Abstract
Summary: The R/Bioconductor package Protein Array Analyzer (PAA) facilitates a flexible analysis of protein microarrays for biomarker discovery (esp., ProtoArrays). It provides a complete data analysis workflow including preprocessing and quality control, uni- and multivariate feature selection as well as several different plots and results tables to outline and evaluate the analysis results. As a main feature, PAA’s multivariate feature selection methods are based on recursive feature elimination (e.g. SVM-recursive feature elimination, SVM-RFE) with stability ensuring strategies such as ensemble feature selection. This enables PAA to detect stable and reliable biomarker candidate panels.
Availability and implementation: PAA is freely available (BSD 3-clause license) from http://www.bioconductor.org/packages/PAA/.
Contact: michael.turewicz@rub.de or martin.eisenacher@rub.de
1 Introduction
Protein microarrays (PMs) such as the ProtoArray by Thermo Fisher Scientific, Waltham, MA, USA, are used for autoimmune antibody screening studies, e.g. to discover biomarker candidate panels in human body fluids to discriminate two groups of samples (e.g. ‘diseased’ and ‘controls’). For ProtoArray data analysis the software Prospector is often used because it provides the advantageous univariate feature ranking approach minimum M statistic (mMs) (Love, 2007) and a ProtoArray-specific robust linear model normalization (rlm) (Sboner et al., 2009). However, since Prospector provides hardly any further functionality for biomarker discovery it is a quite limited tool (Turewicz et al., 2013). Therefore, we have adopted and extended Prospector's key features (mMs, rlm) and implemented PAA which provides a complete data analysis pipeline for ProtoArrays and other single color PMs.
2 PAA workflow
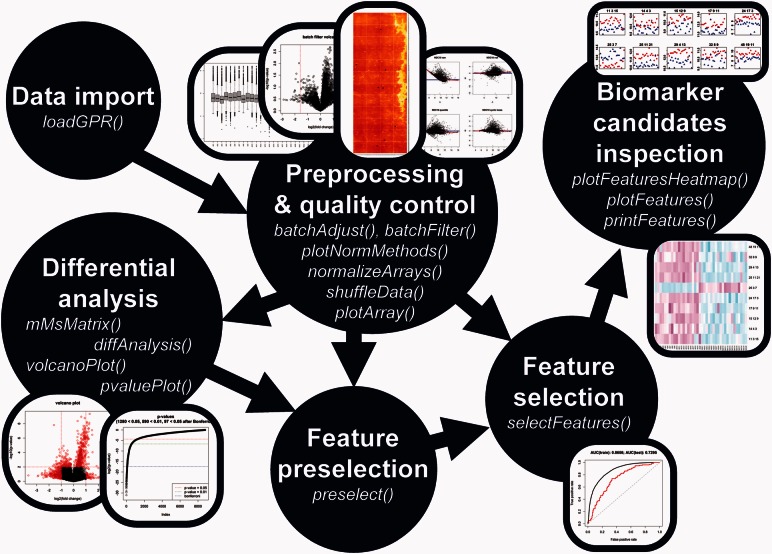
The adaptable PAA workflow consists of six parts (see Fig. 1) which are described in the following subsections.
Fig. 1.
The PAA workflow. The six parts of the PAA workflow including their specific function names and plots are shown. Each analysis begins with ‘data import’ and ends with ‘biomarker candidates inspection’
2.1 Data import
PAA imports microarray data in gpr file format. Therefore, it provides the function loadGPR which imports all needed data into an object of class EListRaw (Expression List). To load the desired files and pass metadata not contained in the gpr files (e.g. mapping between sample IDs and gpr files, batch information, clinical data, etc.) a so called targets file has to be created previously and provided to loadGPR. In case of ProtoArrays, spot duplicates are condensed by taking the smaller value or taking the mean after data import. Besides ProtoArrays, data of all one color microarrays in gpr file format (e.g. other PMs) can be imported.
2.2 Preprocessing and quality control
PAA provides several different preprocessing methods to make all PM intensity values inter- and intra-array-wise comparable. E.g. batch effects must be minimized when PMs from different manufacturing lots are compared in large studies (Turewicz et al., 2013). Therefore, PAA provides the function batchFilter to detect and discard differential features between PM manufacturing lots. Furthermore, the function batchAdjust can be used to adjust for known microarray batches. The function normalizeArrays provides several different normalization methods. E.g. the ProtoArray-specific rlm approach which uses specific control spots has been reimplemented for PAA. Briefly, the model
| (1) |
where is the measured spot signal in log2 scale (of array i, block j, feature k and replicate r), is the array effect, is the block effect, is the actual feature signal and is a random error () is fitted using robust regression to compute the corrected intensities via . Other normalization approaches provided by normalizeArrays are: cyclic loess, quantile and vsn. To assist in choosing an appropriate normalization method, PAA offers two functions: plotMAPlots drawing MA plots and plotNormMethods drawing box plots visualizing differences before and after normalization. For quality control, the function plotArray reconstructs the original spot positions from gpr files to draw a plot mimicking the original scan image and to visualize PMs for which no scan image is available. Then, visual inspection of the spatial intensity pattern can identify strong local tendencies and spatial biases. Moreover, PMs can be inspected after each preprocessing step in order to check the impact of the applied methods.
