Abstract
SUMOylation, the covalent binding of Small Ubiquitin-like Modifier (SUMO) to target proteins, is a posttranslational modification that regulates critical cellular processes in eukaryotes. In insects, SUMOylation has been studied in holometabolous species, particularly in the dipteran Drosophila melanogaster, which contains a single SUMO gene (smt3). This has led to the assumption that insects contain a single SUMO gene. However, the analysis of insect genomes shows that basal insects contain two SUMO genes, orthologous to vertebrate SUMO1 and SUMO2/3. Our phylogenetical analysis reveals that the SUMO gene has been duplicated giving rise to SUMO1 and SUMO2/3 families early in Metazoan evolution, and that later in insect evolution the SUMO1 gene has been lost after the Hymenoptera divergence. To explore the consequences of this loss, we have examined the characteristics and different biological functions of the two SUMO genes (SUMO1 and SUMO3) in the hemimetabolous cockroach Blattella germanica and compared them with those of Drosophila Smt3. Here, we show that the metamorphic role of the SUMO genes is evolutionary conserved in insects, although there has been a regulatory switch from SUMO1 in basal insects to SUMO3 in more derived ones. We also show that, unlike vertebrates, insect SUMO3 proteins cannot form polySUMO chains due to the loss of critical lysine residues within the N-terminal part of the protein. Furthermore, the formation of polySUMO chains by expression of ectopic human SUMO3 has a deleterious effect in Drosophila. These findings contribute to the understanding of the functional consequences of the evolution of SUMO genes.
Keywords: Blattella, Drosophila, insects, SUMO paralogs, polySUMO chains, SUMO, SUMOylation
Introduction
SUMOylation is a posttranslational modification that consists of the covalent and reversible binding of the Small Ubiquitin–like MOdifier, SUMO, to a target protein. This interaction can modify the properties of target proteins affecting their localization, activity, or stability. Protein SUMOylation is involved in many cellular processes, such as cell cycle progression, transcription, nucleocytoplasmic transport, apoptosis, and genome integrity and stability (Geiss-Friedlander and Melchior 2007; Gareau and Lima 2010). As these processes are essential for normal animal development, the study of SUMOylation and its mechanisms of action are of paramount importance.
SUMO proteins are translated as immature precursors and subsequently converted to their mature forms through the activity of sentrin/SUMO-specific proteases (SENPs). Then, mature SUMO is transferred to target proteins through a number of well-defined steps. First, the E1 activating enzyme (SAE1/SAE2) activates SUMO, which can subsequently be recognized and attached to the UBC9 E2-conjugating enzyme. UBC9 mediates covalent interaction of SUMO to the target protein, although the participation of some E3-ligases, which help with substrate recognition, can be also required (Gareau and Lima 2010). The interaction between SUMO and the target protein is a reversible process, as SUMO can be detached through the action of some SENPs. SUMOylation usually occurs at lysine residues in the consensus ΨKxD/E motif, but not all such lysines become SUMOylated and, in some cases, SUMO can also attach to lysine residues outside of this motif (Xu et al. 2008).
The number of SUMO paralogs varies depending on the species. For example, the yeast Saccharomyces cerevisae, the nematode Caenorhabditis elegans and the insect Drosophila melanogaster have only one SUMO gene, whereas plants and vertebrates contain several. Humans have four SUMO paralogs (SUMO1–4). Human SUMO2 and 3 share 97% identity at the amino acid level and comprise the SUMO2/3 subfamily. In contrast, human SUMO1 shares only approximately 50% identity with SUMO2/3. Finally, SUMO4 shares 87% identity with SUMO2, but its activation and conjugation has not been demonstrated (Wilkinson and Henley 2010). Consistent with their sequence differences, SUMO1 and SUMO2/3 usually conjugate to different substrates, but the functional relevance of such specificity remains poorly understood. Vertebrate paralogs also differ in their ability to form polySUMO chains, as SUMO2/3 can be SUMOylated and form chains, whereas SUMO1 cannot and may serve as chain terminator (Matic et al. 2008). PolySUMO chains are functionally important for cellular stress response and degradation of damaged proteins (Saitoh and Hinchey 2000; Castorálová et al. 2012). Because of these key functions, polySUMO chain formation represents an active area of research, although it has only been described in human and yeast.
In insects, SUMOylation has been studied in highly derived holometabolous species, particularly in the dipteran Drosophila melanogaster. In this organism, which has a single SUMO gene (smt3), SUMOylation is required in many developmental processes, such as embryogenesis, wing morphogenesis, nervous system development, immune response, apoptosis, and metamorphosis (Talamillo, Sánchez, and Barrio 2008). Although one might assume that all insects harbor a single SUMO gene, our data and recent genome sequencing efforts reveal that more basal insect species have two SUMO paralogs. In this study, by using the hemimetabolous cockroach Blattella germanica as a model insect with two SUMO genes (SUMO1 and SUMO3), we examine the characteristics and different biological functions of the two Blattella SUMO paralogs to explore the evolutionary consequences of the loss of one SUMO gene during insect evolution. In addition to the study of the paralog-specific functions of SUMO proteins in species with more than one SUMO gene, our work has focused on the requirements and functions for polySUMO chain formation in insects. Here, we report that SUMO1, but not SUMO3, specifically controls different key processes in Blattella metamorphosis, such as cell proliferation, ecdysone biosynthesis and signaling response to this hormone, developmental timing, and proper molting. In contrast, we show that SUMO1 and SUMO3 exert redundant functions in Blattella viability. Finally, we show that, in contrast to vertebrates, insect SUMO proteins cannot form polySUMO chains.
Results and Discussion
Insects Lost the SUMO1 Paralog during Evolution
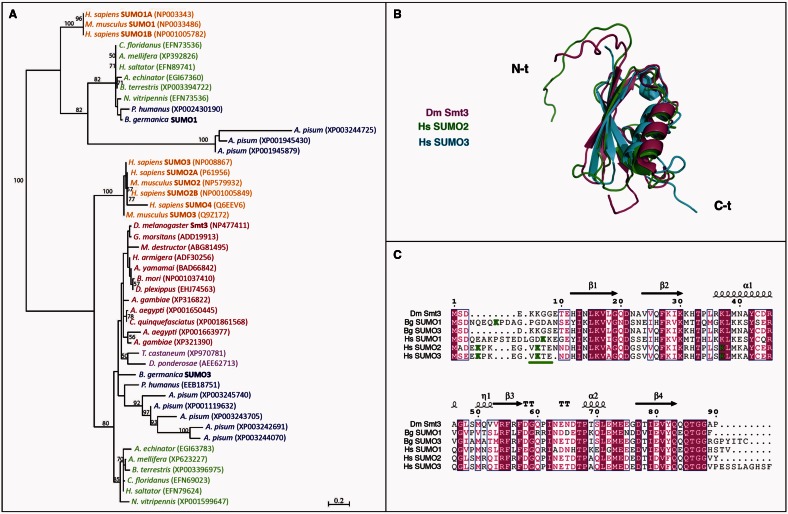
As mentioned above, SUMOylation has been studied in insects that contain a single SUMO gene, particularly Drosophila (Talamillo, Sánchez, and Barrio 2008). However, a detailed analysis of SUMO paralogs in the insect genomes reported in the databases shows that insects present one or two SUMO genes. To determine when, during evolution, insects gained and/or lost SUMO genes, we performed a phylogenetic analysis using all insect SUMO sequences available, as well as those of human and mouse as representatives of the vertebrate group. The multiple sequence alignment and the posterior phylogenetic tree obtained grouped SUMO sequences in two differentiated clusters with a bootstrap support value of 100% (for a total of 100 iterations) (fig. 1A). The upper part of the tree grouped the insect sequences similar to vertebrate SUMO1, whereas the rest of the insect sequences grouped with the vertebrate SUMO2/3 subfamily in the lower branch of the tree. The analysis of the phylogenetic tree shows that Coleopteran, Lepidopteran, and Dipteran species contain only one SUMO gene, orthologous to human SUMO2/3. Conversely, Hymenopteran and all Hemimetabolan species contain two SUMO genes that are orthologous to human SUMO1 and SUMO2/3 genes. Therefore, according to the insect phylogeny currently accepted (Misof et al. 2014), our phylogenetic analysis strongly suggests that insects lost the SUMO1 paralog after the Hymenoptera divergence. Interestingly, it was previously thought that the duplication of the ancestral SUMO gene into two subfamilies, SUMO1 and SUMO2/3, had occurred in the subphylum Vertebrata (Su and Li 2002). However, the presence in basal insects of SUMO1 and SUMO3 paralogs suggests that SUMO gene duplication occurred earlier during animal evolution. Consistent with this hypothesis, a new phylogenetic analysis using SUMO sequences available from species representative of different eukaryotic groups confirmed that the duplication between SUMO1 and SUMO3 occurred at the base of the Metazoa and that later in Metazoan evolution SUMO genes were secondarily lost in different lineages and specifically expanded in vertebrates (supplementary fig. S1, Supplementary Material online).
Fig. 1.
Insects lost the SUMO1 paralog after the hymenopteran divergence. (A) Phylogenetic analysis of insect SUMO proteins. Human and mouse SUMO sequences were included as reference. Branch lengths are proportional to sequence divergence. The bar represents 0.2 substitutions per site. Only bootstrap values ≥50% are shown. EMBL accession number for each sequence is included in parentheses. Color code refers to Hemimetabola (blue), Hymenoptera (green), Coleoptera (purple), Diptera and Lepidoptera (red), and human and mouse (orange). (B) Superposition of the structures of Drosophila Smt3 (magenta), human SUMO2 (green), and human SUMO3 (cyan). The protein backbones are depicted as ribbons showing the different secondary structure elements. The N- and C-terminal ends of the chains, labeled in the figure, were disordered and flexible in solution, and the superposition was based on the well-ordered chain segments. (C) Sequence alignment of the indicated SUMO proteins. The numbers and the secondary structure elements indicated above the sequences correspond to Drosophila Smt3. Arrows are indicative for β-strands, spirals for α-helices (α) and 310 helices (η), and “T” for turns. Conserved regions in all sequences are marked in magenta. Lysine groups being part of SUMOylation consensus sites (underlined) are highlighted in green.
