Abstract
Hydrogels are an important class of biomaterials that have been widely utilized for a variety of biomedical/medical applications. The biological performance of hydrogels, particularly those used as wound dressing could be greatly advanced if imbued with inherent antimicrobial activity capable of staving off colonization of the wound site by opportunistic bacterial pathogens. Possessing such antimicrobial properties would also protect the hydrogel itself from being adversely affected by microbial attachment to its surface. We have previously demonstrated the broad-spectrum antimicrobial activity of supramolecular assemblies of cationic multi-domain peptides (MDPs) in solution. Here, we extend the 1-D soluble supramolecular assembly to 3-D hydrogels to investigate the effect of the supramolecular nanostructure and its rheological properties on the antimicrobial activity of self-assembled hydrogels. Among designed MDPs, the bactericidal activity of peptide hydrogels was found to follow an opposite trend to that in solution. Improved antimicrobial activity of self-assembled peptide hydrogels is dictated by the combined effect of supramolecular surface chemistry and storage modulus of the bulk materials, rather than the ability of individual peptides/peptide assemblies to penetrate bacterial cell membrane as observed in solution. The structure–property–activity relationship developed through this study will provide important guidelines for designing biocompatible peptide hydrogels with built-in antimicrobial activity for various biomedical applications.
Introduction
Hydrogels have been extensively utilized as implantable soft biomaterials for a variety of biomedical applications such as drug delivery, cell scaffolds, wound dressings, and biosensing.1–17 The interface between the implants and biological tissues provides an ideal environment where both chemical and biological functionalities can be modulated to facilitate specific biological processes such as sensing, wound healing and tissue regeneration. However, the hydrogel surface could also be attractive to a variety of microorganisms which may adhere to and grow within the matrix during any step in materials preparation and formulation, cell and tissue culture, in vivo administration and post implantation. Microbe invasion will not only cause malfunction of the implantable hydrogel materials/devices by surface passivation, but can also increase the rate of patient morbidity and mortality as a result of microbial infections. Developing hydrogels that possess inherent antimicrobial activity represents a promising approach that will significantly alleviate any safety concerns associated with microbial contamination of the matrix during and post implantation.6,18–20
Several classes of hydrogels possessing inherent antimicrobial activity have been developed based on natural polymeric hydrogels, synthetic cationic polymeric hydrogels and peptide hydrogels.6,20–32 Amphiphilic peptides consisting of cationic and hydrophobic residues can be rationally designed to mimic the structure of natural antimicrobial peptides (AMPs) and can readily be used as the molecular building blocks for functional supramolecular hydrogels with inherent antimicrobial activity. Several groups have demonstrated the concept of de novo designed amphiphilic peptide hydrogels with broad-spectrum antimicrobial activity while having minimum cytotoxicity to eukaryotic cells.24,27,30,31,33,34 Structure–activity correlations have been thoroughly investigated on the molecular level and the antimicrobial activity was found to be related to the side chain functionality of the charged residues (lysine vs. arginine), charge density, charge accessibility and amphiphilicity.
Recently, by using self-assembled multidomain peptides (MDPs) as a model system, our group for the first time demonstrated the effect of supramolecular structures and interactions on the antimicrobial activity of peptide assemblies in soluble state.35 The antimicrobial activity of the soluble peptide nanofiber is inversely related to its β-sheet content. Supramolecular structures with partially folded β-sheets dramatically enhanced the antimicrobial efficacy compared to those consisting of well-defined β-sheets. While working on the formulation and structure–activity analysis of the soluble self-assembled antimicrobial nanofibers (SAANs) for systemic delivery of AMPs, we observed strong antimicrobial activity of MDPs when being formulated into hydrogels under physiological conditions above 0.5 wt%. MDP-based hydrogels have shown great promise in tissue engineering as stem cell scaffolds and most recently as the extracellular matrix mimics to promote angiogenesis.36,37 Given the biological significance and utility of this new type of material as implantable cell/tissue scaffolds, in the current study, we extended the 1-D soluble supramolecular assembly to 3-D hydrogels to systematically investigate the effect of the supramolecular structure and its rheological properties on the antimicrobial activity of MDP-based peptide hydrogels. Through these studies, we have discovered important information on the supramolecular antimicrobial peptides’ structure–activity relationships at different assembly dimensions that we share herein.
Table 1 shows the four sequences used in the current study for the investigation of the effect of supramolecular nanostructure and their rheological properties on the antimicrobial activity of MDP hydrogels. All peptides contain six repeating units of alternating hydrophilic (Q)–hydrophobic (L) residues that favor the formation of β-sheets. The central (QL) domain was flanked by two charged domains comprised of two or three lysine residues that are used to induce hydrogel formation in phosphate buffer. Tryptophan (W) was incorporated for accurate concentration determination. The molecular and supramolecular structure can be modulated by varying the W position in the flanking charge domain without disrupting the central (QL) domain so as to not significantly alter the driving force for the supramolecular nanofiber formation and stabilization. Positioning W between the (QL) domain and the lysine domain makes an additional hydrophilic (K)–hydrophobic (W) unit on K2W-(QL)6K2 and K3W(QL)6K2 leading to reinforced supramolecular packing compared to their constitutional isomers WK2(QL)6K2 and WK3(QL)6K2, respectively. As demonstrated previously, enhanced supramolecular order compromised the antimicrobial activity of the soluble MDP nanofibers.35 However, when MDPs form hydrogels, the antimicrobial activity increased with optimization of the supramolecular nanostructure and its rheological properties. The structure–activity relationships observed for MDP nanofibers at different dimensions may reflect distinct molecular mechanisms adopted by soluble peptide/polymers (membrane penetration) and hydrogels (anionic sponges) to interact with and disrupt bacterial cell membranes.20,38 Current studies suggest that molecular folding/conformation, assembly states and bulk mechanical properties are among the important design parameters to be considered for future development of more advanced and highly effective antimicrobial hydrogels.
