Abstract
In a recent article, Nelson-Sathi et al. (NS) report that the origins of major archaeal lineages (MAL) correspond to massive group-specific gene acquisitions via HGT from bacteria (Nelson-Sathi et al. 2015. Origins of major archaeal clades correspond to gene acquisitions from bacteria. Nature 517(7532):77-80.). If correct, this would have fundamental implications for the process of diversification in microbes. However, a reexamination of these data and results shows that the methodology used by NS systematically inflates the number of genes acquired at the root of each MAL, and incorrectly assumes bacterial origins for these genes. A reanalysis of their data with appropriate phylogenetic models accounting for the dynamics of gene gain and loss between lineages supports the continuous acquisition of genes over long periods in the evolution of Archaea.
Keywords: Archaea, horizontal gene transfer, ancestral genome reconstruction
Introduction
Reconstructing genome histories is a major challenge in evolutionary biology and the subject of a large body of literature (Maddison 1997; Snel et al. 2002; Mirkin et al. 2003; Hahn 2007; Csűrös 2010; Boussau et al. 2013; Szöllõsi et al. 2013, 2015). In a recent study, Nelson-Sathi et al. (NS) devised an ad hoc method to infer ancestral gene acquisitions in the history of Archaea (Nelson-Sathi et al. 2015). From 134 archaeal proteomes, they built 25,762 protein families of which 2,264 had at least two representatives in a single major archaeal lineages (MAL) (forming a monophyletic group) and at least two bacterial homologs belonging to species (out of 1,847 genomes) from two different phyla. NS concluded that all 2,264 gene families were acquired from Bacteria at the origin of MALs, implying that these acquisitions probably promoted their origin and evolutionary success.
Results and Discussion
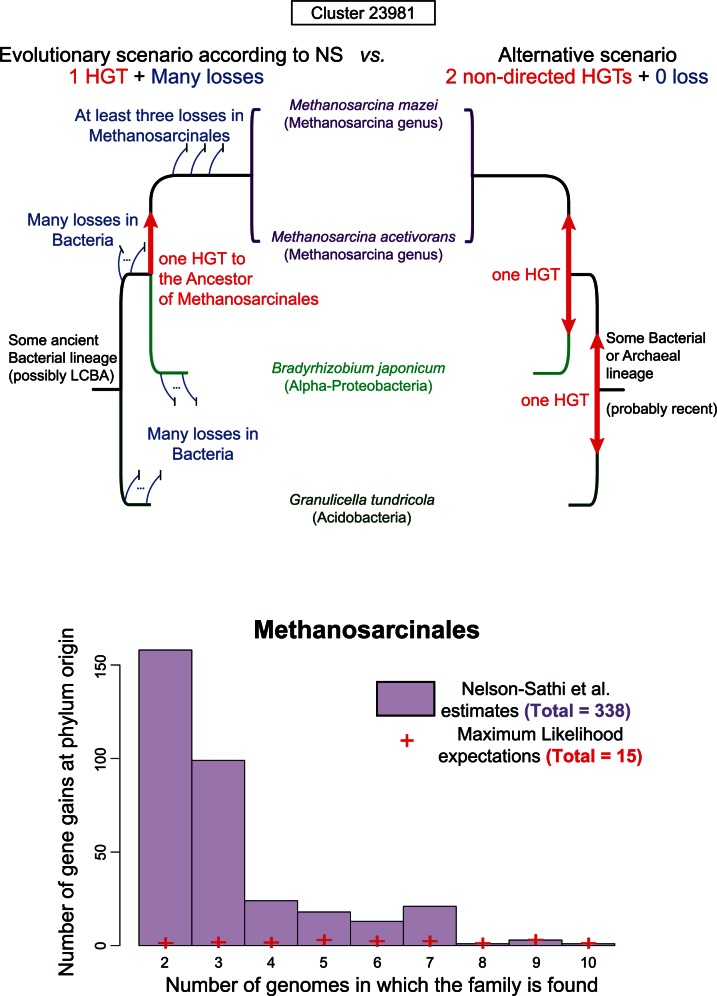
A close look at the results of NS is enough to convince oneself that there are problems with their approach. The set of genes that NS infer to have been acquired at the roots of MAL comprises 2,264 gene “clusters,” which are called “import” clusters. Figure 1a presents the tree reconstructed from one of them, “Cluster 23981,” which we simply sampled from the list of import clusters available as supplementary material accompanying NS (supplementary table S3 from Nelson-Sathi et al. 2015). This gene is found in only two sister species from a single archaeal genus (Methanosarcina), nested in the order Methanosarcinales (one of the 12 MALs) (Petitjean et al. 2015), and two from very distantly related bacteria (Bradyrhizobium japonicum, an alphaproteobacterium and Granulicella tundricola, an acidobacterium), from two different phyla (out of 1,847 bacterial genomes in total). Inclusion of this cluster in the import set implies that, according to NS: 1) the gene was transferred before the origin of Methanosarcinales and 2) that it is “very widespread among diverse bacteria, clearly indicating that [it is an] archaeal acquisition from bacteria, or import” (fig. 1) (Nelson-Sathi et al. 2015). The first is akin to saying that a gene present only in chimpanzee and human necessarily originated in the ancestor of all Vertebrates and has since undergone “systematic” gene loss in “all” other sequenced vertebrate