Insect SUMO3 Paralogs Do Not Form SUMO Chains
The fact that SUMOylation in insects has been only studied in species with a single SUMO gene raises questions about a specific role for the SUMO paralog lost during insect evolution. To address this question, we decided to characterize the SUMOylation process in a hemimetabolous insect model. With a well-characterized postembryonic development, including the metamorphic period, and an efficient and systemic RNA interference (RNAi) response (Martín et al. 2006; Ureña et al. 2014), Blattella germanica is a suitable model to study SUMOylation in an insect with two SUMO genes. To this end, we first cloned the two SUMO sequences of Blattella from a cDNA library synthesized from embryonic cells (UM-BGE-1 cell line). Based on their similarity with the SUMO sequences described, we named them BgSUMO1 (EMBL: LN809887) and BgSUMO3 (EMBL: LN809888) (fig. 1A and supplementary table S1, Supplementary Material online). Sequence alignment of the human, Blattella, and Drosophila SUMO sequences, along with the comparison of the available SUMO structures, reveals high sequence and structure similarities in the central region of the proteins and higher divergence in the N- and C-terminal parts, which are flexible and structurally disorganized (fig. 1B and C). Furthermore, the N-terminal part of BgSUMO1 presents high similarity with human SUMO1, whereas the N-terminal part of BgSUMO3 shows higher similarity with DmSmt3 than with human SUMO3 (fig. 1C).
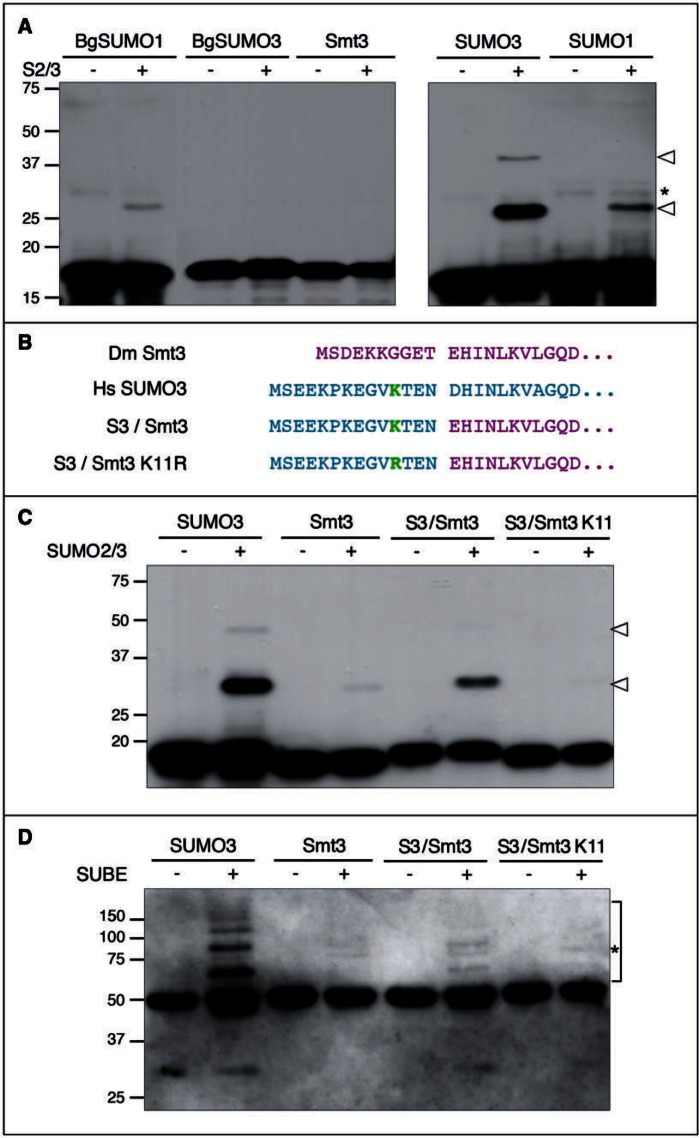
Interestingly, a detailed analysis of the insect SUMO sequences revealed the absence of SUMOylation consensus sites in Drosophila Smt3 and Blattella BgSUMO3 proteins and only a low probability SUMOylation site (QK8PD) in BgSUMO1 (fig. 1C). In contrast, human SUMO2/3 proteins present a SUMOylation consensus site (VK11TE) that is able to form polySUMO chains in vivo, and two nonconsensus lysine residues with lower SUMO affinity (EK5PK and SK42LM) (Knipscheer et al. 2007; Matic et al. 2008; Békés et al. 2011) (fig. 1C). Moreover, human SUMO1 contains one low probability SUMOylation site (DK16KE) that is SUMOylated only when large amounts of the Ubc9 enzyme are added in vitro (Matic et al. 2008) (fig. 1C). Taken together, these data suggest that, in contrast to human proteins, insect SUMO paralogs cannot form polySUMO chains. To test this possibility, we studied the ability of BgSUMO1, BgSUMO3, and Smt3 to be SUMOylated in vitro. As a positive control of strong SUMOylation we used human SUMO3, whereas human SUMO1 was used as a control for residual in vitro SUMOylation. First, we generated hemagglutinin (HA)-tagged forms of the different SUMO proteins and incubated them with recombinant human SUMO2/3 and the proteins of the conjugation machinery (SAE1/2 and Ubc9). These HA-tagged SUMO forms lack the two glycine residues involved in SUMO conjugation and, therefore, can be SUMOylated but not attached to target proteins. Confirming that the system works properly, we detected that human SUMO3 was strongly modified by SUMO2/3 (fig. 2A), as it was previously described (Knipscheer et al. 2007; Matic et al. 2008; Békés et al. 2011). Consistent with our hypothesis, Drosophila Smt3 and BgSUMO3 were not able to form polySUMO chains and, based on the degree of SUMOylation of human SUMO1, only residual SUMOylation was observed for Blattella BgSUMO1 (fig. 2A). As human SUMO1 is not considered to be SUMOylated in vivo despite its modest capacity to be SUMOylated in vitro (Matic et al. 2008), our results indicate that none of the insect SUMO paralogs can form polySUMO chains.
Fig. 2.
Insect SUMO proteins do not form polySUMO chains in vitro. (A) In vitro SUMOylation assay for Blattella BgSUMO1 and BgSUMO3, Drosophila Smt3, and human SUMO1 (negative control) and SUMO3 (positive control). HA-tagged forms of all proteins were incubated in the absence (−) or presence (+) of human SUMO2/3. Arrowheads indicate the modified proteins, whereas asterisk indicates unspecific bands. Molecular weight markers are shown to the left. (B) Residue composition of the chimera S3/Smt3, which has the initial 14 amino acids from human SUMO3 (magenta) and the rest of the protein corresponding to Drosophila Smt3 (cyan). In the chimera S3(K11R)/Smt3, K11 has been mutated to R. (C) In vitro SUMOylation assay for the human SUMO3/Drosophila Smt3 chimeric proteins, either WT (S3/Smt3) or mutant for K11 (S3(K11R)/Smt3. Arrowheads indicate the modified proteins. (D) Detection of SUMO chains formed in vitro through SUBEs. Bracket indicates polySUMO-chains formed and asterisk indicates unspecific bands.
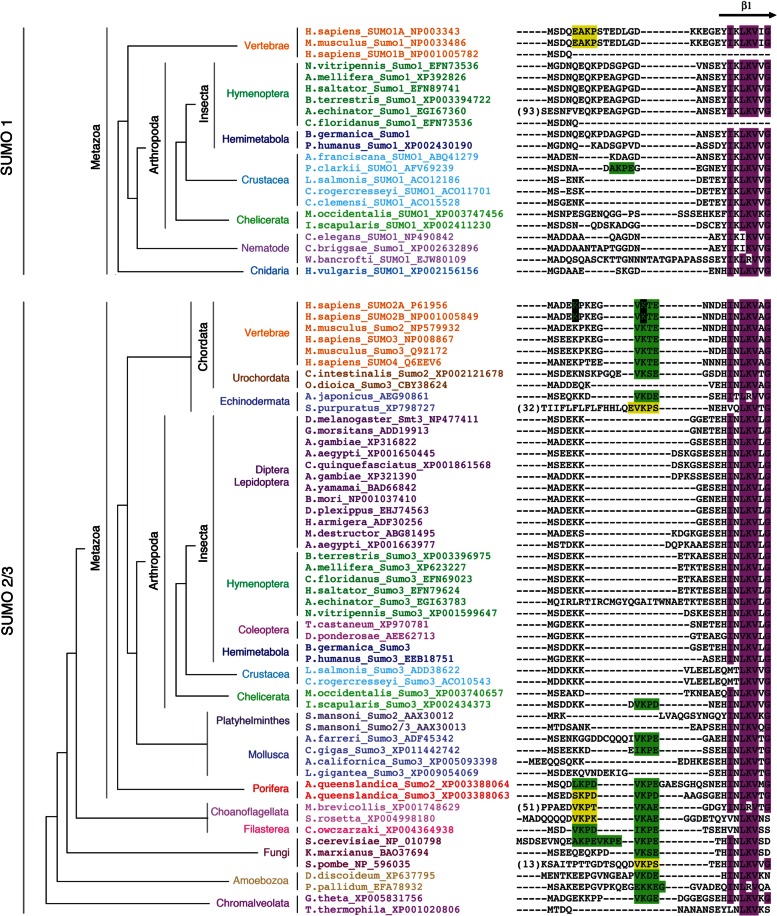
SUMOylation of human SUMO3 in vivo has been preferentially observed on lysine11 (K11) (Matic et al. 2008). In Saccharomyces, Smt3 SUMOylation can also occur in lysine residues located at the N-terminal part of the protein (K11, K15, and K19) (Bylebyl et al. 2003). To further confirm that insect SUMO3 proteins were not SUMOylated by the absence of critical lysine residues in the N-terminal part of their sequences (figs. 1C and 3), we created a chimeric SUMO protein consisting of methionine1-asparagine14 (M1-N14) residues of human SUMO3 (containing K11) and residues glutamic acid11-threonine86 (E11-T86) of Drosophila Smt3 (S3/Smt3) (fig. 2B). Under these conditions, the in vitro analysis confirmed the SUMOylation of the chimeric protein, indicating that the N-terminal region of SUMO proteins is critical to form chains (fig. 2C). In addition, the mutation of K11 to arginine11 (R11) (S3(K11R)/Smt3) completely abolished the polySUMOylation ability of the chimeric protein, further demonstrating the essential role of K11 for SUMO chain formation (fig. 2C). We confirmed these results by using the recently developed SUMO-traps or SUBEs (SUMO-binding entities), which specifically interact with polySUMO chains (Da Silva-Ferrada et al. 2013), to isolate the SUMO chains formed in vitro (fig. 2D). Consistent with the SUMOylation assay, SUBEs recognized chain formation when human SUMO3 or the chimeric S3/Smt3 was used as substrates, but not when Drosophila Smt3 or the chimeric mutant S3(K11R)/Smt3 was used (fig. 2D). Overall, these results confirm that, in contrast to vertebrates, Drosophila and Blattella SUMO3 paralogs have lost the capacity to form polySUMO chains, as they have lost the SUMOylation domains in the N-terminal part of their sequences. Remarkably, a detailed analysis of the N-terminal region of SUMO3 sequences in eukaryote organisms reveals that all insects and crustaceans (Pancrustacea) reported to date have lost the SUMOylation consensus sites (fig. 3). These data suggest that 1) the presence of the SUMOylation domain in the N-terminal part of the SUMO3 protein is an ancestral character of this protein, and 2) this sequence has been lost in Pancrustacea, thus rendering SUMO proteins of insects unable to form polySUMO chains. As all insect SUMO1 proteins do not present SUMOylation domains in their N-terminus either (fig. 3), our result suggests that insect SUMO proteins cannot be SUMOylated.
