Abstract
Background and Aims Agricultural productivity is increasingly being affected by the build-up of salinity in soils and water worldwide. The genetic base of salt-tolerant rice donors being used in breeding is relatively narrow and needs broadening to breed varieties with wider adaptation to salt-affected areas. This study evaluated a large set of rice accessions of diverse origins to identify and characterize novel sources of salt tolerance.
Methods Diversity analysis was performed on 107 germplasm accessions using a genome-wide set of 376 single-nucleotide polymorphism (SNP) markers, along with characterization of allelic diversity at the major quantitative trait locus Saltol. Sixty-nine accessions were further evaluated for physiological traits likely associated with responses to salt stress during the seedling stage.
Key Results Three major clusters corresponding to the indica, aus and aromatic subgroups were identified. The largest group was indica, with the salt-tolerant Pokkali accessions in one sub-cluster, while a set of Bangladeshi landraces, including Akundi, Ashfal, Capsule, Chikirampatnai and Kutipatnai, were in a different sub-cluster. A distinct aus group close to indica contained the salt-tolerant landrace Kalarata, while a separate aromatic group closer to japonica rice contained a number of traditional, but salt-sensitive Bangladeshi landraces. These accessions have different alleles at the Saltol locus. Seven landraces – Akundi, Ashfal, Capsule, Chikirampatnai, Jatai Balam, Kalarata and Kutipatnai – accumulated less Na and relatively more K, maintaining a lower Na/K ratio in leaves. They effectively limit sodium transport to the shoot.
Conclusions New salt-tolerant landraces were identified that are genetically and physiologically distinct from known donors. These landraces can be used to develop better salt-tolerant varieties and could provide new sources of quantitative trait loci/alleles for salt tolerance for use in molecular breeding. The diversity observed within this set and in other donors suggests multiple mechanisms that can be combined for higher salt tolerance.
Keywords: Coastal saline zones, genetic diversity, salinity tolerance, Saltol QTL, single nucleotide polymorphism (SNP)
INTRODUCTION
Salinity is adversely affecting agricultural productivity in >900 000 000 ha of land worldwide. The increasing threat of salinity has become partially linked to the consequences of climate change, especially in low-lying coastal regions. Even in inland areas, salt accumulation is becoming increasingly problematic for crop production because of the build-up of salts as consequences of excessive irrigation with improper drainage and the use of poor-quality water, particularly in arid and semi-arid regions (Ismail et al., 2007, 2010; Munns and Tester, 2008).
Rice constitutes about 43 % of global food grain production, yet rice plants are inherently sensitive to salt stress (Francois and Mass, 1994; Ismail et al., 2007; Singh et al., 2010). Salt stress hinders water uptake and causes leaf damage (Yeo et al., 1990; Noble and Rogers, 1992). It shortens the lifetime of individual leaves due to excessive salt uptake, besides direct effects on carbon assimilation resulting from reduced gas exchange and endo-membrane damage, all of which reduce grain yield (Munns, 2002; Moradi and Ismail, 2007). Despite several studies in the past, our understanding of the mechanisms associated with salt stress responses in rice remain incomplete because of the complexity of the processes involved, which include an ionic component and an osmotic component, triggering numerous morphological, physiological and metabolic changes (Bray, 1993). The water deficit component is considered by some authors to be one of the serious consequences of salinity (Tabaei-Aghdaei et al., 2000; Munns et al., 2006). Salt-tolerant plant species and varieties are reported to accumulate low Na and high K as opposed to sensitive ones, through selective uptake mechanisms (Nobutoshi and Toru, 1991; Khan et al., 1992; Ismail et al., 2007; Platten et al., 2013).
Farmers have selected and grown traditional rice landraces in salt-affected areas for generations, despite their numerous undesirable traits, including longer duration to reach maturity, low yield and poor grain quality, merely because they possess remarkable tolerance to salt stress. Several physiological mechanisms have been suggested to explain the salt tolerance of these landraces, including sodium exclusion, effective sequestration of toxic salts into older leaves and roots, upregulating the antioxidant system during stress (Yeo and Flowers, 1986; Ismail et al., 2007; Moradi and Ismail, 2007) and higher tissue tolerance by compartmenting ions in vacuoles and accumulating compatible solutes in the cytoplasm (Hare et al., 1998; Yeo, 1998; Bohnert et al., 1999). Moreover, tolerance during the seedling stage seems to correlate poorly with tolerance during reproduction in rice, suggesting that different sets of mechanisms are probably involved at each stage (Moradi et al., 2003; Hossain et al., 2015). However, the vast genetic variability reported in rice in response to salinity makes it amenable to genetic manipulation to further enhance its tolerance (Akbar et al., 1972; Flowers and Yeo, 1981). For example, breeders have long made use of the high salinity tolerance of a few landraces, such as Nona Bokra and Pokkali.
Besides characterizing physiological responses to salt stress, advances have been made in identifying quantitative trait loci (QTLs) and genes controlling salinity tolerance traits. For example, several QTLs for salt tolerance have been identified in rice, including a major locus on chromosome 1, containing the major locus Saltol derived from Pokkali and SKC1 (OsHKT1;5) from Nona Bokra. The Saltol locus was reported to be involved in Na/K homeostasis under salt stress (Lin et al., 2004; Ren et al., 2005; Thomson et al., 2010; Platten et al., 2013). As each of the several physiological mechanisms underlying salinity tolerance likely involves several genes, more work is needed to identify additional novel donors and QTLs for salt tolerance, to provide the best functional alleles at these loci or new genes. Single-nucleotide polymorphism (SNP) markers have become the marker system of choice for diversity analysis, genetic mapping, association analysis and marker-assisted selection because of the high abundance of polymorphism and the availability of rapid, high-throughput genotyping systems (Rafalski, 2002; Chagné et al., 2007; Ganal et al., 2009; Thomson, 2014). For example, the Illumina GoldenGate 384-plex SNP genotyping assay has been successfully used for these types of analyses in rice (Chen et al., 2011; Thomson et al., 2012). In addition to genome-wide diversity studies, it is also useful to perform in-depth studies of allelic diversity at key loci for traits of interest, such as the Saltol locus, to determine whether new germplasm accessions have novel alleles at these loci.
Bangladesh is a rich source of diverse rice landraces adapted to its variable rice environments, including the large stretch of salt-affected areas in the southern coastal zones. There are also distinct varieties for the three main production systems: aus, aman and boro (Parsons et al., 1999). Some traditional landraces adapted to salt-affected areas have been identified before, but little is known about their relationships with global rice germplasm or the extent of allelic diversity at any of the loci associated with salinity tolerance that have been mapped before (Lisa et al., 2004). The present study aimed to identify novel sources of tolerance by assessing genetic and phenotypic diversity in a set of rice landraces from Bangladesh, India, Sri Lanka and West Africa together with several reference genotypes. A 384-SNP assay was used to characterize their allelic diversity at the Saltol locus. Moreover, the responses of these accessions to salt stress were evaluated by assessing several physiological traits.
MATERIALS AND METHODS
Plant material
A set of 107 rice genotypes was assembled, including 82 varieties and landraces from Bangladesh, 9 from India, 15 from Sri Lanka and one from the Philippines (Supplementary Data Table S1). Four reference genotypes, representative of the distinct clusters of indica, japonina, aus and aromatic subgroups, were also included for comparison: 93-11 (indica), Nipponbare (japonica), N22 (aus) and Basmati 370 (aromatic). Out of the 82 genotypes originating from Bangladesh, 64 were collected by the Bangladesh Rice Research Institute (BRRI; Gazipur, Bangladesh; http://www.brri.gov.bd/) and 18 were from the T. T. Chang Genetic Resources Center of the International Rice Research Institute (IRRI), Philippines. A set of 18 accessions named ‘Pokkali’ or ‘Pokkalian’ were also requested from IRRI, including 15 accessions originating from Sri Lanka and 3 from India. HanHonKe, Moroberekan and Azucena originated from China, West Africa/Guinea and Philippines, respectively.
Simple sequence repeat and SNP marker genotyping
For the Saltol-linked marker analysis, genomic DNA was extracted from young leaves of 3-week-old plants using a standard miniprep method with Tris/sodium dodecyl sulphate (SDS) extraction buffer (100 mm Tris–HCl pH 8, 50 mm EDTA pH 8, 500 mm NaCl, 1·25 % SDS, 0·38 % sodium bisulphate) and chloroform extraction followed by ethanol precipitation. Five markers were genotyped, including RM1287 (10·8 Mb), AP3206f (11·2 Mb), RM3412b (11·5 Mb) and RM493 (12·2 Mb), which are closely linked to the Saltol locus, and RM7075 (15·1 Mb position) as a flanking marker (Thomson et al., 2010). For the SNP analysis, genomic DNA was extracted from the seedlings of the 107 genotypes and the four reference lines (93-11, Nipponbare, N22 and Basmati 370) using DNeasy Plant Mini Kit (Qiagen, USA) following the manufacturer's protocol (www.qiagen.com). The Illumina GoldenGate assay (Fan et al., 2003) was performed using VeraCode technology on the BeadXpress Reader according to the manufacturer’s protocol. Briefly, about 250 ng of genomic DNA was used to make biotinylated genomic DNA, which then underwent oligonucleotide hybridization to bind the samples to paramagnetic particles, followed by allele-specific extension and ligation, PCR, hybridization to the Veracode Bead Plate, and scanning on the BeadXpress Reader. The analysis employed the VC0011439-OPA set of 384 SNP markers designed to be informative across indica and aus germplasm (Thomson et al., 2012) and was run at the Genotyping Services Laboratory at IRRI (Thomson, 2014; http://gsl.irri.org). Raw hybridization intensity data processing was performed using the genotyping module in the BeadStudio package (Illumina, San Diego, CA, USA), followed by allele calling using ALCHEMY software (Wright et al., 2010). After filtering for low call rates, 376 SNP markers were used in the final analysis.
Molecular marker analysis
For the analysis of molecular markers linked to the Saltol QTL on chromosome 1, the molecular weight of each band was measured using AlphaEaseFC (Alpha Innotech Corporation) version 4.0. Summary statistics, including the number of alleles per locus, major allele frequency, gene diversity and polymorphism information content (PIC) values were determined using POWERMARKER version 3·25 (Liu and Muse, 2005). The PIC values were calculated as previously described (Anderson et al., 1993). Haplotype diversity was determined according to McCartney et al. (2004) and Liu and Anderson (2003). Graphical genotyping and estimation of genetic variability of the 107 accessions were performed using Flapjack software (http://bioinf.scri.ac.uk/flapjack). For the unrooted phylogenetic tree based on the 376-SNP data, the genetic distance was calculated using the C.S. chord (1967) distance matrix (Cavalli-Sforza and Edwards, 1967), followed by phylogeny reconstruction using neighbour-joining as implemented in POWERMARKER.