Table 1.
Primary sequences of MDPs used in the study
| Peptide | N-terminus | Peptide sequencesa | C-terminus | Abbreviation |
|---|---|---|---|---|
| 1 | CH3CO– | KKWQLQLQLQLQLQLKK | CONH2 | K2W(QL)6K2 |
| 2 | CH3CO– | WKKQLQLQLQLQLQLKK | CONH2 | WK2(QL)6K2 |
| 3 | CH3CO– | KKKWQLQLQLQLQLQLKK | CONH2 | K2W(QL)6K2 |
| 4 | CH3CO– | WKKKQLQLQLQLQLQLKK | CONH2 | WK3(QL)6K2 |
Alternating hydrophilic–hydrophobic repeating units are highlighted in bold. Peptides 1 and 3 contain seven repeating units and their constitutional isomer peptides 2 and 4 contain six.
Experimental methods
Peptide synthesis and characterization
All peptides were synthesized on a PS3 peptide synthesizer using MBHA rink amide resin through a standard Fmoc-solid phase peptide synthesis. 20% (v/v) piperidine in DMF was used to deprotect Fmoc groups. HCTU and DIPEA were used as amino acid coupling reagents in a molar ratio of 1 : 1 : 2.5 (amino acid : HCTU : DIPEA). Fmoc protected amino acids were added in 4 equivalents to the resin with double coupling performed. The N-terminus was acetylated in the presence of 50 equivalents of acetic anhydride and 6 equivalents of DIPEA in DMF. Peptides were cleaved from the resin in a mixture of TFA/Tis/water (95/2.5/2.5 by volume) for 3 h. TFA solution was collected and the resin was rinsed twice with neat TFA. After evaporation of the combined TFA solutions, residual peptide solution was triturated with chilled diethyl ether. The resulting precipitate was centrifuged and washed five times with chilled diethyl ether. Crude peptides were dried under vacuum overnight. Due to the exceptional purity of the crude products shown on HPLC (Fig. S1†), we followed the previously reported procedure5,36,37,39–41 for MDP purification through a simple yet extensive dialysis (8 cycles with DI water exchange in every 6 h) of peptides against DI water to remove small molecule impurities during the cleavage reaction. Dialyzed peptides were subsequently lyophilized to yield purified powder. The identity of each peptide was confirmed by MALDI-TOF mass spectrometry (Applied BioSystems Voyager-DE Pro.) using α-cyano-4-hydroxycinnamic acid as the matrix. K2W(QL)6K2, expected [M + H]+ = 2206.7, observed [M + H]+ = 2207.4, WK2(QL)6K2, expected [M + H]+ = 2206.7, observed [M + H]+ = 2207.6, K3W(QL)6K2, expected [M + H]+ = 2334.9, observed [M + H]+ = 2336.5, WK3(QL)6K2, expected [M + H]+ = 2334.9, observed [M + H]+ = 2337.1.
Transmission electron microscopy (TEM)
5 µl of hydrogel (0.5 wt%) was dropped onto a TEM grid. After 2 minutes, excess hydrogel was gently removed with filter paper and the sample was allowed to dry for another minute. 10 µl of negative staining reagent (2 wt% uranyl acetate/water solution) was added onto the grid. The excess staining solution was wicked off by filter paper after 2 minutes. Finally, the sample was allowed to dry overnight before imaging by JEOL 2010 high-resolution TEM.
Oscillatory rheology
The rheological properties of hydrogels were characterized by oscillatory rheology using a discovery hybrid rheometer (TA instruments) with a 12 mm stainless steel parallel plate geometry at 25 °C. 2 wt% hydrogel was prepared by equal volumetric mixing of 4 wt% peptide solution (dissolved in deionized water) and 2× PBS in a syringe, and the hydrogel was equilibrated in the syringe overnight before rheological measurement. ~50 µl of hydrogel was delivered onto the rheometer plate, immediately followed by adjustment of the parallel plate geometry to have a gap height of 350 µm. A dynamic time sweep was performed first for 15 minutes (frequency: 6 rad s−1, strain: 0.2%), followed by a dynamic frequency sweep (frequency range: 1–100 rad s−1, strain: 0.2%). Next, a 1000% strain was applied at a frequency of 6 rad s−1 for 30 seconds to completely disrupt the hydrogel and the storage modulus recovery was monitored by applying another dynamic time sweep (15 minutes, frequency: 6 rad s−1, strain: 0.2%) following the shear deformation. Finally, dynamic strain sweep (strain range: 0.01%–100%, frequency: 6 rad s−1) was performed to determine the yield strain of each hydrogel sample.
Bacterial culture
Staphylococcus aureus (6538) was ordered from Presque Isle Cultures. The bacterium was cultured in Mueller Hinton Broth (23 g L−1 in deionized water) under constant shaking at 100 rpm at 37 °C to reach their mid-exponential growth phase and was used for the evaluation of antimicrobial activity.
