Abstract
In the integrated stress response, phosphorylation of eIF2α (eIF2α-P) reduces protein synthesis while concomitantly promoting preferential translation of specific transcripts associated with stress adaptation. Translation of the glutamyl-prolyl-tRNA synthetase gene EPRS is enhanced in response to eIF2α-P. To identify the underlying mechanism of translation control, we employed biochemical approaches to determine the regulatory features by which upstream ORFs (uORFs) direct downstream translation control and expression of the EPRS coding region. Our findings reveal that translation of two inhibitory uORFs encoded by noncanonical CUG and UUG initiation codons in the EPRS mRNA 5′-leader serve to dampen levels of translation initiation at the EPRS coding region. By a mechanism suggested to involve increased translation initiation stringency during stress-induced eIF2α-P, we observed facilitated ribosome bypass of these uORFs, allowing for increased translation of the EPRS coding region. Importantly, EPRS protein expression is enhanced through this preferential translation mechanism in response to multiple known activators of eIF2α-P and likely serves to facilitate stress adaptation in response to a variety of cellular stresses. The rules presented here for the regulated ribosome bypass of noncanonical initiation codons in the EPRS 5′-leader add complexity into the nature of uORF-mediated translation control mechanisms during eIF2α-P and additionally illustrate the roles that previously unexamined uORFs with noncanonical initiation codons can play in modulating gene expression.
Keywords: aminoacyl tRNA synthetase, endoplasmic reticulum stress (ER stress), eukaryotic initiation factor 2 (eIF2), stress response, translation control, translation initiation, EPRS, integrated stress response, uORF
Introduction
Protein synthesis is modulated in response to a variety of extracellular and intracellular stimuli. Phosphorylation of eukaryotic initiation factor 2 serves as the integration point for these stress signals and regulates the initiation phase of translation (1). During translation initiation, eIF2 combines with initiator Met-tRNAiMet, GTP, and the 40S ribosomal subunit to form the 48S preinitiation complex that facilitates start codon selection. Phosphorylation on the α subunit of eIF2 at serine 51 (eIF2α-P)3 inhibits the exchange of eIF2-GDP for eIF2-GTP, thereby blocking delivery of the initiator Met-tRNAiMet and triggering a global reduction in translation initiation (2). The coincident reduction in protein synthesis allows cells to conserve energy and resources and facilitates the reconfiguration of gene expression to promote the alleviation of stress damage. eIF2α-P facilitates the reprogramming of gene expression by selectively inducing translation of specific transcripts involved in stress adaptation. Because there are multiple mammalian eIF2 kinases that converge on a common substrate, this pathway is referred to as the integrated stress response (ISR) (1, 3). To illustrate, eIF2α is phosphorylated by PERK (protein kinase R-like ER kinase) in response to an accumulation of unfolded protein in the lumen of the ER, whereas amino acid deprivation is sensed by the GCN2 (general control nonderepressible 2) kinase in the cytosol (2, 4).
Included among the downstream ISR induced transcripts are transcription factors ATF4 (CREB2) and CHOP (DDIT3/GADD153) that act to modify gene expression programs to address cellular stress (4–6). GADD34 (PPP1R15A) is also preferentially translated and combines with the catalytic subunit of protein phosphatase 1 to regulate dephosphorylation of eIF2α-P and restoration of protein synthesis after amelioration of the stress damage (7, 8). The 5′-leader of those mRNAs that are preferentially translated contain upstream ORFs (uORFs) that precede the coding sequence (CDS) and are critical for translation control in response to eIF2α-P. ATF4, for example, contains two uORFs that confer its translation control (6, 9). In the “delayed translation reinitiation” model, the 5′-proximal uORF1 in the ATF4 mRNA serves as a positive-acting element that promotes translation reinitiation at downstream ORFs. During cellular stress, delayed delivery of the initiator Met-tRNAiMet to the 40S ribosomal subunit after uORF1 translation allows for scanning ribosomes to surpass an inhibitory uORF2 that overlaps out of frame with the CDS and instead initiate translation at the ATF4 start codon. This model shares features with GCN4 translation control in yeast (10). Preferential translation of both CHOP and GADD34 during eIF2α-P is regulated via a “bypass” mechanism in which an inhibitory uORF is bypassed in part because of a less than optimal start codon context (5, 8).
Although many ISR genes are subject to preferential translation via uORF-mediated mechanisms, the mere presence of an uORF is not sufficient to ensure enhanced translation during eIF2α-P. Genome-wide analyses of changes in translation in response to eIF2α-P suggest that ∼40% of mammalian mRNAs contain uORFs (11, 12). Furthermore, uORFs are suggested to be equally present among those transcripts that are translationally enhanced, repressed, or resistant to eIF2α-P (11). These findings suggest that each uORF contains specific properties that determine whether the 5′-leader of an mRNA serves to activate or repress translation in response to eIF2α-P. Interestingly, recent ribosome profiling analysis has indicated that in addition to uORFs that contain a canonical AUG initiation codon, there are also multitudes of previously unidentified uORFs that begin with noncanonical (CUG, UUG, and GUG) initiation codons (13–15). These observations provide intriguing evidence that the number of uORFs encoded in the genome may be dramatically underestimated in previous studies and suggests that the uORF start codon itself may provide additional information determining how the 5′-leader of a given mRNA regulates translation in response to eIF2α-P.
In this study, we provide an example of translation control mediated via uORFs that contain noncanonical initiation codons and facilitate preferential translation during stress. Translation of the EPRS mRNA encoding glutamyl-prolyl-tRNA synthetase is suggested to be enhanced in response to eIF2α-P (11). The 5′-leader of EPRS contains two UUG initiation codons and three in-frame CUG initiation codons that collectively encode three putative uORFs. Using heterologous reporter systems, we define the regulatory features of the three uORFs that direct translation control of EPRS mRNA. We also show that EPRS protein expression is increased through its preferential translation in response to drugs known to activate PERK and GCN2, implying increased EPRS expression during stress serves to facilitate adaptation in numerous stress contexts. This study provides an understanding of the translation control mechanisms that serves to regulate ISR induced gene transcripts during eIF2α-P but also illustrates the roles that previously unidentified uORFs with noncanonical initiation codons can possess in modulating gene expression.
Experimental Procedures
Cell Culture
Wild-type and A/A MEF cells expressing either a WT version of eIF2α or eIF2α-S51A were cultured in DMEM as previously described (16). GCN2+/+ and GCN2−/− MEF cells were previously described (16) and were cultured in DMEM supplemented with 10% (v/v) fetal bovine serum, 100 units/ml penicillin, 100 μg/ml streptomycin, and 1× nonessential amino acids. For halofuginone treatments, both control and treatment groups were cultured in DMEM supplemented with 10% (v/v) dialyzed fetal bovine serum (Gibco).
Immunoblot Analyses
MEF cells were treated with 1 μm thapsigargin for up to 6 h or left untreated. Alternatively, MEF cells were treated with 20, 50, or 100 nm halofuginone for 6 h or left untreated. Protein lysates were collected followed by quantification and immunoblot analyses as previously described (17). Antibodies used for immunoblot analyses include: EPRS (Abcam catalog no. ab31531), eIF2α-P (Abcam catalog no. ab32157), and β-actin (Sigma catalog no. A5441). Monoclonal antibody measuring total eIF2α was kindly provided by Dr. Scott Kimball (Pennsylvania State University College of Medicine, Hershey, PA). Immunoblots were developed either using chemiluminescence with x-ray film or LI-COR Odyssey (LI-COR Biosciences) imaging.
mRNA Measurement by Quantitative PCR
Total RNA was isolated from MEF cells and polysome fractions with TRIzol reagent (Invitrogen) followed by single-strand cDNA synthesis using a TaqMan reverse transcriptase kit (Applied Biosystems) according to the manufacturer's protocol. Quantitative PCR of transcripts was conducted using SYBR Green (Applied Biosystems) on a Realplex2 Master Cycler (Eppendorf). Primers used for measuring transcripts include: ATF4 forward, 5′-GCCGGTTTAAGTTGTGTGCT-3′; ATF4 reverse, 5′-CTGGATTCGAGGAATGTGCT-3′; EPRS forward, 5′-TGTGGGGAAATTGACTGTGA-3′; EPRS reverse, 5′-AACTCCGACCAAACAAGGTG-3′; Firefly luciferase forward, 5′- CTCACTGAGACTACATCAGC-3′; Firefly luciferase reverse, 5′-TCCAGATCCACAACCTTCGC-3′; β-actin forward, 5′-TGTTACCAACTGGGACGACA-3′; and β-actin reverse, 5′-GGGGTGTTGAAGGTCTCAA-3′.
Polysome Profiling and Sucrose Gradient Ultracentrifugation
MEF cells were left untreated or treated with either 1 μm thapsigargin or 25 nm halofuginone for 6 h. 50 μg/ml cycloheximide was added to culture medium just prior to lysate collection. Lysates were collected, sheared using a 23-gauge needle, and layered onto 10–50% sucrose gradients followed by ultracentrifugation as previously described (11, 18). A piston gradient fractionator (BioComp) and a 254-nm UV monitor with Data Quest Software were used to fractionate sucrose gradients and measure whole cell lysate polysome profiles.