Physiological characterization under salt stress
Out of the 107 rice accessions, 73 landraces collected from the southern coastal region of Bangladesh and others [IR29, a sensitive variety; FL478, a tolerant recombinant inbred line (RIL) from IR29 × Pokkali, commonly used as standard checks; IR64, a popular variety in Asia with intermediate tolerance; and Cheriviruppu and Kalarata, tolerant landraces] were selected for further evaluation under salinity stress at seedling stage. IR29 was used as sensitive check and FL478 and Pokkali were used as tolerant checks. The experiment was conducted in a greenhouse at IRRI, Los Baños, Philippines (14°11ʹ N, 121°15ʹ E, 21 m above sea level) during the 2009 dry season with day/night temperatures of 29/21 °C and 70 % relative humidity. Seeds were incubated for 5 d at 50 °C to break dormancy, then surface-sterilized with fungicide (Vitavax-200; Syngenta) and rinsed several times with distilled water. Sterilized seeds were then placed in Petri dishes with moistened filter paper and incubated at 30 °C for 48 h to germinate. Two pre-germinated seeds were sown per hole on a Styrofoam seedling float as described in Gregorio et al. (1997), suspended on distilled water in 10 L plastic trays for 3 d, then on culture solution (Yoshida et al., 1976) until the plants were 14 d old. The nutrient solution consisted of macronutrients [NH4NO3 (1·43 mm), NaH2PO4.2H2O (0·37 mm), K2SO4 (0·5 mm), CaCl2 (1·00 mm) and MgSO4.7H2O (1·6 mm)] and micronutrients (MnC13.4H20, (NH4)6Mo7024.4H20, ZnS04.7H20, H3B03, CuS04.5H20, FeC13.6H20, C6H807.H20). At 14 d after sowing, NaCl was added to the culture solution to bring its electrical conductivity to 12 dS m−1 (120 mm). Silicon in the form of sodium metasilicate 9 hydrate (16·9 mm) was added to avoid lodging. The pH of the culture solution was adjusted daily to 5 by adding either NaOH or HCl to avoid Fe deficiency (Yoshida et al., 1976) and the solution was renewed every 7 d.
A randomized complete block design was used, with three replications. Three varieties were planted in each tray as checks: IR29, Pokkali (tolerant landrace) and FL478. All entries were monitored and scored based on visual symptoms of salt stress injury as described by Gregorio et al. (1997) using modified Standard Evaluation System for rice (SES; IRRI, 2014) at 7 and 21 d after salinization as the initial and final evaluation, respectively. Final scoring and sampling were accomplished when the sensitive check IR29 scored 7 (SES 1, normal growth; 9, plants are dead). At final scoring, the number of surviving plants of each line was counted to calculate percentage survival. Selected tolerant landraces were then rescued when the final scoring and sampling of the genotypes was completed; they were then transferred to normal nutrient solution for 4 d in a phytotron and subsequently to pots in a net house for seed increase. Seeds of these selected tolerant lines were used for further characterization in replicated trials.
Assessment of physiological traits
To screen for variation in plant vigour, plants were harvested after 14 d on normal culture solution, then shoot fresh and dry weights and average shoot length were recorded. These values were divided by the mean value of the reference check, IR29. The data for each parameter was divided into nine equal class intervals and assigned scores on a 1–9 scale (1 being the most vigorous). The three scores for each variety over replications were averaged to provide the vigour score for each variety.
To measure chlorophyll concentration, freeze-dried leaf samples were weighed and transferred into test tubes. Chlorophyll was extracted with 80 % (v/v) ethanol at 80 °C for 15 min in a water bath. The extracted tissues were cooled at room temperature and the evaporated ethanol was replaced. Optical density of the extract was measured at 663, 652 and 645 nm using a UV/VIS spectrophotometer (DU-800; Beckman Coulter™). Chlorophyll a, b and total chlorophyll (a + b) were calculated in milligrams per gram dry weight (Bruinsma, 1963). To assess Na and K concentrations, samples were collected 21 d after salinization, when the SES score of the susceptible check IR29 reached 7. Ethanol extract was acidified with 1·1 N acetic acid to a final concentration of 0·1 N (1 mL of 1·1 N acetic acid in 10 mL of extract) and heated in a water bath at 90 °C for 2 h. The extracted tissue was then cooled at room temperature, left overnight and filtered using Whatman filter paper no. 40. The extracts were adjusted to volume (10 mL) using 0·1 N acetic acid, to compensate for evaporation during extraction. The Na and K concentrations in the extract were determined using an atomic absorption spectrophotometer (AAnalyst™ 200; Perkin Elmer, USA) and the concentrations of Na and K in leaf tissue were calculated in millimoles per gram dry weight.
To study ion uptake, translocation and distribution in different plant tissues, Na and K concentrations were measured in roots and shoots of different genotypes under salt stress. Total uptake of each ion was calculated based on ion concentration in the tissue and the dry weight of the corresponding tissue. Translocation of Na and K from root to shoot under salt stress was calculated for each genotype based on the total ion uptake in shoots. Translocation of Na from roots to shoots was calculated according to Saqib et al. (2005) using the formula: root-to-shoot Na translocation = shoot Na content (mmol)/root Na content (mmol). In addition, the resulting values of root-to-shoot Na translocation were normalized by dividing them by the values of shoot to root ratio (S/R) of the respective genotypes.
Stomatal conductance (millimoles per square metre per second) was measured on intact youngest fully expanded leaves using an AP4 Leaf Porometer (Dynamax, Houston, TX, USA) 10 d after the imposition of salt stress and four sub-measurements were made in each experimental unit. Leaf relative water content (RWC) was determined following the method of Barrs and Weatherley (1962), using the youngest fully expanded leaf (third leaf from top). Leaf samples were kept in plastic bags placed in an ice chest (around 5 °C) to minimize water loss, and immediately transferred to the laboratory. Leaf samples were weighed to determine their fresh weight (FW), then hydrated to full turgidity by floating them in 25 mL de-ionized water in vials with caps, for 5 h at 4 °C in a cold room. The turgid weight (TW) of the samples was determined after blotting the surface of the leaves dry with filter papers. Samples were then oven-dried at 80 °C for 48 h and weighed (DW). RWC was calculated as: [(FW − DW)/(TW − DW)] × 100.
The membrane stability index (MSI) was evaluated by estimating the electrolyte leakage from fresh leaf tissues into distilled water using the method described by Sairam et al., (2002). About 100 mg of leaf tissue was cut into 5-mm segments and placed in test tubes containing 10 mL of distilled de-ionized water in two sets, with one set placed in a water bath maintained at constant temperature of 40 °C. After 2 h their conductivity (C1) was measured using an electrical conductivity meter (Model CMK-731; Century Instruments). The second set was placed in a boiling water bath at 100 °C for 20 min to completely kill the tissues and release electrolytes. Samples were cooled to 25 °C and conductivity (C2) was measured. The MSI was calculated as (1 − C1/C2) × 100.
Statistical analysis
Data for each parameter were analysed based on a randomized complete block design with three replications. Analysis of variance (ANOVA) and mean comparisons using the least significant difference (LSD) were performed using the Statistical Analysis System package (SAS, USA) and CropStat software version 6.1 (www.irri.org). Associations among characters were examined by Pearson product moment correlation coefficient analysis, which is suitable for quantitative data (Ludbrook, 2002).
RESULTS
Genetic diversity analysis using SNP markers
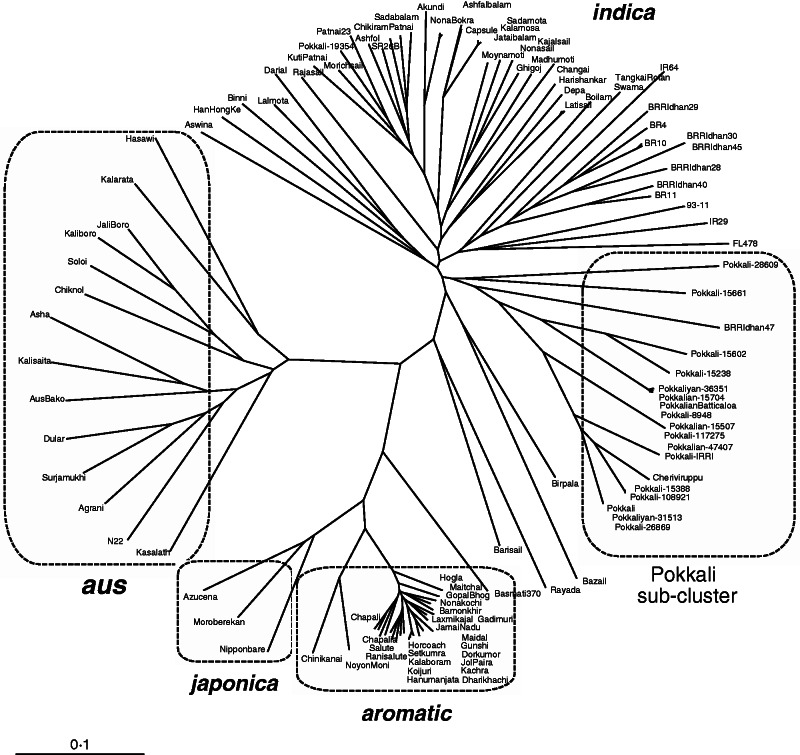
A genetic diversity analysis was performed using 376 SNP markers across the 107 accessions and four reference varieties (Supplementary Data Table S2). The minor allele frequency across SNP markers averaged 35 % with an average of 13·4 % heterozygosity, and PIC values ranging from 0·02 to 0·58 with an average of 0·35. The genetic distance-based clustering revealed four distinct groups in this diverse germplasm, corresponding to japonica, aromatic, aus and indica (Fig. 1 and Supplementary Data Fig. S1). The two tropical japonica varieties, Azucena and Moroberekan, formed a cluster with the reference temperate japonica variety Nipponbare. A distinct aromatic cluster was found near the reference variety Basmati 370, and included Bangladeshi landraces Chinikanai, Noyonmoni, Ranisalute, Gopalbhog, Jamainadu and Bamonkhir, which are popular for their excellent grain quality and aroma. The aus cluster contained the reference variety N22, and included the Bangladeshi landraces Agrani, Kasalath, Surjamukhi, Dular, AusBako, Kalisaita and Jaliboro, along with Kalarata and Hasawi of Indian and Saudi Arabian origins, respectively. The rest of the landraces fell within the largest group corresponding to indica accessions, although four accessions, Birpala, Bazail, Rayada and Barisail, seemed to fall just outside the indica group, between the indica and the aromatic/japonica clusters (Fig. 1 and Supplementary Data Fig. S1). There were several sub-clusters within indica, with all the Pokkali accessions except Pokkali-19354 grouped into one sub-cluster, along with Cheriviruppu and BRRI dhan47, which was distinct from the indica landraces from Bangladesh (Fig. 1).
Fig. 1.
Unrooted neighbour-joining tree showing genetic relationships between the 107 rice accessions and four controls based on 376 SNP markers. The four major groups of Oryza sativa are labelled in bold italics: japonica (Nipponbare as reference), aromatic (Basmati 370 as reference), indica (93-11) and aus (N22 as comparison line). A sub-cluster of Pokkali accessions are shown within the indica group. The Pokkali × IR29-derived RIL FL478 is located near IR29. Most of the salt-tolerant landraces from Bangladesh are within the different indica sub-clusters, apart from the Pokkali accessions.
