Abstract
During 12 recent months of periodic influenza virus surveillance at 9 live poultry markets in Wuxi City China, we identified multiple highly pathogenic H5N6, H5N8, H5N2, and H5N1 avian influenza viruses. The variety of potentially pandemic viruses in this low-risk area is disconcerting and portends an increased pandemic threat.
Keywords: H5N1 influenza A virus, H5N2 influenza A virus, H5N6 influenza A virus, H5N8 influenza A virus, reassortant
Since the first detection of highly pathogenic avian influenza (HPAI) A(H5N1) virus in China in 1996, multiple virus clades [1] have evolved and spread across Asia, Africa, and Europe, causing tremendous economic losses to the poultry industry. Although H5N1 viruses have demonstrated reassortment of internal gene segments [2], major changes were rarely detected before 2013 [3]. However, since 2013, novel H5 viral subtypes (H5N2, H5N6, and H5N8) have emerged and have become globally widespread. These novel subtype viruses have caused multiple outbreaks in wild birds or poultry in Asia, Europe, and North America [4–6]. In China, H5N6 virus has caused 3 human infections in 2014 [7, 8], causing considerable public health concern. In this study, we report recent surveillance for influenza A viruses and the alarming recent detection of multiple H5 subtype viruses from 9 live poultry markets (LPMs) in Wuxi City, China. Wuxi City is a high-socioeconomic area thought to be at low risk of avian influenza virus (AIV) detections.
METHODS
Samples Collection
During July 2013 to June 2014, we conducted periodic active AIV surveillance in Wuxi City, Jiangsu Province, in eastern China. Wuxi City is a prosperous city in China with only 10 H7N9 detections in man since 2015. As permitted by approximately 70% of bird merchants, we collected cloacal and environmental specimens each month from a convenience sample of approximately 10 birds in each of 9 LPMs. This sampling approach yielded 403 cloacal swabs from predominantly healthy chickens, ducks, and geese and 548 environmental samples (water, fecal, and cage swabs). All samples collected from poultry and their environments were kept on ice for up to 4 hours before their preservation −80°C.
Virus Isolation, Subtyping, and Sequencing
All samples were inoculated into 9-day-old, specific pathogen-free embryonated chicken eggs for virus isolation in a biosafety level-3 laboratory. Hemagglutination (HA) assays were performed to detect influenza virus using horse red blood cell. In total, 26 samples were positive for HA activity. The influenza A-positive specimens revealed 5 subtypes: H2, H5, H7, H9, and H11. Nine of the 26 were H5-positive subtypes, and 7 of the H5-positive specimens had evidence of single virus infection: 3 with H5N6 virus (WX-H5N6), 1 with H5N8 virus (WX-H5N8), 2 with H5N1 virus (JS-H5N1), and 1 with H5N2 virus (WX-H5N2) (Table 1); the other 2 H5-positive specimens appeared to be mixed with a H9 subtype virus. Full viral genome sequence studies of the 7 specimens with single H5 virus detections were conducted using Ion Torrent PGM technology (Life Technologies, Grand Island, NY). Sequence data, except for PB2, PB1, and PA of H5N8 (which were not available due to the poor reads), were deposited in Global Initiative on Sharing All Influenza Data (accession nos. EPI_ISL_196571, EPI_ISL_196572, EPI_ISL_196573, EPI_ISL_196591, EPI_ISL_196592, EPI_ISL_196570, EPI_ISL_190702, and PI_ISL_190702). The H2 virus has been previously reported [9].
Table 1.