After fractionation, 10 ng/ml firefly luciferase control RNA (Promega) was added to each fraction to generate polysome shifts for specific transcripts normalized to an exogenous RNA control (11, 18). Fractions were mixed with TRIzol, and RNA isolation and cDNA production were performed as described above. Calculations for percentages of total gene transcript and percentages of individual transcript shifts are as previously described (11). Whole cell lysate polysome profiles and specific transcript polysomes shifts are representative of at least three independent biological experiments.
Plasmid Constructions and Luciferase Assays
A 5′-rapid amplification of cDNA ends (5′-RACE; FirstChoice Ambion) was performed using RNA lysates collected from WT MEF cells left untreated or treated with 1 μm thapsigargin for 6 h to determine the transcriptional start site for EPRS. The cDNA fragment encoding the 5′-leader of EPRS was inserted between SacI and NcoI between the TK promoter and firefly luciferase CDS in a derivative of plasmid pGL3 (6). The resulting PTK-EPRS-Luc contains the mouse EPRS 5′-leader fused to a luciferase reporter. Site-directed mutagenesis and subcloning of synthesized cDNAs was used to generate mutant PTK-EPRS-Luc constructs (Table 1) that were sequenced for verification of nucleotide substitutions. PTK-EPRS-Luc constructs were transiently co-transfected with a Renilla reporter plasmid into WT or A/A MEF cells for 24 h followed by a 6-h 0.1 μm thapsigargin treatment or a 6-h 50 nm halofuginone treatment. Lysates were collected, and firefly and Renilla luciferase activities were measured as described previously (6). Relative values for luciferase measurements are represented with S.D. indicated for at least three independent biological experiments.
TABLE 1.
Description of EPRS mutations used in this study
| Gene construct | Description of mutation |
|---|---|
| EPRS stem loop insertion | Insertion of CTGCAGCCACCACGGCCCCCAAGCTTGGGCCGTGGTGGCTGCAG 3′ of CGTAGCGTGC |
| EPRS uORF1 ΔCUG1 | GGGCTGCGG to GGGAAACGG |
| EPRS uORF1 ΔCUG2 | CACCTGAAC to CACAAAAAC |
| EPRS uORF1 ΔCUG3 | CTCCTGGGA to CTCAAAGGA |
| EPRS uORF2 ΔUUG1 | ACGTTGCAT to ACGAAACAT |
| EPRS uORF3 ΔUUG2 | TCCTTGCTT to TCCAAACTT |
| EPRS uORF1 CUG2 to optimized AUG | CACCTGAAC to ACCATGGAC |
| EPRS uORF2 UUG1 to optimized AUG | ACGTTGCAT to ACCATGGAT |
| EPRS uORF2 UGA to UGG | ACCTGAACC to ACCTGGACC |
The full-length EPRS uORFs were each individually fused in-frame to the luciferase CDS and were transcriptionally expressed from a TK promoter for generation of PTK-CUG123 uORF-Luc, PTK-UUG1 uORF-Luc, and PTK-UUG2 uORF-Luc. Site-directed mutagenesis and subcloning of synthesized cDNAs was used to generate WT and mutant PTK-uORF-Luc constructs (Table 1) that were sequenced for verification of desired nucleotide substitutions. PTK-uORF-Luc constructs were transiently co-transfected with a Renilla reporter plasmid into WT MEF cells for 24 h. Lysates were collected, and firefly and Renilla luciferase activities were measured as described previously (6). Relative values for luciferase measurements are represented with S.D. indicated for at least three independent biological experiments.
Cell Viability Assays
For MTT assays, GCN2+/+ and GCN2−/− MEFs were seeded in 96-well culture plates at 5,000 cells/well 24 h prior to treatment. Cells were treated with 12.5, 25, or 50 nm halofuginone for 6 h, followed by recovery in fresh medium for 18 h. Viability was measured using a CellTiter 96-well nonradioactive cell proliferation assay (Promega). Treatment values were normalized to untreated groups for each respective cell line.
Statistical Analyses
The values indicate the means ± standard deviation and are representative of at least three independent experiments. Calculations of statistical significance were done using the two-tailed Student's t test. One-way analysis of variance and a post hoc Tukey HSD test were used to determine differences between multiple groups. p values less than 0.05 were considered statistically significant and are indicated by asterisks, and treatment groups considered statistically significant from WT control are indicated by pound signs.
Results
EPRS Expression Is Increased in Response to eIF2α-P through Enhanced Translation
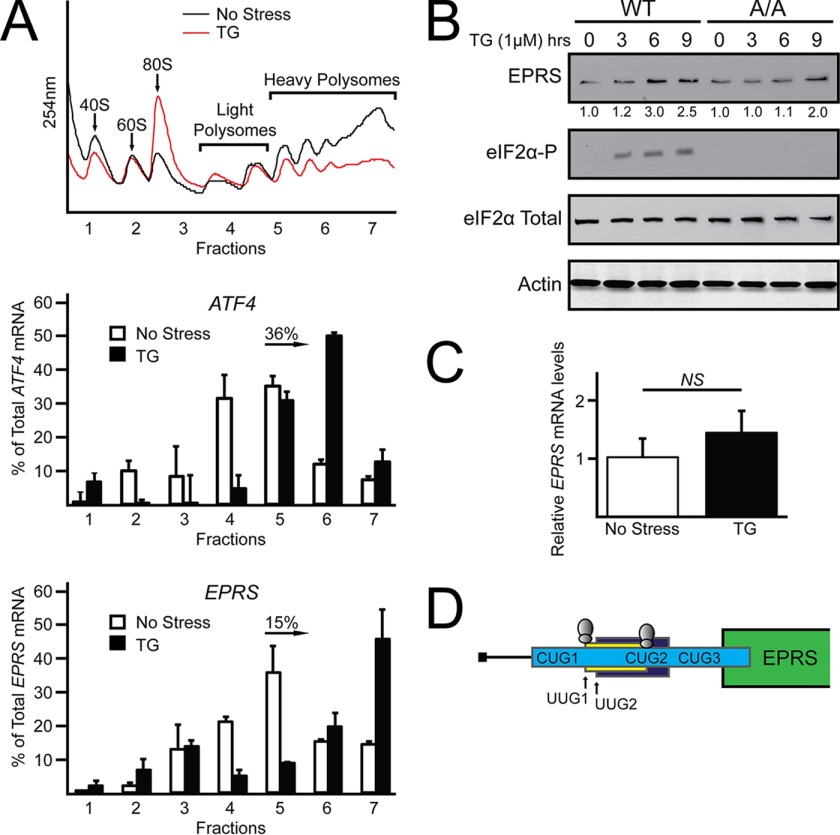
The glutamyl-prolyl-tRNA synthetase gene EPRS was previously identified in a genome-wide analysis of changes in translation as a transcript that has enhanced expression in response to eIF2α-P (11). To further explore the role that eIF2α-P and translation control play in the expression of EPRS, WT mouse embryonic fibroblast (MEF) cells were treated with thapsigargin, an inducer of endoplasmic reticulum stress and PERK activity, and analyzed for changes in translation by sucrose gradient ultracentrifugation. Polysome profiles of cells subjected to ER stress indicated reduced global translation initiation as ascertained by a decrease in heavy polysomes coincident with increased monosomes (Fig. 1A). EPRS and ATF4, a transcript known to be subject to preferential translation (6), were measured for changes in translation by qRT-PCR analysis of polysome fractions. Both EPRS and ATF4 transcripts were predominantly associated with monosomes and light polysomes in the absence of stress. However, upon ER stress induction, EPRS and ATF4 mRNAs significantly shifted to association with heavy polysomes (Fig. 1A). These results suggest that EPRS is preferentially translated in response to ER stress.
FIGURE 1.
EPRS translational expression is increased in response to eIF2α-P. A, WT MEF cells were treated with thapsigargin for 6 h or left untreated. Lysates were collected and separated by sucrose gradient centrifugation followed by analysis of polysome profiles at 254 nm. Total RNA was isolated from sucrose fractions, and the percentage of total ATF4 and EPRS mRNA was determined by qRT-PCR. Profiles and ATF4 and EPRS mRNA polysome shifts are representative of at least three independent biological experiments. B, WT and A/A MEF cells were treated with thapsigargin for up to 9 h or left untreated. Protein lysates were processed, and levels of EPRS, eIF2α-P, eIF2α total, and β-actin were measured by immunoblot. C, total RNA was collected from WT and A/A MEF cells treated with thapsigargin for 6 h or left untreated, and the relative levels of EPRS mRNA were measured using qRT-PCR. D, representation of EPRS 5′-leader. The uORFs in the 5′-leader of the EPRS mRNA are illustrated by the colored boxes, with the initiation codon(s) for each uORF listed. The green box indicates the CDS for EPRS. Ribosomes above UUG1 and CUG2 indicate those start codons that have been suggested to facilitate translation initiation in previous ribosome profiling studies (13, 14). TG, thapsigargin; NS, not significant.