Bacterial live/dead assay
Agar plates were prepared by placing 300 µl of autoclaved mixed solution (50 °C) of agar (15 g L−1 in deionized water) and MHB (23 g L−1 in deionized water) in a confocal dish. The solution was allowed to solidify overnight. 10 µl of the bacterial culture (108 CFU ml−1) was dropped onto the surface of the agar plate and then incubated at 37 °C for 1 h. 40 µl of peptide hydrogel (2 wt%) was delivered through a syringe onto the surface of the agar plate where the bacteria were inoculated. After 16 h of incubation at 37 °C, 25 µl of live/dead assay kit solution was dropped onto the hydrogel. The live/dead assay was performed and the results analyzed by confocal microscopy (Leica, SP2 instrument).
Bacterial cell viability assay
2 µl of bacterial solution (108 CFU ml−1) was dropped on the prepared agar plate as described above. 40 µl of hydrogel was delivered by a syringe to cover the bacteria-inoculated agar surface, followed by incubation for 16 h at 37 °C. Over 16 h of incubation control bacteria (without peptide treatment) grew into opaque colonies with a diameter of <5 mm. The volume of the hydrogel was found to be sufficient to cover the entire bacterial colony, thus eliminating the interference of bacterial growth outside the hydrogel during 16 h of incubation and indeed reflect the peptide’s ability to kill the bacteria underneath. The hydrogel then was gently removed along with the underlying agar on which bacteria were growing and transferred to 10 ml of MHB medium solution (23 g L−1). Thorough vortexing ensured dissolution and suspension of the growing bacteria in MHB medium followed by 10-fold serial dilution of the original bacterial suspension and plating onto agar plates. The bactericidal activity of MDP hydrogels was quantified through bacterial counting on the agar plates and values were normalized to the control group which was not exposed to the hydrogel. Each assay was performed in triplicate in two independent experiments.
Bacterial motility imaging
20 µl of hydrogel (2 wt%) or PBS (control group) was syringe delivered to a confocal dish followed by the addition of 10 µl of S. aureus culture solution (108 CFU mL−1) and 10 µl of bacterial live assay kit solution (SYTO 9, 12 µM in PBS). A glass coverslip was placed on top of the confocal dish and further sealed by a parafilm to prevent solution evaporation and gel drying. After co-incubation for 10 min, the confocal dish was placed under the confocal microscope and the motion of S. aureus was acquired every 3 seconds for a total of 5 min.
Hemolytic activity assay
