Abstract
Late embryogenesis abundant (LEA) proteins are a conserved group of proteins widely distributed in the plant kingdom that participate in the tolerance to water deficit of different plant species. In silico analyses indicate that most LEA proteins are structurally disordered. The structural plasticity of these proteins opens the question of whether water deficit modulates their conformation and whether these possible changes are related to their function. In this work, we characterized the secondary structure of Arabidopsis group 4 LEA proteins. We found that they are disordered in aqueous solution, with high intrinsic potential to fold into α-helix. We demonstrate that complete dehydration is not required for these proteins to sample ordered structures because milder water deficit and macromolecular crowding induce high α-helix levels in vitro, suggesting that prevalent conditions under water deficit modulate their conformation. We also show that the N-terminal region, conserved across all group 4 LEA proteins, is necessary and sufficient for conformational transitions and that their protective function is confined to this region, suggesting that folding into α-helix is required for chaperone-like activity under water limitation. We propose that these proteins can exist as different conformers, favoring functional diversity, a moonlighting property arising from their structural dynamics.
Keywords: Arabidopsis thaliana, intrinsically disordered protein, plant molecular biology, protein folding, protein structure, stress response
Introduction
Low water availability caused by different environmental conditions such as drought, or low temperatures represents a vulnerable situation for many forms of life, particularly for plants. To contend with and to overcome these adverse environments, numerous complex response mechanisms have been selected in the different species of the plant kingdom. One of the most conserved responses is the accumulation of a group of proteins known as late embryogenesis abundant (LEA)3 proteins (1). LEA proteins have been found in all the orthodox dry seeds (embryos) where they have been searched (1, 2), and they also accumulate in response to water limitation in all vegetative tissues (2, 3). Most LEA proteins show high hydrophilicity, high content of small amino acids, and absence or deficit of hydrophobic residues, properties that are extended to a larger set of proteins called hydrophilins, which have been found in species from the three domains of life and that also accumulate under water deficit (2, 4). The composition of these proteins is also characteristic of a group of proteins known as intrinsically disordered proteins (IDPs) (5, 6). Consistent with the predicted structural disorder for most LEA proteins, structural analyses have confirmed this property for some of them in aqueous solution (7–13). Based on their sequence similarity, LEA proteins have been classified in seven groups or families, each one characterized by the presence of specific sequence motifs (2). In Arabidopsis thaliana there are 51 genes encoding LEA proteins from six of the seven families (3). Group 4 LEA (LEA4) proteins are one of the smallest families of LEA proteins in Arabidopsis consisting of only three members: AtLEA4-1 (At1g32560), AtLEA4-2 (At2g35300), and AtLEA4-5 (At5g06760) (3, 14). Group 4 LEA proteins are enriched in charged and small amino acid residues, whereas they lack Cys, Phe, and Trp (2, 3, 14). This group is characterized by an N-terminal region ranging from 74 to 78 amino acid residues, containing conserved amino acid sequence motifs. In silico analysis predicts that this particular region is able to form an amphipathic α-helix structure. The C-terminal region in this protein family is more variable in sequence and length, and it is predicted to be structurally disordered (2, 14). A phylogenetic analysis of group 4 LEA proteins revealed two subclasses in this family (subgroups 4A and 4B) (14). In Arabidopsis, AtLEA4-1 and AtLEA4-2 proteins belong to subgroup 4A, whereas AtLEA4-5 protein fits into subgroup 4B (14).
From the 10 distinctive motifs found in this protein group, the high conservation of motif 2 at the N-terminal region constitutes a signature for this family. The same study also showed that both subgroups emerged from a very early duplication before branching of monocots and dicots, suggesting that this separation gave rise to a subfunctionalization of these subgroups (14). Group 4 LEA proteins and transcripts have been found in dry seeds but also in response to water deficit in vegetative and reproductive tissues (14, 15). Moreover, Arabidopsis mutants deficient in group 4 LEA proteins are sensitive to water deficit, indicating that these proteins participate in the tolerance to this stress condition (14).
Many different functions have been proposed for group 4 LEA proteins such as membrane protectors, sugar or metal binding, radical scavengers, and protein dehydro- and cryo-protectors (16–20). A. thaliana AtLEA4-5 protein was shown to prevent inactivation and conformational changes of reporter enzymes such as lactate dehydrogenase (LDH) and malate dehydrogenase after partial dehydration and freeze/thaw cycles from 1:1 molar ratios, indicating that group 4 LEA proteins have a chaperone-like function to protect other proteins from the effects of water deficit (19, 20).
Studies on animal and bacterial IDPs have shown that these proteins can gain structural order upon binding to a specific partner, interaction that leads to IDP folding (21–25); however, in some other examples, the function of globular chaperones is linked to an order to disorder structural transitions in response to environmental cues such as those imposed by changes in pH or redox state (26, 27). Even though it has been shown that severe dehydration can promote folding of some LEA proteins (8, 11, 13, 28–31), the possibility that the environmental effects caused by mild water deficit in the cell (such as those occurring in vegetative tissues) leads to higher structural order in those IDPs responsive to this stressful environment (e.g. LEA proteins and other hydrophilins) and whether this structural changes could be related to their function are still open questions.
In this work, we demonstrate that even though Arabidopsis members of subgroups 4A (AtLEA4-2) and 4B (AtLEA4-5) LEA proteins are structurally disordered in solution, low osmotic potentials and macromolecular crowding can induce significant levels of α-helix, particularly in the conserved AtLEA4-5 N-terminal region, whereas the C-terminal region displays high structural disorder. We also show that the AtLEA4-5 N-terminal region is necessary and sufficient for the protective effect of this protein on reporter enzyme activities after freeze-thaw cycles and partial dehydration at low molar ratios. Our data support the hypothesis that cellular environment modulates the structural organization of disordered proteins and that these structural changes are related to their functions.
Experimental Procedures
In Silico Analyses
AtLEA4 proteins were aligned using T-Coffee multiple sequence alignment. Secondary structure prediction was determined using AGADIR helical content predictor (32). Intrinsically disordered tendency was predicted using DISpro (33), PONDR (34), and DISOPRED3 (35).
Plasmid Constructions
ORFs of AtLEA4-1, AtLEA4-2, and AtLEA4-5 genes were cloned using cDNA from RNA obtained from Arabidopsis dry seeds. AtLEA4-5 ORF was amplified by PCR using specific primers containing at their ends NcoI (5′-AAACCATGGAGTCGATGAAAGAAAC-3′) and SalI (5′-GCGGTCGACCCGTTTATCCAGTATATCC-3′) restriction sites. This cloning strategy led to a modification in the second codon, which in the recombinant version corresponds to glutamic acid (GAG) instead of glutamine (CAG). The amplicon was cloned into pJET1.2/blunt and subsequently digested with NcoI and SalI for its insertion into pTrc99A vector. To eliminate the Met33 in AtLEA4-5 ORF used in bacterial cells as an alternative translation initiation site and responsible of the production of an additional shorter AtLEA4-5 protein, directed mutagenesis of AtLEA4-5 ORF sequence was conducted using the following overlapping primers to exchange Met33 for a Leu residue: sense (5′-GGAGGAAAAGGCGGAGAAGCTGAAGAC-3′) and antisense (5′-GTCTTCAGCTTCTCCGCCTTTTCCTCC-3′). The modified DNA fragment was inserted into pJET1.2/blunt plasmid vector to produce pJET1.2:AtLEA4-5. For protein production, AtLEA4-5 ORF was transferred to pTrc99A plasmid vector by digesting with NcoI and SalI restriction enzymes. The DNA fragments encoding AtLEA4-5 N- and C-terminal regions were obtained from pJET1.2:AtLEA4-5 plasmid, using the following sense and antisense oligonucleotides: 5′-AAACCATGGAGTCGATGAAAGAAAC-3′ and antisense 5′-CGCGTCGACTCAGGTTCCGGCTCCAGCCGC-3′ and sense 5′-AAACCATGGCCGGTTTAGGTTTGGGGAC-3′ and antisense 5′-GCGGTCGACCCGTTTATCCAGTATATCC-3′, respectively, which were also inserted into pTrc99A to generate pTrc99:AtLEA4-51–77 and pTrc99:AtLEA4-578–158 plasmids, respectively. In all cases, nucleotide sequences were verified accordingly.
