Abstract
Background:
Numerous germline single-nucleotide polymorphisms increase susceptibility to prostate cancer, some lying near genes involved in cellular radiation response. This study investigated whether prostate cancer patients with a high genetic risk have increased toxicity following radiotherapy.
Methods:
The study included 1560 prostate cancer patients from four radiotherapy cohorts: RAPPER (n=533), RADIOGEN (n=597), GenePARE (n=290) and CCI (n=150). Data from genome-wide association studies were imputed with the 1000 Genomes reference panel. Individuals were genetically similar with a European ancestry based on principal component analysis. Genetic risks were quantified using polygenic risk scores. Regression models tested associations between risk scores and 2-year toxicity (overall, urinary frequency, decreased stream, rectal bleeding). Results were combined across studies using standard inverse-variance fixed effects meta-analysis methods.
Results:
A total of 75 variants were genotyped/imputed successfully. Neither non-weighted nor weighted polygenic risk scores were associated with late radiation toxicity in individual studies (P>0.11) or after meta-analysis (P>0.24). No individual variant was associated with 2-year toxicity.
Conclusion:
Patients with a high polygenic susceptibility for prostate cancer have no increased risk for developing late radiotherapy toxicity. These findings suggest that patients with a genetic predisposition for prostate cancer, inferred by common variants, can be safely treated using current standard radiotherapy regimens.
Keywords: prostate cancer, genetic variants, radiotherapy, late toxicity
Prostate carcinoma accounts for a quarter of cancer diagnoses in men in the United Kingdom and is the fourth most common cancer worldwide with an estimated 1.1 million men diagnosed in 2012 (Globocan, 2012; Cancer Research UK, 2014). It is estimated that approximately a third of patients with localised or locally advanced prostate cancer undergo external beam radiotherapy (RT) with curative intent (Foroudi et al, 2003). The use of RT in combination with androgen-deprivation prolongs survival (Martin and D'Amico, 2014), and has contributed to the increase in 5-year survival rate from 30% in the 1970s to 80% in 2009 (Cancer Research UK, 2014). Because of increased cure rates, cancer survivorship and late treatment toxicity have become increasingly important issues in health-care provision (England, 2013).
Late toxicity following irradiation for prostate cancer includes damage to the bladder, bowel and erectile function (Peeters et al, 2005). The median rates of late gastrointestinal (GI) and genitourinary (GU) toxicity are reported to be 15% and 17% respectively (Ohri et al, 2012). The rates of severe GI and GU toxicity are reported to be 2% and 3% respectively (Ohri et al, 2012). There is now supporting evidence that new techniques such as intensity-modulated radiotherapy (IMRT) reduce rates of long-term GI and GU side effects compared with 3D conformal RT, even with dose escalation (Dolezel et al, 2015; Wilkins et al, 2015). Despite these advances, approximately one in five patients will experience some degree of late radiation toxicity (Dolezel et al, 2015).
Studies are attempting to identify the genetic variants that increase an individual's risk of radiation toxicity (Kerns et al, 2013; Barnett et al, 2014; Fachal et al, 2014). This work has highlighted the need to increase the statistical power to identify individual common variants with small effects (Barnett et al, 2012a). To address this need, the Radiogenomics Consortium (RGC) was established in 2009 to facilitate large-scale collaborative research with sufficient power to detect genetic variants that predict a patient's risk of radiation toxicity (West et al, 2010). The RGC groups have undertaken genome-wide association studies (GWASs) and are starting to identify replicated variants that increase a prostate cancer patient's risk of toxicity (Fachal et al, 2014).
In the cancer predisposition field, GWASs have identified 76 common single-nucleotide polymorphisms (SNPs) associated with prostate cancer susceptibility (Eeles et al, 2014). Although the biologic role of these SNPs in the development of prostate cancer is an area of on-going investigation, their proximity to genes that are involved in DNA repair processes suggests that disruption of DNA damage response and repair mechanisms may have a key role (Eeles et al, 2014; Hazelett et al, 2014). If a patient has an inherent compromised ability to repair DNA damage, they may be predisposed to both prostate cancer and toxicity following RT, as the same DNA repair pathways play a central role in cellular response to radiation. In addition, recent epidemiological evidence suggests that radiation exposure increases the risk of developing prostate cancer (Myles et al, 2008; Schmitz-Feuerhake and Pflugbeil, 2011; Kondo et al, 2013). Therefore, the hypothesis underlying this study was that common genetic variants involved in cancer predisposition may have roles in both tumour formation and in the response of normal tissues to radiation-induced DNA damage. The aim of this study was to investigate the association between prostate cancer germline risk SNPs and likelihood of developing late radiation toxicity.
Materials and methods
Patients
This prospective study involved four prostate cancer radiotherapy cohorts: RAPPER (N=533), RADIOGEN (N=597), GenePARE (N=290) and CCI (N=150). Informed consent was obtained from all patients. RAPPER was approved by the Cambridge South Research Ethics Committee (05/Q0108/365). RADIOGEN was approved by the Galician Ethical Committee. GenePARE was approved by the Mount Sinai Medical Center Institutional Review Board. The CCI study was approved by the Health Research Ethics Board of Alberta (Cancer).
The UK RAPPER study (UKCRN1471) recruited patients who received neoadjuvant androgen suppression and external beam radiotherapy (EBRT) from two clinical trials RT01 (ISRCTN47772397) and CHHiP (ISRCTN97182923). A full description of the cohort is available elsewhere (Barnett et al, 2014). The RTO1 study was a randomised dose escalation study using 3D conformal radiotherapy comparing 64 and 74 Gy in the treatment of localised prostate cancer (Dearnaley et al, 2014). The CHHiP study randomised between standard (74 Gy in 37 fractions) and hypofractionated (60 Gy in 20 fractions or 57 Gy in 19 fractions) IMRT (Dearnaley et al, 2012).
RADIOGEN comprised patients who received 3D conformal radical or post-prostatectomy EBRT at the Clinical University Hospital of Santiago de Compostela, Spain. A total of 473 patients had adjuvant hormone therapy. Patients received radical EBRT using doses of between 70 and 76 Gy in 2 Gy per fraction. The adjuvant EBRT doses used were 60–66 Gy in 2 Gy per fraction. A full description of the cohort can be found elsewhere (Fachal et al, 2012).
GenePARE patients received brachytherapy with/without EBRT at the Mount Sinai Hospital, New York. Of the ∼800 patients included in the initial GenePARE study, 290 individuals of European ancestry had high-quality genome-wide SNP data available and were included in the present study. Of these individuals, 147 received adjuvant hormone therapy. The 125I (160 Gy; TG-43) was used in patients undergoing brachytherapy alone and 103Pd (100 Gy) in patients also receiving EBRT. The EBRT regimen was delivered using 3D conformal technique using 24–50 Gy. External beam radiotherapy alone was delivered using IMRT using 66.6–81 Gy, and further full details can be found elsewhere (Kerns et al, 2014b).
The CCI cohort recruited patients from the Cross Cancer Institute in Edmonton and the Tom Baker Cancer Centre in Alberta, Canada. Patients underwent EBRT using a hypofractionated (68 Gy in 25 fractions or 55 Gy in 16 fractions) or conventional (72–82 Gy delivered in 2 Gy per fraction) schedule. Approximately 50% of patients received androgen suppression. Further treatment details can be found elsewhere (Kerns et al, 2013).
Assessment of late radiotherapy toxicity
Late toxicity data were collected prospectively and assessed using standardised scoring systems (Supplementary Table 1). Data collected at 2 years were used as in other RGC studies (Andreassen et al, 2012; Dearnaley et al, 2012; Kerns et al, 2013). For rectal bleeding in GenePARE, a 1–5-year window was allowed, because the scoring system assigns grades based on whether rectal bleeding occurs as a single incident or intermittent symptoms over time.
Decreased stream, urinary frequency and rectal bleeding data were harmonised across the four cohorts to create comparable end points (see Supplementary Table 2). Toxicity end points were analysed as change from baseline rather than actual recorded grade such that the toxicity captured was due to radiotherapy only. Because of the low number of high-grade toxicities (⩾2) it was only possible to analyse toxicities as grade 0 vs ⩾1 (Table 1). Scale-independent Standardised Total Average Toxicity (STAT) scores were derived, as described previously (Barnett et al, 2012b), from a range of individual toxicity end points to provide an overall measure of 2-year toxicity that was comparable across the four cohorts.
