Abstract
Immune-mediated tissue regeneration driven by a biomaterial scaffold is emerging as an innovative regenerative strategy to repair damaged tissues. We investigated how biomaterial scaffolds shape the immune microenvironment in traumatic muscle wounds to improve tissue regeneration. The scaffolds induced a pro-regenerative response, characterized by an mTOR/Rictor-dependent T helper 2 pathway that guides interleukin-4–dependent macrophage polarization, which is critical for functional muscle recovery. Manipulating the adaptive immune system using biomaterials engineering may support the development of therapies that promote both systemic and local pro-regenerative immune responses, ultimately stimulating tissue repair.
Immune homeostasis is indispensable to tissue development, regeneration, and repair (1). Trauma initiates a cascade of local and systemic immune events that trigger the mobilization of cells into the damaged site to initiate host defense and tissue repair. The limited success achieved to date in rebuilding human tissues may be due in part to the tendency for therapeutic strategies to target later processes in wound healing and regeneration, such as stem cell differentiation. Conversely, the immune system is a highly flexible network that serves as a guardian of tissue integrity and is adapted to the nature of the local microenvironment (2). The immune system participates in tissue repair by scavenging debris and dead cells (3), recruiting and supporting the proliferation of tissue progenitor cells (4), and inducing vascularization (5). Previously, immune responses to biomaterials were related to rejection (6–8); however, subsets of innate immune cells have been identified as important mediators of scaffold remodeling (9–11) and can be targeted for immune-mediated tissue regeneration. We explored the role of adaptive immunity in tissue regeneration, identifying TH2 responses as critical in driving the repair of traumatic tissue injury.
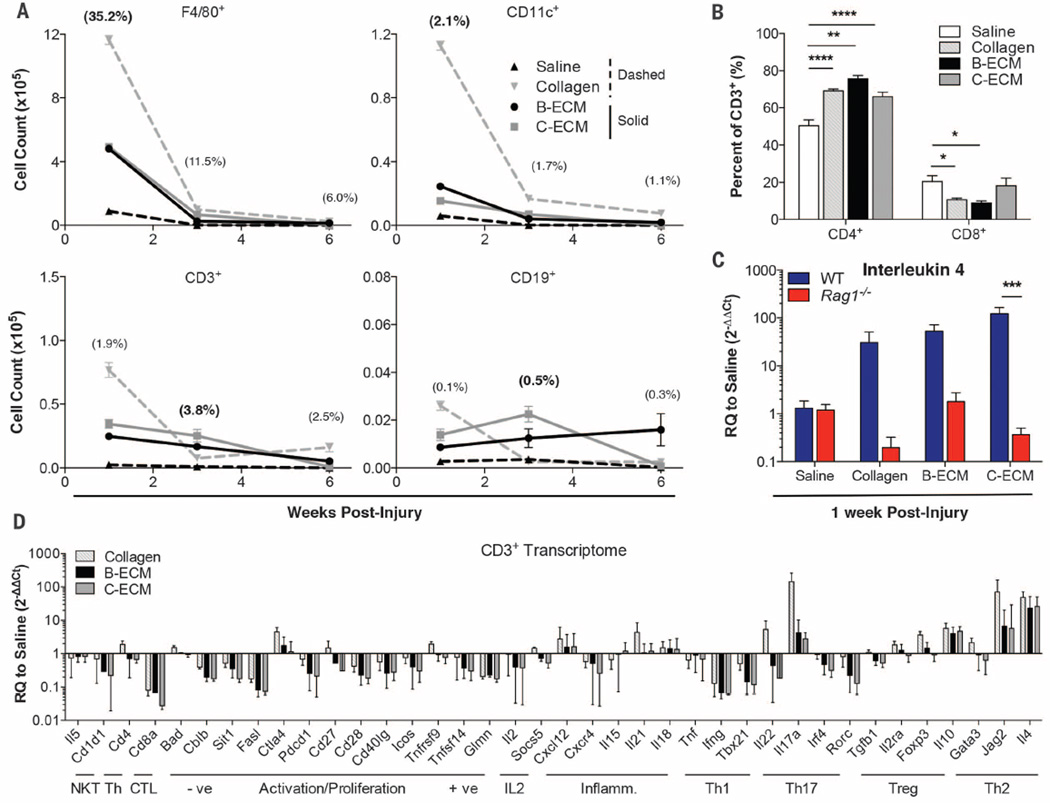
To model a traumatic wound, we surgically excised a portion of the quadriceps muscle group in C57BL/6 mice, provoking an irreversible volumetric muscle loss (VML) injury (12). Based on the regenerative potential and clinical use of tissue-derived extracellular matrix (ECM) scaffolds (10, 11), we screened and selected bone- and cardiac muscle–derived tissue ECM scaffolds (B-ECM and C-ECM) for their immunomodulatory properties [fig. S1 (13)]. The presence of scaffolds in damaged muscle significantly increased the number of myeloid cells (F4/80+ macrophages and CD11c+ dendritic cells; P < 0.0001) and lymphocytes (CD3+ T cells and CD19+ B cells; P < 0.05) present at the injury site as compared to a saline-treated control after 1 and 3 weeks (Fig. 1A and fig. S2). At 1 week, collagen-treated wounds recruited the highest number of immune cells into the defect region (36.0% of total live cells, 13.6 million cells) followed by B-ECM– and C-ECM–treated wounds (39.3%, 5.32 million; and 45.4%, 5.44 million cells), with saline-treated wounds containing significantly fewer cells (36.4%, 