As the Saltol/SKC1 region on chromosome 1 was identified as a major locus for salinity tolerance in rice, the SNP data were analysed to define the genetic relationships based on markers in this region. Out of 376 SNPs, 44 markers covered chromosome 1 and three SNP markers, id1007776, id1008267 and id1008684, were located in the 2-Mb region flanking Saltol/SKC1 between 10·8 and 12·8 Mb (Supplementary Data Fig. S2). The Saltol-linked SNPs were used to compare the graphical genotypes between sets of previously known salt-tolerant genotypes. A comparison with the representative Pokkali-108921 (IRGC 108921) showed that it shared the same alleles at the three Saltol-linked SNPs as Pokkali-15388, Pokkali-26869 and Pokkali-31513 accessions, but was different from the other lines, including FL478, IR29 and the tolerant Bangladeshi landraces such as Capsule (Supplementary Data Fig. S2A). Nona Bokra and FL478 shared similar alleles at the three SNPs with the Bangladeshi landraces Capsule, Akundi and Chikirampatnai, but were different from Kutipatnai, Cheriviruppu and Kalarata (Supplementary Data Fig. S2B, C). Analyses of variation across the entire chromosome 1, using Pokkali-108921 for comparison, showed that the highest variation was for Capsule, Akundi, IR29 and Nona Bokra (56·8–57·8 %), and least variation for Pokkali-15388 (0 %) and Cheriviruppu (9 %) across the 44 SNPs (Supplementary Data Table S3). As Pokkali-108921 and Pokkali-15388 showed no variation even across the entire set of 376 SNPs, it is likely that these came from the same source, while the other Pokkali accessions had variation ranging from 20 to 50 % from Pokkali-108921 for chromosome 1 (Supplementary Data Table S3).
Allelic diversity using Saltol-linked markers
Across the 107 accessions, the responses of rice genotypes to salt stress at the seedling stage ranged from tolerant (SES score 3) to sensitive (score 9) (Supplementary Data Table S4). RIL FL478 and 13 landraces were classified as tolerant (score 3–4) – namely Akundi, Ashfal, Capsule, Chikirampatnai, Jatai Balam and Kutipatnai from Bangladesh and Cheriviruppu, Kalarata, Nona Bokra and four different Pokkali accessions from India – while 44 others were moderately tolerant (score 5–5·7), and the remaining accessions were moderately sensitive to sensitive (score 6–9). Among the 19 Pokkali accessions, five had scores ranging from 3 to 4 (Pokkali-108921, Pokkalian Batticaloa-12092, Pokkali-15388, Pokkali-117275 and Pokkali-IRRI), while Pokkali-8558 (IRGC26869) was moderately tolerant (score 5·3) and the remaining accessions were sensitive to salinity. Out of the five tolerant Pokkali accessions, four were of Sri Lankan origin and one (Pokkali-117275) was from India.
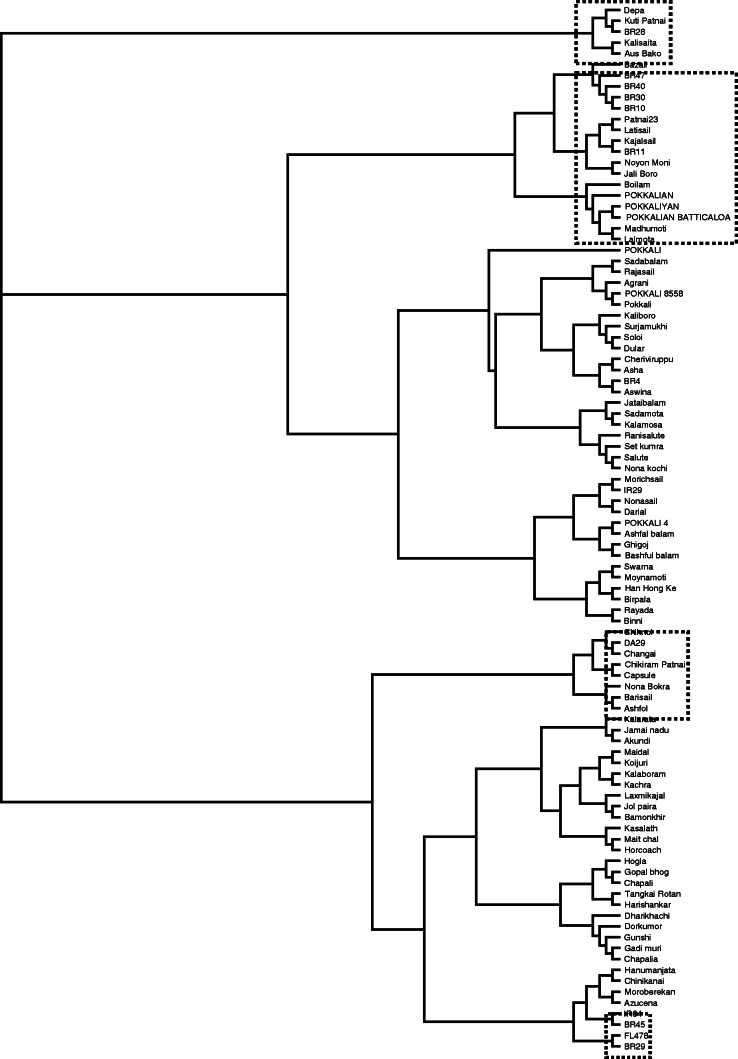
To better characterize allelic diversity across the Saltol/SKC1 region, the set of 107 accessions was genotyped with four simple sequence repeat (SSR) markers across a 1·4-Mb region on chromosome 1 – RM1287 (10·8 Mb), AP3206f (11·2 Mb), RM3412b (11·5 Mb) and RM493 (12·2 Mb) – along with one flanking marker, RM7075 (15·1 Mb). Across the five markers, 3–7 alleles were detected per locus, with an average of 4·6 alleles, while the PIC values ranged from 0·15 (AP3206f) to 0·80 (RM3412b), with an average of 0·55 (Table 1). A genetic diversity analysis using the five markers revealed several clusters (Fig. 2). Depa, Kutipatnai, BR28, Kalisaita and Aus Bako from Bangladesh formed a distinct cluster. Pokkalian, Pokkaliyan and Pokkalian Batticaloa formed a separate cluster with Boilam and a sub-cluster with Jali Boro, Noyonmoni, BR11, Latisail, Kajalsail, BR47, BR40, BR30 and BR10. A number of landraces clustered close to the various Pokkali accessions, whereas Chikirampatnai, Capsule, Changai and DA29 formed a small cluster different from the Pokkali accessions, but near the cluster of Nona Bokra, Barisail and Ashfal (Fig. 2).
Table 1.
Diversity across 107 rice accessions from different sub-groups of rice using five SSR markers linked to the Saltol locus. The panel contained 82 varieties/landraces from Bangladesh, 9 from India, 15 from Sri Lanka and 1 from Philippines
| Marker | Sample size | Major allele frequency | Mean alleles/locus | Mean QTL diversity | PIC |
|---|---|---|---|---|---|
| RM1287 | 107 | 0·60 | 3·00 | 0·56 | 0·49 |
| AP3206f | 107 | 0·92 | 3·00 | 0·16 | 0·15 |
| RM3412b | 107 | 0·27 | 7·00 | 0·82 | 0·80 |
| RM493 | 107 | 0·28 | 6·00 | 0·79 | 0·76 |
| RM7075 | 107 | 0·58 | 4·00 | 0·59 | 0·54 |
| Mean | 107 | 0·53 | 4·60 | 0·58 | 0·55 |
Fig. 2.
An UPGMA (unweighted pair group method with arithmetic mean) cluster dendrogram showing the genetic relationships among 96 rice accessions based on five microsatellite markers (RM1287, AP3206f, RM3412b, RM493 and RM7075) linked to the Saltol locus.
When examined in more detail, a set of five Pokkali accessions shared the same alleles across the five markers: Pokkali-108921, Pokkali-IRRI, Pokkali-8558 (IRGC 26869), and Pokkali-117275. Interestingly, none of the Pokkali accessions in this study carried an allele identical with that of the salt-tolerant RIL FL478 at the Saltol locus. Considering that RM1287, AP3206f and RM3412b were closest to the Saltol/SKC1 locus, there were another five accessions that shared the same alleles as Pokkali-108921 at these three markers: Nona Kochi, Pokkali-Bangladesh, Salute, Set Kumra and Agrani (Supplementary Data Table S4). If Nona Bokra is taken as a comparison line, then none of the other accessions shared the same alleles at all five markers, while four shared the alleles across the first three markers: Changai, Dorkumor, Kutipatnai and DA29 (SR26B). At the same time, many of the other tolerant accessions seemed to carry novel alleles compared with Pokkali-108921 and Nona Bokra, such as the landraces Ashfal, Akundi, Capsule, Chikirampatnai, Kalarata and Kutipatnai (Supplementary Data Table S4).
Characterization of salt stress effects across 73 accessions
A diverse set of 73 accessions was selected from the 107 genotypes by screening under salt stress, with FL478, Pokkali-R and IR29 as checks, to identify new salt-tolerant donors. Most of the accessions in this set have not been explored or characterized before and are of Bangladeshi origin. These 73 genotypes were also included in the SNP and SSR diversity panel. Significant differences were observed for all characters. The standard evaluation system (SES; IRRI, 2014) was used for rating the symptoms of salt stress 7 and 21 d after the imposition of salt stress. The genotypes varied significantly in their visual scores compared with IR29 (sensitive) and the tolerant FL478 and Pokkali. Out of the 73 genotypes, 57 landraces originated from southern Bangladesh, four landraces from India and one japonica genotype from the Philippines. The remaining 11 were modern varieties and breeding lines developed at IRRI, Philippines, and BRRI, Bangladesh (Supplementary Data Table S5). Plants were severely affected by salinity and clearly exhibited symptoms of salt injury, such as leaf burning, chlorosis and stunted growth under salt stress. The sensitive genotypes Azucena, Chapali, Chapalia, Chinikanai, Jamainadu, Kajalsail, Katarangi, Nonakuchi and Soloi and the sensitive IR29 showed the highest symptoms of salt injury, with an SES score of 7·0. The tolerant genotypes, Akundi, Ashfal, Capsule, Chikirampatnai, Cheriviruppu, Jatai Balam, Kalarata and Kutipatnai, showed better performance under salt stress and, together with the tolerant check FL478 and Pokkali, had the lowest average SES score of 3·0. The rest of the genotypes had intermediate SES scores of about 5·0–6·0 (Supplementary Data Table S5).
To account for apparent salinity tolerance due to differences in plant vigour for each genotype, seedling vigour under normal conditions was measured (Yeo et al., 1990). Genotypic differences in plant vigour were highly variable, with Pokkali being the most vigorous (score 1·67) and IR29 the least vigorous (9·0). Across the Bangladeshi landraces, Capsule showed the highest vigour (2·5), followed by Jatai Balam, Kalarata and Kutipatnai, with vigour scores of 3·0, then Cheriviruppu (3·2), Ashfal (3·7), Chikirampatnai (4·0) and Akundi (4·1). The high vigour of these landraces was similar to that of the tolerant checks FL478 and Pokkali. The rest of the sensitive genotypes showed intermediate to low plant vigour (Supplementary Data Table S5).
