAGE followed by the anti-Ub Western blotting analysis.
Whole Cell Extract Preparation
Whole cell extracts were prepared from 6 × 107 cells as described (24). Cells were first washed with buffer A (20 mm Hepes pH 7.5, 5 mm KCl, 0.5 mm MgCl2, 0.1% PMSF, 0.5 mm DTT, 1 μg/ml leupeptin, and 0.2 m sucrose) and lysed in the same buffer without 0.2 m sucrose by passage through a 27-gauge needle. Proteins were precipitated by 65% of ammonium sulfate, collected by centrifugation, resuspended in lysis buffer, followed by dialysis to equilibrium in a buffer containing 20 mm Hepes (pH 7.5), 5 mm KCl, 0.1 mm EDTA, 0.1% PMSF, 0.5 mm DTT, and 1 μg/ml leupeptin.
Heteroduplex Preparation and MMR Assay
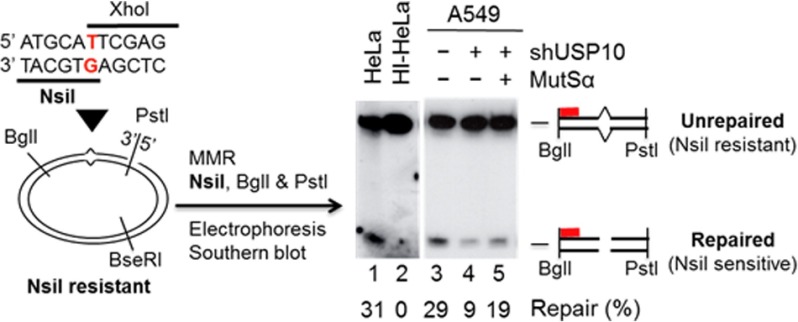
DNA substrates used in this study are circular heteroduplex DNA containing a unique G-T mismatch and a strand break 3′ to the mismatch (Fig. 6). The substrate was prepared from M13mp18-UKY phase series as described previously (25). MMR assays were performed in 20-μl reactions containing 100 ng heteroduplex DNA, 75 μg of whole cell extracts, 10 mm Tris-HCl (pH 7.5), 5 mm MgCl2, 1.5 mm ATP, 0.1 mm dNTPs, 1 mm glutathione and 110 mm KCl at 37 °C for 15 min, as described (24) in the presence or absence of purified human recombinant MutSα (26). Reactions were terminated by addition of Proteinase K. DNA samples were extracted with phenol and recovered by ethanol precipitation. Repair products were digested with PstI, NsiI (repair-scoring enzyme), and BglI, fractionated by polyacrylamide gel electrophoresis, and detected by Southern blot hybridization with a 32P-labeled probe. DNA products were visualized by phosphorimager.
FIGURE 6.
The USP10-knockdown A549 cells display a reduced MMR activity compared with the control cells. DNA substrate (left) used in this study contains a G-T mismatch (red) placed into two overlapping restriction endonucleases and a strand break 172-bp 3′ to the mismatch. Restriction enzymes used for scoring the repair are shown. MMR activity of A549 cells with or without shUSP10 was determined by incubating the circular DNA heteroduplex with 75 μg of whole cell extracts. Repair products were identified by Southern blot analysis using a 32P-labeled oligonucleotide probe (red bar) complementary to the nicked strand at the indicated location. MMR activities in HeLa extracts and heat-inactivated HeLa extracts (HI-HeLa) were used as a positive control and a negative control, respectively. The repaired bands were quantified by densitometry.