0.97 million). The proportion of myeloid cells in the damaged muscle peaked at 1 week after injury, and the T cell fraction, consisting of both CD4+ and CD8+ cells, peaked in all treatment groups at 3 weeks after injury. In the muscle wound, biomaterial scaffolds skewed the ratio of CD4:CD8 T cells toward a higher fraction of CD4+ helper T cells (~70% in scaffold-treated, versus ~50% in saline-treated wounds) at 1 week after injury (Fig. 1B). CD4+FoxP3+ regulatory T cells were also present at low levels and increased over time (fig. S3).
Fig. 1. Biomaterial scaffolds induce a TH2 response in volumetric muscle wounds.
C57BL/6 (WT) and Rag1−/− mice received a critical-size quadriceps muscle injury and were treated immediately with 0.05 ml of saline, particulate collagen, B-ECM, or C-ECM. (A) Proportions of myeloid (F4/80+ macrophages and CD11c+ dendritic cells) and lymphoid (CD3+ T cells and CD19+ B cells) cell populations in the WT wound environment, determined by flow cytometry (% = mean fraction of live cells across all treatments, with peak level shown in bold text). The greatest cell numbers were in scaffold-treated wounds. (B) Proportion of CD3+ T cells that are CD4+ TH cells or CD8+ cytotoxic T lymphocytes at 1 week after injury treated with saline, collagen, B-ECM, or C-ECM by flow cytometry. (C) qRT-PCR analysis of Il4 gene expression in WT and Rag1−/− mice at 1 week after injury. (D) One week after injury, transcriptome of CD3 cells sorted from wounded muscles treated with saline, collagen, B-ECM, or C-ECM, determined by qRT-PCR. Data are displayed as relative quantification (RQ) to saline-treated wounds. Data are means ± SEM, n = 4 mice (2 legs pooled per mouse, representative of at least two independent experiments), analysis of variance (ANOVA): ****P < 0.0001, ***P < 0.001, **P < 0.01, *P < 0.05.
The expression of interleukin 4 (Il4), a gene encoding a canonical type 2 helper T cell (TH2) cytokine that is also important in muscle healing (14–17), increased in the presence of the scaffold (Fig. 1C). Therefore, we sought to understand the role of cells of the adaptive immune system on the formation of the regenerative immune microenvironment. Scaffolds were implanted into B6.129S7-Rag1tm1Mom/J (Rag1−/−) mice, which lack mature T and B cells. In Rag1−/− mice, scaffold-mediated Il4 up-regulation was lost, suggesting a TH2-driven scaffold immune microenvironment. CD3+ cells were sorted out of muscle injuries at 1 week after injury for detailed gene expression analysis (Fig. 1D, fig. S4, and table S1). Scaffolds induced a TH2-type gene expression profile as characterized by increased Il4 expression and decreased expression of Ifng and Tbx21 (TH1 canonical genes). In addition, Jag2, which encodes the Notch ligand Jagged 2, was elevated. Jagged2 helps direct TH differentiation away from TH1 and toward TH2 (18). Il10, which encodes a general anti-inflammatory cytokine that is not TH-specific, was also up-regulated. Other genes that are more selectively expressed by TH1 cells, such as Fasl and Cd28 (the costimulatory receptor for CD86), were likewise down-regulated.
