Abstract
Purpose
Numerous biomarkers for pancreatic cancer have been reported. We determined the extent to which such biomarkers are expressed throughout metastatic progression, including those that effectively predict biologic behavior.
Experimental Design
Biomarker profiling was performed for 35 oncoproteins in matched primary and metastatic pancreatic cancer tissues from 36 rapid autopsy patients. Proteins of significance were validated by immunolabeling in an independent sample set, and functional studies were performed in vitro and in vivo.
Results
Most biomarkers were similarly expressed or lost in expression in most samples analyzed, and the matched primary and metastases from a specific patient were most similar to each other than to other patients. However, a subset of proteins showed extensive inter-patient heterogeneity one of which was p38 MAPK. Strong positive pp38 MAPK immunolabeling was significantly correlated with improved post-resection survival by multivariate analysis (median overall survival 27.9 months, p=0.041). In pancreatic cancer cells, inhibition of functional p38 by SB202190 increased cell proliferation in vitro in both low-serum and low-oxygen conditions. High functional p38 activity in vitro corresponded to lower levels of pJNK protein expression, and p38 inhibition resulted in increased pJNK and pMKK7 by Western blot. Moreover, JNK inhibition by SP600125 or MKK7 siRNA knockdown antagonized the effects of p38 inhibition by SB202190. In vivo, SP600125 significantly decreased growth rates of xenografts with high p38 activity compared to those without p38 expression.
Conclusions
Functional p38 MAPK activity contributes to overall survival through JNK signaling, thus providing a rationale for JNK inhibition in pancreatic cancer management.
Keywords: adjuvant therapy, biomarker, JNK, p38, pancreatic cancer, tissue microarray, metastasis
INTRODUCTION
Despite substantial progress in our understanding of this disease, survival of patients with pancreatic ductal adenocarcinoma remains relatively unchanged over the past 3 decades. Most patients will die of their disease within 5 years. Surgical resection provides the best chance of long-term survival but recurrences occur in most and 5-year survival in this group is only in the range of 20%. The lack of effective population based screening methods, coupled with relatively few treatment options and late stage of diagnosis, all contribute to these statistics (1). Thus, studies to understand the molecular characteristics of pancreatic cancers that impact upon their biological behavior, treatment resistance and ultimately patient survival are needed. Such information would prove invaluable not only for guiding clinical management but for revealing cellular pathways of significance for this disease.
Well documented pathologic markers that correlate with survival in patients with pancreatic cancer include tumor size, level of differentiation and lymph node status (2). Numerous molecular biomarkers for pancreatic cancer have also been described; these can be categorized into diagnostic markers such as Ca19-9, prognostic markers such as Smad4 or MUC1, and predictive markers such as SPARC, HuR or members of the BRCA2 family (3). However, given the wide variety of biomarkers reported and the lack of available pancreatic cancer samples with high-quality outcome data, it remains difficult to stratify these biomarkers into those with the most promise for utility in the clinical setting. Thus, unbiased approaches provide optimal methods to stratify such markers into those most suitable for use in the clinical setting. For example, Winter et al used a survival based tissue microarray of resected pancreatic cancers in association with a set of stringent criteria to identify among more than 2000 potential biomarkers those that were most predictive of patient outcome. This approach identified MUC1 and mesothelin both of which were superior to pathologic features and other putative markers for predicting post operative survival >30 months (4). Such a methodology also has value as it provides clues to the biologic features of pancreatic cancer that underlie the expression of these biomarkers. For example, MUC1 overexpression may enhance HIF-1α stabilization and hypoxia-driven angiogenesis, promote formation of a reactive tumor microenvironment that facilitates metastasis, and induce epithelial to mesenchymal transition (5–7). By contrast, given that mesothelin-specific T-cell responses have been associated with longer survival of pancreatic cancer patients (8), loss of expression of this molecule may signify escape of pancreatic cancer cells from immune surveillance (9).
The goal of this study was to perform biomarker profiling to identify biologic pathways that influence patient survival following surgical resection. To accomplish this goal we used a two well-annotated sets of pancreatic cancer tissues from patients with known patterns of failure at autopsy and from patients who underwent a potentially curative resection (1). Such knowledge is critical for the ultimate goal of identifying the specific features of pancreatic cancer present in diagnostic materials that can be exploited for decision-making during clinical management. We now show that phospho-p38 MAPK expression in pancreatic cancer tissues corresponds to a significant survival advantage in pancreatic cancer patients in the adjuvant setting, and that the mechanism by which functional p38 MAPK promotes survival is due to its ability to inhibit JNK activity at the cellular level.
PATIENTS AND METHODS
Tissues and TMA construction
Paraffin-embedded samples of primary and metastatic pancreatic cancer tissue from 36 patients who underwent an autopsy in association with the Gastrointestinal Cancer Rapid Medical Donation Program (10) were used to construct a tissue microarray (TMA) containing 208 cores (2–4 primary and 2 metastatic tissue cores per patient). All 36 patients died of pancreatic cancer as determined by both gross and histologic analysis. Representative samples of normal pancreas and 12 other normal tissues were also included on the array for orientation under the microscope and as positive or negative controls for labeling by each marker. In addition, we created five TMAs representing 83 resected pancreatic cancer tissues from pancreaticoduodenectomy or distal pancreatectomy specimens at Johns Hopkins Hospital. For each patient two cores each of normal pancreas and the matched pancreatic cancer tissue were included. The Johns Hopkins Institutional Review Board approved collection and use of all autopsy and resected tissues for TMA construction and immunolabeling.
Immunolabeling and Scoring Strategy
Unstained 5-µm sections were cut from each TMA paraffin block and the slides were deparaffinized by routine techniques followed by incubation in 1× sodium citrate buffer (diluted from 10× heat-induced epitope retrieval buffer, Ventana-Bio Tek Solutions, Tucson, AZ) before steaming for 20 minutes at 80°C. Slides were cooled 5 minutes before incubation with each primary antibody (see Supplemental Table 2 for primary antibody details). Immunolabeling was detected per kit instructions (Ventana IVIEW Detection Kits, catalog 760091). Negative controls for each of the antibodies were performed using nonimmune serum instead of the primary antibody. Slides were scored by two of the authors at a two-headed microscope (Y.N. and C.I.D.). For each antibody, the intensity value (scale 0 to 3) and the percent positive cells were multiplied to generate a Histology Score (H-score) with H = % positive cells x intensity of labeling. Using this system H-scores ranged from 0 to 300. H-scores were calculated for each core independently, and the average of all cores for a patient (n=4) or for the primary carcinoma (n=2–4) versus metastasis (n=2) for each patient determined.
