Abstract
Background and Aims Flower colour polymorphism in plants has been used as a classic model for understanding the importance of neutral processes vs. natural selection in population differentiation. However, current explanations for the maintenance of flower colour polymorphism mainly rely on balancing selection, while neutral processes have seldom been championed. Iris lutescens (Iridaceae) is a widespread species in the northern Mediterranean basin, which shows a stable and striking purple–yellow flower colour polymorphism. To evaluate the roles of neutral processes in the spatial variation for flower colour in this species, patterns of neutral genetic variation across its distribution range were quantified, and phenotypic differentiation was compared with neutral genetic differentiation.
Methods Genetic diversity levels and population genetic structure were investigated through the genotyping of a collection of 1120 individuals in 41 populations ranging from Spain to France, using a set of eight newly developed microsatellite markers. In addition, phenotypic differentiation for flower colour was also quantified by counting colour morph frequency in each population, and measuring the reflectance spectra of sampled individuals.
Key Results Populations in Spain present a sharp colour transition from solely purple to solely yellow. The results provide evidence that genetic drift through limited gene flow is important in the evolution of monomorphic populations. In contrast, most populations in France are polymorphic with both phenotypes, and the colour frequencies vary geographically without any spatial gradients observed. A pattern of isolation by distance is detected in France, and gene flow between adjacent populations seems to be an important factor maintaining populations polymorphic.
Conclusions Overall, neutral processes contribute to patterns of spatial variation for flower colour in I. lutescens, but it cannot be excluded that natural selection is also operating. An interaction between neutral processes and natural selection is suggested to explain the spatial variation for flower colour in I. lutescens.
Keywords: Flower colour polymorphism, Iris lutescens, genetic drift, gene flow, isolation by distance
INTRODUCTION
Ecologists have long been intrigued by the emergence and maintenance of polymorphism in floral traits (Weiss, 1995; Schaefer et al., 2004). Indeed, as the pollinator visitation rate is generally correlated with plant fitness in entomogamous species (i.e. 80 % of plant species; Potts et al., 2010), stabilizing selection mediated by associative learning is expected to occur on floral traits, leading to low intra-specific variation (Ashman and Majetic, 2006; Salzmann and Schiestl, 2007; van Kleunen et al., 2008; Dormont et al., 2010). Because flower colour (i.e. colour of the perianth) is one of the most important cues used by pollinators to locate flowers and to learn foraging targets (Menzel and Shmida, 1993; Schoonhoven et al., 2007), any flower colour variation within populations should be rapidly lost due to directional pollinator-mediated selection (Waser and Price, 1981; Levin and Brack, 1995; Campbell et al., 1997; Jones and Reithel, 2001; Chittka and Raine, 2006). This may make the maintenance of flower colour polymorphism surprising, and only a reduced number of species with a genetically controlled flower colour polymorphism within natural populations have been observed (see Rausher, 2008).
From a historical perspective, flower colour polymorphism has made an important contribution to the development of modern evolutionary theory, particularly because Wright (1943) early applied his model of ‘isolation by distance’ (IBD) to investigate spatial patterns of flower colour in Linanthus parryae (Polemoniaceae). He concluded that the spatial distribution of the blue–white morphs in this species was consistent with a random process, and he viewed this species as an example of the drift process required for his shifting balance theory of evolution. In contrast, subsequent observations and experiments led Epling et al. (1960) to conclude that this variation was subject to selection, but Wright (1978) continued to disagree. Only recently has this debate apparently been resolved in Epling’s favour by Schemske and Bierzychudek (2001, 2007). Their results indicate that the flower colour polymorphism in L. parryae is largely a product of temporal and spatial heterogeneity in local selective pressures. So far, this is the only case where the neutrality of flower colour polymorphism has been seriously argued (see also Podolsky and Holtsford, 1995 for various floral traits including stigmate colour and petal colour; Jorgensen et al., 2006 for pollen colour dimorphism).
Indeed, most empirical studies focusing on the maintenance of flower colour polymorphism showed the importance of balancing selection resulting from environmental heterogeneity in time and space (Schemske and Bierzychudek, 2001, 2007; Arista et al., 2013) and antagonistic selection imposed by pollinators and herbivores (Irwin et al., 2003; Strauss et al., 2004; see also Carlson and Holsinger, 2010 for an effect of seed predators). These selective forces are imposed not only by pollinators, but also by the abiotic environment and/or animals with antagonistic effects. This is because the flavonoid/anthocyanin pigments, which are responsible for flower colour in most plants, also have pleiotropic effects on plant survival. For example, these pigments can function in protecting plants against damage caused by UV and visible light, in responses of plants to abiotic stress (e.g. drought and cold) and in resistance to attack by microbes and herbivores (reviewed in Chalker-Scott, 1999; Harborne and Williams, 2000; Winkel-Shirley, 2002; Taylor and Grotewold, 2005; Agati et al., 2012). Whatever the selective pressures involved, spatial segregation among colour morphs is generally observed, and populations are monomorphic, while neutral markers showed little genetic differentiation among morphs (see, for instance, Streisfeld and Kohn, 2005; Sobel and Streisfeld, 2015).
However, some species have a flower colour polymorphism within populations. In food-deceptive species, in particular orchids, negative frequency-dependent selection (NFDS) mediated by learning ability of pollinators is often invoked to maintain such a polymorphism (Gigord et al., 2001). Such a mechanism has only been formally demonstrated once (failed to be detected by Pellegrino et al., 2005a, b; Jersáková et al., 2006; Imbert et al., 2014b for instance), and overall cannot explain variation in flower colour morph proportions among populations (Gigord et al., 2001). For species with a genetic control of flower colour, gene flow among populations could contribute to a transient maintenance of polymorphism. Likewise, any disequilibrium between gene flow and genetic drift should lead a population to become monomorphic.
As neutrality is the null hypothesis, demonstrating its contribution to any phenotypic spatial pattern is a challenge. A classical approach to demonstrate the relative roles of purely neutral processes (i.e. drift in combination with spatially restricted gene flow) and natural selection in spatial differentiation of the heritable phenotypic trait is to study whether the population distribution of the phenotypic character differs from the neutral expectation, i.e. to compare the phenotypic differentiation with the neutral genetic differentiation. In the case of flower colour polymorphism, the spatial flower colour differentiation can be estimated by Euclidean flower colour distance between pairwise populations (e.g. Edh et al., 2007), by quantifying flower colour as a quantitative trait (QST; e.g. Streisfeld and Kohn, 2005) or by calculating the FST at the loci that are responsible for flower colour change (e.g. Schemske and Bierzychudek, 2007; Hopkins et al., 2012); on the other hand, neutral genetic differentiation (FST at neutral loci) has been estimated using neutral markers such as microsatellites (see also Sobel and Streisfeld, 2015). All published studies so far concluded that differentiation in flower colour is driven by selection, rather than neutral processes, with the only exception of the study of Brassica cretica (Brassicaceae), an endemic species in the Aegean island Crete. Indeed, Edh et al. (2007) found that those populations, fixed for different colour morphs, have been heavily influenced by genetic drift. However, in most empirical experiments, the fitness differences among colour morphs cannot be unambiguously ascribed to variation in flower colour genes rather than to variation at linked genes (reviewed in Rausher, 2008). Therefore, we cannot rule out the possibility that neutral processes could contribute to the spatial variation in flower colour, especially in small and isolated populations (Eckhart et al., 2006; Edh et al., 2007).
Here we investigated flower colour polymorphism in a widespread species of the northern Mediterranean basin, Iris lutescens Lam. Iris flowers are among the largest in the Mediterranean flora, and most species are nectarless (Sapir et al., 2002; Rudall et al., 2003). In southern France, the food-deceptive I. lutescens shows a heritable and striking flower colour polymorphism within populations, with both purple- and yellow-flowered individuals growing side by side without any spatial segregation (Fig. 1; Supplementary Data Fig. S1A, B). According to our population survey in consecutive years, the frequency of the yellow morph (FYM) is stable over the years (2009–2013).
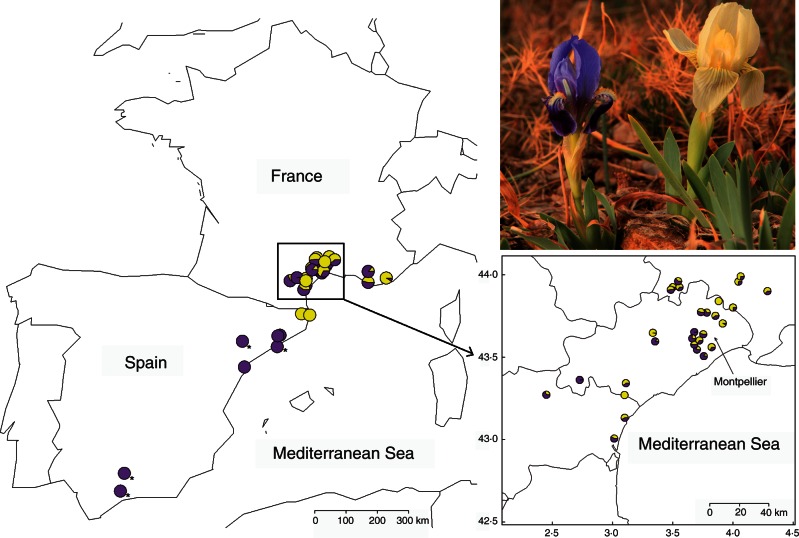
Fig. 1.