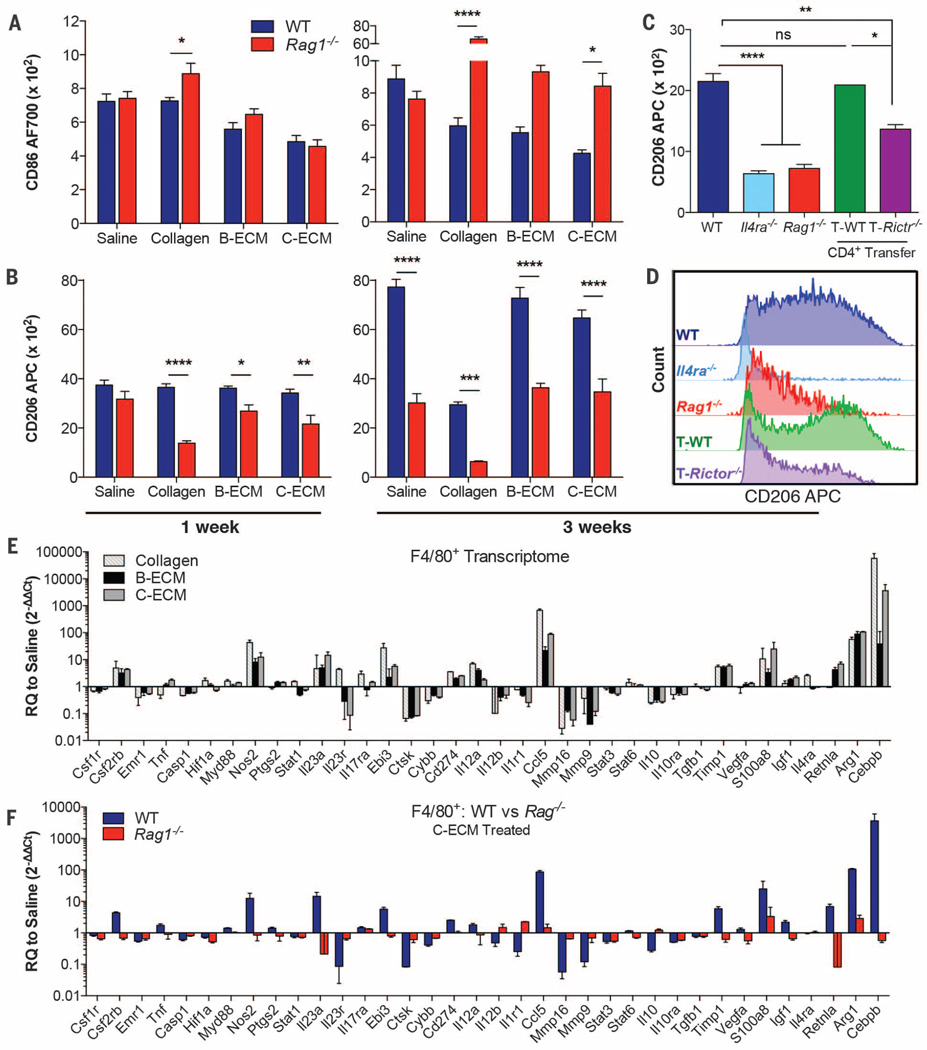
The regenerative outcome of tissue-derived ECM scaffolds in animals and humans is correlated with an immunoregulatory M2 macrophage phenotype during remodeling (9–11). Biomaterials increased the expression of genes associated with a pro-regenerative type 2 immune response, including hallmark genes of M2 myeloid cells, more specifically macrophages that are stimulated by IL-4, known as M(IL-4) macrophages (fig. S5) (19). As with Il4 expression, induction of these M(IL-4) markers was almost completely lost in Rag1−/− mice (fig. S5). In the presence of adaptive immune cells, biomaterial scaffolds inhibited macrophage CD86 up-regulation (a costimulatory molecule expressed at high levels by classical M1 macrophages) at 3 weeks after surgery (Fig. 2A and fig. S6H). In Rag1−/− mice, however, down-regulation of CD86 expression was mitigated, and ECM scaffold–treated wounds returned to a macrophage polarization profile resembling that of saline-treated control animals (Fig. 2A). On the other hand, CD206 expression (a mannose receptor and classical M2 marker) was similar between ECM scaffold–treated and saline-treated mice at 1week after implantation, with increased expression at 3 weeks (Fig. 2B). However, this increase in CD206 was ablated in Rag1−/− mice in both scaffold- and saline-treated wounds, suggesting that the adaptive immune system also has a scaffold-independent role in shaping the wound healing response. Moreover, this CD206 up-regulation was also impaired in B6.129S2-Cd4tm1Mak/J (Cd4−/−) mice, which maintain B cells and CD8+ T cells but lack CD4+ helper T cells (fig. S6A). Additionally, in Cd4−/− mice, the recruitment of B cells (a commonly TH2-driven adaptive effector cell) was diminished (fig. S6G).