Selected Molecular Markers Detected in Avian Influenza A H5 Subtype Viruses in Present Study, China, 2013–2014
| Identified Virus | Collection Data | HA (H3 Numbering) |
PB2 |
NA |
M2 |
PB1-F2 |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cleavage Site | 110 | 158 | 160 | 226 | 228 | 318 | 591 | 627 | 701 | 274 | Stalk Deletion | 31 | 58–90 Truncated | ||
| CK/Jiangsu/927/2013(H5N1) | 18 November 2013 | RERRRKR/G | H | D | A | Q | G | T | Q | E | D | H | 49–68 | S | Yes |
| CK/Jiangsu/2477/2014(H5N1) | 17 June 2014 | RERRRKR/G | H | D | A | Q | G | T | Q | E | D | H | 49–68 | S | Yes |
| DK/Jiangsu/WX156/2013(H5N6) | 22 July 2013 | REKRRKR/G | H | N | A | Q | G | T | Q | E | D | H | No | S | Yes |
| CW/Jiangsu/WX175/2013(H5N6) | 12 July 2013 | REKRRKR/G | H | N | A | Q | G | T | Q | E | D | H | No | S | Yes |
| CK/Jiangsu/WXSB2/2014(H5N6)a | 20 August 2014 | RERRRKR/G | H | N | T | Q | G | T | Q | E | D | H | 58–68 | S | Yes |
| GS/Jiangsu/WX202/2013(H5N8) | 15 February 2014 | REKRRKR/G | H | N | T | Q | G | T | NA | NA | NA | H | No | N | NA |
| DW/Jiangsu/WX1761/2014(H5N2) | 18 April 2014 | REGRRRKR/G | H | N | T | Q | G | T | Q | E | D | H | No | N | No |
Abbreviations: CK, chicken; CW, contaminated water; DK, duck; DW, drinking water; NA, not available due the poor sequence data; WX, Wuxi.
a Identified in a sick chicken.
Phylogenetic Analysis
To understand the molecular epidemiology of the H5 viruses, we first examined 100 or more closely related sequences for each gene in the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/) to infer the overall topology, and then we removed a few extreme outlying sequences from the trees. Maximum-likelihood phylogenetic trees were inferred for all 8 segments of these viruses by using MEGA software, version 6.06 (www.megasoftware.net). To assess the robustness of individual nodes on phylogenetic trees, we used a bootstrap resampling process (1000 replications), the neighbor-joining method, and a best-fit, general, time-reversible model of nucleotide substitution.