Table 1. Distributions of patient characteristics and toxicity.
| RAPPER (N=533) | RADIOGEN (N=597) | GenePARE (N=290) | CCI (N=150) | P-valuea | |
|---|---|---|---|---|---|
|
Age | |||||
| Mean (s.d.) | 67.2 (5.7) | 71.0 (6.5) | 64.0 (7.5) | 66.7 (7.4) | P<0.00005 |
|
Diabetes | |||||
| Yes, n (%) | 39 (7.3) | 144 (24.1) | 16 (5.5) | 24 (16.0) | P<0.00005 |
| No, n (%) | 493 (92.5) | 453 (75.9) | 274 (94.5) | 122 (81.3) | |
| Missing, n (%) | 1 (0.2) | 0 | 0 | 4 (2.7) | |
|
Prior TURP | |||||
| Yes, n (%) | 56 (10.5) | 45 (7.5) | 6 (2.1) | 6 (4.0) | P=0.0002 |
| No, n (%) | 472 ((88.6) | 552 (92.5) | 284 (97.9) | 144 (96.0) | |
| Missing, n (%) | 5 (0.9) | 0 | 0 | 0 | |
|
BED | |||||
| Mean (s.d.) | 120.5 (6.2) | 120.5 (5.6) | 191.9 (22.4) | 125.5 (6.2) | P<0.00005 |
|
STAT 2 years | |||||
| Mean (s.d.) | −0.01 (0.5) | 0.02 (0.8) | 0.12 (0.7) | −0.01 (0.7) | 0.06 |
|
Decreased stream | |||||
| Grade 0, n (%) | 483 (90.6) | 472 (79.1) | 189 (65.2) | NA | P<0.00005 |
| Grade ⩾1, n (%) | 29 (5.5) | 6 (1.0) | 66 (22.7) | NA | |
| Missing, n (%) | 21 (3.9) | 119 (19.9) | 35 (12.1) | NA | |
|
Urine frequency | |||||
| Grade 0, n (%) | 482 (90.5) | 423 (70.9) | 179 (61.7) | 120 (80) | P<0.00005 |
| Grade ⩾1, n (%) | 45 (8.4) | 54 (9.0) | 76 (26.2) | 30 (20) | |
| Missing, n (%) | 6 (1.1) | 120 (20.1) | 35 (12.1) | 0 | |
|
Rectal bleeding | |||||
| Grade 0, n (%) | 446 (83.7) | 522 (87.4) | 208 (71.7) | 110 (73.3) | |
| Grade ⩾1, n (%) | 81 (15.2) | 74 (12.4) | 82 (28.3) | 40 (26.7) | P<0.00005 |
| Missing, n (%) | 6 (1.1) | 1 (0.2) | 0 | 0 | |
Abbreviations: BED=biologically effective dose; NA=not available; STAT=Standardised Total Average Toxicity; TURP=transurethral resection of the prostate.
P-value for test of heterogeneity across cohorts.
Genotyping, quality control and imputation
Samples were genotyped as part of previously completed GWAS (Kerns et al, 2013; Barnett et al, 2014; Fachal et al, 2014). Standard quality control procedures were applied to remove variants that were missing in >5% of samples, had a minor allele frequency (MAF) <1% or displayed genotype frequencies deviating from those expected under Hardy–Weinberg equilibrium (P-value <10−6). Samples that had >3% of all variants missing were removed. Allele frequencies are known to vary by ancestral background, and hence principle component analysis (PCA) was used to identify and exclude individuals with non-European ancestry in order to avoid false positive associations arising from population substructure because of the small number of participants with other ethnicities. Comparable sets of variants were produced through imputation using SHAPEIT (Delaneau et al, 2012) and IMPUTE2 (Howie et al, 2011) with the 1000 Genomes Phase I reference panel (Abecasis et al, 2010). Supplementary Table 3 lists the 76 known prostate cancer susceptibility SNPs. Genotype dosages for the prostate cancer risk alleles were extracted from the imputed data.
Statistical analysis
Polygenic risk scores were created to quantify the patients' genetic risk of prostate cancer. For each patient, genotype dosages for the prostate cancer risk-increasing alleles were calculated and then summed across all the variants. Two types of risk score were calculated:
Non-weighted, for patient i: 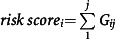
Weighted, for patient i: 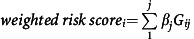
where j=variants 1.76
βj=the per-allele log-odds ratio for risk of prostate cancer associated with variant j
G= risk allele dosage
The log-odds ratios used to weight the risk score were taken from the review paper by Eeles et al (2014).
Within each cohort, logistic regression was used to test the association between each individual toxicity end point and polygenic risk score, adjusted for important nongenetic factors identified by QUANTEC (Bentzen et al, 2010). Total biologically effective dose (BED) was calculated for individuals in all four studies as a measure of radiation dose exposure using an α/β=3. Other nongenetic risk factors included were age at treatment, diabetes (rectal bleeding only), rectal volume (rectal bleeding only), transurethral resection of the prostate (TURP) before radiotherapy (urinary end points only) and baseline toxicity (Table 1). Linear regression was used to test the association between STAT score and polygenic risk score, adjusted for all the nongenetic factors above. Logistic and linear regression was also used to test each genetic variant individually. Regression coefficients and their standard errors were then meta-analysed using standard inverse-variance weighted fixed effects meta-analysis methods.
Power calculations
This study was well powered to detect significant associations between prostate cancer polygenic risk scores and common radiotherapy toxicity end points. Assuming a moderate difference of 0.34 in mean polygenic risk score between prostate cancer patients who experience toxicity and those who do not, with a significance level of α=0.05, the power to detect an association between toxicity (grade ⩾1) and polygenic risk score would be 99% for a toxicity end point with 15% prevalence (grade ⩾1) and 96% for a toxicity end point with 6% prevalence (grade ⩾1). This difference in mean risk would be equivalent to a relative risk of toxicity of 1.4 for the subset of patients with a higher mean polygenic risk of prostate cancer.
Results
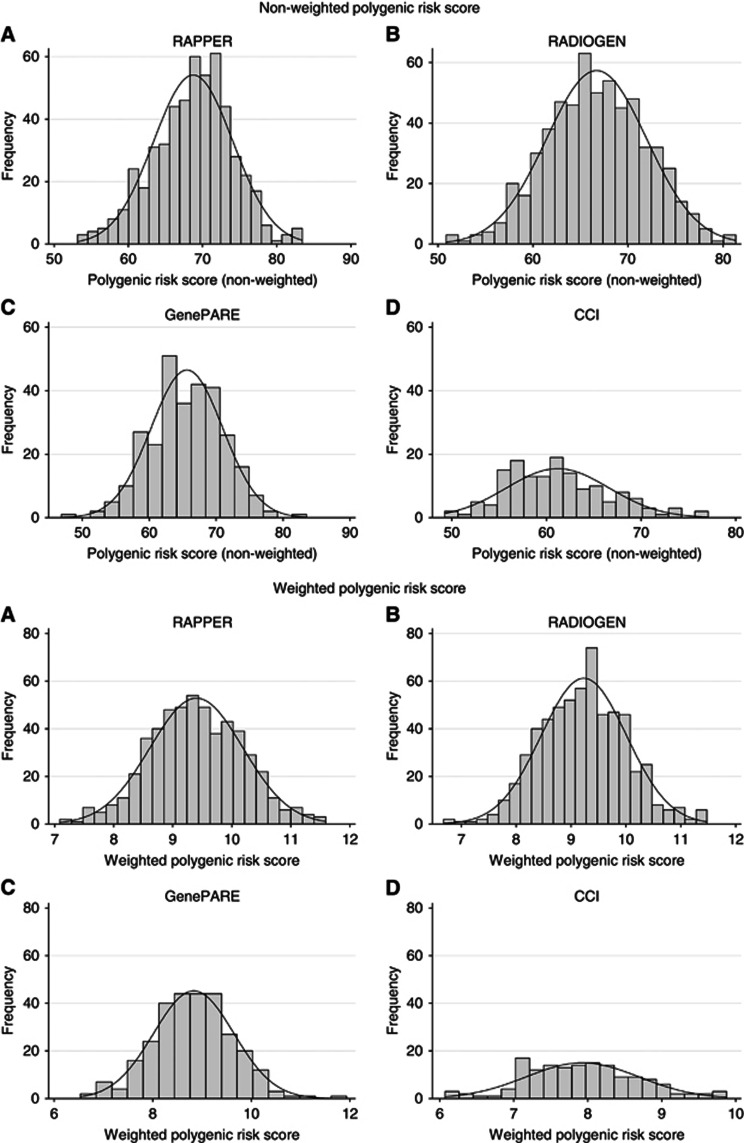
The distributions of patient characteristics, toxicity end points and STAT scores are summarised in Table 1. Of the 76 germline prostate cancer risk SNPs, 75 were genotyped or imputed successfully (R2>0.3; Table 2). Histograms of the polygenic risk scores show an approximate normal distribution within each cohort (Figure 1). Brachytherapy slightly increases urinary toxicity compared with EBRT alone, explaining the higher urinary toxicity in GenePARE (Sutani et al, 2015).