Fig. 2. M(IL-4) pro-regenerative myeloid polarization induced by scaffolds is TH2-dependent.
(A and B) Macrophages in wounded muscle were characterized for CD86 (A) and CD206 (B) expression by flow cytometry at 1 and 3 weeks after injury in the presence of saline or ECM scaffold in WT (blue bars) and Rag1−/− (red bars) mice. The mean of fluorescence is shown. (C) CD206 expression at 3 weeks after injury in C-ECM–treated WT, Il4ra−/−, Rag1−/−, and Rag1−/− mice reconstituted with either WT CD4+ T cells (T-WT, n = 2) or Rictor−/− CD4+ T cells (T-Rictr−/−;TH2-deficient). (D) Representative comparison of CD206 expression between WT, Il4ra−/−, Rag1−/−, and Rag1−/− reconstituted with WT and Rictor−/− CD4+ T cells. (E) qRT-PCR gene expression analysis in cell-sorted macrophages from wounded muscles 1 week after injury and treated with collagen (light gray–striped bars), B-ECM (black solid bars), or C-ECM (gray solid bars) compared to saline control. RQ to saline = 2−ΔΔCt. (F) RQ to saline in WT and Rag1−/− mice when wounds were treated with C-ECM. The figure shows a loss of scaffold-mediated macrophage polarization in Rag1−/− mice. WT, blue bars; Rag1−/−, red bars. Data are means ± SEM, n = 4 mice unless otherwise stated (representative of one or two independent experiments); ANOVA [(A) and (B)] and Student’s t test (D): ****P <0.0001, ***P <0.001, **P <0.01, *P <0.05.
To further elucidate the role of CD4+ T cells, and more specifically TH2 T cells on the polarization of myeloid cells, we evaluated myeloid CD206 expression in Rag1−/− mice that were repopulated with either wild-type (WT) CD4+ T cells or Rictor−/− CD4+ T cells (Fig. 2, C and D, and fig. S7). Rictor is a critical component of the mTORC2 complex that integrates signals from the environment and drives the polarization of TH2 cells (20). Myeloid cells in Rag1−/− mice expressed lower levels of CD206 as compared to WT mice; however, when repopulated with WT CD4+ T cells (T-WT cells), this phenotype was rescued. When mice received TH2-deficient T cells (T-Rictor−/− cells), CD206 expression was not rescued, proving that TH2 T cells, dependent on mTORC2 signaling, are necessary for pro-regenerative myeloid polarization. To confirm the role of IL-4 in TH2-dependent myeloid polarization, we characterized the phenotype of macrophages in BALB/c-Il4ratm1Sz/J (Il4ra−/−) mice that cannot receive signals from IL-4 (Fig. 2, C and D). Compared to WT controls, myeloid cells in Il4ra−/− wounds expressed far lower levels of CD206, suggesting that the macrophage activation was controlled by IL-4, and verifying that the pro-regenerative profile is associated with M(IL-4) cells.
The pleiotropic nature of immune responses typically results in complex expression profiles beyond stereotypical M1 versus M2 “poles” (19). The expression of CD86 and the expression of CD206 on macrophages (classically M1 versus M2) were not mutually exclusive; however, these scaffold-associated macrophages also up-regulated the expression of genes encoding Arg1 and Retnla (encoding Fizz1), similar to the results from quantitative reverse transcription polymerase chain reaction (qRT-PCR) analyses of the whole wound (Fig. 2E and fig. S5). Additionally, Cebpb, and Timp1 were up-regulated, whereas Mmp16 and Mmp9 were down-regulated, further suggesting a pro-regenerative function of the scaffold-associated macrophages (21–23). In Rag1−/− mice, which cannot mount a TH2 immune response, scaffold-associated macrophages lost their pro-regenerative transcriptome (Fig. 2F, fig. S8, and table S2). Several genes directly implicated in muscle regeneration such as Igf1 (insulin-like growth factor-1) (24–26) and Vegfa (vascular endothelial growth factor) (27) decreased significantly in Rag1−/− mice. Gene ontology enrichment analysis of genes differentially expressed in Rag1−/− versus WT macrophages shows enrichment in programs associated with morphogenesis and differentiation, suggesting a reliance on the adaptive immune system for up-regulation of developmentally active immune genes (fig. S9).