Because pTrc99A:AtLEA4-1, pTrc99A:AtLEA4-2, and pTrc99A:AtLEA4-578–158 did not lead to a successful protein expression in bacteria, instead corresponding ORFs were inserted into the pTYB11 vector to obtain them as intein fusion proteins (IMPACT-CN expression system; New England Biolabs Inc.). To this end, AtLEA4-1 and AtLEA4-2 coding sequences were amplified from pJET1.2 intermediary plasmids using specific oligonucleotides containing SapI and PstI restriction sites: 5′-GGTGGTTGCTCTTCCAACATGCAATCGGCGAAACAGAAG-3′ and 5′-GGTGGTCTGCAGTCATTAGTAGTGATGATGATTATGATGTCC-3′ for AtLEA4-1 and, 5′-GGTGGTTGCTCTTCCAACATGCAGTCGGCGAAGG-3′ and 5′-GGTGGTCTGCAGTCATTAGATCTGTCCCGGCG-3′ for AtLEA4-2. To amplify the AtLEA4-578–158 coding sequence, the oligonucleotides used were 5′-GGTGGTTGCTCTTCCAACACCGGTTTAGGTTTGGGGAC-3′ and 5′-GGTGGTCTGCAGTCATTATCCAGTATATCCCCCGC-3′.
Expression and Purification of Recombinant Proteins
Recombinant plasmids derived from pTrc99A or pTYB11 as described above were transformed into Escherichia coli BL21(DE3) pLysS competent cells (Promega). Single colonies were inoculated in fresh LB medium containing 100 μg/ml ampicillin and grown overnight at 37 °C, from which 1 liter of fresh LB medium was inoculated to 0.01 A600 and grown at 37 °C to 0.5–0.8 A600. At this point, protein expression was induced with 1 mm isopropyl β-d-1-thiogalactopyranoside for 6 h at 25 °C. Cell cultures were harvested by centrifugation. For AtLEA4-5 and AtLEA4-51–77 purification, we used a straightforward method designed for nonacidic recombinant unstructured proteins as described by Campos et al. (36). After washing twice with acetone, the protein was resuspended in 10 mm sodium phosphate buffer, pH 7.5, and dialyzed extensively against the same buffer. For intein fused AtLEA4-1, AtLEA4-2, and AtLEA4-578–158, a different purification procedure was followed. Bacterial pellets were resuspended in lysis buffer (20 mm sodium phosphate, pH 8, 500 mm NaCl, 0.1% Triton X-100) containing one tablet of cOmplete protease inhibitor mixture (Roche) per 50 ml of buffer. The cells were lysed by sonication on ice, and the extract was clarified by centrifugation at 20,000 × g for 30 min at 4 °C. To obtain proteins lacking the intein tag, the clarified extract was loaded onto a chitin column following the procedure described by the manufacturer (IMPACTTM-CN kit). The eluted fractions were analyzed for the presence of the recombinant proteins by SDS-PAGE. In contrast to AtLEA4-2, this analysis showed that AtLEA4-1 and AtLEA4-578–158 are most probably protease-susceptible proteins because we were unable to detect the complete corresponding polypeptides, despite using protease-deficient bacterial strains, as is the case for BL21 and RosettaTM 2(DE3) (Merck-Millipore), strains successfully used to purify a number of recombinant proteins from different organisms. Fractions containing AtLEA4-2 were pooled and extensively dialyzed against 10 mm sodium phosphate buffer, pH 7.5. The purity and identity of the different purified proteins were confirmed by SDS-PAGE and by LC-MS. LC-MS was performed by the Proteomic Facility of the Instituto de Biotecnología/Universidad Nacional Autónoma de México following standard methods using LQT-Orbitrap Velos (Thermo-Fisher) mass spectrometer with nanospray ionization system. Proteins were quantified using their molar extinction coefficient at 280 nm (ϵ = 2,980 m−1 cm−1). Because AtLEA4-51–77 lacks aromatic amino acid residues, its concentration was determined by Bradford assay and verified by SDS-PAGE comparing with known concentrations of AtLEA4-5. Purified proteins were conserved by lyophilization or in aliquots at −80 °C until used. Because it was not possible to obtain whole AtLEA4-578–158, the complete chemically synthesized polypeptide corresponding to this truncated protein was purchased from Biomatik (Cambridge, Canada) and extensively dialyzed before use. This polypeptide was verified by mass spectrometry and HPLC.
Far UV Circular Dichroism Spectroscopy
AtLEA4-2, AtLEA4-5, and AtLEA4-51–77 recombinant proteins, as well as AtLEA4-578–158 polypeptide were diluted to 0.3 mg/ml and far UV CD spectra were recorded using a Jasco J-715 CD spectropolarimeter (JASCO Analytical Instruments) on a 0.1-cm-path length cell from 190 to 250 nm. Desired temperature was regulated with a Peltier temperature-controlled cell holder (PTC-4235; JASCO). Three spectra were averaged and smoothed to reduce noise. Each spectrum was acquired every 1 nm with 2-s average time per point and 1-nm band pass. Secondary structure estimation was calculated using Dichroweb software (37, 38). The CDSSTR algorithm was used with 4, 7, and SP175 data sets. These assays were reproduced using proteins from at least three independent purification batches.
In Vitro Freeze-Thaw Assay
Freeze-thaw in vitro assays were carried out as previously described by Reyes et al. (19) with small modifications. Briefly, LDH from rabbit muscle (Roche) was diluted to a final concentration of 250 nm (monomer) with or without the corresponding test protein (AtLEA4-2, AtLEA4-5, AtLEA4-51–77, AtLEA4-578–158, or lysozyme) in buffer 25 mm Tris-HCl, pH 7.5. The different test proteins were set to the desired molar ratio from 0.5:1 to 20:1 (test protein:LDH), considering 250 nm as 1:1 molar ratio. Mixtures in a final volume of 100 μl were frozen for 30 s in liquid N2 and subsequently thawed at 25 °C in a thermomixer (Eppendorf). This procedure constituted one freeze-thaw cycle, which was repeated up to seven times. After the treatment, LDH activity was measured as reported (19). Enzyme activities for each sample were measured in at least three independent tests (each one with three technical replicates). These experiments were reproduced with proteins from at least three independent purification batches.
In Vitro Partial Dehydration Assay
Partial dehydration in vitro assays were performed as previously described by Reyes et al. (20) with small modifications. LDH from rabbit muscle (Roche) was diluted to 250 nm (monomer) as the final concentration, in the presence or absence of the corresponding test protein (AtLEA4-2, AtLEA4-5, AtLEA4-51–77, or AtLEA4-578–158) in buffer of 25 mm Tris-HCl, pH 7.5. The different test proteins were set to 5:1 molar ratio (1.25 μm). Mixtures in a final volume of 25 μl were placed in a SpeedVac concentrator (Savant Instruments), and water was evaporated to achieve ≥98% water loss, keeping constant temperature. The percentage of partial water loss was defined as the amount of water evaporated from the samples by means of weight. Partially dehydrated samples were rehydrated to the initial weight with water, assuring that all solutes were completely resuspended. LDH activity was measured as described above. These assays were reproduced using protein samples from at least three independent purification batches.
Statistical Analyses
Statistical analyses were carried out using one-way analysis of variance test. Significant differences were calculated with Tukey's multiple comparison post-test (p < 0.01).
Results
A. thaliana Group 4 Late Embryogenesis Abundant Proteins Are Intrinsically Disordered Proteins in Aqueous Solution
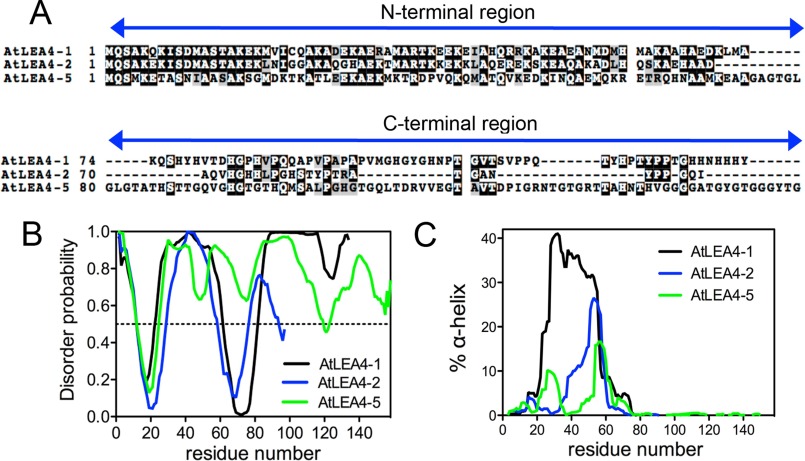
In A. thaliana, there are three genes encoding group 4 LEA proteins: AtLEA4-1, AtLEA4-2, and AtLEA4-5 (3, 14). These proteins have low molecular masses between 10.5 and 16.2 kDa and pI values between 8.95 and 9.67. All of them present a 74–78-amino acid long N-terminal domain, highly conserved across all group 4 LEA proteins described so far in the plant kingdom, which contains distinctive motifs for this protein family (Fig. 1A). Because of the amino acid composition of this group of LEA proteins, it was proposed that they are IDPs (2, 12). In silico analyses using PONDR (VLXT) (34) (Fig. 1B), DISpro (33), and DISOPRED3 (35) (data not shown) indicated that, despite their sequence similarity, they possess different levels of disorder.