species. Similarly, a “widespread” distribution (2) signifies transfer from a bacterial donor only if it is interpreted as a sign of antiquity in Bacteria, again implying extensive gene losses, this time in Bacteria. A much more parsimonious scenario (fig. 1) requires only two transfers, and avoids massive convergent losses in Archaea and Bacteria. In this scenario, the gene is acquired at the origin of the genus Methanosarcina (but not the phylum Methanosarcinales), and the direction of horizontal gene transfer (HGT) is unknown.
Fig. 1.
Forcing HGTs to the origins of archaeal phyla. (a) Competing evolutionary scenarios for « Cluster 23981 » from NS’s import set. Left: the gene is ancient in Bacteria and was subsequently transferred (red arrow) to the ancestor of Methanosarcinales. A large number of losses (blue lines) in Bacteria and Methanosarcinales is necessary to explain the narrow pattern of presence in extant species. Right: the gene is not ancient in Bacteria and was absent at the origin of Methanosarcinales. It has been transferred twice from Bacteria or Archaea and among Bacteria, and no loss is necessary. (b) Distribution of gene gains at the origin of Methanosarcinales (for other phyla, see the supplementary figure S1, Supplementary Material online). NS estimates are represented in purple. The distribution is very skewed toward sparsely distributed genes. ML expectations of gains on the corresponding set of genes are represented by red crosses.
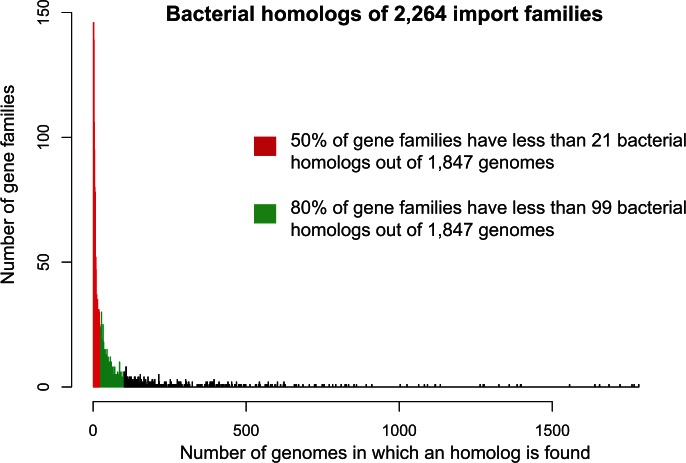
We emphasize that this cluster is not an exception but is representative of the import set (figs. 1b and 2 and supplementary fig. S1, Supplementary Material online). In fact, most of the genes reported by NS as acquired at the origins of a MAL are present in very few species in Archaea and Bacteria. More precisely, 52% (1,171/2,264 import clusters) are represented in only two or three archaeal species, strongly suggesting that these genes have been acquired during the diversification of MALs rather than at their root (see below). Furthermore, the definition of import genes by NS requires that they have homologs in bacterial species from at least two phyla, of which they claim one has to be the donor (Nelson-Sathi et al. 2015). Although this is not explicit in their paper, it can only mean, as we argue above, that NS consider that if a gene has representatives in two different bacterial phyla, it is “ancient” (i.e., it was present in the common ancestor of these two phyla) and hence of bacterial origin. Yet, these import genes could be more recent and instead could have been transferred among Bacterial phyla. In support of this hypothesis, we observe that these genes have a very narrow and patchy distribution: half of these import genes have homologs in less than 1.2% of the bacterial genomes considered (21/1,847) (fig. 2). Because these genomes are from at least two phyla, such a patchy distribution is consistent with, and strongly suggests, recent HGTs within the bacterial domain. Their presence in an ancestral bacterial genome (at least as old as the common ancestor of a MAL) cannot be assumed. Instead, these genes appear to have very complex evolutionary histories, and NS’s assumption that they were transferred from Bacteria to Archaea rather than the reverse is unfounded.