Peptide hydrogel (2 wt%) was prepared in a 96-well plate as follows: 50 µL PBS (2×) was placed in each well, followed by the addition of 50 µL of 4 wt% peptide stock solution. The peptide hydrogel was equilibrated in a shaker for 3 h at 37 °C. Afterwards, 200 µl PBS (1×) was gently added onto the surface of the hydrogel and the peptide hydrogel was allowed to incubate overnight in the shaker at 37 °C. 200 µl PBS (1×) was gently removed before cell seeding. Human red blood cells (hRBCs) donated from healthy volunteers were washed three times with equal volumes of 1× PBS followed by centrifugation for 10 min at 3460 rpm. The supernatant was removed after centrifugation. 100 µL of hRBCs were resuspended in 20 mL PBS buffer resulting in a 0.5% (v/v) stock suspension. 200 µL of the hRBCs stock suspension was introduced into the hydrogels and a control surface. Negative and positive controls were included by adding 100 µl PBS (1×) and 100 µl 1% Triton X-100 solution to hRBCs on the control surface, respectively. Samples were incubated at 37 °C on a shaker for 1 h. The solution on the top of the surfaces was removed and centrifuged at 14 000 rpm for 10 min. 100 µl of the supernatant was transferred to another 96-well plate and the released hemoglobin was calculated based upon the absorbance at 405 nm. Each assay was performed in triplicate. The percentage of intact hRBCs was calculated using the following equation:
Results and discussion
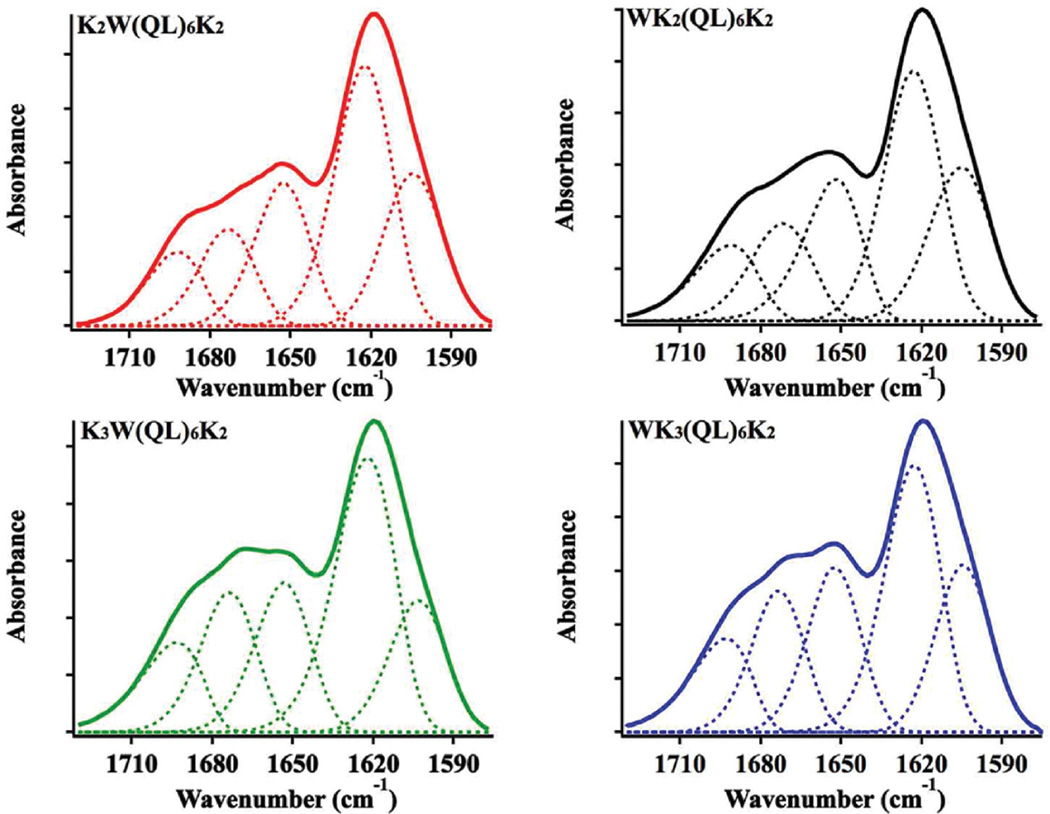
Upon charge neutralization under physiological buffer conditions, all four peptides exhibited the ability to self-assemble into hydrogels above a critical concentration at 0.5 wt%. The secondary structures of peptide hydrogels at 2 wt% were determined by FT-IR spectroscopy (Fig. 1), showing a predominant β-sheet secondary structure as characterized by the strong absorption at 1625 cm−1 in the amide I region for all four peptides. The shoulder peak at 1695 cm−1 is indicative of an antiparallel packing of β-sheets, in agreement with previous experimental results and molecular dynamic simulation.42,43 Absorbance at 1673 cm−1 could be due to residual TFA which was found to be critical to facilitate the dissolution of peptides at a sufficiently high concentration (4 wt%) in water. We attempted to remove TFA through various desalting procedures, but the materials were found to aggregate in water stock solution within seconds before gelation is triggered through the addition of exogenous salts. Therefore, throughout the study, further efforts to remove TFA were not conducted and the results based on TFA containing MDPs are consistent using several different synthesis batches of peptides. The IR peaks at 1655 cm−1 and 1602 cm−1 were attributed to the absorption of the glutamine side chain and the indole group on tryptophan respectively.44 Circular dichroism (CD) spectroscopy (Fig. S2†) confirmed the formation of β-sheets as the predominant protein secondary structure for all MDPs which showed a minimum absorption at 216 nm as a result of n–π* transition of the peptide bonds. The difference in terms of peak intensity is largely due to different solution viscosity and turbidity exhibited by each MDP hydrogel. The translucent and highly viscous peptide hydrogels may induce light scattering to varying degrees so that the amplitude of CD absorption varies among different samples and/or peptides.45,46 However, the position of the minimum CD absorption suggested that at a relatively high solution concentration all four peptides self-assembled into predominantly β-sheets.