FIGURE 1.
In silico structure prediction of Arabidopsis group 4 LEA proteins. A, sequence alignment of Arabidopsis group 4 LEA proteins showing N-terminal conserved and C-terminal variable regions. B, AtLEA4-1 (black), AtLEA4-2 (blue), and AtLEA4-5 (green) structural disorder levels using PONDR predictor. According to this algorithm, proteins with disorder probability values above 0.5 are considered highly disordered. C, percentage of α-helix predicted using AGADIR for AtLEA4-1 (black), AtLEA4-2 (blue), and AtLEA4-5 (green).
To characterize the LEA4 proteins structural features, we expressed and purified the three recombinant proteins in E. coli. AtLEA4-2 and AtLEA4-5 were obtained with more than 95% purity (data not shown). Because AtLE4–1 was mostly degraded during different expression and purification procedures, the rest of the experiments were performed only with AtLEA4-2 and AtLEA4-5, representing the two LEA4 subgroups, 4A and 4B, respectively (14).
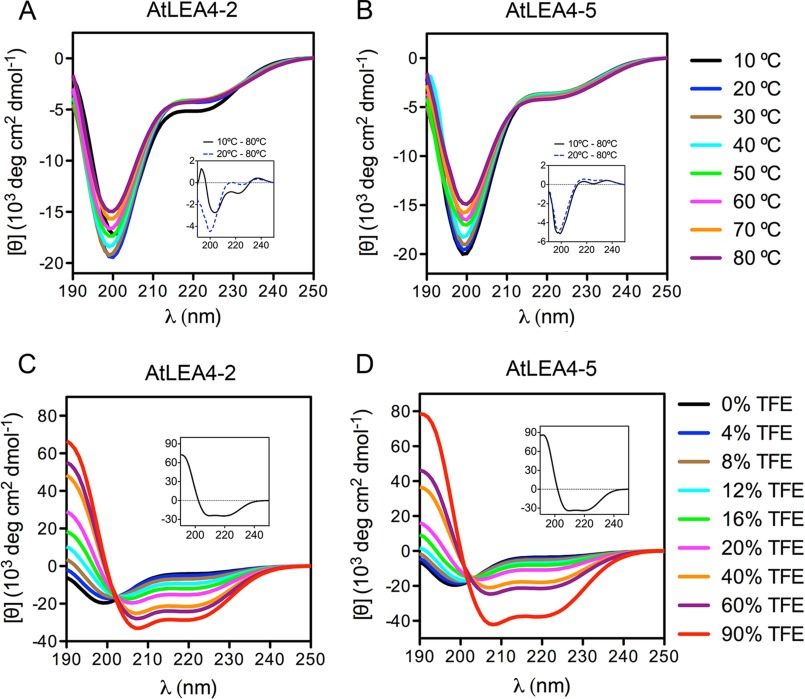
To determine the secondary structure of AtLEA4-2 and AtLEA4-5 in solution, the purified proteins were analyzed by far UV CD. The results obtained corroborated that both proteins are mostly disordered in solution over a wide range of temperatures (Fig. 2, A and B) and pH (data not shown), with the characteristic negative band for random coil structures around 198 nm. Both spectra showed a significant negative signal around 222 nm (typical of α-helix), suggesting that these proteins possess residual α-helix structure. The comparative analysis of the AtLEA4-2 CD difference spectra obtained at different temperatures (Δ10–80 °C and Δ20–80 °C) revealed that this protein is able to form α-helix structures at low temperatures (10 °C), whereas at higher temperatures the protein is mostly disordered (Fig. 2A, inset). By contrast, a similar analysis indicated that AtLEA4-5 is mostly disordered under all temperatures tested (Fig. 2B, inset). We did not find evidence of extended helical conformations (e.g. poly-l-proline II) for any of these proteins, such as those found in LEA groups 1, 2, and 6 (10, 39). Altogether, these data demonstrate that AtLEA4 proteins are IDPs with residual α-helix structure in aqueous solution.
FIGURE 2.
Far UV CD spectra of AtLEA4-2 and AtLEA4-5 in aqueous solution at various temperatures and in different TFE concentrations. A and B, far UV CD spectra of AtLEA4-2 (A) and AtLEA4-5 (B) at 10 °C (black), 20 °C (blue), 30 °C (brown), 40 °C (cyan), 50 °C (green), 60 °C (magenta), 70 °C (orange), and 80 °C (purple). Insets in A and B show Δ10–80 °C (continuous black line) and Δ20–80 °C (dashed blue line) difference spectra. C and D, AtLEA4-2 (C) and AtLEA4-5 (D) in TFE/water mixtures at 0% (black), 4% (blue), 8% (brown), 12% (cyan), 16% (green), 20% (magenta), 40% (orange), 60% (purple), and 90% (red) TFE. The difference spectra (Δ90–0% TFE) are shown as insets in C and D. These results were reproduced at least four times in autonomous experiments, using three independent purification batches of both proteins.
AtLEA4-2 and AtLEA4-5 Have the Potential to Acquire High Levels of Ordered Structure
To determine the intrinsic ability of AtLEA4-2 and AtLEA4-5 to attain helicity, CD analyses were performed in the presence of different concentrations of 2,2,2-trifluoroethanol (TFE), a well known α-helix inducer (40, 41). Increasing TFE concentrations promote α-helix formation in both proteins, as revealed by the progressive increase in [θ]198 toward positive values and the transition of the minimum at [θ]222 onward more negative values (Fig. 2, C and D). Difference spectra showed that both proteins are able to gain high helicity levels; however, Δ90–0% TFE indicates that AtLEA4-5 reaches higher α-helix percentage than AtLEA4-2 (Fig. 2, C and D, insets). Assessment of protein secondary structure using Dichroweb (37) indicated that AtLEA4-2 adopts 7% of α-helix in 4% TFE, and it reaches up to 80% α-helix in 90% TFE (Table 1), showing a decrease in its structural disorder from 61% in aqueous solution to 16% in 90% TFE. The α-helix content for AtLEA4-5 increased from 6% to 88% in 4% to 90% ΤFΕ, whereas its disordered structure decreased from 61% to 10% (Table 1). Both AtLEA4-2 and AtLEA4-5 CD spectra obtained with 4% to 90% TFE showed isodichroic points (Fig. 2, C and D), indicating that these proteins are able to adopt two structural conformations in equilibrium under these conditions: one mostly unstructured favored in aqueous solution (U4–2 and U4–5) and a second one with higher helicity promoted by increasing TFE concentrations (F4–2 and F4–5). The fitted straight lines obtained from transition diagrams for AtLEA4-2 and AtLEA4-5 support the formation of two conformers for both proteins (Fig. 3, E and F), as was indicated by the presence of isodichroic points. These data demonstrate that AtLEA4-2 and AtLEA4-5 possess an intrinsic ability to form α-helical species, which under some conditions are in equilibrium with unfolded conformations (U4–2 ↔ F4–2; U4–5 ↔ F4–5).
TABLE 1.
Percentage of helix, strand, and unordered structures in AtLEA4-2 and AtLEA4-5
Secondary structure content in AtLEA4-2 and AtLEA4-5 proteins was obtained by far UV CD spectrometry and calculated with Dichroweb server.
