Abstract
Background & Aims
Ubiquitination is a reversible protein modification involved in the major cellular processes that define cell phenotype and behaviour. Ubiquitin modifications are removed by a large family of proteases named deubiquitinases. The role of deubiquitinases in hepatic stellate cell (HSC) activation and their contribution to fibrogenesis are poorly defined. We have identified that the deubiquitinase ubiquitin C-terminal hydrolase 1 (UCHL1) is highly induced following HSC activation, determined its function in activated HSC and its potential as a therapeutic target for fibrosis.
Methods
Deubiquitinase expression was determined in day 0 and day 10 HSC. Increased UCHL1 expression was confirmed in human HSC and in an alcoholic liver disease (ALD) patient liver. The importance of UCHL1 in hepatic fibrosis was investigated in CCl4 and bile duct ligation injured mice using a pharmacological inhibitor (LDN 57444). The effects of UCHL1 inhibition on HSC proliferation were confirmed by Western blot and 3H thymidine incorporation.
Results
Here we report that pharmacological inhibition of UCHL1 blocks progression of established fibrosis in CCl4 injured mice. UCHL1 siRNA knockdown, LDN 57444 treatment, or HSC isolated from UCHL1–/– mice show attenuated proliferation in response to the mitogen, platelet-derived growth factor. Additionally, we observed changes in the phosphorylation of the cell cycle regulator retinoblastoma protein (Rb) in the absence of UCHL1 highlighting a potential mechanism for the reduced proliferative response.
Conclusions
UCHL1 expression is highly upregulated upon HSC activation and is involved in the regulation of HSC proliferation. This study highlights therapeutic opportunities for pharmacological targeting of UCHL1 in chronic liver disease.
Keywords: Deubiquitinase, UCHL1, HSC, Liver fibrosis, LDN57444
Introduction
Activation or transdifferentiation of quiescent hepatic stellate cells (HSC) to a myofibroblast state is central to the fibrogenic process in acute hepatic wound-repair and chronic liver disease [1,2]. HSC transdifferentiation is an epigenetically regulated process that induces genome-wide changes in gene expression that enable the cell to adopt its profibrogenic functions including proliferation, migration and the expression and secretion of large quantities of extracellular matrix proteins. To acquire the capability for these functions the transdifferentiated HSC must also undergo fundamental alterations in protein turnover and in the post-translational regulatory mechanisms that control the localisation and function of proteins. The major post-translational processes that control cell phenotype and behaviour include the addition of small chemical groups (e.g. by phosphorylation, acetylation, methylation and hydroxylation), alterations in the chemical nature of amino acids (e.g. by citrullination), addition of functional molecules (e.g. lipids or sugars) or the covalent linkage of proteins to other protein or peptide modules (e.g. by ubiquitination, sumoylation, and neddylation) [3]. While there have been many studies describing the role of protein phosphorylation in HSC, principally concerning signal transduction events implicated in fibrogenesis, there has been relatively little attention to the contributions of other forms of protein modifications towards HSC activation and fibrosis.
Ubiquitin (Ub) is a small and highly conserved protein comprised of 76 amino acids that is expressed throughout the eukaryotic kingdom. The addition of Ub to proteins is controlled by the coordinated activity of three types of enzymes; Ub activating enzyme (E1), Ub conjugating enzyme (E2) and Ub ligase (E3). Ub is conjugated either as a monomer or end-to-end polymer chain, but more commonly as a branched polymer where seven different lysine (K) residues (K6, K11, K27, K29, K33, K48, and K63) within the Ub peptide can serve as points of Ub-Ub conjugation. The best-characterised poly-Ub chains are the K48-linked polymers, which target their associated proteins for processing or degradation by the 26S proteasome [4]. By contrast, K63-linked chains avoid the 26S proteasome and instead are more akin to phosphorylation in that they regulate a wide variety of signalling events such as activation of NF-κB [5]. Ubiquitination is a reversible modification, important for termination of signalling events, editing of Ub-protein modifications and recycling of Ub [6]. Deubiquitination is catalysed by a large family of over 80 different deubiquitinases (DUBs), which can be subdivided into five distinct super-families, the ubiquitin specific proteases (USPs), the ubiquitin C-terminal hydrolases (UCHs), the Machado-Josephin domain proteases (MJDs), the ovarian tumour proteases (OTU) and the JAB1/MPN/Mov34 (JAMM) domain metalloproteinases [7]. DUB specificity is also determined by cellular location, specific binding interactions and the type of ubiquitin chain linkage and removal of these ubiquitin adducts regulates either the proteins stability or its activity [8]. Noteworthy is that several DUBs have been identified with polymorphisms that are linked to a range of human diseases including cancers and neurological disorders [9–12].
Given the important regulatory role of DUBs in ubiquitination we hypothesised that HSC transdifferentiation will be associated with stable alterations in the expression of the DUBs and will be associated with specific functional requirements of the activated HSC. By testing this hypothesis we hoped to identify one or more DUBs that may be targeted to attenuate the fibrogenic activities of HSC. Here, we present data demonstrating that ubiquitin C-terminal hydrolase 1 (UCHL1 or PGP9.5) is highly induced with HSC activation and plays a role as a regulator of HSC proliferation. Moreover, we show that pharmacological inhibition of UCHL1 blocks HSC proliferation and when administered in vivo acts in a therapeutic way to block progression of established fibrosis despite continued liver injury.
Materials and methods
Human samples
Alcoholic liver disease (ALD) and human control liver, normal and diseased lung (idiopathic pulmonary fibrosis, IPF) and kidney tissue samples for histology were taken under full ethical approval with patient consent (REC references 10/H0906/41, 11/NE/0291, and 13/EM/0311).
Animals
A description of the UCHL1–/– mice including details of the knockout strategy appear in Coulombe et al., [13]. Authors hold appropriate licences for work relating to all experiments and animal procedures were approved by local ethical review committee and the UK Home Office.
Chronic carbon tetrachloride (CCl4) liver injury model
Wild-type (WT) male mice (25–30 g) were intraperitoneal injected twice weekly for 8 weeks with CCl4 at 2 μl/g body weight (CCl4:olive oil at 1:3[vol:vol]) or olive oil vehicle. From week 4 mice received an additional bi-weekly injection intraperitoneal of UCHL1 inhibitor LDN 57444 (0.4 mg/kg) or vehicle (DMSO). 24 h after the final CCl4 injection mice were humanely killed and liver and serum samples prepared.
Bile duct ligation
Bile duct ligation (BDL) was performed as previously described under buprenorphine pain relief [14]. Mice were allowed to develop cholestatic disease and fibrosis over a period of 14 days. Alzet osmotic minipumps were implanted subcutaneously at day 0 delivering (0.4 mg/kg/day) LDN 57444 or (DMSO 50%, PEG 50%) vehicle control. Animals were humanely killed and liver and serum samples prepared.