Shoot Na and K concentrations across the 73 accessions
The lowest shoot Na concentration was that of Capsule (0·10 mmol g−1 d.wt) and the highest was Azucena (0·67 mmol g−1 d.wt). Sodium concentration in the sensitive check IR29 (0·61 mmol g−1 d.wt) was much higher than that of the tolerant landraces Akundi, Ashfal, Capsule, Chikirampatnai, Jatai Balam, Kalarata and Kutipatnai (Supplementary Data Table S5). Shoot K concentration in the tolerant genotypes Akundi (0·16 mmol g−1 d.wt), Ashfal (0·20 mmol g−1 d.wt), Chikirampatnai (0·18 mmol g−1 d.wt), Jatai Balam (0·24 mmol g−1 d.wt), Kalarata (0·22 mmol g−1 d.wt) and Kutipatnai (0·19 mmol g−1 d.wt) was greater than that of the sensitive check IR29 (0·16 mmol g−1 d.wt) but the tolerant genotype Capsule had a lower concentration (0·12 mmol g−1 d.wt) (Supplementary Data Table S5). The tolerant genotypes Capsule, Chikirampatnai and Kalarata maintained lower Na/K ratios (0·8, 0·9 and 0·9, respectively) than the sensitive genotypes Hanumanjata, IR29 and Jamainadu, which had the highest Na/K ratios in leaves, of 4·6, 3·8 and 3·7, respectively.
The SES scores correlated with shoot Na concentration and plant vigour, (Supplementary Data Figs S3 and S4). The correlation between shoot Na concentration and chlorophyll concentration in leaves was also significant, but negative (−0·13). Based on average SES scores, plant vigour, shoot Na transport, Na/K ratio and chlorophyll concentration, the genotypes Akundi, Ashfal, Capsule, Cheriviruppu, Chikirampatnai, Jatai Balam, Kalarata and Kutipatnai can be classified as salt-tolerant and potentially provide alternative sources of new genes for breeding. Ashfal Balam, which showed relatively moderate or higher tolerance, was also included in this analysis. These nine landraces were further characterized to confirm their tolerance. Pokkali-108921 and FL478 were used as tolerant checks and IR29 as sensitive.
Characterization of the tolerant landraces for Na and K uptake and distribution
The tolerant landraces Akundi, Ashfal, Capsule, Kalarata Chikirampatnai, Jatai Balam and Kutipatnai showed higher salt tolerance than the sensitive check IR29 based on visual (SES) scores. The sensitive check IR29 had the highest shoot Na concentration (0·88 mmol g−1 d.wt), about 4-fold that of the tolerant landraces Ashfal, Capsule, Kalarata and Pokkali and about 3-fold that of Chikirampatnai, Jatai Balam, Kutipatnai and FL478 (Table 2). Genotypic differences in shoot K concentration were also significant. The highest shoot K concentration was in Pokkali (0·67 mmol g−1 d.wt), Ashfal Balam (0·60 mmol g−1d.wt) and Cheriviruppu (0·59 mmol g−1 d.wt) under salt stress. Most of the landraces had significantly higher shoot K concentration than IR29 (0·39 mmol g−1 d.wt), except for Capsule and Kalarata (0·50 mmol g−1 d.wt) and Kutipatnai (0·48 mmol g−1d.wt), the differences being non-significant (Table 2). These three landraces also did not show significant variation in shoot K concentration compared with FL478 (0·53 mmol g−1d.wt.), the tolerant check.
Table 2.
Responses of nine rice salt-tolerant landraces and checks to salt stress (electrical conductivity of 12 dS m−1) at seedling stage under phytotron conditions. Data are means of four replications with three sub-samples per replication
| Genotype | SES score | Shoot Na (mmol g−1 d.wt.) | Shoot K (mmol g−1 d.wt.) | Total uptake (mg−1 plant) |
Root Na (mmol g−1 d.wt.) | Root K (mmol g−1 d.wt.) | Root-to-shoot Na translocation |
RWC (%) | MSI (%) | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Shoot Na | Shoot K | Absolute | Normalized | ||||||||
| Akundi | 3·5bc | 0·26cde | 0·50cd | 2·72cd | 5·23b | 0·86ab | 0·08b | 0·32de | 0·32 | 62·4e | 57·1g |
| Ashfal Balam | 4·0b | 0·40b | 0·60ab | 4·25b | 6·38a | 0·82abc | 0·09b | 0·48b | 0·46 | 62·3e | 66·6f |
| Ashfal | 3·0c | 0·21e | 0·53bc | 2·26d | 5·71b | 0·89a | 0·08b | 0·24e | 0·23 | 70·1c | 82·3b |
| Capsule | 3·0c | 0·23de | 0·50cd | 2·39d | 5·20b | 0·80abcd | 0·09b | 0·25e | 0·25 | 70·3c | 69·6e |
| Chikirampatnai | 3·0c | 0·32bc | 0·52bc | 3·29c | 5·34b | 0·71bcd | 0·09b | 0·43bc | 0·44 | 80·6a | 84·1a |
| Cheriviruppu | 3·0c | 0·41b | 0·59ab | 4·31b | 6·20a | 0·71bcd | 0·23a | 0·57b | 0·55 | 72·9b | 75·6c |
| Jatai Balam | 3·5bc | 0·32bc | 0·55bc | 3·38c | 5·82b | 0·67de | 0·05b | 0·47b | 0·45 | 61·6e | 56·8g |
| Kalarata | 3·0c | 0·23cde | 0·50cd | 2·33d | 5·08bc | 0·81abcd | 0·13b | 0·27e | 0·28 | 66·1d | 65·4f |
| Kutipatnai | 3·0c | 0·31cd | 0·48cd | 3·24c | 5·02c | 0·87a | 0·07b | 0·36cd | 0·35 | 69·5c | 84·1a |
| IR29 (S. ck) | 7·0a | 0·88 a | 0·39d | 9·14a | 4·04c | 0·65e | 0·10b | 1·49a | 1·48 | 44·2f | 49·5h |
| FL478 (R. ck) | 3·0c | 0·32bc | 0·53bc | 3·29c | 5·45b | 0·72bcde | 0·10b | 0·44bc | 0·45 | 62·6e | 73·2d |
| Pokkali (R. ck) | 3·0c | 0·22de | 0·67a | 2·29d | 6·98a | 0·84abc | 0·11b | 0·29de | 0·30 | 65·8d | 76·1c |
| Significance | |||||||||||
| Genotype | *** | *** | *** | ** | * | *** | ns | *** | – | *** | *** |
| LSD P < 0·05 | 0·14 | 0·09 | 0·09 | 1·01 | 1·10 | 0·15 | 0·08 | 0·08 | – | 1·01 | 1·63 |
| LSD P < 0·01 | 0·50 | 0·12 | 0·12 | – | 1·51 | 0·20 | – | 0·11 | – | 1·32 | 2·40 |
| CV (%) | 14·2 | 18·3 | 11·3 | 12·4 | 15·3 | 13·2 | 10·1 | 11·7 | – | 1·07 | 1·62 |
Different letters (a–h) within a column denote significance based on LSD test (P < 0·05 and P < 0·01).
Root-to-shoot Na translocation was calculated as: shoot Na content (mmol)/root Na content (mmol) (Saqib et al., 2005).
Normalized root-to-shoot Na translocation was obtained by dividing root-to-shoot Na translocation values by the values of root:shoot ratio of the respective genotypes.
*P < 0·05; **P < 0·01; ***P < 0·001; ns, not significant; S. ck, sensitive check; R. ck, tolerant check; CV, coefficient of variation.
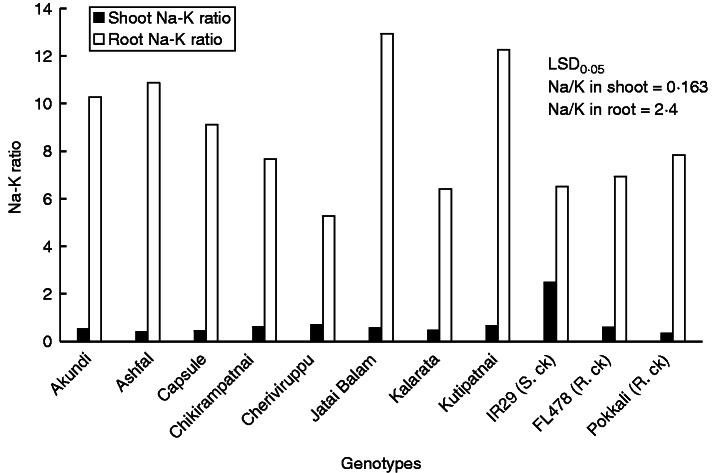
The salt-tolerant landraces maintained much lower Na/K ratios in shoots than the sensitive IR29, which showed the highest shoot ratio (2·48) (Fig. 3). Pokkali had the lowest (0·33) followed by Ashfal (0·39), Capsule (0·45), Kalarata (0·47) and Jatai Balam (0·57). The other genotypes, like Chikirampatnai (0·61), Cheriviruppu (0·69) and Kutipatnai (0·64), did not show significant variation among them and compared with the tolerant check FL478 (0·60), though their shoot Na/K ratio was considerably lower than that of IR29.
Fig. 3.
Na/K ratio in shoots and roots of selected landraces and checks under salt stress. Data are mean values of four replications, with three sub-samples per replication.
Root Na concentration was highest in Ashfal (0·89 mmol g−1d.wt) and Kutipatnai (0·86 mmol g−1 d.wt) followed by Akundi, Kalarata, Capsule, Chikirampatnai, Cheriviruppu and Jataibalam (Table 2). The lowest Na concentration (0·65 mmol g−1 d.wt) in roots was found in the sensitive check IR29, while the tolerant checks FL478 and Pokkali had root Na concentrations within the range of the tolerant Bangladeshi landraces. No significant differences were observed among genotypes in root K concentration except for Cheriviruppu (0·23 mmol g−1 d.wt), which was high under salt stress. Tolerant landraces apparently accumulated higher amounts of Na in their roots, resulting in higher Na/K ratios than in IR29, but markedly lower Na concentrations and lower Na/K ratios in their shoots than IR29 (Fig. 3). The highest root Na/K ratios were in Jatai Balam (12·93), Kutipatnai (12·27) and Ashfal (11·07), which were much higher than those of IR29 (6·5) and the tolerant FL478 (6·94) and Pokkali (7·84). This shows that under salt stress most of the salt-tolerant landraces maintain higher Na/K ratios in roots than the sensitive genotypes (except Cheriviruppu and Kalarata). Variation in this trait among tolerant genotypes is interesting and should be further evaluated.
The tolerant landraces showed a lower Na uptake that was similar to the tolerant checks Pokkali and FL478. Conversely, the sensitive IR29 had the lowest shoot K (4·04 mg plant−1) uptake, while the tolerant landrace Ashfal Balam had the highest (6·38 mg plant−1), followed by Cheriviruppu (6·20 mg plant−1), which was similar to Pokkali (6·98 mg plant−1) (Table 2). On average, shoot K uptake of the sensitive check IR29 was about 25–50 % lower than that of the tolerant landraces Ashfal, Capsule, Kalarata and Pokkali.
IR29 translocated the highest amount of Na from roots to shoots (Table 2). Significantly lower amounts were translocated by Ashfal, Capsule, Kalarata and Akundi, followed by Kutipatnai, Chikirampatnai and Jatai Balam.
