Abstract
Mitochondrial DNA (mtDNA) is strictly maternally inherited in metazoans. The major exception to this rule has been found in many bivalve species which allow the presence of different sex-linked mtDNA molecules. This mechanism, named doubly uniparental inheritance (DUI), is characterized by the presence of two mtDNAs: The female mtDNA is found in somatic tissue and female gonads, whereas the male mtDNA is usually found in male gonads and sperm. In this study we highlight the existence of two divergent mitochondrial haplotypes with a low genetic difference around 6–8% in Arctica islandica, a long-lived clam belonging to the Arcticidae, a sister group to the Veneridae in which DUI has been found. Phylogenetic analysis on cytochrome b and 16S sequences from somatic and gonadic tissues of clams belonging to different populations reveals the presence of the “divergent” type in male gonads only and the “normal” type in somatic tissues and female gonads. This peculiar segregation of divergent mtDNA types speaks for the occurrence of the DUI mechanism in A. islandica. This example also highlights the difficulties to assess the presence of such particular mitochondrial inheritance system and underlines the possible misinterpretations in phylogeographic and phylogenetic studies of bivalve species linked to the presence of two poorly differentiated mitochondrial genomes.
Keywords: doubly uniparental inheritance, mitochondrial DNA, Arcticidae, Arctica islandica, long-lived clam
Introduction
Mitochondria are the main generators of cellular energy in eukaryotes, and each mitochondrion possesses multiple copies of species-specific mitochondrial genome. Mitochondrial DNA (mtDNA), albeit circular, double-stranded and short (≈16.5 kb), is of crucial importance because genetic variation within the encoded sequence of mitochondrial protein subunits can have significant consequences for whole animal metabolism and fitness (Blier et al. 2001). mtDNA is maternally inherited and shows a high mutation rate, rendering it particularly interesting for population genetic and phylogeographic studies.
The doubly uniparental inheritance (DUI) system, described in more than 40 bivalve species, constitutes an exception to the commonly accepted rule of maternal mtDNA inheritance in animals. It is characterized by the existence of two distinct sex-associated mitochondrial genomes: The female (F-) mtDNA which is transmitted through the eggs to all offspring, and the male (M-) mtDNA which is exclusively present in the sperm. In female embryos, M-mtDNA is selectively degraded whereas in males this genome aggregates in one blastomere from the first division, destined to produce the male germline (Ghiselli et al. 2011; Milani et al. 2011). In the latter case, no M-genome degradation will occur as a male-specific open-reading frame (ORF) prevents the recognition of the M-type by the degradation machinery (Milani, Ghiselli, Maurizii, et al. 2014), or by the presence of a protein complex binding the M-mtDNA to protect it from degradation (Kyriakou et al. 2015). Thus, in adults, F-mtDNA is usually present in all tissues of either sex, except in sperm where the M-type-mtDNA remains predominant, leading to the generation of male heteroplasmic individuals (Zouros 2013; Kyriakou et al. 2015).
As the detection of DUI remains difficult, the probability of unrecognized cases remains high, obscuring the clear taxonomic classification of DUI across invertebrate clades. DUI is currently known from approximately 40 species in some but not all parts of the bivalve tree. Although a systematic study is still missing, DUI is not uncommon in the Mytiloidea with additional cases in Unionoidea, Veneroidea, and Nuculanoidea (Theologidis et al. 2008; Doucet-Beaupré et al. 2010; Boyle and Etter 2013; Zouros 2013; Plazzi et al. 2015). DUI is notably absent in hermaphroditic bivalves and may thus be incompatible with this reproductive strategy (Breton et al. 2011).
It is currently unclear whether the scattered appearance of DUI is the result of a single evolutionary event at the base of the bivalve tree, followed by its reduction in several branches, or whether it evolved independently many times (Doucet-Beaupré et al. 2010; Zouros 2013). Recent evidence provides plausible explanations for both scenarios. “Masculinization” events, that is, replacement of the M-mtDNA by the F-genome even in male gonads, have been observed in hybrids between Mytilus species and can explain the apparent repeated loss of the male mitochondrial genome in independent lineages (Zouros 2013). Recent evidence for the existence of DUI-regulating mitochondrial ORFs of viral origin supports the recurrent initiation of DUI (Milani, Ghiselli, Maurizii, et al. 2014, Plazzi et al. 2015).
Arctica islandica (Linnaeus 1767) belongs to the Heterodonta, order Veneroida and is the sole living representative of the Arcticidae family (Morton 2011). This species, known as the longest-lived non colonial species with a maximum reported life span of 507 years, is of great interest for studies of aging in marine non model species (Strahl and Abele 2010; Morton 2011; Strahl et al. 2011; Abele and Philipp 2013; Gruber et al. 2015). The different populations are widely distributed in the Northern hemisphere (from east coast of North America to the European North Atlantic around Iceland, north of Norway and into the White Sea). The confirmed range of distribution further extends south into the Dutch and German North Sea (NS) and the Western Baltic Sea (BS), with populations showing a broad spectrum of environmental adaptations in terms of salinity (20–35 PSU), temperature (0–19 °C), and oxygen concentrations (Dahlgren et al. 2000; Begum et al. 2010; Basova et al. 2012).