| Treatment | AtLEA4-2 |
AtLEA4-5 |
||||
|---|---|---|---|---|---|---|
| Helix | Strand | Unordered | Helix | Strand | Unordered | |
| % | % | |||||
| Aqueous solution | 5 | 33 | 61 | 5 | 34 | 61 |
| 4% TFE | 7 | 32 | 61 | 6 | 33 | 61 |
| 8% TFE | 16 | 25 | 58 | 6 | 32 | 61 |
| 12% TFE | 21 | 23 | 56 | 17 | 23 | 59 |
| 16% TFE | 35 | 15 | 51 | 18 | 25 | 55 |
| 20% TFE | 42 | 15 | 44 | 32 | 17 | 51 |
| 40% TFE | 57 | 9 | 33 | 49 | 13 | 39 |
| 60% TFE | 62 | 9 | 30 | 57 | 8 | 35 |
| 90% TFE | 80 | 3 | 16 | 88 | 2 | 10 |
| 10% glycerol | 5 | 34 | 60 | 4 | 34 | 61 |
| 20% glycerol | 8 | 32 | 61 | 7 | 36 | 57 |
| 30% glycerol | 18 | 22 | 60 | 7 | 34 | 59 |
| 40% glycerol | 21 | 23 | 57 | 18 | 24 | 57 |
| 50% glycerol | 33 | 12 | 53 | 18 | 23 | 59 |
| 60% glycerol | 43 | 14 | 43 | 34 | 15 | 51 |
| 70% glycerol | 49 | 9 | 41 | 42 | 11 | 48 |
| 80% glycerol | 54 | 12 | 33 | 46 | 9 | 45 |
| 45% PEG 5000 | 37 | 11 | 52 | 39 | 9 | 53 |
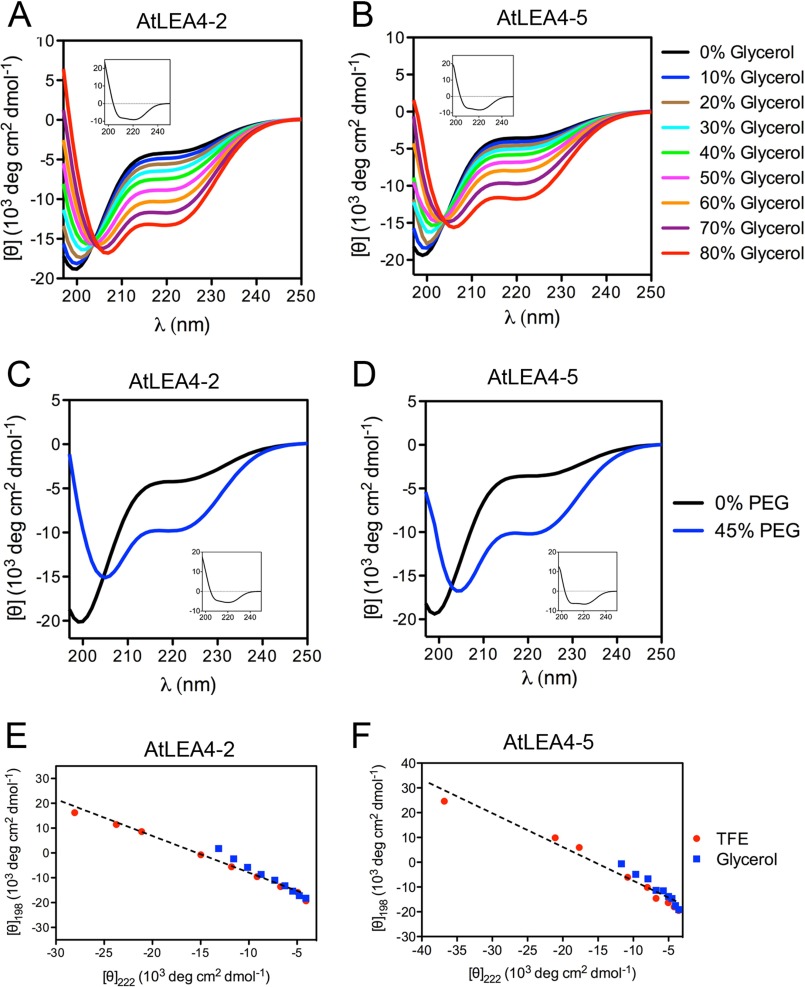
FIGURE 3.
Far UV CD spectra of AtLEA4-2 and AtLEA4-5 in different concentrations of glycerol or PEG. A and B, far UV CD spectra of AtLEA4-2 (A) and AtLEA4-5 (B) in glycerol/water mixtures at 0% (black), 10% (blue), 20% (brown), 30% (cyan), 40% (green), 50% (magenta), 60% (orange), 70% (purple), and 80% (red) glycerol. Difference spectra (Δ80–0% glycerol) are shown as insets in A and B. C and D, AtLEA4-2 (C) and AtLEA4-5 (D) in aqueous solution (black line) and in PEG/water mixture at 45% PEG 5000 (blue line). Difference spectra (Δ45–0% PEG) are shown as insets in C and D. E and F, transition diagrams for AtLEA4-2 (E) and AtLEA4-5 (F) were obtained using the ellipticity values at 198 and 222 nm from TFE and glycerol titrations. Dashed lines represent the linear fits of the data. These results were reproduced at least four times in autonomous experiments, using three independent purification batches of both proteins.
AtLEA4-2 and AtLEA4-5 Fold to α-Helix in Response Low Water Availability and Macromolecular Crowding Induced in Vitro
Previous studies have shown that recombinant LEA proteins from different groups acquire secondary structure, mostly α-helix, when subjected to complete dehydration (8, 11, 13, 28–31). Given the intrinsic potential of AtLEA4-2 and AtLEA4-5 to gain α-helix conformation and because these proteins accumulate even under mild water limitation, we hypothesized that in vitro conditions limiting water availability could induce changes in their secondary structure. Addition of increasing glycerol concentrations led to a notorious progressive gain in α-helix structure in both proteins, as shown by the [θ]198 change toward positive values and a deeper minimum at [θ]222 (Fig. 3, A and B). Analysis using Dichroweb estimated a small difference in α-helix content between these two proteins: 54% α-helix for AtLEA4-2 and 46% for AtLEA4-5 at the highest glycerol concentration (80%) (Table 1). We observed the presence of isodichroic points in both cases (Fig. 3, A and B), which was also supported by their corresponding transition diagrams (Fig. 3, E and F), that together with those obtained from TFE treatments showed that AtLEA4-2 and AtLEA4-5 seem to follow the same folding pathway to α-helix under both treatments (Fig. 3, E and F).
Inside living cells, macromolecules are present at very high concentrations (∼400 g/liter) (42–44), a condition that is typically known as macromolecular crowding (45, 46). This state is further exacerbated in cells under water deficit, reaching macromolecular concentrations up to ∼900 g/liter upon severe dehydration (42). PEG was used to simulate a crowded environment in vitro. The addition of 45% PEG 5000 to AtLEA4-2 or AtLEA4-5 solutions clearly induced changes in their structural conformations (Fig. 3, C and D). Dichroweb estimations indicate 37 and 39% α-helix gains for AtLEA4-2 and AtLEA4-5, respectively (Table 1). Together, these data indicate that AtLEA4-2 and AtLEA4-5 can acquire secondary structure under low water availability or macromolecular crowding in vitro, possibly reflecting what occurs in plant cells under water deficit.
The N-terminal Region of AtLEA4 Proteins Is Necessary and Sufficient for the Conformational Changes Induced by Water Deficit
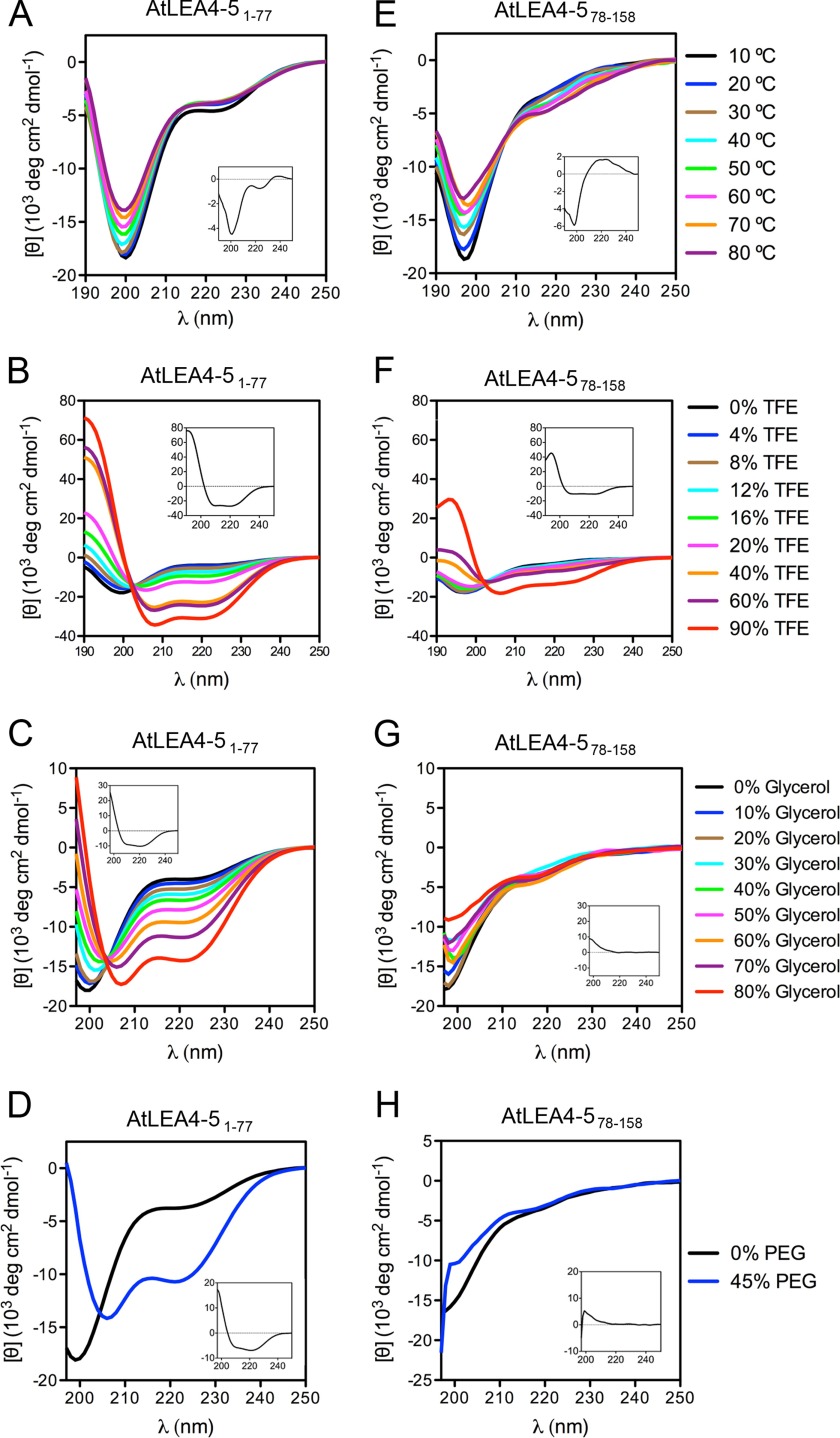
Plant group 4 LEA proteins are characterized by the presence of conserved motifs at their N-terminal region (2, 14). In silico analysis predicts that this region of 70, 74, and 77 residues in AtLEA4-1, AtLEA4-2, and AtLEA4-5, respectively, has a higher propensity to adopt α-helical conformations than the C-terminal region (Fig. 1C). To test this prediction, we performed far UV CD experiments using AtLEA4-5 truncated versions: one containing the first 77 amino acids, named AtLEA4-51–77, and a second one corresponding to the 81 amino acid C-terminal region, from residues 78 to 158 (AtLEA4-578–158) (47).