RESULTS
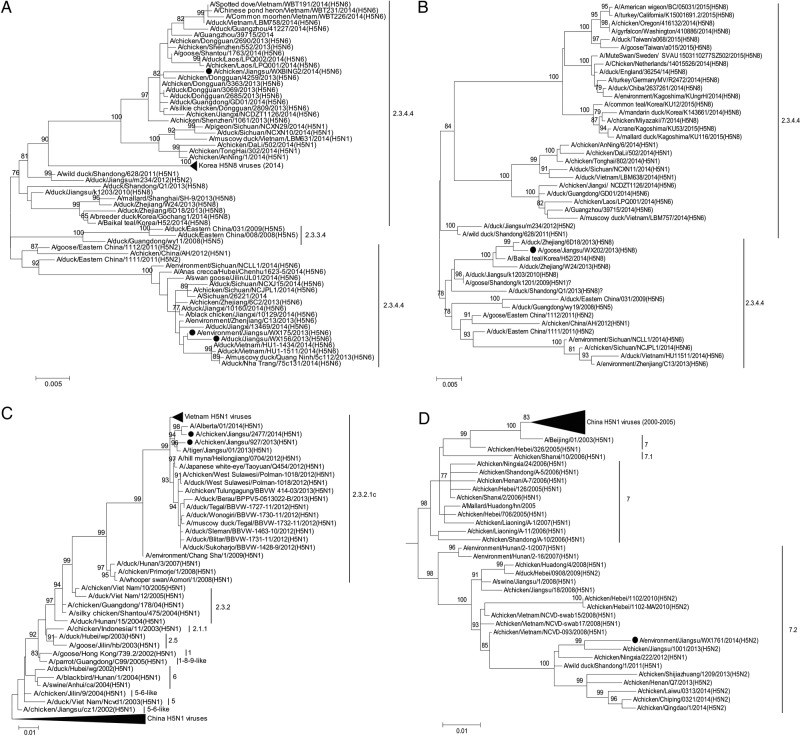
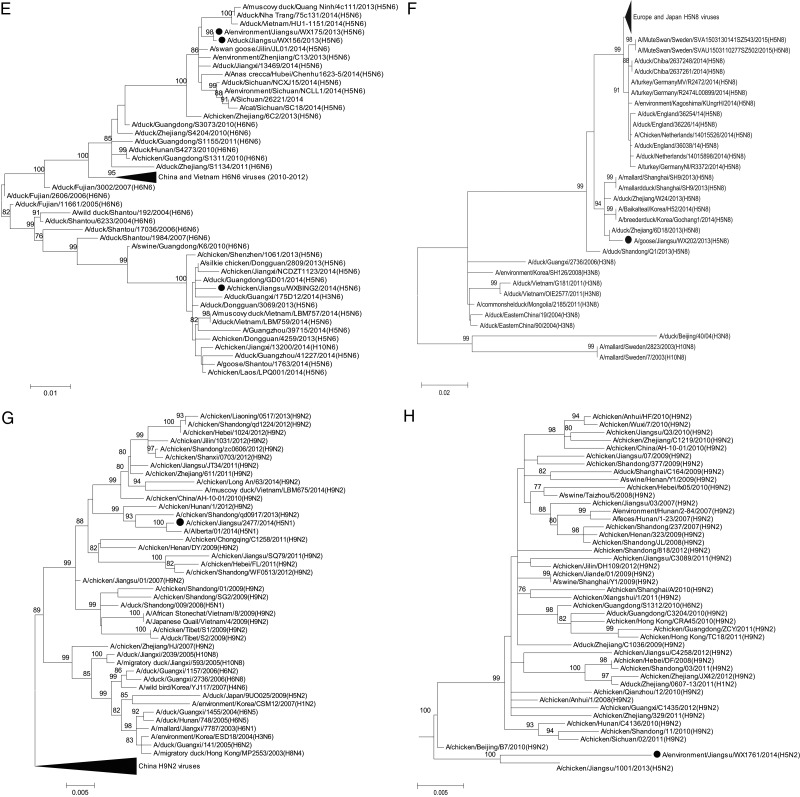
The HA gene phylogeny showed that the WXS-H5N6 virus was closely related to A/chicken/Dongguan/4259/2013(H5N6) from Guangdong Province, China of the H5N6 Jiangxi lineage, clade 2.3.4.4. The 2 WX-H5N6 viruses were closely related to A/environment/Zhejiang/C13/2013(H5N6) of the Sichuan lineage [10], which also belonged to clade 2.3.4.4. These 3 H5N6 viruses likely originated from H5 subtype HPAI viruses with N1, N2, and N8 neuraminidases (NAs) detected in poultry in China since 2010 and from viruses detected in Vietnam in 2014 (Figure 1A). The NA genes of these 3 H5N6 viruses also clustered into Sichuan and Jiangxi lineages and likely originated from H6N6 AIV circulating in the Fujian and Guangdong Provinces of China (Figure 1E). The 6 internal genes of 3 H5N6 viruses were found to cluster with H5N6 strains identified in China and Vietnam (Supplementary Figure 1).
Figure 1.
Maximum-likelihood phylogenetic tree for the hemagglutinin genes of highly pathogenic avian influenza A(H5N6) (A), A(H5N8) (B), A(H5N1) (C), and A(H5N2) (D) strains and the neuraminidase of highly pathogenic avian influenza A(H5N6) (E), A(H5N8) (F), and A(H5N2) (H) strains and for the basic polymerase 2 genes of A(H5N1/H9N2 7:1) (G) strain from Wuxi City, Jiangsu Province, China. Bootstrap values (n = 1000) at key nodes are indicated. Values lower than 75 were not shown. Scale bar indicates evolutionary distance (nucleotide substitutions per site). The black dots indicate the H5 subtype viruses reported in this study.