Recent sequencing of the entire mitochondrial genome did not report either the presence or the absence of DUI in A. islandica (Glöckner et al. 2013). A phylogeographic study on A. islandica mentioned one individual (out of 83 individuals tested) with at least two sharply distinct cytochrome b haplotype groups (haploype X in Dahlgren et al. 2000), which could be a first indicator of DUI in this species. Indeed the Arcticidae is a sister group to the Veneridae (Glöckner et al. 2013), a family in which several DUI species have been reported (Venerupis philippinarum, Meretrix lamarckii) whereas it is supposed to be absent in others (Callista chione, Mercenaria mercenaria, Ruditapes deccussatus, Venus verrucosa) (Passamonti and Scali 2001; Plazzi et al. 2015).
In this article, we report the discovery of two distinct mitochondrial haplotype groups with DUI-like inheritance with respect to sex and tissue specificity in NS and BS populations of the long-lived clam A. islandica.
Results
Both cytochrome b and 16S primers successfully amplified fragments from almost all the 117 DNA extracts (115 sequences for cytochrome b and 114 sequences for 16S) (supplementary table S1, Supplementary Material online) yielding fragment lengths of 274-bp sequences for cytochrome b and 624- or 498-bp sequences for 16S.
The alignments revealed the presence of two different sequence groups in both loci, which will be referred to as “normal” and “divergent” forms in this article, with the haplotype group closest to the already published sequences being assigned “normal” whereas sequences resembling the haplotype X (approximately 6% different from the other sequences) of Dahlgren et al. (2000) were termed “divergent.”
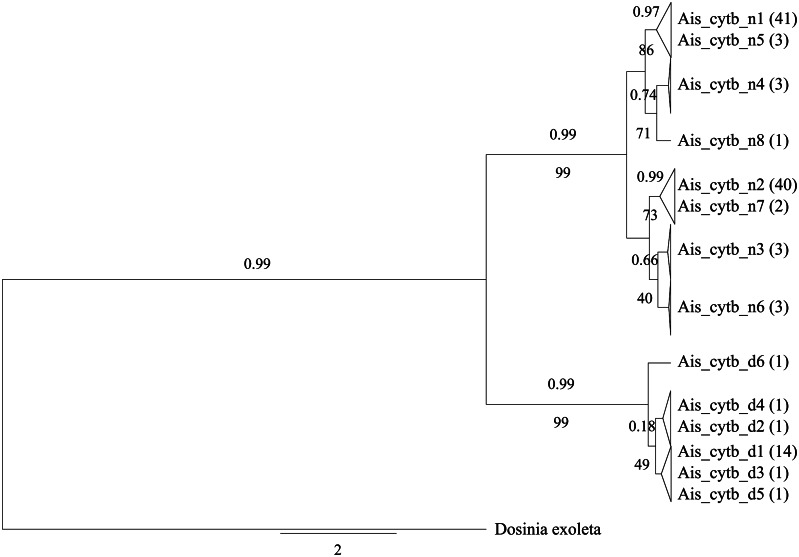
The “divergent” cytochrome b group consisted of 6 haplotypes clearly separated by 18 nucleotide substitutions from the other 8 haplotypes representing the “normal” group (fig. 1) and the replacement of 4 amino acids with no stop codons (table 1). The p-distance between the two groups (normal vs. divergent = 0.079 ± 0.014) was higher than the intragroup distances (normal = 0.011 ± 0.004; divergent = 0.003 ± 0.001).
Fig. 1.
Phylogenetic tree based on cytochrome b partial sequences (274 bp). Sequences on the same branch with a p-distance <0.005 are collapsed together and the name of the corresponding haplotype is indicated. Numbers above the branches indicate the posterior probability determined from the Bayesian Inference analysis, and numbers below the branches refer to the bootstrap value determined from the maximum-likelihood phylogeny. Dosinia exoleta cytochrome b sequence was used to root the tree.
Table 1.
Variable Positions between 12 Cytochrome b Haplotypes Sequenced from Different Tissues of Arctica islandica (six “divergent” and eight “normal” haplotypes).
| haplotypes | n | 3 | 12 | 33 | 42 | 48 | 57 | 60 | 64 | 69 | 75 | 90 | 111 | 114 | 123 | 132 | 144 | 154 | 157 | 159 | 160 | 165 | 171 | 172 | 198 | 201 | 240 | 241 | 243 | 255 | 273 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ais_cytb_n1 | 41 | A | A | T | T | G | T | C | G | G | G | G | C | T | C | T | T | T | C | G | G | G | T | A | A | T | C | C | G | A | T |
| Ais_cytb_n2 | 40 | . | . | C | . | . | . | . | . | . | . | . | T | . | . | . | C | . | T | . | . | A | . | . | . | C | . | . | . | . | . |
| Ais_cytb_n3 | 3 | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | A | . | . | . | C | . | . | . | . | . |
| Ais_cytb_n4 | 3 | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . | A | . | . |
| Ais_cytb_n5 | 3 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . |
| Ais_cytb_n6 | 3 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | A | A | . | . | C | . | . | . | . | . |
| Ais_cytb_n7 | 2 | . | . | C | . | . | . | . | . | . | . | . | T | . | . | . | C | . | T | . | . | A | . | . | . | . | . | . | . | . | . |
| Ais_cytb_n8 | 1 | . | . | C | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | A | . | . | . | . | . | . | . | . | . |
| Ais_cytb_d1 | 14 | G | . | . | C | A | C | . | A | A | A | T | . | . | T | C | C | C | G | A | A | . | . | G | G | . | T | T | . | . | C |
| Ais_cytb_d2 | 1 | G | . | . | C | A | C | T | A | A | A | T | . | . | T | C | C | C | G | A | A | . | . | G | G | . | T | T | . | . | C |
| Ais_cytb_d3 | 1 | G | . | . | G | A | C | . | A | A | A | T | . | . | T | C | C | C | G | A | A | . | . | G | G | . | T | T | . | . | C |
| Ais_cytb_d4 | 1 | G | . | . | C | A | C | T | A | A | A | T | . | . | T | C | G | C | G | A | A | . | . | G | G | . | T | T | A | . | C |
| Ais_cytb_d5 | 1 | G | . | . | C | A | C | . | A | A | A | A | . | . | T | C | C | C | G | A | A | . | . | G | G | . | T | T | . | . | C |
| Ais_cytb_d6 | 1 | G | G | . | C | A | . | . | A | A | A | T | . | . | T | C | C | C | G | A | A | . | . | G | G | . | T | T | . | . | C |
Note.—Numbers indicate the position along the 274-bp fragment. Numbers in bold indicate the first codon positions, the others are the third codon positions. Nucleotides in bold induce amino acids changes on the predicted protein.