In contrast to in silico predictions, AtLEA4-51–77 behaved as a disordered protein in aqueous solution under all temperatures tested (Fig. 4A), with a difference spectrum profile (Δ10–80 °C) similar to that obtained for the complete AtLEA4-5 (Figs. 2B and 4A, insets). However, the addition of increasing TFE concentrations progressively induced α-helix formation in the truncated protein containing the N-terminal region (Fig. 4B). Likewise, treatments with glycerol and PEG led to the same behavior in this protein (Fig. 4, C and D), reaching up to 55 and 42% α-helix at the highest glycerol and PEG concentrations, respectively (Table 2). Also complete AtLEA4-5 and AtLEA4-2, far UV CD spectra from AtLEA4-51–77 showed isodichroic points when treated with progressively increasing TFE or glycerol concentrations, indicating that this protein is in equilibrium between two states in these conditions: disordered and α-helix conformations (Fig. 4, B and C). The high linear correlation of transition diagrams confirmed this observation (data not shown).
FIGURE 4.
N-terminal conserved region of AtLEA4-5 is necessary and sufficient to induce folding to α-helix under the different conditions tested. Far UV CD spectra of AtLEA4-51–77 (A–D) and AtLEA4-578–158 (E–H) under different temperatures (A and E), different TFE (B and F) and glycerol concentrations (C and G), and under 45% PEG 5,000 (D and H). Difference spectra (Δ10–80 °C; Δ90–0% TFE; Δ80–0% glycerol; Δ45–0% PEG) are shown as insets in each graph. These results were reproduced at least four times in autonomous experiments, using three independent purification batches of AtLEA4-51–77 protein and one batch for AtLEA4-578–158 polypeptide.
TABLE 2.
Percentage of helix, strand, and unordered structures in AtLEA4-51–77 and AtLEA4-578–158
Secondary structure content in AtLEA4-2 and AtLEA4-5 proteins was obtained by far UV CD spectrometry and calculated with Dichroweb server. NA, not available.
| Treatment | AtLEA4-51–77 |
AtLEA4-578–158 |
||||
|---|---|---|---|---|---|---|
| Helix | Strand | Unordered | Helix | Strand | Unordered | |
| % | % | |||||
| Aqueous solution | 6 | 32 | 61 | 3 | 38 | 58 |
| 4% TFE | 6 | 33 | 59 | 4 | 37 | 58 |
| 8% TFE | 7 | 32 | 60 | 4 | 37 | 58 |
| 12% TFE | 18 | 25 | 57 | 5 | 36 | 59 |
| 16% TFE | 22 | 24 | 53 | 5 | 35 | 59 |
| 20% TFE | 36 | 16 | 48 | 5 | 36 | 59 |
| 40% TFE | 59 | 9 | 31 | 8 | 33 | 58 |
| 60% TFE | 65 | 7 | 27 | 19 | 28 | 54 |
| 90% TFE | 80 | 3 | 16 | 49 | 15 | 35 |
| 10% glycerol | 6 | 33 | 60 | NA | NA | NA |
| 20% glycerol | 16 | 23 | 62 | NA | NA | NA |
| 30% glycerol | 19 | 24 | 58 | 6 | 45 | 51 |
| 40% glycerol | 19 | 23 | 57 | 0 | 53 | 45 |
| 50% glycerol | 24 | 18 | 57 | 6 | 36 | 56 |
| 60% glycerol | 39 | 12 | 48 | 4 | 41 | 55 |
| 70% glycerol | 45 | 9 | 46 | NA | NA | NA |
| 80% glycerol | 55 | 8 | 35 | 5 | 35 | 58 |
| 45% PEG 5000 | 42 | 9 | 50 | NA | NA | NA |
Using the chemically synthesized peptide corresponding to the AtLEA4-5C-terminal region (AtLEA4-578–158), far UV CD analysis confirmed that AtLEA4-578–158 is highly disordered in aqueous solution at different temperatures (Fig. 4E). Unlike AtLEA4-2, AtLEA4-5, or AtLEA4-51–77, the difference spectrum Δ10–80 °C of AtLEA4-578–158 indicated the possible formation of poly-l-proline-like structures (Fig. 4E, inset). In contrast to the N-terminal region of these proteins, addition of up to 60% TFE had only minor effects on the structure of AtLEA4-578–158 (Fig. 4F), inducing 4% to 19% α-helix (Table 2). It was not until a high TFE concentration (90%) was reached that AtLEA4-578–158 became helical (Fig. 4F), indicating a low intrinsic competence to acquire ordered structures. This finding was further supported by the results obtained from the addition of glycerol or PEG to AtLEA4-578–158 solutions, which showed no effect on its structure (Fig. 4, G and H), given that the negative band at [θ]222 did not show any change and that only a slight increase in the [θ]198 signal was detected. Under the highest glycerol (80%) and PEG (45%) concentrations, AtLEA4-578–158 only showed 5% α-helix formation (Table 2). Comparison between the difference spectra for AtLEA4-51–77 and AtLEA4-578–158 in TFE, glycerol and PEG showed the magnitude of the AtLEA4-51–77 folding to α-helix, compared with AtLEA4-578–158 (Fig. 4, A–H, insets). In accordance, the transition diagram for glycerol titration of AtLEA4-578–158 showed no linear behavior (data not shown). Together, these data demonstrate that the AtLEA4-5 N-terminal region (AtLEA4-51–77) is necessary and sufficient to drive α-helix conformations in this protein under low water availability or macromolecular crowding conditions.