To determine the role that eIF2α-P plays in the expression of EPRS, we measured changes in EPRS protein levels in WT MEF cells and mutant MEF cells (A/A) that express eIF2α-S51A that cannot be phosphorylated. As expected, eIF2α-P was detectable only in WT cells treated with thapsigargin. EPRS protein expression was increased 3-fold by 6 h of thapsigargin treatment in WT MEFs, whereas A/A MEFs presented with reduced and delayed induction in EPRS expression (Fig. 1B). EPRS mRNA expression did not significantly change in response to ER stress (Fig. 1C), suggesting that EPRS is not subject to transcriptional regulation in this cell line, but is preferentially translated upon ER stress and eIF2α-P.
Preferential Translation of EPRS Features Two uORFs with Noncanonical Initiation Codons
The advent of ribosome profiling has resulted in the genome-wide identification of previously uncharacterized translation initiation sites including those for amino-terminal protein extensions, protein truncations, and uORFs with noncanonical (non-AUG) initiation codons (13–15). Intriguingly, the 5′-leader of EPRS contains three putative uORFs with five noncanonical initiation codons, two of which have been identified in ribosome profiling studies as functional initiation codons (Fig. 1D) (13).
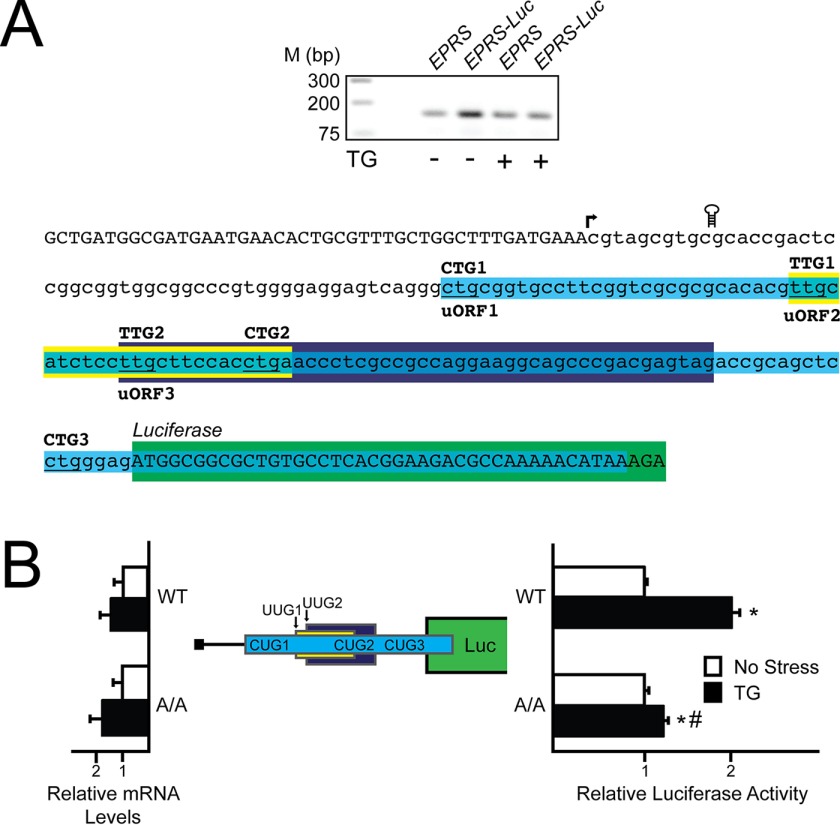
Multiple mechanisms of preferential translation rely on uORF-mediated translation control (5, 6, 8, 19). To determine the role of the EPRS 5′-leader in its preferential translation, we next conducted a 5′-RACE to define the transcriptional start site of the mouse EPRS gene (Fig. 2A). A cDNA segment encoding the 155-nucleotide EPRS 5′-leader was cloned in between a minimal TK promoter and the firefly luciferase CDS, producing PTK-EPRS-Luc. This construct and the subsequent ones that follow feature in-frame replacement of the firefly luciferase initiation codon with the initiation codon of the EPRS CDS. Luciferase activity from PTK-EPRS-Luc was induced 2-fold in WT MEF cells treated with thapsigargin compared with minimal induction in A/A cells (Fig. 2B). In both these and the following reporter measurements, there was no significant change in EPRS-Luc mRNA levels, suggesting that the changes in luciferase activity are the result of translation control. These results suggest that the EPRS 5′-leader is sufficient to direct preferential translation of EPRS in response to eIF2α-P.
FIGURE 2.
The 5′-leader of the EPRS mRNA directs preferential translation. A, top panel, a 5′-RACE was conducted for EPRS using WT MEFs treated with thapsigargin for 6 h or left untreated. Total RNA and cDNA were prepared, and DNA products were separated by gel electrophoresis, with markers for the indicted base pair sizes listed on the left. Bottom panel, nucleotide representation of the EPRS 5′-leader in lowercase letters, with uppercase letters representing the 5′-linker added during the 5′-RACE procedure and the beginning of the CDS of the EPRS-Luc fusion. Colored boxes represent the EPRS uORFs, with uORF1 in blue, uORF2 in yellow, and uORF3 in purple. Start codons for each uORF are indicated above the colored boxes. The coding region of the EPRS-Luc fusion is illustrated by the green box. The transcription start site is indicated with an arrow, and the location of the stem loop insertion is illustrated. B, the PTK-EPRS-Luc construct and a Renilla luciferase reporter were co-transfected into WT or A/A MEFs and treated with thapsigargin for 6 h or left untreated. EPRS 5′-leader mediated translation control was measured via Dual-Luciferase assay, and the corresponding EPRS-Luc mRNA was measured by qRT-PCR. The PTK-EPRS-Luc construct contains the cDNA sequence corresponding to the EPRS 5′-leader fused to the luciferase reporter gene with both the EPRS uORFs and the CDS of the EPRS-Luc fusion indicated with colored boxes that are the analogues to those indicated in Fig. 2A. TG, thapsigargin; Luc, luciferase.
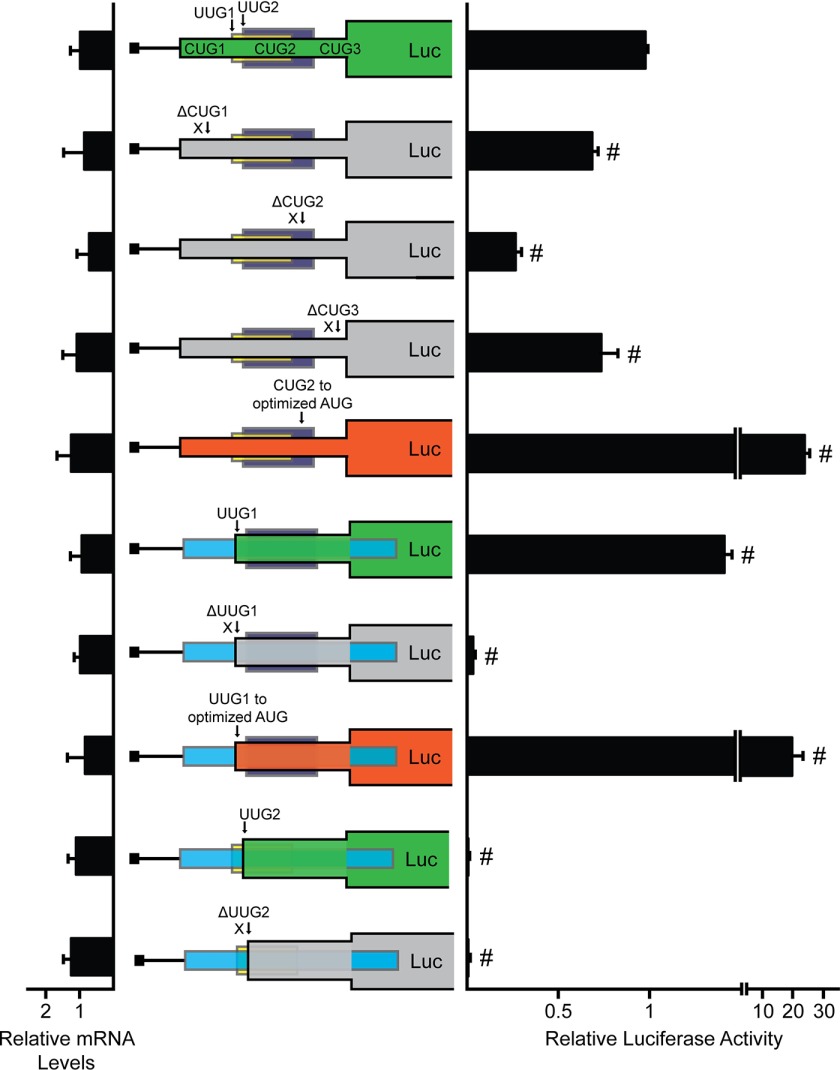
To determine which, if any, of the five noncanonical initiation codons can serve as functional sites of translation initiation, we constructed in-frame fusions of each uORF with the firefly luciferase CDS. These constructs featured deletion of the luciferase AUG, ensuring that any measureable luciferase activity was the product of translation initiation at the in-frame uORF. Fusion of the first uORF resulted in the in-frame fusion of three CUG initiation codons, here denoted CUG1, CUG2, and CUG3. Fusion of uORF1 with the firefly luciferase CDS had measurable luciferase activity, indicative of at least one functional initiation codon (Fig. 3). Individual deletion of CUG1 and CUG3 each resulted in slight decrease in luciferase activity, whereas deletion of CUG2 resulted in a 75% decrease in luciferase activity. This indicates that although all three CUGs can serve as functional initiation codons, CUG2 is the dominant site of translation initiation in uORF1. Optimization of CUG2 to an AUG with strong Kozak context (ACCAUGG) resulted in a 24-fold increase in luciferase activity as compared with fusion of uORF1 with all three CUG initiation codons intact. Together, these results indicate that CUG2 is the dominant initiation codon in uORF1 and that optimization of CUG2 to an AUG with strong Kozak context can further facilitate translation initiation at this site.