Fig. 1.
Amide I region of FTIR spectra of peptide hydrogels (2 wt%) formed by K2W(QL)6K2, WK2(QL)6K2, K3W(QL)6K2 and WK3(QL)6K2 after complete drying. Solid curve: original IR spectra. Dashed line: deconvolution of the original spectra.
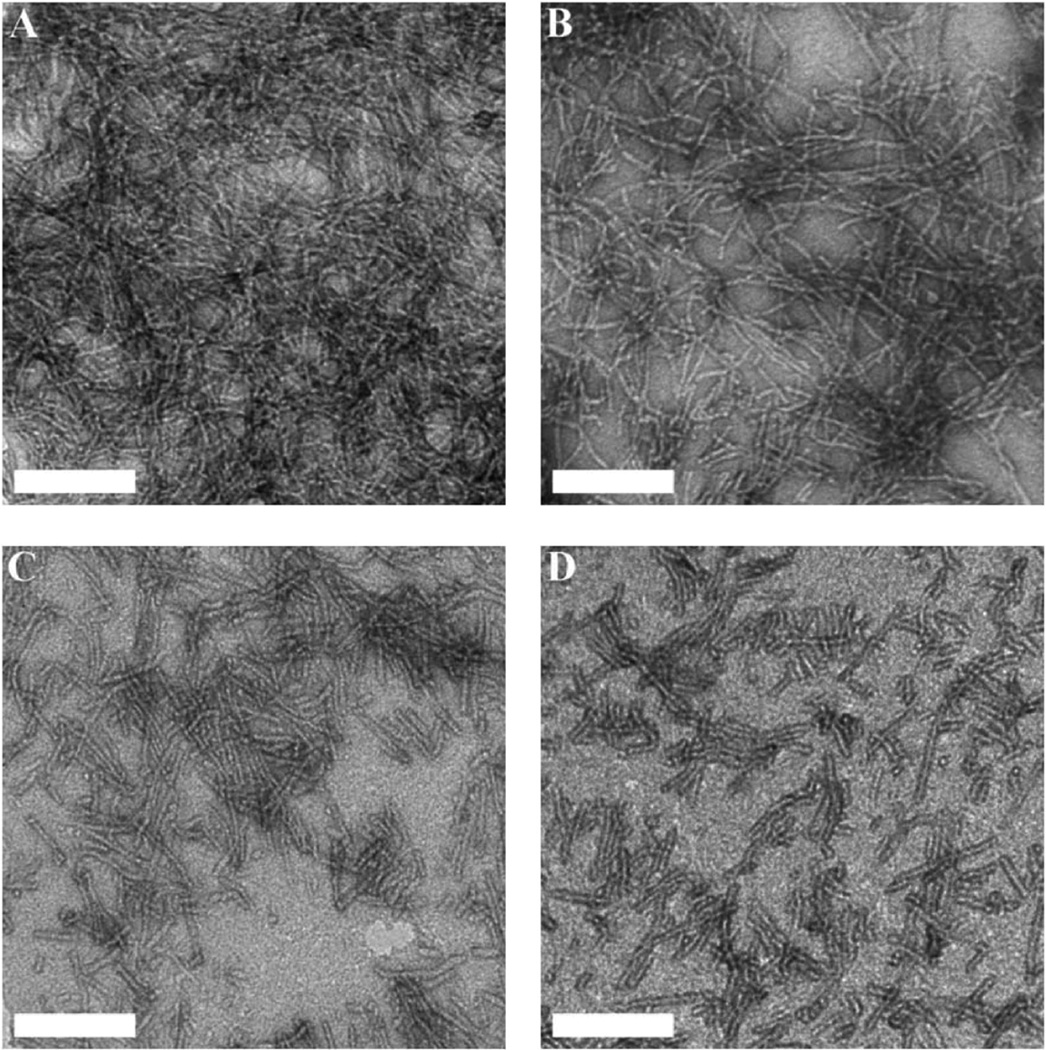
All hydrogels displayed nanostructured fiber morphology as a result of the packing of β-sheets driven by the attractive molecular interaction between the (QL) repeating units (Fig. 2). However, the degree of the supramolecular polymerization varies and is largely dictated by peptide primary structures featuring different ratios of core/terminal domain. K2W(QL)6K2 contains a core domain of seven hydrophilic–hydrophobic repeating units, i.e. one “KW” + six “QL” while its constitutional isomer WK2(QL)6K2 consists of six “QL” repeating units and the same number of lysine residues. The additional KW unit allows K2W(QL)6K2 to shift the balance toward supramolecular assembly leading to elongated nanofibers compared to WK2(QL)6K2 (Fig. 2).
Fig. 2.
TEM images of dried peptide hydrogels (0.5 wt%) formed by (A) K2W(QL)6K2, (B) WK2(QL)6K2, (C) K3W(QL)6K2 and (D) WK3(QL)6K2. Scale bar: 100 nm.
Densely packed nanofiber networks are visible on the negatively-stained TEM image of K2W(QL)6K2 hydrogels. WK3(QL)6K2 with a suboptimal balance of attractive/repulsive forces displays individually dispersed short nanofibers. K3W(QL)6K2 of the same chemical composition, but with an additional KW unit to drive the supramolecular assembly, showed nanofibers of increasing length that can potentially form physical crosslinks. The supramolecular structural morphologies correlate well with the storage moduli displayed by the four peptide hydrogels as discussed below.
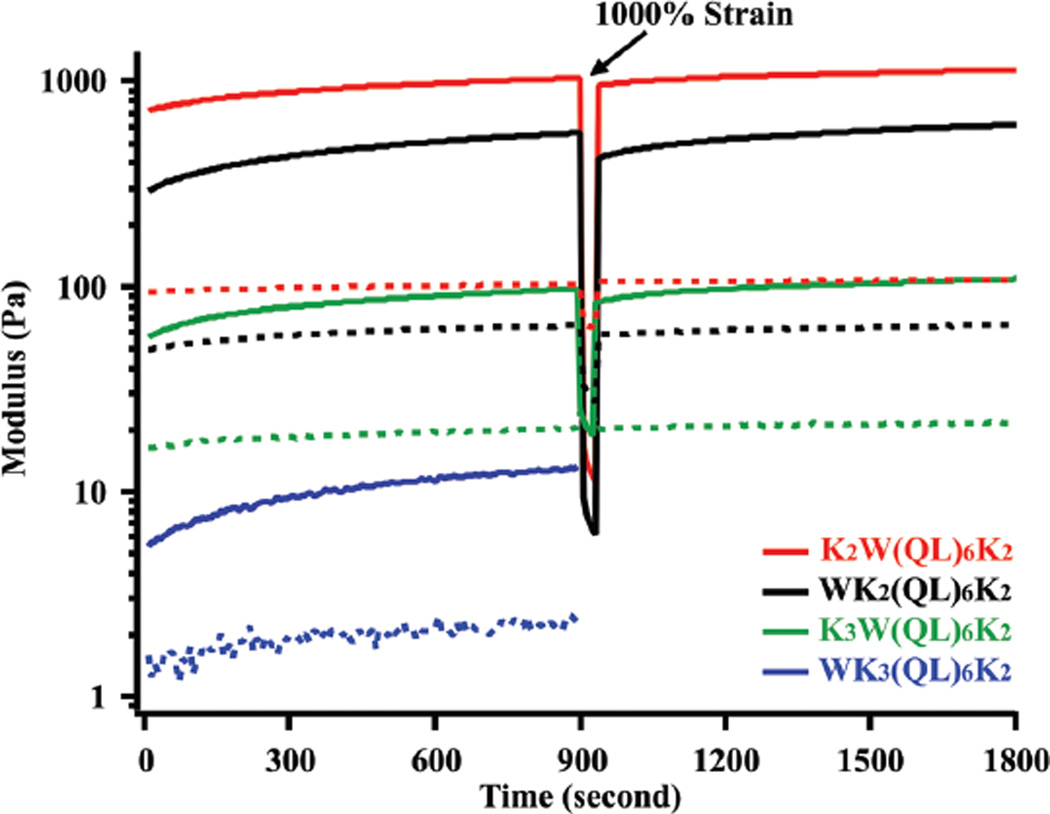
The rheological properties of peptide hydrogels were characterized by oscillatory rheology and the results are shown in Fig. 3, S3 and S4.† For all peptide hydrogels at 2 wt%, the storage modulus (G′) is significantly larger than the loss modulus (G″) within the relevant linear viscoelastic region, confirming the formation of elastic hydrogels. The hydrogels, however, demonstrated storage moduli across two orders of magnitude. K2W(QL)6K2 formed self-supporting, very rigid hydrogels with the highest storage modulus at ~1200 Pa, followed by WK2(QL)6K2 at ~500 Pa, K3W(QL)6K2 at ~100 Pa and WK3(QL)6K2 at ~10 Pa. The different rheological properties reflect the internal molecular order of peptide building blocks within the nanofibers and are strongly correlated with the supramolecular nanostructures observed by TEM.