Variation in stomatal conductance, leaf water content and membrane stability
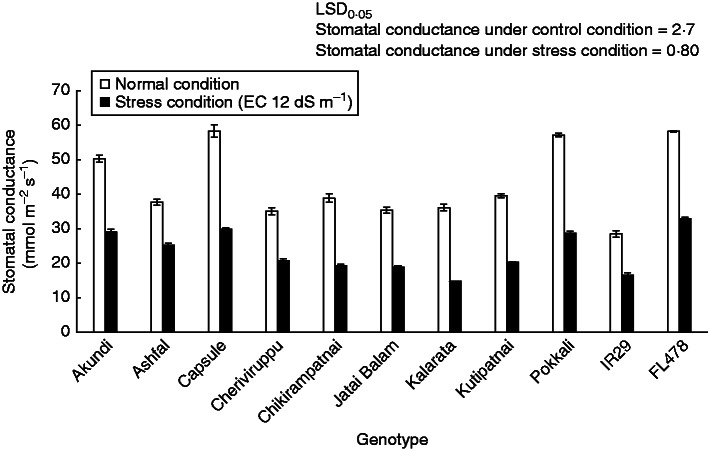
Salt stress caused a significant reduction in stomatal conductance; on average conductance decreased by about 50 % compared to that under control conditions (Fig. 4). The highest stomatal conductance was observed in Capsule under both control and stress conditions. Akundi, Ashfal and Capsule showed trends similar to those of the tolerant FL478 and Pokkali under salt stress.
Fig. 4.
Stomatal conductance of rice landraces and checks under control and salt stress conditions. Data are mean values of four replications, with three sub-samples per replication. Vertical bars indicate standard error of the mean.
Relative water content was significantly higher in Chikirampatnai (80 %), followed by Capsule, Ashfal (70 %), Kutipatnai (69·5 %) and Kalarata (66·1 %), and was lowest in IR29 (44 %; Table 2). Membrane stability index decreased under salt stress and was lowest in the sensitive IR29 (49·5 %) (Table 2). The tolerant landraces had significantly higher MSI than IR29; the highest was in Chikirampatnai and Kutipanai (84 %), followed by Ashfal (82 %) and Cheriviruppu (75 %).
Correlations among physiological traits
The SES scores correlated negatively with shoot K (r = −0·56, P < 0·01), root Na (r = −0·47, P < 0·01), RWC (r = −0·84, P < 0·001), MSI (r = −0·69, P < 0·01) and stomatal conductance (r = −0·37, P < 0·05) (Table 3). SES scores also correlated positively with shoot Na (r = 0·95, P < 0·001), shoot Na/K ratio (r = 0·96, P < 0·001) and root-to-shoot Na translocation (r = 0·94, P < 0·001), suggesting that the amount and concentration of Na in shoots constitute the main cause of the symptoms of salt injury reflected by SES scores. The correlation between shoot Na and shoot K was negative (r = −0·55, P < 0·01). Potassium concentration in the shoot also correlated negatively with root Na, shoot Na/K ratio and root-to-shoot translocation, and positively with MSI and RWC (Table 3). These correlations confirmed the importance of regulating Na uptake and its translocation to sensitive plant tissue under salt stress. Root-to-shoot Na translocation correlated positively and strongly with Na concentration (r = 0·99, P < 0·001) and Na/K ratio in shoots (r = 0·98, P < 0·001), but negatively with root Na concentration (r = −0·66, P < 0·01). RWC correlated negatively (r = −73, P < 0·01) with root-to-shoot Na translocation. However, since the selected accessions represent a set of tolerant lines, the scatters mostly tended to cluster for the tolerant lines versus values for the only sensitive IR29 (Supplementary Data Fig. S5)
Table 3.
Correlation coefficients for the association among Na and K concentrations in shoots, Na/K ratio in shoots, root Na concentration, root K concentration, Na/K ratio in roots, root-to-shoot translocation, RWC and MSI under salt stress in diverse landraces and checks
| Trait | SES score | Shoot Na (mmol g−1 d.wt.) | Shoot K (mmol g−1 d.wt.) | Na/K ratio in shoots | Root Na (mmol g−1 d.wt.) | Root K (mmol g−1 d.wt.) | Na/K ratio in root | Root-to-shoot translocation | RWC (%) | MSI (%) |
|---|---|---|---|---|---|---|---|---|---|---|
| Shoot Na (mmol g−1 d.wt.) | 0·95*** | |||||||||
| Shoot K (mmol g−1 d.wt.) | −0·56** | −0·55** | ||||||||
| Na/K ratio in shoots | 0·96*** | 0·99*** | −0·65** | |||||||
| Root Na (mmol g−1 d.wt.) | −0·47** | −0·61** | 0·24ns | −0·56** | ||||||
| Root K (mmol g−1 d.wt.) | −0·10ns | 0·11ns | 0·26ns | 0·03ns | −0·21ns | |||||
| Na/K ratio in roots | −0·17ns | −0·33* | −0·08ns | −0·28ns | 0·39* | −0·82*** | ||||
| Root-to-shoot translocation | 0·94*** | 0·99*** | −0·53** | 0·98*** | −0·66** | 0·11ns | −0·33* | |||
| RWC (%) | −0·84*** | −0·72*** | 0·41** | −0·75*** | 0·30* | 0·20ns | 0·02ns | −0·73*** | ||
| MSI (%) | −0·69** | −0·54** | 0·37* | −0·56** | 0·41** | 0·14ns | 0·02ns | −0·56** | 0·82*** | |
| Stomatal conductance | −0·37* | −0·44** | 0·28ns | −0·41** | 0·33* | −0·20ns | 0·12ns | −0·42** | 0·14ns | 0·21ns |
*P < 0·05; **P < 0·01; ***P < 0·001; ns, not significant.
DISCUSSION
Considerable genetic variability in tolerance of salt stress is available in rice. The availability of diverse sources of tolerance for breeding and gene discovery will help widen the genetic base and increase the degree of tolerance by combining superior alleles from different sources. We identified new potential donors for salt tolerance: Akundi, Ashfal, Capsule, Cheriviruppu, Chikirampatnai, Jatai Balam, Kalarata and Kutipatnai. These new sources show similar tolerance to the previously identified donors but vary in some of the traits associated with tolerance compared with the commonly used Pokkali group and Nona Bokra. They also showed allelic variation at the known Saltol locus and are genetically distinct, suggesting they could provide new alleles for tolerance. These landraces are currently being grown by farmers in the salt-affected areas of the coastal region in South Bangladesh, despite their inferior grain quality and lower yields. They could therefore be useful for use in cultivar improvement using standard breeding tools as well as sources of new QTLs/genes for use in molecular breeding. Developing new varieties with higher salt tolerance will be of significant benefit to farmers in the region who depend on rice for food security.
Bangladesh landraces are genetically distinct from common salt tolerance donors
The genome-wide 376-SNP marker analyses revealed that only Cheriviruppu and BRRI dhan47 clustered with the well-known Pokkali, the traditional donor of salinity tolerance used in most rice breeding programmes. The other landraces from Bangladesh formed separate clusters genetically distant from Pokkali accessions (Fig. 1). Although several of the Bangladeshi landraces clustered near the previously known donor Nona Bokra, allelic diversity analysis showed that they had different alleles at the SKC1 locus (Supplementary Data Table S4), which is a major determinant of salinity tolerance in Nona Bokra (Ren et al., 2005). Moreover, most of the accessions had different Saltol alleles from that of the reference Pokkali accession (IRGC-108921). Only Cheriviruppu shared the same allele as Pokkali across four out of the five markers, but differed from it at RM3412b. The fact that Akundi, Ashfal, Capsule, Chikirampatnai, Kalarata and Kutipatnai diverged from Pokkali in both SSR and SNP diversity analyses, despite their high tolerance of salt stress, makes them potential novel salt tolerance donors.
A previous report showed considerable genetic variation among different accessions of Pokkali (Thomson et al., 2010). The current study included 18 Pokkali accessions, out of which only four accessions were tolerant of salinity: Pokkali-IRRI (SES = 3·0), Pokkali-108921 (reference line, SES = 3·0), Pokkali-15388 (SES = 4·0) and Pokkali-117275 (SES = 4·0). Two accessions were moderately tolerant and the remaining 12 Pokkali accessions were sensitive. The SNP analysis showed that most of the Pokkali accessions clustered together, except for Pokkali-19354. In addition, the analysis of five Saltol-linked markers showed that the tolerant Pokkali accessions shared the same Saltol allele (Supplementary Data Table S4). This information is crucial, and suggests that only the four tolerant Pokkali accessions should be used in breeding salt-tolerant varieties, particularly for Saltol introgression.
Considering that the Bangladeshi landraces Akundi, Ashfal, Capsule, Chikirampatnai, Jatai Balam, Kalarata and Kutipatnai have different alleles from Pokkali at Saltol, but still have high salinity tolerance, suggests that these landraces are potential novel donors with new tolerance mechanisms and will likely provide alternative sources of salt tolerance genes. These novel donors are currently being grown by farmers in the salt-affected coastal region of Southern Bangladesh. The diversity observed within this set of accessions and with previously identified donors suggests the existence of multiple tolerance mechanisms that could be combined to further enhance salt tolerance of rice beyond the current level attained using the traditional donor Pokkali and its derivatives, such as FL478. Using these new donors will help widen the genetic base of salt tolerance in breeding and could allow the combination of several alleles for independent traits, but with additive effects (Ismail et al., 2007). However, further studies are needed to identify these genes or alleles and to evaluate the tolerance associated with the alternative Saltol alleles they possess. The ideal combination of particular alleles on different genetic backgrounds also needs to be established.
Physiological responses of tolerant landraces to salt stress at seedling stage
Plant vigour or faster growth and the extent of Na uptake and transport to the shoot determine Na concentration in the shoot, which is the major determinant of salt tolerance in rice (Platten et al., 2013). When the shoot grows faster, the concentration resulting from the same amount of transported Na will be lower than in slowly growing shoots. Several studies suggested faster growth under salinity as an indicator of the extent of salt tolerance of a genotype, as it reduces the toxic effects through dilution (Yeo and Flowers, 1986; Flowers et al., 1989; Yeo et al., 1990). In the current study, the salinity-tolerant Bangladeshi landraces tended to have high seedling vigour, ranging from 2·5 for Capsule to 5·4 for Jatai Balam, on a scale of 1–9, with the sensitive IR29 scoring 9. This suggests vigour as one of the mechanisms likely involved in salt stress tolerance in these landraces.
Shoot Na and K homeostasis is important for salinity tolerance in these landraces
On average, salt stress increased shoot sodium concentration by about 2-fold and root Na concentration 4-fold, with substantial variation between genotypes. The tolerant landraces Akundi, Ashfal, Capsule, Chikirampatnai, Jatai Balam, Kalarata and Kutipatnai had significantly lower Na concentration in their leaves, averaging less than a third of that accumulated by the sensitive IR29, and with Capsule maintaining the lowest concentration (Supplementary Data Table S5). Sodium concentration in seedlings of the seven tolerant landraces (2·85 mmol g−1 d.wt) was only about 30 % of that in the sensitive genotype IR29 (9·14 mmol g−1 d.wt), suggesting that these genotypes strongly exclude sodium from entering roots. However, the mechanism by which this is accomplished in rice awaits further analyses.