The N-terminal Region of AtLEA4 Proteins Is Necessary and Sufficient to Prevent Inactivation of Lactate Dehydrogenase Caused by Freeze-Thaw Cycles and Partial Dehydration
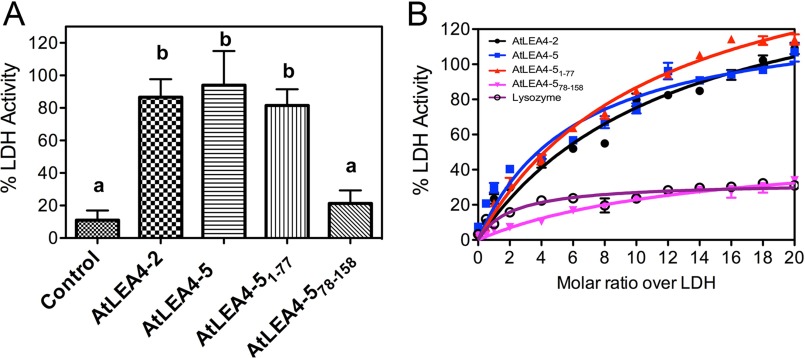
Because the motif conservation and ability to fold into α-helical conformations of the AtLEA4-5 are preferentially located at the N-terminal region as compared with its C-terminal region, we asked about the competence of these individual segments to protect the reporter enzyme LDH from deleterious effects caused by freeze-thaw cycles and partial dehydration, as was previously shown for the complete protein (19, 20). For this purpose, LDH in the presence or absence of AtLEA4-5, AtLEA4-2, AtLEA4-51–77, or AtLEA4-578–158 was subjected to in vitro freeze-thaw cycles and partial dehydration treatments. AtLEA4-2, the smallest protein of this group, mostly consisting of the N-terminal region (Fig. 1A), showed a similar protective effect on LDH activity as AtLEA4-5 after partial dehydration (Fig. 5A). Interestingly, a comparable protection was produced by AtLEA4-51–77, contrasting with the negligible protective levels showed by the AtLEA4-5 C-terminal region (AtLEA4-578–158), whose values were rather close to those shown by lysozyme, an unrelated globular protein (Fig. 5, A and B). A similar conclusion can be drawn when different molar ratios of these proteins were used in freeze-thaw in vitro assays, where it is evident that the various proteins containing the N-terminal region showed an equivalent protective trend, in opposition to the C-terminal region represented by AtLEA4-578–158 or to lysozyme (Fig. 5B). This analysis revealed the highest and lowest protective efficiencies for AtLEA4-5 and AtLEA4-578–158, respectively, among the different proteins tested (Fig. 5B). Because the hydrophilicity index and length are similar between the N-terminal and C-terminal regions, these data are consistent with the hypothesis that the protective activity of these proteins is rather dependent on the conserved motifs present in the N-terminal region and/or on its ability to fold into α-helical conformations.
FIGURE 5.
The N-terminal conserved region of AtLEA4-5 is necessary and sufficient to prevent inactivation of LDH after partial dehydration and freeze/thaw cycles in vitro. A, remaining LDH enzymatic activity after ≥98% water loss induced in vitro of samples containing LDH without any additive (control) or with AtLEA4-2, AtLEA4-5, AtLEA4-51–77, and AtLEA4-578–158 in a 5:1 molar ratio (additive:LDH). B, remaining LDH enzymatic activity after seven freeze-thaw cycles of LDH without any additive or with increasing molar amounts (from 0:1 to 20:1 molar ratio additive:LDH) of AtLEA4-2 (black line with circles), AtLEA4-5 (blue line with squares), AtLEA4-51–77 (red line with up triangles), AtLEA4-578–158 (magenta line with inverted triangles), and lysozyme (purple line with open circles). Error bars indicate S.E. of three independent tests (with three internal repetitions). Letters indicate significant differences calculated with Tukey's multiple comparison post-test (p < 0.01). The data in B were fit to a hyperbola curve, and significant differences were subsequently calculated with Tukey's multiple comparison post-test (p < 0.01). These results were reproduced at least four times in autonomous experiments, using three independent purification batches of AtLEA4-2, AtLEA4-5, and AtLEA4-51–77 proteins, and one batch for AtLEA4-578–158 polypeptide.
Discussion
During the last nearly 20 years, research on the so-called IDPs has challenged the classical structure-function paradigm (48). IDPs that either completely lack a well defined three-dimensional structure or contain short regions of disorder, known as intrinsically disordered regions (IDRs) within folded domains are highly abundant in eukaryotic proteomes and perform important functions (6). In plants, IDPs participate in developmental control, light perception, transcriptional regulation, and response to abiotic stress (12). LEA proteins are the plant protein family with the highest number of proteins either predicted or characterized as IDPs (12). There is experimental evidence showing that LEA proteins from different groups acquire α-helix after complete drying (8, 11, 13, 28–31). These findings suggest that these proteins form α-helix in the dry seed, but little is known about LEA proteins structural behavior under less severe conditions, such as those present in vegetative tissues under drought. Mouillon et al. (49) showed that three Arabidopsis group 2 LEA proteins (Cor47, Lti29, and Lti30) are IDPs in solution and that they remain disordered when subjected to low water potential or macromolecular crowding simulated in vitro with glycerol and PEG, respectively. Even though Cor47 showed a slight folding into α-helix under high concentrations of glycerol and PEG, the authors propose that dehydrins have evolved to stay disordered under cellular conditions and that disorder might be required to properly fulfill their function (49). A similar effect was also observed experimentally for α-casein, MAP2c, and p21Cip1, three different animal IDPs that show no conformational change when subjected to macromolecular crowding, supporting the idea that the physiological state of these IDPs is also disordered (50).
Even though, most LEA proteins can be considered as IDPs, the analysis of their amino acid sequences indicates variety in their potential to attain different levels of secondary structure (2). Because LEA proteins accumulate not only under severe dehydration, such as that occurring in the dry seed, but also in different tissues under a wide range of water limitation, we investigated the possibility that these proteins could adopt secondary structures under conditions prevailing upon water loss, not necessarily as extreme as those occurring in dry seeds. Group 4 LEA proteins in Arabidopsis represented a suitable set of proteins for this purpose, given that there are two protein subtypes (subgroups 4A and 4B): a long variant conformed by two distinctive N-terminal and C-terminal regions (AtLEA4-5, representative of subgroup 4B) and a short one with only the conserved N-terminal region (AtLEA4-2, representative of subgroup 4A) (14). Furthermore, in silico analysis showed that in contrast to the C-terminal region, the N-terminal region sequence has the potential to fold into α-helix conformations offering an appropriate system to experimentally compare their structural properties under different environments. In this work, we demonstrate not only that the N-terminal regions of AtLEA4-2 and AtLEA4-5 have an intrinsic capacity to fold into α-helix as shown by the far UV CD spectra in the presence of TFE but also that they gain significant helicity in low water potential and macromolecular crowded solutions. Interestingly, the AtLEA4-5 C-terminal region, which we show is disordered in aqueous solution as predicted, remains disordered under all conditions tested. These results indicate the presence of two functional domains in this LEA protein group. The N-terminal region can become ordered under conditions of water restriction, short of absolute dryness, whereas the persistent disorder of the C-terminal region could indicate a requirement to expose some amino acid residues for a more effective or additional function. Such possibility could be related to the abundance of His residues in this region, which seems to be involved in their binding to metal ions (18), suggesting multifunctionality in some LEA protein families.
All LEA proteins studied to date from groups 1, 2, 3, 4, and 6 show structural disorder in aqueous solution, but most of them also have an intrinsic potential to acquire helical conformations in the presence of TFE (12). These results suggest that there are conditions where LEA proteins exhibit such structural transformation. For most of these LEA proteins, folding has been detected after extreme dehydration (8, 11, 13, 28–31), but extreme conditions may not be necessary in all cases to induce structural transformation. We show in this work that group 4 LEA proteins reach up to 54 and 39% α-helix under less severe environments regarding water availability and macromolecular crowding, respectively. The levels of α-helix formation for group 4 LEA proteins are significantly higher than those estimated for LEA proteins from other groups under similar treatments (12, 39), indicating that the ability to fold is not necessarily the same for different IDPs or intrinsically disordered regions. This is also supported by the wide range of α-helix formation observed among different LEA proteins upon comparable treatments (9, 12, 39, 49, 51, 52). Other proteins seem to be unable to gain ordered conformations such as Rab18, a group 2 LEA protein (9). This information indicates diversity in their structural plasticity, action mechanisms, and/or in their function.
The results from the protection activity assays demonstrate that the protective role of LEA 4 proteins on LDH under low water availability and/or molecular crowding is confined to the conserved N-terminal region of this protein family. There is no apparent participation of the C-terminal region, which completely lacks this safeguard function. These observations imply that the ability of a LEA4 N-terminal domain to gain helicity is related to this chaperone-like activity, particularly under conditions prevailing in water-deficit environments. Based on in vitro evidence, it has been proposed that one mechanism for this activity involves protein-protein interactions (20, 53–55). A comprehensive view brings into consideration the existence of different LEA4 conformers in equilibrium (partially folded or unfolded) under crowded or water-deficit environments, which supports the existence of preformed secondary structural elements, denominated as prestructured motifs that could be implicated in the recognition of different and specific binding partners (56). Our findings strongly suggest that water deficit leads to the stabilization of particular conformations in group 4 LEA proteins that may allow the exposure of different motifs necessary for the binding of their target molecules, hence supporting the idea that LEA proteins of this group function as a structural ensemble, whose dynamism can be modulated by environmental conditions (57–60). This proposed mode of action also exhibits possible binding promiscuity, in consonance with their role as chaperone-like molecules needed during water scarcity. At this point, we cannot discount the possibility that different conformers in these proteins could favor different functions such as protection of proteins and/or membranes, as well as metal binding, a moonlighting property arising from their structural dynamics.