Fig. 3.
Insect SUMO proteins lack SUMOylation consensus motifs. Sequence alignment of the N-terminal region of SUMO proteins. Conserved regions in all sequences are marked in magenta. High- and low-probability SUMOylation consensus sites are highlighted in green or yellow, respectively. K5 and K11 residues within the SUMOylation consensus sites in human SUMO2/3 sequences are highlighted in black.
SUMOylation Is Essential for Blattella Metamorphosis
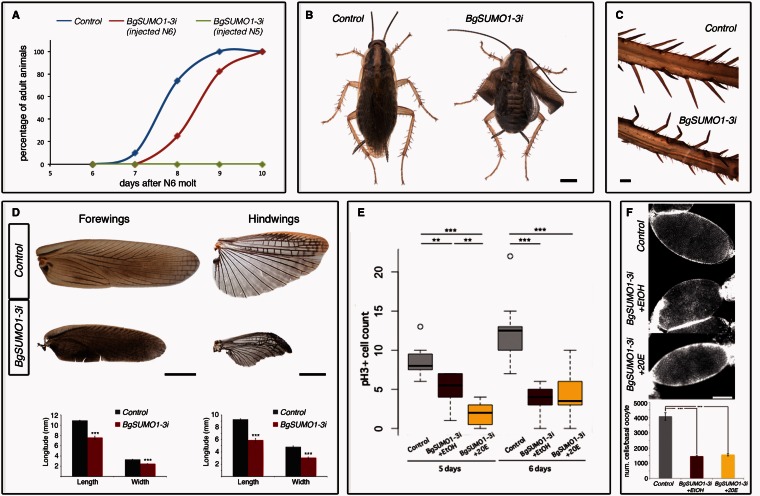
To study the biological functions of the two SUMO paralogs in Blattella, we first analyzed their spatial and developmental expression patterns. Both BgSUMO mRNAs were present in all tissues analyzed, namely the fat body, epidermis, ovary, brain, corpora allata, digestive tract, and prothoracic glands (supplementary fig. S2A, Supplementary Material online). Given that both genes are ubiquitously expressed, we analyzed the mRNA levels of BgSUMO1 and BgSUMO3 in the prothoracic gland, as a model tissue, and found that both were expressed throughout development (supplementary fig. S2B, Supplementary Material online). Next, we used systemic RNAi to examine the role of SUMO genes in Blattella. We focused our functional analysis on the metamorphosis of Blattella, as it has been shown that Drosophila Smt3 has essential roles in the metamorphic process (Talamillo, Sánchez, Cantera, et al. 2008). In Blattella, metamorphosis occurs during the last nymphal instar stage (N6) and is mainly characterized by the appearance of functional wings (Mané-Padrós et al. 2010). First, we lowered the expression of both BgSUMO genes by injecting simultaneously two dsRNAs targeting each BgSUMO transcript in newly emerged N5 female nymphs (herein called BgSUMO1-3i animals) to ensure the depletion of both BgSUMOs during the metamorphic stage (supplementary fig. S2C–E, Supplementary Material online). Specimens injected with dsMock were used as negative controls (Control animals). All BgSUMO1-3i nymphs molted into N6 six days after injection, as the Control animals (supplementary table S2, Supplementary Material online). However, BgSUMO1-3i nymphs died 1–4 days after the N6 molt, thus precluding the functional analysis of SUMO proteins during metamorphosis (fig. 4A and supplementary table S2, Supplementary Material online). To circumvent this problem, we injected the dsRNA into newly emerged N6 female nymphs. Under this condition, the majority of BgSUMO1-3i nymphs molted into adults although showing a significant delay of approximately 1 day compared with Control animals (fig. 4A and supplementary table S3, Supplementary Material online). As before, all BgSUMO1-3i adults died 1–4 days after the imaginal molt, confirming that SUMOylation is essential for Blattella viability. A similar role in viability has been described in a wide range of species, such as Drosophila, Saccharomyces, Caenorhabditis, Danio rerio, or Mus musculus (Dohmen et al. 1995; Johnson et al. 1997; Jones et al. 2001; Nacerddine et al. 2005; Nowak and Hammerschmidt 2006; Huang et al. 2011). Interestingly, BgSUMO1-3i adults presented clear deficiencies in the extension of the wings and malformations in the leg bristles, indicative of molting problems (fig. 4B and C). They also showed a significant reduction in the size of the fore- and hindwings (fig. 4D), suggesting impairment of cell proliferation during metamorphosis. To further study whether SUMOylation is required for cell proliferation during metamorphosis, we analyzed the increase of follicular epithelial cells that surround basal oocytes. These cells proliferate intensively during the metamorphic nymphal stage from approximately 500 cells in newly ecdysed N6 nymphs to approximately 3,500 cells just before the imaginal molt (Mané-Padrós et al. 2008). As figure 4E shows, depletion of both BgSUMOs dramatically impaired the proliferative activity of follicular cells as revealed by a temporal analysis of cell division by phospho-Histone H3 (pH3) staining. We also analyzed whether the absence of BgSUMOs induces cell death by immunostaining the follicular cells with anti-activated-caspase-3 antibody. We did not detect any increase in cell death in the BgSUMO1-3i animals (supplementary fig. S5, Supplementary Material online). As a result, the number of the follicular cells was significantly reduced in BgSUMO1-3i animals, confirming that SUMOylation is required for cell proliferation during Blattella metamorphosis (fig. 4F). The role of SUMOylation in cell cycle progression and tissue proliferation has been demonstrated in other eukaryotic organisms, indicating its conserved function (Nacerddine et al. 2005; Poulin et al. 2005; Nowak and Hammerschmidt 2006; Liao et al. 2010; Terada and Furukawa 2010; Kanakousaki and Gibson 2012). In Drosophila, for example, many proteins involved in cell division are SUMOylated, such as PCNA, RFC2, Topoisomerase I, Topoisomerase II, and Polo (Nie et al. 2009). In addition to the proliferation impairment, the nuclei of the follicular cells of BgSUMO1-3i animals were bigger and presented an abnormal shape compared with Control nuclei (fig. 4F). Similarly, SUMOylation-deficient mice and zebrafish embryos also present abnormal nucleus morphology due to aberrant sister chromatid separation, hypocondensation, and polyploidy (Nacerddine et al. 2005; Nowak and Hammerschmidt 2006).
Fig. 4.
SUMOylation controls Blattella metamorphosis and developmental timing. (A) Representation of the timing of imaginal molt in Control (blue), BgSUMO1-3i animals injected in N5 (green), and BgSUMO3i animals injected in N6 (red). (B) Dorsal view of a Control specimen 1 day after the imaginal molt, showing a normal winged adult shape (left), and a BgSUMO1-3i adult showing defects on wing extension in both the forewings and hindwings (right). Scale bar: 2 mm. (C) Detail of the adult bristles in Control and BgSUMO1-3i individuals. Scale bar: 0.2 mm. (D) Forewings (left) and hindwings (right) in newly molted Control and BgSUMO1-3i adults. Scale bars: 2 mm. Means of maximum length and width of forewings (bottom left) and hindwings (bottom right) in Control and BgSUMO1-3i adults. Error bars represent SEM (n = 5). Asterisks indicate differences statistically significant as follows: ***P ≤ 0.0005 (t-test). (E) Graphical representation of the mitotic capacity of follicular cells in 5- and 6-day-old Control (gray bars), BgSUMO1-3i (red bars), and BgSUMO1-3i N6 nymphs treated with the edysteroid hormone 20E (yellow bars), represented by the number of nuclei marked by the mitotic marker PH3. Asterisks indicate differences statistically significant as follow: **P < 0.001; ***P < 0.0001 (n = 25–30). (F) DAPI staining and follicular cell number from basal oocytes in newly molted Control, BgSUMO1-3i, and BgSUMO1-3i adults treated with 20E. Scale bar: 5 μm. Error bars represent SEM (n = 5–15). Asterisks indicate differences statistically significant as follows: ***P ≤ 0.0005 (t-test).
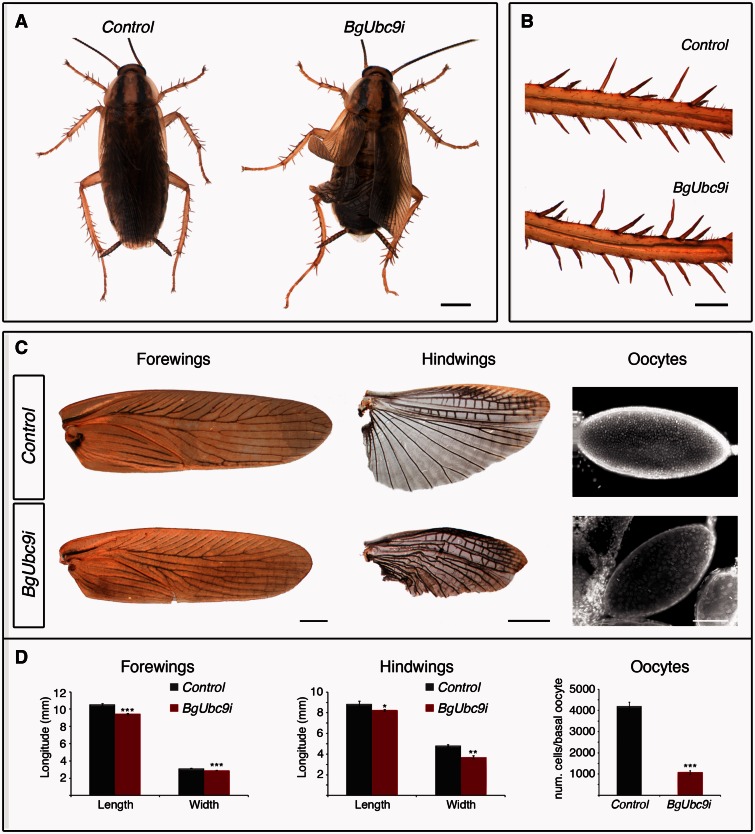
To further confirm that the phenotype of BgSUMO1-3i animals was due to the impairment of SUMOylation, we cloned and depleted by RNAi the SUMO-conjugating enzyme BgUbc9 (BgUbc9i animals) (supplementary fig. S3B, Supplementary Material online). As both BgSUMOs, BgUbc9 mRNA was present throughout development (supplementary fig. S3A, Supplementary Material online). Like BgSUMO1-3i animals, BgUbc9-depleted nymphs molted into adults showing clear morphological deficiencies in the legs and the spreading of the wings (fig. 5A and B). They also showed a significant reduction in the size of the fore- and hindwings and in the number of follicle cells (fig. 5C and D). Likewise, BgUbc9i adults also died 1–4 days after the imaginal molt. The similar phenotypes between BgSUMO1-3i and BgUbc9i animals thus confirm that the defects observed in BgSUMO1-3i animals were due to the block of the SUMOylation process. Overall, these results show that SUMOylation has essential roles on Blattella viability and metamorphosis, particularly in the control of cell proliferation, developmental timing, and proper molting.