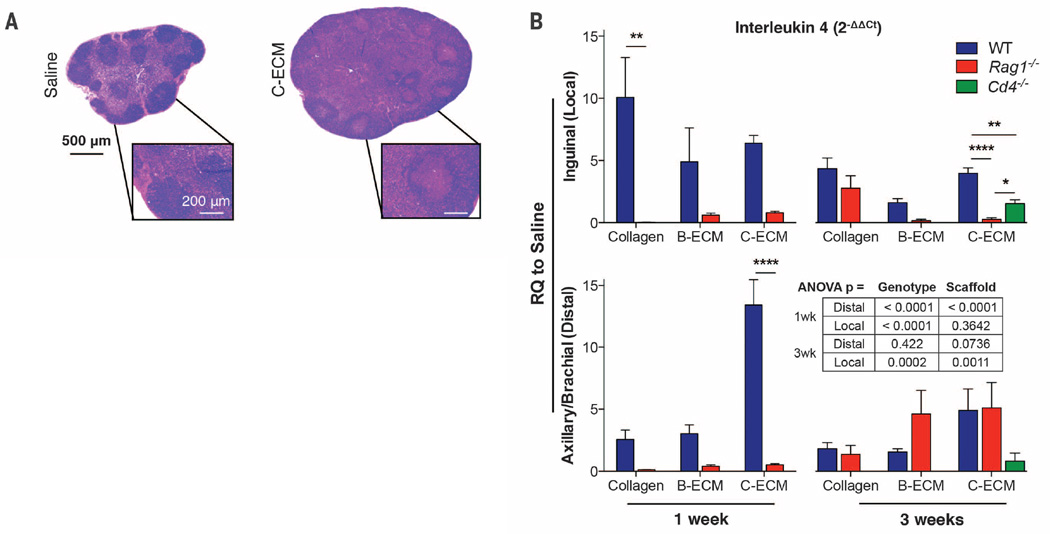
The detection of a local scaffold-associated TH2 polarization led us to investigate the potential systemic T cell response (28). Subcutaneous scaffold implants produce a systemic TH2-like response in the bloodstream, but the connection to wound healing and regeneration is unknown (28, 29) (Fig. 3 and fig. S10). Scaffold treatment induced hypertrophy of local draining lymph nodes (Fig. 3A), which accompanied a robust increase in Il4 expression (Fig. 3B and fig. S10). This Il4 induction was absent at 1 week after injury in Rag1−/− mice but present after 3 weeks, suggesting an early adaptive immune–dependent Il4 up-regulation followed by an innate immune–driven Il4 up-regulation later in the wound healing and regeneration processes. Additionally, Cd4−/− mice displayed a significant decrease in scaffold-mediated Il4 up-regulation in inguinal lymph nodes at 3 weeks after injury in C-ECM–treated animals (Fig. 3B). This Il4 expression level was higher than that in Rag1−/− mice, demonstrating an important role of CD4+ T cells in scaffold-induced systemic type 2 immunity, but with potential further contributions by B cells or CD8+ T cells.
Fig. 3. Systemic immune homeostasis is modified by application of biomaterial scaffolds.
(A) Inguinal lymph node morphology at 1 week after injury in saline- (left) and C-ECM– (right) treated WT animals. Hematoxylin and eosin (H&E) staining is shown. (B) qRT-PCR analysis of Il4 gene expression in local draining lymph nodes (inguinal, top bar graphs) and distal lymph nodes (axillary/brachial, bottom bar graphs) in WT, Rag1−/−, and Cd4−/− mice at 1 and 3 weeks after wound treatment with collagen, B-ECM, or C-ECM. RQ to saline is 2−ΔΔCt. Data are means ± SEM, n = 4 mice (representative of at least two independent experiments), ANOVA: ****P < 0.0001, **P < 0.01, *P < 0.05.