Isolation of primary HSC
Primary human HSCs were isolated from normal margins of surgically resected liver. Rat HSCs were isolated from normal livers of Sprague-Dawley rats. Mouse HSC were isolated from normal livers of UCHL1–/– or WT littermate controls. Liver tissue was digested with pronase and collagenase B (Roche) and the cell suspension was subsequently separated by an 11.5% Optiprep gradient (Sigma). HSCs were seeded onto plastic (Corning), cultured in Dulbecco’s modified Eagle’s medium (Life Technologies) supplemented with 16% fetal bovine serum, pyruvate, glutamine, penicillin, and streptomycin (Life Technologies) and maintained in an incubator at 37 °C with 5% CO2. Freshly isolated (day 0) cells were considered quiescent and (day 10) cultures regarded as activated unless specifically stated otherwise.
Histology
Liver tissue sections stained with Haematoxylin-eosin were evaluated by an experienced liver pathologist (DGT) for inflammatory cell infiltrate, severity of necroinflammation including extent and topography of parenchymal necrosis, hepatocyte ballooning and apoptosis, and type and extent of steatosis. A necroinflammatory score (range 0–6) was devised based on the semi-quantification of confluent necrosis by Ishak et al. [15]. Staging of fibrosis was assessed according to Ishak et al. [15] on sections stained with Sirius red as previously described [16].
Immunohistochemistry
Immunohistochemistry for alpha smooth muscle actin (αSMA) was performed on 4 μm thick formalin-fixed paraffin-embedded sections as previously described [14]. For UCHL1, slides were deparaffinised, rehydrated through graded alcohols, endogenous peroxidase was blocked in hydrogen peroxide/methanol and antigen retrieval was performed using citric saline for 20 min in the microwave, followed by incubation in trypsin for 10 min at 37 °C. Sections were blocked using the Vector avidin/biotin blocking kit and 20% swine serum, prior to overnight incubation at 4 °C with primary antibody UCHL1 (Serotec) at a 1:100 dilution. Next day slides were washed in PBS and incubated with biotinylated rabbit anti-mouse at 1:2000 (Vector labs) for 2 h. After PBS washing, slides were incubated with streptavidin biotin-peroxidase complex (Vector Labs) and incubated at room temperature for 45 min. UCHL1-positive cells were visualized by 3,3-diaminobenzidine tetrahydrochloride (DAB) and counterstained with haematoxylin.
Hydroxyproline assay
The collagen content of liver tissue was measured using the hydroxyproline method, as previously described [17]. Briefly, liver samples were hydrolysed for 18 h at 110 °C in 1 ml of 6 N HCl. Samples were then neutralised in 10 N NaOH before colourisation with Ehrlich’s reagent. A standard curve comprised of dilutions of 400 μM hydroxyproline was used for quantification.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting
Total protein was fractionated by 12% SDS-PAGE and transferred to nitrocellulose membrane. Blots were blocked with TBS/Tween 20 (0.1% T-TBS) containing 5% milk or 5% BSA protein before overnight incubation with primary antibodies. Primary antibodies raised against UCHL1 (#3524 Cell signalling), USP44 (Santa Cruz 79330), αSMA (A5228 Sigma), p27 (Santa cruz 528), pRb (Cell signalling #9301), total Rb (Santa cruz 74562), and GAPDH (ab22555 Abcam) were used at 1:1000 dilution. Membranes were washed in T-TBS and incubated with anti-rabbit (#7074S, Cell Signalling), horseradish peroxidise (HRP)-conjugate antibodies at 1:2000 dilution for 1 h. Blots were washed and antigen detected by ECL (Amersham Biosciences).
RNA isolation and quantitative reverse transcriptase-polymerase (qRT-PCR)
Total RNA was purified from isolated cells using RNeasy Mini Kit (Qiagen) following the manufacturer’s instructions. Primer sequences are listed in (Supplementary Table 1). qRT-PCR was performed using SYBR-Green jumpstart Taq Readymix (Sigma) according to manufacturer’s instructions. Relative transcriptional differences were calculated using the (1/2A)_100 calculation.
Tritiated thymidine incorporation assay
Control, LDN 57444 treated or siRNA transfected rat/human HSC or UCHL1–/– and WT mouse HSC were serum starved (0.5%) for 18 h prior to treatment with/without mouse or human platelet-derived growth factor BB (PDGFBB) (eBioscience) 20 ng/ml or 0.5% media control and pulsed with 18_5 kBq methyl-3[H]thymidine (74 GBq/mmol; PerkinElmer, Cambridge, UK) for 24 h as described previously [15]. Cells were trypsinised and radioactivity was quantified using a liquid scintillation counter (PerkinElmer) and results were presented as mean counts/min ± SEM.
Statistical analysis
P values were calculated using one way ANOVA for multiple comparisons followed by Tukey’s post-hoc tests or a two-tailed unpaired student’s t test where *p <0.05, **p <0.01 or ***p <0.001 was considered significant.
Results
DUB mRNA analysis reveals a significant increase in UCHL1 and a decrease in USP44 with HSC activation
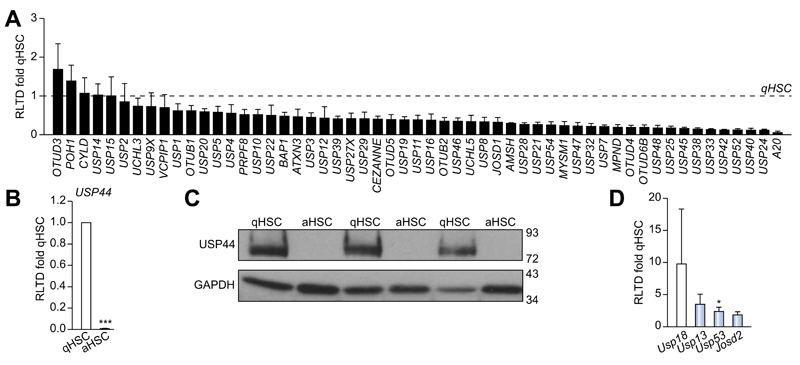