Results
USP10 Interacts with MSH2
We previously identified HDAC6 as an ubiquitin E3 ligase of MSH2 (8). To identify novel MSH2-interacting proteins associated with the ubiquitin-proteasome pathway, we overexpressed HA-MSH2 in 293T cells for 48 h followed by treatment with 50 μm MG132 for 4 h. The HA-MSH2 protein was immunoprecipitated (IP-ed) with anti-HA agarose beads, and the unique bands existing in the anti-HA, but not in the control anti-IgG, immunoprecipitate were excised and analyzed by the liquid chromatography-tandem mass spectrometry (LC-MS/MS). The mass spectrometry analysis identified 14 peptide sequences of USP10, a ubiquitin-specific peptidase (data not shown), suggesting that USP10 exists in the anti-MSH2 immunoprecipitate. Reciprocally, endogenous USP10 was IP-ed by anti-USP10 antibodies in HeLa S3 cells. The immunoprecipitate was resolved by SDS-PAGE, and the bands were excised and examined by LC-MS/MS. MSH2 was identified in the USP10 immunoprecipitate (data not shown), suggesting that USP10 is associated with MSH2.
We next confirmed the interaction between USP10 and MSH2 by performing co-IP assays with anti-USP10 antibodies using 293T and mouse embryonic fibroblasts (MEFs) cell extracts. As shown in Fig. 1A, MSH2 was only detected in the anti-USP10 immunoprecipitates (lanes 3 and 6), but not in the anti-IgG controls (lanes 2 and 5). In a reciprocal fashion, the anti-MSH2 antibody, but not anti-IgG, specifically IP-ed USP10 in HeLa and MEFs (Fig. 1B). Therefore, USP10 and MSH2 indeed interact with each other in vivo.
FIGURE 1.
USP10 interacts with MSH2. A and B, endogenous USP10 and MSH2 interact with each other. A, 293T cells and MEFs were lysed and immunoprecipitated (IP-ed) with either anti-IgG or anti-USP10 antibodies followed by an anti-MSH2 Western blotting analysis (upper panel). The blot was stripped and reprobed with anti-USP10 antibodies (lower panel). B, HeLa cells and MEFs were lysed and IP-ed with either anti-IgG or anti-MSH2 antibodies followed by an anti-USP10 Western blotting analysis (upper panel). The blot was stripped and reprobed with anti-MSH2 antibodies (lower panel). C, diagrams of the domain structure of MSH2 and the deletion constructs of MSH2. D, USP10 physically interacts with MSH2, and the N terminus of MSH2 binds to USP10. Escherichia coli lysates harboring His-USP10 were incubated with bacterially-purified GST, GST-MSH2, GST-MSH2-(1–378), GST-MSH2-(200–700), or GST-MSH2-(624–934). GST pull-down was performed followed by an anti-His Western blotting analysis (upper panel). Protein expression of GST, GST-MSH2, GST-MSH2-(1–378), GST-MSH2-(200–700), and GST-MSH2-(624–934) was shown by Coomassie Blue staining (lower panel). E, diagrams of USP10 domain structure and USP10 deletion constructs. The numbers indicate the amino acids. UCH stands for the ubiquitin C-terminal hydrolase domain. F, C terminus of USP10 binds MSH2. Both wild type and catalytically-dead mutant of USP10 bind MSH2. Bacterially purified GST-MSH2 was incubated with 293T cell lysates transfected with Flag-empty vector (F-vector) or indicated USP10 constructs. The GST pull-down analyses were performed followed by an anti-Flag Western blotting analysis (upper panel). The input of Flag-USP10 deletion proteins and wild type (WT) and catalytically-dead mutant (C424A) of USP10 proteins was detected by an anti-Flag Western blotting analysis (middle panel). The amount of GST-MSH2 used for GST pull-down assays was examined by an anti-GST Western blotting analysis (lower panel).
To determine whether USP10 directly interacts with MSH2 or through other associated proteins, the in vitro GST pull-down assay was carried out. As shown in Fig. 1D, GST-MSH2, but not GST, was able to pull down His-USP10 (lanes 1 and 2). This result strongly indicates a direct interaction between USP10 and MSH2.
We next attempted to map which region of MSH2 binds to USP10. MSH2 can be divided into five domains according to its crystal structure, namely mismatch binding, connector, level, clamp, and ATPase (Fig. 1C) (27). As shown in Fig. 1D, the N terminus of MSH2 (1–378), but not the middle region (200–700) or C terminus of MSH2-(624–934), binds to USP10 directly (lanes 3, 4, and 5). Thus, the major region responsible for the interaction with USP10 is located in the mismatch binding and connector domains of MSH2.