Map of the western Mediterranean basin showing locations of 41 sampled populations of Iris lutescens. Each pie chart indicates the proportion of yellow and purple individuals in each population in 2013. The top right inset shows purple- and yellow-flowered individuals of I. lutescens. Photographs was taken by Bruce Anderson near Montpellier, Languedoc-Roussillon region, France. The bottom right inset shows 29 populations around Montpellier in southern France. An asterisk indicates a population only involved in neutral genetic analyses (FST calculated), and no information for flower colour reflectance and flower size is available (no phenotypic distance calculated).
The purple–yellow flower colour difference in this species is due to anthocyanins, mainly delphanin, present in extremely higher concentrations in purple flowers than in yellow flowers, indicating a regulatory change in the flavonoid/anthocyanin biosynthetic pathway (Wang et al., 2013). However, the locus(i) responsible for this regulation change has not yet been identified. The two colour morphs of I. lutescens do not differ in phenology, floral scent, pollen/ovule production or any vegetative characteristics (Wang et al., 2013; Imbert et al., 2014a). So far, it has only been shown that purple flowers are larger than the yellow ones (Imbert et al., 2014b; also see the Results). According to our previous studies, the two colour morphs are pollinated by similar kinds of pollinators (an assortment of bees, including bumble-bees), and it does not seem that pollinators have an over-riding, innate preference for either morph (Imbert et al., 2014a). The fitness survey in natural populations of southern France shows that female reproductive success of both morphs is positively correlated with FYM, at both the population scale and the local scale, which is in contradiction to the NFDS hypothesis in food-deceptive species (Imbert et al., 2014b). Therefore, the intriguing question of how these two colour morphs are maintained in natural populations remains unanswered.
In this study, we compared phenotypic distribution and neutral genetic variation to assess the importance of neutral processes in explaining the spatial variation of flower colour in I. lutescens. First, we conducted an extensive population survey in France and Spain to document the spatial distribution of flower colour in I. lutescens. We measured floral morphological traits and quantified flower colour in the populations we visited to determine the phenotypic differentiation among populations. We thus specifically tested for spatial segregation of FYM. Next, we genotyped individuals sampled from natural populations at multiple microsatellite loci to examine how neutral genetic variation is structured within and among populations and colour morphs. We then compared the extent of population differentiation in flower colour (as well as flower size) with that of neutral loci.
MATERIALS AND METHODS
The study species
Iris lutescens Lam. (Iridaceae) is a perennial rhizomatous species, with a distribution range that extends from Spain through France to Italy. The species occurs in open and dry places in the Mediterranean region of these countries. It grows 10–30 cm tall, with erect, sword-shaped leaves and one showy flower at the apex of each shoot (Fig. 1). Each flower has three pendent, bearded sepals (also referred to as falls), alternating with three erect petals (standards). Because of the vegetative reproduction of the underground rhizome, a genet can produce several flowering stems (Supplementary Data Fig. S1B) that cannot be distinguished.
Flowers of I. lutescens are hermaphroditic but self-incompatible, and thus are totally pollinator dependent for sexual reproduction. Pollination is probably achieved by food deception since the species does not produce nectar, and pollinators have not been observed to forage for pollen. Flowering occurs in early spring (March to early May), and no co-flowering species is likely to represent a model in a putative mimicry system (Imbert et al., 2014a). It is most likely that the food deception is due to exploitation of insect perceptual biases, a model based on innate cognitive biases of insects in particular traits (Schaefer and Ruxton, 2009), and on exploitation of newly emerged naive insects. The main pollinators of I. lutescens are honey-bees (Apis mellifera), bumble-bees (Bombus spp.), and solitary bee species of several genera (Apidae; e.g. Anthophora, Eucera and Xylocopa). A florivorous beetle (Tropinota hirta, Cetoniidae) is also commonly observed on flowers. Observations in natural populations have failed to show any preference for a particular morph in either Apoids or beetles (Imbert et al., 2014a). Leaf herbivory is infrequent (Imbert et al., 2014a). Neither fruit predation nor pre-dispersal seed predation has been documented so far in natural populations. Note that apart from the yellow and purple morphs, some intermediate or extreme phenotypes (e.g. white or blue) can also be found in natural populations, but they are very rare (see Imbert et al., 2014a).
Population survey and sampling
A total of 41 populations were surveyed (Fig. 1; Supplementary Data Table S1). In May 2012, we visited two populations in southern Spain, designated ‘Cabra’ and ‘Antequera’. From March to May of 2013, we conducted a census of 39 populations in Spain and France. For each population, we recorded its latitude, longitude and altitude, surveyed population size and flower colour proportions, and sampled individuals for floral morphological measurements, flower colour phenotyping and genotyping.
Flower colour proportions were quantified with non-linear transects, and were always done by the same experimenter (E.I.) to reduce sampling errors. Counting points were located at least 4 m apart along the transect, and at each point the number of yellow and purple flowers was counted within a 2 m radius. The sampling effort, i.e. the number of points per population, depended on both population density and size. This survey was used to estimate population size (total number of flowering stems without information about the number of genets), and the frequency of the yellow morph (FYM) and the purple morph (1 – FYM) (see also Imbert et al., 2014b).
In each population, we haphazardly chose 30 fully blooming flowers for floral morphological measurements. Effort was made to distribute the sampling throughout the populations, and no neighbouring plants (minimum distance between 1 and 5 m, depending on the size of the population) were chosen in order to reduce the probability of sampling the same genet. In some small populations, or populations at the very early/late flowering period when visited, fewer than 30 individuals were sampled; for four monomorphic purple populations in Spain, only leaves were collected for genotyping (see Fig. 1; Supplementary Data Table S1). In polymorphic populations, we sampled an equal number of individuals for each colour morph, while for some populations with extremely low or high FYM (e.g. Liausson and Croix), we sampled the rare phenotype as much as we could (Supplementary Data Table S1). For each individual, four morphological traits were measured (Supplementary Data Fig. S1C): (1) flower height – from the ground to the top of the flower; (2) flower length – distance from the bottom of the fall to the top of the erect petal; (3) sepal width – at the widest part of the sepal; and (4) petal width – at the widest part of the petal. Measurements were always done by the same experimenter (H.W.) to reduce sampling errors. Subsequently, one petal and one leaf were collected for quantifying the flower colour and genotyping, respectively. The fresh petals and leaves were immediately desiccated in silica gel and stored in the dark at room temperature.
Measurements and analyses of flower colour as perceived by pollinators
The reflectance spectra of the dry petals were measured using a JAZ-PX spectrophotometer with a pulsed-xenon light source (Ocean Optics, Inc., Dunedin, FL, USA). Each spectrum was recorded between 300 and 700 nm, covering the spectral sensitivity range for most insects (Briscoe and Chittka, 2001). The spectrometric data were then analysed with AVICOL v.6 (Gomez, 2006). As we were interested in insect pollinator responses to colour, we characterized petal colour using measurements based on the properties of insect vision. Most insects have receptors with peak sensitivities near 350 nm (UV receptor), 440 nm (blue receptor) and 530 nm (green receptor) (Briscoe and Chittka, 2001). Both purple and yellow flowers of I. lutescens have very low reflection in the UV wavelengths (Fig. 2), while the reflection differs more strongly in the blue and green. We used the shape model to extract the reflectance value at 440 and 530 nm, respectively, and calculated the ratio between them (R440/R530), which is referred to as the reflectance ratio (Frey, 2004; Campbell et al., 2012). This reflectance ratio captures much of the variation between flowers in the shape of the reflectance curve, and thus we used it as an index of flower colour.
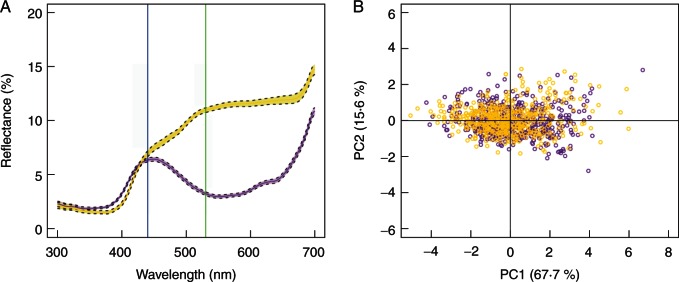
Fig. 2.