Fig. 5.
Depletion of Blattella BgUbc9 phenocopies the absence of BgSUMO1 and BgSUMO3. (A) Dorsal view of newly molted Control (left) and BgUbc9i (right) adults. Scale bar: 2 mm. (B) Details of the adult bristles in Control and BgUbc9i individuals. Scale bar: 0.5 mm. (C) Phenotypic analysis of the forewings (left column), hindwings (central column), and DAPI staining of basal oocytes (right column) of newly molted Control and BgUbc9i adults. Scale bars: 1 mm (forewings), 2 mm (hindwings), and 5 μm (oocytes). (D) Means of maximum length and width of forewings and hindwings, and follicular cell number from basal oocytes of newly molted Control and BgUbc9i adults. Error bars represent SEM (n = 5). Asterisks indicate differences statistically significant as follows: *P ≤ 0.05; **P ≤ 0.001; ***P ≤ 0.0005 (t-test).
Functional Differences between Blattella SUMO Paralogs
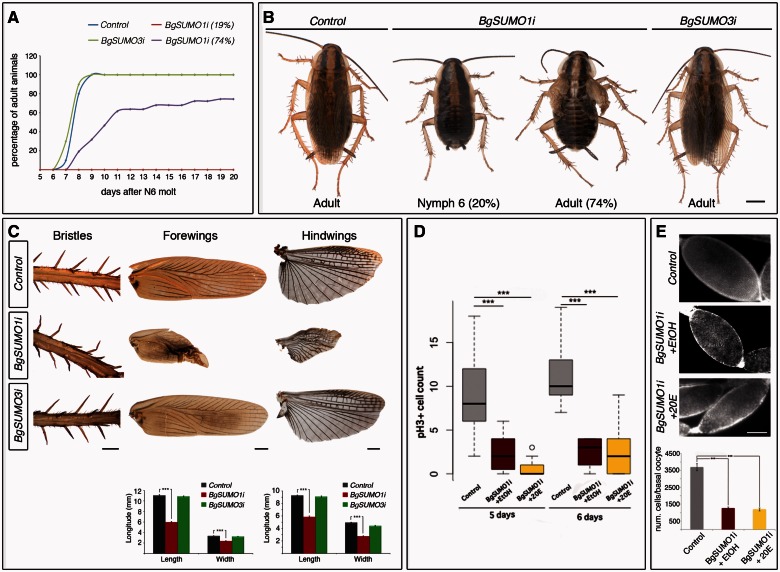
To assess the contribution of each BgSUMO paralog to the observed phenotype, dsRNAs targeting each isoform were injected separately into newly emerged penultimate-N5 instar nymphs (BgSUMO1i and BgSUMO3i animals) (see supplementary fig. S2C, Supplementary Material online). These treatments resulted in a remarkable decrease in the corresponding transcript without affecting the expression of the other paralog (supplementary fig. S4, Supplementary Material online). All the Control, BgSUMO1i, and BgSUMO3i animals molted properly to the last-N6 nymphal instar (supplementary table S4, Supplementary Material online). In contrast to BgSUMO1-3i animals injected in N5, all the Control, BgSUMO1i and BgSUMO3i N6 nymphs survived during the metamorphic N6 stage. After that, all the Control and BgSUMO3i nymphs completed the imaginal molt at the same time (fig. 6A and B and supplementary table S4, Supplementary Material online). Likewise, the majority of BgSUMO1i animals (74.5%) also reached adulthood although showing a significant developmental delay (mean average for N6 length in Control animals: 8.1 ± 0.1 days; in BgSUMO3i animals: 7.8 ± 0.14 days; in BgSUMO1i animals: 10.46 ± 0.46 days, P ≤ 0.0005, Log-Rank test) (fig. 6A and B and supplementary table S4, Supplementary Material online). In contrast, approximately 19% of BgSUMO1i animals never reached the adult stage and stayed as nymphs until they died (fig. 6A and B and supplementary table S4, Supplementary Material online). Remarkably, although BgSUMO3i adults did not show any morphological defects, the phenotype of BgSUMO1i animals that molted into adults was identical to that observed in BgSUMO1-3i and BgUbc9i adults (fig. 6B). The leg bristles in BgSUMO1i adults were twisted, indicating molting problems, and the fore- and hindwings were abnormally unfolded and their size was significantly reduced (fig. 6B and C). Likewise, the follicular epithelia of BgSUMO1i basal oocytes presented a significant impairment of cell division without any increase in cell death, which resulted in a clear reduction in the number of cells (fig. 6D and E and supplementary fig. S5, Supplementary Material online). Importantly, BgSUMO1i and BgSUMO3i adults were fully viable, in contrast to BgSUMO1-3i adults, which died soon after the imaginal molt. Altogether, these results show that 1) BgSUMO1 specifically controls SUMO-dependent processes in the metamorphosis and developmental timing of Blattella, whereas BgSUMO3 is dispensable in these processes; and 2) both paralogs are redundant for essential SUMOylation-dependent functions related to organism viability.
Fig. 6.
BgSUMO1 is the essential paralog in Blattella metamorphosis and developmental timing. (A) Representation of the timing of imaginal molt in Control (blue), BgSUMO3i (green), and BgSUMO1i animals. BgSUMO1i specimens are separated into those that molt to the adult stage (purple) or remained as nymphs (red). (B) Dorsal views of Control and BgSUMO3i specimens 1 day after the imaginal molt, showing normal winged adult shapes (Control, left; BgSUMO3i, right) and BgSUMO1i specimens that remained as nymphs after 14 days (BgSUMO1i Nymph 6) or that molt into adults showing defects of wing extension in both the forewings and hindwings (BgSUMO1i Adult). Scale bar: 2 mm. (C) Phenotypic analysis of the bristles, forewings and hindwings in newly molted Control, BgSUMO1i and BgSUMO3i adults. Scale bars: 0.5 mm (bristles) and 1 mm (forewings and hindwings). Means of maximum length and width of forewings and hindwings in newly molted Control, BgSUMO1i, and BgSUMO3i adults (lower panels). Error bars represent SEM (n = 5). Asterisks indicate differences statistically significant as follows: ***P ≤ 0.0005 (t-test). (D) Graphical representation of the mitotic capacity of follicular cells in 5- and 6-day-old Control (gray bars), BgSUMO1i (red bars) and BgSUMO1i N6 nymphs treated with 20E (yellow bars), represented by the number of nuclei marked by the mitotic marker PH3. Asterisks indicate differences statistically significant as follows: ***P < 0.0001. (n = 25–30). (E) DAPI staining and follicular cell number from basal oocytes in newly molted Control, BgSUMO1i, and BgSUMO1i adults treated with 20E. Scale bar: 5 μm. Error bars represent SEM (n = 5–15). Asterisks indicate differences statistically significant as follows: ***P ≤ 0.0005 (t-test).
BgSUMO1 Is Required for Proper Ecdysteroid Synthesis
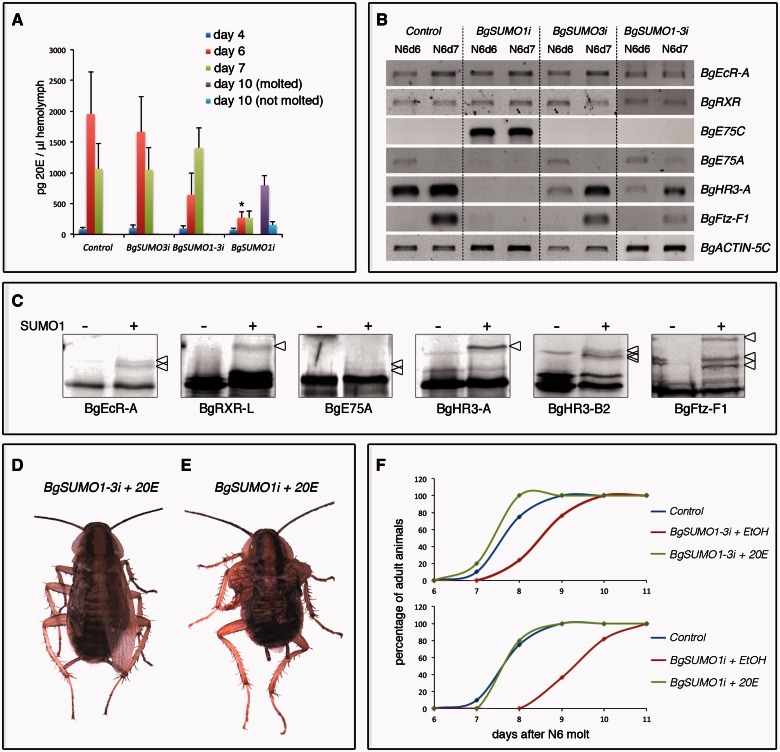
In insects, periodic pulses of the ecdysteroid hormone 20-hydroxyecdysone (20E) at the end of each instar promote the transition to the next stage (Yamanaka et al. 2013). The significant delay in the majority of BgSUMO1i nymphs to reach the adult stage, along with the approximately 19% of BgSUMO1i nymphs that never reach adulthood, suggested that the biosynthesis of 20E may be defective in these animals. To test this possibility, we measured the levels of 20E in the hemolymph of Control and BgSUMO1i, as well as in BgSUMO3i and BgSUMO1-3i animals, during the N6 instar. As figure 7A shows, the levels of circulating 20E in Control animals presented the expected peak at the end of the instar. Remarkably, consistent with the strong developmental delay of BgSUMO1i nymphs, the circulating 20E titer of these animals was abnormally low compared with Control animals (fig. 7A). The levels of 20E eventually increased after a prolonged N6 instar in the BgSUMO1i animals that were able to molt into adults (75%) (fig. 7A, purple bar), whereas the increase in 20E levels was not detected in the BgSUMO1i nymphs that did not reach adulthood (19%) (fig. 7A, light blue bar). This difference in 20E levels between the two BgSUMO1i groups of animals is most likely the result of different RNAi efficiency. Nevertheless, these results show that BgSUMO1 is required for the biosynthesis of 20E in Blattella. On the other hand, no differences in 20E levels were found between Control and BgSUMO3i animals, whereas the increase of circulating 20E in BgSUMO1-3i nymphs showed an approximately 1-day delay, also correlating with the developmental delay of these animals (fig. 7A).