The 16S was characterized by 9 “normal” and 10 “divergent” haplotypes, which were clearly separated by 27 fully diagnostic sites and the presence of a 127-bp indel. The Ais_16S_d4 haplotype from male BS animals deviated from the nucleotide substitution pattern shared by the other divergent haplotypes at position 11. Two “normal” haplotypes, Ais_16S_n4 and Ais_16S_n5 from female animals (from BS and NS, respectively), had a “divergent”-like substitution at positions 369 and 396, respectively (table 2). The “divergent” and “normal” 16S groups have an inter group p-distance of 0.059 ± 0.009, higher than the intra group distance (normal = 0.006 ± 0.002; divergent = 0.002 ± 0.0008). The cytochrome b and 16S sequences showed an average value of 65.4% and 64.4% A+T, respectively. Transition/transversion bias was R = 5.75 for cytochrome b and R = 7.38 for 16S.
Table 2.
Variable Positions between 19 16S Haplotypes Sequenced from Different Tissues of Arctica islandica (ten “divergent” and nine “normal” haplotypes).
| haplotypes | n | 4 | 6 | 7 | 11 | 32 | 37 | 47 | 65 | 70 | 135 | 144 | 155 | 168 | 185 | 232 | 237 | 240-366 | 368 | 369 | 374 | 376 | 377 | 383 | 388 | 394 | 395 | 396 | 397 | 399 | 400 | 402 | 406 | 409 | 410 | 435 | 439 | 443 | 445 | 446 | 450 | 467 | 485 | 525 | 536 | 597 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ais_16S_n1 | 39 | A | T | A | G | T | C | T | G | G | G | G | G | T | G | A | A | - | T | C | T | A | T | G | A | G | A | C | T | C | T | A | A | A | A | A | T | G | G | A | G | A | A | T | G | A |
| Ais_16S_n2 | 37 | . | C | . | . | . | . | . | . | . | . | . | . | C | . | . | . | - | . | . | . | . | . | . | G | . | . | . | . | . | . | G | . | . | . | . | C | . | . | . | . | . | . | C | . | . |
| Ais_16S_n3 | 6 | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | - | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . |
| Ais_16S_n4 | 3 | . | C | . | . | . | . | . | . | . | . | . | . | C | . | . | . | - | . | T | . | . | . | . | G | . | . | . | . | . | . | G | . | . | . | . | C | . | . | . | . | . | . | C | . | . |
| Ais_16S_n5 | 3 | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | - | . | . | . | . | . | . | G | . | . | T | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . |
| Ais_16S_n6 | 3 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | - | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | . | . | . | . | . |
| Ais_16S_n7 | 2 | . | C | . | . | . | . | . | . | . | . | . | . | C | . | . | . | - | . | . | C | . | . | . | G | . | . | . | . | . | . | G | . | . | . | . | C | . | . | . | . | . | . | C | . | . |
| Ais_16S_n8 | 2 | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | - | . | . | . | . | . | . | G | . | . | . | . | . | . | G | . | . | . | . | C | . | . | . | . | . | . | C | . | . |
| Ais_16S_n9 | 1 | . | . | G | . | . | . | . | . | . | . | . | . | C | . | . | . | - | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | C | . | . |
| Ais_16S_d1 | 7 | G | . | . | A | G | T | A | A | A | . | . | A | C | A | . | . | 127 bp indel | . | T | . | G | A | A | G | A | C | T | G | T | . | G | - | G | G | G | C | . | A | G | . | G | G | C | A | . |
| Ais_16S_d2 | 3 | G | . | . | A | G | T | A | A | A | A | . | A | C | A | . | . | 127 bp indel | . | T | . | G | A | A | G | A | C | T | G | T | . | G | - | G | G | G | C | . | A | G | . | G | G | C | A | . |
| Ais_16S_d3 | 1 | G | . | . | A | G | T | A | A | A | A | . | A | C | A | . | . | 127 bp indel (pos 344: G) | . | T | . | G | A | A | G | A | C | T | G | T | . | G | - | G | G | G | C | . | A | G | . | G | G | C | A | G |
| Ais_16S_d4 | 1 | G | . | . | . | G | T | A | A | A | . | . | A | C | A | . | . | 127 bp indel | . | T | . | G | A | A | G | A | C | T | G | T | . | G | - | G | G | G | C | . | A | G | . | G | G | C | A | . |
| Ais_16S_d5 | 1 | G | . | . | A | G | T | A | A | A | . | . | A | C | A | . | . | 127 bp indel | . | T | . | G | A | A | G | A | C | T | G | T | C | G | - | G | G | G | C | . | A | G | . | G | G | C | A | . |
| Ais_16S_d6 | 1 | G | . | . | A | G | T | A | A | A | A | . | A | C | A | . | . | 127 bp indel | . | T | . | G | A | A | G | A | C | T | G | T | . | G | - | G | G | G | C | A | A | G | . | G | G | C | A | . |
| Ais_16S_d7 | 1 | G | . | . | A | G | T | A | A | A | . | A | A | C | A | G | G | 127 bp indel | . | T | . | G | A | A | G | A | C | T | G | T | . | G | - | G | G | G | C | . | A | G | . | G | G | C | A | . |
| Ais_16S_d8 | 1 | T | . | . | A | G | T | A | A | A | . | . | A | C | A | . | . | 127 bp indel | . | T | . | G | A | A | G | A | C | T | G | T | . | G | - | G | G | G | C | . | A | G | . | G | G | C | A | . |
| Ais_16S_d9 | 1 | G | . | . | A | G | T | A | A | A | A | . | A | C | A | . | . | 127 bp indel | C | T | . | G | A | A | G | A | C | T | G | T | . | G | - | G | G | G | C | . | A | G | . | G | G | C | A | . |