The HA gene of WX-H5N8 virus clustered with clade 2.3.4.4 H5N8 viruses that have been detected in China and South Korea since 2013. The HA sequence was most closely related to a duck isolate [A/duck/Zhejiang/6D18/2013(H5N8)] detected in Zhejiang Province, China (Figure 1B). However, this HA gene was notably different from H5N8 isolates detected in Japan, Europe, and North America (Figure 1B). Studies of the NA and 3 internal genes confirmed the clustering observed for HA gene (Figure 1F and Supplementary Figure 2). All 5 genes with available sequences data of WX-H5N8 were closely related to other H5 subtype viruses, such as A/duck/Zhejiang/6D18/2013(H5N8), A/Baikal teal/Korea/2014(H5N8), A/duck/Zhejiang/W24/2013(H5N8), and A/breeder duck/Korea/Gochang1/2014(H5N8), which circulated in China and South Korea.
Phylogenetic analyses of HA genes of the 2 JS-H5N1 viruses demonstrated that they both fell within clade 2.3.2.1c and were similar to H5N1 viruses detected in China and Vietnam identified during 2012 to 2013 (Figure 1C). The HA gene of JS2477/14 and JS927/13 were most closely related to the sequence of A/Alberta/01/2014(H5N1) virus that was isolated from the first human infection with H5N1 in Canada [11] and also closely related to A/tiger/Jiangsu/1/2013(H5N1), respectively (Figure 1C). They had 99.5% and 99.4% nucleotide (nt) similarity with A/Alberta/01/2014(/H5N1) and A/tiger/Jiangsu/1/2013(H5N1), respectively. The NA and internal genes, except for the PB2 gene of JS927/14, confirmed the clustering observed for HA gene, clustering with strains identified from China, Vietnam, and Indonesia (Supplementary Figure 3). The PB2 gene showed discordant clustering with H9N2 viruses identified from China and Vietnam during 2007–2013. It had the highest nt similarity (98.5%–99.6%) with Alberta/14, A/chicken/Hunan/1/2012(H9N2), and A/chicken/Shandong/qd0917/2013(H9N2), suggesting a reassortment with an H9N2-subtyping lineage PB2 gene (Figure 1G).
The HA gene of WX-H5N2 virus clustered with H5 subtype HPAI viruses with N1 and N2 subtypes (clade 7.2) circulating in Vietnam and China, and it had the highest nt similarity (97.3%−97.9%) with A/chicken/Jiangsu/1001/2013(H5N2) and A/chicken/Ningxia/222/2012(H5N1) (Figure 1D). The NA gene clustered with H9N2 viruses identified in China and had the same highest nt similarity (97.7%) with A/chicken/Jiangsu/1001/2013(H5N2) and A/chicken/Beijing/B7/2010(H9N2) but showed a distinct lineage (Figure 1H). In particular, the 6 internal genes also clustered with H9N2 subtype viruses circulating in China. All were closely related to A/chicken/Hebei/YT/2010(/H9N2), A/chicken/Beijing/B7/2010(H9N2), and A/chicken/Jiangsu/1001/2013(H5N2) (Supplementary Figure 4), indicating a reassortment with H9N2 viruses circulating in China.
The HA protein of the 7 subtype H5 viruses possessed the polybasic amino acid (aa) cleavage site, indicating all were of the highly pathogenic phenotype (Table 1). In addition, the HA proteins of these viruses had the aas Q226 and G228 (H3 numbering), indicating that the viruses preferentially bind to avian-like receptors [12]. The NA stalk of these H5N1 viruses possessed a 20-aa deletion (positions 49–68), which might enhance virus adaptation to domestic fowl and increase virulence in mammals [13, 14]. The 2 WXS-H5N6 viruses had a deletion of 11 aa residues at positions 58–68 (N6 numbering) in the NA stalk region, which suggested that these viruses might have different adaptation and virulence characteristics in poultry and mammals [10]. Moreover, these H5 subtype viruses contained a truncated PB1-F2 protein of 57 aa in length (except for WX-H5N2), which might influence their virulence in mammals [15]. Amino acids Q591, E627, and D701 were observed in the PB2 protein, suggesting that these novel H5N1 viruses had not adapted to infect mammals [16]. No drug resistance-associated mutations were observed in our detected viruses isolates except that mutation 31N in M2 protein was found in the WX-H5N8 and WX-H5N2 strains.