Table 2. SNPs associated with prostate cancer.
|
RAPPER |
RADIOGEN |
GenePARE |
CCI |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Chromosome | Position | Alleles major/minor | MAF | R2 a | MAF | R2 a | MAF | R2 a | MAF | R2 a |
| rs1218582 | 1 | 154834183 | A/G | 0.45 | 0.94 | 0.50 | 0.87 | 0.43 | 0.96 | 0.48 | 0.96 |
| rs4245739 | 1 | 204518842 | A/C | 0.24 | 0.99 | 0.30 | 0.99 | 0.28 | 1 | 0.23 | 1 |
| rs11902236 | 2 | 10117868 | G/A | 0.27 | 0.93 | 0.29 | 1 | 0.27 | 0.91 | 0.28 | 1 |
| rs13385191 | 2 | 20888265 | G/Ab | 0.25 | 0.99 | 0.24 | 1 | 0.27 | 1 | 0.26 | 1 |
| rs1465618 | 2 | 43553949 | G/A | 0.22 | 0.98 | 0.26 | 0.98 | 0.23 | 0.98 | 0.22 | 1 |
| rs721048 | 2 | 63131731 | G/A | 0.22 | 1 | 0.22 | 1 | 0.18 | 0.94 | 0.17 | 0.93 |
| rs10187424 | 2 | 85794297 | A/G | 0.37 | 1 | 0.44 | 0.99 | 0.42 | 0.99 | 0.42 | 1 |
| rs12621278 | 2 | 173311553 | A/G | 0.04 | 0.97 | 0.04 | 1 | — | — | — | — |
| rs2292884 | 2 | 238443226 | A/G | 0.28 | 1 | 0.23 | 0.97 | 0.24 | 0.99 | 0.22 | 0.99 |
| rs3771570 | 2 | 242382864 | G/A | 0.18 | 0.99 | 0.16 | 0.96 | 0.14 | 1 | 0.15 | 1 |
| rs2660753 | 3 | 87110674 | C/T | 0.11 | 1 | 0.16 | 1 | 0.19 | 0.96 | 0.08 | 0.98 |
| rs2055109 | 3 | 87467332 | C/Tb | 0.26 | 1 | 0.28 | 0.99 | 0.24 | 0.86 | 0.22 | 0.99 |
| rs7611694 | 3 | 113275624 | A/C | 0.41 | 1 | 0.37 | 0.98 | 0.36 | 1 | 0.39 | 1 |
| rs10934853 | 3 | 128038373 | C/A | 0.29 | 0.98 | 0.29 | 0.94 | 0.32 | 1 | 0.34 | 1 |
| rs6763931 | 3 | 141102833 | C/T | 0.45 | 0.98 | 0.39 | 1 | 0.40 | 0.96 | 0.39 | 1 |
| rs10936632 | 3 | 170130102 | A/C | 0.49 | 0.94 | 0.45 | 0.95 | 0.46 | 0.94 | 0.47 | 0.95 |
| rs1894292 | 4 | 74349158 | G/A | 0.47 | 0.86 | 0.47 | 1 | 0.40 | 0.91 | 0.44 | 1 |
| rs12500426 | 4 | 95514609 | C/A | 0.48 | 0.96 | 0.48 | 0.98 | 0.49 | 0.98 | 0.49 | 0.99 |
| rs17021918 | 4 | 95562877 | C/T | 0.31 | 0.99 | 0.31 | 0.99 | 0.35 | 1 | 0.37 | 1 |
| rs7679673 | 4 | 106061534 | C/A | 0.34 | 1 | 0.38 | 0.99 | 0.49 | 0.99 | 0.38 | 1 |
| rs2242652 | 5 | 1280028 | G/A | 0.19 | 0.91 | 0.15 | 0.45 | 0.22 | 0.62 | 0.19 | 0.62 |
| rs12653946 | 5 | 1895829 | C/T | 0.45 | 1 | 0.46 | 0.58 | 0.48 | 0.90 | 0.44 | 0.90 |
| rs2121875 | 5 | 44365545 | T/G | 0.35 | 1 | 0.37 | 1 | 0.41 | 0.99 | 0.34 | 1 |
| rs6869841 | 5 | 172939426 | G/A | 0.23 | 0.98 | 0.21 | 0.99 | 0.24 | 0.99 | 0.23 | 0.99 |
| rs130067c | 6 | 31118511 | T/G | 0.21 | 1 | 0.19 | 1 | NA | NA | NA | NA |
| rs3096702d | 6 | 32192331 | G/A | 0.42 | 0.97 | 0.28 | 1 | NA | NA | NA | NA |
| rs1983891 | 6 | 41536427 | C/T | 0.29 | 0.95 | 0.35 | 0.98 | 0.31 | 1 | 0.31 | 1 |
| rs2273669 | 6 | 109285189 | A/G | 0.15 | 0.99 | 0.16 | 0.97 | 0.12 | 0.97 | 0.14 | 0.99 |
| rs339331 | 6 | 117210052 | T/Cb | 0.27 | 0.97 | 0.25 | 1 | 0.19 | 1 | 0.32 | 1 |
| rs1933488 | 6 | 153441079 | A/G | 0.38 | 0.99 | 0.41 | 1 | 0.44 | 1 | 0.41 | 1 |
| rs9364554 | 6 | 160833664 | C/T | 0.33 | 0.99 | 0.25 | 1 | 0.25 | 1 | 0.29 | 1 |
| rs12155172 | 7 | 20994491 | G/A | 0.22 | 0.82 | 0.21 | 1 | 0.21 | 0.97 | 0.22 | 0.97 |
| rs10486567 | 7 | 27976563 | G/Ab | 0.19 | 0.97 | 0.20 | 1 | 0.25 | 0.99 | 0.21 | 0.98 |
| rs6465657 | 7 | 97816327 | T/C | 0.49 | 1 | 0.47 | 1 | 0.41 | 1 | 0.49 | 1 |
| rs2928679 | 8 | 23438975 | C/T | 0.48 | 1 | 0.45 | 0.99 | 0.49 | 0.97 | 0.46 | 1 |
| rs1512268 | 8 | 23526463 | G/A | 0.45 | 0.99 | 0.50 | 0.98 | 0.49 | 0.99 | 0.42 | 1 |
| rs11135910 | 8 | 25892142 | G/A | 0.2 | 0.93 | 0.15 | 1 | 0.16 | 0.97 | 0.18 | 0.99 |
| rs12543663 | 8 | 127924659 | A/C | 0.35 | 0.94 | 0.26 | 0.97 | 0.31 | 0.94 | 0.33 | 0.94 |
| rs10086908 | 8 | 128011937 | T/C | 0.27 | 1 | 0.31 | 1 | 0.26 | 1 | 0.26 | 0.99 |
| rs16901979 | 8 | 128124916 | C/A | 0.05 | 0.99 | 0.05 | 1 | 0.05 | 1 | 0.05 | 1 |
| rs620861 | 8 | 128335673 | C/T | 0.34 | 1 | 0.36 | 0.99 | 0.36 | 0.99 | 0.31 | 0.99 |
| rs6983267 | 8 | 128413305 | G/Tb | 0.43 | 0.95 | 0.40 | 1 | 0.48 | 0.98 | 0.45 | 1 |
| rs1447295 | 8 | 128485038 | C/A | 0.14 | 0.97 | 0.07 | 1 | 0.09 | 0.99 | 0.10 | 0.99 |
| rs817826 | 9 | 110156300 | T/C | 0.17 | 0.64 | 0.17 | 0.94 | 0.21 | 1 | 0.12 | 1 |
| rs1571801 | 9 | 124427373 | C/A | 0.28 | 0.76 | 0.23 | 1 | 0.22 | 1 | 0.32 | 1 |
| rs10993994 | 10 | 51549496 | C/T | 0.46 | 0.79 | 0.43 | 1 | 0.47 | 0.92 | 0.43 | 0.92 |
| rs3850699 | 10 | 104414221 | A/G | 0.28 | 0.99 | 0.28 | 0.96 | 0.29 | 0.97 | 0.26 | 0.97 |
| rs2252004 | 10 | 122844709 | G/Tb | 0.09 | 0.99 | 0.09 | 0.99 | 0.11 | 1 | 0.09 | 1 |
| rs4962416 | 10 | 126696872 | T/C | 0.28 | 0.93 | 0.31 | 1 | 0.31 | 0.98 | 0.24 | 0.98 |
| rs7127900 | 11 | 2233574 | G/A | 0.22 | 0.98 | 0.23 | 0.91 | 0.27 | 0.59 | 0.22 | 0.96 |
| rs1938781 | 11 | 58915110 | T/C | 0.21 | 1 | 0.21 | 1 | 0.20 | 0.99 | 0.19 | 1 |
| rs7931342 | 11 | 68994497 | G/T | 0.46 | 1 | 0.41 | 1 | 0.36 | 1 | 0.41 | 1 |
| rs11568818 | 11 | 102401661 | A/G | 0.44 | 0.89 | 0.46 | 0.99 | 0.42 | 0.91 | 0.42 | 0.93 |
| rs902774 | 12 | 53273904 | G/A | 0.16 | 0.98 | 0.14 | 1 | 0.17 | 0.99 | 0.18 | 1 |
| rs1270884 | 12 | 114685571 | G/A | 0.48 | 0.93 | 0.49 | 0.98 | 0.48 | 0.98 | 0.48 | 1 |
| rs10875943 | 12 | 49676010 | T/C | 0.32 | 0.86 | 0.28 | 1 | 0.32 | 0.93 | 0.29 | 0.95 |
| rs9600079 | 13 | 73728139 | G/T | 0.46 | 0.86 | 0.47 | 1 | 0.43 | 0.89 | 0.46 | 0.95 |