FIGURE 3.
Preferential translation of EPRS features two uORFs with noncanonical initiation codons. The full-length EPRS uORFs were each individually fused in-frame to the luciferase CDS and were transcriptionally expressed from a TK promoter for generation of PTK-CUG123 uORF-Luc, PTK-UUG1 uORF-Luc, and PTK-UUG2 uORF-Luc. WT and mutant versions of the uORF-Luc fusions were transfected into WT MEF cells, and uORF translation control was measured by Dual-Luciferase assay, and the corresponding CUG123 uORF-Luc, UUG1 uORF-Luc, and UUG2 uORF-Luc mRNA levels were measured by qRT-PCR. WT versions of each luciferase fusion are illustrated by the green boxes. Mutant versions of PTK-CUG123 uORF-Luc include mutations of the CUG initiation codons, as represented by ΔCUG1, ΔCUG2, and ΔCUG3, and optimization of the start codon for CUG2 to an AUG in strong Kozak consensus sequence (ACCAUGG), as represented by CUG2 to optimized AUG. Mutant versions of PTK-UUG1 uORF-Luc include mutation of the UUG initiation codon, as represented by ΔUUG1, and optimization of the UUG start codon to an AUG in a strong Kozak consensus sequence (ACCAUGG), as represented by UUG1 to optimized AUG. Mutation of PTK-UUG2 uORF-Luc includes mutation of the UUG initiation codon, as represented by ΔUUG2. Loss of the indicated initiation codon in the uORF-Luc fusion is illustrated by the gray boxes, and the optimized initiation codon in the uORF-Luc fusion is illustrated in orange. Relative values are represented as histograms for each with the S.D. indicated. Luc, luciferase.
Fusion of the second uORF that contains a UUG initiation codon resulted in a 1.5-fold increase in luciferase activity as compared with fusion of uORF1 (Fig. 3). Deletion of the UUG initiation codon, denoted UUG1, by mutating it to AAA resulted in no appreciable luciferase activity caused by the lack of any additional initiation codons in the uORF2 fusion. Mutation of the UUG1 initiation codon to an AUG in optimal context (ACCAUGG) resulted in nearly a 14-fold increase in luciferase activity as compared with the WT UUG1 fusion. These results suggest that UUG1 serves as the functional site of translation initiation in uORF2 and that translation initiation at this site can be further increased with the introduction of an AUG initiation codon in Kozak context.
Fusion of the third uORF also with a UUG initiation codon, denoted UUG2, did not result in any appreciable luciferase activity (Fig. 3). Furthermore, deletion of UUG2 by mutation to AAA did not result in any change in luciferase activity, indicating that there are no functional initiation sites in the uORF3 fusion. Consistent with a previous ribosome profiling study, these combined results suggest that CUG2 and UUG1 can both serve as functional sites of translation initiation in the EPRS 5′-leader (Fig. 1D) (13) and that the 5′-leader of EPRS can direct EPRS preferential translation in response to eIF2α-P.
EPRS Translation Control Involves Bypass of Two uORFs with Noncanonical Initiation Codons
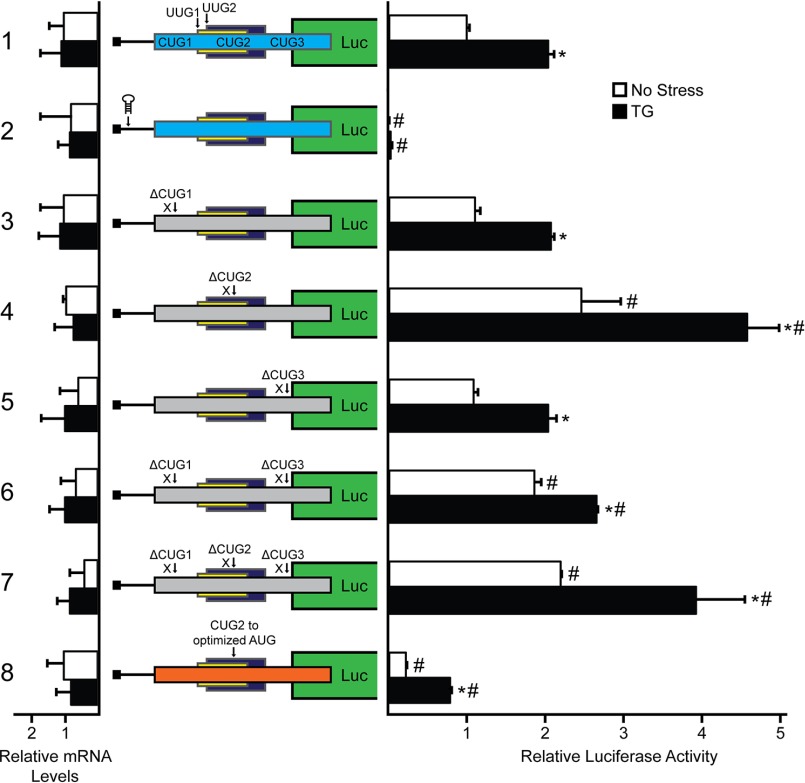
To determine whether preferential translation of EPRS occurs through ribosome scanning, a stem loop structure with a predicted free energy of ΔG = −41 kcal/mol was inserted 10 nucleotides downstream of the 5′ cap of the EPRS-Luc transcript (Figs. 2A and 4, construct 2). Introduction of this palindromic sequence to the EPRS-Luc mRNA significantly reduced luciferase activity independent of stress, suggesting that EPRS preferential translation is mediated by ribosome scanning beginning from the 5′-end of the EPRS transcript.
FIGURE 4.
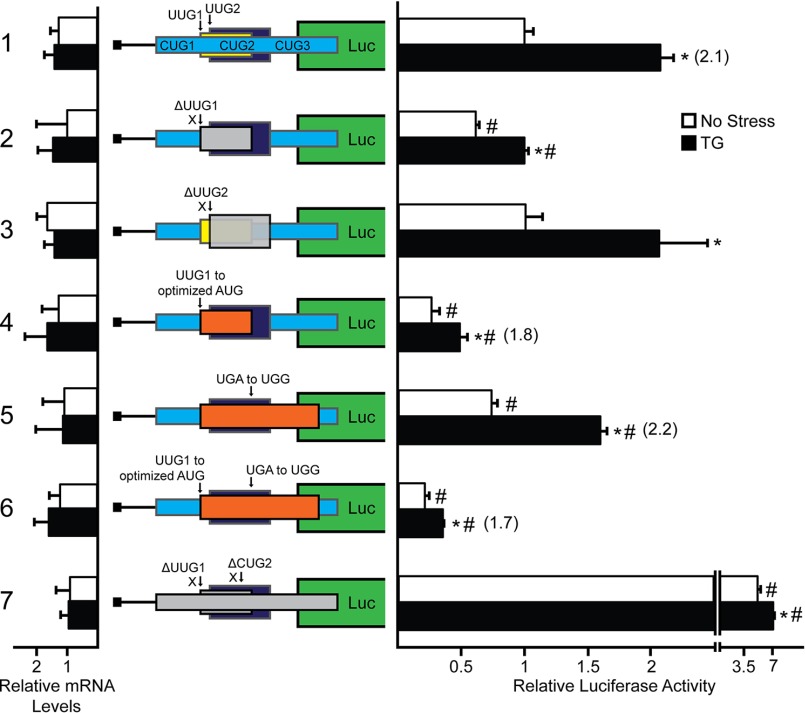
EPRS translation control involves bypass of an uORF with a noncanonical CUG initiation codon. WT and mutant versions of PTK-EPRS-Luc constructs were transfected into WT MEF cells, treated for 6 h or left untreated, and measured using a Dual-Luciferase assay, and the relative levels of the corresponding EPRS-Luc mRNAs were measured by qRT-PCR. Mutant versions of PTK-EPRS-Luc include a stem loop insertion and mutation of the CUG initiation codons individually or together, as represented by ΔCUG1, ΔCUG2, and ΔCUG3. Losses of the initiation codons CUG1, CUG2, and CUG3 are indicated in the EPRS-Luc fusion that is indicated by the gray boxes. Optimization of the CUG2 initiation codon to an AUG in optimal Kozak consensus sequence (ACCAUGG) is represented as CUG to optimized AUG (orange box). Relative values are represented as histograms for each with the S.D. indicated. TG, thapsigargin; Luc, luciferase.