DISCUSSION
In our limited surveillance effort, we identified 3 H5N6 viruses, 1 H5N8 virus, 1 H5N2 virus, and 2 H5N1 viruses from 9 LPMs from a high-socioeconomic area of eastern China during a 12-month period in 2013−2014. Genetic evidence suggested that these viruses were HPAI and the progeny of multiple H5, H6, and H9N2 AIV strains previously detected in Asia. In addition, the H7, H9, and H11 viruses were also identified, which will be reported in a separate work.
Phylogenetic and antigenic analyses suggest that 3 H5N6 viruses belong to clade 2.3.4.4 and have evolved into 2 subclades (Jiangxi and Sichuan) with altered antigenicity. Moreover, a very close genetic relationship between the human and avian H5N6 viruses was observed, suggesting that direct contact with poultry contributed to the human infection. Furthermore, these H5N6 viruses have distinct evolutionary characteristics, such as the A160 mutation in the HA protein and an 11-aa deletion in the NA stalk, which may enhance their adaptation and infectivity in mammals, including humans. Although all 11 human infections in China had recent history of direct contact with poultry, the potential for infection, outbreaks, and pandemic in humans should be carefully monitored. Similar to these H5N6 viruses in this study, 1 WX-H5N8 virus also belonged to clade 2.3.4.4, but its genes were derived from different subtype viruses. The WX-H5N8 virus was very closely related to the H5N8 virus isolated in China and South Korea, but it was slightly different from the H5N8 viruses detected in European, Japan, and North America (Figure 1B and 1F), which may reflect the virus evolution among migratory wild birds through overlapping flyways. Fortunately, genetic analyses demonstrated that our WX-H5N8 virus had no molecular evidence of mammalian adaption. However, with continued evolutionary pressure, the virus could pose future health threats to humans.
Our phylogenetic analysis suggested that the WX-H5N2 virus in this study possessed an HA gene from the clade 7.2 H5N1 virus. However, the remaining 7 genes of the virus were likely from A/chicken/Hebei/YT/2010-like and A/chicken/Beijing/B7/2010-like H9N2 viruses. The WX-H5N2 strain possessed the characteristic sequence of the HPAI virus at the cleavage site, suggesting that the H5N2 virus inherited a highly pathogenic phenotype from the reassortment involved in HPAI H5N1 viruses. It is also interesting to note that a novel reassortant JS-H5N1/2477 virus (H5N1/H9N2 7:1 PB2; clade 2.3.2.1c) in this study was similar to a H5N1 virus identified in a man in Canada in January of 2013 [11] and in a tiger found deceased in a zoo in China in 2013 [17]. Considering the widespread cocirculation of H5N1 and H9N2 viruses with other subtypes of AIVs in China, the emergence of a variety of potentially pandemic H5 subtype viruses is alarming. Although it is hard to identify (1) where and when the reassortment event occurred in our strain or (2) where future such reassortant or recombinant events will be identified, it seems probable that our H5N2 strain's relationship with H5N1 and H9N2 viruses identified in China, Vietnam, and Indonesia could be due to the cross-border movement of viruses between China and other countries.
CONCLUSIONS
In conclusion, we are concerned that these HPAI H5 viruses may be more widespread than we have recognized and that they may portend increased H5 pandemic risk. It now seems extremely important to increase surveillance for novel influenza virus in poultry in China and to conduct interventions to stop their spread.
Supplementary Data
Supplementary material is available online at Open Forum Infectious Diseases online (http://OpenForumInfectiousDiseases.oxfordjournals.org/).
Acknowledgments
Author contributions. M.-J. M. and W.-C. C. conceived the study design. M.-J. M., S.-H. C., T. Z., Y.-H. Q., G.-L. W., M.-N. W., Y. L., and B. L. performed the laboratory analysis. M.-J. M., G.-L. W., M.-N. W., and Y. L. performed sequences data analysis. M.-J. M. drafted the manuscript. All authors critically reviewed the manuscript. All authors read and approved the final manuscript.
Disclaimers. The funders had no role in study design, data collection and analysis, preparation of the manuscript, or the decision to publish.