Fig. 7.
20E biosynthesis and the 20E-signaling response are impaired in BgSUMO1i nymphs. (A) Ecdysteroid titers in the hemolymph of Control, BgSUMO1i, and BgSUMO3i N6 nymphs determined by ELISA. In BgSUMO1i animals, 20E levels are also measured in specimens that molted into adults after a prolonged N6 instar (day 10 molted) and in specimens that remained as nymphs (day 10 not molted). Results are expressed as ng/μl of 20E equivalents. Vertical bars indicate the SEM (n = 4). Asterisks indicate differences statistically significant at *P ≤ 0.005 (t-test). (B) Expression levels of the indicated NRs in Control, BgSUMO1i, BgSUMO3i, and BgSUMO1-3i nymphs. mRNA levels were analyzed by RT-PCR from prothoracic glands of 6- and 7-day-old last instar nymphs. BgACTIN-5C levels were measured as reference. The results shown are representative of five replicates. (C) In vitro SUMOylation assay for Blattella nuclear receptors BgEcR-A, BgRXR-L, BgE75-A, BgHR3-A, BgHR3-B2, and BgFtz-F1. Proteins were incubated in the absence (−) or presence (+) of human SUMO1. Arrowheads indicate the SUMOylated proteins. Dorsal views of a 20E-treated BgSUMO1-3i adult (D) and a 20E-treated BgSUMO1i adult (E) showing defects on wing extension in both the forewings and hindwings. Scale bar: 2 mm. (F) Effect of 20E administration to BgSUMO1-3i (upper panel) and BgSUMO1i animals (lower panel) on the timing of imaginal molt of these animals, compared with Control animals. BgSUMO1i animals that remained as nymphs were not included in the analysis.
The very low levels of circulating 20E in BgSUMO1i nymphs suggest that these animals may have affected the proper temporal progression of the 20E-controlled hierarchy of nuclear hormone receptors (NRs) that directs the transition to the adult stage in Blattella (Maestro et al. 2005; Cruz et al. 2006, 2007, 2008; Martín et al. 2006; Mané-Padrós et al. 2008, 2012). To test this possibility, we measured the expression of the 20E-dependent NRs in the prothoracic gland of 6- and 7-day-old N6 Control, BgSUMO1i, BgSUMO3i and BgSUMO1-3i nymphs, just when the levels of 20E are higher in Control animals. As figure 7B shows, mRNA levels of BgEcR-A and BgRXR, which form the 20E heterodimeric receptor, were normal in BgSUMO1i, BgSUMO3i, and BgSUMO1-3i nymphs. In contrast, the sequential expression of three NRs that are induced by the increasing levels of 20E, namely BgE75A, BgHR3-A and BgFTZ-F1, was completely halted in BgSUMO1i nymphs when compared with Control, BgSUMO3i, and BgSUMO1-3i animals. Accordingly, the transcript levels of BgE75C, an NR that is expressed at low levels of 20E and repressed at high levels of this hormone, were very high in BgSUMO1i nymphs, whereas it was undetectable in Control, BgSUMO3i, and BgSUMO1-3i nymphs (fig. 7B). It is interesting to note that the lower expression levels of BgHR3-A and BgFTZ-F1 in BgSUMO1-3i nymphs are consistent with the delay in the increase of 20E levels observed in these animals. Taken together, these results show that BgSUMO1 is required for the proper synthesis of 20E and, therefore, for the correct transduction of the hormonal signaling. In Drosophila, Smt3 is also essential for 20E biosynthesis and signaling in the prothoracic gland during the last larval stage (Talamillo, Sánchez, Cantera, et al. 2008). In this gland, Smt3 controls the expression and transcriptional activity of Ftz-f1 and regulates cholesterol intake through controlling the expression of Scavenger receptors (SR-BI) (Talamillo et al. 2013). Furthermore, Smt3 is essential for maintaining the Ultraspiracle (USP) protein levels (the RXR ortholog in Drosophila) and USP SUMOylation is required for full 20E-induced activity in the fly (Wang et al. 2014). Therefore, it is plausible that, in addition to the 20E-biosynthesis defect, the missregulation of the 20E-signaling cascade observed in BgSUMO1i animals is also partially provoked by deficient SUMOylation of the NRs that transduce the hormonal signal. Supporting this possibility, we have found that the Blattella 20E-dependent NRs analyzed, including BgRXR and BgEcR, are SUMOylated in vitro (fig. 7C).
To further determine to what extent the metamorphic defects, developmental retardation and viability impairment in BgSUMO-1i and BgSUMO1-3i animals are caused by defective 20E biosynthesis, we injected 20E to 5-day-old BgSUMO-depleted nymphs, just when the levels of 20E start to increase in Control nymphs. This procedure is an established approach used to examine the role of 20E in 20E-defective Blattella nymphs (Mané-Padrós et al. 2008). Interestingly, the adult morphological defects of BgSUMO1i and BgSUMO1-3i animals were not rescued (fig. 7D and E). 20E also failed to rescue the proliferation capacity of the follicular cells of BgSUMO1i and BgSUMO1-3i animals, nor increased the cell death of these cells (figs. 4E, 4F, 6D, and 6E and supplementary fig. S5, Supplementary Material online). Likewise, supplying 20E did not rescue the viability of BgSUMO1-3i adults, as they also died 1–4 days after the imaginal molt. In contrast, supplement of 20E completely rescued the developmental delay in the N6-adult transition in BgSUMO1i and BgSUMO1-3i animals (fig. 7F). Altogether, these results show that BgSUMO1 not only exerts critical regulatory activities through the regulation of 20E biosynthesis (developmental timing) but also acts in a 20E-independent manner (cell proliferation, molting, and adult viability).
In summary, our paralog-specific RNAi analyses indicate that BgSUMO1 is essential for Blattella metamorphosis, particularly in cell proliferation, 20E biosynthesis and signaling response, developmental timing and molting, whereas BgSUMO3 is dispensable in these processes. In vertebrates, most of the SUMOylation substrates are uniquely or preferentially modified by a specific paralog in normal conditions and only a certain overlap (∼15%) has been demonstrated (Manza et al. 2004; Rosas-Acosta et al. 2005; Vertegaal et al. 2006). Despite this specificity, the lack of SUMO1 promotes compensatory SUMOylation with SUMO2/3, suggesting certain redundancy in their functions (Yuan et al. 2010; Citro and Chiocca 2013). Consistent with this observation, SUMO1 is dispensable in zebrafish and mice development (Zhang et al. 2008; Yuan et al. 2010). In contrast, our results demonstrate that BgSUMO3 cannot compensate the absence of BgSUMO1 during Blattella metamorphosis, uncovering the occurrence of paralog-specific functions in insects with two SUMO paralogs.
Blattella BgSUMO3 Paralog Can Substitute for Drosophila Smt3
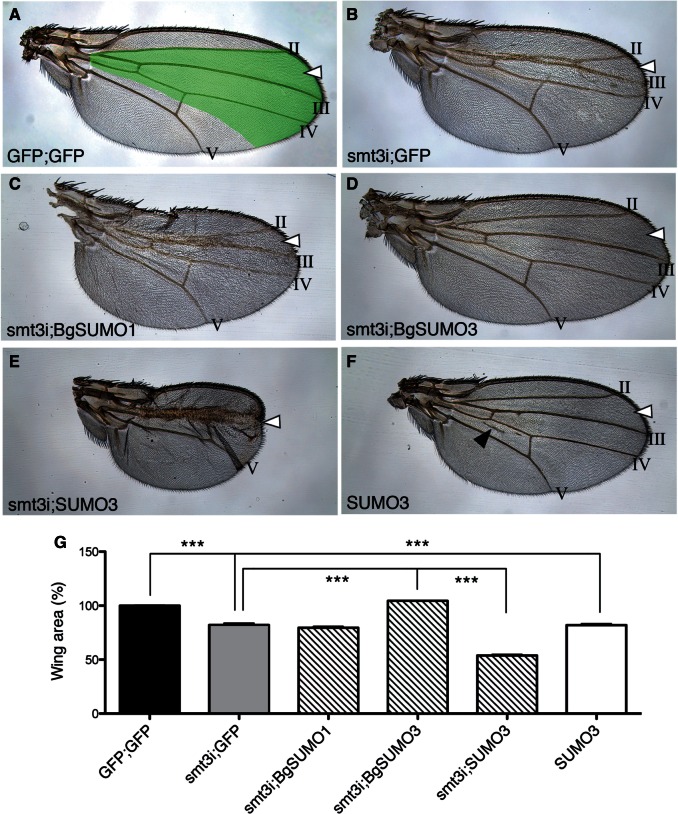
The results above indicate that, although the role of SUMOylation in insect metamorphosis is evolutionarily conserved, this function has switched from SUMO1 in basal orders of insects (Blattella) to SUMO3 in more derived holometabolous insects (Drosophila). To further characterize this evolutionary switch, we asked whether Blattella BgSUMO1 could functionally substitute for Drosophila Smt3 in the control of a SUMO-dependent process, namely the development of Drosophila wing. Growth and patterning in Drosophila wings are highly stereotypical processes and any variation in the size and the location of the longitudinal veins is easily detectable (fig. 8A). Thus, the depletion of Smt3 from the central part of the wing (smt3i wings) by using the SalEPv-GAL4 driver provoked smaller wings and affected the formation of the intervein region between LII and LIII veins (fig. 8B and G). In this smt3i background, the overexpression of Blattella BgSUMO1 did not rescue the wing deficiencies (fig. 8C and G), whereas the overexpression of BgSUMO3 restored perfectly the wing size and the integrity of the LII–LIII intervein region (fig. 8D and G). To confirm these results, we turned to the prothoracic gland as a model tissue. The downregulation of smt3 in the prothoracic gland by using the phantom-GAL4 driver prevents larvae from entering pupariation (table 1), due to the inability to produce the 20E pulse that induces metamorphosis (Talamillo, Sánchez, Cantera, et al. 2008). Consistent with the results obtained in the wings, the overexpression of BgSUMO1 in smt3-depleted prothoracic glands was not able to rescue the smt3i phenotype, whereas the overexpression of BgSUMO3 completely rescued the pupariation phenotype, resulting in normal adult flies (table 1). Altogether, these results indicate that although BgSUMO1 is the crucial SUMO paralog in the metamorphosis of basal insects, it cannot substitute for Smt3 in Drosophila development, most probably due to the low sequence similarity. Conversely, BgSUMO3, which is dispensable in Blattella metamorphosis, contains all the elements necessary to function properly in Drosophila.