Hierarchical Cluster Analysis
Agglomorative hierarchical clustering based on Euclidean distance and using Ward’s method with complete linkage was used to cluster H-score data over both genes and samples(11). Multivariate logistic regression was used to evaluate the predictive power of candidate biomarkers of metastatic burden. The response variable was metastatic burden with samples classified as low (MB≤10) or high (MB>10), and 12 markers found to have significant variation across samples were used as predictors. To mitigate the effects of overfitting, performance was evaluated with leave-one-out cross-validation. Specifically, each sample was set aside in turn, while the coefficients of the logistic regression model were estimated using the remaining samples. The resulting model was applied to the left out sample in order to predict the metastatic burden. Both continuous predictions (the estimated probability of high MB) and binary predictions (estimated probability of high MB >0.5) were reported.
Cell Lines
The Panc 5.04 and Panc 2.5 cell lines were obtained from Dr. Elizabeth Jaffee (Johns Hopkins). The HPNE-E6/E7 cell line was obtained from the ATCC, and the HPDE line was provided by Dr. Michael Goggins (Johns Hopkins). The A2.1, A6L, A10.7, A38.44 and A13D cell lines were created in our own laboratory. All cell lines were confirmed mycoplasma free and authenticated at the time of creation by the Johns Hopkins Cell Core.
In vitro Cell Proliferation
All tumor cell lines were cultured in DMEM supplemented with 10% FBS, 100 units/ml penicillin, 100 µg/ml streptomycin and 2 mmol/L L-glutamine at 37°C and 5% CO2. Cells were treated with p38 inhibitors (SB202190, SB203580 and SB239063) at a variety of concentrations (5 µM, 10 µM or 20 µM), or treated with p38 inhibitors and 10 µM of gemcitabine (GEM) then analyzed for cell proliferation at 1, 24, 48 and 72 hour points using cell counting Kit-8 (Dojindo molecular technologies, Kumamoto, Japan). As for JNK inhibitor treatment experiments, 10 µM of SP600125 was used. Each assay experiment was performed in triplicate and data was derived from at least three independent experiments. A solvent tolerance test was performed to consider the solvent toxicity by setting up 4 different concentrations (0 %, 0.05 %, 0.1 % and 0.2 %) of DMSO (Supplemental Figure 5). Based on these results 0.1 % of DMSO was used as a vehicle treated control for all experiments.
Western blots
Equal amounts of proteins were separated on SDS-polyacrylamide gel and transferred onto PVDF membranes (DuPont NEN, Boston, MA). Each membrane was hybridized with primary antibody for two hours followed by horseradish peroxidase (HRP)-linked IgG and visualized by the enhanced chemiluminescence (ECL) system (Amersham). Expression of β-actin was used as an internal control. All primary antibodies were purchased from Cell Signaling and used at the recommended dilutions (phospho-ATF2, #9221; AKT, #9272; Phospho-Akt (Ser473), #9271; JNK, #9252; phosphor-JNK, #4668; p38, #9212; phospho-p38, #9216; MKK7, #4172; phosphor-MKK7, #4171). Films were scanned into JPEG format using a Canon MG7100 scanner, and images were analyzed and quantitated in Image J following the method outlined at http://www.lukemiller.org/journal/2007/08/quantifying-western-blots-without.html.
p38 MAP Kinase Assay
The p38 MAP Kinase assay kit was used to assess functional p38 activity (Cell Signaling Technology, Cat# 9820). Cell lysates were prepared by suspending cell pellets in RIPA buffer (20 mM Tris, 0.1% SDS, 1% Triton X-100, 1% sodium deoxycholate, pH 7.4) supplemented with protease inhibitor cocktail tablets (Roche Diagnostics GmbH, Mannheim, Germany). Next, 20 µl of immobilized phospho-p38 MAPK (Thr180/Tyr182) mAb primary antibody was added to 400 µg of cellular protein in a 200 µl-volume and incubated at 4°C overnight. The IP products were washed two times with 1x cell lysis buffer and two times with 1x kinase buffer followed by resuspension in 50 µl of 1x kinase buffer containing 200 µM ATP and 1 µl of kinase substrate. After incubation at 4°C for 30 minutes the reaction was terminated with 25 µL 3X SDS sample buffer. Western blot analysis was performed as described above.
Hypoxia chambers
Cell proliferation after p38 inhibitors and gemcitabine (GEM) treatment was investigated for evaluation of p38 MAPK’s role under normoxia and hypoxia conditions using Hypoxia chambers (Billups-Rothenberg Inc, Del Mar, CA). Cells cultivated in 96-well plates or 10 cm cell culture dishes were placed in a hypoxia chamber. The chamber was flushed with 1% O2/5% CO2/93% N2 gas mixture for 5 min, sealed and kept at 37°C. For longer incubation periods the chamber was refilled after 24 hours to ensure constant oxygen concentrations.
siRNA knockdowns
Panc5.04, A10.7 and A38.44 cells were cultured in Dulbecco's modified medium containing 10%FBS. Cells were transfected with validated SignalSilence® MKK7 siRNA II (#6323, Cell signaling) or SignalSilence® Control siRNA (#6568, Cell signaling) using Lipofectamine® RNAiMAX reagent (Cat. #. 13778-075, Invitrogen) following the Reverse transfection protocol. Inhibition of gene expression by siRNA was determined after 24 hours by Western blot analysis. As for cell proliferation assay, the control and MKK7 siRNA-transfected cells were treated with 10 µM of SB202190, and then subjected to cell proliferation assay using cell counting Kit-8 in following days. Each transfection experiment was performed in triplicate and data was derived from at least two independent experiments.
Xenograft assays
For in vivo tumor growth analysis, 4×106 of tumor cells was subcutaneously injected into 6 week old male CD1−/− (nude) mice. Starting from the next day, the mice (n=3 of each group) were intraperitoneally injected 200 µl of 0.1% solvent (v/v) or SB202190 (2.5mg/kg/day) every day for 3 weeks. Mice were sacrificed 3 weeks post-injection and size of tumors was measured. As for SP600125 treatment, the mice (n=3 of each group) were intraperitoneally injected 200 µl of SP600125 (40mg/kg/day) once daily for 5 consecutive days starting on the next day of implantation, then followed for 3 weeks. Data was represented as mean ± SEM.
Statistics
Summary data are presented as the median value and/or mean ± S.D. Parametric distributions were compared by a Student’s T-test for two groups, or a one-way analysis of variance in the event of multiple comparisons, and nonparametric distributions by χ2 test or Fisher exact test in the event of frequency values <5. Disease specific survival analyses were performed using the Kaplan Meier method with individual arms compared by a log rank test. Multivariate analyses were performed using the Cox proportional hazards model. P values ≤0.05 were considered significant.
RESULTS
Biomarker Profiling of Pancreatic Cancer
A discovery set of 36 patients’ pancreatic cancer tissues was used to create a tissue microarray for biomarker profiling (Figure 1A). For 30 of these patients the two cores each of the primary carcinoma and one matched metastasis were included, and for six patients no metastatic disease was found at autopsy and thus four cores of the primary carcinoma were included. The clinicopathologic features of these patients are shown in Supplemental Table 1.