| rs8008270 | 14 | 53372330 | G/A | 0.15 | 1 | 0.19 | 0.99 | 0.19 | 1 | 0.16 | 1 |
| rs7141529 | 14 | 69126744 | G/Ab | 0.47 | 1 | 0.45 | 1 | 0.46 | 0.99 | 0.49 | 0.99 |
| rs4430796 | 17 | 36098040 | G/A | 0.47 | 0.94 | 0.49 | 0.88 | 0.49 | 0.90 | 0.45 | 0.93 |
| rs7210100e | 17 | 47436749 | A/G | — | — | — | — | — | — | — | — |
| rs11649743 | 17 | 36074979 | G/Ab | 0.2 | 1 | 0.19 | 1 | 0.15 | 1 | 0.16 | 1 |
| rs11650494 | 17 | 47345186 | G/A | 0.09 | 0.99 | 0.08 | 0.93 | 0.13 | 0.99 | 0.09 | 0.99 |
| rs684232 | 17 | 618965 | A/G | 0.35 | 0.99 | 0.33 | 0.98 | 0.34 | 0.98 | 0.39 | 0.99 |
| rs1859962 | 17 | 69108753 | T/G | 0.5 | 0.99 | 0.45 | 1 | 0.43 | 1 | 0.47 | 1 |
| rs7241993 | 18 | 76773973 | G/A | 0.28 | 0.94 | 0.28 | 0.91 | 0.32 | 0.51 | 0.32 | 0.56 |
| rs8102476 | 19 | 38735613 | C/Tb | 0.42 | 0.99 | 0.34 | 0.96 | 0.38 | 0.97 | 0.43 | 0.97 |
| rs11672691 | 19 | 41985587 | A/Gb | 0.24 | 0.87 | 0.21 | 0.93 | 0.23 | 0.93 | 0.27 | 0.93 |
| rs103294f | 19 | 54797848 | T/C | 0.23 | 1 | 0.20 | 1 | 0.22 | 0.30 | — | — |
| rs2735839 | 19 | 51364623 | G/A | 0.12 | 1 | 0.15 | 1 | 0.17 | 0.93 | 0.16 | 0.97 |
| rs2427345 | 20 | 61015611 | G/A | 0.35 | 0.99 | 0.35 | 0.37 | 0.39 | 0.91 | 0.40 | 0.96 |
| rs6062509 | 20 | 62362563 | A/C | 0.32 | 1 | 0.26 | 0.99 | 0.27 | 0.98 | 0.28 | 0.98 |
| rs5759167 | 22 | 43500212 | G/T | 0.47 | 1 | 0.45 | 0.90 | 0.46 | 0.80 | 0.47 | 1 |
| rs2405942 | 23 | 9814135 | A/G | 0.17 | 1 | 0.20 | 0.95 | 0.21 | 0.94 | 0.23 | 0.93 |
| rs5919432 | 23 | 67021550 | A/G | 0.14 | 0.98 | 0.18 | 1 | 0.21 | 1 | 0.18 | 1 |
| rs5945619 | 23 | 51241672 | T/C | 0.41 | 0.92 | 0.44 | 1 | 0.38 | 1 | 0.33 | 1 |
Abbreviations: MAF=minor allele frequency; NA=not available; SNP=single-nucleotide polymorphism.
R2 refers to the ‘imputation info' metric produced by IMPUTE2 that represents the certainty with which the SNP has been imputed and lies between 0 (no certainty) and 1 (high certainty; R2=1 for genotyped SNPs).
Major allele is associated with increased risk of prostate cancer.
Merged SNP rs115664826.
Merged SNP rs114376585.
The rs7210100 MAF=0, R2=0, excluded from RAPPER analyses; RADIOGEN R2=0.005; not imputed in GenePARE or CCI data sets.
rs103294 poorly imputed in CCI.
Figure 1.
Histograms showing the approximate normal distributions for the non-weighted and the weighted polygenic risk scores in (A) the RAPPER cohort, (B) the RADIOGEN cohort, (C) the GenePARE cohort and (D) the CCI cohort.
The results of the association analyses are shown in Table 3 and Supplementary Tables 4–7. Neither the non-weighted nor the weighted polygenic risk score was associated with any late radiotherapy individual toxicity end points or STAT score in any of the individual studies or on meta-analysis (meta-analysis P>0.35 and P>0.33 for non-weighted and weighted scores respectively; Table 3). None of the individual SNPs were associated with late radiation toxicity at 2 years at the prespecified significance level of P-value <5 × 10−4 in any of the individual studies or on meta-analysis (Supplementary Tables 4–7). There was no statistical evidence of heterogeneity between studies for any individual SNPs or the polygenic risk score.
Table 3. Polygenic risk score analysis results.
|
RAPPER |
RADIOGEN |
GenePARE |
CCI |
Meta-analysis |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| β (95% CI) | Pa | β (95% CI) | P | β (95% CI) | P | β (95% CI) | P | β (95% CI) | P | Q, P-hetb | |
|
STAT score | |||||||||||
| Non-weighted risk score | 0.003 (−0.006, 0.01) | 0.49 | 0.003 (−0.008, 0.01) | 0.61 | −0.01 (−0.03, 0.01) | 0.15 | −0.01 (−0.04, 0.01) | 0.25 | 0.00002 (−0.01, 0.01) | 0.99 | 3.88, 0.28 |
| Weighted risk score | 0.04 (−0.02, 0.10) | 0.19 | 0.03 (−0.05, 0.10) | 0.44 | −0.06 (−0.17, 0.05) | 0.28 | −0.09 (−0.26, 0.08) | 0.29 | 0.01 (−0.03, 0.06) | 0.49 | 4.15, 0.25 |
|
Decreased stream | |||||||||||
| Non-weighted risk score | 0.01 (−0.06, 0.08) | 0.78 | 0001 (−0.001, 0.003) | 0.36 | 0.004 (−0.06, 0.06) | 0.91 | NA | NA | 0.001 (−0.001, 0.003) | 0.35 | 0.08, 0.96 |
| Weighted risk score | 0.34 (−0.15, 0.83) | 0.17 | 0.01 (−0.01, 0.02) | 0.36 | 0.04 (−0.35, 0.43) | 0.86 | NA | NA | 0.01 (−0.01, 0.02) | 0.33 | 1.84, 0.40 |
|
Urine frequency | |||||||||||
| Non-weighted risk score | −0.004 (−0.06, 0.06) | 0.90 | −0.001 (−0.01, 0.004) | 0.66 | −0.04 (−0.10, 0.02) | 0.18 | −0.03 (−0.12, 0.05) | 0.45 | −0.002 (−0.007, 0.004) | 0.54 | 2.19, 0.53 |
| Weighted risk score | −0.003 (−0.40, 0.40) | 0.99 | −0.01 (−0.05, 0.02) | 0.50 | −0.31 (−0.72, 0.08) | 0.12 | 0.04 (−0.56, 0.62) | 0.89 | −0.01 (−0.05, 0.02) | 0.42 | 2.18, 0.54 |
|
Rectal bleeding | |||||||||||
| Non-weighted risk score | 0.04 (−0.009, 0.08) | 0.11 | −0.002 (−0.01, 0.003) | 0.47 | −0.02 (−0.08, 0.04) | 0.48 | −0.0003 (−0.07, 0.07) | 0.99 | −0.002 (−0.01, 0.003) | 0.55 | 3.22, 0.36 |
| Weighted risk score | 0.28 (−0.03, 0.59) | 0.08 | −0.01 (−0.04, 0.03) | 0.78 | −0.06 (−0.45, 0.32) | 0.75 | −0.05 (−0.57, 0.47) | 0.86 | −0.002 (−0.04, 0.03) | 0.90 | 3.32, 0.34 |
Abbreviations: CI=confidence interval; NA=not available; STAT=Standardised Total Average Toxicity.