| Ais_16S_d10 | 1 | G | . | . | A | G | T | A | A | A | . | . | A | C | A | . | . | 127 bp indel (pos 354: A) | . | T | . | G | A | A | G | A | C | T | G | T | . | G | - | G | G | G | C | . | A | G | . | G | G | C | A | . |
Note.—Numbers indicate the position along the longest fragment (624 bp).
Phylogenetic analysis of partially sequenced cytochrome b and 16S was conducted using Maximum-Likelihood and Bayesian Inference methods and showed a similar overall topology (figs. 1 and 2; supplementary figs. S1–S4, Supplementary Material online). Briefly, “divergent” and “normal” haplotypes formed different intraspecific clades for both genes, strongly supported by a bootstrap value of 99 and a posterior probability of 0.99 (figs. 1 and 2). For 16S, these phylogenetic analyses were repeated on the same sequences with the 127-bp indel replaced by a single nucleotide to treat the indel as a single insertion/deletion event rather than an accumulation of events affecting single nucleotides. In both cases, the inferred tree topology using both methods was similar, with a strong separation between both haplotype groups (supplementary figs. S5 and S6, Supplementary Material online).
Fig. 2.
Phylogenetic tree based on 16S partial sequences (498–624 bp). Sequences on the same branch with a p-distance <0.005 are collapsed together and the corresponding haplotype name is indicated. Numbers above the branches indicate the posterior probability determined from the Bayesian Inference analysis, and numbers below the branches refer to the bootstrap value estimated from the maximum-likelihood phylogeny. Dosinia exoleta was used as outgroup.
“Divergent” cytochrome b and 16S haplotypes always co/occurred in the same DNA extracts. These “divergent” forms were only present in male gonadic tissue (supplementary table S1, Supplementary Material online). An attentive analysis of the sequences amplified from the male gonadic tissue DNA extracts also showed rare cases of co-amplification of the “divergent” and the “normal” forms (cloning results; data not shown). Somatic tissues of both sexes and female gonads exclusively carried “normal” cytochrome b and 16S forms. Except for one female, all animals featured the same “normal” haplotype for the two studied genes in all somatic tissue within each individual (supplementary table S1, Supplementary Material online). Finally, both “normal” and “divergent” groups for cytochrome b and 16S partial sequences were present in BS and NS populations. The presence of one or the other genome type is only linked to sex and tissue specificity, refuting the possibility that our data indicate existence of cryptic species (supplementary fig. S7 and table S1, Supplementary Material online).
Discussion
The existence of the DUI pathway for mitochondrial genomes can only be proved by 1/ the detection of heteroplasmic individuals carrying male and female mtDNA forms (Stewart et al. 1995; Passamonti and Scali 2001; Theologidis et al. 2008; Boyle and Etter 2013; Zouros 2013, and references therein) and, 2/ the in vivo localization of M-mtDNA in the male germline due to the aggregation of the male mitochondria during the embryonic development (Milani et al. 2011). Our phylogenetic and distance/based analyses of two partial mitochondrial sequences stemming from three different tissues highlight the coexistence of distinct mtDNA forms in A. islandica named “normal” and “divergent.” Our data demonstrate that the allelic linkage of the two loci is absolute, that is, the cytochrome b “divergent” allele is always associated with the “divergent” allele in 16S and likewise for the “normal” alleles in the two loci. It suggests that the two mitochondrial genomes are evolving as independent entities with no recombination acting to break the tight allelic association. Females possess only the “normal” mitochondrial genome across all tissue types whereas males also feature a “divergent” mtDNA exclusively in the gonads, genomes that can be linked to the F- and M-types mtDNA, respectively. The co-amplification of both forms in male gonadic tissue (detected by the presence of multiple sequencing peaks and the amplification of both sequence types in our cloning results) can be explained by the presence of nonreproductive cells in male gonads that may express the F-mtDNA (Passamonti and Scali 2001; Theologidis et al. 2008). Thus, our data are the first evidence for the existence of two distinct and sex-linked mitochondrial genomes that are inherited in a DUI fashion in the long-lived clam A. islandica in NS and BS (supplementary fig. S7 and table S1, Supplementary Material online). This study highlights another species from the order Veneroida inside the bivalves in which the DUI system is revealed (Passamonti and Scali 2001; Plazzi et al. 2015). It also underlines the difficulties to ascertain the presence of this particular sex-related mitochondrial genome inheritance within the bivalve evolutionary tree, which is still poorly sampled with respect to DUI (Theologidis et al. 2008; Plazzi et al. 2015).