Phenotypic variation in flower colour and flower size. (A) Mean reflectance (full line) and standard error of the mean (dotted lines) for typical spectra curves of purple and yellow petals. The blue line and green line represent the positions of 440 nm and 530 nm, respectively. (B) Flower size differentiation of all purple and yellow individuals measured. Points represent principal component scores on the first two principal component axes.
Analyses of floral morphological traits: flower size and flower colour
First, the variation of colour among populations was examined by performing generalized linear mixed models (GLMMs) on the four morphological variables, with colour as a fixed effect and population as a random factor. Because the four floral morphological traits are correlated with each other, principal component analysis (PCA) was performed on these traits to extract the multivariate index of overall flower size. The four variables were scaled prior to analyses. Linear models were then performed on the first two principle components (PC1 and PC2). The same linear model was also used to analyse the reflectance ratio of flower colour spectra.
DNA extraction and genotyping
In total, 1120 individuals from 41 populations were used to evaluate population genetic structure. Genomic DNA was extracted from leaf tissue using the cetyltrimethyl ammonium bromide (CTAB) method (Doyle and Doyle, 1990). Each individual was genotyped at eight microsatellite loci specifically designed for this study (Table 1).
Table 1.
Microsatellite loci information, including information for PCR amplification
| Locus | Primer sequence (5'–3') | Repeat | Size range | Multiplex | Dye | [μM] | Genbank accession | N | A | Na | He | Ho | FIS |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Icpm01 | F: AAAACGGAGTCGTCAAATGG | (CT)13 | 206–258 | IcMix01 | 6-FAM | 0·1 | KR259393 | 842 | 25 | 7·7 (2·4) | 0·81 (0·07) | 0·30 (0·16) | 0·61 |
| R: GGTAGAAGATTCTGGATGAACAAA | |||||||||||||
| pmIc01 | F: CCGTTGCTCGAACCTACAAT | (CA)24 | 118–184 | IcMix01 | 6-FAM | 0·2 | KR259394 | 981 | 31 | 13·2 (3·9) | 0·87 (0·09) | 0·81 (0·14) | 0·06 |
| R: AGCATAAGATTAATCACGACAACA | |||||||||||||
| pmIc17 | F: TTGAAGGGTTGACATGAAACA | (CA)13 | 88–138 | IcMix01 | VIC | 0·2 | KR259365 | 910 | 27 | 6·9 (3·1) | 0·67 (0·15) | 0·35 (0·21) | 0·54 |
| R: CCCTTCAAGACAAGGTTTCC | |||||||||||||
| pmIc24 | F: TTCAGAGATGACGAAGAACCAA | (GAA)11 | 278–314 | IcMix01 | PET | 0·4 | KR259396 | 693 | 12 | 5·4 (1·7) | 0·72 (0·13) | 0·41 (0·22) | 0·38 |
| R: AAGTGAAGGACGTCTGTCCG | |||||||||||||
| pmIc34 | F: TCTAGAGTCTTGACATGTGGAAAAG | (GA)11 | 111–161 | IcMix01 | PET | 0·1 | KR259397 | 988 | 31 | 9·9 (3·4) | 0·78 (0·09) | 0·76 (0·13) | 0·02 |
| R: GCTGGTTTTGGTCTACCCCT | |||||||||||||
| pmIc05 | F: CATGGGTATGGTGGCCTAGA | (GT)16 | 86–154 | IcMix02 | 6-FAM | 0·2 | KR259398 | 1017 | 30 | 11·9 (3·4) | 0·86 (0·07) | 0·81 (0·11) | 0·06 |
| R: TCCTTTACGTCACACTTGCAT | |||||||||||||
| pmIc33 | F: GCTCACCCAAATACAAAGGG | (AC)12 | 99–135 | IcMix02 | PET | 0·2 | KR259399 | 998 | 20 | 7·6 (1·9) | 0·78 (0·09) | 0·73 (0·14) | 0·07 |
| R: CGGAAAATGAAGGTGACAAG | |||||||||||||
| pmIc41 | F: TCGCATCTCAAGATTTTCTCTT | (TC)11 | 71–125 | IcMix02 | VIC | 0·2 | KR259400 | 911 | 42 | 10·6 (3·4) | 0·83 (0·10) | 0·76 (0·16) | 0·07 |
| R: CAGGCTCACCAACCAGTTTC |
Size range in bp. Annealing temperature is 60 °C for all primers.
N, total number of individuals with amplification; A, total number of alleles; Na, He, Ho, FIS: the number of alleles, the expected heterozygosity, the observed heterozygosity and the fixation index at each locus averaged across all 41 populations, respectively.
For Na, He and Ho, values are mean and standard deviation (s.d.).
The library of microsatellite markers was produced at Genoscreen (Lille, France) by coupling multiplex microsatellite enrichment and next-generation sequencing on 454 GS-FLX Titanium platforms (Malausa et al., 2011). The QDD pipeline (Meglécz et al., 2010) was used to select and analyse 504 sequences and design primers. Finally eight primer pairs (Table 1) produced amplicons of the expected size and showed polymorphism, and therefore were retained for genotyping. PCR amplifications were performed separately for each locus and then multiplexed for two sub-sets of loci (IcMix01, IcMix02). In each multiplex, the primers were directly labelled using different fluorescent dyes (Applied Biosystems, Table 1). Microsatellites were amplified using the Qiagen multiplex PCR kit (Qiagen) in a 10 μL reaction volume containing 0·1–0·4 μm of each primer (Table 1) and 1 μL of genomic DNA. PCRs were conducted using Mastercycler pro (Eppendorf) under the following conditions: an initial denaturation step at 95 °C for 15 min; 35 cycles consisting of 30 s at 95 °C, 90 s at 60 °C and 60 s at 72 °C; finally, a supplementary extension step of 30 min at 60 °C. A 3 μL aliquot of the diluted PCR products (1/100) was pooled in 15 μL of HI-DI™ formamide with 0·125 μL of GeneScan-500 LIZ size standard (all Applied Biosystems). Products were analysed in an ABI PRISM 3130XL DNA Analyzer (Applied Biosystems) at the LabEx CeMEB sequencing platform (Montpellier, France). Fragment analyses and scoring were carried out using GeneMapper version 5.0 software (Applied Biosystems).
Analyses of microsatellite polymorphism and genetic diversity
We first tested the genotypic linkage disequilibrium between each pair of loci within each population, using the G-test available in GENEPOP 4.2 (Rousset, 2008). Multiple tests were then corrected using Benjamini–Hochberg correction (Benjamini and Hochberg, 1995). We also used exact tests implemented by GENEPOP software to test for departure from Hardy–Weinberg equilibrium (Rousset and Raymond, 1995). For all the tests performed in GENEPOP, we used the default settings. Several multilocus genetic diversity parameters for each population, including mean number of alleles (Na), observed heterozygosity (Ho), unbiased expected heterozygosity (He) and FIS, were computed using GENETIX 4.05 (Belkhir et al., 1996–2004). From these basic analyses (summarized in Table 1), it appears that three loci (Icpm01, pmIc17 and pmIc24) are likely to have null alleles (reduced number of successful amplifications, low number of alleles per population, high values of FIS; Table 1). Therefore, all the following analyses have been performed with and without these three loci. While the results remained unchanged qualitatively, here we only presented the results of the statistical analyses performed with five loci.
Genetic differentiation and isolation by distance
Global and pairwise FST estimates were calculated following Weir and Cockerham (1984) in GENEPOP, and tests of the genotypic differentiation among populations were performed using the exact G test provided by GENEPOP (Raymond and Rousset, 1995). We then tested for IBD using a Mantel test with 9999 permutations. The two-dimensional pairwise geographic distance matrix was created from the GPS co-ordinates using the ‘spDists’ function available in the ‘sp’ package in R (Bivand et al., 2013). Distances were log-transformed for all statistical analyses following Raymond and Rousset (1995). The pairwise FST/(1 – FST) genetic distance matrix was created by GENEPOP.
In order to test whether flower colour is a barrier to gene flow, genetic differentiation between the two colour groups was further investigated by comparing within- and between-group IBD patterns (Rousset, 1999). We chose populations with both purple and yellow individuals growing completely in sympatry (model assumption in Rousset, 1999), i.e. the 30 polymorphic populations, and artificially separated the sampled purple and yellow individuals within each polymorphic population into two sub-populations. Pairwise FST estimates among sub-populations were calculated, and three IBD patterns were compared: among purple sub-populations (within purple group), among yellow sub-populations (within yellow group) and between purple and yellow sub-populations (between groups). If flower colour is not a barrier to gene flow, then IBD should be observed between groups, and the differentiation between groups should be intermediate between the genetic differentiations within each group (Rousset, 1999); if colour is a barrier to gene flow, then the differentiation between groups should be independent of the distance, and the differentiation between groups should exceed within-group differentiation (e.g. Martel et al., 2003).