P-value corresponding to β-estimate.
P-value for test of heterogeneity between studies.
Discussion
This study found no evidence that prostate cancer patients with a high polygenic risk score for susceptibility to the disease have an increased risk of developing late toxicity following RT. The study was well powered to detect an association between prostate cancer polygenic risk and radiotherapy toxicity end points with a prevalence ⩾6% and a moderate effect of RR=1.4. There was also no evidence for individual SNPs to be associated with risk of toxicity, although the study was not sufficiently powered to detect associations with individual SNPs that are each likely to carry a very small risk for radiotherapy toxicity. Rare, highly penetrant variants like BRCA1, BRCA2 and HOXB13 were not included in this analysis as they require sequencing in a much larger number of patients and different statistical analysis methods.
The biggest nongenetic determinant of radiotherapy toxicity is known to be dose (Kerns et al, 2015). In this study we calculated BED to allow comparison across cohorts receiving external beam therapy only (RAPPER, RADIOGEN and CCI) and those receiving brachytherapy as well (GenePARE). Other important nongenetic factors such as age and comorbidities were also adjusted for. Tests for heterogeneity in these factors across the cohorts were highly significant, suggesting that the cohorts are not homogeneous. However, none of the meta-analysis P-values for heterogeneity were statistically significant. Thus, although the heterogeneity of the cohorts may have reduced the power of the meta-analysis, it is unlikely to have biased the results for the SNPs.
The prostate cancer risk SNPs are mostly located in intronic regions and the functional target genes through which they increase prostate cancer risk are not known. However, some SNPs associated with prostate cancer risk reside near genes that may influence the DNA repair process. The SNP rs817826, identified in a Han Chinese population, lies in an intergenic region between RAD23B and KLF4 (Xu et al, 2012). RAD23B is a key protein involved in the nucleotide excision repair pathway that functions to repair single-strand DNA breaks from ionising radiation (Clement et al, 2010). Defects in this pathway have been associated with photosensitive conditions such as xeroderma pigmentosa (XP) and increase the likelihood of double-strand breaks and late radiation toxicity (Feltes and Bonatto, 2015). Another SNP, rs1938781, found on chromosome 11q12 lies very close to FAM111A and FAM111B (Akamatsu et al, 2012). Mutations in FAM111B have been associated with the development of hereditary fibrosing poililoderma with pulmonary fibrosis, tendon contracture and myopathy (Mercier et al, 2013). The underlying mechanism in which FAM111B causes the above abnormalities is not known. One of the most interesting SNPs, rs7141529 on chromosome 14q24, is an intronic SNP in the DNA repair gene RAD51B (Eeles et al, 2013). Though the functional effect of this SNP is unknown, RAD51B is involved in homologous recombination repair induced by double-strand DNA breaks such as those caused by RT. The SNPs in the TERT locus of 5p15 have been shown to affect prostate cancer risk by interfering with TERT expression (Amin Al Olama et al, 2013). The TERT gene functions by adding telomere repeat sequences at the end of chromosomes that prevent cells undergoing telomere-dependent senescence (Kote-Jarai et al, 2013). A number of proteins have been identified that are involved in telomere maintenance as well as being involved in repair of DNA double-strand breaks by homologous recombination (Huda et al, 2009). Another SNP that has an association with aggressive prostate cancer is rs4245739 that is located in the 3′ untranslated region (UTR) of MDM4 on chromosome 1q32 (Eeles et al, 2013). When functioning normally, MDM4 is a critical negative regulator of the tumour suppressor gene TP53. MDM4 is frequently overexpressed in many cancers that have wild-type TP53 (Wynendaele et al, 2010). TP53 is involved in DNA repair (Merino and Malkin, 2014).
Studies investigating genetic variation in relation to risk of radiotherapy toxicity focused initially on ATM, because individuals with homozygote mutations are extremely sensitive to radiation. The first SNP studies were reported at the start of twenty-first century, and the most widely studied genes encoded proteins associated with DNA repair (e.g., ATM), the development of fibrosis (e.g., TGFB1) and scavenging of reactive oxygen species (e.g., SOD2). Although significant associations were reported, replication was often unsuccessful (Andreassen et al, 2012; Barnett et al, 2012a). Since the establishment of the RGC, replicated associations have been found in both large candidate gene (Talbot et al, 2012; Seibold et al, 2015) and genome-wide association (Fachal et al, 2014) studies. It is interesting to note that the SNPs being identified through GWAS fall in or near genes associated with the function of the tissue irradiated (Fachal et al, 2014; Kerns et al, 2014a, 2014c, 2015; Rosenstein et al, 2014). Although DNA damage response gene products have a clear role in cancer eradication, other pathways are clearly important in the pathogenesis of late radiotherapy toxicity (Bentzen, 2006).
The study reported here had a number of limitations. First, the findings are limited to prostate cancer risk conferred by common variants only – many thousands of participants will need to be studied to assess a role for rare variants. Second, our analysis was limited to men who were genetically of European ancestry and therefore the conclusions may not be generalisable to men of other ethnicities. Third, many genes that predispose to prostate cancer have not yet been identified. Fourth, there are likely to be unrecorded toxicities in patients because underreporting is a known issue of data collection in radiotherapy studies (Bentzen et al, 2010). For example, it was not possible to analyse sexual dysfunction as no data were available for two of the cohorts.
Only 33% of common germline variants that predict the familial risk of developing prostate cancer have so far been discovered (Eeles et al, 2014). The top 1% of the risk distribution have a 4.7 times increased risk of developing prostate cancer than the average population being profiled (Eeles et al, 2013). The National Institute of Health-funded GAME-ON initiative is a cross-cancer genotyping project that will include 100 000 prostate cancer patient samples on a genotyping array of 500 000 SNPs. Through this expanded genotyping effort, additional risk SNPs for prostate cancer susceptibility are expected to be identified. Approximately 5000 samples from the RGC are included in the OncoArray genotyping initiative, and can be used to test associations between SNPs and radiotherapy toxicity in a future larger study with more SNPs covering a larger percentage of the familial risk. The larger sample size should allow for better testing of individual SNPs.
In summary, this work showed that there is no association between genetic susceptibility to developing prostate cancer and the development of late radiation toxicity. The implication of this finding is that standard RT for prostate cancer can be given to patients with an increased genetic burden for prostate cancer without the risk of increased late radiotherapy toxicity.