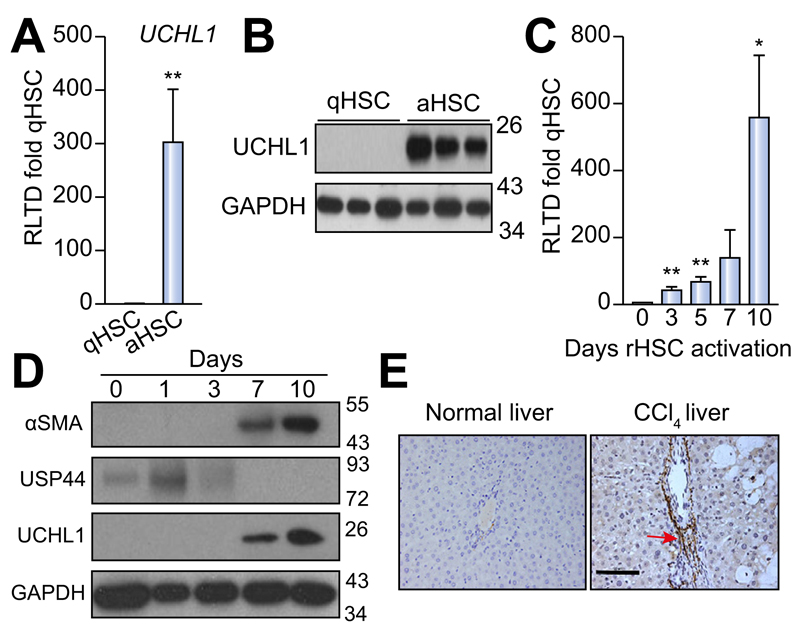
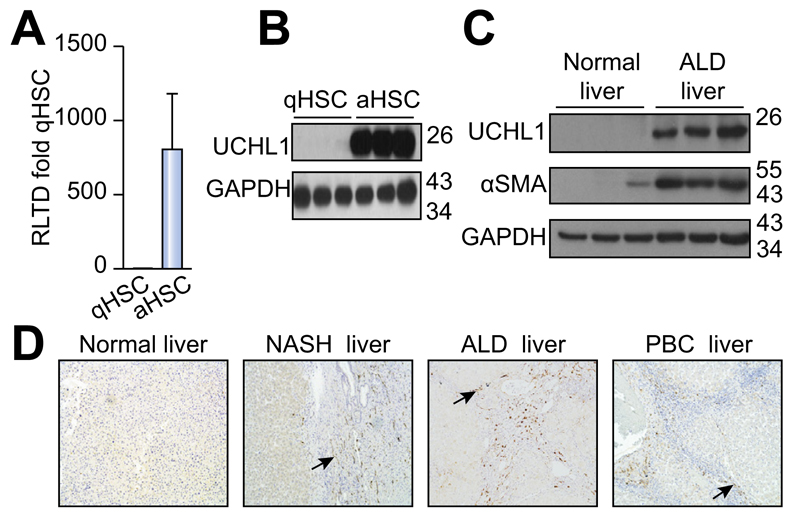
To determine DUB expression levels and evaluate changes upon HSC activation we designed a panel of rat DUB qRT-PCR primers (Supplementary Table 1) and analysed mRNA expression in primary quiescent (qHSC) and culture-activated (aHSC) rat HSC. We discovered either no significant differences or a reduction in mRNA expression upon HSC activation in the 55 DUBs examined (Fig. 1A). USP44 is a DUB involved in the negative regulation of histone 2B lys20 ubiquitinylation and embryonic stem cell differentiation [18]. USP44 was notable in that it was highly expressed in qHSC but was undetectable at mRNA and protein level in aHSC (Fig. 1B, C). By contrast four DUBs displayed a modest increase in aHSC including three ubiquitin specific proteases; USP18, USP13, USP53 and the josephin domain containing protein JOSD2 (Fig. 1D). However, UCHL1 was of particular interest as it was undetectable in qHSC and dramatically induced to very high levels in aHSC, with an estimated 300 fold induction (Fig. 2A). Western blot analysis confirmed a substantial increase in UCHL1 protein expression upon HSC activation (Fig. 2B). These observations encouraged us to further investigate the expression of UCHL1 in diseased liver and to determine if it makes an important functional contribution to the aHSC. A detailed time course for HSC activation revealed that UCHL1 is induced during the so-called transitionary-phase of HSC activation concomitant with induction of αSMA, and thereafter expression levels continued to rise with each subsequent day of culture as the HSC become fully differentiated to the myofibroblast phenotype (Fig. 2C, D). By contrast, the expression of USP44 was inversely correlated with HSC activation and completely absent at the protein level by day 7 (Fig. 2D). To investigate whether expression levels of UCHL1 increased following liver injury, Sprague-Dawley rats were subjected to repeated 4-week CCl4 injury to induce in vivo HSC activation and fibrosis. UCHL1 expression was undetectable by immunohistochemistry in sham olive oil treated rats (Fig. 2E left panel). However, UCHL1 expression was substantially upregulated in CCl4 injured rats and was selectively localised to activated myofibroblast-like cells present within the fibrotic tissue (Fig. 2E right panel). To determine whether UCHL1 expression is also increased during activation of human HSC we examined both mRNA and protein levels in human day 0 (quiescent) and day 10 (activated) HSC. Results confirm that UCHL1 expression is absent in human quiescent HSC and dramatically increased upon activation in concurrence with our findings with rat HSC (Fig. 3A, B). Activated myofibroblasts are found associated with fibrotic lesions in various liver diseases including ALD. By Western blot we discovered that UCHL1 protein expression is increased in ALD in correlation with elevated αSMA levels (Fig. 3C). In addition, we also examined the location of UCHL1 in human liver by immunohistochemistry. In the absence of obvious fibrosis we failed to detect UCHL1 in any cell type (Fig. 3D left panel), by contrast myofibroblast-like cells were stained positive for UCHL1 in fibrotic tissue in the diseased livers of NASH, ALD, and PBC patients (Fig. 3D).
Fig. 1. Comparison of deubiquitinase expression in quiescent (qHSC) vs. activated (aHSC) rat HSC.
Deubiquitinase mRNA expression levels were quantified by qRT-PCR and expressed as relative level of transcriptional difference (RLTD) of activated rat HSC (day 10) compared with quiescent (day 0) n = minimum of 3 animals per group (A, B, and D). (C) Western blots for USP44 expression in quiescent (n = 3) and day 10 activated rat HSC (n = 3). Statistical analysis between quiescent and activated HSC were compared using two-tailed unpaired students t test *p <0.05; ***p <0.001.
Fig. 2. UCHL1 expression is significantly elevated in activated (aHSC) vs. quiescent (qHSC) rat stellate cells.
(A) UCHL1 mRNA expression levels were quantified by qRT-PCR and expressed as relative level of transcriptional difference (RLTD) of activated rat HSC (day 10) compared with quiescent (day 0) n = minimum of 3 animals per group. (B) Western blots for UCHL1 expression in quiescent (n = 3) and day 10 activated rat HSC (n = 3). (C) UCHL1 mRNA expression in rat HSC (rHSC) harvested at days 0–10 of culture and results are expressed as RLTD (day 0 HSC). (D) Western blot for αSMA, USP44, UCHL1, and GAPDH expression in rat HSC harvested at days 0–10 of culture. Molecular weight markers shown on right panel. (E) Representative photomicrographs of UCHL1 stained liver sections from rats treated with olive oil vehicle (left panel) or CCl4 injured for 4 weeks (right panel), n = 3 independent experiments. Statistical analysis between quiescent and activated HSC were compared using two-tailed unpaired students t test *p <0.05; **p <0.01.
Fig. 3. UCHL1 expression is elevated in human HSC in vitro and in vivo in ALD livers.