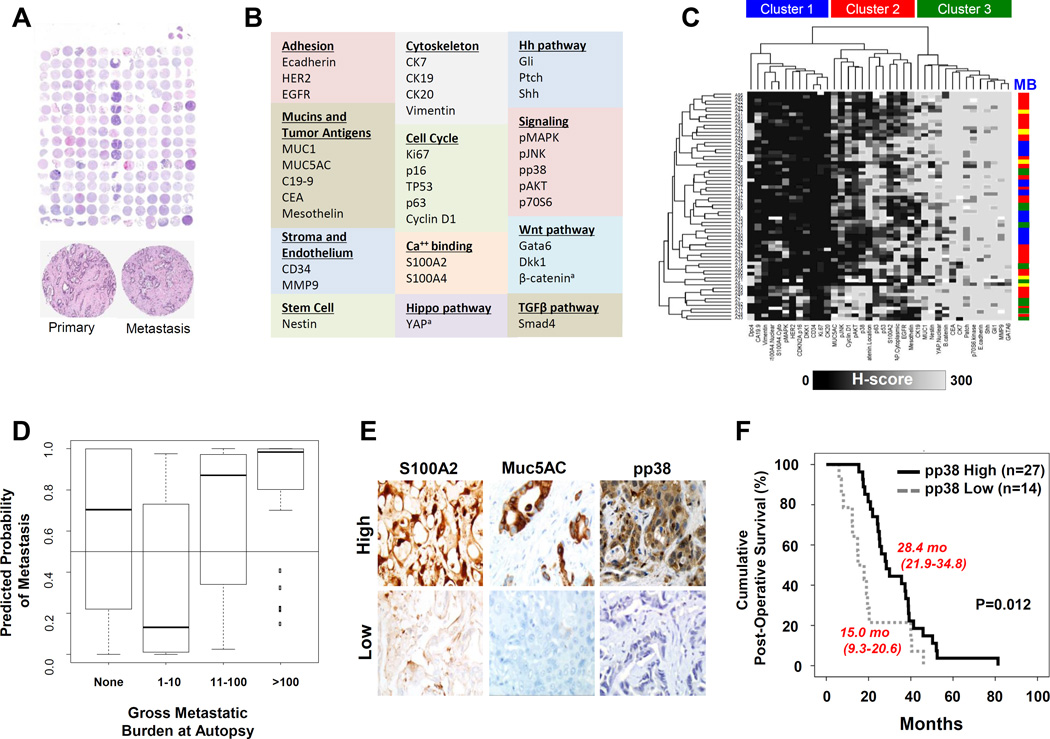
Figure 1. Biomarker Profiling of a Matched Primary and Metastasis Tissue Microarray.
(A). Scanning power view of a hematoxylin and eosin stained section of the TMA constructed for biomarker profiling. An example of cores of the matched primary and liver metastasis from a single patient are shown at 10x magnification. (B) Proteins and evaluated by biomarker profiling. (C) Heatmap generated by unbiased Hierarchical Cluster Analysis of expression data. Dendrograms indicate the presence of three distinct clusters based on patterns of protein expression. The relationship of protein expression to the gross metastatic burdens (MB) of each patient at autopsy are shown to the right of the heat map, with the colors corresponding to no metastases (yellow), 1–10 metastases (green), 11–100 metastases (blue) and >100 metastases (red). For each patient labeling patterns for both the primary carcinoma and matched metastasis are independently plotted on their own row. (D) Box plot illustrating the predicted probability of having a high metastatic burden (>10 metastases). (E) Examples of high (H-score ≥ 150) and low (H-score <150) protein expression for pp38, MUC5AC and S100A2 in the neoplastic epithelium of resected pancreatic cancer tissues. (F) Kaplan Meier survival plot of patients who completed adjuvant chemoradiation with high versus low pp38 immunolabeling of their resected tissues.
Immunolabeling was performed on recuts of this TMA for 35 different proteins representing a variety of cellular pathways, desmoplastic stromal constituents or oncoproteins reported to play a role, either directly or indirectly, in pancreatic cancer biology (Figure 1B and Supplemental Table 2). High quality labeling was achieved for 94% of the total 5184 cores analyzed, corresponding to a total of 4889 individual datapoints of immunolabeling data. For proteins such as β-catenin or Yes-associated protein (YAP), in which pattern of expression is important, both the intensity of labeling and the location of labeling were recorded. In order to identify expression patterns that represented similar biological behavior, the dataset was next analyzed by unsupervised hierarchical cluster analysis (Figure 1C). In general, the primary and metastases from a specific patient were most similar to each other than to other patients. A tendency to clump patients with similar metastatic burdens was also noted, although there were many such clusters that were all relatively small and interspersed with one another. Boxplots where then created for each protein organized by primary versus metastasis, and by metastatic burden (Supplemental Figure 1). This again showed little variation across primary versus metastatic site, and across categories of metastatic burden. However, some proteins such as Smad4 did show a relationship with metastatic burden, consistent with prior reports for this gene product (12,13).
Closer review of the data indicated the presence of three clusters using this set of markers (Figure 1C). The first cluster (Cluster 1) corresponded to those biomarkers with consistently abnormal patterns of immunolabeling compared to that of normal epithelial cells within the same sections. Oncoproteins in this cluster corresponded to those with either frequent loss of expression (i.e. CDKN2A) or overexpression compared to normal ductal epithelium (i.e. Ca19-9) in the majority of cancer tissues studied. Such patterns are consistent with the general dysregulation of gene expression that accompanies pancreatic carcinogenesis (14–17). This cluster also included cytoskeletal and matrix proteins that were consistently negative in both normal and neoplastic samples (i.e. CK20). By contrast, Cluster 3 corresponded to those proteins with expression patterns that were similar in intensity or cellular localization to that of normal epithelial cells in the majority of these samples. These proteins included those such as CK7, a marker of pancreaticobiliary epithelium, and the transcription factor Gata6. Finally, Cluster 2 corresponded to tissue biomarkers with significant heterogeneity of expression across the samples and patients analyzed, and this cluster of proteins was further explored.
Identification of p38 MAPK as a Biomarker of Patient Outcome
Cluster 2 corresponded to 12 (33%) of the 35 proteins analyzed. These 12 proteins were highly meaningful for pancreatic cancer biology and included MUC5AC, cyclin D1, p53, p63, β-catenin, pp38, pJNK, pAKT, S100A2, YAP, EGFR and mesothelin. β-catenin and YAP alterations in this cluster corresponded specifically to their altered cellular localization (membranous and cytoplasmic, respectively) compared to the patterns expected for normal epithelial cells (18).