In conclusion, the findings reported here indicate that the in vitro chaperone-like function of the intrinsically disordered group 4 LEA proteins is closely associated to their ability to adopt ordered structural conformations under prevailing conditions in water-deficit environments. The high correlation found between accumulation under water deficit and intrinsic structural disorder, common features in typical LEA proteins and other hydrophilins (4), suggests a functional advantage for this attribute throughout evolution, not only to maintain structural plasticity that could avoid undesirable consequences under stressful environments, such as dramatic structural modifications leading to a functional breakdown, but also to gain functional and mechanistic diversity given their conformational freedom.
Author Contributions
C. L. C.-V. and A. A. C. designed the experimental strategy. C. L. C.-V. conducted cloning, protein expression, and protein purification. C. L. C.-V. and G. S.-R. performed circular dichroism spectroscopy experiments. C. L. C.-V. and J. L. R. carried out in vitro protection assays. A. A. C. supervised research. C. L. C.-V., G. S.-R., and A. A. C. analyzed data. C. L. C.-V. and A. A. C. wrote the article, which was read and approved by all authors.
Acknowledgments
We thank H. Jane Dyson for critical reviewing this manuscript, Rosa M. Solórzano for technical assistance in protein purification, and Francisco Campos for the gift of pTrc99A/AtLEA4-51–77 plasmid. We are also grateful to the proteomics (Unidad de Proteómica) and DNA sequencing (Unidad de Síntesis y Secuenciación de ADN) core facilities at the Instituto de Biotecnología of the Universidad Nacional Autónoma de México.
This work was partially supported by Grants IN208212 from the Dirección General de Apoyo al Personal Académico/Programa de Apoyo a Proyectos de Investigación e Innovación Tecnológica PAPIIT, Universidad Nacional Autónoma de México and Grants 132258 and 221448 from the Consejo Nacional de Ciencia y Tecnología. The authors declare that they have no conflicts of interest with the contents of this article.
The nucleotide sequence(s) reported in this paper has been submitted to the DDBJ/GenBankTM/EBI Data Bank with accession number(s) AEE31503.1, AEC09091.1, and AED91062.1.
- LEA
- late embryogenesis abundant
- IDP
- intrinsically disordered proteins
- LDH
- lactate dehydrogenase
- TFE
- 2,2,2-trifluoroethanol.
References
- 1. Bray E. A. (1997) Plant responses to water deficit. Trends Plant Sci. 2, 48–54 [Google Scholar]
- 2. Battaglia M., Olvera-Carrillo Y., Garciarrubio A., Campos F., and Covarrubias A. A. (2008) The enigmatic LEA proteins and other hydrophilins. Plant Physiol. 148, 6–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hundertmark M., and Hincha D. K. (2008) LEA (late embryogenesis abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genomics 9, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Garay-Arroyo A., Colmenero-Flores J. M., Garciarrubio A., and Covarrubias A. A. (2000) Highly hydrophilic proteins in prokaryotes and eukaryotes are common during conditions of water deficit. J. Biol. Chem. 275, 5668–5674 [DOI] [PubMed] [Google Scholar]
- 5. Dyson H. J., and Wright P. E. (2005) Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 6, 197–208 [DOI] [PubMed] [Google Scholar]
- 6. Tompa P. (2012) Intrinsically disordered proteins: a 10-year recap. Trends Biochem. Sci. 37, 509–516 [DOI] [PubMed] [Google Scholar]
- 7. Hughes S., and Graether S. P. (2011) Cryoprotective mechanism of a small intrinsically disordered dehydrin protein. Protein Sci. 20, 42–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hundertmark M., Popova A. V., Rausch S., Seckler R., and Hincha D. K. (2012) Influence of drying on the secondary structure of intrinsically disordered and globular proteins. Biochem. Biophys. Res. Commun. 417, 122–128 [DOI] [PubMed] [Google Scholar]
- 9. Mouillon J. M., Gustafsson P., and Harryson P. (2006) Structural investigation of disordered stress proteins. Comparison of full-length dehydrins with isolated peptides of their conserved segments. Plant Physiol. 141, 638–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rivera-Najera L. Y., Saab-Rincón G., Battaglia M., Amero C., Pulido N. O., García-Hernández E., Solórzano R. M., Reyes J. L., and Covarrubias A. A. (2014) A group 6 late embryogenesis abundant protein from common bean is a disordered protein with extended helical structure and oligomer-forming properties. J. Biol. Chem. 289, 31995–32009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shih M. D., Hsieh T. Y., Lin T. P., Hsing Y. I., and Hoekstra F. A. (2010) Characterization of two soybean (Glycine max L.) LEA IV proteins by circular dichroism and Fourier transform infrared spectrometry. Plant Cell Physiol. 51, 395–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sun X., Rikkerink E. H., Jones W. T., and Uversky V. N. (2013) Multifarious roles of intrinsic disorder in proteins illustrate its broad impact on plant biology. Plant Cell 25, 38–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tolleter D., Jaquinod M., Mangavel C., Passirani C., Saulnier P., Manon S., Teyssier E., Payet N., Avelange-Macherel M. H., and Macherel D. (2007) Structure and function of a mitochondrial late embryogenesis abundant protein are revealed by desiccation. Plant Cell 19, 1580–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Olvera-Carrillo Y., Campos F., Reyes J. L., Garciarrubio A., and Covarrubias A. A. (2010) Functional analysis of the group 4 late embryogenesis abundant proteins reveals their relevance in the adaptive response during water deficit in Arabidopsis. Plant Physiol. 154, 373–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schmid M., Davison T. S., Henz S. R., Pape U. J., Demar M., Vingron M., Schölkopf B., Weigel D., and Lohmann J. U. (2005) A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37, 501–506 [DOI] [PubMed] [Google Scholar]
- 16. Dang N. X., Popova A. V., Hundertmark M., and Hincha D. K. (2014) Functional characterization of selected LEA proteins from Arabidopsis thaliana in yeast and in vitro. Planta 240, 325–336 [DOI] [PubMed] [Google Scholar]
- 17. Hundertmark M., Dimova R., Lengefeld J., Seckler R., and Hincha D. K. (2011) The intrinsically disordered late embryogenesis abundant protein LEA18 from Arabidopsis thaliana modulates membrane stability through binding and folding. Biochim. Biophys. Acta 1808, 446–453 [DOI] [PubMed] [Google Scholar]
- 18. Liu G., Xu H., Zhang L., and Zheng Y. (2011) Fe binding properties of two soybean (Glycine max L.) LEA4 proteins associated with antioxidant activity. Plant Cell Physiol. 52, 994–1002 [DOI] [PubMed] [Google Scholar]
- 19. Reyes J. L., Campos F., Wei H., Arora R., Yang Y., Karlson D. T., and Covarrubias A. A. (2008) Functional dissection of hydrophilins during in vitro freeze protection. Plant Cell Environ. 31, 1781–1790 [DOI] [PubMed] [Google Scholar]
- 20. Reyes J. L., Rodrigo M. J., Colmenero-Flores J. M., Gil J. V., Garay-Arroyo A., Campos F., Salamini F., Bartels D., and Covarrubias A. A. (2005) Hydrophilins from distant organisms can protect enzymatic activities from water limitation effects in vitro. Plant Cell Environ. 28, 709–718 [Google Scholar]
- 21. Borcherds W., Theillet F. X., Katzer A., Finzel A., Mishall K. M., Powell A. T., Wu H., Manieri W., Dieterich C., Selenko P., Loewer A., and Daughdrill G. W. (2014) Disorder and residual helicity alter p53-Mdm2 binding affinity and signaling in cells. Nat. Chem. Biol. 10, 1000–1002 [DOI] [PubMed] [Google Scholar]
- 22. Dyson H. J., and Wright P. E. (2002) Coupling of folding and binding for unstructured proteins. Curr. Opin. Struct. Biol. 12, 54–60 [DOI] [PubMed] [Google Scholar]
- 23. Radhakrishnan I., Pérez-Alvarado G. C., Parker D., Dyson H. J., Montminy M. R., and Wright P. E. (1997) Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: a model for activator:coactivator interactions. Cell 91, 741–752 [DOI] [PubMed] [Google Scholar]