(A) UCHL1 mRNA levels in human quiescent (day 0) and activated (day 10) HSC were quantified by qRT-PCR and results are expressed as RLTD (quiescent). (B) Western blot for UCHL1 and GAPDH expression in human HSC harvested at day 0 (quiescent) and day 10 (activated). (C) Western blot for UCHL1, αSMA, and GAPDH expression in control and alcoholic liver disease (ALD) liver samples. (D) Representative photomicrographs of UCHL1 stained human liver sections from normal human liver (left panel) and NASH, ALD and PBC patient liver tissue (right panels). Data is representative of n = 3 independent experiments.
Fibrosis is a pathological process that is associated with chronic disease in a number of different organs where it is postulated that myofibroblasts are generated by similar events to those described in the liver and stimulate fibrogenesis by the secretion and deposition of extracellular matrix proteins [19]. We were therefore interested to discover if UCHL1 expression is a common (core) feature of fibrosis-associated myofibroblasts. We failed to detect UCHL1 in normal healthy human lung. However, in lung sections from patients suffering IPF with evident fibrotic foci there were clear UCHL1 positive myofibroblasts, and additionally we also detected positively stained immune cells (Supplementary Fig. 1). In sections of kidney there was UCHL1 staining in podocytes and tubules, which was more evident in the nephrectomy samples with evidence of tubulointerstitial fibrosis and tubular atrophy. In addition, there was clear UCHL1 positive staining in the interstitial fibroblasts associated with fibrotic regions (Supplementary Fig. 1). We conclude that abundant UCHL1 expression is characteristic of fibrosis-associated human myofibroblasts and as such, is likely to contribute to the fibrogenic process.
UCHL1 inhibition reduces liver fibrogenesis
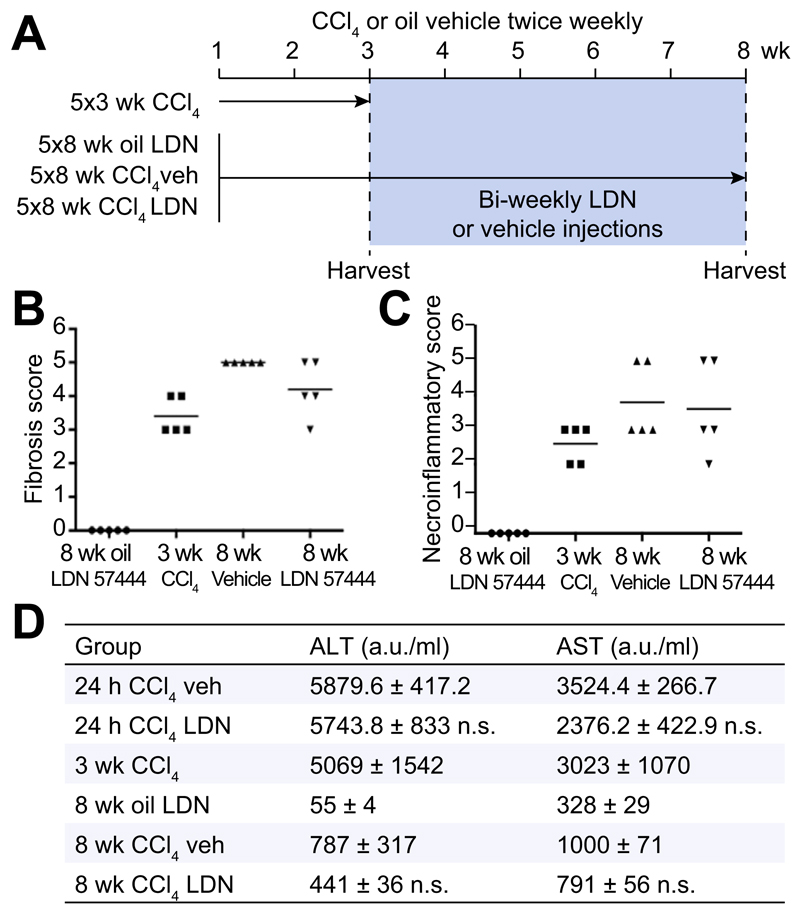
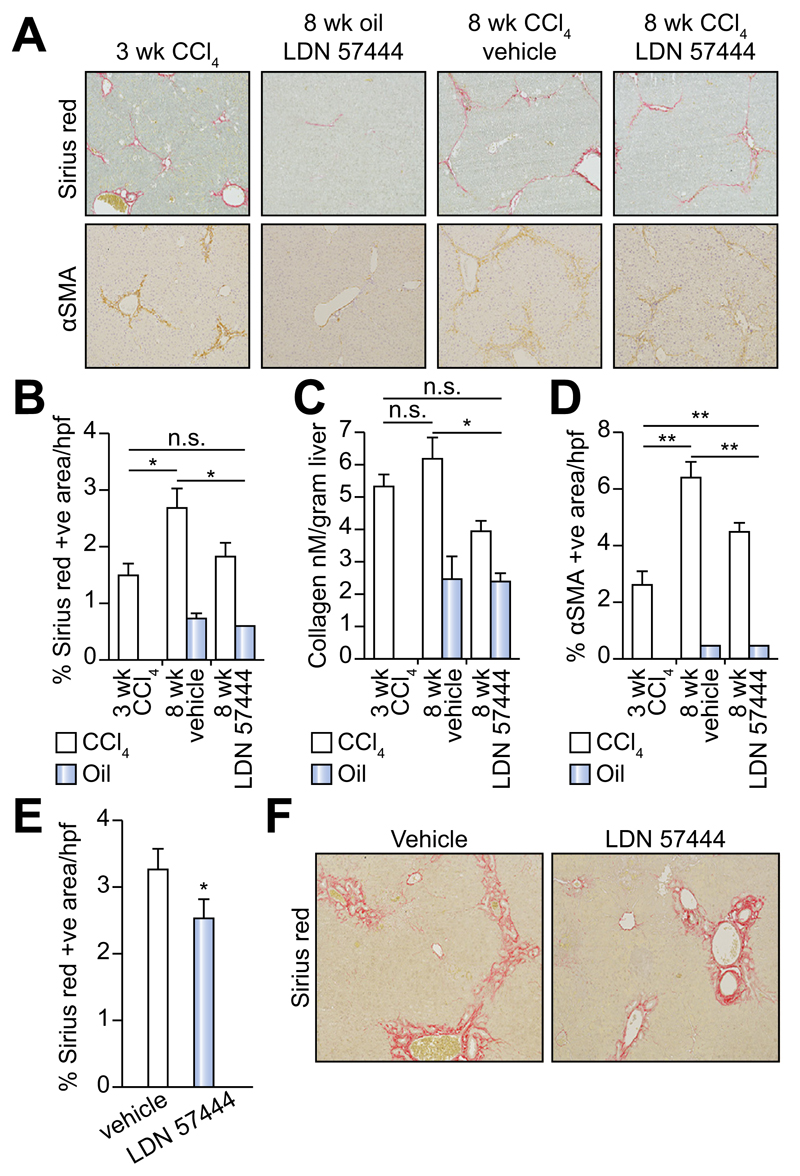
LDN 57444 is a reversible, competitive, active-site directed inhibitor of UCHL1 [20]. To investigate whether the inhibition of UCHL1 would reduce liver fibrogenesis we established an experimental therapeutic regime in mice (Fig. 4A). In this model, the animals were repeatedly administered CCl4 over a period of 3-weeks which as expected resulted in a mild fibrotic reaction (Fig. 4A, B). With other groups of these mice we then continued CCl4 injury over a further 5-weeks during which time we either administered the UCHL1 inhibitor LDN 57444 or control (DMSO). Mice treated with LDN 57444 showed a significant improvement in the Ishak fibrosis pathology score (Fig. 4B) indicative of a therapeutic blockade of fibrosis progression. We did not observe any difference in the necroinflammatory score between the drug and vehicle treated groups (Fig. 4C). However, we did observe a slight but not significant improvement in liver function with LDN 57444 treatment as assessed by reduced levels of the liver transaminases ALT and AST following either an acute or chronic dose of CCl4 (Fig. 4D). Collagen deposition was determined by Sirius red and hydroxyproline assay (Fig. 5A–C) and aHSC quantification by αSMA staining (Fig. 5A and D). Collagen deposition was detected at high levels 3 weeks post-CCl4 treatment and increased further at the 8 week CCl4 time-point. This increase was significantly reduced by LDN 57444 treatment p <0.05 (Fig. 5A–C). In addition, the observed reduction in collagen content correlated with reduced levels of HSC determined by αSMA staining p <0.01 (Fig. 5D). To further assess the anti-fibrotic potential of LDN 57444 we determined its effects in the BDL model of cholestatic liver disease. Immediately following BDL surgery, mice were implanted with an osmotic minipump supplying 0.4 mg/kg/day LDN 57444 or vehicle control, animals were then left for 14 days to develop liver disease and sacrificed to assess liver damage. Although we did not detect a significant reduction in αSMA positive cells in this model the animals developed significantly less fibrosis demonstrated by Sirius red staining (Fig. 5E, F). We conclude that LDN 57444 or similar compounds that might target UCHL1 have potential for development as anti-fibrotics.