To determine the potential relevance of this subset of markers, we first used a logistic regression model of these 12 proteins to predict met burden> 10, in a leave-one-out cross validation. This correctly predicted 30 of 40 pancreatic cancers as having high burden and 10 of 16 as having low burden, further suggesting the biologic significance of this set of proteins (Figure 1D). Next, we immunolabeled a validation set of 83 surgically resected pancreatic cancers for 11 of these proteins and correlated their expression patterns to patient outcome. Clinical features of these 83 patients are shown in Supplemental Table 3. Consistent with prior reports, S100A2 and MUC5AC overexpression were each significantly related to worse overall postsurgical survival by univariate analysis (19,20)(Figure 1E and Supplemental Figures 2A, 2B). Of interest, cytoplasmic phospho-p38 MAPK (pp38) overexpression within the neoplastic epithelium corresponded to an improved overall survival in these same patients (p=0.0155), with a median overall survival of 24.4 months (95% CI: 19.7–29.1 months) for carcinomas with high expression (H-score >150) compared to 14.7 months (95% CI: 7.6–21.8 months) for those with low expression (H-score ≤150)(Figure 1E and Supplemental Figure 2C). High pp38 expression also corresponded to a higher likelihood of a lymph node ratio <0.4 (p=0.031) (Supplemental Table 4). There was no difference in the rates of pp38 overexpression in these 83 resected carcinomas (53 of 83, 64%) compared to that of the 36 patients with end stage disease in the discovery set of patients (28 of 36, 78%) (p=0.1345), suggesting loss of pp38 does not occur to a greater extent with disease progression.
Given that a subset of these patients did not complete adjuvant chemoradiation, a reflection of poor performance status or disease progression (21), or details of adjuvant data were not available, we repeated the survival analysis of pp38 expression solely among the 41 patients confirmed to have completed adjuvant therapy. The vast majority of these patients received 1–2 cycles of gemcitabine followed by adjuvant 5-FU based chemotherapy and radiation to 50–54 Gy followed by another 2–4 months of gemcitabine as per the Radiation Therapy Oncology Group Trial (RTOG) 97-04 (22).. Some patients received adjuvant therapy closer to home; while confirmed to have completed therapy the full details were not available. This analysis maintained a significant relationship between pp38 expression and overall survival, with patients with high expression having a median overall survival of 28.4 months (95% CI: 21.9–38.4 months) compared to those with low expression having a median overall survival of 15.0 months (95% CI: 9.3–20.6 months) (p=0.012), an almost two-fold difference (Figure 1F). We further evaluated pp38 overexpression in a multivariate model in the 26 patients similarly treated per RTOG 97-04, indicating that pp38 overexpression was an independent predictor of improved patient survival in this cohort (Table 1 and Supplemental Figure 2D).
Table 1.
Multivariate Analysis of 26 Pancreatic Cancer Patients who Completed Adjuvant Chemoradiation per the RTOG 97-04 protocol.
| Variable | Hazard Ratio |
95.0% Confidence Interval |
P Value |
|---|---|---|---|
| Age | 1.049 | .952–1.156 | .331 |
| Vascular Invasion |
28.779 | 1.917–432.070 | .015 |
| Perineural Invasion |
.297 | .009–10.252 | .501 |
| Tumor Differentiation |
.128 | .022−.748 | .022 |
| Node Ratio >0.4 | .439 | .021–8.994 | .593 |
| Surgical Margin | 2.471 | .884–6.910 | .085 |
| Clinical Stage | 1.280 | .199–8.229 | .795 |
| pp38 MAPK expression |
.236 | .059−.942 | .041 |
No relationships with overall survival were found for the remaining proteins; this does not rule out the significance of these proteins for pancreatic cancer biology, only that their expression patterns were not a reliable predictive marker of patient outcome in our validation set. Given that the role of p38 MAPK in pancreatic cancer outcome has not been explored in detail, we sought to determine the significance and mechanisms of action of this signaling pathway in pancreatic cancer.
Functional p38 acts by restraining pancreatic cancer cell growth
To determine the extent to which p38 is functionally expressed in pancreatic cancer, we performed western blotting for total and phosphorylated forms of p38α as this is the predominant isoform expressed in most cell types (23) in two immortalized normal and seven pancreatic cancer cell lines (Figure 2A). Despite detectable total p38 protein in six of seven cancer cell lines, only three (Panc5.04, A10.7 and A38.44) expressed significant levels of phosphorylated p38 suggesting endogenous activation of the pathway in these cells. To confirm pp38 was functional, we determined the abundance of phospho-ATF2 in each cell line by western blot, a known protein target of pp38 (24). Levels of pATF2 paralleled levels of pp38 protein, further indicating that this subset of pancreatic cancer cell lines has functional p38 expression.
Figure 2. Effects of p38 MAPK inhibitor SB202190 on pancreatic cell proliferation.
(A) Western blotting for total and phospho-p38 (pp38) MAPK expression in immortalized normal (HPNE, HPDE) and pancreatic cancer cell lines. Expression of actin was used as a loading control. The ratio of phosphorylated to total p38 is shown for each cell line, indicating high levels of phosphorylation for Panc5.04, A10.7 and A38.44 cells. Functional p38 MAPK activity as determined by immunoprecipitation of phosphorylated ATF2, a known protein target of p38, is also shown. (B) Immunohistochemical staining for pp38 MAPK in cancer tissues of patients A2 (source of cell line A2.1) and A6 (source of cell line A6L) indicating low levels of pp38 expression. Cancer cells are indicated by arrowheads and scale bars correspond to 50 microns. (C) Cell lines Panc5.04, A10.7 and A38.44 with high p38 MAPK activity were treated with 10 µM of the p38 MAPK inhibitor SB202190 and subjected to cell proliferation assay. (D) Functional p38 MAPK activity levels as reflected by pATF2 levels in Panc5.04, A10.7 and A38.44 cells after treatment with increasing concentrations of SB202190. (E) A2.1 and A6L cells with low levels of p38 MAPK activity were treated with 10 µM of SB202190 and subjected to cell proliferation assay. 0.1% (v/v) of DMSO was used as a vehicle treated control. *, p<0.05; **, p<0.01; ***, p<0.001 (treated group versus untreated control).
To determine the general mechanism by which p38 functional activity may contribute to pancreatic cancer survival in patients, we used three different pharmacologic inhibitors of p38 (SB202190, SB203580 and SB239063) to assess the growth in vitro of cell lines Panc5.04, A10.7 and A38.44. All three inhibitors of p38 resulted in increased growth in vitro in a dose dependent manner indicating functional p38 expression restrains the growth of these cells (Figure 2C and Supplemental Figure 3). However, as off targets effects were seen at concentrations greater than 10 um of inhibitor, and all three inhibitors behaved similarly, all remaining experiments were performed with SB202190 at a 10 um concentration. Immunoprecipitation of pATF2 in Panc5.04, A10.7 and A38.44 cells after treatment with SB202190 (Figure 2D) further confirmed the specificity of p38 inhibition in these cells at this concentration. Finally and as an additional confirmation these effects were specific to p38 activity, we repeated the cell proliferation assays in cell lines A2.1 and A6L that do not express high levels of pp38 (Figure 2A). Modest decreases in growth were seen in these cell lines after addition of SB202190, consistent with off-target effects in these cells (Figure 2E). Thus, we conclude that functional p38 expression in pancreatic cancer cells acts by restraining cell growth, consistent with data indicating p38 negatively regulates the cell cycle at the G1/S and G2M transitions (25). Note that for A2.1 and A6L cells we considered the possibility that lack of functional p38 may reflect growth in culture conditions and not the microenvironment that these cells arose in. To address this possibility we immunolabeled the matched paraffin embedded tissues that these cell lines were originally derived from for pp38 expression. Weak or no expression was seen for pp38 (Figure 2B) indicating that their lack of pp38 expression in vitro is an accurate reflection of their in vivo status for this phosphoprotein.