Ribosomes scanning the EPRS mRNA would encounter the three uORFs located in the EPRS 5′-leader (Fig. 2A). To determine the contribution of each uORF and noncanonical initiation codon to the preferential translation of the EPRS CDS, we began by mutating the three CUG initiation codons located in uORF1 to AAA, as indicated by ΔCUG in the figure (Fig. 4). Deletion of CUG1 and CUG3 individually resulted in no significant difference in basal luciferase activity and were induced 2-fold with ER stress treatment, a similar result as the WT PTK-EPRS-Luc construct (Fig. 4 constructs 1, 3, and 5). Combined deletion of CUG1 and CUG3 resulted in a slight increase in luciferase activity both basally and with ER stress treatment (Fig. 4, construct 6). This result is consistent with the observation that CUG1 and CUG3 incur low amounts of translation initiation (Fig. 3) and serve as mild dampeners of downstream translation. Deletion of CUG2, however, resulted in an almost 2.5-fold increase in basal luciferase activity that was stress-inducible (Fig. 4, construct 4). Furthermore, deletion of all three CUGs resulted in similar luciferase activity as deletion of CUG2 alone, supporting the role of CUG2 as the dominant regulatory initiation codon in uORF1 (Fig. 4, constructs 4 and 7).
Multiple preferentially translated mRNAs rely on ribosome bypass of uORFs with poor start codon context for optimal expression during cellular stress (5, 8). To determine whether noncanonical initiation codons function in a similar manner by allowing for ribosome bypass, we next mutated CUG2 to an AUG with the optimal Kozak consensus sequence (ACCAUGG). Introduction of the optimized AUG reduced luciferase activity over 60% (Fig. 4, construct 8). Because uORF1 overlaps out of frame with the EPRS CDS, the observed decrease in luciferase activity after substitution of the AUG for CUG2 suggests that although CUG2 serves as the dominant site of initiation in uORF1, it can be bypassed, in part because of a noncanonical initiation codon, to facilitate translation at the downstream EPRS CDS.
To determine the contribution of the remaining two uORFs, we next mutated the UUG initiation codon for uORF2 to AAA, as indicated by ΔUUG1 (Fig. 5, construct 2). Deletion of UUG1 resulted in lower basal luciferase activity as compared with WT PTK-EPRS-Luc and a decreased induction ratio upon thapsigargin treatment, suggesting that UUG1 can act as a positive element that facilitates initiation at the downstream CDS. Deletion of the third uORF by substituting the UUG initiation codon for AAA, denoted ΔUUG2, resulted in no difference in luciferase activity as compared with the WT PTK-EPRS-Luc construct (Fig. 5, constructs 1 and 3). This finding is consistent with the low levels of luciferase activity observed for the UUG2 in-frame fusion (Fig. 3) and indicates that the uORF3 does not serve a regulatory role in EPRS translation.
FIGURE 5.
EPRS translational control involves bypass of uORFs with noncanonical initiation codons. WT and mutant versions of PTK-EPRS-Luc constructs were transfected into WT MEF cells, treated for 6 h or left untreated, and measured using a Dual-Luciferase assay, and the corresponding EPRS-Luc mRNA levels were measured by qRT-PCR. Mutant versions of PTK-EPRS-Luc include mutation of the UUG1 and UUG2 initiation codons (ΔUUG1 and ΔUUG2), optimization of the UUG1 initiation codon to an AUG with optimal Kozak consensus sequence (UUG1 to optimized AUG), mutation of the stop codon of the UUG1 uORF to generate an overlapping out of frame uORF (UGA to UGG), combined mutation of UUG1 to an AUG with optimal Kozak consensus sequence (ACCAUGG) with the stop codon mutation (UUG1 to optimized AUG and UGA to UGG), and combined mutation of the initiation codons for UUG1 and CUG2 (ΔUUG1 and ΔCUG2). Loss of the indicated initiation codon in the EPRS-Luc fusion is illustrated by a gray box, and those involving optimization of the initiation codon or extension of the uORF are indicated by orange boxes. Relative values are represented as histograms for each with the S.D. indicated. TG, thapsigargin; Luc, luciferase.
To further dissect the role of uORF2 in EPRS translation control, we next mutated the UUG initiation codon to an AUG in optimal context, as indicated by UUG1 to optimize AUG in the figure (Fig. 5, construct 4). Mutation of UUG1 to an optimized AUG resulted in a decrease in both the basal luciferase activity and the luciferase induction ratio upon ER stress treatment. This result suggests that the ability of uORF2 to allow for initiation at the downstream CDS relies upon its noncanonical UUG initiation codon.
In addition to bypass of uORFs, downstream translation initiation can also be regulated by ribosome reinitiation (6, 10, 19). To determine the contribution of ribosome reinitiation after uORF2 translation, the uORF2 stop codon was mutated from TGA to TGG to generate an overlapping out of frame uORF. Mutation of the uORF2 stop codon resulted in a 25% decrease in basal luciferase activity that was still induced 2-fold upon thapsigargin treatment. The basal reduction in luciferase activity suggests that after translation initiation at uORF2 UUG1 in the WT PTK-EPRS-Luc construct, a certain amount of translating ribosomes terminate and later reinitiate at the EPRS CDS. UUG1 is 5′ proximal to CUG2 and likely plays a positive-acting role in the EPRS translational control scheme by precluding a small number of scanning ribosomes from initiating translation at the inhibitory, overlapping out of frame CUG2. Instead UUG1 would allow for a measurable amount of ribosome reinitiation 3′ to the predominant CUG2 initiation codon in the uORF1. Combined mutation of UUG1 to an optimized AUG with the TGA stop codon substituted for TGG resulted in a further decrease in basal luciferase activity that was only induced 1.7-fold upon stress treatment. Overall, these results suggest that although a portion of the ribosomes that translate uORF2 can reinitiate downstream, scanning ribosomes also bypass uORF2 because of its noncanonical UUG1 initiation codon and initiate translation at the downstream CDS.
Because CUG2 and UUG1 are suggested to be the dominant initiation codons for uORF1 and uORF2, respectively, in the EPRS 5′-leader, we finally generated their combined deletion by mutating each to AAA (Fig. 5, construct 7). Combined deletion of both UUG1 and CUG2 resulted in 5-fold increase in luciferase activity independent of stress, further supporting the roles of UUG1 and CUG2 as overall repressing elements in EPRS translation control. A model for EPRS translation control and its broader medical implications will be expanded upon under “Discussion.”
Translation Control of EPRS during Treatment with the Drug Halofuginone
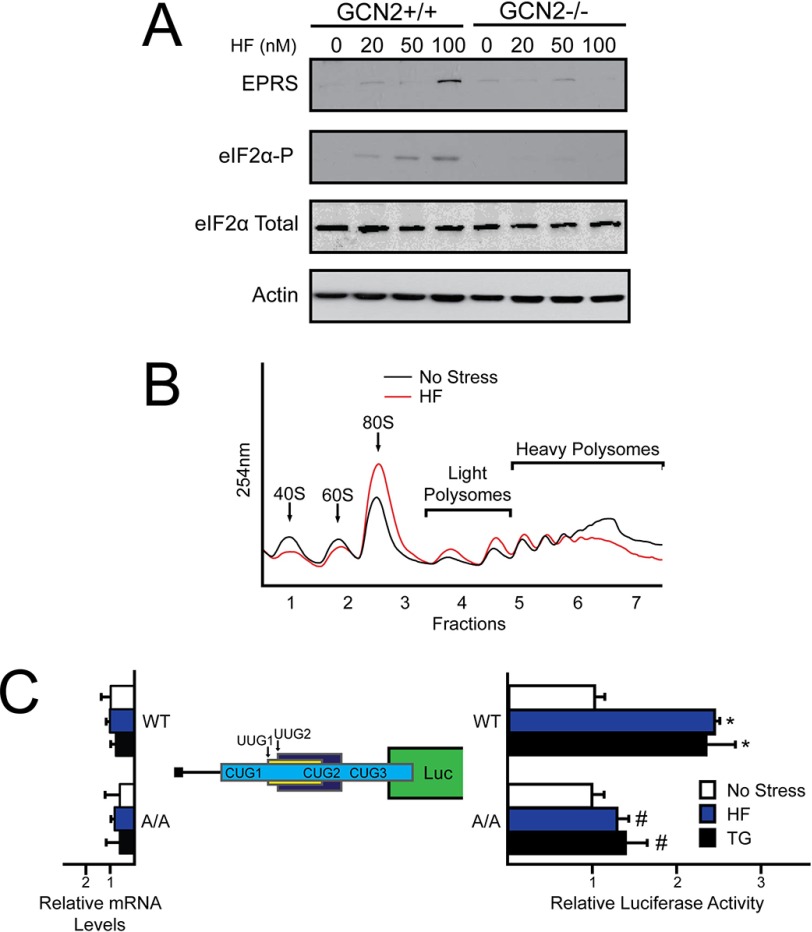
Halofuginone, a drug currently in phase II clinical trials for the treatment of fibrotic disease and solid tumors (Clinical Trial NCT00064142), was shown to confer surgical stress resistance in an animal model by a mechanism requiring the eIF2α kinase GCN2 (20). Halofuginone competes with proline for the active site of EPRS, leading to an accumulation of uncharged tRNAPro and activation of the GCN2/eIF2α-P pathway (20–22). Dietary restriction has been associated with an improved clinical outcome prior to an ischemic event in both animal and clinical models. The pharmacological induction of the GCN2/eIF2α-P pathway by halofuginone offers the exciting potential of conferring presurgical stress resistance using a pharmaceutical. We determined that EPRS mRNA is preferentially associated with heavy polysomes upon ER stress and that the 5′-leader of the EPRS gene transcript confers translational control in luciferase reporter constructs, suggesting preferential translation of EPRS in response to eIF2α-P. As a result, we proposed that halofuginone treatment would lead to the enhanced expression of its target substrate EPRS. To first examine the impact of halofuginone treatment on eIF2α-P and EPRS expression, WT and GCN2−/− MEF cells were treated with increasing concentrations of halofuginone for 6 h (Fig. 6A). As expected, eIF2α-P was detectable only in WT cells treated with halofuginone. EPRS protein expression was increased in a dose-dependent manner with halofuginone treatment in WT MEFs, whereas GCN2−/− MEFs displayed minimal induction in EPRS expression.