The two mitochondrial lineages highlighted in A. islandica show a relatively low level of sequence divergence, between 6% and 8% for 16S and cytochrome b, respectively, compared with the value obtained in other DUI species belonging to other bivalves superfamilies, for example, in Mytilus galloprovincialis (22%; Mizi et al. 2005; Zouros 2013), Ledella ultima (27%; Boyle and Etter 2013), Tapes philippinarum (15–33%; Passamonti et al. 2003), Donax truculus (37%; Theologidis et al. 2008; Boyle and Etter 2013), or Inversidens japanensis (50%; Doucet-Beaupré et al. 2010). This divergence between sequences is in part caused by the presence of a 127-bp-long indel in 16S, a situation already described in Unionids with the 120-bp extension of the cox2 genes at the 3′-end (review in Zouros 2013), and the existence of specific synonymous and nonsynonymous mutations in cytochrome b, leading to the substitution of four amino acids in the predicted protein. Arctica thus represents the lowest divergence between M- (divergent) and F- (normal) mtDNAs among DUI species known to date. Possible explanations for the comparatively low differentiation are: 1/ a “masculinization” event replacing the male genome by a female one erasing all previous records of the “divergent” genome (Hoeh et al. 1996; Zouros 2013); 2/ a recent separation of “divergent” and “normal” lineages, implying a DUI origin event particular to A. islandica, and therefore the occurrence of several independent DUI origins within the Heterondonta subclass (Plazzi et al. 2015); or 3/ a reduced mutation rate in A. islandica which can be due to a co-adaptation between the nuclear and the mitochondrial genomes (Blier et al. 2001) required to allow this long-lived bivalve to deal with contrasting environmental conditions in the NS and the BS (Dahlgren et al. 2000; Begum et al. 2010).
The “divergent” M-type was already found in A. islandica in adductor muscle from two individuals from Northern Scotia and Iceland. Dahlgren et al. (2000) described the “X” cytochrome b haplotype, which was the most distantly related haplotype in their study (6.8% compared with the nearest neighbor and identical to the one named “Ais_cytb_d1” in our study; see table 1) but did not discuss this haploype further in their article. This example highlights that a co-amplification of the M-genome can be misinterpreted as a highly differentiated ortholog marker and potentially misleads phylogeographic and phylogenetic studies. This problem is exacerbated when the two genomes are poorly differentiated or if for some reason the “normal” F-genome fails to amplify (Theologidis et al. 2008; Plazzi et al. 2015).
The “normal” F-mtDNAs variability tends to be higher compared with the “divergent” M-mtDNAs, which differs with most of the previous studies on DUI system (Zouros 2013, and references therein). Passamonti (2007) described the same situation in Musculista senhousia and proposed, among other hypotheses, that the M-genome can experience a strongest selection pressure. It contrasts to the common statement suggesting that the M-genome is under a relaxed selection, leading to a higher diversity compared with the F-genome (Stewart et al. 1996). The F-mtDNA ensures energy production in a lot of different tissues (all tissues in female and somatic in male), whereas the M-type influence is restricted to the sperm function, probably only during late stage of spermatogenesis (Zouros 2013; Kyriakou et al. 2015). In this study, the divergence within groups was estimated using partial sequences, leading to the reduction of the number of rare variants detected, and so, an unavoidable underestimation of the real mtDNA variability, which can only be estimated by sequencing the entire mitochondrial genome (Theologidis et al. 2008; Ghiselli et al. 2013; Plazzi et al. 2015). Considering this technical limitation, the difference of variability within each sex has to be taken with caution, and the common statement of a relaxed selection on the M-genome reconsidered (Passamonti 2007; Plazzi et al. 2015). A M-type under relaxed selection might affect sperm energetic production and motility (Jha et al. 2008; Breton et al. 2009), which might then reduce male fertility and endanger the viability of the population (Ghiselli et al. 2013).
So far, the DUI of mitochondrial genome remains exclusive to the bivalve clade. However, the technical limitations for DUI detection (see above), associated with a poorly sampled but highly diverse evolutionary tree, do not permit to ascertain the occurrence, the origin (unique or multiple), or the mechanisms controlling this particular mtDNA inheritance system within the bivalves (Theologidis et al. 2008; Zouros 2013; Milani, Ghiselli, Maurizii, et al. 2014; Kyriakou et al. 2015; Plazzi et al. 2015). A coupling between the presence of the M-genome and maleness is clearly established even if the characteristic of the link, causative or associative, remains under debate (Milani et al. 2013; Zouros 2013; Kyriakou et al. 2015). Thus, Breton et al. (2011) suggested that DUI is absent in hermaphroditic Unionidae and Margaritiferidae, these species carrying an F-like mitochondrial genome only. However, rare cases of hermaphroditism were revealed in natural populations of A. islandica (Mann 1982; Thorarinsdottir 2000) and sex change (sequential hermaphroditism) was proposed to support high longevity (Abele and Philipp 2013). It suggests that the close link between sex determination and M-mtDNA makes the conservation of DUI in hermaphrodite individuals difficult but not impossible (Zouros 2013). Closer investigations on these particular hermaphroditic individuals belonging to a DUI species could be promising to unravel the function of the M- and F-genomes with respect to sex determination. Furthermore, previous studies showed that the M-genome strongly influences the motility (Jha et al. 2008) and the COX activity of Mytilus sperm (Breton et al. 2009). The M-genome can also be detected and transcribed in somatic tissue of R. philippinarum (Passamonti and Scali 2001; Milani, Ghiselli, Iannello, et al. 2014) suggesting that it might impact the cellular energetic homeostasis and physiology. The M-genome may therefore affect metabolism outside the sperm cell in some circumstances, thus creating a unique opportunity to study the compatibility between nucleus and mitochondrial genomes and the implication of mtDNA change occurring under natural conditions on bivalve physiology and ecology.