Fig. 4. UCHL1 inhibition reduces liver fibrosis in vivo.
(A) Experimental plan: Mice were administered with CCl4 or olive oil for 3 or 8 weeks with the addition of bi-weekly injections of LDN 57444 (LDN drug) or DMSO (vehicle) from week 4. (B) Fibrosis scores expressed as individual data and mean mice according to the Ishak fibrosis scale (n = 5). (C) Necroinflammatory scores expressed as individual data and mean mice according to the Ishak necroinflammation scale (n = 5). (D) Serum plasma levels of the liver enzymes ALT and AST (arbitrary units a.u./ml) were compared between the acute injured mice groups (24 h CCl4) and chronic (3 and 8 week oil/CCl4), n = 5 mice per group. Acute CCl4 groups were compared using two-tailed unpaired students t test; not significant (n.s) p >0.05. Chronic CCl4 multicomparison analysis was performed using two-way repeated measures ANOVA between all groups (p = 0.0002 ALT, p = 0.004 AST); followed by Tukeys post-tests between 8 week CCl4 veh and drug groups (n.s) p >0.05.
Fig. 5. Inhibition of UCHL1 reduces fibrogenesis in two liver fibrosis models.
(A) Representative photomicrographs of Sirius red and αSMA stained liver sections from 3 week CCl4, 8 week oil/LDN 57444, 8 week CCl4/vehicle and CCl4/LDN 57444 treated mice. (B) Fibrosis densitometry of Sirius red stained liver sections. (C) Liver collagen content as assessed by hydroxyproline assay. (D) Average mean% area of αSMA stained liver sections (n = 5 per group). (E) Fibrosis densitometry and (F) representative photomicrographs of Sirius red stained liver sections from BDL control or LDN 57444 treated mice (n = 8 control and 7 LDN 57444). Multicomparison analysis was performed using two-way repeated measures ANOVA between all groups p <0.05 considered significant; followed by Tukeys post-tests between individual groups where *p <0.05; **p <0.01.
Proliferation is significantly reduced in UCHL1–/– and siRNA knockdown HSC
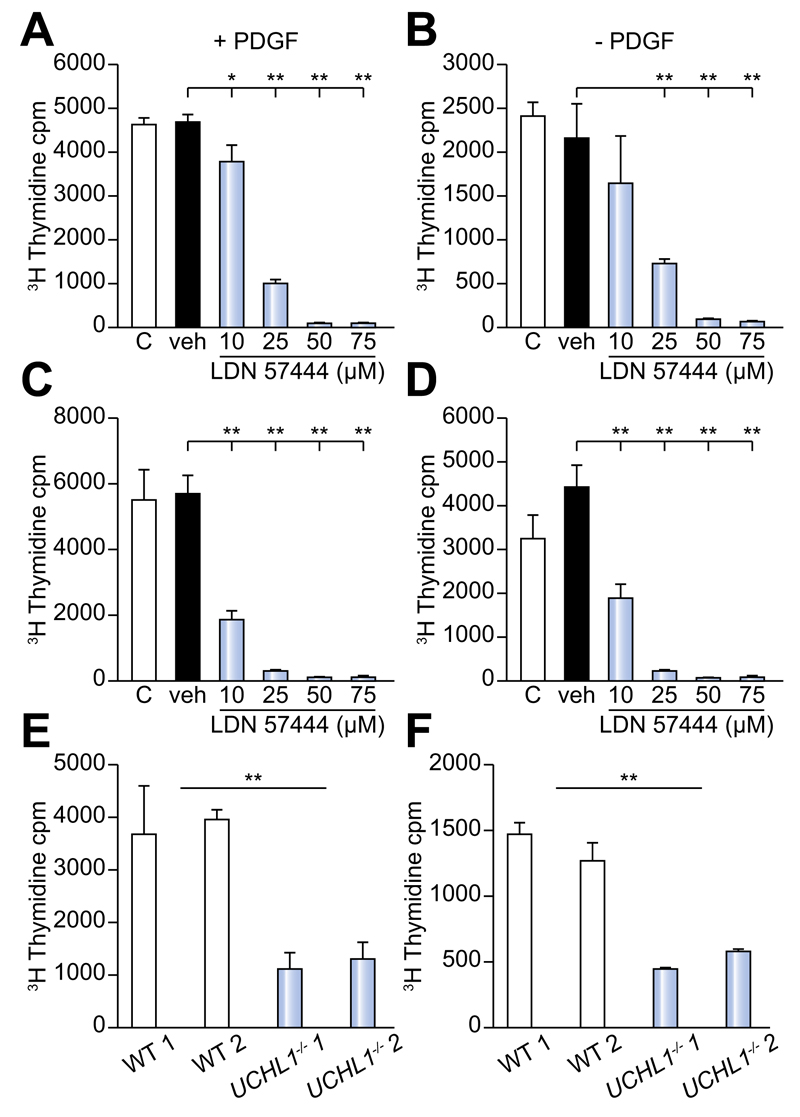
PDGFBB promotes proliferative and migratory responses of aHSC [21]. To investigate a function for UCHL1 in HSC proliferation, day 10 activated rat HSC were pre-treated with either transfection reagent alone, scrambled control siRNA or UCHL1 specific siRNAs and subsequently cultured for 24 h ± 20 ng/ml PDGFBB as a proliferative stimulus. Tritiated thymidine incorporation was used to measure proliferation and siRNA treated HSC were also examined by Western blotting to confirm knockdown of the UCHL1 protein (Supplementary Fig. 2A–C). Results confirm that following siRNA treatment UCHL1 was undetectable at the protein level (Supplementary Fig. 2A). UCHL1 specific siRNA knockdown lowered the proliferative response of HSC to the PDGFBB stimulus in comparison with the HSC treated with siRNA negative control (Supplementary Fig. 2B). We next wanted to examine the proliferative capacity of both rat and human activated HSC following UCHL1 inhibition using incremental doses of LDN 57444 (Fig. 6A–D). LDN 57444 treatment at doses of >10 μm (rat HSC + PDGFBB) and >10 μm (human ± PDGFBB) significantly reduced HSC proliferation in comparison with the DMSO vehicle control treated cells (Fig. 6A–D). This reduction in proliferative capacity was not due to drug toxicity as only doses of >75 μM caused apoptosis in isolated HSC (Supplementary Fig. 3A, B). In addition, we did not observe any toxicity <75 μM in isolated or in vivo hepatocytes (Supplementary Fig. 3C, D). Furthermore, while UCHL1–/– HSC showed no significant difference in basal levels of apoptosis or reduced levels of fibrogenic and inflammatory gene expression in comparison to WT (Supplementary Fig. 4), they displayed significantly reduced proliferative responses ± PDGFBB (Fig. 6E, F). Of note, due to their progressive neurodegeneration and short lifespan we were unable to confirm the in vivo function of UCHL1 with UCHL1–/– mice [13].