FIGURE 6.
EPRS translation control is regulated in response to halofuginone treatment. A, GCN2+/+ and GCN2−/− MEF cells were treated with increasing concentrations of halofuginone for 6 h. Protein lysates were processed, and the levels of EPRS, eIF2α-P, eIF2α total, and β-actin were measured by immunoblot. B, WT MEF cells were treated with halofuginone for 6 h or left untreated. Lysates were collected, sheared using a 23-gauge needle, and layered on to 10–50% sucrose gradients followed by ultracentrifugation and analysis of whole lysate polysome profiles at 254 nm. C, the PTK-EPRS-Luc construct and a Renilla luciferase reporter were co-transfected into WT or A/A MEFs and treated with thapsigargin or halofuginone for 6 h or left untreated. EPRS 5′-leader mediated translation control was measured via Dual-Luciferase assay and corresponding EPRS-Luc mRNA values were measured by qRT-PCR. The relative values are represented as histograms for each with the S.D. indicated. HF, halofuginone; TG, thapsigargin.
To determine the outcome of increased eIF2α-P with halofuginone treatment on global translation initiation, WT MEF cells were treated with halofuginone for 6 h or left untreated. Halofuginone treatment substantially reduced heavy polysomes with an accumulation of the 80S monosome peak, indicative of an eIF2α-P induced defect in global translation initiation (Fig. 6B). The biological implication of this during preconditioning is that, upon ischemic reperfusion, halofuginone would induce eIF2α-P providing the benefits of target UPR genes important for stress remediation. Coincident with the induction of the UPR, the increase in EPRS protein levels would quickly alleviate the toxicity associated with the drug treatment.
To further address whether EPRS mRNA is subject to translational control during halofuginone treatment, WT and A/A MEF cells were transfected with the PTK-EPRS-Luc reporter followed by either halofuginone treatment or no treatment. Both cell types were also treated with thapsigargin as a positive control for preferential translation of EPRS during eIF2α-P. In the WT MEF cells, both halofuginone and thapsigargin treatment resulted in a 2.5-fold induction of EPRS-Luc expression (Fig. 6C). Importantly, this increase in EPRS-Luc mRNA translation was absent in the alanine mutant (Fig. 6C). We conclude that translation of EPRS is enhanced in response to different stress conditions, including that triggered by halofuginone.
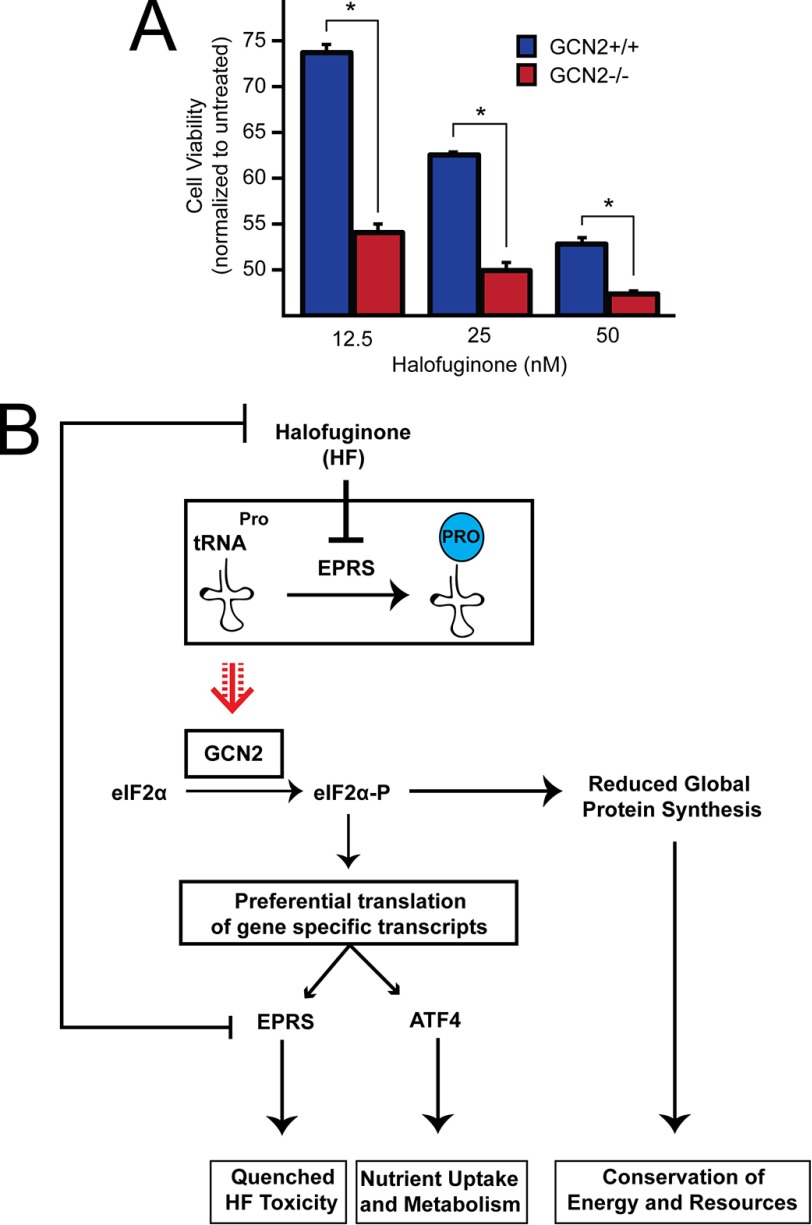
To examine the role of the ISR on cell fate, we treated WT and GCN2−/− MEF cells with increasing doses of halofuginone for 6 h and then allowed the cells to recover for 18 h in fresh media prior to measuring viability via MTT assay. From this analysis, we observed a sharp decrease in viability in the GCN2−/− cells compared with their WT counterparts. This difference was most notable at the 12.5 nm treatment, at which we observed an over 20% decrease in viability in the GCN2−/− cells compared with WT (Fig. 7A). Collectively, these results suggest that GCN2 activation and subsequent EPRS translational control are paramount to cell survival during halofuginone treatment (Fig. 7B).
FIGURE 7.
GCN2 confers protection against halofuginone-induced toxicity. A, equal numbers of GCN2+/+ and GCN2−/− MEFs were seeded in 96-well plates, cultured for 24 h, and treated with 12.5, 25, or 50 nm halofuginone for 6 h, followed by recovery in fresh media for 18 h. MTT activity was measured by the conversion of tetrazolium to formazan. B, model depicting gene regulation downstream of the eIF2 kinase GCN2 during halofuginone treatment. With the accumulation of uncharged tRNAPro during halofuginone treatment, activated GCN2 phosphorylates eIF2α and decreases global mRNA translation initiation. Coincident with a decrease in overall translation, mRNA encoding ATF4 is subject to preferential translation, ultimately leading to an increase in ATF4 downstream targets central to stress remediation. Also subject to preferential translation during eIF2α-P is mRNA encoding EPRS. During halofuginone treatment, EPRS is preferentially translated, and the resulting increase in its expression is suggested to quench chronic drug toxicity. HF, halofuginone.