Fig. 3.
Rheological properties of peptide hydrogels (2 wt%) formed by K2W(QL)6K2, WK2(QL)6K2, K3W(QL)6K2 and WK3(QL)6K2 during dynamic time sweep (frequency: 6 rad s−1, strain: 0.2%). 1000% strain was applied at the 900th second to completely disrupt the hydrogel and the recovery of storage modulus was continually measured afterwards. Solid line: storage modulus, dashed line: loss modulus.
The intermolecular interaction among K2W(QL)6K2 building units is more favorable to drive the packing of β-sheets into supramolecular nanofibers compared to WK2(QL)6K2 which has a shorter core domain. K3W(QL)6K2, despite the expanded core domain, showed a surprisingly low storage modulus. Dynamic frequency sweep tests showed K2W(QL)6K2 and WK2(QL)6K2 hydrogels could withstand oscillatory frequencies up to 25 rad s−1, whereas K3W(QL)6K2 hydrogels deform at 10 rad s−1 (Fig. S3†). K2W(QL)6K2 and K3W(QL)6K2 differ by one lysine residue at the N-terminus. The molecular conformation of the charged domain seems to be a critical determinant of the supramolecular packing and structure and bulk mechanical properties of the nanofibers upon charge neutralization in phosphate buffer. CD and FT-IR may not be sufficient to provide accurate structural information for specific peptide regimes. NMR spectroscopy using site-specific isotope labeling will enable deeper understanding of the effect of local peptide secondary structure on the supramolecular packing and rheological properties. Related work has been initiated in the lab and the results will be reported separately in future.
Regulating non-covalent interactions among peptide subunits is a powerful strategy to tune the morphology of supramolecular nanostructures and thus control the rheological properties of hydrogels. MDPs consisting of QL repeating units have been shown to form weak hydrogels of low storage modulus at ~100 Pa under physiological conditions.44 Replacing glutamine with serine was found to slightly improve the rigidity of the hydrogels presumably due to the formation of hydrogen bonding between the serine residues.39,40,44 The approach detailed herein offers an alternative route to manipulate both the nanostructure and mechanical properties of the MDP hydrogels through simple variation of the terminal domain without significantly affecting the supramolecular packing of the core domain. Moreover, the storage moduli of K2W(QL)6K2, WK2(QL)6K2, and K3W(QL)6K2 were recovered to ~90%, ~75%, and ~80% of their original values within 10 seconds after shear strain at 1000%. The ability of shear thinning and rapid recovery is a key characteristic of dynamic hydrogels, providing significant promise to using these hydrogels for future implantable biomaterials development.
The ability of MDP hydrogels to inhibit bacterial growth was evaluated in a commonly used Gram-positive bacterial pathogen, S. aureus. WK3(QL)6K2 formed a very fragile hydrogel and thus was excluded for the activity measurement due to the difficulty of removing them from the agar surface cultured bacterial colonies for quantitative viability measurements. Hydrogels based on MDPs 1–3 provided a sufficiently broad range of storage moduli for us to investigate the effect of supramolecular nanostructure and its rheological properties on their antimicrobial activity. Before conducting the aforementioned activity tests, the antimicrobial activities of MDPs 1–3 first were evaluated in solution by comparing their minimum inhibitory concentrations (MICs) and the results are shown in Table 2.
Table 2.