Because p38 coordinates signals related to environmental and inflammatory stresses (1,23), we nonetheless repeated these assays in the setting of low serum and low oxygen conditions to mimic features of the microenvironment typical of human pancreatic cancer (2,26–28) (Supplemental Figure 4). A physiologic response to hypoxia in all cells was confirmed by upregulation of the HIF-1α target genes ADM and NDRG-1 (Supplemental Figure 4B)(3,29). Growth of each cell line in 1% FBS or 1% O2 ((Supplemental Figure 4A,4C) for up to 96 hours led to a reduction in cell proliferation, whereas p38 inhibition in the setting of low serum or oxygen abolished this effect, indicating that functional p38 restrains cell growth in conditions of cellular stress typical of the pancreatic cancer microenvironment.
p38 exerts its growth inhibitory effects via JNK
p38 has been shown to act in concert with JNK to coordinate responses to environmental stimuli or growth signals. However, whereas in some tissue types JNK activation has been shown to result in cytostasis, in others JNK may promote cell growth (4,23). To determine if JNK mediates the observed inhibition of cell growth by functional p38 in pancreatic cells, we first determined the extent to which JNK is active in these same cells by Western blot (Figure 3A). We found activated JNK (pJNK) in all cell lines, with exception of A2.1 and A6L. Of interest, Panc 2.5 and A13D cells that do not have functional p38 also expressed high levels of pJNK indicating activation of JNK may occur by more than one mechanism in pancreatic cancer, such as direct upstream activation by MAP2Ks or by AKT (5–7,30,31). However, pAKT was not expressed to any great extent in these pancreatic cancer cell lines indicating that this is not a common mechanism for JNK activation in this tumor type.
Figure 3. Effects of functional p38 MAPK on cell proliferation through the JNK pathway in pancreatic cancer cells.
(A) Western blotting for total and phosphorylated JNK and AKT expression in immortalized normal and pancreatic cancer cells. (B) Western blotting for total and phosphorylated JNK, MKK4 and MKK7 expression levels in Panc5.04, A10.7 and A38.44 cells after treatment with 10 µM of SB202190 for up to 2 days. Quantification of pJNK, pMKK4 and pMKK7 expression was shown as a ratio of normalized phosphorylated protein level in the SB202190 treated group to that of vehicle control. (C) Cell proliferation assays of Panc5.04, A10.7 and A38.44 cells treated with 10 µM of p38 MAPK inhibitor SB202190 alone or in combination with 10 µM of the JNK inhibitor SP600125. 0.1% (v/v) of DMSO was used as a vehicle treated control. *, p<0.05; **, p<0.01; ***, p<0.001 (treated group versus untreated control).
To determine if p38 inhibition influences JNK activity in pancreatic cancer cells, Panc 5.04, A10.7 and A38.44 cells were incubated with SB202190 and levels of pJNK determined. Shown in Figure 3B, inhibition of p38 resulted in increased pJNK levels in Panc 5.04, A10.7 and A38.44 cells by 24 hours of treatment, leading us to next assess the ability of JNK inhibition to counteract the effects of functional p38. Shown in Figures 3C and Supplemental Figure 5, pharmacologic inhibition of JNK by SP600125 in Panc 5.04, A10.7 and A38.44 cells neutralized the effects of p38 inhibition, supporting JNK as the mediator of p38 effects in pancreatic cancer cells. Because p38 may inhibit JNK though MKK4 and/or MKK7 (8,32), we next determined the expression levels of these two proteins after p38 inhibition. MKK7 phosphorylation levels increased in all three cell lines in association with p38 inhibition, whereas MKK4 levels only increased in Panc 5.04 and A38.44(Figure 3B). To confirm this possibility, we used an siRNA to knock down MKK7 expression in each cell line and repeated each MTT assay in the presence of the p38 inhibitor (Supplemental Figure 6). This indicated that MKK7 knockdown effectively abolished the effects of p38 inhibition in each cell line, similar to that seen for pharmacologic JNK inhibition (Figure 3C).
Finally, we determined the relationships of p38 and JNK expression to growth using a subcutaneous xenograft model (Figure 4). Similar to our findings in vitro, growth of xenografted Panc 5.04 or A10.7 cells with functional p38 expression was enhanced in mice treated with SB202190 (Figure 4A, 4B). By contrast, growth of these same xenografts was decreased in mice treated with SP600125. No change in growth was seen with either inhibitor in A2.1 or A6L cells consistent with their minimal levels of expression of pp38 and pJNK. Immunolabeling for pJNK in harvested xenografts revealed upregulation of pJNK in A10.7 cells treated with SP600125, whereas no differences in labeling were seen in A2.1 or A6L cells (Figure 5). Of interest, no viable tumor cells were found in the SP600125 treated Panc 5.04 xenograft tissues despite morphologic review of several step sections of these tissues. We cannot rule out that viable cells are present, only that they were likely few in number and could not easily be recognized in the setting of extensive fibrosis and inflammation.
Figure 4. Effects of p38 and JNK inhibition on pancreatic cancer cells in vivo.
(A) Representative subcutaneous xenografts of Panc 5.04, A10.7, A2.1 or A6L cell lines harvested from mice treated with vehicle (Veh), SB202190 or SP600125 for three weeks. (B) Summary data of xenograft tumor volumes at the end of the experiment. Values shown are the mean ± S.D. of six unique xenografts per cell line per condition. * p<0.05 (C). Western blots of total protein extracted from A10.7 xenografts after treatment with vehicle, SB202190 or SP600125. Quantification of pJNK, and p-p38 MAPK expression was shown as a ratio of normalized phosphorylated protein in the treated group to that of the vehicle control.
Figure 5. Histologic features of treated xenografts.
Hematoxylin and eosin or pJNK immunolabeling of representative xenografts from Panc 5.04, A10.7, A2.1 and A6L cells after treatment with vehicle or SB202190 or SP600125. Positive nuclear labeling is seen in Panc 5.04 and A10.7 cells treated with vehicle or SB202190 (images 600x). By contrast, abundant inflammation was seen in SP600125 treated Panc 5.04 cells (100x), and loss of nuclear labeling was seen in A10.7 (600x) Minimal to no pJNK expression was seen in A2.1 and A6L cells (400x). However, arrows in A2.1 indicate rare pJNK positive labeling nuclei.