Financial support. This work was supported by the Program of International Science & Technology Cooperation of China (2013DFA30800), the National Natural Science Foundation of China (81402730), the US National Institute for Allergy and Infection Diseases (R01AI108993-01A1; to G. C. G.), the Program of Jiangsu Provincial Science & Technology (H201448), and the Major Project of Wuxi Health Bureau (Z201404).
Potential conflicts of interest. All authors: No reported conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
References
- 1.Toward a unified nomenclature system for highly pathogenic avian influenza virus (H5N1). Emerg Infect Dis 2008; 14:e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lei F, Shi W. Prospective of genomics in revealing transmission, reassortment and evolution of wildlife-borne avian influenza A (H5N1) viruses. Curr Genomics 2011; 12:466–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao K, Gu M, Zhong L et al. Characterization of three H5N5 and one H5N8 highly pathogenic avian influenza viruses in China. Vet Microbiol 2013; 163:51–7. [DOI] [PubMed] [Google Scholar]
- 4.World Health Organization. Antigenic and genetic characteristics of zoonotic influenza viruses and development of candidate vaccine viruses for pandemic preparedness. Available at: http://www.who.int/influenza/vaccines/virus/201502_zoonotic_vaccinevirusupdate.pdf?ua=1 Accessed 26 February 2015.
- 5.European Food Safety Authority. Highly pathogenic avian influenza A subtype H5N8. Parma, Italy: EFSA, 2014. Available at: http://www.efsa.europa.eu/en/efsajournal/doc/3941.pdf Accessed 5 December 2014.
- 6.Jhung MA, Nelson DI. Outbreaks of avian influenza A (H5N2), (H5N8), and (H5N1) among birds--United States, December 2014-January 2015. MMWR Morb Mortal Wkly Rep 2015; 64:111. [PMC free article] [PubMed] [Google Scholar]
- 7.Pan M, Gao R, Lu Q et al. Human infection with a novel highly pathogenic avian influenza A (H5N6) virus: virological and clinical findings. J Infect 2016; 72:52–9. [DOI] [PubMed] [Google Scholar]
- 8.Yang ZF, Mok CK, Peiris JS, Zhong NS. Human infection with a novel avian influenza A (H5N6) virus. N Engl J Med 2015; 373:487–9. [DOI] [PubMed] [Google Scholar]
- 9.Ma MJ, Yang XX, Qian YH et al. Characterization of a novel reassortant influenza A virus (H2N2) from a domestic duck in Eastern China. Sci Rep 2014; 4:7588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bi Y, Mei K, Shi W et al. Two novel reassortants of avian influenza A (H5N6) virus in China. J Gen Virol 2015; 96:975–81. [DOI] [PubMed] [Google Scholar]
- 11.Pabbaraju K, Tellier R, Wong S et al. Full-genome analysis of avian influenza A(H5N1) virus from a human, North America, 2013. Emerg Infect Dis 2014; 20:887–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stevens J, Blixt O, Tumpey TM et al. Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science 2006; 312:404–10. [DOI] [PubMed] [Google Scholar]
- 13.Matsuoka Y, Swayne DE, Thomas C et al. Neuraminidase stalk length and additional glycosylation of the hemagglutinin influence the virulence of influenza H5N1 viruses for mice. J Virol 2009; 83:4704–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou H, Yu Z, Hu Y et al. The special neuraminidase stalk-motif responsible for increased virulence and pathogenesis of H5N1 influenza A virus. PLoS One 2009; 4:e6277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zamarin D, Ortigoza MB, Palese P. Influenza A virus PB1-F2 protein contributes to viral pathogenesis in mice. J Virol 2006; 80:7976–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Z, Chen H, Jiao P et al. Molecular basis of replication of duck H5N1 influenza viruses in a mammalian mouse model. J Virol 2005; 79:12058–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He S, Shi J, Qi X et al. Lethal infection by a novel reassortant H5N1 avian influenza A virus in a zoo-housed tiger. Microbes Infect 2015; 17:54–61. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.