Discussion
In this study, we address the mechanisms by which uORFs with noncanonical initiation codons modulate gene expression in response to eIF2α-P. Previously, identification of uORFs was largely dependent on the presence of an AUG initiation codon in the 5′-leader of a given mRNA. Recent ribosome profiling evidence has suggested that this mechanism of uORF identification has vastly underestimated the number of functional uORFs and that uORFs with noncanonical initiation codons can also be translated and serve regulatory roles in gene expression (13–15). The 5′-leader of the glutamyl-prolyl-tRNA synthetase gene EPRS contains five noncanonical initiation codons that are divided between three uORFs. We determined here the regulatory features by which the uORFs direct translation control of EPRS and promote increased EPRS protein expression in response to diverse cellular stresses.
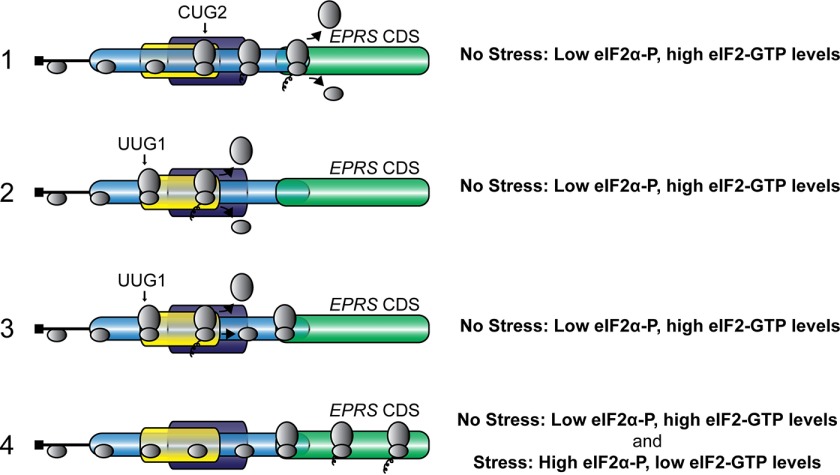
As illustrated in the model presented in Fig. 8, translation initiation at either CUG2, encoded in uORF1, or the uORF2 UUG reduce basal translation initiation at the EPRS CDS. uORF1 overlaps out of frame with the EPRS CDS, and translation of uORF1 results in translation termination 3′ of the start codon for EPRS (Fig. 8). Additionally, only ∼25% of the ribosomes that translate uORF2 reinitiate at the downstream EPRS initiation codon, thereby dampening basal EPRS expression (Fig. 5). Central to the EPRS mechanism of translation control are the noncanonical initiation codons for the two functional uORFs. Replacement of either CUG2 or UUG1 with an AUG initiation codon resulted in over a 60% decrease in luciferase activity (Figs. 4 and 5). The presence of the CUG and UUG initiation codons allows for a portion of the scanning ribosomes to bypass the uORFs, at least in part because of their noncanonical initiation codons, and instead initiate translation at the EPRS CDS during basal conditions. eIF2α-P further facilitates bypass of the uORFs and allows for an increase in EPRS expression in response to stress. This modulation of ribosome bypass ensures appropriate expression of EPRS protein to perform its function as a dual function aminoacyl tRNA synthetase and allows for increased EPRS expression upon cellular stress and changes in demand for appropriately charged aminoacyl-tRNAs.
FIGURE 8.
Model for EPRS translational control. EPRS translation control involves bypass of two inhibitory uORFs with noncanonical initiation codons. In the absence of stress, low levels of eIF2α-P, and high eIF2-GTP, ribosomes scan the 5′-leader of the EPRS mRNA and initiate translation at CUG2, encoded in uORF1, or UUG1, encoded in uORF2. uORF1 overlaps out of frame with the EPRS CDS, and translation of uORF1 results in translation termination 3′ of the start codon for EPRS. A portion of the ribosomes that translate uORF2, encoded by UUG1, terminate and are released from the EPRS mRNA. Alternatively, ribosomes can reinitiate at the downstream EPRS CDS after uORF2 translation. The presence of the CUG and UUG initiation codons allows for a portion of the scanning ribosomes to bypass the uORFs, at least in part because of their noncanonical initiation codons and instead initiate translation at the EPRS CDS during basal conditions. In the presence of stress, high levels of eIF2α-P and diminished eIF2-GTP levels are suggested to further facilitate bypass of the uORFs and allow for an increase in EPRS CDS translation and subsequent protein expression.
Expression of EPRS Is Induced during Diverse Cellular Stresses
EPRS is responsible for charging of glutamyl and prolyl tRNAs with their cognate amino acids. Levels of aminoacyl tRNA synthetases are critical for translation through charging of tRNAs and modulating the available aminoacyl-tRNA pool (23, 24). We show here that EPRS protein expression is enhanced through an uORF-mediated translation mechanism in response to ER stress and to treatment with halofuginone, which is suggested to cause a decrease in the charged prolyl-tRNA pool (21). Lowered global protein synthesis by eIF2α-P would help to conserve resources and allow cells to reconfigure gene expression to alleviate these stress conditions. In the case of halofuginone treatment, increased amounts of uncharged prolyl-tRNAs are suggested to directly activate GCN2 phosphorylation of eIF2α-P (Fig. 7B). The eIF2α-P would then lead to preferential translation of key ISR genes including ATF4, which would facilitate nutrient uptake and alter metabolism to better manage the change in tRNA charging. Of importance, eIF2α-P would also lead to increased EPRS expression that would serve to diminish the toxicity of the drug (Fig. 7B). This model is central to the surgical stress resistance concept whereby nutrient depletion prior to surgery provides for a boost in expression of genes that would enable for subsequent protection from ischemic damage occurring during surgical procedures (20). GCN2 induction of protective patterns of gene expression is suggested not just to be restricted to nutrient depletion. For example, budding yeast exposure to high salinity results in a transient decrease in the charging of several different tRNAs, most likely because of changes in amino acid transport and/or aminoacyl tRNA synthetase expression or activity (25–27). Therefore, stresses not directly linked to starvation for amino acids can change the status of tRNA charging and activate GCN2 and the ISR.
Halofuginone directly inhibits the prolyl-tRNA charging function of EPRS, and expression of EPRS in the ISR is suggested to diminish the toxicity of halofuginone treatment. However, expression of EPRS and other aminoacyl-tRNA synthetases are induced by eIF2α-P and the ISR in response to diverse environmental stresses (28). One benefit of enhanced levels of aminoacyl tRNA synthetases would be to rapidly restore translation in the feedback regulation of the ISR. In addition to their role in regulating translation through tRNA charging, aminoacyl tRNA synthetases are also suggested to serve a role in proofreading to prevent tRNA charging with damaged amino acids, thereby ensuring translation fidelity and proper protein folding and function (29, 30). Finally, aminoacyl tRNA synthetases can serve other functions unrelated to tRNA charging. EPRS, for example, functions in the GAIT complex (γ-interferon-activated inhibitor of translation) to repress translation of a class of inflammatory mRNAs in immune cells (31).
Specific Features of uORFs Function to Regulate Translation Control
Although many ISR genes are subject to preferential translation via uORF-mediated mechanisms, the mere presence of an uORF is not sufficient to ensure enhanced translation during eIF2α-P. Indeed, uORFs are suggested to be equally present among those transcripts that are translationally enhanced, repressed, or resistant to eIF2α-P (11). This finding suggests that each uORF contains specific features that determine whether the 5′-leader of an mRNA serves to activate or repress translation in response to eIF2α-P. Our study suggests that the use of a noncanonical start codon for an uORF allows for basal ribosome bypass that can be further enhanced during eIF2α-P. Because glutamyl- and prolyl-tRNA charging by EPRS is required for cellular homeostasis, bypass of uORFs with noncanonical initiation codons allows for appropriate levels of the aminoacyl-tRNA pool basally that can be fine-tuned during disruptions to the cellular environment.
The EPRS mechanism of translation control contains features of both the ATF4 and CHOP preferential translation mechanisms. The ATF4 mechanism of “delayed translation reinitiation” relies upon translation of an uORF that overlaps out of frame with the ATF4 CDS to thwart ATF4 protein production during basal conditions (6). EPRS translation control similarly utilizes an overlapping out of frame uORF to dampen basal EPRS expression. During cellular stress and eIF2α-P, however, the overlapping uORF1 and an additional inhibitory 5′ proximal uORF2 in the EPRS 5′-leader can be bypassed through the use of noncanonical initiation codons in the inhibitory uORFs (Fig. 8). This is reminiscent of a feature from the CHOP and GADD34 mechanisms of preferential translation in which ribosomes bypass an inhibitory uORF in part because of its poor Kozak start codon context (5, 8).
In addition to EPRS, a recent study illustrated that BiP (GRP78/HSPA5) is an example of another gene that is required for protein homeostasis whose translational expression is regulated by uORFs with a noncanonical initiation codon (32). Interestingly, BiP appears to incorporate these uORFs in a mechanism of translational control that involves an internal ribosome entry sequence that would allow for BiP translation to efficiently occur despite robust eIF2α-P during environmental stress (32–34). Additionally, amino acid starvation results in increased GADD45G expression through a mechanism involving an overlapping out of frame uORF with a CUG initiation codon (15). The rules for regulation of ribosome bypass examined herein for EPRS provide new insight into the uORF-mediated translation control mechanisms that serve to regulate ISR-induced gene transcripts during eIF2α-P but also illustrate the role that previously unexamined uORFs with noncanonical initiation codons can possess in modulating gene expression. These findings have implications for the genome-wide assessment of uORF prevalence and provides an additional uORF feature that can be used to predict the basic translation control properties for an mRNA during basal conditions and those that induce eIF2α-P.