Comparison between phenotypic differentiation and genetic differentiation
From the basic expectation that neutral processes contribute to spatial differentiation in flower colour, we predict a significant non-linear relationship between FYM and genetic diversity, the well-mixed populations (0·25 ≤ FYM ≤ 0·75) being more diverse than the yellow/purple dominant populations (0 ≤ FYM < 0·25 or 0·75 < FYM ≤ 1). In other words, populations with a low value for FYM or a high value for FYM should present lower values for diversity parameters, such as mean number of alleles (Na) or unbiased expected heterozygosity (He), therefore an inverted U-shaped relationship should be observed between the FYM and multilocus genetic diversity estimates. Furthermore, we also expected that genetic diversity increases with population size, as usually observed because of the coalescence within populations. We thus fitted a polynomial model, with population size, FYM and FYM2 as explanatory variables, and the multilocus genetic diversity parameters as response variables. The significant contribution of each explanatory variable was tested using the stepwise multiple regression procedure.
The QST/FST approach is a long used method to compare phenotypic differentiation and neutral genetic differentiation (Whitlock, 2008). Because I. lutescens is a perennial species with a very low germination rate ( < 10 %) and a non-controlled flowering stage, measuring the phenotypes in controlled conditions for individuals of known pedigrees and next estimating QST was not possible. Therefore, comparisons between phenotypic differentiation and genotypic differences were based upon Euclidean distances.
The measurement of flower colour distance between pairs of populations was calculated as the Euclidean distance of flower colour, i.e. the Euclidean distance between the weight-mean reflectance ratio [mean purple × (1 – FYM) + mean yellow × FYM] of pairwise populations. In addition to flower colour, we also used the PC1 as a quantitative measure of flower size. To avoid the confounding effect of flower colour, we removed the colour effect by computing (purple PC1 – mean purple PC1) and (yellow PC1 – mean yellow PC1). Using this flower size estimator, we calculated the pairwise phenotypic distance for flower size as well.
To determine whether the neutral processes contributed to the observed spatial distribution of flower colour and flower size, we tested whether phenotypic distance and neutral genetic distance (FST) increase in parallel with geographic distance. If the observed cline in trait values is solely attributable to IBD, phenotypic distance and FST for neutral markers should exhibit concordant patterns of increase with geographic distance (Storz, 2002; Streisfeld and Kohn, 2005). Following Guillot and Rousset (2013), we did not perform partial Mantel tests. Therefore, in addition to testing the correlation between FST and geographic distance, we performed simple Mantel tests to examine whether phenotypic distance is correlated with geographic distance. We also tested for a correlation between phenotypic distance and FST. The degree of correlation can inform us about the importance of genetic drift and selection in population differentiation for colour morph frequencies (Takahashi et al., 2014).
All of the statistical analyses were performed using R software version 3.1.0 (R Core Team, 2014). PCA was performed using the ‘FactoMineR’ library (Husson et al., 2014). Linear models were fitted using the ‘glmer’ function from the ‘lme4’ library (Bates et al., 2014). Mantel tests were performed with the ‘ecodist’ library (Goslee and Urban, 2007).
RESULTS
Spatial pattern of flower colour distribution
All nine populations found in Spain were monomorphic (Fig. 1), seven of which were purple and the two populations close to France were yellow. In contrast, among all the 32 populations visited in France, only two were yellow monomorphic, and the remaining 30 populations were polymorphic, with both purple and yellow individuals growing side by side within each population (Fig. 1; Supplementary Data Fig. S1A, B). The FYM in the polymorphic populations ranged from 0·02 to 0·98 (Supplementary Data Table S1), and the mean value was 0·49.
Among all of the 41 populations surveyed, a significant positive relationship was found between the difference in FYM (Euclidean distance) and the geographical distance (Mantel test based on Spearman’s rank correlation, r = 0·28, P = 0·0001). Significant correlations were also observed between the FYM and latitude (Spearman’s rank test, r = 0·47, P = 0·002) and longitude (r = 0·49, P = 0·001), but this pattern is largely due to the clear separation of purple and yellow populations in Spain (Fig. 1). When analysing populations in France alone, none of the correlations was significant (P > 0·10).
Phenotypic variation of flower colour and flower size
Purple- and yellow-flowered individuals strongly differed in their reflectance ratio (Table 2; for typical reflectance curves of the two colour morphs see Fig. 2A), and variation among populations accounted for only 9·5 % of the total variance, confirming that flower colour is mainly genetically controlled. Regarding the PCA for floral morphological traits (Table 2; Fig. 2B), PC1 and PC2 represented 67·7 and 15·6 % of the total variance, respectively. Co-ordinates on PC1 were highly and positively correlated with all four measured morphological traits (Pearson’s correlation test, r = 0·71 – 0·87, P < 0·0001), and among-population variation accounted for a non-negligible part of the total variance (44·6 %), indicating an important contribution of environmental factors to phenotypic values of flower size. Co-ordinates on PC2 were highly and positively correlated with flower height (r = 0·70, P < 0·0001), but negatively correlated with sepal width and petal width (r = –0·29 and –0·30, respectively, P < 0·0001). The variance among populations accounted for 29·4 % of the total variance. The colour effect was significant for both of the two axes (Table 2), and purple flowers were larger overall. As PC1 explained 67·7 % of the total variance in floral morphology, and factor loadings for all the four traits measured were uniformly high and positive, it is interpretable as an overall flower size vector.
Table 2.
Summary statistics for floral traits in yellow and purple individuals sampled from the 37 natural populations
| Purple morph, mean (s.d., range) | Yellow morph, mean (s.d., range) | P-value | Population (%) | |
|---|---|---|---|---|
| (A) Flower colour | ||||
| Reflectance ratio | 2·06 (0·45, 0·58–3·45) | 0·64 (0·11, 0·30–0·96) | <0·0001 | 9·5 |
| (B) Flower size | ||||
| Flower height (cm) | 14·06 (4·16, 6·0–35·5) | 14·23 (4·54, 6·5–33·0) | 0·07 | 50·6 |
| Flower length (mm) | 61·33 (10·49, 29·89–1 07·12) | 58·79 (10·37, 5·89–101·80) | <0·001 | 32·5 |
| Sepal width (mm) | 23·90 (3·70, 13·37–35·29) | 22·88 (3·73, 10·59–37·41) | <0·0001 | 27·6 |
| Petal width (mm) | 26·55 (3·91, 15·10–41·19) | 26·38 (3·63, 16·23–37·41) | <0·05 | 35·2 |
| PC1 (67·7 %) | 0·14 (1·61, –4·23–6·68) | –0·14 (1·67, –5·03–5·96) | <0·01 | 44·6 |
| PC2 (15·6 %) | –0·07 (0·80, –2·79–2·78) | 0·07 (0·78, –1·82–2·84) | <0·0001 | 29·4 |
P-values indicate the significance of the differences between colour (fixed effect) in GLMMs. Percentage variance explained by the factor ‘population’ is given.
The ‘Reflectance ratio’ and ‘PC1’ were used to calculate phenotypic distances for flower colour and flower size, respectively.
Microsatellite polymorphism and genetic diversity
The number of successfully scored individuals ranged from 693 for pmIc24 to 1017 for pmIc05 (Table 1). In total we obtained 561 individuals with complete scoring (i.e. amplification for all of the eight loci). The total number of alleles ranged from 12 (pmIc24) to 42 (pmIc41), and the average number of alleles (Na) per population varied from 5·4 (pmIc24) to 13·2 (pmIc01) (Table 1). Thirteen genotypic disequilibria out of 410 (3·2 % were significant at the 5 % level after Benjamini–Hochberg correction, mainly occurring in small and monomorphic populations (CapDeCreus, Amposta, Lauret and Lleida). We therefore treated all loci as independent.
The unbiased expected heterozygosity (He) per population was generally high (Table 1). Three loci, Icpm01, pmIc17 and pmIc24, showed significant heterozygote deficiency in more than two-thirds of the populations (P < 0·05), while the other five loci were in Hardy–Weinberg equilibrium in most populations. Overall, 40 out of 41 populations (excluding population Llers) showed significant heterozygote deficiency (Supplementary Data Table S2). After removing those three loci, only 17 populations showed departure from Hardy–Weinberg equilibrium. For the following analyses, we used data excluding the three loci with suspected null alleles (Icpm01, pmIc17 and pmIc24).