Fig. 2.
The sporadic distribution of the 2,264 import gene families in Bacteria. The distribution of the 2,264 gene families in the 1,847 bacterial genomes is represented. The distribution is very skewed: half of the gene families have fewer than 21 bacterial homologs out of the 1,847 genomes (1.1%), and a vast majority of them (80%) are present in fewer than 99 bacterial genomes (5%). Because for each family the bacterial homologs are from at least two different phyla, this distribution is highly suggestive of recent HGTs among bacteria and of complex evolutionary scenarios for these families, preventing the inference of a direction of transfer.
How did NS come to their conclusions about the origins of these genes? To assess whether the 2,264 gene acquisitions correspond to the origins of MAL, they employed an ad hoc phylogenetic test, which compares distributions of splits in the “import” and “recipient” set of gene trees. The recipient set is comprised of gene families only present in a single MAL, whereas members of the import set, discussed above, also have (typically sparse) homologs in Bacterial species. NS show that the import and recipient sets exhibit similar distributions of splits for 6 out of 13 MALs. They interpret this result as evidence that the import set of genes has been vertically inherited after a single acquisition at the root of the corresponding MAL. In reality, this result only shows that tree distributions are not statistically different between these two—arbitrary—sets of genes. This similarity “does” imply that the pattern of presence/absence of genes and their transfer rates are similar between the two sets. It “does not” imply that either set was predominantly acquired before the root of a particular MAL. This would only be the case if genes of one of the sets (e.g., the recipient set) were predominantly acquired before the root of the given MAL. In reality, both recipient and import genes have a very skewed distribution: respectively 59% and 52% of these genes are present in fewer than four species of a given MAL. Furthermore, this test is only applicable to families with four or more genes (see the supplementary methods of NS), which only represent 48% of the 2,264 families in the import set. Nevertheless, NS extend their conclusions to all genes in the import set. Finally, while 7 out of 13 MALs do not pass their congruence test for a lack of statistical signal, NS nonetheless argue that all 2,264 import genes were acquired at the origins of 13 MALs (as clearly stated, for instance in the abstract or the caption of figure 3 in Nelson-Sathi et al. [2015]).
Fig. 3.
Most of the 2,264 import gene families were acquired after phyla origination in Archaea. We used the tree reconstructed by NS to represent the points of acquisition of genes of the import set in the evolution of Archaea. Colors on branches represent the expected number of gains per branch, relatively to the number of phylum-specific families. Branches with a gain expectation less than 1 are colored in grey. On the left, NS estimations are represented. All phylum-specific families were acquired at the origin of each phylum, and no subsequent gains are inferred. On the right, ML estimates show that gene family gains are spread over the history of diversification of each phyla and that most of the families were acquired after the origination of phyla.
In fact, with the method used by NS, no gene acquisition is possible after the ancestor of a MAL because the relationships among species within MALs are ignored. In order to assess how many of the genes specific to MALs have been acquired more recently, it is necessary to analyze the data in a phylogenetic framework. Ideally, it would be necessary to apply a method that simultaneously infers the species tree and the scenarios of gene evolution for each cluster based on the corresponding gene trees (Szöllõsi et al. 2012). However, these methods require extensive computation and are currently limited in the number of species that they can efficiently analyze. We hence used Count (Csűrös 2010), using a maximum likelihood (ML) approach with an evolutionary model of gene gain and loss (see Materials and Methods) on the phylogenetic (presence/absence) profiles of each gene cluster. We estimated lineage-specific rates of gain per gene cluster and computed the expected lineage-specific number of gains along the reference tree reconstructed by Nelson-Sathi et al. (2015). This approach is of course a simplification, because we assume that the tree of species within MALs is correct and we ignore the phylogenetic signal contained in individual gene alignments. Yet, it is conservative for our comparison here because errors in the relationships of species within a MAL and ignoring the phylogenetic conflict between a gene tree and the species tree will both tend to yield a higher probability for genes to be present at the origins of MALs (Szöllõsi et al. 2015). Among the 2,264 import genes, the great majority (75%) appears during the diversification of each MAL (fig. 3 and supplementary fig. S1, Supplementary Material online). In other words, the acquisitions of the import genes defined by NS are spread over long periods in the evolution of MALs, and only a minority (25%) are inferred to have been acquired at their origins. For instance, the number of gene acquisitions at the root of Methanosarcinales and Thermoproteales is now very small (15 instead of 338 and 5 instead of 59, respectively). These estimates are consistent using ML or parsimony with a variety of parameter values (see Materials and Methods and supplementary fig. S2, Supplementary Material online). Interestingly, we observe a strong, positive and significant correlation between the number of ancestrally acquired import genes and the branch lengths of the reference species tree (ρ = 0.47, P < 10−7, see supplementary fig. S3, Supplementary Material online). This correlation is actually also observed with the recipient set (ρ = 0.54 P < 10−9). Although the number of substitutions per site is only a rough estimate of time, this suggests that gene acquisition is a continual, rather than a punctuated process in archaeal evolution in both the import and recipient gene sets. Therefore, as MALs are, by definition, well separated from other lineages, their root branches tend to have slightly higher gains. However, figure 3 suggests that in most MALs (Haloarchaea, Methanosarcinales, Methanomicrobiales, Thermoplasmatales, Methanobacteriales, Desulfurococcales, and Thermoproteales), some internal branches experienced more gains than their root branch leading to their last common ancestor (fig. 3).