Author Contributions
S. K. Y. conceived the study, designed, performed and analyzed experiments, and wrote the manuscript. T. D. B. conceived the study, designed, performed and analyzed experiments, and wrote the manuscript. R. C. W. conceived and coordinated the study, designed and analyzed experiments, and wrote the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank members of the Wek laboratory for helpful discussions, and Christopher Davis who was instrumental in earlier pilot studies.
This work was supported by National Institutes of Health Grant GM049164 (to R. C. W.) and the Ralph W. and Grace M. Showalter Research Trust Fund. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- eIF2α-P
- phosphorylation of eIF2α
- ISR
- integrated stress response
- uORF
- upstream ORF
- ER
- endoplasmic reticulum
- CDS
- coding sequence
- RACE
- rapid amplification of cDNA ends
- MEF
- mouse embryonic fibroblast
- qRT-PCR
- quantitative RT-PCR
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- PERK
- protein kinase R-like ER kinase.
References
- 1. Hinnebusch A. G. (2014) The scanning mechanism of eukaryotic translation initiation. Annu. Rev. Biochem. 83, 779–812 [DOI] [PubMed] [Google Scholar]
- 2. Baird T. D., and Wek R. C. (2012) Eukaryotic initiation factor 2 phosphorylation and translational control in metabolism. Adv. Nutr. 3, 307–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kozak M. (1984) Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucleic Acids Res. 12, 857–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Walter P., and Ron D. (2011) The unfolded protein response: from stress pathway to homeostatic regulation. Science 334, 1081–1086 [DOI] [PubMed] [Google Scholar]
- 5. Palam L. R., Baird T. D., and Wek R. C. (2011) Phosphorylation of eIF2 facilitates ribosomal bypass of an inhibitory upstream ORF to enhance CHOP translation. J. Biol. Chem. 286, 10939–10949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vattem K. M., and Wek R. C. (2004) Reinitiation involving upstream open reading frames regulates ATF4 mRNA translation in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 101, 11269–11274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Novoa I., Zeng H., Harding H. P., and Ron D. (2001) Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2alpha. J. Cell Biol. 153, 1011–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Young S. K., Willy J. A., Wu C., Sachs M. S., and Wek R. C. (2015) Ribosome reinitiation directs gene-specific translation and regulates the integrated stress response. J. Biol. Chem. 290, 28257–28271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Harding H. P., Novoa I., Zhang Y., Zeng H., Wek R., Schapira M., and Ron D. (2000) Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell 6, 1099–1108 [DOI] [PubMed] [Google Scholar]
- 10. Hinnebusch A. G. (2005) Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 59, 407–450 [DOI] [PubMed] [Google Scholar]
- 11. Baird T. D., Palam L. R., Fusakio M. E., Willy J. A., Davis C. M., McClintick J. N., Anthony T. G., and Wek R. C. (2014) Selective mRNA translation during eIF2 phosphorylation induces expression of IBTKalpha. Mol. Biol. Cell 25, 1686–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Iacono M., Mignone F., and Pesole G. (2005) uAUG and uORFs in human and rodent 5′untranslated mRNAs. Gene 349, 97–105 [DOI] [PubMed] [Google Scholar]
- 13. Ingolia N. T., Lareau L. F., and Weissman J. S. (2011) Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147, 789–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lee S., Liu B., Lee S., Huang S. X., Shen B., and Qian S. B. (2012) Global mapping of translation initiation sites in mammalian cells at single-nucleotide resolution. Proc. Natl. Acad. Sci. U.S.A. 109, E2424–2432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gao X., Wan J., Liu B., Ma M., Shen B., and Qian S. B. (2015) Quantitative profiling of initiating ribosomes in vivo. Nat. Methods 12, 147–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jiang H. Y., Wek S. A., McGrath B. C., Lu D., Hai T., Harding H. P., Wang X., Ron D., Cavener D. R., and Wek R. C. (2004) Activating transcription factor 3 (ATF3) is integral to the eIF2 kinase stress response. Mol. Cell. Biol. 24, 1365–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Teske B. F., Fusakio M. E., Zhou D., Shan J., McClintick J. N., Kilberg M. S., and Wek R. C. (2013) CHOP induces activating transcription factor 5 (ATF5) to trigger apoptosis in response to perturbations in protein homeostasis. Mol. Biol. Cell 24, 2477–2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Teske B. F., Baird T. D., and Wek R. C. (2011) Methods for analyzing eIF2 kinases and translational control in the unfolded protein response. Methods Enzymol. 490, 333–356 [DOI] [PubMed] [Google Scholar]
- 19. Zhou D., Palam L. R., Jiang L., Narasimhan J., Staschke K. A., and Wek R. C. (2008) Phosphorylation of eIF2 directs ATF5 translational control in response to diverse stress conditions. J. Biol. Chem. 283, 7064–7073 [DOI] [PubMed] [Google Scholar]
- 20. Peng W., Robertson L., Gallinetti J., Mejia P., Vose S., Charlip A., Chu T., and Mitchell J. R. (2012) Surgical stress resistance induced by single amino acid deprivation requires Gcn2 in mice. Sci. Transl. Med. 4, 118ra111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Keller T. L., Zocco D., Sundrud M. S., Hendrick M., Edenius M., Yum J., Kim Y. J., Lee H. K., Cortese J. F., Wirth D. F., Dignam J. D., Rao A., Yeo C. Y., Mazitschek R., and Whitman M. (2012) Halofuginone and other febrifugine derivatives inhibit prolyl-tRNA synthetase. Nat. Chem. Biol. 8, 311–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhou H., Sun L., Yang X. L., and Schimmel P. (2013) ATP-directed capture of bioactive herbal-based medicine on human tRNA synthetase. Nature 494, 121–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ibba M., and Soll D. (2000) Aminoacyl-tRNA synthesis. Annu. Rev. Biochem. 69, 617–650 [DOI] [PubMed] [Google Scholar]
- 24. Ling J., Reynolds N., and Ibba M. (2009) Aminoacyl-tRNA synthesis and translational quality control. Annu. Rev. Microbiol. 63, 61–78 [DOI] [PubMed] [Google Scholar]
- 25. Zaborske J. M., Narasimhan J., Jiang L., Wek S. A., Dittmar K. A., Freimoser F., Pan T., and Wek R. C. (2009) Genome-wide analysis of tRNA charging and activation of the eIF2 kinase Gcn2p. J. Biol. Chem. 284, 25254–25267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bernales S., Papa F. R., and Walter P. (2006) Intracellular signaling by the unfolded protein response. Annu. Rev. Cell Dev. Biol. 22, 487–508 [DOI] [PubMed] [Google Scholar]
- 27. Narasimhan J., Staschke K. A., and Wek R. C. (2004) Dimerization is required for activation of eIF2 kinase Gcn2 in response to diverse environmental stress conditions. J. Biol. Chem. 279, 22820–22832 [DOI] [PubMed] [Google Scholar]
- 28. Harding H. P., Zhang Y., Zeng H., Novoa I., Lu P. D., Calfon M., Sadri N., Yun C., Popko B., Paules R., Stojdl D. F., Bell J. C., Hettmann T., Leiden J. M., and Ron D. (2003) An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell 11, 619–633 [DOI] [PubMed] [Google Scholar]
- 29. Bullwinkle T. J., Reynolds N. M., Raina M., Moghal A., Matsa E., Rajkovic A., Kayadibi H., Fazlollahi F., Ryan C., Howitz N., Faull K. F., Lazazzera B. A., and Ibba M. (2014) Oxidation of cellular amino acid pools leads to cytotoxic mistranslation of the genetic code. eLife 3, 02501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bullwinkle T., Lazazzera B., and Ibba M. (2014) Quality control and infiltration of translation by amino acids outside of the genetic code. Annu. Rev. Genet. 48, 149–166 [DOI] [PubMed] [Google Scholar]
- 31. Mukhopadhyay R., Jia J., Arif A., Ray P. S., and Fox P. L. (2009) The GAIT system: a gatekeeper of inflammatory gene expression. Trends Biochem. Sci. 34, 324–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Starck S. R., Tsai J. C., Chen K., Shodiya M., Wang L., Yahiro K., Martins-Green M., Shastri N., and Walter P. (2016) Translation from the 5′ untranslated region shapes the integrated stress response. Science 351, aad3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cho S., Park S. M., Kim T. D., Kim J. H., Kim K. T., and Jang S. K. (2007) BiP internal ribosomal entry site activity is controlled by heat-induced interaction of NSAP1. Mol. Cell. Biol. 27, 368–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kim Y. K., and Jang S. K. (2002) Continuous heat shock enhances translational initiation directed by internal ribosomal entry site. Biochem. Biophys. Res. Commun. 297, 224–231 [DOI] [PubMed] [Google Scholar]