Fig. 8.
Functional analysis of Blattella and human SUMOs in Drosophila. (A) Control wing. Green area represents SalEPv-Gal4 expression domain. LII–LIII intervein region (arrowhead) and longitudinal veins II–V are indicated. (B) Wing silenced for smt3 (smt3i), showing surface reduction on LII–LIII intervein region. (C) Wing silenced for smt3 that simultaneously overexpresses Blattella BgSUMO1, which cannot rescue Smt3 absence. (D) Wing silenced for smt3 that simultaneously overexpresses Blattella BgSUMO3, which rescues the absence of Smt3 and recuperates the LII–LIII intervein region. (E) Wing silenced for smt3 that simultaneously overexpresses human SUMO3, showing stronger deficiencies than smt3i wings (B). (F) Wing overexpressing human SUMO3 in a WT background. Wings are smaller than control and present ectopic veins (black arrowhead), confirming the adverse effect of human SUMO3 on wing development. (G) Analysis of the wing area under the aforementioned conditions. Error bars represent SEM (n = at least 15 wings). Asterisks indicate differences statistically significant as follows: ***P ≤ 0.0005 (t-test).
Table 1.
Phenotypic Effect of BgSUMO1 or BgSUMO3 Overexpression in Prothoracic Glands of smt3i Drosophila Larvae.
| Adults (%) | Larval Arrest (%) | |
|---|---|---|
| +;+ | 100 | — |
| smt3i;+ | — | 100 |
| smt3i;BgSUMO1 | — | 100 |
| smt3i;BgSUMO3 | 100 | — |
If sequence similarity between BgSUMO3 and Drosophila Smt3 is important to allow proper SUMOylation, then it would be expected that human SUMO3, with a sequence that is also highly similar to Smt3 (fig. 1B and C), might be able to rescue the smt3i phenotypes in Drosophila. However, contrary to Drosophila Smt3 and Blattella SUMO3, human SUMO3 protein can form polySUMO chains efficiently and differs from the others in its N-terminal sequence (figs. 1C and 2A). This difference allows us to analyze in insects the functional implication of the N-terminal region of the SUMOs. Toward this aim, we performed rescue experiments overexpressing human SUMO3 in smt3-depleted wings (smt3i;SUMO3). Interestingly, we observed that human SUMO3 did not rescue the absence of Drosophila Smt3 despite their high sequence homology (fig. 8E and G). In fact, the phenotype observed in smt3i;SUMO3 wings was stronger than that observed in smt3i animals, indicating that human SUMO3 has a deleterious effect in Drosophila. Consistently, the overexpression of human SUMO3 in a wild-type background (GFP;SUMO3) resulted in smaller wings with ectopic veins (fig. 8F and G). Altogether, our results suggest that the N-terminal region of human SUMO3 is detrimental in insects. This feature represents an important evolutionary difference with vertebrates, including the capacity to form polySUMO chains, which are essential for several key processes, such as cellular stress response and ubiquitin-dependent degradation of damaged proteins by proteasome (Saitoh and Hinchey 2000; Castorálová et al. 2012). PolySUMOylated proteins are recognized by the SUMO-targeted ubiquitin ligase RNF4, which in turn ubiquitylates the substrate and targets it for proteasome degradation (Tatham et al. 2008). The RNF4 ortholog in Drosophila, Degringolade, can recognize and ubiquitylate SUMOylated and non-SUMOylated proteins, targeting them for degradation (Abed et al. 2011). However, consistent with the insect inability to form SUMO chains, Degringolade null mutants are viable (Barry et al. 2011). Interestingly, SUMO chain formation is neither essential in yeast, as mutation of the SUMOylable lysines in Saccharomyces Smt3 (K11, K15, and K19) impairs formation of chains but does not affect viability, growth or stress sensitivity (Bylebyl et al. 2003). These data suggest that the essential roles controlled by polySUMO chains could be an innovative feature of vertebrates, although SUMO orthologs from other animal species should be studied to confirm this hypothesis.
Finally, based on our results we conclude the following: 1) The duplication of the ancestral SUMO gene occurred at the base of Metazoan evolution. Hence, basal insects possess two SUMO genes, encoding for SUMO1 and SUMO3 paralogs. However, during insect evolution the SUMO1 gene has been lost after the hymenopteran divergence, and, as a result, Coleopteran, Lepidopteran and Dipteran species have only the SUMO3 gene. 2) Despite the difference in the number of SUMO genes, the role of SUMOylation in the control of metamorphosis is conserved in hemimetabolous and holometabolous insects. 3) The processes regulated by SUMOylation in Blattella metamorphosis (cell proliferation, ecdysone signaling response, and proper molting) are specifically controlled by the SUMO1 paralog. In contrast, SUMO1 and SUMO3 exert redundant functions in Blattella viability. 4) During insect evolution, derived holometabolous insects (Drosophila) have co-opted SUMO3 for all the metamorphic functions that are controlled specifically by the SUMO1 paralog in more basal insects (Blattella). This switch has probably allowed the loss of SUMO1 gene in the derived holometabolan species. 5) The sequence and structure differences between SUMO1 and SUMO3 paralogs are crucial for their biological roles, as Blattella SUMO1 protein cannot functionally substitute for Drosophila Smt3 in the control of SUMO-dependent processes during the metamorphosis of the fly, despite being the critical paralog in Blattella metamorphosis. In contrast, Blattella SUMO3 paralog, which is dispensable in the cockroach metamorphosis, contains all the elements necessary to substitute Drosophila Smt3. 6) Unlike SUMO3 from vertebrates, insect SUMO3 proteins have lost the capacity to form polySUMO chains due to the loss of key lysine residues within their flexible and disorganized N-terminal part. In addition, and given that Blattella SUMO1 protein is only residually polySUMOylated in vitro, we conclude that insect SUMO proteins cannot form polySUMO chains.
Materials and Methods
Insects
Blattella specimens were reared in the dark at 30 ± 1°C and 60–70% relative humidity. Drosophila were raised on standard medium at 25°C. Knockdown and overexpression experiments were performed using the GAL4/UAS system (Brand and Perrimon 1993). phantom-Gal4,UAS-mCD8::GFP/TM6B,Tb strain was obtained from P. Leopold and C. Mirth (Colombani et al. 2005; Mirth et al. 2005). Other strains used were SalEPv-Gal4 (Barrio and de Celis 2004) and UAS-RNAi line UAS-smt3i (Talamillo, Sánchez, Cantera, et al. 2008). UAS-BgSUMO1, UAS-BgSUMO3, and UAS-SUMO3 strains were generated as described below.
Plasmid Construction and Generation of Transgenic Strains
BgSUMO1 (EMBL: LN809887), BgSUMO3 (EMBL: LN809888), and human SUMO3 (EMBL: NM006936) coding regions were amplified using the specific primers described in supplementary table S5, Supplementary Material online. Fragments obtained were subsequently cloned into the EcoRI–XhoI sites (BgSUMO1 and BgSUMO3) or into the BglII–XhoI sites (human SUMO3) of pUASTattb vector (Bischof et al. 2007). Transgenic lines were generated following standard transformation procedures (Spradling and Rubin 1982).
Cloning of Blattella BgSUMO1, BgSUMO3, and BgUbc9 cDNAs
The two full length cDNA clones corresponding to BgSUMO1 and BgSUMO3 were obtained from a suppression subtractive hybridization library that was carried out using the polymerase chain reaction (PCR)-selected cDNA Subtraction Kit (Clontech), following the manufacturer’s protocols. The tester library was prepared with 1 μg of polyA + mRNA from UM-BGE-1 embryonic cells from Blattella treated with 20E during 10 h. The driver library was prepared with the same amount of polyA + mRNA from untreated UM-BGE-1 cells. The BgUbc9 cDNA clone was isolated by PCR using cDNA template from prothoracic glands of Blattella last instar nymphs. Total RNA was isolated using GenElute Mammalian Total RNA kit (Sigma) and DNase-treated with RQ1 RNase-Free DNase (Promega). The cDNA was generated by reverse transcription with Transcriptor First Strand cDNA Synthesis Kit (Roche) as previously described (Maestro et al. 2005; Cruz et al. 2006). The sequence of BgUbc9 homolog was obtained by PCR using degenerate primers: Fw 5′-TGGMGNAARGANCAYCC-3′ and Rv 5′-ACNCKYTTNTCRTAYTC-3′. A 398-bp fragment was amplified, subcloned into the pSTBlue-1 vector (Novagen), and sequenced. This was followed by 5′- and 3′-RACE Rapid amplification of cDNA ends (RACE) (5′- and 3′-RACE System Version 2.0; Invitrogen) to obtain the full BgUbc9 coding region, using the specific primers described in supplementary table S6, Supplementary Material online. All PCR products were subcloned into pSTBlue-1 vector (Novagen) and sequenced in both directions. A final 654-bp fragment of BgUbc9 gene covering full coding region was obtained (EMBL: LN809889).
Semiquantitative Reverse Transcriptase PCR
Total RNA was extracted from different tissues using the GenElute Mammalian Total RNA Kit (Sigma), and equivalent amounts of RNA were used for cDNA synthesis as described above. Reverse transcriptase (RT)-PCR without reverse transcription was used to confirm the absence of genomic contamination. All PCR amplifications were carried out using GoTaq DNA Polymerase (Promega) following manufacturer’s instructions. As a reference, Blattella BgActin-5C expression levels were measured in each cDNA sample. Primer sequences used for RT-PCR are described in supplementary table S7, Supplementary Material online.
Quantitative Real-Time RT-PCR
Total RNA from three staged prothoracic glands was isolated and retrotranscribed as described above. Relative transcript levels were determined using Power SYBR Green PCR Mastermix (Applied Biosystems). To standardize the quantitative real-time RT-PCR (qPCR) inputs, a master mix that contained Power SYBR Green PCR Mastermix and forward and reverse primers was prepared to a final concentration of 100 nM for each primer. The qPCR experiments were conducted with the same quantity of organ equivalent input for all treatments, and each sample was run in duplicate using 2 µl of cDNA per reaction. All the samples were analyzed on the iCycler iQReal Time PCR Detection System (Bio-Rad). For each standard curve, one reference DNA sample was diluted serially. Primer sequences used for qPCR are described in supplementary table S8, Supplementary Material online.