We then examined which region of USP10 binds to MSH2. Human USP10 contains an ubiquitin C-terminal hydrolase domain (412–792) for its deubiquitination activities (Fig. 1E). The N terminus of USP10 has been reported to interact with p53 (15) and G3BP (18). By contrast, as shown in Fig. 1F, the C terminus of USP10 interacts with MSH2 (lane 3).
We also examined whether USP10's enzymatic activity influences its binding to MSH2. As shown in Fig. 1F, the USP10 catalytically dead mutant (18), USP10 (C424A), interacted with MSH2 to the same extent as the wild type did (lanes 4 and 5), suggesting that the interaction between USP10 and MSH2 is independent of USP10 enzymatic activity.
USP10 Stabilizes MSH2
We previously showed that HDAC6 serves as an ubiquitin E3 ligase to promote MSH2's degradation (8). Here we explored whether USP10 can counteract HDAC6 to stabilize MSH2. As shown in Fig. 2A, in 293T cells, the level of HA-MSH2 increased significantly by overexpressing USP10; in MEFs, the level of endogenous MSH2 increased dramatically by overexpressing of USP10, as well. To verify whether USP10 regulates the level of MSH2, we knocked down USP10 by shRNA against USP10 in A549 and H1299 cells. As shown in Fig. 2B, the level of MSH2 was significantly decreased in the USP10 knockdown A549 and H1299 cells compared with that of MSH2 in the control cells. We then examined whether USP10 influences the stability of MSH2 by measuring the half-life of MSH2. As shown in Fig. 2, C and D, MSH2 half-life in the USP10-knockdown A549 cells is around 6 h, while MSH2 half-life in control cells is more than 12 h, suggesting that USP10 can prolong MSH2's half-life. We next used a USP10 inhibitor, known as specific and potent autophagy inhibitior-1 (Spautin-1) to treat A549 cells. As shown in Fig. 2E, Spautin-1 was able to decrease the level of MSH2, but not USP10, in a time-dependent manner, suggesting that suppressing the deubiquitinating enzyme activity of USP10 reduces the protein level of MSH2.
We next surveyed a panel of lung cancer cell lines to determine whether there is a positive correlation between USP10 and MSH2 in these cell lines. As shown in Fig. 2, F and G, the expression of MSH2 is positively correlated to the expression of USP10, indicating that USP10 stabilizes MSH2 in lung cancer cells.
USP10 Deubiquitinates MSH2
We then determined whether MSH2 is a substrate of USP10. As shown in Fig. 3A, overexpression of USP10 reduced the ubiquitination of F-MSH2 in A549 and H1299 cells. Conversely, knockdown of USP10 significantly increased the ubiquitination of MSH2 in A549 and H1299 cell lines (Fig. 3B).
To directly examine the deubiquitination activity of USP10 toward MSH2, we utilized a cell-free system. We prepared ubiquitinated GST-MSH2 (Ub-GST-MSH2) as described in “Experimental Procedures” and then performed the deubiquitination assay. As shown in Fig. 3C, F-USP10 efficiently deubiquitinated MSH2 in vitro (compare lanes 2 and 3).
We then tested whether USP10 is able to deubiquitinate MSH2 in vivo. As shown in Fig. 3D, USP10 WT, but not USP10 (C424A), reduced His-MSH2 ubiquitination. To ensure that the ubiquitination signal was indeed from His-MSH2 and not from its associated proteins, His-MSH2 was washed under the denaturing conditions and the ubiquitination status was examined by the anti-Ub Western blotting analysis. Overall, we demonstrated that MSH2 is a substrate of USP10.