The approach used by NS was first applied in a previous study that found that massive gene transfers from bacteria had occurred at the origin of Haloarchaea (Nelson-Sathi et al. 2012). These conclusions have since been shown to be unfounded in a recent study using a more comprehensive taxonomic sampling in Haloarchaea (Becker et al. 2014). Gene transfer seems to have occurred continuously across the tree of life (Ochman et al. 2000; Gogarten and Townsend 2005; Abby et al. 2012; Szöllõsi et al. 2012), and probably contributed to the greatest as well as the smallest innovations. Among the genes that have been acquired at the origins of MALs, some have probably promoted metabolic innovations and played a decisive role in their evolutionary success. But both the quantification and the argument for a systematic bacterial origin defended by NS are erroneous. Actually, many genes acquired throughout the evolution of MALs have no bacterial homologs (the recipient set) and have therefore been largely overlooked by NS. Understanding the evolutionary history of adaptations in relation to gene transfer requires and deserves more accurate analysis of the data in an integrated phylogenetic framework.
Materials and Methods
The data used in Nelson-Sathi et al. (2015) were kindly provided by the authors. Statistical analyses were performed in R (R Core Team 2013) and ancestral gene repertoire reconstructions were performed with the Count program (Csűrös 2010). Gene clusters were first coded into phylogenetic (presence/absence) profiles depending on their presence or absence in archaeal genomes. Gene clusters are considered independent of each other. We used a probabilistic birth–death model of gain and loss of genes to model the evolutionary dynamic of each gene cluster and to compute probabilities of presence/absence at each internal node of the reference tree (Csűrös and Miklós 2006; Csűrös 2010). The model defines two rate parameters: a gain parameter κ and a loss parameter µ. We used a version of the model in which κ and µ are allowed to vary across branches of the reference tree. All rates were optimized by maximizing the likelihood. We estimated the rates from the 25,762 protein families, along the archaeal reference species tree reconstructed by Nelson-Sathi et al. (2015). After optimization of the branch-specific parameters, ancestral gene repertoire reconstructions were carried out for each of the 2,264 families, by computing their branch-specific posterior probabilities of evolutionary events. For instance, each gene has a posterior probability of being gained on each branch of the reference tree. At a given branch, the expected total number of gene acquisitions across all families was computed by summing all family specific gene gain posterior probabilities.
Parsimonious reconstructions were also done in Count with the Wagner parsimony algorithm (Farris 1970; Csűrös 2008), which penalizes gains and losses differently. For each gene cluster, the scenario of ancestral presence/absence that minimizes the total cost of gain and loss is retained. A large range of gain and loss cost combinations was tested.
All parsimony and ML estimations gave very similar results (See supplementary fig. S2, Supplementary Material online). A Count session file that will allow users to reproduce our results is available at ftp://pbil.univ-lyon1.fr/pub/datasets/DAUBIN/HGT_Archaea/.
Supplementary Material
Supplementary material is available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors are thankful to Eric Alm, Greg Fournier, Andrew Roger and Eric Tannier for their advice and for fruitful discussions. The authors also thank Nelson-Sathi et al. (2015) for promptly making their data available. B.B., V.D., C.B.-A., and M.G. are supported by the Ancestrome Project ANR-10-BINF-01–01.