To conclude, we provide strong evidence of the existence of the DUI system in A. islandica with the description of a new “divergent” male genome specifically to male gonadic tissue. Because DUI was found in A. islandica, the sole representative of a family with a fossil history going back to the Cretaceous (Morton 2011), and in other species of the super class Autolamellibranchia, it suggests that this particular mtDNA inheritance system may represent an ancient mechanism that appeared more than 400 Ma (review in Zouros 2013). This study also highlights the difficulties to determine the presence of such particular but still not completely decrypted mitochondrial inheritance mechanisms, yet essential for the reproduction of these species. Further investigations of the DUI mechanism will permit a better understanding of the crucial role played by the mtDNA with respect to cellular physiology and organisms’ adaptive capacities.
Materials and Methods
Origin of Samples
Mature A. islandica individuals were collected in July 2014 in the BS in Kiel Bay and in August 2014 in NS around Helgoland (see Begum et al. 2010 for the sample location map). They were transported in cooling boxes at 10 °C to the Alfred Wegener Institute Helmholtz Center for Polar and Marine Research in Bremerhaven and held in 60-l aquaria with recirculating water and weekly fed with Nannochloropsis algae (PhytoMaxx, Germany). Water parameters were adjusted to match the natural conditions at the collection site, that is, 7 °C and 20 PSU for the BS individuals and 7 °C and 33 PSU for the NS clams. Each individual was sexed through direct light microscopic inspection of gonads for the presence of mature eggs or sperm. The animals were dissected (BS animals n = 13; NS animals n = 26) and tissues (mantle, foot, gonads) frozen in liquid nitrogen.
DNA Extraction, Polymerase Chain Reaction Amplification, and Sequencing
Total DNA was extracted from gonads, mantle, and foot using the Qiagen DNeasy Blood & Tissue Kit, quantified using Nanodrop D-1000 and stored at −80 °C.
Partial cytochrome b sequence amplifications were carried out in 15 µl reaction volume comprising 1.5 µl 10× Taq buffer (5Prime), 0.3 µl dNTP mix (10 mM), 0.075 µl of each primer (100 µM, forward primer: 5′-TAA TAA TTG CTT TCA CTG G-3′, reverse primer: 5′-CTA TTA TAA TAA AAA TTC CAA CTA G-3′), 0.09 µl Hot Master Taq DNA polymerase (5 U/µl; 5Prime, Hamburg, Germany), and 2 µl DNA extract (10 ng/µl). Reactions were performed on Eppendorf MasterCycler with the following polymerase chain reaction (PCR) conditions: 2 min at 94 °C initial denaturation, 38 cycles of denaturation for 20 s at 94 °C, annealing for 20 s at 52 °C, and extension for 50 s at 65 °C. The amplification was completed with a final extension (65 °C, 8 min) according to the manufacturer’s instructions.
The universal 16S primers 16SAR (5′-CGCCTGTTTATCAAAAACAT-3′) and 16SAB (5′-CCGGTCTGAACTCAGATCACG-3′) yielded different fragment from total DNA extraction (Palumbi 1996). PCR was conducted in 15 µl reaction volume comprising 1.5 µl 10× Taq buffer (5Prime), 0.3 µl dNTP mix (10 mM), 0.075 µl of each primer (100 µM), 0.15 µl betain (5 M), 0.09 µl Hot Master Taq DNA polymerase (5 U/µl; 5Prime), and 2 µl DNA extract (10 ng/µl). Reactions were performed on Eppendorf MasterCycler following the PCR conditions: 2 min at 94 °C, followed by 36 cycles of denaturation for 20 s at 94 °C, annealing for 10 s at 52 °C, and extension for 50 s at 65 °C. The amplification was completed with a final extension (65 °C, 8 min) according to the manufacturer’s instructions.
All PCR products were sent to Eurofins Genomics GmbH (Ebersberg, Germany) to be sequenced on both strands using the primers that were used during amplification.
DNA Cloning and Sequencing
The amplified products from male gonads that revealed a co-amplification of different variants were cloned using the TOPO TA Cloning Kit (Invitrogen) to confirm the presence of each form. Recombinant clones were sent to Eurofins Genomics GmbH to be sequenced using M13 universal primers (M13 Forward: 5′-GTAAAACGACGGCCAG-3′; M13 reverse: 5′-CAGGAAACAGCTATGAC-3′).