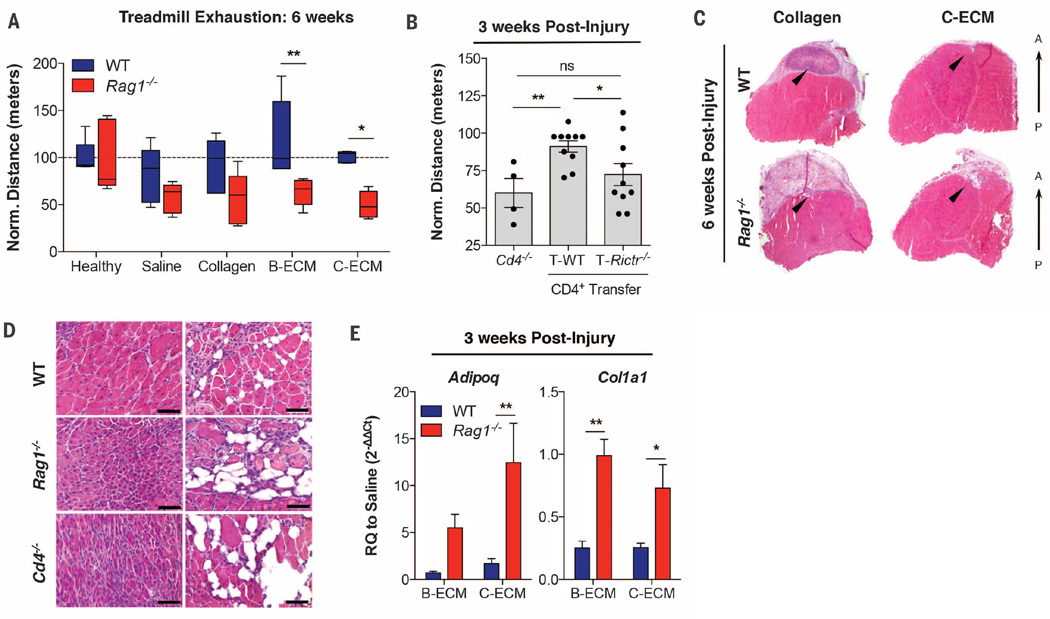
Functionally, WT animals recovered to be able to run distances similar to those of healthy uninjured counterparts after 6 weeks (Fig. 4A). However, this restoration of running capacity was ablated in the absence of T and B cells (Rag1−/−) in ECM scaffold–treated wounds. At 3 weeks after injury, repopulation of Rag1−/− mice with WTT cells rescued their functional capacity, and the animals could run greater distances as compared to mice lacking the CD4 subset (Fig. 4B; 91.11 ± 3.83 versus 60.06 ± 9.69, P = 0.0032). Furthermore, Rag1−/− mice repopulated with WT CD4 T cells performed better than those repopulated with Rictor−/− CD4+ T cells (72.31 ± 7.40, P = 0.0368), confirming the role of TH2 CD4+ T cells in functional muscle regeneration.
Fig. 4. TH2/M(IL-4) responses to biomaterial-treated muscle wound promote functional tissue regeneration.
(A) Treadmill exhaustion assay of mice at 6 weeks after injury to test muscle function in WT (blue bars) and Rag1−/− (red bars) mice. Results are normalized to the distance run by an uninjured control (100 m). n = 5 mice per condition and genotype. (B) Treadmill exhaustion at 3 weeks in Cd4−/−, and Rag1−/− mice repopulated with WT (T-WT) or Rictor−/− (T-Rictr−/−; TH2 deficient) CD4+ T cells. n = 4 mice (Cd4−/−) or n = 10 mice (T-WT and T-Rictr−/−) (C) Transverse section of quadriceps muscle at 6 weeks after injury in collagen- and C-ECM–treated WT and Rag1−/− mice. The black arrowheads indicate the injury/treatment area. A, anterior, P, posterior, with H&E staining shown. (D) C-ECM–treated VML at 3 weeks after injury in WT, Rag1−/−, and Cd4−/− mice stained with H&E. Small muscle fibers and ectopic adipogenesis are present in Rag1−/− and Cd4−/− wounds. Scale bars, 50 µm. (E) Gene expression (qRT-PCR) of Adipoq (adipose marker) and Col1a1 (collagen I) showing increased adipose gene expression in Rag1−/− as well as increased collagen gene expression, suggesting alterations in connective tissue deposition and possible scarring. n = 4 mice unless otherwise stated (representative of at least two independent experiments). Data are means ± SEM; ANOVA [(A) and (D)] and Student’s t test (E): ****P <0.0001, ***P < 0.001, **P < 0.01, *P < 0.05.