RNAi Synthesis and Injection
RNAi in vivo in nymphs of Blattella was performed as previously described (Martín et al. 2006; Cruz et al. 2007). A volume of 1 µl of each dsRNA solution (4 µg/µl) was injected into the abdomen of newly emerged female nymphs. In case of coinjection of two dsRNAs (dsBgSUMO1 and dsBgSUMO3), 0.5 μl of each solution was applied in a single injection of 1 μl (2 μg/μl each dsRNA). In case of injections to newly emerged fifth-instar nymphs, the animals were reinjected just after molting to the subsequent nymphal instar. Control dsRNA (dsMock) consisted of a noncoding sequence from the pSTBlue-1 vector (Cruz et al. 2006). Primers used to generate construct templates for dsRNA synthesis are described in supplementary table S9, Supplementary Material online.
Treatments In Vivo with 20E
N6 nymphs were injected at day 5 with 20 μg of 20E (Sigma) per specimen in 1 μl of Ringer saline with 10% ethanol. Controls received the same volume of solvent.
Quantification of Circulating Ecdysteroids
Hemolymph ecdysteroids were quantified by enzyme-linked immunosorbent assay (ELISA) as previously described (Cruz et al. 2003). 20E (Sigma) and 20E-acetylcholinesterase (Cayman Chemical, Ann Arbor, MI) were used as a standard and an enzymatic tracer, respectively. The antiserum (Cayman Chemical) was used at a dilution of 1:50,000. Absorbances were read at 450 nm, using a Multiscan Plus II Spectrophotometer (Labsystems, Madrid, Spain). The ecdysteroid antiserum used has the same affinity for ecdysone and 20E, but as the standard curve was obtained with the later compound, results are expressed as 20E equivalents.
Microscopy and Immunocytochemistry
All dissections of Blattella nymphal and adult tissues were carried out in Ringer’s saline on carbon dioxide-anesthetized specimens. Ovaries were fixed in 4% paraformaldehyde, permeabilized in Phosphate-buffered saline (PBS)–0.2% Tween (PBT), and incubated for 10 min in 1 μg/ml DAPI4', 6'-Diamidino-2-phenylindole (DAPI) in PBT. After two washes with PBT, ovaries were mounted in Mowiol 4-88 (Calbiochem). For immunocytochemistry analysis, anti-pH3 (Ser10) and anti-Caspase 3 antibodies (Cell Signaling Technologies) were used to analyze cell proliferation and cell death, respectively. Ovaries were fixed in 4% paraformaldehyde, permeabilized in PBS–0.1% Triton 30 min, and incubated 2 h with anti-pH3 (dilution 1:100) or anti-Caspase 3 (dilution 1:250) antibodies. After washing, ovaries were incubated 2 h with antirabbit secondary antibody, counterstained with DAPI, and mounted in Vectashield Mounting Medium for Fluorescence H-1000 (Vector Laboratories). All samples were examined with an AxioImager.Z1 (ApoTome 213 System; Zeiss) microscope and images were subsequently processed using Adobe Photoshop. To calculate the number of follicular cells, basal oocytes stained with DAPI were used. The oocyte surface was measured with the formula: Oocyte surface = 4π · (length/2) · (width/2). Total number of follicular cells (N) was determined after counting the number of nucleus present in a square (n) and extrapolating the value to the entire oocyte through the formula: N = (oocyte surface · n)/square surface. Maximum length and width of forewings and hindwings were measured in Adobe Photoshop from images taken with Zeiss SteREO Discovery.V8 and AxioCam MRc5.
In Vitro SUMOylation and SUBE Assays
Drosophila, Blattella, and human SUMOs were amplified using the primers described in supplementary table S10, Supplementary Material online. Each primer contains the T7 promoter sequence in the 5′-end followed by the sequence of HA tag. For SUMOylation of the Blattella nuclear receptors, the following cDNAs were used: BgEcR-A (EMBL: AM039690), BgRXR-L (EMBL: AJ854490), BgE75-A (EMBL: AM238653), BgHR3-A (EMBL: AM259128), BgHR3-B2 (EMBL: AM259130), and BgFtz-F1 (EMBL: FM163377). The products of amplification, or the cDNAs used as templates, were translated using TNT Wheat Germ extract (Promega) and, in the case of Blattella nuclear receptors, 35S-methionine (Amersham Biosciences and Pierce). All proteins were incubated in a buffer containing an ATP regenerating system (50 mM Tris pH 7.5, 10 mM MgCl2, 2 mM ATP, 10 mM creatine phosphate [Sigma], 3.5 U/ml of creatine kinase [Sigma], and 0.6 U/ml of inorganic pyrophosphatase [Sigma]), SUMO1 (for nuclear receptors) or SUMO2 and SUMO3 (for SUMO proteins) (5 µg each), Ubc9 (0.325 µg), and purified SAE1/2 (0.8 µg; ENZO Life Sciences). Reactions were incubated at 30°C for 2 h, resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and exposed.
For SUBE pulldown assays, one-tenth of input was saved and the rest of the reaction was incubated with 50 µl of Glutathione S-Transferase (GST)-agarose beads containing 50 µg of SUBEs or GST (data not shown) and 1 mM Dithiothreitol (DTT) for 2 h at 4 °C. After incubation, beads were pulled down by centrifugation and one-tenth of the unbound fraction was kept for western analysis. Subsequently, the same beads were washed with the binding buffer (50 mM Tris pH 8.5; 50 mM NaCl, 5 mM ethylenediaminetetraacetic acid, and 1% Igepal) and were resuspended in Laemmli Buffer before being loaded on a 12% SDS-PAGE.
Phylogenetic Analysis
For the phylogenetic analyses, we used the sequences detailed in supplementary file S1, Supplementary Material online. Protein sequences were aligned using ClustalX (Thompson et al. 1997) or MAFFT with default parameters (Katoh et al. 2002) and alignment-ambiguous regions were removed. The resulting alignment (99 positions) was analyzed by the PhyML program (Guindon and Gascuel 2003), based on the maximum-likelihood principle with the model of amino acid substitution. The data sets were bootstrapped for 100 replicates using PhyML. In the phylogenetic analysis with all eukaryote SUMO sequences, the tree was rooted by the plants.
Structural Analysis
Among the different three-dimensional structures available at the Protein Data Bank (PDB), those corresponding to the isolated SUMO proteins (not bound to other proteins) and spanning the longest amino acid sequences were chosen for comparison: HsSUMO2 (PDB: 2AWT), HsSUMO3 (PDB: 1U4A), and DmSmt3 (PDB: 2K1F). All three are solution Nuclear magnetic resonance (NMR) structures represented by ensembles of 20 models, and one model from each ensemble was used for the superposition shown in figure 1B. Because the N- and C-terminal ends were found disordered and flexible, superposition was based only on the well-defined regions (residues 18–90 of HsSUMO2, residues 17–89 of HsSUMO3, and residues 13–85 of DmSmt3) as seen in the ensembles. The superposition and the figure 1B were made with PyMol. The alignment with the secondary structure elements of DmSmt3 on top of it was prepared with ESPRIPT (Gouet et al. 2003).
Statistical Analysis
pH3 data were analyzed by using the R environment (R Development Core Team 2014). The normal distributions were performed using the Shapiro–Wilk test (R function Shapiro.test). The statistical significance of values between three groups was evaluated by ANOVA (analysis of variance) test followed by Bonferroni’s Honestly Significant Difference post hoc test. pH3 graphs were generated in R using a custom script based on the base boxplot function superimposed with individual data points plotted with the beeswarm function (package beeswarm).
Supplementary Material
Supplementary file S1, figures S1–S5, and tables S1–S10 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors thank Dr Jose Castresana and Dr Jesús Gómez-Zurita for advice and assistance during phylogenetic analysis, and Xavier Franch-Marro for helpful suggestions on the manuscript. E.U. was supported by a predoctoral fellowship from Consejo Superior de Investigaciones Cientificas. This work was funded by the Spanish Ministerio de Ciencia e Innovacion (Projects BFU2009-10571 to D.M. and BFU2011-25986 to R.B.). R.B. acknowledges the Consolider program (CSD2007-008-25120), the Departments of Education and Industry of the Basque Government (PI2012/42), and the Bizkaia County. This article is based upon work from COST Action (PROTEOSTASIS BM1307), supported by COST (European Cooperation in Science and Technology). The research has also benefited from FEDER.