Acknowledgments
This work was supported by Cancer Research UK (C1094/A11728 to CMLW and Neil Burnet for the RAPPER study, C26900/A8740 to Gil Barnett and C5047A17528 to Ros Eeles), the Royal College of Radiologists (to Gil Barnett), Prostate Cancer UK (P2012148 to Ros Eeles), The ELLIPSE Consortium on behalf of the GAME-ON Network, The National Institute for Health Research (to Gil Barnett), Addenbrooke's Charitable Trust (to Gil Barnett), NIHR support to the Biomedical Research Centre at The Institute of Cancer Research and Royal Marsden NHS Foundation Trust, The National Institute for Health Research Cambridge Biomedical Research Centre (to Neil Burnet), UK Medical Research Council (LD), the Experimental Cancer Medicine Centre (CMLW), the Royal Marsden NHS Foundation Trust (to DPD), the United States National Institutes of Health (1R01CA134444 to BSR), the American Cancer Society (RSGT-05-200-01-CCE to BSR), the United States Department of Defense (PC074201 to BSR), Mount Sinai Tisch Cancer Institute Developmental Fund Award (to BSR), the Instituto de Salud Carlos III (FIS PI10/00164 and PI13/02030 to AV), Fondo Europeo de Desarrollo Regional (FEDER 2007-2013 to AV), Xunta de Galicia and the European Social Fund (POS-A/2013/034 to LF), and the Alberta Cancer Board Research Initiative Program (103.0393.71760001404 to MP). Laboratory infrastructure for the RAPPER study was funded by Cancer Research UK (C8197/A10123). DPD acknowledges support from the National Institute for Health Research RM/ICR Biomedical Research Centre and all the researchers at the Royal Marsden Hospital and the Institute of Cancer Research. The RAPPER cohort comprises patients and data recruited into the RT01 and CHHiP UK radiotherapy trials. The RT01 trial was supported by the UK Medical Research Council. The CHHiP trial (CRUK/06/016) was supported by the Department of Health and Cancer Research UK (C8262/A7253); trial recruitment was facilitated within centers by the National Institute for Health Research Cancer Research Network. We acknowledge the participants who agreed to be a part of this study.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
Supplementary Material
References
- Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA, Hurles ME, Mcvean GA (2010) A map of human genome variation from population-scale sequencing. Nature 467: 1061–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akamatsu S, Takata R, Haiman CA, Takahashi A, Inoue T, Kubo M, Furihata M, Kamatani N, Inazawa J, Chen GK, Le Marchand L, Kolonel LN, Katoh T, Yamano Y, Yamakado M, Takahashi H, Yamada H, Egawa S, Fujioka T, Henderson BE, Habuchi T, Ogawa O, Nakamura Y, Nakagawa H (2012) Common variants at 11q12, 10q26 and 3p11.2 are associated with prostate cancer susceptibility in Japanese. Nat Genet 44(426-9): s1. [DOI] [PubMed] [Google Scholar]
- Amin Al Olama A, Kote-Jarai Z, Schumacher FR, Wiklund F, Berndt SI, Benlloch S, Giles GG, Severi G, Neal DE, Hamdy FC, Donovan JL, Hunter DJ, Henderson BE, Thun MJ, Gaziano M, Giovannucci EL, Siddiq A, Travis RC, Cox DG, Canzian F, Riboli E, Key TJ, Andriole G, Albanes D, Hayes RB, Schleutker J, Auvinen A, Tammela TL, Weischer M, Stanford JL, Ostrander EA, Cybulski C, Lubinski J, Thibodeau SN, Schaid DJ, Sorensen KD, Batra J, Clements JA, Chambers S, Aitken J, Gardiner RA, Maier C, Vogel W, Dork T, Brenner H, Habuchi T, Ingles S, John EM, Dickinson JL, Cannon-Albright L, Teixeira MR, Kaneva R, Zhang HW, Lu YJ, Park JY, Cooney KA, Muir KR, Leongamornlert DA, Saunders E, Tymrakiewicz M, Mahmud N, Guy M, Govindasami K, O'brien LT, Wilkinson RA, Hall AL, Sawyer EJ, Dadaev T, Morrison J, Dearnaley DP, Horwich A, Huddart RA, Khoo VS, Parker CC, Van As N, Woodhouse CJ, Thompson A, Dudderidge T, Ogden C, Cooper CS, Lophatonanon A, Southey MC, Hopper JL, English D, Virtamo J, Le Marchand L, Campa D, Kaaks R, Lindstrom S, Diver WR, Gapstur S, Yeager M, Cox A, Stern MC, Corral R, Aly M, Isaacs W, Adolfsson J, Xu J, Zheng SL, Wahlfors T, Taari K, Kujala P, Klarskov P, Nordestgaard BG, Røder MA, Frikke-Schmidt R, Bojesen SE, FitzGerald LM, Kolb S, Kwon EM, Karyadi DM, Orntoft TF, Borre M, Rinckleb A, Luedeke M, Herkommer K, Meyer A, Serth J, Marthick JR, Patterson B, Wokolorczyk D, Spurdle A, Lose F, McDonnell SK, Joshi AD, Shahabi A, Pinto P, Santos J, Ray A, Sellers TA, Lin HY, Stephenson RA, Teerlink C, Muller H, Rothenbacher D, Tsuchiya N, Narita S, Cao GW, Slavov C, Mitev V UK Genetic Prostate Cancer Study Collaborators/British Association of Urological Surgeons' Section of Oncology; UK ProtecT Study Collaborators; Australian Prostate Cancer Bioresource; PRACTICAL Consortium Chanock S, Gronberg H, Haiman CA, Kraft P, Easton DF, Eeles RA (2013) A meta-analysis of genome-wide association studies to identify prostate cancer susceptibility loci associated with aggressive and non-aggressive disease. Hum Mol Genet 22: 408–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreassen CN, Barnett GC, Langendijk JA, Alsner J, De Ruysscher D, Krause M, Bentzen SM, Haviland JS, Griffin C, Poortmans P, Yarnold JR (2012) Conducting radiogenomic research—do not forget careful consideration of the clinical data. Radiother Oncol 105: 337–340. [DOI] [PubMed] [Google Scholar]
- Barnett GC, Coles CE, Elliott RM, Baynes C, Luccarini C, Conroy D, Wilkinson JS, Tyrer J, Misra V, Platte R, Gulliford SL, Sydes MR, Hall E, Bentzen SM, Dearnaley DP, Burnet NG, Pharoah PD, Dunning AM, West CM (2012. a) Independent validation of genes and polymorphisms reported to be associated with radiation toxicity: a prospective analysis study. Lancet Oncol 13: 65–77. [DOI] [PubMed] [Google Scholar]
- Barnett GC, Thompson D, Fachal L, Kerns S, Talbot C, Elliott RM, Dorling L, Coles CE, Dearnaley DP, Rosenstein BS, Vega A, Symonds P, Yarnold J, Baynes C, Michailidou K, Dennis J, Tyrer JP, Wilkinson JS, Gomez-Caamano A, Tanteles GA, Platte R, Mayes R, Conroy D, Maranian M, Luccarini C, Gulliford SL, Sydes MR, Hall E, Haviland J, Misra V, Titley J, Bentzen SM, Pharoah PD, Burnet NG, Dunning AM, West CM (2014) A genome wide association study (GWAS) providing evidence of an association between common genetic variants and late radiotherapy toxicity. Radiother Oncol 111: 178–185. [DOI] [PubMed] [Google Scholar]
- Barnett GC, West CM, Coles CE, Pharoah PD, Talbot CJ, Elliott RM, Tanteles GA, Symonds RP, Wilkinson JS, Dunning AM, Burnet NG, Bentzen SM (2012. b) Standardized total average toxicity score: a scale- and grade-independent measure of late radiotherapy toxicity to facilitate pooling of data from different studies. Int J Radiat Oncol Biol Phys 82: 1065–1074. [DOI] [PubMed] [Google Scholar]
- Bentzen SM (2006) Preventing or reducing late side effects of radiation therapy: radiobiology meets molecular pathology. Nat Rev Cancer 6: 702–713. [DOI] [PubMed] [Google Scholar]
- Bentzen SM, Constine LS, Deasy JO, Eisbruch A, Jackson A, Marks LB, Ten Haken RK, Yorke ED (2010) Quantitative Analyses of Normal Tissue Effects in the Clinic (QUANTEC): an introduction to the scientific issues. Int J Radiat Oncol Biol Phys 76: S3–S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Research UK (2014) Prostate Cancer Key Stats [Online]. Available at http://www.cancerresearchuk.org/cancer-info/cancerstats/keyfacts/prostate-cancer/ (accessed February 2015).