Muscle structure correlated with the differences in functional capacity. Histologically, at 6 weeks after injury, the quadriceps muscle treated with the C-ECM scaffold appeared similar to that of healthy controls, with minimal scaffold visible and repair tissue fully integrated within the surrounding musculature. A large region of fibrous tissue with active inflammation was present in muscles treated with the collagen scaffold (Fig. 4C and fig. S11). Rag1−/− mice displayed increased adipose deposition, fibrosis, scaffold persistence, and smaller-diameter muscle fibers than their WT counterparts. At 3 weeks after injury, centrally nucleated muscle fibers, which are indicative of active regeneration or recovery from injury, were present within the biomaterial scaffold and around the defect site (Fig. 4D and fig. S12). WT mice produced muscle with larger, more rounded fibers, whereas Rag1−/− mice muscles contained smaller, irregularly shaped fibers, indicating a defect in muscle regeneration. In addition, the pathologic Rag1−/− histomorphology was recapitulated in Cd4−/− mice, confirming the role of CD4+ T cells in fibro-adipogenic lineage commitment (Fig. 4D). Increased gene expression of Adipoq (adiponectin) confirmed ectopic adipogenesis in Rag1−/− whole muscle. Similarly, the expression of Col1a1 (type I collagen) increased in Rag1−/− mice muscles, highlighting increased fibrosis (Fig. 4E and fig. S13). Although scaffold treatment reduced fibro- and adipogenesis markers in WT animals, this benefit was lost in Rag1−/− mice.
We have demonstrated that tissue-derived biomaterial scaffolds enhance the development of a pro-regenerative immune environment and have implicated adaptive immune cells, specifically mTORC2-dependent CD4+ TH2 T cells, in the process of functional tissue restoration (fig. S14). Just as cancer research has made great strides in T cell therapies, these concepts can be translated to biomaterials design to improve tissue repair and regeneration (30–33).
Supplementary Material
Acknowledgments
The authors acknowledge the Sidney Kimmel Comprehensive Cancer Center Flow Cytometry Research Core and L. Blosser for their help on flow cytometric studies; A. Anderson for assistance with adipogenesis studies; S. Sommerfeld and X. Wang for assistance with the clinical product testing; A. Rittenbach for assistance with computed tomography imaging; M. de Palma for generous donation of the iBMM macrophage cell line; M. Swartz and the Swartz lab for assistance in the pilot studies and J.H.E. sabbatical hosting; A. Ewald for helpful discussions; and J. Schneck for critical review. The data presented in this manuscript are tabulated in the main paper and in the supplementary materials. K.S., F.H., D.P., and J.H.E. are inventors on provisional patent application no. 48317-502P01US, filed by Johns Hopkins University (JHU) related to regenerative immunology. J.H.E. holds equity in Aegeria Soft Tissue, a company that has licensed JHU intellectual property not directly related to the materials used in this study but similar enough that it may benefit from the results. The conflict is being managed by the Johns Hopkins Office of Policy Coordination. This work was funded by the Maryland Stem Cell Research Fund (MSCRF), grant 113345; the Armed Forces Institute for Regenerative Medicine; and U.S. Department of Defense grant W81XWH-11-2-0022 (awarded to J.H.E.) and the Jules Stein Professorship from the Research to Prevent Blindness Foundation. M.T.W. was supported by a postdoctoral fellowship from the Hartwell Foundation. D.M.P., J.D.P., B.S.L. and H.W. were supported by Cancer Center Core grant P30CA006973. J.D.P. was supported by a grant from the National Institutes of Health, R01AI077610. J.D.P, J.H.E., and D.M.P acknowledge support from the Bloomberg-Kimmel Institute for Cancer Immunotherapy.