Salt exclusion functions to reduce the rate at which salt accumulates in roots and transpiring organs. In rice, it is believed that most of the salt enters the plant roots through passive pathways, via bypass flow (membrane leakage or apoplastic leakage). However, for translocation from roots to shoots it is estimated that close to 95 % of the volume flow is under the control of membrane selectivity before reaching the shoot (Yeo et al., 1987). Salt-tolerant rice landraces identified in this study are probably more efficient in exclusion of Na resulting in reduced Na uptake, and this has been identified as a major trait associated with salt tolerance in rice (Yeo et al., 1987; Ismail et al., 2007; Munns and Tester, 2008; Rajendran et al., 2009; Platten et al., 2013). Besides, these genotypes are efficient in restricting salt movement from root to shoot, where, on average, they translocate only about 22 % of that translocated by IR29. This will protect active tissue against toxic salt accumulation. The mechanisms by which salt is confined to the roots in these tolerant genotypes and candidate genes for these controlling mechanisms need to be identified.
The shoot Na/K ratio was higher under salt stress, mainly because of an increase in Na uptake relative to K. This result was similar to that reported by Khan et al. (1997), Asch et al. (2000) and Moradi and Ismail (2007). These authors postulated that salt-tolerant rice genotypes accumulate less Na and more K in comparison with susceptible ones. In this study, the most sensitive genotypes, Hanumanjata, IR29 and Jamainadu, had the highest Na/K ratio (4·6, 3·8 and 3·7, respectively) in leaves, while this ratio was much lower in the tolerant landraces Capsule, Chikirampatnai and Kalarata. Variation in Na/K ratio could be one of the most important determinants of salt tolerance in rice as a measure of the ability to sustain the ionic homeostasis necessary for metabolic function. However, the substantially higher correlation of Na/K ratio with Na than with K concentration in shoot suggests that this ratio is mainly driven by Na uptake and translocation to shoots. Pires et al. (2015) also made similar observations.
The correlation between chlorophyll and sodium concentrations in leaf tissue is negative (r = −0·123), indicating direct damage to photosynthetic apparatus caused by higher salt accumulation. The greater biomass produced by the tolerant landraces under salt stress could reduce Na toxicity by dilution and enhance chlorophyll retention (Moradi and Ismail, 2007).
In contrast to the shoot, root Na concentration was significantly higher in the tolerant landraces Ashfal and Kutipatnai, followed by Akundi, Ashfalbalam, Kalarata, Capsule, Chikirampatnai, Cheriviruppu and Jataibalam, but was lowest in the sensitive IR29. Accumulation of salt in roots could therefore be adaptive by restricting its translocation to shoots, possibly through selective loading into xylem tissue via HKT1;5. Compartmenting higher amounts of Na in roots under salt stress dramatically increased the Na/K ratio in roots. The ability of the tolerant landraces to restrict this increase in the amount of Na in their roots is an indication of higher tissue tolerance and might increase root osmotic potential and enhance water uptake from saline soil solution, while protecting the photosynthetic and actively growing tissues of the shoot.
Interestingly, no significant variations were found among these landraces and IR29 in root K concentration, contrary to the observations of Flores (2004) and Shannon et al. (1998), who reported higher K concentrations and lower Na/K ratios in roots of some salt-tolerant rice genotypes. The overall genetic variability in Na concentration and in Na/K ratio in roots observed here has also been reported in different sets of rice germplasm (Flowers et al., 1985; Yeo et al., 1990; Moradi et al., 2003). The variation in either shoot or root sodium concentration could in fact depend on one or more of the mechanisms that lead to low Na concentration in functional tissues and favourable ion homeostasis (Platten et al., 2013). Gregorio and Senadhira (1993) also observed a positive relation between low Na/K ratios in the shoot with salinity tolerance in rice.
Several studies using different sets of rice accessions reported similar genetic variability (Yeo and Flowers, 1983; Gregorio and Senadhira, 1993; Senadhira et al., 1996; Moradi et al., 2003; Ismail et al., 2007; Sexcion et al., 2009; Horie et al., 2012; Kumar et al., 2013). Tolerant landraces apparently accumulated higher Na in their roots, resulting in higher Na/K ratios but lower Na concentrations, and Na/K ratios in their shoots. This might reflect the importance of maintaining K homeostasis in shoots rather than in roots under saline conditions. Lee et al. (2003) reported that tolerant indica rice varieties had greater ability to absorb K and maintain a lower Na/K ratio in shoots when experiencing salt stress. Adequate K concentration in shoot and root cells is crucial for continued growth, as K is needed not only for cell turgor maintenance to drive cell expansion, but also as a co-factor for numerous enzymes (Leigh and Jones, 1984).
To determine the extent of salt exclusion by roots of individual genotypes, total ion uptake in roots and shoots was calculated. Under salt stress, the tolerant landrace Ashfal had the lowest total Na uptake, while FL478 showed the highest uptake, which could partially be due to their differences in shoot biomass as well as in compartmentation into roots (Ashfal and FL478) versus translocation to shoots. Significant differences in total uptake were also observed between tolerant genotypes, ranging from 2·33 to 4·31 mg per plant compared with IR29 (9·14 mg per plant).
The seven most tolerant landraces translocated significantly lower amounts of Na from roots to shoots than IR29. Also, root-to-shoot Na translocation was lower in these tolerant landraces compared with FL478, but was similar to that of Pokkali. Ability to retain more Na in roots is probably associated with salt stress tolerance in these genotypes. Interestingly, the absolute and normalized values for root-to-shoot Na translocation are similar among tolerant genotypes, suggesting similar translocation rates and probably similar mechanisms for exclusion from the shoot or recirculation (James et al., 2006). Tsuchiya et al. (1994) suggested some possible mechanisms that could explain the higher ability of salt-tolerant rice cultivars to restrict Na flow from root to shoot. Shi and Zhu (2002) and Hasegawa et al., (2000) argued that plasma membrane transporters like Na/H+ antiporters could have important roles in excluding Na from the shoot during xylem loading and long-distance Na transport in plants.
Several physiological parameters are important for salinity tolerance in Bangladeshi rice landraces
Salinity inhibits plant growth by a range of mechanisms, including direct ion toxicity and interference with the uptake of nutrients, particularly K (Leigh and Jones, 1984; Zhu et al., 1998). In this study, the strong negative correlation between K and Na concentration (Table 3) could be attributed to the fact that Na and K are sufficiently similar in size, charge, geometry and electronic configuration to compete for similar sites of absorption, adsorption and active transport in the plant root surface and within plant tissues. Zhu (2007) reported that Na has more damaging effects on low-affinity K transporters than high-affinity transporters, because low-affinity K transporters have lower K/Na selectivity. However, high-affinity K transporters may also act as low-affinity Na transporters in some plants under high salinity (Rubio et al., 1995; Gorham et al., 1997). The tolerant genotypes are good sodium excluders during uptake by roots. The mechanisms by which salt absorption into roots is regulated in these tolerant landraces and candidate genes for these controlling mechanisms need to be identified. The considerably stronger correlation of Na/K ratio with Na than with K concentration in shoots suggests that this ratio is mainly dependent on Na uptake and translocation to the shoot.
Plants with lower ratios of Na to K or those with more efficient mechanisms for K/Na discrimination can better tolerate salt stress. The level of inhibition of K uptake by sodium depends on how much K is present and a lower Na/K ratio is reported to be protective against the toxic effects of Na (Zhu, 2007). Ren et al. (2005) reported that the rice OsHKT1;5 transporter, which controls the SKC1 QTL, is Na-selective and is involved in Na retrieval from the xylem. In general, HKT-type transporters appear to play key roles in Na accumulation and salt sensitivity in plants (Davenport et al., 2007). Identification of the OsHKT1;5 gene suggests the existence of additional highly effective exclusion mechanisms in rice (Genc et al., 2010; Cotsaftis et al., 2011, 2012; Platten et al., 2013) and some members of the HKT family function as low-affinity Na transporters at high Na concentrations and could play important roles in regulating Na transport from roots to shoots in rice, as observed in this study (Laurie et al., 2002; Ren et al., 2005; Davenport et al., 2007; Rodríguez-Navarro and Rubio, 2006). AtHKT1 is likely important in regulating Na+ and K+ homeostasis in Arabidopsis, as mutations that disrupt its function alter the transport of Na from the root to the shoot, and the Na/K ratio in the root (Davenport et al., 2007). In addition, the SOS1 (salt overly sensitive 1; AtNHX7) locus, an Na+/H+ antiporter on the plasma membrane, expressed in root cells Shi and Zhu, 2002), is essential for Na+ and K+ homeostasis, and sos1 mutations render plants more sensitive to growth inhibition by high Na and low K environments in Arabidopsis thaliana (Shi et al., 2000). SOS1 effluxes Na from cells and may be important in Na extrusion from roots into the external medium (Munns 2005).
All tolerant landraces had higher RWC, ranging from 61 to 81 %, compared with the sensitive IR29 (44 %), suggesting that these lines were able to maintain the relatively high turgidity required for leaf function. Since sensitive genotypes usually transfer larger amounts of Na from roots to shoots, this could result in higher (less negative) osmotic potential in their roots and less water uptake from saline soil solution. However, the role of this in tolerance and how this excessive salt is compartmented in roots warrant further studies. Sensitive genotypes are also known to have less control over their stomata when subjected to salt stress, resulting in higher transpiration and greater water loss, both of which could be reflected in lower values of leaf RWC and consequent cellular dehydration (Gadallah, 1999; Sairam et al., 2002; Moradi and Ismail, 2007; Qin et al., 2010). Reduction in stomatal conductance under salt stress, however, has been observed before (Brugnoli and Lauteri, 1991; Moradi and Ismail, 2007), and will presumably reduce photosynthesis and plant growth.
The MSI was significantly higher in tolerant genotypes (56·8–84·1 %) and lower in IR29 (49·5 %). MSI is indicative of salt tolerance as it measures the extent of cell membrane injury under stress, as observed previously for heat (Ismail and Hall, 1999), cold (Ismail et al., 1997) and salt stress (Bhattacharjee and Mukherjee 1996; Farooq and Azam, 2006). We also observed significant negative correlations of MSI with shoot Na (r = −0·54, P < 0·01), SES score (r = −0·69, P < 0·01), Na/K ratio in shoots (r = −0·56, P < 0·01) and root-to-shoot translocation (r = −0·56, P < 0·01) (Table 3). MSI also showed a significant positive correlation with shoot K, root Na and leaf RWC. Farooq and Azam (2006) observed that MSI correlates negatively with Na and positively with K in rice shoots.
The genome-wide 376-SNP marker analysis showed that only Cheriviruppu and BRRI dhan47 (Pokkali origin) clustered with the well-known Pokkali, the traditional donor of salinity tolerance in most breeding programmes. The other six landraces (Akundi, Ashfal, Capsule, Chikirampatnai, Jatai Balam and Kutipatnai) formed a cluster with the indica group and Kalarata fell into the aus group. These tolerant genotypes had significantly lower shoot Na concentration, and control of root-to-shoot Na translocation appears to be important for salt tolerance in these genotypes. These landraces will constitute alternative sources of salt tolerance genes in rice and could help in developing varieties combining functional alleles of several tolerance traits.