Genetic differentiation and isolation by distance
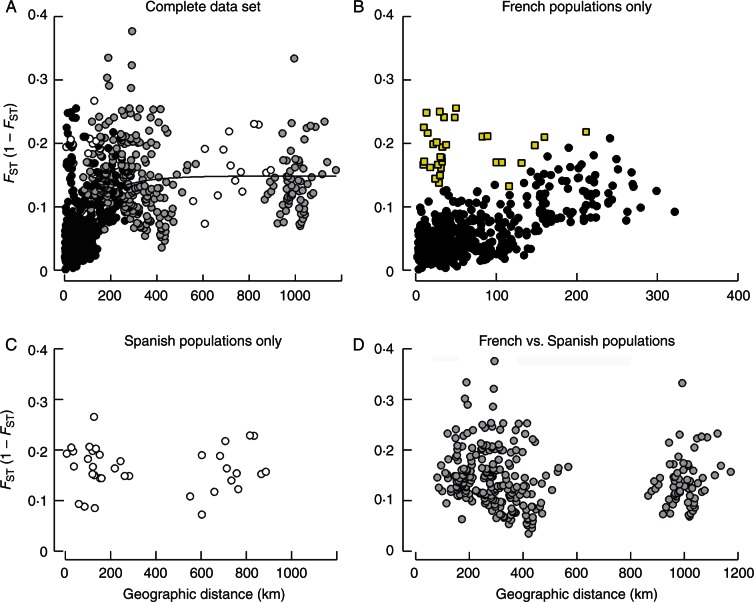
Microsatellite FST between populations averaged 0·08 (significantly different from 0, P < 0·0001). Pairwise FST (41 populations, 820 pairs) ranged from 0·002 to 0·27: 818 P-values were significantly different from 0 at the 5 % level (exact G test). There was no genotypic differentiation between two pairs of polymorphic populations in France: Blandas and Montdardier (FST = 0·002, P = 0·42, 4·4 km apart), and Vacquerolles and Saint-Paul (FST = 0·01, P = 0·06, 59·4 km apart); whereas the largest differentiation was between the purple monomorphic population Teix in Spain and the yellow monomorphic population Lauret in France (FST = 0·27, P < 0·0001, 293 km apart).
The Mantel test indicated a significant correlation between pairwise FST/(1 – FST) and geographic distance (r = 0·41, P = 0·004; Fig. 3A). However, the patterns were different in Spain and France. In France, where most populations are polymorphic, a significant IBD was observed (r = 0·51, P = 0·001; Fig. 3B), and the FST between populations averaged 0·06 (s.d. = 0·04). In particular, most of the largest FST values were associated with a single population: the yellow monomorphic population Lauret (FST = 0·16 ± 0·02 mean ± s.d.; Fig. 3B), indicating a genetic discontinuity between this population and the other French populations. However, this pattern was not observed for the second yellow monomorphic French population (Nissan, FST = 0·06 ± 0·03). In Spain, where only monomorphic populations were found, the neutral markers were relatively highly differentiated (FST = 0·14 ± 0·03), even among geographically close populations (Fig. 3C). Consistently, no IBD was detected at the between-population level (P = 0·41). Finally, the FST values between French and Spanish populations were also large (FST = 0·12 ± 0·04, Fig. 3D). Overall, this pattern created a saturated curve for the correlation between pairwise FST/(1 – FST) and geographic distance (Fig. 3A).
Fig. 3.
Scatterplots of pairwise FST/(1 – FST) against geographic distances (km). (A) Pairwise combinations of 41 populations sampled. (B) Pairwise combinations of 32 populations in France, yellow square symbols represent the pairwise combinations between Lauret, a yellow monomorphic population, and the other populations in France. (C) Pairwise combinations of nine populations in Spain and (D) pairwise combinations between populations in France and Spain. Statistical analyses have been performed with ln (geographical distance), but here we present non-transformed distances to illustrate the saturated shape.
Genetic differentiation between the two colour groups in polymorphic populations was further investigated by comparing within- and between-group IBD patterns (Fig. 4). The IBD pattern was significant within the purple group (slope = 0·017, 95 % confidence interval (CI) 0·009–0·025), within the yellow group (slope = 0·025, 95 % CI 0·014–0·041) and between colour groups (slope = 0·021, 95 % CI 0·012–0·032). The three slopes were not significantly different from each other as their 95 % CIs overlapped. The differentiation between the two colour groups was intermediate between the genetic differentiations within each colour group (Fig. 4).
Fig. 4.
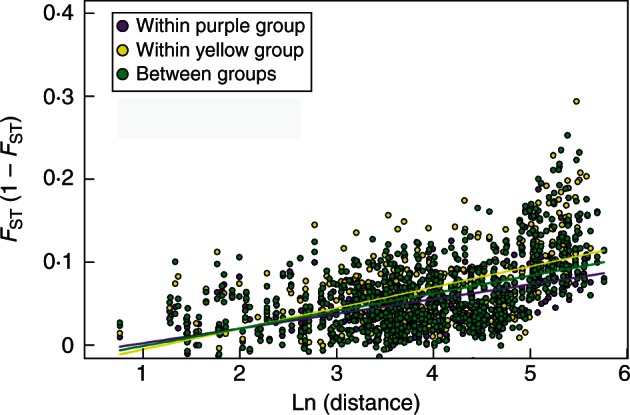
Three regressions of FST/(1 – FST) against ln (geographic distance) (km) for within and between colour in polymorphic populations. Purple, among purple sub-populations (within purple group); yellow, among yellow sub-populations (within yellow group); green, between yellow and purple sub-populations (between groups).
Comparison between phenotypic differentiation and genetic differentiation
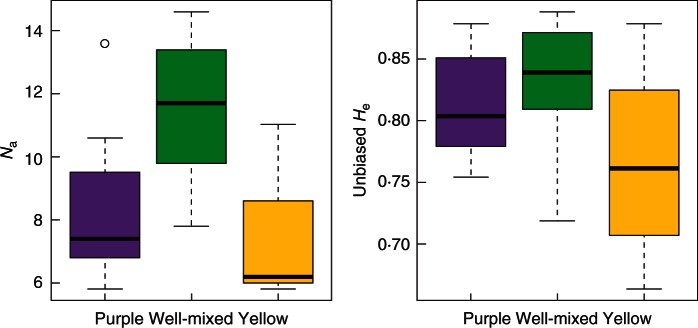
We observed an inverted-U relationship between the mean number of alleles (Na) and FYM2, and also between unbiased expected heterozygosity (He) and FYM2 (P < 0·0001 for both parameters, Fig. 5). In other words, Na and unbiased He in well-mixed populations (0·25 ≤ FYM ≤ 0·75) were larger than those in the purple/yellow dominant populations (Fig. 5). Note that the same relationship was still upheld when analysing French populations alone. However, we did not observe this relationship for the observed heterozygosity (Ho) and FIS (P > 0·40). Population size did not contribute to explain the Na and unbiased He (P > 0·40), and there was no confounding effect between the FYM and population size (P > 0·49, Spearman coefficient of correlation).
Fig. 5.
Boxplot for the estimates of genetic diversity according to the colour type of populations. Na, mean number of alleles per population; unbiased He, unbiased expected heterozygosity per population. Purple dominant: 0 ≤ FYM < 0·25; well-mixed: 0·25 ≤ FYM ≤ 0·75; yellow dominant: 0·75 < FYM ≤ 1. For both parameters, the relationship with FYM2 is significantly different from 0 (P < 0·0001, see text for details).
The Mantel test indicated that pairwise distances for flower colour were significantly correlated with both geographic distances and neutral genetic distances (FST, Table 3). For populations in France, a significant correlation was observed between distances for flower colour and FST (r = 0·20, P = 0·03; Table 3), but not between colour distance and geographic distance. In contrast, for populations in Spain, where IBD was not detected, colour distance was significantly correlated with geographic distance (r = 0·64, P = 0·008; Table 3), but not correlated with FST (Table 3). For the tests concerning flower size, the phenotypic distance was correlated neither with geographic distance nor with FST (Table 3).
Table 3.
Results of Mantel tests (r and P-value) for pairwise populations with four distances calculated: neutral genetic distance (FST), geographic distance (ln-transformed), flower colour distance (Euclidean distance of the weight-mean reflectance ratio) and flower size distance (Euclidean distance of the PC1 co-ordinates, see text for details)
| Distance 1 | Distance 2 | All of the populations (37 populations) |
French populations only (32 populations) |
Spanish populations only (five populations) |
|||
|---|---|---|---|---|---|---|---|
| r | P-value | r | P-value | r | P-value | ||
| Genetic | Geographic | 0·51 | <0·0001 | 0·35 | 0·008 | –0·36 | 0·74 |
| Flower colour | Geographic | 0·29 | <0·001 | 0·13 | 0·07 | 0·64 | 0·008 |
| Flower colour | Genetic | 0·29 | 0·001 | 0·20 | 0·03 | 0·10 | 0·31 |
| Flower size | Geographic | 0·05 | 0·29 | 0·15 | 0·08 | –0·37 | 0·79 |
| Flower size | Genetic | 0·09 | 0·21 | 0·12 | 0·15 | 0·19 | 0·27 |
Significance is based on 9999 randomized permutations.