Depletion of USP10 Decreases Cellular Sensitivity to MNNG and 6-TG and Decreases DNA Mismatch Repair Activities in A549 Cells
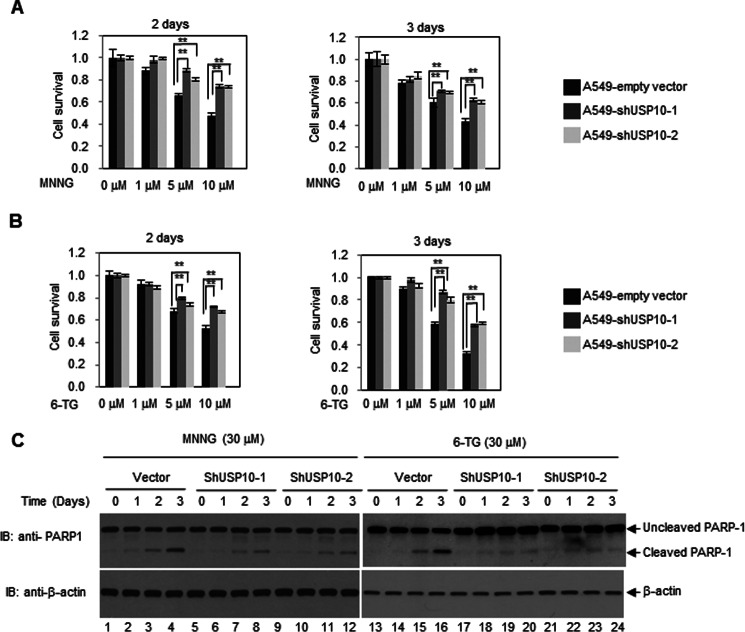
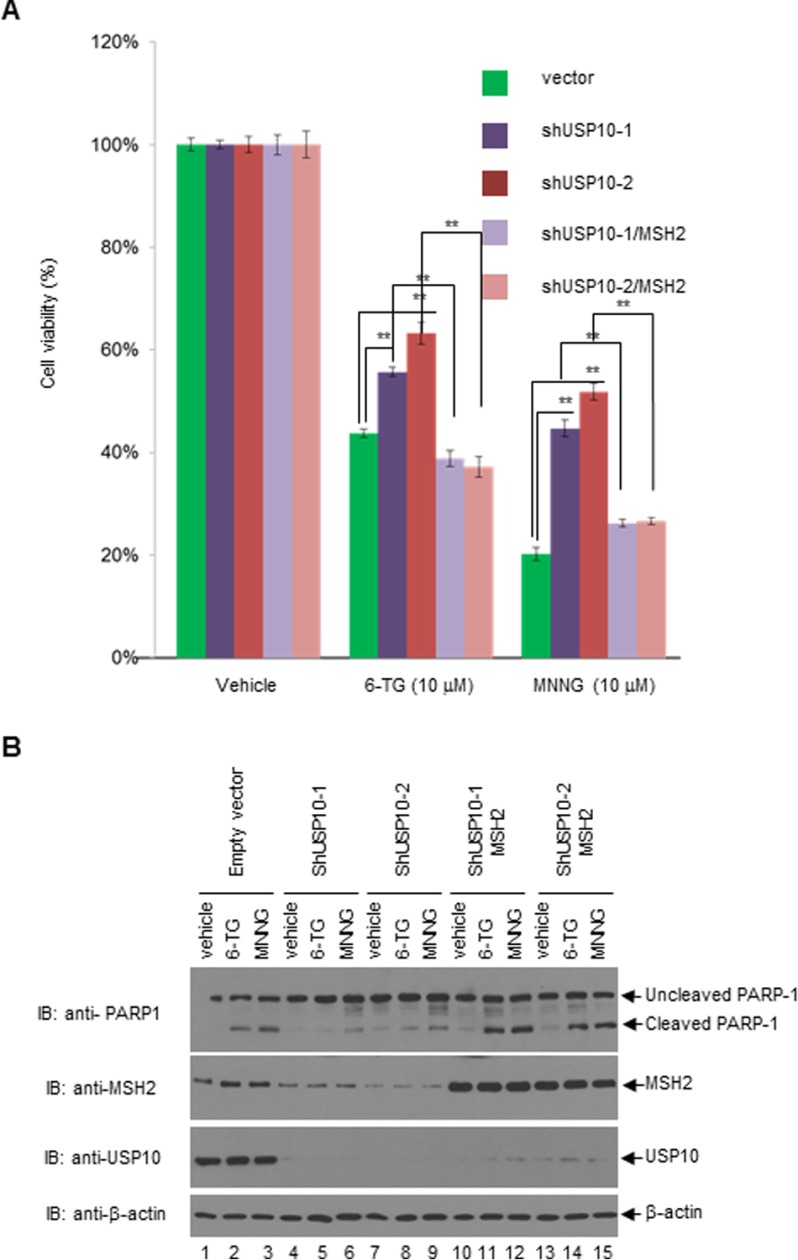
The MSH2-MSH6 heterodimers recognize DNA-damaging agents, such as 6-TG, MNNG, cisplatin, carboplatin, doxorubicin, and etoposide, which form DNA adducts that cannot be removed by MMR (5). It has been well documented that the levels of MSH2 are inversely correlated with 6-TG or MNNG resistance (5). Because USP10 is able to up-regulate the level of MSH2, we set out to test whether USP10 plays a role in regulating MNNG- or 6-TG-mediated cell killing. We depleted the expression of USP10 using two different shRNAs (shUSP10–1 and shUSP10–2) in A549 cells and measured the cytotoxicity of MNNG and 6-TG by MTT assays. As shown in Fig. 4, A and B, knockdown of USP10 using both shUSP10–1 and shUSP10–2 in A549 cells significantly increased cell survival compared with the control cells. To determine whether this increased survival is due to the decrease of apoptosis, we detected the cleavage of poly ADP-ribose polymerase 1 (PARP-1) by Western blotting analyses. As shown in Fig. 1C, knockdown of USP10 with either shUSP10–1 or shUSP10–2 decreased the PARP-1 cleavage upon MNNG and 6-TG treatment compared with the control. To ensure that the increased cell survival is due to the reduction of MSH2, HA-MSH2 was introduced to the A549 pools transduced by shUSP10–1 or shUSP10–2 lentiviruses, and the resulting cells displayed decreased cell survival and increased apoptosis upon 6-TG or MNNG treatment (Fig. 5). Therefore, our data suggest a USP10-MSH2 pathway governing cellular sensitivity to 6-TG and MNNG. In addition, we explored whether depletion of USP10 affects DNA MMR activities. As shown in Fig. 6, knockdown of USP10 in A549 cells significantly reduces the MMR activities, while the addition of the MutSα complex partially restores MMR activity. This result indicates that USP10 regulates the cellular MMR activities via modulating the level of MSH2.