Conclusions
The SNP diversity analysis revealed three major clusters corresponding to the indica, aus and aromatic subgroups. The largest group was indica, with the salt-tolerant Pokkali accessions in one sub-cluster, while the set of Bangladeshi landraces including Akundi, Ashfal, Capsule, Chikirampatnai and Kutipatnai were in a different sub-cluster. A distinct aus group close to indica contained the salt-tolerant landrace Kalarata, while a separate aromatic group closer to japonica rice contained a number of traditional but salt-sensitive Bangladeshi varieties. Moreover, these landraces have different new alleles at the previously characterized Saltol QTL. The seven most tolerant landraces maintained a substantially lower shoot Na concentration through reduced uptake and translocation to shoots, probably resulting in more favourable Na–K homeostasis in functional leaves and actively growing tissues. These tolerant genotypes are promising sources for sodium exclusion from roots and restricted transport to shoots. However, the control mechanisms in these tolerant landraces and the genes involved need to be identified. These accessions are novel donors for salinity tolerance and for identifying new salt tolerance genes, and will be used to develop high yielding salt-tolerant rice varieties using conventional and genome-assisted breeding strategies.
SUPPLEMENTARY DATA
Supplementary data are available online at www.aob.oxfordjournals.org and consist of the following. Table S1: list of 111 accessions used in the study and their origins. Table S2: molecular marker data from 376 SNPs across 111 accessions. Table S3: origin, pedigree, SES score and genetic variability of different Pokkali accessions and selected salt-tolerant landraces using 376 SNP markers to define genetic diversity. Table S4: SSR allele distribution in a diverse collection of rice genotypes differing in reaction to salt stress. Table S5: variation among a diverse set of rice genotypes including 62 landraces and 11 modern varieties, in physiological traits associated with tolerance of salt stress using the Standard Evaluation System. Figure S1: an UPGMA (unweighted pair group method with arithmetic mean) cluster dendrogram showing the genetic relationships between the 107 rice accessions and the four reference lines Nipponbare, 93-11, N22 and Basmati 370 based on 376 SNP markers. Figure S2: graphical genotypes showing the diversity between (A) Pokkali-108921 as control line for comparison and other Pokkali accessions including selected salt-tolerant landraces, (B) Nona Bokra (comparison line) and different salt-tolerant landraces with Pokkali-108921 (C) FL478 (comparison line) and different salt-tolerant landraces including Pokkali and Nona Bokra in the SNP alleles on chromosome 1 using Flapjack graphical genotyping software. Figure S3: scatter diagram for SES scores and shoot Na concentration. Figure S4: scatter diagram for SES scores and plant vigour. Figure S5. Scatter diagram for association of SES scores with (A) shoot Na, (B) shoot Na- K Ratio and (C) root-to-shoot translocation.
ACKNOWLEDGEMENTS
We thank Rochelle Zantua for technical assistance with marker genotyping and Macario del Valle and Ricardo Eugenio for assistance in phenotyping. This work was funded in part by the CGIAR Challenge Program on Water and Food (Competitive Project number 7) and the German Federal Ministry for Economic Cooperation and Development (BMZ).
LITERATURE CITED
- Akbar M, Yabuno T, Nakao S. 1972. Breeding for saline resistant varieties of rice. I. Variability for salt-tolerance among some rice varieties. Japanese Journal of Breeding 22: 277–284. [Google Scholar]
- Anderson JA, Churchill GA, Autrique JE, Tanksley SD, Sorrells ME. 1993. Optimizing parental selection for genetic linkage maps. Genome 36: 181–186. [DOI] [PubMed] [Google Scholar]
- Asch F, Dingkuhn M, Dorffling K, Miezan K. 2000. Leaf K-Na ratio predicts salinity induced yield loss in irrigated rice. Euphytica 113: 109–118. [Google Scholar]
- Barrs HD, Weatherley PE. 1962. A re-examination of the relative turgidity technique for estimating water deficit in leaves. Australian Journal of Biological Sciences 15: 413–428. [Google Scholar]
- Bhattacharjee S, Mukherjee AK. 1996. Ethylene evolution and membrane lipid peroxidation as indicators of salt injury in leaf tissues of Amaranthus lividus seedlings. Indian Journal of Experimental Biology 34: 279–281. [Google Scholar]
- Bohnert HJ, Su H, Shen B. 1999. Molecular mechanisms of salinity tolerance. In: Shinozaki K, Yamaguchi-Shinozaki K, eds. Molecular responses to cold, drought, heat and salt stress in higher plants. Austin: R. G. Landes, 29–60. [Google Scholar]
- Bray EA. 1993. Molecular responses to water deficit. Plant Physiology 103: 1035–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugnoli E, Lauteri M. 1991. Effects of salinity on stomatal conductance, photosynthetic capacity, and carbon isotope discrimination of salttolerant (Gossypium hirsutum L.) and salt-sensitive (Phaseolus vulgaris L.) C3 non-halophytes. Plant Physiology 95: 628–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruinsma J. 1963. The quantitative analysis of chlorophylls a and b in plant extracts. Photochemistry and Photobiology 2: 241–249. [Google Scholar]
- Cavalli-Sforza LL, Edwards AWF. 1967. Phylogenetic analysis. Models and estimation procedures. American Journal of Human Genetics 19: 233–257. [PMC free article] [PubMed] [Google Scholar]
- Chagné D, Batley J, Edwards D, Forster JW. 2007. Single nucleotide polymorphism genotyping in plants. In: Oraguzie NC, Rikkerink EHA, Gardiner SE, De Silva HN. eds. Association mapping in plants. Berlin: Springer, 77–94. [Google Scholar]
- Chen H, He H, Zou Y, et al. 2011. Development and application of a set of breeder-friendly SNP markers for genetic analyses and molecular breeding of rice (Oryza sativa L.). Theoretical and Applied Genetics 123: 869–879. [DOI] [PubMed] [Google Scholar]
- Cotsaftis O, Pletta D, Johnsona AAT, et al. 2011. Root-specific transcript profiling of contrasting rice genotypes in response to salinity stress. Molecular Plant 4: 25–41. [DOI] [PubMed] [Google Scholar]
- Cotsaftis O, Plett D, Shirley N, Tester M, Hrmova M. 2012. A Two-staged model of Na+ exclusion in rice explained by 3D modeling of HKT transporters and alternative splicing. PLoS One 7: e39865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport RJ, Muñoz-Mayor A, Jha D, Essah PA, Rus A, Tester M. 2007. The transporter AtHKT1;1 controls retrieval of Na+ from the xylem in Arabidopsis . Plant, Cell & Environment 30: 497–507. [DOI] [PubMed] [Google Scholar]
- Fan JB, Oliphant A, Shen R, et al. 2003. Highly parallel SNP genotyping. Cold Spring Harbor Symposia on Quantitative Biology 68: 69–78. [DOI] [PubMed] [Google Scholar]
- Farooq S, Azam F. 2006. The use of cell membrane stability (CMS) technique to screen for salt tolerant wheat varieties. Journal of Plant Physiology 163: 629–637. [DOI] [PubMed] [Google Scholar]
- Flores NRL. 2004. Role of root signals in rice (Oryza sativa L.) responses to salinity stress. MSc Thesis, University of the Philippines. [Google Scholar]
- Flowers TJ, Yeo AR. 1981. Variability in the resistance of sodium chloride salinity within rice (Oryza sativa L.) varieties. New Phytologist 88: 363–373. [Google Scholar]
- Flowers TJ, Duque E, Hajibagheri MA, McGonigle TP, Yeo AR. 1985. The effect of salinity on leaf ultrastructure and net photosynthesis of two varieties of rice: further evidence of cellular component of salt-resistance. New Phytologist 100: 37–43. [Google Scholar]
- Flowers TJ, Salama FM, Yeo AR. 1989. Water use efficiency in rice (Oryza sativa L.) in relation to resistance to salinity. Plant, Cell & Environment 11: 453–459. [Google Scholar]
- Francois LB, Mass EV. 1994. Crop response and management on salt affected soils. In: Pessarakli M, ed. Handbook of plant and crop stress. New York: Marcel Dekker, 149–181. [Google Scholar]
- Gadallah MAA. 1999. Effects of proline and glycinebetaine on Vicia faba response to salt stress. Biologia Plantarum 42: 249–257. [Google Scholar]
- Ganal MW, Altmann T, Roder MS. 2009. SNP identification in crop plants. Current Opinion in Plant Biology 12: 1–7. [DOI] [PubMed] [Google Scholar]
- Genc Y, Oldach K, Verbyla AP, et al. 2010. Sodium exclusion QTL associated with improved seedling growth in bread wheat under salinity stress. Theoretical and Applied Genetics 121: 877–894. [DOI] [PubMed] [Google Scholar]
- Gorham J, Bridges J, Dubcovsky J, et al. 1997. Genetic analysis and physiology of a trait for enhanced K+/Na+ discrimination in wheat. New Phytologist 137: 109–116. [Google Scholar]
- Gregorio GB, Senadhira D. 1993. Genetic analysis of salinity tolerance in rice. Theoretical and Applied Genetics 86: 333–338. [DOI] [PubMed] [Google Scholar]
- Gregorio GB, Senadhira D, Mendoza RD. 1997. Screening rice for salinity tolerance. Manila: International Rice Research Institute; Discussion Paper Series No. 22. [Google Scholar]
- Hare PD, Cress WA, Van Staden J. 1998. Dissecting roles of osmolyte accumulation during stress. Plant, Cell & Environment 21: 535–553. [Google Scholar]
- Hasegawa PM, Bressan RA, Zhu JK, Bohnert HJ. 2000. Plant cellular and molecular responses to high salinity. Annual Review of Plant Physiology and Plant Molecular Biology 51: 463–499. [DOI] [PubMed] [Google Scholar]
- Horie T, Karahara I, Katsuhara M. 2012. Salinity tolerance mechanisms in glycophytes: an overview with the central focus on rice plants. Rice 5: 11 http://www.thericejournal.com/content/5/1/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain H, Rahman MA, Alam MS, Singh RK. 2015. Mapping of quantitative trait loci associated with reproductive-stage salt tolerance in rice. Journal of Agronomy and Crop Science 201: 17–31. [Google Scholar]
- IRRI. 2014. Standard evaluation system for rice (SES), 5th edn. Los Banos, Philippines: International Rice Research Institute. [Google Scholar]
- Ismail AM, Hall AE. 1999. Reproductive-stage heat-tolerance, leaf membrane thermostability and plant morphology in cowpea. Crop Science 39: 1762–1768. [Google Scholar]
- Ismail AM, Hall AE, Close T J. 1997. Chilling tolerance during emergence of cowpea associated with a dehydrin and slow electrolyte leakage. Crop Science 37: 1270–1277. [Google Scholar]
- Ismail AM, Heuer S, Thomson MJ, Wissuwa M. 2007. Genetic and genomic approaches to develop rice germplasm for problem soils. Plant Molecular Biology 65: 547–570. [DOI] [PubMed] [Google Scholar]