References
- Abed M, Barry KC, Kenyagin D, Koltun B, Phippen TM, Delrow JJ, Parkhurst SM, Orian A. 2011. Degringolade, a SUMO-targeted ubiquitin ligase, inhibits Hairy/Groucho-mediated repression. EMBO J. 30:1289–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrio R, de Celis JF. 2004. Regulation of spalt expression in the Drosophila wing blade in response to the Decapentaplegic signaling pathway. Proc Natl Acad Sci U S A. 101:6021–6026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry KC, Abed M, Kenyagin D, Werwie TR, Boico O, Orian A, Parkhurst SM. 2011. The Drosophila STUbL protein Degringolade limits HES functions during embryogenesis. Development 138:1759–1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Békés M, Prudden J, Srikumar T, Raught B, Boddy MN, Salvesen GS. 2011. The dynamics and mechanism of SUMO chain deconjugation by SUMO-specific proteases. J Biol Chem. 286:10238–10247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof J, Maeda RK, Hediger M, Karch F, Basler K. 2007. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc Natl Acad Sci U S A. 104:3312–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118:401–415. [DOI] [PubMed] [Google Scholar]
- Bylebyl GR, Belichenko I, Johnson ES. 2003. The SUMO isopeptidase Ulp2 prevents accumulation of SUMO chains in yeast. J Biol Chem. 278:44113–44120. [DOI] [PubMed] [Google Scholar]
- Castorálová M, Březinová D, Svéda M, Lipov J, Ruml T, Knejzlík Z. 2012. SUMO-2/3 conjugates accumulating under heat shock or MG132 treatment result largely from new protein synthesis. Biochim Biophys Acta. 1823:911–919. [DOI] [PubMed] [Google Scholar]
- Citro S, Chiocca S. 2013. Sumo paralogs: redundancy and divergencies. Front Biosci (Schol Ed). 5:544–553. [DOI] [PubMed] [Google Scholar]
- Colombani J, Bianchini L, Layalle S, Pondeville E, Dauphin-Villemant C, Antoniewski C, Carré C, Noselli S, Léopold P. 2005. Antagonistic actions of ecdysone and insulins determine final size in Drosophila. Science 310:667–670. [DOI] [PubMed] [Google Scholar]
- Cruz J, Mané-Padrós D, Bellés X, Martín D. 2006. Functions of the ecdysone receptor isoform-A in the hemimetabolous insect Blattella germanica revealed by systemic RNAi in vivo. Dev Biol. 297:158–171. [DOI] [PubMed] [Google Scholar]
- Cruz J, Martín D, Bellés X. 2007. Redundant ecdysis regulatory functions of three nuclear receptor HR3 isoforms in the direct-developing insect Blattella germanica. Mech Dev. 124:180–189. [DOI] [PubMed] [Google Scholar]
- Cruz J, Martín D, Pascual N, Maestro J, Piulachs M, Bellés X. 2003. Quantity does matter. Juvenile hormone and the onset of vitellogenesis in the German cockroach. Insect Biochem Mol Biol. 33:1219–1225. [DOI] [PubMed] [Google Scholar]
- Cruz J, Nieva C, Mané-Padrós D, Martín D, Bellés X. 2008. Nuclear receptor BgFTZ-F1 regulates molting and the timing of ecdysteroid production during nymphal development in the hemimetabolous insect Blattella germanica. Dev Dyn. 237:3179–3191. [DOI] [PubMed] [Google Scholar]
- Da Silva-Ferrada E, Xolalpa W, Lang V, Aillet F, Martin-Ruiz I, de la Cruz-Herrera CF, Lopitz-Otsoa F, Carracedo A, Goldenberg SJ, Rivas C, et al. 2013. Analysis of SUMOylated proteins using SUMO-traps. Sci Rep. 3:1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohmen RJ, Stappen R, McGrath JP, Forrová H, Kolarov J, Goffeau A, Varshavsky A. 1995. An essential yeast gene encoding a homolog of ubiquitin-activating enzyme. J Biol Chem. 279:18099–18109. [DOI] [PubMed] [Google Scholar]
- Gareau JR, Lima CD. 2010. The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat Rev Mol Cell Biol. 11:861–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiss-Friedlander R, Melchior F. 2007. Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol. 8:947–956. [DOI] [PubMed] [Google Scholar]
- Gouet P, Robert X, Courcelle E. 2003. ESPript/ENDscript: extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 31:3320–3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 52:696–704. [DOI] [PubMed] [Google Scholar]
- Huang H, Du G, Chen H, Liang X, Li C, Zhu N, Xue L, Ma J, Jiao R. 2011. Drosophila Smt3 negatively regulates JNK signaling through sequestering Hipk in the nucleus. Development 138:2477–2485. [DOI] [PubMed] [Google Scholar]
- Johnson ES, Schwienhorst I, Dohmen RJ, Blobel G. 1997. The ubiquitin-like protein Smt3p is activated for conjugation to other proteins by an Aos1p/Uba2p heterodimer. EMBO J. 16:5509–5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D, Crowe E, Stevens TA, Candido EPM. 2001. Functional and phylogenetic analysis of the ubiquitylation system in Caenorhabditis elegans: ubiquitin-conjugating enzymes, ubiquitin-activating enzymes, and ubiquitin-like proteins. Genome Biol. 1:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanakousaki K, Gibson MC. 2012. A differential requirement for SUMOylation in proliferating and non-proliferating cells during Drosophila development. Development 139:2751–2762. [DOI] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knipscheer P, van Dijk WJ, Olsen JV, Mann M, Sixma TK. 2007. Noncovalent interaction between Ubc9 and SUMO promotes SUMO chain formation. EMBO J. 26:2797–2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao S, Wang T, Fan K, Tu X. 2010. The small ubiquitin-like modifier (SUMO) is essential in cell cycle regulation in Trypanosoma brucei. Exp Cell Res. 316:704–715. [DOI] [PubMed] [Google Scholar]
- Maestro O, Cruz J, Pascual N, Martín D, Bellés X. 2005. Differential expression of two RXR/ultraspiracle isoforms during the life cycle of the hemimetabolous insect Blattella germanica (Dictyoptera, Blattellidae). Mol Cell Endocrinol. 238:27–37. [DOI] [PubMed] [Google Scholar]
- Mané-Padrós D, Borràs-Castells F, Belles X, Martín D. 2012. Nuclear receptor HR4 plays an essential role in the ecdysteroid-triggered gene cascade in the development of the hemimetabolous insect Blattella germanica. Mol Cell Endocrinol. 348:322–330. [DOI] [PubMed] [Google Scholar]
- Mané-Padrós D, Cruz J, Vilaplana L, Nieva C, Ureña E, Bellés X, Martín D. 2010. The hormonal pathway controlling cell death during metamorphosis in a hemimetabolous insect. Dev Biol. 346:150–160. [DOI] [PubMed] [Google Scholar]
- Mané-Padrós D, Cruz J, Vilaplana L, Pascual N, Bellés X, Martín D. 2008. The nuclear hormone receptor BgE75 links molting and developmental progression in the direct-developing insect Blattella germanica. Dev Biol. 315:147–160. [DOI] [PubMed] [Google Scholar]
- Manza LL, Codreanu SG, Stamer SL, Smith DL, Wells KS, Roberts RL, Liebler DC. 2004. Global shifts in protein sumoylation in response to electrophile and oxidative stress. Chem Res Toxicol. 17:1706–1715. [DOI] [PubMed] [Google Scholar]
- Martín D, Maestro O, Cruz J, Mané-Padrós D, Bellés X. 2006. RNAi studies reveal a conserved role for RXR in molting in the cockroach Blattella germanica. J Insect Physiol. 52:410–416. [DOI] [PubMed] [Google Scholar]
- Matic I, van Hagen M, Schimmel J, Macek B, Ogg SC, Tatham MH, Hay RT, Lamond AI, Mann M, Vertegaal ACO. 2008. In vivo identification of human small ubiquitin-like modifier polymerization sites by high accuracy mass spectrometry and an in vitro to in vivo strategy. Mol Cell Proteomics. 7:132–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirth C, Truman JW, Riddiford LM. 2005. The role of the prothoracic gland in determining critical weight for metamorphosis in Drosophila melanogaster. Curr Biol. 15:1796–1807. [DOI] [PubMed] [Google Scholar]
- Misof B, Liu S, Meusemann K, Peters RS, Donath A, Mayer C, Frandsen PB, Ware J, Flouri T, Beutel RG, et al. 2014. Phylogenomics resolves the timing and pattern of insect evolution. Science (80-) 346:763–767. [DOI] [PubMed] [Google Scholar]
- Nacerddine K, Lehembre F, Bhaumik M, Artus J, Cohen-Tannoudji M, Babinet C, Pandolfi PP, Dejean A. 2005. The SUMO pathway is essential for nuclear integrity and chromosome segregation in mice. Dev Cell. 9:769–779. [DOI] [PubMed] [Google Scholar]
- Nie M, Xie Y, Loo JA, Courey AJ. 2009. Genetic and proteomic evidence for roles of Drosophila SUMO in cell cycle control, Ras signaling, and early pattern formation. PLoS One 4:e5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak M, Hammerschmidt M. 2006. Ubc9 regulates mitosis and cell survival during zebrafish development. Mol Biol Cell. 17:5324–5336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulin G, Dong Y, Fraser AG, Hopper NA, Ahringer J. 2005. Chromatin regulation and sumoylation in the inhibition of Ras-induced vulval development in Caenorhabditis elegans. EMBO J. 24:2613–2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. 2014. R: a language and environment for statistical computing. Vienna (Austria): R Foundation for Statistical Computing; Available from: http://www.R-project.org. [Google Scholar]
- Rosas-Acosta G, Russell WK, Deyrieux A, Russell DH, Wilson VG. 2005. A universal strategy for proteomic studies of SUMO and other ubiquitin-like modifiers. Mol Cell Proteomics. 4:56–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh H, Hinchey J. 2000. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J Biol Chem. 275:6252–6258. [DOI] [PubMed] [Google Scholar]
- Spradling AC, Rubin GM. 1982. Transposition of cloned P elements into Drosophila germ line chromosomes. Science 218:341–347. [DOI] [PubMed] [Google Scholar]
- Su H-L, Li SS-L. 2002. Molecular features of human ubiquitin-like SUMO genes and their encoded proteins. Gene 296:65–73. [DOI] [PubMed] [Google Scholar]
- Talamillo A, Herboso L, Pirone L, Pérez C, González M, Sánchez J, Mayor U, Lopitz-Otsoa F, Rodriguez MS, Sutherland JD, et al. 2013. Scavenger receptors mediate the role of SUMO and Ftz-f1 in Drosophila steroidogenesis. PLoS Genet. 9:e1003473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talamillo A, Sánchez J, Barrio R. 2008. Functional analysis of the SUMOylation pathway in Drosophila. Biochem Soc Trans. 36:868–873. [DOI] [PubMed] [Google Scholar]
- Talamillo A, Sánchez J, Cantera R, Pérez C, Martín D, Caminero E, Barrio R. 2008. Smt3 is required for Drosophila melanogaster metamorphosis. Development 135:1659–1668. [DOI] [PubMed] [Google Scholar]
- Tatham MH, Geoffroy M-C, Shen L, Plechanovova A, Hattersley N, Jaffray EG, Palvimo JJ, Hay RT. 2008. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat Cell Biol. 10:538–546. [DOI] [PubMed] [Google Scholar]
- Terada K, Furukawa T. 2010. Sumoylation controls retinal progenitor proliferation by repressing cell cycle exit in Xenopus laevis. Dev Biol. 347:180–194. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ureña E, Manjón C, Franch-Marro X, Martín D. 2014. Transcription factor E93 specifies adult metamorphosis in hemimetabolous and holometabolous insects. Proc Natl Acad Sci U S A. 111:7024–7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vertegaal ACO, Andersen JS, Ogg SC, Hay RT, Mann M, Lamond AI. 2006. Distinct and overlapping sets of SUMO-1 and SUMO-2 target proteins revealed by quantitative proteomics. Mol Cell Proteomics. 5:2298–2310. [DOI] [PubMed] [Google Scholar]
- Wang J, Wang S, Li S. 2014. Sumoylation modulates 20-hydroxyecdysone signaling by maintaining USP protein levels in Drosophila. Insect Biochem Mol Biol. 54C:80–88. [DOI] [PubMed] [Google Scholar]
- Wilkinson KA, Henley JM. 2010. Mechanisms, regulation and consequences of protein SUMOylation. Biochem J. 428:133–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, He Y, Qiang B, Yuan J, Peng X, Pan X-M. 2008. A novel method for high accuracy sumoylation site prediction from protein sequences. BMC Bioinformatics 9:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka N, Rewitz KF, O’Connor MB. 2013. Ecdysone control of developmental transitions: lessons from Drosophila research. Annu Rev Entomol. 58:497–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H, Zhou J, Deng M, Liu X, Le Bras M, de The H, Chen SJ, Chen Z, Liu TX, Zhu J. 2010. Small ubiquitin-related modifier paralogs are indispensable but functionally redundant during early development of zebrafish. Cell Res. 20:185–196. [DOI] [PubMed] [Google Scholar]
- Zhang F-P, Mikkonen L, Toppari J, Palvimo JJ, Thesleff I, Jänne OA. 2008. Sumo-1 function is dispensable in normal mouse development. Mol Cell Biol. 28:5381–5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.