DISCUSSION
Geographic variation among populations in phenotypic traits provides key insights into the relative roles of neutral processes and natural selection to population differentiation (McKay and Latta, 2002; Takahashi et al., 2014). Therefore, a critical issue is to determine to what extent neutral processes interact with natural selection to explain among-population differentiation (Eroukhmanoff et al., 2009; Runemark et al., 2010). In the present study, we investigated the spatial variation for flower colour in the Mediterranean species Iris lutescens, quantified patterns of neutral genetic variation across its distribution range and compared the phenotypic differentiation with neutral genetic differentiation. Our results provide empirical evidence that neutral processes contribute to the flower colour polymorphism in I. lutescens, and the overall spatial variation of flower colour in natural populations is possibly achieved through the interaction between neutral processes and certain selective forces.
The striking flower colour polymorphism of I. lutescens seems to be a peculiar, and thus intriguing case that provides insights into the relative roles of neutral processes and natural selection in population differentiation. In most of the published investigations of species with flower colour polymorphism, selection is found to play a predominant role in flower colour divergence, and most populations are monomorphic and have sharp geographical clines for flower colour transition (e.g. Streisfeld and Kohn, 2005; Hopkins et al., 2012; Arista et al., 2013), sometimes at a reduced spatial scale (Schemske and Bierzychudek, 2007). Among the 41 natural populations of I. lutescens surveyed, the spatial distribution of flower colour appeared as a combination of two opposing patterns: similarly to other studies cited above, all Spanish populations are monomorphic (most populations are purple while only two are yellow); but in France most populations are polymorphic, with a wide range of FYM, and no detectable spatial gradient. Maintenance of flower colour polymorphism within populations has been tentatively explained by negative frequency-dependsnt selection resulting from pollinator behaviour in one species of orchid (Gigord et al., 2001). However, in I. lutescens, as observed in other species, we did not detect such pollinator-mediated selection (Imbert et al., 2014a, b). Furthermore, such a process could hardly explain important spatial variation for morph proportions, as we observed in I. lutescens. From the present study, comparing the phenotypic differentiation with neutral genetic differentiation, we provide evidence that neutral processes might be partly responsible for the spatial variation in the proportions of flower colour morph.
In this study, we considered flower colour as a quantitative trait and used the reflectance ratio as an index of flower colour. Although both yellow and purple morphs display a fair amount of variation in colour intensity, the reflectance ratios differ strongly between them, while population origin makes a trivial contribution to the total variance. In addition, the measurements of reflectance spectra in field-collected plants presented in this study showed no significant difference compared with those of the common-garden plants (Wang et al., 2013). Both results indicate that flower colour difference in this species is mainly genetically controlled, consistent with investigations in various plant species showing that colour transition is due to modification in the regulatory network of the flavonoid/anthocyanin biosynthetic pathway (Rausher, 2008). Unfortunately, the locus(i) responsible for the genetic segregation of colour in this species is still unknown. Furthermore, consistent with our pollinator observations (Imbert et al., 2014a), we also observed that this flower colour differentiation does not act as a barrier to gene flow. Therefore, it makes sense to consider that the frequency of the yellow morph, and thus the frequency of the purple morph, can be influenced by gene flow among populations. We also have to interpret our data considering that different mutations in the biosynthetic pathway can lead to a similar change in colour phenotype (e.g. Quattrocchio et al., 1999).
As often observed in plant species, we also observed IBD in I. lutescens, since the Mantel test indicated a significant correlation between differentiation for neutral markers and geographic distances across the 41 sampled populations. However, the respective roles of gene flow and drift differ between the French polymorphic populations and the Spanish monomorphic populations. Indeed, in France, a significant pattern of IBD is detected at neutral loci, indicating that these populations have reached a regional equilibrium between gene flow and drift. It is worth noting that many pairwise combinations of populations (80 %; 397 out of 496 pairs) are geographically close (<200 km apart) with relatively low FST values (FST < 0·10), indicating that gene flow occurs regularly between adjacent populations. In contrast, we did not detect IBD for the Spanish populations, and these populations are highly differentiated, even though some of them are geographically very close (<30 km apart). This result suggests that gene flow is limited and these populations are somehow isolated from each other, giving a potential important role to genetic drift. We note that the Spanish populations included in this study are of reduced population size, compared with that of the French populations (see Supplementary Data Table S1). This probably results from habitat fragmentation, but it remains to be fully investigated. Restricted gene flow is also detected for the yellow monomorphic population Lauret, which exists as a genetic discontinuity in France. However, Spanish and French populations are not genetically separated, as the genetic differentiation between Spanish and French populations was of the same amplitude as that among Spanish populations only.
Compared with the polymorphic populations, the flower colour monomorphic populations showed increased pairwise FST estimates, and decreased genetic diversity (Na and He). This again supports the results from IBD analyses showing that the monomorphic populations are isolated from other populations and can drift independently, so flower colour polymorphism within populations seems unlikely to occur. Edh et al. (2007) has also reported a similar pattern in Brassica cretica: nearby monomorphic populations show high differentiation at neutral markers without any spatial segregation for flower colour. Our conclusion here is in good agreement with the expectation that random genetic drift within populations should lead to the fixation of one colour morph (Gray and Mckinnon, 2007). However, if genetic drift is the only mechanism explaining a population being monomorphic, a random process should lead to similar numbers of yellow and purple monomorphic populations. Nonetheless, we clearly observed a bias towards being purple monomorphic in Spain. In addition to the present study, 23 more populations were visited across Spain in 2014, and all of them are purple monomorphic (E. Imbert et al., unpubl. res). In contrast, the Mantel test indicated that the phenotypic differentiation for flower colour between Spanish populations is correlated with geographic distance, but not with differentiation for neutral markers. This pattern could be explained by repetitions of founder events, as observed during range expansion (Slatkin and Excoffier, 2012). To detect founder effects, we need to know the historical phylogeography of these populations, but the results presented in the current study do not provide sufficient information.
Despite the fact that our results suggest a regional equilibrium between gene flow and drift among the French populations, this does not fully explain why they remain highly polymorphic with a wide range of FYM. First, gene flow should lead to a convergence for the frequency of yellow morph at the regional scale, and at a long-term scale to the fixation of one colour morph (Gray and McKinnon, 2007). However, it is worth noting that vegetative reproduction via rhizomes could contribute to retarding the convergence in FYM between adjacent populations. Also, convergence in FYM is also dependent on the number of loci determining the colour phenotypes. Furthermore, our previous survey in the French populations revealed that female fitness is positively correlated with FYM at both the population scale and the local scale, although the female fitness of the yellow morph does not differ from that of the purple morph (Imbert et al., 2014b). In addition to gene flow, factors favouring purple morphs, which might balance yellow morph advantage as well, should be investigated. For example, so far flower size, apart from flower colour, is the only trait that we have found differentiated between morphs (Imbert et al., 2014b; this study). A recent study on another Mediterranean Iris species, Iris atropurpurea, documented an effect of flower size on pollinator attractiveness and thus on plant fitness (Lavi and Sapir, 2015). However, such an effect has not been observed in Iris haynei (Lavi and Sapir, 2015) and Iris tuberosa (Pellegrino, 2015). We also have to consider the benefits of high anthocyanin concentration of the purple morph in extreme environmental conditions, such as drought (e.g. Schemske and Bierzychudek, 2001, 2007; Arista et al., 2013).
Taken together, neutral processes contribute to the spatial variation for proportions of flower colour morph in I. lutescens, although we cannot exclude that natural selection is also operating. Interestingly, our results imply that the Spanish and French populations are experiencing two different mechanisms. In Spain, gene flow is limited and genetic drift seems influential. Therefore, the populations are somehow isolated from each other, and flower colour polymorphism within populations seems unlikely to occur. In France, where most populations are polymorphic with both purple and yellow phenotypes, a pattern of IBD has been detected, and gene flow between adjacent populations is an essential factor maintaining populations polymorphic. We propose two directions for future work: first, it is necessary to investigate the historic phylogeography of this species, which could help to illustrate the contribution of founder effects to the determination of flower colour in small monomorphic populations and flower colour variation in small polymorphic populations; secondly, it is necessary to investigate the environmental heterogeneity across the distribution range of this species. Local adaptation might be an explanation for the sharp flower colour cline in Spain (see Arista et al., 2013), and abiotic factors favouring the purple morph, in addition to gene flow, might form balancing selection to maintain flower colour polymorphism in France.