2.3 Differential analysis
PAA offers univariate biomarker discovery with fold change and P-value calculation via the functions diffAnalysis, pvaluePlot and volcanoPlot.
2.4 Biomarker candidate selection
Biomarker candidate selection via feature selection methods is the central task in computational biomarker discovery. Multivariate approaches based on embedded classifier algorithms model feature interdependencies, interact with the classifier and result in more accurate classifications than simpler strategies (Saeys et al., 2007). Hence, PAA comes with three recursive feature elimination (RFE) algorithms: (i) a reimplementation of SVM-RFE (Guyon et al., 2002) which utilizes the weights of linear SVMs; (ii) a similar RFE approach using Random Forests (RFs) (Jiang et al., 2004) called RF-RFE; (iii) an interface to RJ-RFE, the RFE method of the C ++ package Random Jungle (RJ) (Schwarz et al., 2010) which is a fast RF reimplementation. All three variants of RFE can be called via the function selectFeatures and are embedded in frequency-based feature selection (FFS) (Baek et al., 2009) and ensemble feature selection (EFS) (Abeel et al., 2010) which are strategies that ensure stable and reliable biomarker panels.
2.5 Feature preselection
Because RFE embedded in FFS or EFS are computationally expensive multivariate methods for large datasets (e.g. group sizes >30 each) it is often beneficial to reduce the number of variables beforehand. Therefore, PAA provides several univariate preselection methods via the function preselect. The default method is mMs (implemented in C ++ to improve run times) which provides a P-value based on an urn model (similar approach to the hypergeometric test). Besides mMs, PAA provides t test- and MRMR-based (Peng et al., 2005) preselection.
2.6 Biomarker candidates inspection
PAA returns various output for results evaluation. E.g. the plots returned by pvaluePlot and volcanoPlot visualize differential features from the univariate perspective. ROC curves and results files outlining the classification performance can be returned by selectFeatures. After feature selection the resulting biomarker candidate panel can be inspected. Therefore, PAA comes with three functions: (i) plotFeatures plots the fluorescence intensities of the selected biomarker candidates in group specific colors (one sub-figure per candidate) in order to visualize the differences; (ii) the selected panel and all related protein information can be saved via printFeatures into a txt file suitable for analysis with external tools (e.g. annotation); (iii) a heat map of the candidate panel can be plotted by plotFeaturesHeatmap as an alternative to plotFeatures.
3 Conclusion
PAA provides a comprehensive toolbox and an adaptable workflow for PM data analysis. It comprises the most important methods of Prospector and goes far beyond. Especially the multivariate feature selection based on RFE embedded in FFS or EFS, which is a cutting edge strategy for biomarker discovery, enables PAA to identify stable and reliable feature panels. Finally, PAA is flexible since the R/Bioconductor framework facilitates workflow extension and customization.
Funding
This work was supported by P.U.R.E., a project of Nordrhein-Westfalen, a federal state of Germany; and de.NBI, a project of the German Federal Ministry of Education and Research (BMBF) [grant number FKZ 031 A 534A].
Conflict of Interest: none declared.
References
- Abeel T. et al. (2010) Robust biomarker identification for cancer diagnosis with ensemble feature selection methods. Bioinformatics, 26, 392–398. [DOI] [PubMed] [Google Scholar]
- Baek S. et al. (2009) Development of biomarker classifiers from high-dimensional data. Brief Bioinform., 10, 537–546. [DOI] [PubMed] [Google Scholar]
- Guyon I. et al. (2002) Gene selection for cancer classification using support vector machines. Mach. Learn., 46, 389–422. [Google Scholar]
- Jiang H. et al. (2004) Joint analysis of two microarray gene-expression data sets to select lung adenocarcinoma marker genes. BMC Bioinformatics, 5, 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love B. (2007) The analysis of protein arrays In: Predki P.F. (ed). Functional Protein Microarrays in Drug Discovery. CRC Press, Boca Raton, pp. 381–402. [Google Scholar]
- Peng H. et al. (2005) Feature selection based on mutual information: criteria of max-dependency, max-relevance, and min-redundancy. IEEE Trans. Pattern Anal. Mach. Intell., 27, 1226–1238. [DOI] [PubMed] [Google Scholar]
- Saeys Y. et al. (2007) A review of feature selection techniques in bioinformatics. Bioinformatics, 23, 2507–2517. [DOI] [PubMed] [Google Scholar]
- Sboner A. et al. (2009) Robust-linear-model normalization to reduce technical variability in functional protein microarrays. J. Proteome Res., 8, 5451–5464. [DOI] [PubMed] [Google Scholar]
- Schwarz D.F. et al. (2010) On safari to Random Jungle: a fast implementation of Random Forests for high-dimensional data. Bioinformatics, 26, 1752–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turewicz M. et al. (2013) Improving the default data analysis workflow for large autoimmune biomarker discovery studies with protoarrays. Proteomics, 13, 2083–2087. [DOI] [PMC free article] [PubMed] [Google Scholar]