- Ismail AM, Thomson MJ, Vergara GV, et al. 2010. Designing resilient rice varieties for coastal deltas using modern breeding tools. In: Hoanh CT, Szuster BW, Pheng KS, Ismail AM, Nobel AD, eds. Tropical deltas and coastal zones: food production, communities and environment at the land-water interface. Wallingford: CAB, 154–165. [Google Scholar]
- James RA, Davenport RJ, Munns R. 2006. Physiological characterization of two genes for Na+ exclusion in durum wheat, Nax1 and Nax2 . Plant Physiology 142: 1537–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan AH, Ashraf MY, Azmi AR. 1992. Osmotic adjustment in sorghum under NaCl stress, Physiologia Plantarum 14: 159–164. [Google Scholar]
- Khan MSA, Hamid A, Karim MA. 1997. Effect of sodium chloride on germination and seedling characters of different types of rice (Oryza sativa L.). Journal of Agronomy and Crop Science 179: 163–169. [Google Scholar]
- Kumar K, Kumar M, Kim SR, Ryu H, Cho YG. 2013. Insights into genomics of salt stress response in rice. Rice 6: 27 http://www.thericejournal.com/content/6/1/27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie S, Feeney KA, Maathuis FJM, Heard PJ, Brown SJ, Leigh RA. 2002. A role for HKT1 in sodium uptake by wheat roots. The Plant Journal 32: 139–149. [DOI] [PubMed] [Google Scholar]
- Lee K-S, Choi W-Y, Ko J-C, Kim T-S, Gregorio GB. 2003. Salinity tolerance of japonica and indica rice (Oryza sativa L.) at the seedling stage. Planta 216: 1043–1046. [DOI] [PubMed] [Google Scholar]
- Leigh RA, Jones RGW. 1984. A hypothesis relating critical potassium concentration for growth to the distribution and functions of this ion in the plant cell. New Phytologist 97:1–13. [Google Scholar]
- Lin HX, Zhu MZ, Yano M, et al. 2004. QTLs for Na+ and K+ uptake of the shoots and roots controlling rice salt tolerance. Theoretical and Applied Genetics 108: 253–260. [DOI] [PubMed] [Google Scholar]
- Lisa LA, Seraj ZI, Elahi CMF, et al. 2004. Genetic variation in microsatellite DNA, physiology and morphology of coastal saline rice (Oryza sativa L.) landraces of Bangladesh. Plant and Soil 263: 213–228. [Google Scholar]
- Liu K, Muse SV. 2005. PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21: 2128–2129. [DOI] [PubMed] [Google Scholar]
- Liu S, Anderson JA. 2003. Targeted molecular mapping of major wheat QTL for Fusarium head blight resistance using wheat ESTs and synteny with rice. Genome 46: 817–823. [DOI] [PubMed] [Google Scholar]
- Ludbrook J. 2002. Statistical techniques for comparing measures and methods of measurement: A critical review. Clinical and Experimental Pharmacology and Physiology 29: 527–536. [DOI] [PubMed] [Google Scholar]
- McCartney CA, Sommers DJ, Fedak G, Cao W. 2004. Haplotype diversity at fusarium head blight resistance QTLs in wheat. Theoretical and Applied Genetics 109: 261–271. [DOI] [PubMed] [Google Scholar]
- Moradi F, Ismail AM. 2007. Reponses of photosynthesis, chlorophyll fluorescence and ROS-scavenging systems to salt stress during seedling and reproductive stages in rice. Annals of Botany 99: 1161–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moradi F, Ismail AM, Gregorio GB, Egdane JA. 2003. Salinity tolerance of rice during reproductive development and association with tolerance at the seedling stage. Indian Journal of Plant Physiology 8: 105–116. [Google Scholar]
- Munns R. 2002. Comparative physiology of salt and water stress. Plant, Cell & Environment 25: 239–250. [DOI] [PubMed] [Google Scholar]
- Munns R. 2005. Genes and salt tolerance: bringing them together. New Phytologist 167: 645–663. [DOI] [PubMed] [Google Scholar]
- Munns R, Tester M. 2008. Mechanisms of salinity tolerance. Annual Review of Plant Biology 59: 651–681. [DOI] [PubMed] [Google Scholar]
- Munns R, James RA, Läuchli A. 2006. Approaches to increasing the salt tolerance of wheat and other cereals. Journal of Experimental Botany 57: 1025–1043. [DOI] [PubMed] [Google Scholar]
- Noble CL, Rogers NE. 1992. Arguments for the use of physiological criteria for improving the salt tolerance in crops. Plant and Soil 146: 99–107. [Google Scholar]
- Nobutoshi M, Toru M. 1991. Characterization of Na+ exclusion mechanism of salt- tolerant reed plants in comparison with salt sensitive rice plants. Physiologia Plantarum 83: 170–176. [Google Scholar]
- Parsons BJ, Newbury HJ, Jackson MT. 1999. The genetic structure and conservation of aus, aman and boro rices from Bangladesh. Genetic Resources and Crop Evolution 46: 587–598. [Google Scholar]
- Pires PS, Negrã S, Oliveira MM, Purugganan MD. 2015. Comprehensive phenotypic analysis of rice (Oryza sativa) response to salinity stress. Physiologia Plantarum 155: 43–54. [DOI] [PubMed] [Google Scholar]
- Platten JD, Egdane JA, Ismail AM. 2013. Salinity tolerance, Na+ exclusion and allele mining of HKT1;5 in Oryza sativa and O. glaberrima: many sources, many genes, one mechanism? BMC Plant Biology 13: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Dong WY, He KN, et al. 2010. NaCl salinity-induced changes in water status, ion contents and photosynthetic properties of Shepherdia argentea (Pursh) Nutt seedlings. Plant, Soil and Environment 56: 325–332. [Google Scholar]
- Rafalski A. 2002. Applications of single nucleotide polymorphisms in crop genetics. Current Opinion in Plant Biology 5: 94–100. [DOI] [PubMed] [Google Scholar]
- Rajendran K, Tester M, Roy SJ. 2009. Quantifying the three main components of salinity tolerance in cereals. Plant, Cell & Environment 32: 237–249. [DOI] [PubMed] [Google Scholar]
- Ren ZH, Gao JP, Li LG, et al. 2005. A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nature Genetics 37: 1141–1146. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Navarro A, Rubio F. 2006. High-affinity potassium and sodium transport systems in plants. Journal of Experimental Botany 57: 1149–1160. [DOI] [PubMed] [Google Scholar]
- Rubio F, Gassmann W, Schroeder JI. 1995. Sodium driven potassium uptake by the plant potassium transporter HKT1 and mutations conferring salt tolerance. Science 270: 1660–1663. [DOI] [PubMed] [Google Scholar]
- Sairam RK, Rao KV, Srivastava GC. 2002. Differential response of wheat genotypes to long term salinity stress in relation to oxidative stress, antioxidant activity and osmolyte concentration. Plant Science 163: 1037–1046. [Google Scholar]
- Saqib M, Zörb C, Rengel Z, Schubert S. 2005. The expression of the endogenous vacuolar Na+/H+ antiporters in roots and shoots correlates positively with the salt resistance of wheat (Triticum aestivum L.). Plant Science 169: 959–965. [Google Scholar]
- Senadhira D, Gregorio GB, Roxas JP, Mendoza RD. 1996. Challenges and opportunities in breeding for salinity tolerance in rice. In: Proceedings, Workshop on Soil Salinity in Rice, 24–26 January, PhilRice, Muñoz, Philippines, 31–40. [Google Scholar]
- Sexcion PSH, Egdane JA, Ismail AM, Dionisio-Sese ML. 2009. Morpho-physiological traits associated with tolerance of salinity during seedling stage in rice (Oryza sativa L.). Philippines Journal of Crop Science 34: 27–37. [Google Scholar]
- Shannon MC, Rhodes JD, Draper JH, Scardaci SC, Spyres MD. 1998. Assessment of salt tolerance in rice cultivars in response to salinity problems in California. Crop Science 38: 394–398. [Google Scholar]
- Shi H, Zhu J-K. 2002. Regulation of expression of the vacuolar Na+/H+ antiporter gene AtNHX1 by salt stress and abscisic acid. Plant Molecular Biology 50: 543–550. [DOI] [PubMed] [Google Scholar]
- Shi H, Ishitani M, Kim C, Zhu JK. 2000. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proceedings of the National Academy of Sciences of the USA 97: 6896–6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh RK, Redoña ED, Refuerzo L. 2010. Varietal improvement for abiotic stress tolerance in crop plants: special reference to salinity in rice. In: Pareek A, Sopory SK, Bohnert HJ, Govindjee, eds. Abiotic stress adaptation in plants: physiological, molecular and genomic foundation. New York: Springer, 387–415. [Google Scholar]
- Tabaei-Aghdaei S, Harrison P, Pearee RS. 2000. Expression of dehydration-stress related genes in crown of wheat grass species having contrasting acclimation to salt, cold and drought. Plant, Cell & Environment 23:561–71. [Google Scholar]
- Thomson MJ. 2014. High-Throughput SNP genotyping to accelerate crop improvement. Plant Breeding and Biotechnology 2:195–212. [Google Scholar]
- Thomson MJ, de Ocampo M, Egdane J, et al. 2010. Characterizing the Saltol quantitative trait locus for salinity tolerance in rice. Rice 3:148–160. [Google Scholar]
- Thomson MJ, Zhao K, Wright M, et al. 2012. High-throughput single nucleotide polymorphism genotyping for breeding applications in rice using the BeadXpress platform. Molecular Breeding 29: 875–886. [Google Scholar]
- Tsuchiya M, Miyake M, Naito H. 1994. Physiological response to salinity in rice plant. III. A possible mechanism for Na+ exclusion in rice root under NaCl-stress conditions. Japanese Journal of Crop Science 63: 326–332. [Google Scholar]
- Wright MH, Tung CW, Zhao K, Reynolds A, McCouch SR, Bustamante CD. 2010. ALCHEMY: a reliable method for automated SNP genotype calling for small batch sizes and highly homozygous populations. Bioinformatics 26: 2952–2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo A. 1998. Molecular biology of salt tolerance in the context of whole plant physiology. Journal of Experimental Botany 49: 915–923. [Google Scholar]
- Yeo AR, Flowers TJ. 1983. Varietal differences in the toxicity of sodium ions in rice leaves. Physiologia Plantarum 59: 189–195. [Google Scholar]
- Yeo AR, Flowers TJ. 1986. Salinity resistance in rice (Oryza sativa L.) and a pyramiding approach to breeding varieties for saline soils. Australian Journal of Plant Physiology 13:161–173. [Google Scholar]
- Yeo AR, Yeo ME, Flowers TJ. 1987. The contribution of an apoplastic pathway to sodium uptake by rice roots in saline conditions. Journal of Experimental Botany 38: 1141–1153. [Google Scholar]
- Yeo AR, Yeo ME, Flowers SA, Flowers TJ. 1990. Screening of rice (Oryza sativa L.) genotypes for physiological characters contributing to salinity resistance, and their relationship to overall performance. Theoretical and Applied Genetics 79: 377–384. [DOI] [PubMed] [Google Scholar]
- Yoshida S, Forno DA, Cock JH, Gomez KA. 1976. Laboratory manual for physiological studies of rice. Philippines: IRRI. [Google Scholar]
- Zhu J-K. 2007. Plant salt stress. In: Encyclopedia of Life Sciences. Wiley Online Library, doi:10.1002/9780470015902.a0001300.pub2. [Google Scholar]
- Zhu J-K, Liu J, Xiong L. 1998. Genetic analysis of salt tolerance in Arabidopsis thaliana: evidence of a critical role for potassium nutrition. Plant Cell 10: 1181–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.