SUPPLEMENTARY DATA
Supplementary data are available online at www.aob.oxfordjournals.org and consist of the following. Table S1: characteristics of the 41 populations sampled. Table S2: estimates of multilocus genetic diversity for the 41 populations sampled. Figure S1: photographs showing polymorphic populations in southern France; and the four floral morphological traits (flower size) measured in this study.
ACKNOWLEDGEMENTS
We thank Nicolas Siro and Julliette Pouzadoux for their help with the DNA extraction and microsatellite genotyping. Genotyping data used in this work were partly produced through the technical facilities of the platform GenSeq (génotypage-séquençage) in LabEx CeMEB (Centre Méditerranéen de l’Environnement et de la Biodiversité), Montpellier, France. We also thank Guillaume Martin, François Rousset and Charles B. Fenster for insightful discussions and comments on early versions of this paper. This work was supported by a PhD grant of the China Scholarship Council to H.W., FEDER Funds of the Spanish Government (MINCYT CGL2009-08257 and MINECO CGL2012-33270) to M.T., an Erasmus Mundus scholarship to Y.M. and the French CNRS funds PICS-6196 to E.I.
Author contributions: H.W. designed the experiment, collected phenotypic data in natural populations, genotyped the individuals, analysed data and wrote the paper. This work was a part of her PhD thesis. M.T. participated in data collection, in particular for Spanish populations, and participated in writing the paper. Y.M. participated in data collection and writing the paper. E.F. designed the microsatellite primers and supervised all the molecular work. E.I. supervised the entire study and participated in experimental design, data collection, data analyses and redaction.
LITERATURE CITED
- Agati G, Azzarello E, Pollastri S, Tattini M. 2012. Flavonoids as antioxidants in plants: location and functional significance. Plant Science 196: 67–76. [DOI] [PubMed] [Google Scholar]
- Arista M, Talavera M, Berjano R, Ortiz PL. 2013. Abiotic factors may explain the geographical distribution of flower colour morphs and the maintenance of colour polymorphism in the scarlet pimpernel. Journal of Ecology 101: 1613–1622. [Google Scholar]
- Ashman TL, Majetic CJ. 2006. Genetic constraints on floral evolution: a review and evaluation of patterns. Heredity 96: 343–352. [DOI] [PubMed] [Google Scholar]
- Bates D, Maechler M, Bolker B, Walker S. 2014. lme4: linear mixed-effects models using Eigen and S4. R package version 1.1-6. http://CRAN.R-project.org/package=lme4. [Google Scholar]
- Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F. 1996–2004. GENETIX 4.05, logiciel sous Windows™ pour la génétique des populations. Montpellier, France: Laboratoire Génome, Populations, Interactions, CNRS UMR 5171, Université de Montpellier II. [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society, Series B 57: 289–300. [Google Scholar]
- Bivand RS, Pebesma E, Gómez-Rubio V. 2013. Applied spatial data analysis with R, 2nd edn. New York: Springer; http://www.asdar-book.org/. [Google Scholar]
- Briscoe AD, Chittka L. 2001. The evolution of color vision in insects. Annual Review of Entomology 46: 471–510. [DOI] [PubMed] [Google Scholar]
- Campbell DR, Waser NM, Melendez-Ackerman EJ. 1997. Analyzing pollinator-mediated selection in a plant hybrid zone: hummingbird visitation patterns on three spatial scales. American Naturalist 149: 295–315. [Google Scholar]
- Campbell DR, Bischoff M, Lord JM, Robertson AW. 2012. Where have all the blue flowers gone: pollinator responses and selection on flower colour in New Zealand Wahlenbergia albomarginata. Journal of Evolutionary Biology 25: 352–364. [DOI] [PubMed] [Google Scholar]
- Carlson JE, Holsinger KE. 2010. Natural selection on inflorescence color polymorphisms in wild Protea populations: the role of pollinators, seed predators, and intertrait correlations. American Journal of Botany 97: 934–944. [DOI] [PubMed] [Google Scholar]
- Chalker-Scott L. 1999. Environmental significance of anthocyanins in plant stress responses. Photochemistry and Photobiology 70: 1–9. [Google Scholar]
- Chittka L, Raine NE. 2006. Recognition of flowers by pollinators. Current Opinion in Plant Biology 9: 428–435. [DOI] [PubMed] [Google Scholar]
- Dormont L, Delle-Vedove R, Bessière JM, Hossaert-McKey M, Schatz B. 2010. Rare white-flowered morphs increase the reproductive success of common purple morphs in a food-deceptive orchid. New Phytologist 185: 300–310. [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. 1990. Isolation of plant DNA from fresh tissue. Focus 12: 13–15. [Google Scholar]
- Eckhart VM, Rushing NS, Hart GM, Hansen JD. 2006. Frequency-dependent pollinator foraging in polymorphic Clarkia xantiana ssp. xantiana populations: implications for flower colour evolution and pollinator interactions. Oikos 112: 412–421. [Google Scholar]
- Edh K, Widén B, Ceplitis ALF. 2007. Nuclear and chloroplast microsatellites reveal extreme population differentiation and limited gene flow in the Aegean endemic Brassica cretica (Brassicaceae). Molecular Ecology 16: 4972–4983. [DOI] [PubMed] [Google Scholar]
- Epling C, Lewis H, Ball FM. 1960. The breeding group and seed storage: a study in population dynamics. Evolution 14: 238–255. [Google Scholar]
- Eroukhmanoff F, Hargeby A, Arnberg NN, Hellgren O, Bensch S, Svensson EI. 2009. Parallelism and historical contingency during rapid ecotype divergence in an isopod. Journal of Evolutionary Biology 22: 1098–1110. [DOI] [PubMed] [Google Scholar]
- Frey FM. 2004. Opposing natural selection from herbivores and pathogens may maintain floral-colour variation in Claytonia virginica (Portulacaceae). Evolution 58: 2426–2437. [DOI] [PubMed] [Google Scholar]
- Gigord LD, Macnair MR, Smithson A. 2001. Negative frequency-dependent selection maintains a dramatic flower color polymorphism in the rewardless orchid Dactylorhiza sambucina (L.) Soò. Proceedings of the National Academy of Sciences, USA 98: 6253–6255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez D. 2006. AVICOL, a program to analyse spectrometric data. Last update October 2011. Free executable available at http://sites.google.com/site/avicolprogram/ or from the author at dodogomez@yahoo.fr. [Google Scholar]
- Goslee SC, Urban DL. 2007. The ecodist package for dissimilarity-based analysis of ecological data. Journal of Statistical Software 22: 1–19. [Google Scholar]
- Gray SM, McKinnon JS. 2007. Linking color polymorphism maintenance and speciation. Trends in Ecology and Evolution 22: 71–79. [DOI] [PubMed] [Google Scholar]
- Guillot G, Rousset F. 2013. Dismantling the Mantel tests. Methods in Ecology and Evolution 4: 336–344. [Google Scholar]
- Harborne JB, Williams CA. 2000. Advances in flavonoid research since 1992. Phytochemistry 55: 481–504. [DOI] [PubMed] [Google Scholar]
- Hopkins R, Levin DA, Rausher MD. 2012. Molecular signatures of selection on reproductive character displacement of flower color in Phlox drummondii. Evolution 66: 469–485. [DOI] [PubMed] [Google Scholar]
- Husson F, Josse J, Lê S, Mazet J. 2014. FactoMineR: multivariate exploratory data analysis and data mining with R. R package version 1.26. http://CRAN.R-project.org/package= FactoMineR. [Google Scholar]
- Imbert E, Wang H, Anderson B, Hervouet B, Talavera M, Schatz B. 2014a. Reproductive biology and colour polymorphism in the food-deceptive Iris lutescens (Iridaceae). Acta Botanica Gallica 161: 117–127. [Google Scholar]
- Imbert E, Wang H, Conchou L, Vincent H, Talavera M, Schatz B. 2014b. Positive effect of the yellow morph on female reproductive success in the flower colour polymorphic Iris lutescens (Iridaceae), a deceptive species. Journal of Evolutionary Biology 27: 1965–1974. [DOI] [PubMed] [Google Scholar]
- Irwin RE, Strauss SY, Storz S, Emerson A, Guibert G. 2003. The role of herbivores in the maintenance of a flower color polymorphism in wild radish. Ecology 84: 1733–1743. [Google Scholar]
- Jersáková J, Kindlmann P, Renner SS. 2006. Is the colour dimorphism in Dactylorhiza sambucina maintained by differential seed viability instead of frequency-dependent selection? Folia Geobotanica 41: 61–76. [Google Scholar]
- Jones KN, Reithel JS. 2001. Pollinator-mediated selection on a flower color polymorphism in experimental populations of Antirrhinum (Scrophulariaceae). American Journal of Botany 88: 447–454. [PubMed] [Google Scholar]