- Clement FC, Camenisch U, Fei J, Kaczmarek N, Mathieu N, Naegeli H (2010) Dynamic two-stage mechanism of versatile DNA damage recognition by xeroderma pigmentosum group C protein. Mutat Res 685: 21–28. [DOI] [PubMed] [Google Scholar]
- Dearnaley D, Syndikus I, Sumo G, Bidmead M, Bloomfield D, Clark C, Gao A, Hassan S, Horwich A, Huddart R, Khoo V, Kirkbride P, Mayles H, Mayles P, Naismith O, Parker C, Patterson H, Russell M, Scrase C, South C, Staffurth J, Hall E (2012) Conventional versus hypofractionated high-dose intensity-modulated radiotherapy for prostate cancer: preliminary safety results from the CHHiP randomised controlled trial. Lancet Oncol 13: 43–54. [DOI] [PubMed] [Google Scholar]
- Dearnaley DP, Jovic G, Syndikus I, Khoo V, Cowan RA, Graham JD, Aird EG, Bottomley D, Huddart RA, Jose CC, Matthews JH, Millar JL, Murphy C, Russell JM, Scrase CD, Parmar MK, Sydes MR (2014) Escalated-dose versus control-dose conformal radiotherapy for prostate cancer: long-term results from the MRC RT01 randomised controlled trial. Lancet Oncol 15: 464–473. [DOI] [PubMed] [Google Scholar]
- Delaneau O, Marchini J, Zagury JF (2012) A linear complexity phasing method for thousands of genomes. Nat Methods 9: 179–181. [DOI] [PubMed] [Google Scholar]
- Dolezel M, Odrazka K, Zouhar M, Vaculikova M, Sefrova J, Jansa J, Paluska P, Kohlova T, Vanasek J, Kovarik J (2015) Comparing morbidity and cancer control after 3D-conformal (70/74 Gy) and intensity modulated radiotherapy (78/82 Gy) for prostate cancer. Strahlenther Onkol 191(4): 338–346. [DOI] [PubMed] [Google Scholar]
- Eeles R, Goh C, Castro E, Bancroft E, Guy M, Al Olama AA, Easton D, Kote-Jarai Z (2014) The genetic epidemiology of prostate cancer and its clinical implications. Nat Rev Urol 11: 18–31. [DOI] [PubMed] [Google Scholar]
- Eeles RA, Olama AA, Benlloch S, Saunders EJ, Leongamornlert DA, Tymrakiewicz M, Ghoussaini M, Luccarini C, Dennis J, Jugurnauth-Little S, Dadaev T, Neal DE, Hamdy FC, Donovan JL, Muir K, Giles GG, Severi G, Wiklund F, Gronberg H, Haiman CA, Schumacher F, Henderson BE, Le Marchand L, Lindstrom S, Kraft P, Hunter DJ, Gapstur S, Chanock SJ, Berndt SI, Albanes D, Andriole G, Schleutker J, Weischer M, Canzian F, Riboli E, Key TJ, Travis RC, Campa D, Ingles SA, John EM, Hayes RB, Pharoah PD, Pashayan N, Khaw KT, Stanford JL, Ostrander EA, Signorello LB, Thibodeau SN, Schaid D, Maier C, Vogel W, Kibel AS, Cybulski C, Lubinski J, Cannon-Albright L, Brenner H, Park JY, Kaneva R, Batra J, Spurdle AB, Clements JA, Teixeira MR, Dicks E, Lee A, Dunning AM, Baynes C, Conroy D, Maranian MJ, Ahmed S, Govindasami K, Guy M, Wilkinson RA, Sawyer EJ, Morgan A, Dearnaley DP, Horwich A, Huddart RA, Khoo VS, Parker CC, Van As NJ, Woodhouse CJ, Thompson A, Dudderidge T, Ogden C, Cooper CS, Lophatananon A, Cox A, Southey MC, Hopper JL, English DR, Aly M, Adolfsson J, Xu J, Zheng SL, Yeager M, Kaaks R, Diver WR, Gaudet MM, Stern MC, Corral R, Joshi AD, Shahabi A, Wahlfors T, Tammela TL, Auvinen A, Virtamo J, Klarskov P, Nordestgaard BG, Røder MA, Nielsen SF, Bojesen SE, Siddiq A, Fitzgerald LM, Kolb S, Kwon EM, Karyadi DM, Blot WJ, Zheng W, Cai Q, McDonnell SK, Rinckleb AE, Drake B, Colditz G, Wokolorczyk D, Stephenson RA, Teerlink C, Muller H, Rothenbacher D, Sellers TA, Lin HY, Slavov C, Mitev V, Lose F, Srinivasan S, Maia S, Paulo P, Lange E, Cooney KA, Antoniou AC, Vincent D, Bacot F, Tessier DC COGS–Cancer Research UK GWAS–ELLIPSE (part of GAME-ON) Initiative; Australian Prostate Cancer Bioresource; UK Genetic Prostate Cancer Study Collaborators/British Association of Urological Surgeons' Section of Oncology; UK ProtecT (Prostate testing for cancer and Treatment) Study Collaborators; PRACTICAL (Prostate Cancer Association Group to Investigate Cancer-Associated Alterations in the Genome) Consortium Kote-Jarai Z, Easton DF (2013) Identification of 23 new prostate cancer susceptibility loci using the iCOGS custom genotyping array. Nat Genet 45(385-391): 391e1–392e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- England N (2013) National Cancer Survivorship Initiative [Online]. Available at http://www.ncsi.org.uk (accessed 12 January 2015).
- Fachal L, Gomez-Caamano A, Barnett GC, Peleteiro P, Carballo AM, Calvo-Crespo P, Kerns SL, Sanchez-Garcia M, Lobato-Busto R, Dorling L, Elliott RM, Dearnaley DP, Sydes MR, Hall E, Burnet NG, Carracedo A, Rosenstein BS, West CM, Dunning AM, Vega A (2014) A three-stage genome-wide association study identifies a susceptibility locus for late radiotherapy toxicity at 2q24.1. Nat Genet 46: 891–894. [DOI] [PubMed] [Google Scholar]
- Fachal L, Gomez-Caamano A, Sanchez-Garcia M, Carballo A, Peleteiro P, Lobato-Busto R, Carracedo A, Vega A (2012) TGFbeta1 SNPs and radio-induced toxicity in prostate cancer patients. Radiother Oncol 103: 206–209. [DOI] [PubMed] [Google Scholar]
- Feltes BC, Bonatto D (2015) Overview of xeroderma pigmentosum proteins architecture, mutations and post-translational modifications. Mutat Res Rev Mutat Res 763: 306–320. [DOI] [PubMed] [Google Scholar]
- Foroudi F, Tyldesley S, Barbera L, Huang J, Mackillop WJ (2003) Evidence-based estimate of appropriate radiotherapy utilization rate for prostate cancer. Int J Radiat Oncol Biol Phys 55: 51–63. [DOI] [PubMed] [Google Scholar]
- Globocan (2012) Prostate Cancer: Estimated Incidence, Mortality and Prevalence Worldwide in 2012 [Online]. Available at http://globocan.iarc.fr/old/FactSheets/cancers/prostate-new.asp (accessed November 2015).
- Hazelett DJ, Rhie SK, Gaddis M, Yan C, Lakeland DL, Coetzee SG, Henderson BE, Noushmehr H, Cozen W, Kote-Jarai Z, Eeles RA, Easton DF, Haiman CA, Lu W, Farnham PJ, Coetzee GA (2014) Comprehensive functional annotation of 77 prostate cancer risk loci. PLoS Genet 10: e1004102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie B, Marchini J, Stephens M (2011) Genotype imputation with thousands of genomes. G3 (Bethesda) 1: 457–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huda N, Tanaka H, Mendonca MS, Gilley D (2009) DNA damage-induced phosphorylation of TRF2 is required for the fast pathway of DNA double-strand break repair. Mol Cell Biol 29: 3597–3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerns SL, Kundu S, Oh JH, Singhal SK, Janelsins M, Travis LB, Deasy JO, Janssens AC, Ostrer H, Parliament M, Usmani N, Rosenstein BS (2015) The prediction of radiotherapy toxicity using single nucleotide polymorphism-based models: a step toward prevention. Semin Radiat Oncol 25: 281–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerns SL, Ostrer H, Rosenstein BS (2014. a) Radiogenomics: using genetics to identify cancer patients at risk for development of adverse effects following radiotherapy. Cancer Discov 4: 155–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerns SL, De Ruysscher D, Andreassen CN, Azria D, Barnett GC, Chang-Claude J, Davidson S, Deasy JO, Dunning AM, Ostrer H, Rosenstein BS, West CM, Bentzen SM (2014. b) STROGAR - strengthening the reporting of genetic association studies in radiogenomics. Radiother Oncol 110: 182–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerns SL, Stock RG, Stone NN, Blacksburg SR, Rath L, Vega A, Fachal L, Gomez-Caamano A, De Ruysscher D, Lammering G, Parliament M, Blackshaw M, Sia M, Cesaretti J, Terk M, Hixson R, Rosenstein BS, Ostrer H (2013) Genome-wide association study identifies a region on chromosome 11q14.3 associated with late rectal bleeding following radiation therapy for prostate cancer. Radiother Oncol 107: 372–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerns SL, West CM, Andreassen CN, Barnett GC, Bentzen SM, Burnet NG, Dekker A, De Ruysscher D, Dunning A, Parliament M, Talbot C, Vega A, Rosenstein BS (2014. c) Radiogenomics: the search for genetic predictors of radiotherapy response. Future Oncol 10: 2391–2406. [DOI] [PubMed] [Google Scholar]