MIC results of K2W(QL)6K2, WK2(QL)6K2, and K3W(QL)6K2 in solution against S. aureus
| Peptides | Sequences | MIC (µM) |
|---|---|---|
| 1 | K2W(QL)6K2 | >160 |
| 2 | WK2(QL)6K2 | 160 |
| 3 | K3W(QL)6K2 | 20 |
In solution, K3W(QL)6K2 showed the most effective bactericidal activity followed by WK2(QL)6K2 and K2W(QL)6K2. As demonstrated in our previous work, the different antimicrobial activities of self-assembled MDPs in solution are related to the intermolecular interactions between their peptide subunits.35 K3W(QL)6K2 due to increased electrostatic repulsion, led to nanofibers with weaker intermolecular interactions among the building blocks compared to those formed by WK2(QL)6K2 and K2W(QL)6K2. Loosely packed supramolecular nanofibers will allow for the individual peptide chains or fragments of the chain to intercalate into and disrupt bacterial cell membranes. In a recent report, the effect of a peptide’s intermolecular interactions on mammalian cell toxicity was discussed.47 Our work represents another example of using de novo designed peptides to generate supramolecular nanostructures with tunable intermolecular interactions within the assembly to modulate and optimize their bioactivity, in this case the inherent antimicrobial properties of hydrogels.
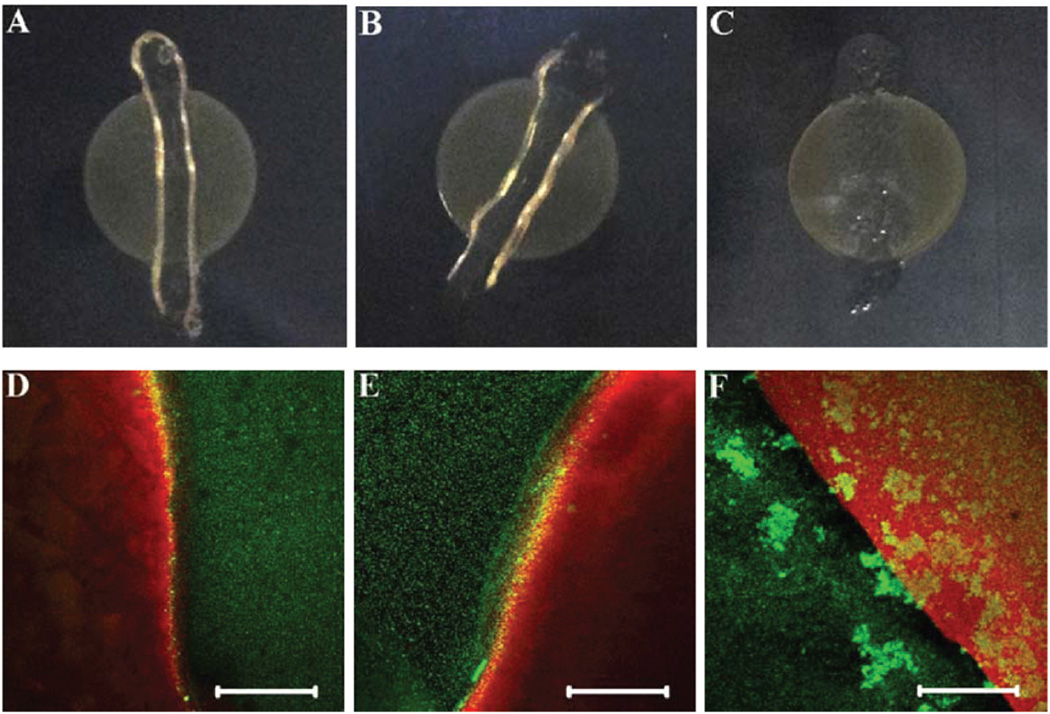
Interestingly, the antimicrobial activity of MDP hydrogels was found to follow an opposite trend to that observed in solution, which may suggest a distinct bactericidal mechanism for hydrogel-based antimicrobial materials. Here the evaluation of the bactericidal activity of MDP hydrogels followed an established protocol in the literature.27,30 In the present study, bacteria were first cultured on agar plates for 1 h at 37 °C, followed by deposition of the same amount of peptide hydrogels (40 µl) on the inoculated surface and incubation for another 16 h at 37 °C. The top panel in Fig. 4 presents the optical images of S. aureus cultured on agar plates in the presence of peptide hydrogels for 16 h.
Fig. 4.
Optical (top panel) and confocal (bottom panel) images of S. aureus cultured on agar plates treated by K2W(QL)6K2 (A, D), WK2(QL)6K2 (B, E) and K3W(QL)6K2 (C, F) hydrogels after 16 h. Scale bar: 120 µm.
It is noticeable that the hydrogels formed by K2W(QL)6K2 and WK2(QL)6K2 are more robust and remain highly hydrated, whereas the hydrogels assembled from K3W(QL)6K2 tend to flatten and collapse after 16 h of incubation. Visual inspection of the optical images suggested that hydrogels formed by K2W-(QL)6K2 and WK2(QL)6K2 are more efficient at inhibiting the growth of bacteria as demonstrated by the absence of bacterial colonies underneath the hydrogel (Fig. 4A and B). K3W(QL)6K2 was found to be the least effective to inhibit the growth of S. aureus as shown by the invasion of the bacteria to the hydrogel- treated areas (Fig. 4C). Bactericidal activity of different MDP hydrogels was further confirmed by live/dead staining assay. Confocal fluorescence images were photographed at the hydrogel–bacteria interface and the results are shown in the bottom panels of Fig. 4D–F. The red fluorescence is attributed to PI staining as an indicator of bacterial cell membrane disruption or death and the green fluorescence arises from nucleic acid staining of live bacteria with SYTO 9. Consistent with the optical imaging results, K2W(QL)6K2 and WK2(QL)6K2 hydrogels with higher storage moduli were found to be more effective in inhibiting bacterial growth as demonstrated by the clear boundary between the gel-treated and non-treated agar surface than K3W(QL)6K2 which showed large quantities of live bacterial cells on the hydrogel treated agar surface.