Fig. 6. Proliferation is significantly reduced following UCHL1 knockdown, inhibition and in UCHL1–/– HSC.
(A) 3H Thymidine incorporation in (A and B) rat and (C and D) human activated HSC in ± the addition of 20 ng/ml PDGFBB cultured in media (c), DMSO vehicle control (veh) or incremental doses of LDN 57444. (E and F) 3H Thymidine incorporation in 2 separate lines of HSC from UCHL–/– mice or WT littermates ± the addition of 20 ng/ml PDGFBB. 3H thymidine incorporation results are expressed as mean counts/min (cpm) values from triplicate wells (±SEM). Data is representative of 2 (WT and UCHL1–/–) and 3 (human and rat HSC) independent experiments. Multicomparison analysis was performed using two-way repeated measures ANOVA between all groups p <0.05 considered significant; followed by Tukeys post-tests between individual groups where *p <0.05; **p <0.01.
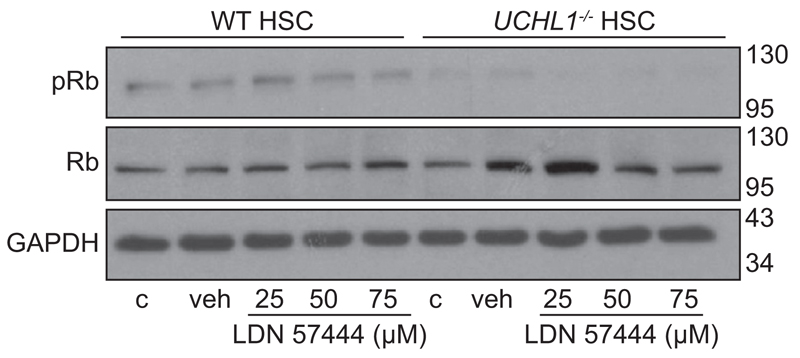
A role for UCHL1 as a potentiator of cyclin-dependent kinases (CDK) in nerve and cancer cell lines has recently been described [22]. CDK4 is able to phosphorylate cell cycle regulators including retinoblastoma protein (Rb). This in turn leads to the release of Rb bound E2F transcription factors, ubiquitination and degradation of the CDK inhibitor p27 and the promotion of cell cycle progression from G1-S phase [23]. However, the endogenous role of UCHL1 in Rb phosphorylation in non-cancerous cells such as HSC is not known. Therefore, to determine whether inhibition and/or loss of UCHL1 in isolated mouse HSC would lead to a reduction in Rb phosphorylation or a change in levels of G1-S cell cycle regulators, isolated UCHL1–/– and WT HSC were treated with incremental doses of LDN 57444 for 24 h, then pRb and total Rb were measured by Western blot. Results confirm that levels of pRb were decreased in UCHL1–/– in comparison with WT HSC yet; the effect of the inhibitor on WT HSC was negligible (Fig. 7). Of note, the G1-S inhibitor p27 which is usually degraded following Rb phosphorylation was also increased in WT mice following LDN 57444 treatment and in the UCHL1–/– mice n = 1 experiment (Supplementary Fig. 5). Taken together with the requirement of UCHL1 expression and activity for optimal proliferative responses to PDGFBB, we conclude that one mechanism by which UCHL1 regulates HSC function and fibrosis is via the post-translational control of cell cycle proteins.
Fig. 7. UCHL1 knockdown and pharmacological inhibition decreases Rb phosphorylation.
(A) Western blot for phosphorylated Rb (pRb), total Rb and GAPDH in isolated WT and UCHL1 –/– mouse HSC treated with media (c), DMSO (veh), or incremental doses of LDN 57444. Molecular weight markers shown on right side. Data is representative of 2 independent experiments.