- Kondo H, Soda M, Mine M, Yokota K (2013) Effects of radiation on the incidence of prostate cancer among Nagasaki atomic bomb survivors. Cancer Sci 104: 1368–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kote-Jarai Z, Saunders EJ, Leongamornlert DA, Tymrakiewicz M, Dadaev T, Jugurnauth-Little S, Ross-Adams H, Al Olama AA, Benlloch S, Halim S, Russell R, Dunning AM, Luccarini C, Dennis J, Neal DE, Hamdy FC, Donovan JL, Muir K, Giles GG, Severi G, Wiklund F, Gronberg H, Haiman CA, Schumacher F, Henderson BE, Le Marchand L, Lindstrom S, Kraft P, Hunter DJ, Gapstur S, Chanock S, Berndt SI, Albanes D, Andriole G, Schleutker J, Weischer M, Canzian F, Riboli E, Key TJ, Travis RC, Campa D, Ingles SA, John EM, Hayes RB, Pharoah P, Khaw KT, Stanford JL, Ostrander EA, Signorello LB, Thibodeau SN, Schaid D, Maier C, Vogel W, Kibel AS, Cybulski C, Lubinski J, Cannon-Albright L, Brenner H, Park JY, Kaneva R, Batra J, Spurdle A, Clements JA, Teixeira MR, Govindasami K, Guy M, Wilkinson RA, Sawyer EJ, Morgan A, Dicks E, Baynes C, Conroy D, Bojesen SE, Kaaks R, Vincent D, Bacot F, Tessier DC, Easton DF, Eeles RA (2013) Fine-mapping identifies multiple prostate cancer risk loci at 5p15, one of which associates with TERT expression. Hum Mol Genet 22: 2520–2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin NE, D'Amico AV (2014) Progress and controversies: radiation therapy for prostate cancer. CA Cancer J Clin 64: 389–407. [DOI] [PubMed] [Google Scholar]
- Mercier S, Kury S, Shaboodien G, Houniet DT, Khumalo NP, Bou-Hanna C, Bodak N, Cormier-Daire V, David A, Faivre L, Figarella-Branger D, Gherardi RK, Glen E, Hamel A, Laboisse C, Le Caignec C, Lindenbaum P, Magot A, Munnich A, Mussini JM, Pillay K, Rahman T, Redon R, Salort-Campana E, Santibanez-Koref M, Thauvin C, Barbarot S, Keavney B, Bezieau S, Mayosi BM (2013) Mutations in FAM111B cause hereditary fibrosing poikiloderma with tendon contracture, myopathy, and pulmonary fibrosis. Am J Hum Genet 93: 1100–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merino D, Malkin D (2014) p53 and hereditary cancer. Subcell Biochem 85: 1–16. [DOI] [PubMed] [Google Scholar]
- Myles P, Evans S, Lophatananon A, Dimitropoulou P, Easton D, Key T, Pocock R, Dearnaley D, Guy M, Edwards S, O'brien L, Gehr-Swain B, Hall A, Wilkinson R, Eeles R, Muir K (2008) Diagnostic radiation procedures and risk of prostate cancer. Br J Cancer 98: 1852–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohri N, Dicker AP, Showalter TN (2012) Late toxicity rates following definitive radiotherapy for prostate cancer. Can J Urol 19: 6373–6380. [PMC free article] [PubMed] [Google Scholar]
- Peeters ST, Heemsbergen WD, Van Putten WL, Slot A, Tabak H, Mens JW, Lebesque JV, Koper PC (2005) Acute and late complications after radiotherapy for prostate cancer: results of a multicenter randomized trial comparing 68 Gy to 78 Gy. Int J Radiat Oncol Biol Phys 61: 1019–1034. [DOI] [PubMed] [Google Scholar]
- Rosenstein BS, West CM, Bentzen SM, Alsner J, Andreassen CN, Azria D, Barnett GC, Baumann M, Burnet N, Chang-Claude J, Chuang EY, Coles CE, Dekker A, De Ruyck K, De Ruysscher D, Drumea K, Dunning AM, Easton D, Eeles R, Fachal L, Gutierrez-Enriquez S, Haustermans K, Henriquez-Hernandez LA, Imai T, Jones GD, Kerns SL, Liao Z, Onel K, Ostrer H, Parliament M, Pharoah PD, Rebbeck TR, Talbot CJ, Thierens H, Vega A, Witte JS, Wong P, Zenhausern F (2014) Radiogenomics: radiobiology enters the era of big data and team science. Int J Radiat Oncol Biol Phys 89: 709–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz-Feuerhake I, Pflugbeil S (2011) 'Lifestyle' and cancer rates in former East and West Germany: the possible contribution of diagnostic radiation exposures. Radiat Prot Dosimetry 147: 310–313. [DOI] [PubMed] [Google Scholar]
- Seibold P, Behrens S, Schmezer P, Helmbold I, Barnett G, Coles C, Yarnold J, Talbot CJ, Imai T, Azria D, Koch CA, Dunning AM, Burnet N, Bliss JM, Symonds RP, Rattay T, Suga T, Kerns SL, Bourgier C, Vallis KA, Sautter-Bihl ML, Claβen J, Debus J, Schnabel T, Rosenstein BS, Wenz F, West CM, Popanda O, Chang-Claude J (2015) XRCC1 Polymorphism Associated With Late Toxicity After Radiation Therapy in Breast Cancer PatientsExternal Web Site Policy. Int J Radiat Oncol Biol Phys 92(5): 1084–1092. [DOI] [PubMed] [Google Scholar]
- Sutani S, Ohashi T, Sakayori M, Kaneda T, Yamashita S, Momma T, Hanada T, Shiraishi Y, Fukada J, Oya M, Shigematsu N (2015) Comparison of genitourinary and gastrointestinal toxicity among four radiotherapy modalities for prostate cancer: conventional radiotherapy, intensity-modulated radiotherapy, and permanent iodine-125 implantation with or without external beam radiotherapy. Radiother Oncol 117: 270–276. [DOI] [PubMed] [Google Scholar]
- Talbot CJ, Tanteles GA, Barnett GC, Burnet NG, Chang-Claude J, Coles CE, Davidson S, Dunning AM, Mills J, Murray RJ, Popanda O, Seibold P, West CM, Yarnold JR, Symonds RP (2012) A replicated association between polymorphisms near TNFα and risk for adverse reactions to radiotherapyExternal Web Site Policy. Br J Cancer 107(4): 748–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West C, Rosenstein BS, Alsner J, Azria D, Barnett G, Begg A, Bentzen S, Burnet N, Chang-Claude J, Chuang E, Coles C, De Ruyck K, De Ruysscher D, Dunning A, Elliott R, Fachal L, Hall J, Haustermans K, Herskind C, Hoelscher T, Imai T, Iwakawa M, Jones D, Kulich C, Langendijk JH, O'neils P, Ozsahin M, Parliament M, Polanski A, Rosenstein B, Seminara D, Symonds P, Talbot C, Thierens H, Vega A, West C, Yarnold J (2010) Establishment of a radiogenomics consortium. Int J Radiat Oncol Biol Phys 76: 1295–1296. [DOI] [PubMed] [Google Scholar]
- Wilkins A, Mossop H, Syndikus I, Khoo V, Bloomfield D, Parker C, Logue J, Scrase C, Patterson H, Birtle A, Staffurth J, Malik Z, Panades M, Eswar C, Graham J, Russell M, Kirkbride P, O'sullivan JM, Gao A, Cruickshank C, Griffin C, Dearnaley D, Hall E (2015) Hypofractionated radiotherapy versus conventionally fractionated radiotherapy for patients with intermediate-risk localised prostate cancer: 2-year patient-reported outcomes of the randomised, non-inferiority, phase 3 CHHiP trial. Lancet Oncol 16:: 1605–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynendaele J, Bohnke A, Leucci E, Nielsen SJ, Lambertz I, Hammer S, Sbrzesny N, Kubitza D, Wolf A, Gradhand E, Balschun K, Braicu I, Sehouli J, Darb-Esfahani S, Denkert C, Thomssen C, Hauptmann S, Lund A, Marine JC, Bartel F (2010) An illegitimate microRNA target site within the 3' UTR of MDM4 affects ovarian cancer progression and chemosensitivity. Cancer Res 70: 9641–9649. [DOI] [PubMed] [Google Scholar]
- Xu J, Mo Z, Ye D, Wang M, Liu F, Jin G, Xu C, Wang X, Shao Q, Chen Z, Tao Z, Qi J, Zhou F, Wang Z, Fu Y, He D, Wei Q, Guo J, Wu D, Gao X, Yuan J, Wang G, Xu Y, Wang G, Yao H, Dong P, Jiao Y, Shen M, Yang J, Ou-Yang J, Jiang H, Zhu Y, Ren S, Zhang Z, Yin C, Gao X, Dai B, Hu Z, Yang Y, Wu Q, Chen H, Peng P, Zheng Y, Zheng X, Xiang Y, Long J, Gong J, Na R, Lin X, Yu H, Wang Z, Tao S, Feng J, Sun J, Liu W, Hsing A, Rao J, Ding Q, Wiklund F, Gronberg H, Shu XO, Zheng W, Shen H, Jin L, Shi R, Lu D, Zhang X, Sun J, Zheng SL, Sun Y (2012) Genome-wide association study in Chinese men identifies two new prostate cancer risk loci at 9q31.2 and 19q13.4. Nat Genet 44: 1231–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.