FIGURE 4.
Depletion of USP10 in A549 cells confers MNNG and 6-TG resistance. A, knockdown of USP10 increases cell viability to MNNG. A549 cells transduced with lentiviruses containing empty vectors, shUSP10–1 or shUSP10–2 were treated with MNNG of the indicated concentration for 2 days and 3 days. MTT assays were performed. B, knockdown of USP10 increases cell viability to 6-TG. A549 cells transduced with lentiviruses containing empty vectors, shUSP10–1 or shUSP10–2 were treated with 6-TG. MTT assays were performed. C, knockdown of USP10 reduces apoptosis upon treatment of MNNG and 6-TG. A549 cells transduced with lentiviruses containing empty vectors, shUSP10–1 or shUSP10–2 were treated with MNNG or 6-TG at indicated times and concentrations. Cells were lysed, and the anti-PARP1 and anti-β-actin Western blotting analyses were performed. For A and B, the error bars stand for a standard error (S.E.). **, denotes p < 0.001 by a two-tailed Student's t test (n = 6).
FIGURE 5.
Overexpression of MSH2 in USP10-knockdown A549 cells restores the 6-TG- and MNNG-mediated growth inhibition and apoptosis. A, A549 stable pools were transduced with vectors, shUSP10–1 and shUSP10–2, and the latter two pools were transfected with HA-MSH2. Then the above five groups of cells were treated with vehicle, 10 μm 6-TG or 10 μm MNNG for 3 days. MTT assays were performed. B, five groups of cells as shown in 5A were treated with vehicle, 30 μm 6-TG, or 30 μm MNNG for 3 days. Cells were then harvested and lysed. The anti-PARP1, anti-MSH2, anti-USP10, and anti-β-actin Western blotting analyses were performed. For A, the error bars stand for S.E. **, denotes p < 0.001 by a two-tailed Student's t test (n = 6).