DISCUSSION
In the future, comprehensive characterizations that merge clinical, pathologic and molecular data at the time of diagnosis of a pancreatic neoplasm will provide the greatest benefit to patients by allowing a personalization of their care (9,33). This study thus aims to fulfill that purpose by surveying a large series of biomarkers in a set of well-annotated discovery set of patient specimens. It is important to note that this biomarker list was not meant to be all inclusive but rather to consider major pathways and oncoproteins of significance for solid tumors including pancreatic cancer, a list that continues to grow (1,3). Our sample set is also not fully representative of all patients with this disease as it was based on postmortem tissues from patients at the extremes of metastatic failure (none to >100) encountered at autopsy. However, Winter et al used a similar approach based on a tissue microarray representing the survival extremes of resected patients which was nonetheless fruitful in identifying biomarkers of significance for this disease (4,10), as we now show for the p38-JNK pathway. Our approach also illustrates that biomarker studies of pancreatic cancer progression can be segregated into at least two categories: those that are consistently changed in expression with tumor progression (either gained or lost), and those that are heterogeneous in expression thereby highlighting subsets of pancreatic cancer with inherent differences in biology.
The significance of p38 and JNK signaling in cancer is highly dependent on the cellular context and the tissue of origin (11,23). Moreover, while the significance of this pathway has been elucidated in several human tumor types, data are limited in pancreatic cancer as most studies have typically focused on mechanistic studies of only one or two cell lines and have shown opposite effects (12,13,34–36). Handra-Luca used immunolabeling for pp38 in resected pancreatic cancer tissues, although in that study pp38 overexpression was associated with a worse recurrence free survival (14–17,37). Our findings now serve to clarify the role of p38 in pancreatic cancer by indicating it fulfills a tumor suppressive-type function through JNK inhibition, similar to that found in hematopoietic cells, fibroblasts and hepatocytes (18,32).
In a whole exome sequencing survey of 24 pancreatic cancers, Jones et al identified 12 core pathways that were recurrently targeted by somatic alteration in the majority of cancers analyzed, one of which was the JNK pathway (19,20,38). Thus, our data based on protein expression in an independent set of pancreatic cancer tissues not only validate the relevance of this pathway, but also indicate that the significance of p38 expression may be for its effects on JNK. However, while the genetic alterations observed by Jones et al served to identify the JNK pathway, the functional consequences of these genes on JNK signaling were not defined. For example, short term activation of JNK may promote cell survival (21,39), whereas long term activation may lead to increased apoptosis such as through the E3 ubiquitin ligase ITCH that promotes c-Jun degradation (23,40,41). Of interest, USP9x has recently been identified as a tumor suppressor in pancreatic cancer that also exerts it effects though regulation of ITCH expression (24,42), lending support to the notion that ITCH-dependent JNK signaling that leads to apoptosis, a consequence of chronic JNK activation by ongoing DNA damage (25,43), may be one of the cellular functions targeted.
Differential pp38 expression in pancreatic cancer may be due to underlying changes in expression of total p38, or to relative changes in activation (44). However, most cases in the current study robustly expressed total p38 by immunolabeling analyses of tissue or Western blot analyses of cell line extracts, suggesting phosphorylation levels as the underlying mechanism of p38 activity. p38 may be activated by the canonical MAP2Ks MKK3 or MKK6, both of which are expressed in pancreatic cancer, or by MKK4 that also activates JNK (44); it may undergo autophosphorylation as well although this mechanism appears specific to activated T-lymphocytes (45). However, p38 may also be dephosphorylated by a number of phosphatases thus providing an alternative mechanism for control of p38 activity and by extension JNK (44). This possibility is perhaps most relevant given reports of frequent loss of DUSP6 and MKP1 expression in pancreatic cancer cells and tissues (46,47).
Ultimately, elucidation of the role of p38 and JNK signaling in pancreatic cancer is most relevant for its potential value in the clinical setting. We now show that functional p38 signaling in pancreatic cancer is a predictive marker of outcome in resected pancreatic cancer. This is consistent with the findings by Koizumi et al who noted the necessity of p38 activity for gemcitabine-induced toxicity of pancreatic cancer cells (48), and that JNK inhibition suppressed pancreatic cancer cell growth in both in vitro assays and in a genetically engineered mouse model of pancreatic cancer (49), lending support to in human studies of JNK inhibition in the adjuvant setting and potentially in patients receiving first line therapy for newly diagnosed advanced stage disease as well (50).
Supplementary Material
STATEMENT OF TRANSLATIONAL RELEVENCE.
Pancreatic cancer is a highly lethal tumor type for which few therapeutic options exist, thus identification of pathways for targeting remains paramount. We used biomarker profiling of pancreatic cancer tissues to identify p38 MAPK as a predictive biomarker of longer overall survival in patients who undergo surgical resection, and validated this finding in in vitro and in vivo models. A potential mechanism by which p38 confers a survival advantage was identified as inhibition of MKK7 and its downstream target JNK. These results identify p38 MAPK and JNK signaling as a viable therapeutic target in pancreatic cancer for immediate use in clinical trial design.
Acknowledgments
Supported by National Institutes of Health grants CA140599, CA101955 and CA62924, The International Research Fund for Subsidy of Kyushu University School of Medicine Alumni, The Alfredo Scatena Memorial, The George Rubis Endowment for Pancreatic Cancer Research, The Michael Rolfe Pancreatic Cancer Foundation, Sigma Beta Sorority, The Joseph C. Monastra Foundation, The Gloria Swan Pancreatic Cancer Foundation, The Skip Viragh Pancreatic Cancer Center and The Patty Boshell Pancreas Cancer Foundation.
Footnotes
The authors have no financial conflicts of interest related to this work.