References
- Abby SS, Tannier E, Gouy M, Daubin V. 2012. Lateral gene transfer as a support for the tree of life. Proc Natl Acad Sci U S A. 109:4962–4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker EA, Seitzer PM, Tritt A, Larsen D, Krusor M, Yao AI, Wu D, Madern D, Eisen JA, Darling AE, et al. 2014. Phylogenetically driven sequencing of extremely halophilic archaea reveals strategies for static and dynamic osmo-response. PLoS Genet. 10:e1004784–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boussau B, Szöllõsi GJ, Duret L, Gouy M, Tannier E, Daubin V. 2013. Genome-scale coestimation of species and gene trees. Genome Res. 23:323–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csűrös M. 2008. Ancestral reconstruction by asymmetric Wagner parsimony over continuous characters and squared parsimony over distributions. In: Comparative genomics. Vol. 5267 Lecture Notes in Computer Science. Berlin, Heidelberg: Springer Berlin Heidelberg. p. 72–86. [Google Scholar]
- Csűrös M. 2010. Count: evolutionary analysis of phylogenetic profiles with parsimony and likelihood. Bioinformatics 26:1910–1912. [DOI] [PubMed] [Google Scholar]
- Csűrös M, Miklós I. 2006. A probabilistic model for gene content evolution with duplication, loss, and horizontal transfer. In: Research in computational molecular biology. Vol. 3909 Lecture Notes in Computer Science. Berlin, Heidelberg: Springer Berlin Heidelberg. p. 206–220. [Google Scholar]
- Farris JS. 1970. Methods for computing Wagner trees. Syst Biol. 19:83–92. [Google Scholar]
- Gogarten JP, Townsend JP. 2005. Horizontal gene transfer, genome innovation and evolution. Nat Rev Microbiol. 3:679–687. [DOI] [PubMed] [Google Scholar]
- Hahn MW. 2007. Bias in phylogenetic tree reconciliation methods: implications for vertebrate genome evolution. Genome Biol. 8:R141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison WP. 1997. Gene trees in species trees. Syst Biol. 46:523–536. [Google Scholar]
- Mirkin BG, Fenner TI, Galperin MY, Koonin EV. 2003. Algorithms for computing parsimonious evolutionary scenarios for genome evolution, the last universal common ancestor and dominance of horizontal gene transfer in the evolution of prokaryotes. BMC Evol Biol. 3:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson-Sathi S, Dagan T, Landan G, Janssen A, Steel M, McInerney JO, Deppenmeier U, Martin WF. 2012. Acquisition of 1,000 eubacterial genes physiologically transformed a methanogen at the origin of Haloarchaea. Proc Natl Acad Sci U S A. 109:20537–20542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson-Sathi S, Sousa FL, Roettger M, Lozada-Chávez N, Thiergart T, Janssen A, Bryant D, Landan G, Schönheit P, Siebers B, et al. 2015. Origins of major archaeal clades correspond to gene acquisitions from bacteria. Nature 517:77–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H, Lawrence JG, Groisman EA. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299–304. [DOI] [PubMed] [Google Scholar]
- Petitjean C, Deschamps P, López-García P, Moreira D, Brochier-Armanet C. 2015. Extending the conserved phylogenetic core of archaea disentangles the evolution of the third domain of life. Mol Biol Evol. 32:1242–1254. [DOI] [PubMed] [Google Scholar]
- Snel B, Bork P, Huynen MA. 2002. Genomes in flux: the evolution of archaeal and proteobacterial gene content. Genome Res. 12:17–25. [DOI] [PubMed] [Google Scholar]
- Szöllõsi GJ, Boussau B, Abby SS, Tannier E, Daubin V. 2012. Phylogenetic modeling of lateral gene transfer reconstructs the pattern and relative timing of speciations. Proc Natl Acad Sci U S A. 109:17513–17518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szöllõsi GJ, Davín AA, Tannier E, Daubin V, Boussau B. 2015. Genome-scale phylogenetic analysis finds extensive gene transfer among fungi. Philos Trans R Soc Lond B Biol Sci. 370(1678): 20140335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szöllõsi GJ, Rosikiewicz W, Boussau B, Tannier E, Daubin V. 2013. Efficient exploration of the space of reconciled gene trees. Syst Biol. 62:901–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2013. R: a language and environment for statistical computing. R Foundation for Statistical Computing; Vienna (Austria): https://www.R-project.org. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.