Discussion
In this study, we have identified a novel MSH2-interacting protein, USP10, which stabilizes and deubiquitinates MSH2 in vitro and in vivo. The USP10-MSH2 axis regulates the MSH2 and MutSα homeostasis and cellular sensitivity to DNA damage. We previously demonstrated an unexpected E3 ligase activity of HDAC6, which regulates MSH2 proteasome-dependent degradation (8). We have now identified a deubiquitinating enzyme USP10, which counteracts HDAC6's activity. However, how HDAC6 and USP10 work in concert to regulate MSH2 stability is not clear. Based on our domain mapping data, HDAC6 and USP10 bind to different domains of MSH2. HDAC6 binds to MSH2 C-terminal region (8), while USP10 binds to MSH2 N-terminal region. Therefore, HDAC6 and USP10 may not compete with each other to bind to MSH2.
Unlike one of USP10 substrates, p53, which has a very short half-life, MSH2 has a much longer half-life. We suspect that USP10-mediated deubiquitination of MSH2 may partially account for MSH2 stability. We previously identified four C-terminal lysines (Lys-845, Lys-847, Lys-871, and Lys-892) in MSH2, which can be either acetylated or ubiquitinated (8). We proposed that HDAC6 sequentially deacetylates at these four sites to dissemble the MSH2-MSH6 heterodimers and ubiquitinates the monomer of MSH2. Thus, USP10 may promote deubiquitination of the MSH2 monomer to facilitate MSH2 acetylation and the MSH2-MSH6 dimer formation. We are currently testing this hypothesis in the laboratory.
We also found that USP10 interacts with MSH6 in vivo by the immunoprecipitation assay (data not shown). However, we failed to show that USP10 physically interacts with MSH6. So it is possible that USP10 influences MSH6's function through a direct interaction with MSH2.
We previously showed that upon 6-TG and MNNG treatment, the level of MSH2 ubiquitination was decreased (data not shown and Ref. 8). Thus, it is likely that USP10 is more activated under the 6-TG or MNNG treatment and deubiquitinates MSH2 more efficiently. Yuan et al., reported that under stress conditions, such as ionizing radiation (IR), USP10 can be phosphorylated by ATM and translocate to the nucleus to stabilize p53 (15). We recently showed that USP10 phosphorylation was increased upon MNNG treatment. A MAPK/CDKs family kinase might be responsible for USP10 phosphorylation (data not shown). Further studies are needed to elucidate how MNNG-induced USP10 phosphorylation affects its enzymatic activity to regulate MSH2 stability.
To explore whether USP10 and MSH2 can serve as biomarkers for the sensitivity of chemotherapeutic drugs in lung cancer patients, we examined the mRNA levels of USP10 and MSH2 by qRT-PCR in a cohort of non-small cell lung cancer patients under clinical trials (28). We found that there was a positive correlation between USP10 and MSH2.4 Future studies will determine the protein levels of USP10 and MSH2 in these patients to examine whether USP10 also stabilizes MSH2 in lung cancer patients.
Author Contributions
M. Z. and C. H. performed most of the experiments. D. T. performed the MMR experiment. M. Z. and X. Z. designed the study and wrote the paper. S. X. did the statistical analysis. K. W. revised the manuscript. W. B., G. M. L., and G. B. provided the critical reagents and helped design the experiments. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank H. Lee Moffitt Cancer Center Proteomics facility for the mass spectrometry analysis, Dr. Zhenkun Lou for some USP10 reagents and critical reading of the manuscript, Agnes Malysa for assistance, and Joshua Haakenson for proofreading the manuscript.