Discussion
The Ub system is an important post-translational regulator of many cellular processes including protein turnover and cell signalling, that has also been mechanistically implicated in many human diseases including cancers in which inhibitors of Ub processing are used in clinical trials. Fibrosis has many similarities to cancer including alterations in cell phenotype and dysregulation of the cell cycle, which promotes the proliferation of myofibroblasts and spread of fibrosis [24–26]. Hence, it is surprising that to date there has been little attention focused on the Ub system and its regulators in fibrosis. In the present study we have began to examine the case for further study of Ub and its regulatory enzymes by focusing on the DUBs. Using the well-characterised model of culture-induced HSC activation (rodent and human) with confirmation of observations in situ with diseased tissues we have shown that there are discreet but profound changes in the expression of a small number of DUBs. Of particular interest was the dramatic induction of UCHL1. UCHL1 was first identified as an abundant protein found in the brain and testes (1–2% of soluble brain protein) and is predominantly described as a neuronal marker, however, later studies have demonstrated that its expression is not limited to the neuronal compartment [27]. The role of UCHL1 is extremely diverse as it is able to function as both a Ub ligase and a DUB, however it can also interact and stabilize cell proteins such as β catenin, and activate kinases such as CDK4 without the requirement for its enzymatic activity [22,28]. Therefore, the function of UCHL1 is likely to vary widely in a cell and environment specific manner. Indeed, diverse roles are described in kidney disease, neurodegeneration and cancer where UCHL1 exerts its effects by influencing levels of free proteins such as cellular Ub, glutathione and by regulation of the cell cycle [13,29–33]. In accordance with this, following our expression analysis we went on to show that UCHL1 is involved in the post-translational control of the cell cycle protein Rb and in the proliferative response of aHSC to PDGFBB. In addition, we also observed an increase in the levels of p27 with LDN 57444 >50 μM in WT mice and in the UCHL1–/–. However, there was also a slight decrease in the p27 levels in the UCHL1–/– mice with >50 μM suggesting potential off-target effects of the inhibitor in the absence of the UCHL1 protein potentially due to effects on the UCHL1 systemic isoform UCHL3 which is ubiquitously expressed and may have opposing roles. A role for UCHL1 in proliferation is consistent with the pro-oncogenic functions of UCHL1 in various cancers [34] and the findings that in lymphoma cells UCHL1 knockdown decreases cellular growth and viability [30]. Although we observe reduced proliferation in UCHL1–/– HSC their viability was not reduced in comparison with WT HSC (Supplementary Fig. 4). In addition, neither HSC nor primary mouse hepatocytes showed increased levels of apoptosis in the presence of the UCHL1 inhibitor at doses <75 μM. However, we did see a slight reduction in transaminase levels and hepatocyte apoptosis in vivo after drug treatment and therefore cannot rule out the potential for mild hepatoprotective effects of the drug. This hepatoprotection, is unlikely to be a direct effect of the drug on hepatocytes as we have no evidence of UCHL1 expression in hepatocytes either in vitro or in vivo. UCHL1 plays various documented roles in cellular function not limited to cellular proliferation but also including cellular migration and invasion both relative to HSC survival and motility [35]. Although, we did not observe any significant difference in the migration of UCHL1–/– HSC in comparison to WT using an in vitro scratch assay (data not shown), the role of UCHL1 in the motility of HSC in vivo has not yet been explored.
Perhaps our most exciting discovery was that a pharmacological inhibitor (LDN 57444) with specificity for UCHL1 was able to block the further progression of pre-established liver fibrosis despite ongoing liver damage. There are now other UCHL1 targeting inhibitors commercially available as UCHL1 expression appears to be restricted to myofibroblasts in the diseased liver, these data highlight interesting therapeutic opportunities for pharmacological targeting of the DUB in chronic liver disease that warrant further attention. We also suggest that it is now time for a wider examination of the role of the Ub system and its regulatory enzymes in myofibroblast biology where there are likely to be important contributions to tissue fibrosis and cancer.
Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.jhep.2015.07.034.
Acknowledgements
We would like to thank Rachel Howarth for assistance with the animal studies.
Abbreviations
- HSC
hepatic stellate cells
- UCHL1
ubiquitin c-terminal hydrolase
- Ub
ubiquitin
- DUB
deubiquitinase
- USP
ubiquitin specific protease
- IPF
idiopathic pulmonary fibrosis
- CCl4
carbon tetrachloride
- BDL
bile duct ligation
- qHSC
quiescent hepatic stellate cells
- aHSC
activated hepatic stellate cells
- αSMA
alpha smooth muscle actin
- ALD
alcoholic liver disease
- PDGFBB
platelet-derived growth factor BB
- CDK
cyclin-dependent kinase
- Rb
retinoblastoma protein
Footnotes
Conflict of interest
The authors who have taken part in this study declared that they do not have anything to disclose regarding funding or conflict of interest with respect to this manuscript.
Authors’ contributions
C.L.W. performed the majority of the laboratory based and in vivo experiments and the data analyses. L.M. performed a proportion of the immunohistochemistry and analyses. J.L. performed and analysed hydroxyproline assay. S.K. and J.F. provided access to human liver and samples. C.R.F. carried out minipump implant surgery. N.S.S. and A.F. provided kidney and lung samples and provided expert analyses of immunohistochemistry. J.H.R. assisted in the tritiated thymidine proliferation experiments. D.G.T. carried out the morphological analyses of the tissue sections from the CCl4 model and scoring of liver pathology. D.A.G. provided us with the UCHL1–/– mice. C.W., F.O., and D.M. designed the experiments and wrote the manuscript. All authors discussed the paper and commented on the manuscript.
This work was funded by the UK Medical Research Council (Grant G0700890 and MR/K0019494/1 to D.A.M.) and the Wellcome Trust (WT086755MA to D.A.M.).
References
- 1.Friedman SL. Hepatic stellate cells: protean, multifunctional, and enigmatic cells of the liver. Physiol Rev. 2008;88:125–172. doi: 10.1152/physrev.00013.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mann J, Mann DA. Transcriptional regulation of hepatic stellate cells. Adv Drug Deliv Rev. 2009;61:497–512. doi: 10.1016/j.addr.2009.03.011. [DOI] [PubMed] [Google Scholar]
- 3.Mann M, Jensen ON. Proteomic analysis of post-translational modifications. Nat Biotechnol. 2003;21:255–261. doi: 10.1038/nbt0303-255. [DOI] [PubMed] [Google Scholar]
- 4.Heride C, Urbe S, Clague MJ. Ubiquitin code assembly and disassembly. Curr Biol. 2014;24:R215–R220. doi: 10.1016/j.cub.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 5.Chen ZJ, Sun LJ. Nonproteolytic functions of ubiquitin in cell signaling. Mol Cell. 2009;33:275–286. doi: 10.1016/j.molcel.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 6.Kulathu Y, Komander D. Atypical ubiquitylation - The unexplored world of polyubiquitin beyond Lys48 and Lys63 linkages. Nat Rev Mol Cell Biol. 2012;13:508–523. doi: 10.1038/nrm3394. [DOI] [PubMed] [Google Scholar]