Sequence Analysis
Cytochrome b and 16S sequences were aligned using the ClustalW algorithm of CodonCode Aligner program (version 4.2.5; CodonCode Corporation, Dedham, MA). Amino acid composition for cytochrome b was deduced using the invertebrate mtDNA genetic code. MEGA 6 (version6.06; Tamura et al. 2013) was used to calculate p-distance between sequences (with bootstrap analysis 1,000 replicates) and estimate the transition/transversion ratio. Maximum likelihood (using MEGA 6) with bootstrap analysis (1,000 replicates) and Bayesian phylogenies (using BEAST, version 1.8.2; Drummond and Rambaut 2007) were performed on cytochrome b and 16S sequences with Dosinia exoleta (Bivalvia, Veneridae, GenBank accession number: cytochrome b—GQ166609.1; 16S—JF808184.1) as outgroup (closest sequence according to BLAST [Basic Local Alignment Search Tool] search) using Hasegawa–Kishino–Yano with Invariable site nucleotide substitution model as determined by jModelTest2 (version 1.6; Guindon and Gascuel 2003; Darriba et al. 2012). FigTree (version 1.4.2; Rambaut and Drummond 2012) was used to edit the phylogenetic tree for better readability. Sequences on the same branch with a p-distance <0.005 were collapsed into a single group. DNA haplotype networks for both genes were realized using Haplotype Viewer (Ewing 2012).
Supplementary Material
Supplementary files S1 and S2, figures S1–S7, and table S1 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors thank Dr Stephanie Ismar and Michael Janke for providing Arctica islandica specimens and Andrea Eschbach for her technical support. This study was financially supported by the Alexander von Humboldt foundation (http://www.humboldt-foundation.de) to C.D.
References
- Abele D, Philipp E. 2013. Environmental control and control of the environment: the basis of longevity in bivalves. Gerontology 59(3):261–266. [DOI] [PubMed] [Google Scholar]
- Basova L, Begum S, Strahl J, Sukhotin A, Brey T, Philipp E, Abele D. 2012. Age dependent patterns of antioxidants in Arctica islandica from six regionally separate populations with different life spans. Aquat Biol. 14:141–152. [Google Scholar]
- Begum S, Basova L, Heilmayer O, Philipp EE, Abele D, Brey T. 2010. Growth and energy budget models of the bivalve Arctica islandica at six different sites in the Northeast Atlantic realm. J Shellfish Res. 29(1):107–115. [Google Scholar]
- Blier PU, Dufresne F, Burton RS. 2001. Natural selection and the evolution of mtDNA-encoded peptides: evidence for intergenomic co-adaptation. Trends Genet. 17(7):400–406. [DOI] [PubMed] [Google Scholar]
- Boyle EE, Etter RJ. 2013. Heteroplasmy in a deep-sea protobranch bivalve suggests an ancient origin of doubly uniparental inheritance of mitochondria in Bivalvia. Mar Biol. 160(2):413–422. [Google Scholar]
- Breton S, Stewart DT, Blier PU. 2009. Role-reversal of gender-associated mitochondrial DNA affects mitochondrial function in Mytilus edulis (Bivalvia: Mytilidae). J Exp Zool B Mol Dev Evol. 312(2):108–117. [DOI] [PubMed] [Google Scholar]
- Breton S, Stewart DT, Shepardson S, Trdan RJ, Bogan AE, Chapman EG, Ruminas AJ, Piontkivska H, Hoeh WR. 2011. Novel protein genes in animal mtDNA: a new sex determination system in freshwater mussels (Bivalvia: Unionoida)? Mol Biol Evol. 28(5):1645–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlgren TG, Weinberg JR, Halanych KM. 2000. Phylogeography of the ocean quahog (Arctica islandica): influences of paleoclimate on genetic diversity and species range. Mar Biol. 137(3):487–495. [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9(8):772–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucet-Beaupré H, Breton S, Chapman EG, Blier PU, Bogan AE, Stewart DT, Hoeh WR. 2010. Mitochondrial phylogenomics of the Bivalvia (Mollusca): searching for the origin and mitogenomic correlates of doubly uniparental inheritance of mtDNA. BMC Evol Biol. 10(1):50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 7(1):214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing GB. 2012. Haplotype viewer. Vienna (Austria): Center for Integrative Bioinformatics Vienna [accessed 2015 Apr]. Available from: http://www.cibiv.at/∼greg/haploviewer [Google Scholar]
- Ghiselli F, Milani L, Guerra D, Chang PL, Breton S, Nuzhdin SV, Passamonti M. 2013. Structure, transcription, and variability of metazoan mitochondrial genome: perspectives from an unusual mitochondrial inheritance system. Genome Biol Evol. 5(8):1535–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiselli F, Milani L, Passamonti M. 2011. Strict sex-specific mtDNA segregation in the germ line of the DUI species Venerupis philippinarum (Bivalvia: Veneridae). Mol Biol Evol. 28(2):949–961. [DOI] [PubMed] [Google Scholar]
- Glöckner G, Heinze I, Platzer M, Held C, Abele D. 2013. The mitochondrial genome of Arctica islandica; Phylogeny and variation. Plos One 8(12):e82857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber H, Wessels W, Boynton P, Xu J, Wohlgemuth S, Leeuwenburgh C, Qi W, Austad SN, Schaible R, Philipp EE. 2015. Age-related cellular changes in the long-lived bivalve Arctica islandica. Age 37(5):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 52(5):696–704. [DOI] [PubMed] [Google Scholar]