This work was supported in part by National Institutes of Health Grant R01CA164147, a New Investigator Grant 09KN-17 from Florida James and Esther King Biomedical Program, a Liz Tilberis award from Ovarian Cancer Research Fund, and the Karmanos Cancer Institute start-up funds (to X. Z.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
G. Bepler, unpublished data.
- MMR
- mismatch repair
- USP10
- ubiquitin-specific peptidase 10
- DDR
- DNA damage response
- 6-TG
- 6-thioguanine
- MNNG
- N-methyl-N′-nitro-N-nitrosoguanidine
- DUB
- deubiquitinating enzyme
- USP
- ubiquitin-specific peptidase
- MSH
- MutS homolog.
References
- 1. Kunkel T. A., and Erie D. A. (2005) DNA mismatch repair. Annu. Rev. Biochem. 74, 681–710 [DOI] [PubMed] [Google Scholar]
- 2. Drummond J. T., Li G. M., Longley M. J., and Modrich P. (1995) Isolation of an hMSH2-p160 heterodimer that restores DNA mismatch repair to tumor cells. Science 268, 1909–1912 [DOI] [PubMed] [Google Scholar]
- 3. Genschel J., Littman S. J., Drummond J. T., and Modrich P. (1998) Isolation of MutSbeta from human cells and comparison of the mismatch repair specificities of MutSβ and MutSα. J. Biol. Chem. 273, 19895–19901 [DOI] [PubMed] [Google Scholar]
- 4. Kolodner R. D. (1995) Mismatch repair: mechanisms and relationship to cancer susceptibility. Trends Biochem. Sci. 20, 397–401 [DOI] [PubMed] [Google Scholar]
- 5. Fink D., Aebi S., and Howell S. B. (1998) The role of DNA mismatch repair in drug resistance. Clin. Cancer Res. 4, 1–6 [PubMed] [Google Scholar]
- 6. Scherer S. J., Maier S. M., Seifert M., Hanselmann R. G., Zang K. D., Muller-Hermelink H. K., Angel P., Welter C., and Schartl M. (2000) p53 and c-Jun functionally synergize in the regulation of the DNA repair gene hMSH2 in response to UV. J. Biol. Chem. 275, 37469–37473 [DOI] [PubMed] [Google Scholar]
- 7. Humbert O., Achour I., Lautier D., Laurent G., and Salles B. (2003) hMSH2 expression is driven by AP1-dependent regulation through phorbol-ester exposure. Nucleic Acids Res. 31, 5627–5634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang M., Xiang S., Joo H. Y., Wang L., Williams K. A., Liu W., Hu C., Tong D., Haakenson J., Wang C., Zhang S., Pavlovicz R. E., Jones A., Schmidt K. H., Tang J., et al. (2014) HDAC6 deacetylates and ubiquitinates MSH2 to maintain proper levels of MutSα. Mol. Cell 55, 31–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Humbert O., Hermine T., Hernandez H., Bouget T., Selves J., Laurent G., Salles B., and Lautier D. (2002) Implication of protein kinase C in the regulation of DNA mismatch repair protein expression and function. J. Biol. Chem. 277, 18061–18068 [DOI] [PubMed] [Google Scholar]
- 10. Hicke L., and Dunn R. (2003) Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu. Rev. Cell Dev. Biol. 19, 141–172 [DOI] [PubMed] [Google Scholar]
- 11. Clague M. J., and Urbé S. (2006) Endocytosis: the DUB version. Trends Cell Biol. 16, 551–559 [DOI] [PubMed] [Google Scholar]
- 12. Mukhopadhyay D., and Riezman H. (2007) Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science 315, 201–205 [DOI] [PubMed] [Google Scholar]
- 13. Love K. R., Catic A., Schlieker C., and Ploegh H. L. (2007) Mechanisms, biology and inhibitors of deubiquitinating enzymes. Nat. Chem. Biol. 3, 697–705 [DOI] [PubMed] [Google Scholar]