- 7.Komander D, Clague MJ, Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10:550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 8.Clague MJ, Coulson JM, Urbe S. Cellular functions of the DUBs. J Cell Sci. 2012;125:277–286. doi: 10.1242/jcs.090985. [DOI] [PubMed] [Google Scholar]
- 9.Elbaz A, Levecque C, Clavel J, Vidal JS, Richard F, Correze JR, et al. S18Y polymorphism in the UCH-L1 gene and Parkinson’s disease: evidence for an age-dependent relationship. Mov Disord. 2003;18:130–137. doi: 10.1002/mds.10326. [DOI] [PubMed] [Google Scholar]
- 10.Luise C, Capra M, Donzelli M, Mazzarol G, Jodice MG, Nuciforo P, et al. An atlas of altered expression of deubiquitinating enzymes in human cancer. PLoS One. 2011;6:e15891. doi: 10.1371/journal.pone.0015891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hellerbrand C, Bumes E, Bataille F, Dietmaier W, Massoumi R, Bosserhoff AK. Reduced expression of CYLD in human colon and hepatocellular carcinomas. Carcinogenesis. 2007;28:21–27. doi: 10.1093/carcin/bgl081. [DOI] [PubMed] [Google Scholar]
- 12.Todi SV, Paulson HL. Balancing act: deubiquitinating enzymes in the nervous system. Trends Neurosci. 2011;34:370–382. doi: 10.1016/j.tins.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coulombe J, Gamage P, Gray MT, Zhang M, Tang MY, Woulfe J, et al. Loss of UCHL1 promotes age-related degenerative changes in the enteric nervous system. Front Aging Neurosci. 2014;6:129. doi: 10.3389/fnagi.2014.00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gieling RG, Elsharkawy AM, Caamano JH, Cowie DE, Wright MC, Ebrahimkhani MR, et al. The c-Rel subunit of nuclear factor-kappaB regulates murine liver inflammation, wound-healing, and hepatocyte proliferation. Hepatology. 2010;51:922–931. doi: 10.1002/hep.23385. [DOI] [PubMed] [Google Scholar]
- 15.Ishak K, Baptista A, Bianchi L, Callea F, De Groote J, Gudat F, et al. Histological grading and staging of chronic hepatitis. J Hepatol. 1995;22:696–699. doi: 10.1016/0168-8278(95)80226-6. [DOI] [PubMed] [Google Scholar]
- 16.Oakley F, Mann J, Nailard S, Smart DE, Mungalsingh N, Constandinou C, et al. Nuclear factor-kappaB1 (p50) limits the inflammatory and fibrogenic responses to chronic injury. Am J Pathol. 2005;166:695–708. doi: 10.1016/s0002-9440(10)62291-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wynn TA, Barron L, Thompson RW, Madala SK, Wilson MS, Cheever AW, et al. Quantitative assessment of macrophage functions in repair and fibrosis. Current protocols in immunology / edited by John E Coligan [et al] 2011;Chapter 14:Unit14 22. doi: 10.1002/0471142735.im1422s93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fuchs G, Shema E, Vesterman R, Kotler E, Wolchinsky Z, Wilder S, et al. RNF20 and USP44 regulate stem cell differentiation by modulating H2B monoubiquitylation. Mol Cell. 2012;46:662–673. doi: 10.1016/j.molcel.2012.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wynn TA. Cellular and molecular mechanisms of fibrosis. J Pathol. 2008;214:199–210. doi: 10.1002/path.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y, Lashuel HA, Choi S, Xing X, Case A, Ni J, et al. Discovery of inhibitors that elucidate the role of UCH-L1 activity in the H1299 lung cancer cell line. Chem Biol. 2003;10:837–846. doi: 10.1016/j.chembiol.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 21.Lechuga CG, Hernandez-Nazara ZH, Hernandez E, Bustamante M, Desierto G, Cotty A, et al. PI3K is involved in PDGF-beta receptor upregulation post-PDGF-BB treatment in mouse HSC. Am J Physiol Gastrointest Liver Physiol. 2006;291:G1051–G1061. doi: 10.1152/ajpgi.00058.2005. [DOI] [PubMed] [Google Scholar]
- 22.Kabuta T, Mitsui T, Takahashi M, Fujiwara Y, Kabuta C, Konya C, et al. Ubiquitin C-terminal hydrolase L1 (UCH-L1) acts as a novel potentiator of cyclin-dependent kinases to enhance cell proliferation independently of its hydrolase activity. J Biol Chem. 2013;288:12615–12626. doi: 10.1074/jbc.M112.435701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dick FA, Rubin SM. Molecular mechanisms underlying RB protein function. Nat Rev Mol Cell Biol. 2013;14:297–306. doi: 10.1038/nrm3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hewitson TD, Becker GJ. Interstitial myofibroblasts in IgA glomerulonephritis. Am J Nephrol. 1995;15:111–117. doi: 10.1159/000168813. [DOI] [PubMed] [Google Scholar]
- 25.Grotendorst GR, Rahmanie H, Duncan MR. Combinatorial signaling pathways determine fibroblast proliferation and myofibroblast differentiation. FASEB J. 2004;18:469–479. doi: 10.1096/fj.03-0699com. [DOI] [PubMed] [Google Scholar]
- 26.Duffield JS, Lupher M, Thannickal VJ, Wynn TA. Host responses in tissue repair and fibrosis. Annu Rev Pathol. 2013;8:241–276. doi: 10.1146/annurev-pathol-020712-163930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Day IN, Thompson RJ. UCHL1 (PGP 9.5): neuronal biomarker and ubiquitin system protein. Prog Neurobiol. 2010;90:327–362. doi: 10.1016/j.pneurobio.2009.10.020. [DOI] [PubMed] [Google Scholar]
- 28.Bheda A, Yue W, Gullapalli A, Whitehurst C, Liu R, Pagano JS, et al. Positive reciprocal regulation of ubiquitin C-terminal hydrolase L1 and beta-catenin/TCF signaling. PLoS One. 2009;4:e5955. doi: 10.1371/journal.pone.0005955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoglinger GU, Breunig JJ, Depboylu C, Rouaux C, Michel PP, Alvarez-Fischer D, et al. The pRb/E2F cell-cycle pathway mediates cell death in Parkinson’s disease. Proc Natl Acad Sci U S A. 2007;104:3585–3590. doi: 10.1073/pnas.0611671104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hussain S, Foreman O, Perkins SL, Witzig TE, Miles RR, van Deursen J, et al. The de-ubiquitinase UCH-L1 is an oncogene that drives the development of lymphoma in vivo by deregulating PHLPP1 and Akt signaling. Leukemia. 2010;24:1641–1655. doi: 10.1038/leu.2010.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leroy E, Boyer R, Auburger G, Leube B, Ulm G, Mezey E, et al. The ubiquitin pathway in Parkinson’s disease. Nature. 1998;395:451–452. doi: 10.1038/26652. [DOI] [PubMed] [Google Scholar]
- 32.Meyer-Schwesinger C, Meyer TN, Sievert H, Hoxha E, Sachs M, Klupp EM, et al. Ubiquitin C-terminal hydrolase-l1 activity induces polyubiquitin accumulation in podocytes and increases proteinuria in rat membranous nephropathy. Am J Pathol. 2011;178:2044–2057. doi: 10.1016/j.ajpath.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Read NC, Gutsol A, Holterman CE, Carter A, Coulombe J, Gray DA, et al. Ubiquitin C-terminal hydrolase L1 deletion ameliorates glomerular injury in mice with ACTN4-associated focal segmental glomerulosclerosis. Biochim Biophys Acta. 2014;1842:1028–1040. doi: 10.1016/j.bbadis.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 34.Hurst-Kennedy J, Chin LS, Li L. Ubiquitin C-terminal hydrolase l1 in tumorigenesis. Biochem Res Int. 2012;2012:123706. doi: 10.1155/2012/123706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim HJ, Kim YM, Lim S, Nam YK, Jeong J, Kim HJ, et al. Ubiquitin C-terminal hydrolase-L1 is a key regulator of tumor cell invasion and metastasis. Oncogene. 2009;28:117–127. doi: 10.1038/onc.2008.364. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.