Footnotes
SUPPLEMENTARY MATERIALS
www.sciencemag.org/content/352/6283/366/suppl/DC1
Materials and Methods
Figs. S1 to S14
Tables S1 and S2
References (34, 35)
REFERENCES AND NOTES
- 1.Wynn TA, Chawla A, Pollard JW. Nature. 2013;496:445–455. doi: 10.1038/nature12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matzinger P, Kamala T. Nat. Rev. Immunol. 2011;11:221–230. doi: 10.1038/nri2940. [DOI] [PubMed] [Google Scholar]
- 3.Peng Y, et al. J Autoimmun. 2007;29:303–309. doi: 10.1016/j.jaut.2007.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Otis JS, et al. PLOS One. 2014;9:e92363. doi: 10.1371/journal.pone.0092363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frantz S, Vincent KA, Feron O, Kelly RA. Circ. Res. 2005;96:15–26. doi: 10.1161/01.RES.0000153188.68898.ac. [DOI] [PubMed] [Google Scholar]
- 6.Anderson JM. Annu. Rev. Mater. Res. 2001;31:81–110. [Google Scholar]
- 7.Anderson JM, Miller KM. Biomaterials. 1984;5:5–10. doi: 10.1016/0142-9612(84)90060-7. [DOI] [PubMed] [Google Scholar]
- 8.Anderson JM. ASAIO Trans. 1988;34:101–107. doi: 10.1097/00002480-198804000-00005. [DOI] [PubMed] [Google Scholar]
- 9.Sicari BM, et al. Sci. Transl. Med. 2014;6:234ra58. doi: 10.1126/scitranslmed.3008085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mase VJ, Jr, et al. Orthopedics. 2010;33:511. doi: 10.3928/01477447-20100526-24. [DOI] [PubMed] [Google Scholar]
- 11.Brown BN, et al. Acta Biomater. 2012;8:978–987. doi: 10.1016/j.actbio.2011.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sicari BM, et al. Tissue Eng. Part A. 2012;18:1941–1948. doi: 10.1089/ten.tea.2012.0475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beachley VZ, et al. Nat. Methods. 2015;12:1197–1204. doi: 10.1038/nmeth.3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salmon-Ehr V, et al. Lab. Invest. J. Tech. Methods Pathol. 2000;80:1337–1343. doi: 10.1038/labinvest.3780141. [DOI] [PubMed] [Google Scholar]
- 15.Gause WC, Wynn TA, Allen JE. Nat. Rev. Immunol. 2013;13:607–614. doi: 10.1038/nri3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heredia JE, et al. Cell. 2013;153:376–388. doi: 10.1016/j.cell.2013.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horsley V, Jansen KM, Mills ST, Pavlath GK. Cell. 2003;113:483–494. doi: 10.1016/s0092-8674(03)00319-2. [DOI] [PubMed] [Google Scholar]
- 18.Fang TC, et al. Immunity. 2007;27:100–110. doi: 10.1016/j.immuni.2007.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murray PJ, et al. Immunity. 2014;41:14–20. doi: 10.1016/j.immuni.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delgoffe GM, et al. Nat. Immunol. 2011;12:295–303. doi: 10.1038/ni.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruffell D, et al. Proc. Natl. Acad. Sci. U.S.A. 2009;106:17475–17480. doi: 10.1073/pnas.0908641106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blackwell J, et al. J Physiol. Sci. 2015;65:145–150. doi: 10.1007/s12576-014-0350-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davis ME, Gumucio JP, Sugg KB, Bedi A, Mendias CL. J Appl. Physiol. 2013;115:884–891. doi: 10.1152/japplphysiol.00137.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mourkioti F, Rosenthal N. Trends Immunol. 2005;26:535–542. doi: 10.1016/j.it.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 25.Musarò A, et al. Nat. Genet. 2001;27:195–200. doi: 10.1038/84839. [DOI] [PubMed] [Google Scholar]
- 26.Liu JP, Baker J, Perkins AS, Robertson EJ, Efstratiadis A. Cell. 1993;75:59–72. [PubMed] [Google Scholar]
- 27.Arsic N, et al. Molecular Therapy: J. Am. Soc. Gene Therapy. 2004;10:844–854. doi: 10.1016/j.ymthe.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 28.Allman AJ, et al. Transplantation. 2001;71:1631–1640. doi: 10.1097/00007890-200106150-00024. [DOI] [PubMed] [Google Scholar]
- 29.Allman AJ, McPherson TB, Merrill LC, Badylak SF, Metzger DW. Tissue Eng. 2002;8:53–62. doi: 10.1089/107632702753503054. [DOI] [PubMed] [Google Scholar]
- 30.Rosenberg SA, Yang JC, Restifo NP. Nat. Med. 2004;10:909–915. doi: 10.1038/nm1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ali OA, et al. Cancer Res. 2014;74:1670–1681. doi: 10.1158/0008-5472.CAN-13-0777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim J, Mooney DJ. Nano Today. 2011;6:466–477. doi: 10.1016/j.nantod.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ali OA, Tayalia P, Shvartsman D, Lewin S, Mooney DJ. Adv. Funct. Mater. 2013;23:4621–4628. doi: 10.1002/adfm.201203859. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.