- 14. Millard S. M., and Wood S. A. (2006) Riding the DUBway: regulation of protein trafficking by deubiquitylating enzymes. J. Cell Biol. 173, 463–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yuan J., Luo K., Zhang L., Cheville J. C., and Lou Z. (2010) USP10 regulates p53 localization and stability by deubiquitinating p53. Cell 140, 384–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jochemsen A. G., and Shiloh Y. (2010) USP10: friend and foe. Cell 140, 308–310 [DOI] [PubMed] [Google Scholar]
- 17. Liu J., Xia H., Kim M., Xu L., Li Y., Zhang L., Cai Y., Norberg H. V., Zhang T., Furuya T., Jin M., Zhu Z., Wang H., Yu J., Hao Y., et al. (2011) Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP13. Cell 147, 223–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Soncini C., Berdo I., and Draetta G. (2001) Ras-GAP SH3 domain binding protein (G3BP) is a modulator of USP10, a novel human ubiquitin specific protease. Oncogene 20, 3869–3879 [DOI] [PubMed] [Google Scholar]
- 19. Bomberger J. M., Barnaby R. L., and Stanton B. A. (2009) The deubiquitinating enzyme USP10 regulates the post-endocytic sorting of cystic fibrosis transmembrane conductance regulator in airway epithelial cells. J. Biol. Chem. 284, 18778–18789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bomberger J. M., Barnaby R. L., and Stanton B. A. (2010) The deubiquitinating enzyme USP10 regulates the endocytic recycling of CFTR in airway epithelial cells. Channels 4, 150–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Draker R., Sarcinella E., and Cheung P. (2011) USP10 deubiquitylates the histone variant H2A.Z and both are required for androgen receptor-mediated gene activation. Nucleic Acids Res. 39, 3529–3542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang Y., Zhang M., Dong H., Yong S., Li X., Olashaw N., Kruk P. A., Cheng J. Q., Bai W., Chen J., Nicosia S. V., and Zhang X. (2009) Deacetylation of cortactin by SIRT1 promotes cell migration. Oncogene 28, 445–460 [DOI] [PubMed] [Google Scholar]
- 23. Fu W., Ma Q., Chen L., Li P., Zhang M., Ramamoorthy S., Nawaz Z., Shimojima T., Wang H., Yang Y., Shen Z., Zhang Y., Zhang X., Nicosia S. V., Pledger J. W., et al. (2009) MDM2 acts downstream of p53 as an E3 ligase to promote FOXO ubiquitination and degradation. J. Biol. Chem. 284, 13987–14000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tong D., Ortega J., Kim C., Huang J., Gu L., and Li G. M. (2015) Arsenic inhibits DNA mismatch repair by promoting EGFR expression and PCNA phosphorylation. J. Biol. Chem. 290, 14536–14541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Guo S., Presnell S. R., Yuan F., Zhang Y., Gu L., and Li G. M. (2004) Differential requirement for proliferating cell nuclear antigen in 5′ and 3′ nick-directed excision in human mismatch repair. J. Biol. Chem. 279, 16912–16917 [DOI] [PubMed] [Google Scholar]
- 26. Zhang Y., Yuan F., Presnell S. R., Tian K., Gao Y., Tomkinson A. E., Gu L., and Li G. M. (2005) Reconstitution of 5′-directed human mismatch repair in a purified system. Cell 122, 693–705 [DOI] [PubMed] [Google Scholar]
- 27. Warren J. J., Pohlhaus T. J., Changela A., Iyer R. R., Modrich P. L., and Beese L. S. (2007) Structure of the human MutSα DNA lesion recognition complex. Mol. Cell 26, 579–592 [DOI] [PubMed] [Google Scholar]
- 28. Bepler G., Williams C., Schell M. J., Chen W., Zheng Z., Simon G., Gadgeel S., Zhao X., Schreiber F., Brahmer J., Chiappori A., Tanvetyanon T., Pinder-Schenck M., Gray J., Haura E., et al. (2013) Randomized international phase III trial of ERCC1 and RRM1 expression-based chemotherapy versus gemcitabine/carboplatin in advanced non-small-cell lung cancer. J. Clin. Oncol. 31, 2404–2412 [DOI] [PMC free article] [PubMed] [Google Scholar]