REFERENCES CITED
- 1.Stathis A, Moore MJ. Advanced pancreatic carcinoma: current treatment and future challenges. Nat Rev Clin Oncol. 2010;7:163–172. doi: 10.1038/nrclinonc.2009.236. [DOI] [PubMed] [Google Scholar]
- 2.Hidalgo M. Pancreatic cancer. N Engl J Med. 2010;362:1605–1617. doi: 10.1056/NEJMra0901557. [DOI] [PubMed] [Google Scholar]
- 3.Fong ZV, Winter JM. Biomarkers in pancreatic cancer: diagnostic, prognostic, and predictive. Cancer J. 2012;18:530–538. doi: 10.1097/PPO.0b013e31827654ea. [DOI] [PubMed] [Google Scholar]
- 4.Winter JM, Tang LH, Klimstra DS, Brennan MF, Brody JR, Rocha FG, et al. A novel survival-based tissue microarray of pancreatic cancer validates MUC1 and mesothelin as biomarkers. PLoS ONE. 2012;7:e40157. doi: 10.1371/journal.pone.0040157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kitamoto S, Yokoyama S, Higashi M, Yamada N, Takao S, Yonezawa S. MUC1 enhances hypoxia-driven angiogenesis through the regulation of multiple proangiogenic factors. Oncogene. 2012 doi: 10.1038/onc.2012.478. [DOI] [PubMed] [Google Scholar]
- 6.Behrens ME, Grandgenett PM, Bailey JM, Singh PK, Yi C-H, Yu F, et al. The reactive tumor microenvironment: MUC1 signaling directly reprograms transcription of CTGF. Oncogene. 2010;29:5667–5677. doi: 10.1038/onc.2010.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roy LD, Sahraei M, Subramani DB, Besmer D, Nath S, Tinder TL, et al. MUC1 enhances invasiveness of pancreatic cancer cells by inducing epithelial to mesenchymal transition. Oncogene. 2011;30:1449–1459. doi: 10.1038/onc.2010.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thomas AM, Santarsiero LM, Lutz ER, Armstrong TD, Chen Y-C, Huang L-Q, et al. Mesothelin-specific CD8(+) T cell responses provide evidence of in vivo cross-priming by antigen-presenting cells in vaccinated pancreatic cancer patients. J Exp Med. 2004;200:297–306. doi: 10.1084/jem.20031435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leao IC, Ganesan P, Armstrong TD, Jaffee EM. Effective depletion of regulatory T cells allows the recruitment of mesothelin-specific CD8 T cells to the antitumor immune response against a mesothelin-expressing mouse pancreatic adenocarcinoma. Clin Transl Sci. 2008;1:228–239. doi: 10.1111/j.1752-8062.2008.00070.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Embuscado EE, Laheru D, Ricci F, Yun KJ, de Boom Witzel S, Seigel A, et al. Immortalizing the complexity of cancer metastasis: genetic features of lethal metastatic pancreatic cancer obtained from rapid autopsy. Cancer Biol Ther. 2005;4:548–554. doi: 10.4161/cbt.4.5.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning. 2nd. New York: Springer Publishers; 2009. [Google Scholar]
- 12.Iacobuzio-Donahue CA, Fu B, Yachida S, Luo M, Abe H, Henderson CM, et al. DPC4 gene status of the primary carcinoma correlates with patterns of failure in patients with pancreatic cancer. J Clin Oncol. 2009;27:1806–1813. doi: 10.1200/JCO.2008.17.7188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yachida S, White C, Naito Y, Zhong Y, Brosnan JA, Macgregor-Das AM, et al. Clinical Significance of the Genetic Landscape of Pancreatic Cancer and Implications for Identification of Potential Long Term Survivors. Clin Cancer Res. 2012 doi: 10.1158/1078-0432.CCR-12-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Logsdon CD, Simeone DM, Binkley C, Arumugam T, Greenson JK, Giordano TJ, et al. Molecular profiling of pancreatic adenocarcinoma and chronic pancreatitis identifies multiple genes differentially regulated in pancreatic cancer. Cancer Res. 2003;63:2649–2657. [PubMed] [Google Scholar]
- 15.Iacobuzio-Donahue CA, Maitra A, Shen-Ong GL, van Heek T, Ashfaq R, Meyer R, et al. Discovery of novel tumor markers of pancreatic cancer using global gene expression technology. Am J Pathol. 2002;160:1239–1249. doi: 10.1016/S0002-9440(10)62551-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crnogorac-Jurcevic T, Efthimiou E, Nielsen T, Loader J, Terris B, Stamp G, et al. Expression profiling of microdissected pancreatic adenocarcinomas. Oncogene. 2002;21:4587–4594. doi: 10.1038/sj.onc.1205570. [DOI] [PubMed] [Google Scholar]
- 17.Han H, Bearss DJ, Browne LW, Calaluce R, Nagle RB, Hoff Von DD. Identification of differentially expressed genes in pancreatic cancer cells using cDNA microarray. Cancer Res. 2002;62:2890–2896. [PubMed] [Google Scholar]
- 18.Steinhardt AA, Gayyed MF, Klein AP, Dong J, Maitra A, Pan D, et al. Expression of Yes-associated protein in common solid tumors. Hum Pathol. 2008;39:1582–1589. doi: 10.1016/j.humpath.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Biankin AV, Kench JG, Colvin EK, Segara D, Scarlett CJ, Nguyen NQ, et al. Expression of S100A2 calcium-binding protein predicts response to pancreatectomy for pancreatic cancer. Gastroenterology. 2009;137:558-68-568.e1–558-68-568.e11. doi: 10.1053/j.gastro.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 20.Aloysius MM, Zaitoun AM, Awad S, Ilyas M, Rowlands BJ, Lobo DN. Mucins and CD56 as markers of tumour invasion and prognosis in periampullary cancer. Br J Surg. 2010;97:1269–1278. doi: 10.1002/bjs.7107. [DOI] [PubMed] [Google Scholar]
- 21.Mullinax JE, Hernandez JM, Toomey P, Villadolid D, Bowers C, Cooper J, et al. Survival after pancreatectomy for pancreatic adenocarcinoma is not impacted by performance status. Am J Surg. 2012;204:704–708. doi: 10.1016/j.amjsurg.2012.01.016. [DOI] [PubMed] [Google Scholar]
- 22.Regine WF, Winter KA, Abrams RA, Safran H, Hoffman JP, Konski A, et al. Fluorouracil vs gemcitabine chemotherapy before and after fluorouracil-based chemoradiation following resection of pancreatic adenocarcinoma: a randomized controlled trial. JAMA. 2008;299:1019–1026. doi: 10.1001/jama.299.9.1019. [DOI] [PubMed] [Google Scholar]
- 23.Wagner EF, Nebreda AR. Signal integration by JNK and p38 MAPK pathways in cancer development. Nat Rev Cancer. 2009;9:537–549. doi: 10.1038/nrc2694. [DOI] [PubMed] [Google Scholar]
- 24.Ono K, Han J. The p38 signal transduction pathway: activation and function. Cell Signal. 2000;12:1–13. doi: 10.1016/s0898-6568(99)00071-6. [DOI] [PubMed] [Google Scholar]
- 25.Ambrosino C, Nebreda AR. Cell cycle regulation by p38 MAP kinases. Biol Cell. 2001;93:47–51. doi: 10.1016/s0248-4900(01)01124-8. [DOI] [PubMed] [Google Scholar]