- 24. Rogers J. M., Wong C. T., and Clarke J. (2014) Coupled folding and binding of the disordered protein PUMA does not require particular residual structure. J. Am. Chem. Soc. 136, 5197–5200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sugase K., Dyson H. J., and Wright P. E. (2007) Mechanism of coupled folding and binding of an intrinsically disordered protein. Nature 447, 1021–1025 [DOI] [PubMed] [Google Scholar]
- 26. Reichmann D., Xu Y., Cremers C. M., Ilbert M., Mittelman R., Fitzgerald M. C., and Jakob U. (2012) Order out of disorder: working cycle of an intrinsically unfolded chaperone. Cell 148, 947–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tapley T. L., Körner J. L., Barge M. T., Hupfeld J., Schauerte J. A., Gafni A., Jakob U., and Bardwell J. C. (2009) Structural plasticity of an acid-activated chaperone allows promiscuous substrate binding. Proc. Natl. Acad. Sci. U.S.A. 106, 5557–5562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Goyal K., Tisi L., Basran A., Browne J., Burnell A., Zurdo J., and Tunnacliffe A. (2003) Transition from natively unfolded to folded state induced by desiccation in an anhydrobiotic nematode protein. J. Biol. Chem. 278, 12977–12984 [DOI] [PubMed] [Google Scholar]
- 29. Popova A. V., Hundertmark M., Seckler R., and Hincha D. K. (2011) Structural transitions in the intrinsically disordered plant dehydration stress protein LEA7 upon drying are modulated by the presence of membranes. Biochim. Biophys. Acta 1808, 1879–1887 [DOI] [PubMed] [Google Scholar]
- 30. Shih M. D., Hsieh T. Y., Jian W. T., Wu M. T., Yang S. J., Hoekstra F. A., and Hsing Y. I. (2012) Functional studies of soybean (Glycine max L.) seed LEA proteins GmPM6, GmPM11, and GmPM30 by CD and FTIR spectroscopy. Plant Sci. 196, 152–159 [DOI] [PubMed] [Google Scholar]
- 31. Shimizu T., Kanamori Y., Furuki T., Kikawada T., Okuda T., Takahashi T., Mihara H., and Sakurai M. (2010) Desiccation-induced structuralization and glass formation of group 3 late embryogenesis abundant protein model peptides. Biochemistry 49, 1093–1104 [DOI] [PubMed] [Google Scholar]
- 32. Muñoz V., and Serrano L. (1994) Elucidating the folding problem of helical peptides using empirical parameters. Nat. Struct. Biol. 1, 399–409 [DOI] [PubMed] [Google Scholar]
- 33. Cheng J. L., Sweredoski M. J., and Baldi P. (2005) Accurate prediction of protein disordered regions by mining protein structure data. Data Min. Knowl. Disc. 11, 213–222 [Google Scholar]
- 34. Romero P., Obradovic Z., Li X., Garner E. C., Brown C. J., and Dunker A. K. (2001) Sequence complexity of disordered protein. Proteins 42, 38–48 [DOI] [PubMed] [Google Scholar]
- 35. Jones D. T., and Cozzetto D. (2015) DISOPRED3: precise disordered region predictions with annotated protein-binding activity. Bioinformatics 31, 857–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Campos F., Guillén G., Reyes J. L., and Covarrubias A. A. (2011) A general method of protein purification for recombinant unstructured non-acidic proteins. Protein Expr. Purif. 80, 47–51 [DOI] [PubMed] [Google Scholar]
- 37. Lobley A., Whitmore L., and Wallace B. A. (2002) DICHROWEB: an interactive website for the analysis of protein secondary structure from circular dichroism spectra. Bioinformatics 18, 211–212 [DOI] [PubMed] [Google Scholar]
- 38. Whitmore L., and Wallace B. A. (2004) DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 32, W668–W673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Soulages J. L., Kim K., Walters C., and Cushman J. C. (2002) Temperature-induced extended helix/random coil transitions in a group 1 late embryogenesis-abundant protein from soybean. Plant Physiol. 128, 822–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Buck M. (1998) Trifluoroethanol and colleagues: cosolvents come of age: recent studies with peptides and proteins. Q. Rev. Biophys. 31, 297–355 [DOI] [PubMed] [Google Scholar]
- 41. Luo P.., and Baldwin R. L. (1997) Mechanism of helix induction by trifluoroethanol: a framework for extrapolating the helix-forming properties of peptides from trifluoroethanol/water mixtures back to water. Biochemistry 36, 8413–8421 [DOI] [PubMed] [Google Scholar]
- 42. Ellis R. J. (2001) Macromolecular crowding: an important but neglected aspect of the intracellular environment. Curr. Opin. Struct. Biol. 11, 114–119 [DOI] [PubMed] [Google Scholar]
- 43. Zimmerman S. B., and Minton A. P. (1993) Macromolecular crowding: biochemical, biophysical, and physiological consequences. Annu. Rev. Biophys. Biomed. Struct. 22, 27–65 [DOI] [PubMed] [Google Scholar]
- 44. Zimmerman S. B., and Trach S. O. (1991) Estimation of macromolecule concentrations and excluded volume effects for the cytoplasm of Escherichia coli. J. Mol. Biol. 222, 599–620 [DOI] [PubMed] [Google Scholar]
- 45. Minton A. P. (2000) Implications of macromolecular crowding for protein assembly. Curr. Opin. Struct. Biol. 10, 34–39 [DOI] [PubMed] [Google Scholar]
- 46. Minton A. P. (1997) Influence of excluded volume upon macromolecular structure and associations in “crowded” media. Curr. Opin. Biotech. 8, 65–69 [DOI] [PubMed] [Google Scholar]
- 47. Campos F., Zamudio F., and Covarrubias A. A. (2006) Two different late embryogenesis abundant proteins from Arabidopsis thaliana contain specific domains that inhibit Escherichia coli growth. Biochem. Biophys. Res. Commun. 342, 406–413 [DOI] [PubMed] [Google Scholar]
- 48. Wright P. E., and Dyson H. J. (2015) Intrinsically disordered proteins in cellular signalling and regulation. Nat. Rev. Mol. Cell Biol. 16, 18–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mouillon J. M., Eriksson S. K., and Harryson P. (2008) Mimicking the plant cell interior under water stress by macromolecular crowding: disordered dehydrin proteins are highly resistant to structural collapse. Plant Physiol. 148, 1925–1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Szasz C. S., Alexa A., Toth K., Rakacs M., Langowski J., and Tompa P. (2011) Protein disorder prevails under crowded conditions. Biochemistry 50, 5834–5844 [DOI] [PubMed] [Google Scholar]
- 51. Haaning S., Radutoiu S., Hoffmann S. V., Dittmer J., Giehm L., Otzen D. E., and Stougaard J. (2008) An unusual intrinsically disordered protein from the model legume Lotus japonicus stabilizes proteins in vitro. J. Biol. Chem. 283, 31142–31152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mccubbin W. D., Kay C. M., and Lane B. G. (1985) Hydrodynamic and optical-properties of the wheat-germ Em protein. Can. J. Biochem. Cell Biol. 63, 803–811 [Google Scholar]
- 53. Cuevas-Velazquez C. L., Rendón-Luna D. F., and Covarrubias A. A. (2014) Dissecting the cryoprotection mechanisms for dehydrins. Front. Plant Sci. 5, 583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Olvera-Carrillo Y., Luis Reyes J., and Covarrubias A. A. (2011) Late embryogenesis abundant proteins: versatile players in the plant adaptation to water limiting environments. Plant Signal. Behav. 6, 586–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tompa P., and Kovacs D. (2010) Intrinsically disordered chaperones in plants and animals. Biochem. Cell Biol. 88, 167–174 [DOI] [PubMed] [Google Scholar]
- 56. Lee S. H., Kim D. H., Han J. J., Cha E. J., Lim J. E., Cho Y. J., Lee C., and Han K. H. (2012) Understanding pre-structured motifs (PreSMos) in intrinsically unfolded proteins. Curr. Protein Pept. Sci. 13, 34–54 [DOI] [PubMed] [Google Scholar]
- 57. Cino E. A., Karttunen M., and Choy W. Y. (2012) Effects of molecular crowding on the dynamics of intrinsically disordered proteins. PLoS One 7, e49876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mittag T., Kay L. E., and Forman-Kay J. D. (2010) Protein dynamics and conformational disorder in molecular recognition. J. Mol. Recognit. 23, 105–116 [DOI] [PubMed] [Google Scholar]
- 59. Mittag T., Orlicky S., Choy W. Y., Tang X., Lin H., Sicheri F., Kay L. E., Tyers M., and Forman-Kay J. D. (2008) Dynamic equilibrium engagement of a polyvalent ligand with a single-site receptor. Proc. Natl. Acad. Sci. U.S.A. 105, 17772–17777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Uversky V. N. (2013) Unusual biophysics of intrinsically disordered proteins. Biochim. Biophys. Acta 1834, 932–951 [DOI] [PubMed] [Google Scholar]