- Hoeh WR, Stewart DT, Sutherland BW, Zouros E. 1996. Multiple origins of gender-associated mitochondrial DNA lineages in bivalves (Mollusca: Bivalvia). Evolution 50(6):2276–2286. [DOI] [PubMed] [Google Scholar]
- Jha M, Côté J, Hoeh WR, Blier PU, Stewart DT. 2008. Sperm motility in Mytilus edulis in relation to mitochondrial DNA polymorphisms: implications for the evolution of doubly uniparental inheritance in bivalves. Evolution 62(1):99–106. [DOI] [PubMed] [Google Scholar]
- Kyriakou E, Kravariti L, Vasilopoulos T, Zouros E, Rodakis GC. 2015. A protein binding site in the M mitochondrial genome of Mytilus galloprovincialis may be responsible for its paternal transmission. Gene 562(1):83–94. [DOI] [PubMed] [Google Scholar]
- Mann R. 1982. The seasonal cycle of gonadal development in Arctica islandica from the southern New England shelf. Fish Bull. 80(2):315–326. [Google Scholar]
- Milani L, Ghiselli F, Maurizii MG, Passamonti M. 2011. Doubly uniparental inheritance of mitochondria as a model system for studying germ line formation. PLoS One 6(11):e28194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani L, Ghiselli F, Nuzhdin SV, Passamonti M. 2013. Nuclear genes with sex bias in Ruditapes philippinarum (Bivalvia, Veneridae): mitochondrial inheritance and sex determination in DUI species. J Exp Zool B Mol Dev Evol. 320(7):442–454. [DOI] [PubMed] [Google Scholar]
- Milani L, Ghiselli F, Iannello M, Passamonti M. 2014. Evidence for somatic transcription of male-transmitted mitochondrial genome in the DUI species Ruditapes philippinarum (Bivalvia: Veneridae). Curr Genet. 60(3):163–173. [DOI] [PubMed] [Google Scholar]
- Milani L, Ghiselli F, Maurizii MG, Nuzhdin SV, Passamonti M. 2014. Paternally transmitted mitochondria express a new gene of potential viral origin. Genome Biol Evol. 6(2):391–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizi A, Zouros E, Moschonas N, Rodakis GC. 2005. The complete maternal and paternal mitochondrial genomes of the Mediterranean mussel Mytilus galloprovincialis: implications for the doubly uniparental inheritance mode of mtDNA. Mol Biol Evol. 22(4):952–967. [DOI] [PubMed] [Google Scholar]
- Morton B. 2011. The biology and functional morphology of Arctica islandica (Bivalvia: Arcticidae)—a gerontophilic living fossil. Mar Biol Res. 7(6):540–553. [Google Scholar]
- Palumbi SR. 1996. Nucleic acids II: the polymerase chain reaction. In: Hillis DM, Moritz C, Mable BK, editors. Molecular Systematics. p. 205–247. [Google Scholar]
- Passamonti M. 2007. An unusual case of gender-associated mitochondrial DNA heteroplasmy: the mytilid Musculista senhousia (Mollusca Bivalvia). BMC Evol Biol. 7(Suppl. 2):S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passamonti M, Boore JL, Scali V. 2003. Molecular evolution and recombination in gender-associated mitochondrial DNAs of the Manila clam Tapes philippinarum. Genetics 164(2):603–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passamonti M, Scali V. 2001. Gender-associated mitochondrial DNA heteroplasmy in the venerid clam Tapes philippinarum (Mollusca Bivalvia). Curr Genet. 39(2):117–124. [DOI] [PubMed] [Google Scholar]
- Plazzi F, Cassano A, Passamonti M. 2015. The quest for Doubly Uniparental Inheritance in heterodont bivalves and its detection in Meretrix lamarckii (Veneridae: Meretricinae). J Zool Syst Evol Res. 53(1):87–94. [Google Scholar]
- Rambaut A, Drummond AJ. 2012. FigTree version 1.4. Edinburgh (United Kingdom): University of Edinburgh. [Google Scholar]
- Stewart DT, Kenchington ER, Singh RK, Zouros E. 1996. Degree of selective constraint as an explanation of the different rates of evolution of gender-specific mitochondrial DNA lineages in the mussel Mytilus. Genetics 143(3):1349–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart DT, Saavedra C, Stanwood RR, Ball AO, Zouros E. 1995. Male and female mitochondrial DNA lineages in the blue mussel (Mytilus edulis) species group. Mol Biol Evol. 12(5):735–747. [DOI] [PubMed] [Google Scholar]
- Strahl J, Abele D. 2010. Cell turnover in tissues of the long-lived ocean quahog Arctica islandica and the short-lived scallop Aequipecten opercularis. Mar Biol. 157(6):1283–1292. [Google Scholar]
- Strahl J, Brey T, Philipp EE, Thorarinsdóttir G, Fischer N, Wessels W, Abele D. 2011. Physiological responses to self-induced burrowing and metabolic rate depression in the ocean quahog Arctica islandica. J Exp Biol. 214(24):4223–4233. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30(12):2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theologidis I, Fodelianakis S, Gaspar MB, Zouros E. 2008. Doubly uniparental inheritance (DUI) of mitochondrial DNA in Donax trunculus (Bivalvia: Donacidae) and the problem of its sporadic detection in Bivalvia. Evolution 62(4):959–970. [DOI] [PubMed] [Google Scholar]
- Thorarinsdottir GG. 2000. Annual gametogenic cycle in ocean quahog, Arctica islandica from north-western Iceland. J Mar Biol Assoc UK. 80(04):661–666. [Google Scholar]
- Zouros E. 2013. Biparental inheritance through uniparental transmission: the doubly uniparental inheritance (DUI) of mitochondrial DNA. Evol Biol. 40(1):1–31 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.