- 26.Olive KP, Jacobetz MA, Davidson CJ, Gopinathan A, McIntyre D, Honess D, et al. Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science. 2009;324:1457–1461. doi: 10.1126/science.1171362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Provenzano PP, Cuevas C, Chang AE, Goel VK, Hoff Von DD, Hingorani SR. Enzymatic targeting of the stroma ablates physical barriers to treatment of pancreatic ductal adenocarcinoma. Cancer Cell. 2012;21:418–429. doi: 10.1016/j.ccr.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jacobetz MA, Chan DS, Neesse A, Bapiro TE, Cook N, Frese KK, et al. Hyaluronan impairs vascular function and drug delivery in a mouse model of pancreatic cancer. Gut. 2013;62:112–120. doi: 10.1136/gutjnl-2012-302529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Semenza GL. Defining the role of hypoxia-inducible factor 1 in cancer biology and therapeutics. Oncogene. 2010;29:625–634. doi: 10.1038/onc.2009.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haeusgen W, Herdegen T, Waetzig V. The bottleneck of JNK signaling: molecular and functional characteristics of MKK4 and MKK7. Eur J Cell Biol. 2011;90:536–544. doi: 10.1016/j.ejcb.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 31.Vivanco I, Palaskas N, Tran C, Finn SP, Getz G, Kennedy NJ, et al. Identification of the JNK signaling pathway as a functional target of the tumor suppressor PTEN. Cancer Cell. 2007;11:555–569. doi: 10.1016/j.ccr.2007.04.021. [DOI] [PubMed] [Google Scholar]
- 32.Hui L, Bakiri L, Mairhorfer A, Schweifer N, Haslinger C, Kenner L, et al. p38alpha suppresses normal and cancer cell proliferation by antagonizing the JNK-c-Jun pathway. Nat Genet. 2007;39:741–749. doi: 10.1038/ng2033. [DOI] [PubMed] [Google Scholar]
- 33.Crane CH, Iacobuzio-Donahue CA. Keys to Personalized Care in Pancreatic Oncology. J Clin Oncol. 2012 doi: 10.1200/JCO.2012.45.1799. [DOI] [PubMed] [Google Scholar]
- 34.Ding XZ, Adrian TE. MEK/ERK-mediated proliferation is negatively regulated by P38 map kinase in the human pancreatic cancer cell line, PANC-1. Biochem Biophys Res Commun. 2001;282:447–453. doi: 10.1006/bbrc.2001.4595. [DOI] [PubMed] [Google Scholar]
- 35.Habiro A, Tanno S, Koizumi K, Izawa T, Nakano Y, Osanai M, et al. Involvement of p38 mitogen-activated protein kinase in gemcitabine-induced apoptosis in human pancreatic cancer cells. Biochem Biophys Res Commun. 2004;316:71–77. doi: 10.1016/j.bbrc.2004.02.017. [DOI] [PubMed] [Google Scholar]
- 36.Dreissigacker U, Mueller MS, Unger M, Siegert P, Genze F, Gierschik P, et al. Oncogenic K-Ras down-regulates Rac1 and RhoA activity and enhances migration and invasion of pancreatic carcinoma cells through activation of p38. Cell Signal. 2006;18:1156–1168. doi: 10.1016/j.cellsig.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 37.Handra-Luca A, Lesty C, Hammel P, Sauvanet A, Rebours V, Martin A, et al. Biological and prognostic relevance of mitogen-activated protein kinases in pancreatic adenocarcinoma. Pancreas. 2012;41:416–421. doi: 10.1097/MPA.0b013e318238379d. [DOI] [PubMed] [Google Scholar]
- 38.Jones S, Zhang X, Parsons DW, Lin JC-H, Leary RJ, Angenendt P, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ventura J-J, Hübner A, Zhang C, Flavell RA, Shokat KM, Davis RJ. Chemical genetic analysis of the time course of signal transduction by JNK. Mol Cell. 2006;21:701–710. doi: 10.1016/j.molcel.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 40.Chang L, Kamata H, Solinas G, Luo J-L, Maeda S, Venuprasad K, et al. The E3 ubiquitin ligase itch couples JNK activation to TNFalpha-induced cell death by inducing c-FLIP(L) turnover. Cell. 2006;124:601–613. doi: 10.1016/j.cell.2006.01.021. [DOI] [PubMed] [Google Scholar]
- 41.Gao M, Labuda T, Xia Y, Gallagher E, Fang D, Liu Y-C, et al. Jun turnover is controlled through JNK-dependent phosphorylation of the E3 ligase Itch. Science. 2004;306:271–275. doi: 10.1126/science.1099414. [DOI] [PubMed] [Google Scholar]
- 42.Perez-Mancera PA, Rust AG, van der Weyden L, Kristiansen G, Li A, Sarver AL, et al. The deubiquitinase USP9X suppresses pancreatic ductal adenocarcinoma. Nature. 2012;486:266–270. doi: 10.1038/nature11114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koorstra J-BM, Hong S-M, Shi C, Meeker AK, Ryu JK, Offerhaus GJA, et al. Widespread activation of the DNA damage response in human pancreatic intraepithelial neoplasia. Mod Pathol. 2009;22:1439–1445. doi: 10.1038/modpathol.2009.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cuadrado A, Nebreda AR. Mechanisms and functions of p38 MAPK signalling. Biochem J. 2010;429:403–417. doi: 10.1042/BJ20100323. [DOI] [PubMed] [Google Scholar]
- 45.Lajevic MD, Suleiman S, Cohen RL, Chambers DA. Activation of p38 mitogen-activated protein kinase by norepinephrine in T-lineage cells. Immunology. 2011;132:197–208. doi: 10.1111/j.1365-2567.2010.03354.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Furukawa T, Sunamura M, Motoi F, Matsuno S, Horii A. Potential tumor suppressive pathway involving DUSP6/MKP-3 in pancreatic cancer. Am J Pathol. 2003;162:1807–1815. doi: 10.1016/S0002-9440(10)64315-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu S, Furukawa T, Kanai N, Sunamura M, Horii A. Abrogation of DUSP6 by hypermethylation in human pancreatic cancer. J Hum Genet. 2005;50:159–167. doi: 10.1007/s10038-005-0235-y. [DOI] [PubMed] [Google Scholar]
- 48.Koizumi K, Tanno S, Nakano Y, Habiro A, Izawa T, Mizukami Y, et al. Activation of p38 mitogen-activated protein kinase is necessary for gemcitabine-induced cytotoxicity in human pancreatic cancer cells. Anticancer Res. 2005;25:3347–3353. [PubMed] [Google Scholar]
- 49.Takahashi R, Hirata Y, Sakitani K, Nakata W, Kinoshita H, Hayakawa Y, et al. Therapeutic effect of c-Jun N-terminal kinase inhibition on pancreatic cancer. Cancer Sci. 2012 doi: 10.1111/cas.12080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Plantevin Krenitsky V, Nadolny L, Delgado M, Ayala L, Clareen SS, Hilgraf R, et al. Discovery of CC-930, an orally active anti-fibrotic JNK inhibitor. Bioorg Med Chem Lett. 2012;22:1433–1438. doi: 10.1016/j.bmcl.2011.12.027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.