- Jorgensen TH, Richardson DS, Andersson S. 2006. Comparative analyses of population structure in two subspecies of Nigella degenii: evidence for diversifying selection on pollen-color dimorphisms. Evolution 60: 518–528. [PubMed] [Google Scholar]
- van Kleunen M, Meier A, Saxenhofer M, Fischer M. 2008. Support of the predictions of the pollinator-mediated stabilizing selection hypothesis. Journal of Plant Ecology 1: 173–178. [Google Scholar]
- Lavi R, Sapir Y. 2015. Are pollinator the agents of selection for the extreme large size and dark color in Oncocyclus irises? New Phytologist 205: 369–377. [DOI] [PubMed] [Google Scholar]
- Levin DA, Brack ET. 1995. Natural selection against white petals in Phlox. Evolution 49: 1017–1022. [DOI] [PubMed] [Google Scholar]
- Malausa T, Gilles A, Meglecz E, et al. 2011. High-throughput microsatellite isolation through 454 GS-FLX Titanium pyrosequencing of enriched DNA libraries. Molecular Ecology Resources 11: 638–644. [DOI] [PubMed] [Google Scholar]
- Martel C, Réjasse A, Rousset F, Bethenod MT, Bourguet D. 2003. Host-plant-associated genetic differentiation in Northern French populations of the European corn borer. Heredity 90: 141–149. [DOI] [PubMed] [Google Scholar]
- McKay JK, Latta RG. 2002. Adaptive population divergence: markers, QTL and traits. Trends in Ecology and Evolution 17: 285–291. [Google Scholar]
- Meglécz E, Costedoat C, Dubut V, et al. 2010. QDD: a user-friendly program to select microsatellite markers and design primers from large sequencing projects. Bioinformatics 26: 403–404. [DOI] [PubMed] [Google Scholar]
- Menzel R, Shmida A. 1993. The ecology of flower colours and the natural colour vision of insect pollinators: the Israeli flora as a study case. Biological Reviews 68: 81–120. [Google Scholar]
- Pellegrino G. 2015. Pollinator limitation on reproductive success in Iris tuberosa. AoB PLANTS 7: plu089; doi:10.1093/aobpla/plu089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrino G, Bellusci F, Musacchio A. 2005a. Evidence of post-pollination barriers among three colour morphs of the deceptive orchid Dactylorhiza sambucina (L.) Soó. Sexual Plant Reproduction 18: 179–185. [Google Scholar]
- Pellegrino G, Caimi D, Noce ME, Musacchio A. 2005b. Effects of local density and flower colour polymorphism on pollination and reproduction in the rewardless orchid Dactylorhiza sambucina (L.) Soó. Plant Systematics and Evolution 251: 119–129. [Google Scholar]
- Podolsky RH, Holtsford TP. 1995. Population structure of morphological traits in Clarkia dudleyana. I. Comparison of FST between allozymes and morphological traits. Genetics 140: 733–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts SG, Biesmeijer JC, Kremen C, Neumann P, Schweiger O, Kunin WE. 2010. Global pollinator declines: trends, impacts and drivers. Trends in Ecology and Evolution 25: 345–353. [DOI] [PubMed] [Google Scholar]
- Quattrocchio F, Wing J, van der Woude K, et al. 1999. Molecular analysis of the anthocyanin2 gene of petunia and its role in the evolution of flower color. The Plant Cell 11: 1433–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausher MD. 2008. Evolutionary transitions in floral color. International Journal of Plant Sciences 169: 7–21. [Google Scholar]
- Raymond M, Rousset F. 1995. An exact test for population differentiation. Evolution 49: 1280–1283. [DOI] [PubMed] [Google Scholar]
- R Core Team. 2014. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: URL http://www.R-project.org/. [Google Scholar]
- Rousset F. 1999. Genetic differentiation within and between two habitats. Genetics 151: 397–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousset F. 2008. Genepop’007: a complete re-implementation of the genepop software for Windows and Linux. Molecular Ecology Resources 8: 103–106. [DOI] [PubMed] [Google Scholar]
- Rousset F, Raymond M. 1995. Testing heterozygote excess and deficiency. Genetics 140: 1413–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudall PJ, Manning JC, Goldblatt P. 2003. Evolution of floral nectaries in Iridaceae. Annals of the Missouri Botanical Garden 90: 613–631. [Google Scholar]
- Runemark A, Hansson B, Pafilis P, Valakos ED, Svensson EI. 2010. Island biology and morphological divergence of the Skyros wall lizard Podarcis gaigeae: a combined role for local selection and genetic drift on color morph frequency? BMC Evolutionary Biology 10: 269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzmann CC, Schiestl FP. 2007. Odour and colour polymorphism in the food-deceptive orchid Dactylorhiza romana. Plant Systematics and Evolution 267: 37–45. [Google Scholar]
- Sapir Y, Shmida AVI, Fragman ORI, Comes HP. 2002. Morphological variation of the Oncocyclus irises (Iris: Iridaceae) in the southern Levant. Botanical Journal of the Linnean Society 139: 369–382. [Google Scholar]
- Schaefer HM, Ruxton GD. 2009. Deception in plants: mimicry or perceptual exploitation? Trends in Ecology and Evolution 24: 676–685. [DOI] [PubMed] [Google Scholar]
- Schaefer HM, Schaefer V, Levey DJ. 2004. How plant–animal interactions signal new insights in communication. Trends in Ecology and Evolution 19: 577–584. [Google Scholar]
- Schemske DW, Bierzychudek P. 2001. Perspective: evolution of flower color in the desert annual Linanthus parryae: Wright revisited. Evolution 55: 1269–1282. [DOI] [PubMed] [Google Scholar]
- Schemske DW, Bierzychudek P. 2007. Spatial differentiation for flower color in the desert annual Linanthus parryae: was Wright right? Evolution 61: 2528–2543. [DOI] [PubMed] [Google Scholar]
- Schoonhoven LM, van Loon JJA, Dicke M. 2007. Insect–plant biology, 2nd edn. Oxford: Oxford University Press. [Google Scholar]
- Slatkin M, Excoffier L. 2012. Serial founder effects during range expansion: a spatial analog of genetic drift. Genetics 191: 171–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobel JM, Streisfeld MA. 2015. Strong premating reproductive isolation drives incipient speciation in Mimulus aurantiacus. Evolution 69: 447–461. [DOI] [PubMed] [Google Scholar]
- Storz JF. 2002. Contrasting patterns of divergence in quantitative traits and neutral DNA markers: analysis of clinal variation. Molecular Ecology 11: 2537–2551. [DOI] [PubMed] [Google Scholar]
- Strauss SY, Irwin RE, Lambrix VM. 2004. Optimal defence theory and flower petal colour predict variation in the secondary chemistry of wild radish. Journal of Ecology 92: 132–141. [Google Scholar]
- Streisfeld MA, Kohn JR. 2005. Contrasting patterns of floral and molecular variation across a cline in Mimulus aurantiacus. Evolution 59: 2548–2559. [PubMed] [Google Scholar]
- Takahashi Y, Nagata N, Kawata M. 2014. Antagonistic selection factors induce a continuous population divergence in a polymorphism. Heredity 112: 391–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor LP, Grotewold E. 2005. Flavonoids as developmental regulators. Current Opinion in Plant Biology 8: 317–323. [DOI] [PubMed] [Google Scholar]
- Wang H, Conchou L, Bessière JM, Cazals G, Schatz B, Imbert E. 2013. Flower color polymorphism in Iris lutescens (Iridaceae): biochemical analyses in light of plant–insect interactions. Phytochemistry 94: 123–134. [DOI] [PubMed] [Google Scholar]
- Waser NM, Price MV. 1981. Pollinator choice and stabilizing selection for flower color in Delphinium nelsonii. Evolution 35: 376–390. [DOI] [PubMed] [Google Scholar]
- Weir BS, Cockerham CC. 1984. Estimating F-statistics for the analysis of population structure. Evolution 38: 1358–1370. [DOI] [PubMed] [Google Scholar]
- Weiss MR. 1995. Floral color change: a widespread functional convergence. American Journal of Botany 82: 167–185. [Google Scholar]
- Whitlock MC. 2008. Evolutionary inference from QST. Molecular Ecology 17: 1885–1896. [DOI] [PubMed] [Google Scholar]
- Winkel-Shirley B. 2002. Biosynthesis of flavonoids and effects of stress. Current Opinion in Plant Biology 5: 218–223. [DOI] [PubMed] [Google Scholar]
- Wright S. 1943. An analysis of local variability of flower color in Linanthus parryae. Genetics 28: 139–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. 1978. Variability within and among natural populations. Volume 4 in Evolution and the genetics of populations. Chicago: University of Chicago Press. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.