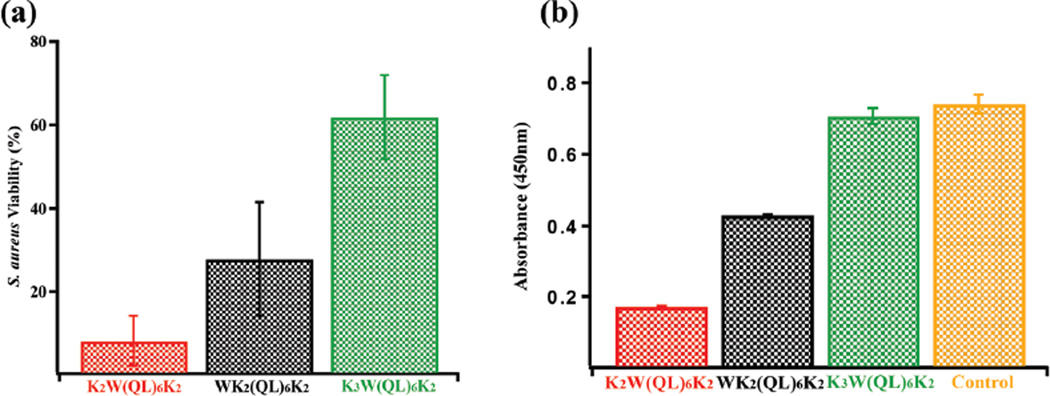
The bactericidal activity of the three hydrogels was further evaluated quantitatively and compared using a cell viability assay. After 16 h of coincubation of gels with S. aureus cultured on thin agar plates (diameter: 5 mm, depth: 2 mm), hydrogels were gently removed along with the underlying bacteria growing on the agar surface and were transferred to 10 ml of culture medium followed by cell resuspension, serial dilution, plating and colony counting. All the procedures were performed with extreme care to prevent unintentional loss of bacterial cells during gel removal and transfer to broth culture medium. Notably, small amounts of agar did not cause problems with resuspension and quantification of the isolated bacteria that were originally attached to the agar surface. The assay has been validated thoroughly against both peptidetreated and non-treated agar surface-cultured bacteria as a control. As shown in Fig. 5a, K2W(QL)6K2 showed the highest growth inhibitory effect with 8% cell viability compared to the untreated bacterial culture, followed by WK2(QL)6K2 at 28% and K3W(QL)6K2 at 62%. Colorimetric XTT viability assay results confirm the differing antimicrobial activities among the three peptides. The UV absorbance taken at 450 nm (Fig. 5b) is proportional to the number of live bacteria and thus is inversely related to the peptides’ antimicrobial activity. K2W(QL)6K2 with the lowest UV absorbance exhibited the most potent antimicrobial activity followed by WK2(QL)6K2 and K3W(QL)6K2.
Fig. 5.
Viability of S. aureus quantified by (a) bacterial colony counting assay and (b) XTT-menadione assay following exposure of S. aureus for 16 h to different peptide hydrogels.
Intriguingly, K3W(QL)6K2 did not show appreciable antimicrobial activity when self-assembled into hydrogels, however it was found to be the most effective at killing S. aureus in solution (Table 2). This may suggest that it is not the soluble peptide/nanofibers that prevent the growth of the bacteria on the agar plate and a different antimicrobial mechanism may be used by self-assembled nanofiber hydrogels.48 We postulate that the antimicrobial activity displayed by hydrogels may be largely attributed to the surface chemistry of the supramolecular hydrogel nanofibers and their bulk rheological properties. Due to the formation of large quantities of elongated peptide nanofibers, excessive charge density is possible to trap the bacterial cell on the hydrogel surface and greatly reduces their motility. Further and tighter adherence of the bacteria to the matrix could be facilitated by a combined effect of the local charge density on the hydrogel surface and the porous network of the densely crosslinked nanofibers as shown by SEM (Fig. S5†).20 To investigate the effect of storage modulus of MDP hydrogels on bacterial motility and further link such an effect on their antimicrobial activities, the motion of S. aureus was monitored under a confocal microscope in cell culture medium with 50% PBS, culture medium with 50% K2W(QL)6K2 and culture medium with 50% K3W(QL)6K2. The results firmly confirmed the effect of storage moduli on bacterial motility. While S. aureus was rapidly moving in cell culture medium, the motility was dramatically reduced in both hydrogels. Through visual inspection, minimal movement was observed in K2W(QL)6K2 hydrogels which has the highest storage modulus. The reduced motility could be a result of a strong suctioning force from the nanofibrous hydrogels which may vary depending on the supramolecular nanostructure and their rheological properties. Membrane disruption may occur locally through such combined chemical and mechanical interactions. Since the process occurs entirely at the surface of the hydrogels, the positive charge density on the matrix surface is perhaps the most crucial parameter to be considered for antimicrobial hydrogel design. Rheological properties, in this case storage modulus also appears to play an important role by providing mechanical support for individual nanofibers and fibrous networks to direct their desirable chemical and biological functionalities against the restrained bacteria. The supramolecular assembly of MDPs described herein offers a versatile platform where both the surface chemistry of the supramolecular nanofibers and their rheological properties can be readily tuned by incorporating various cationic moieties, not limited to natural cationic amino acids to enhance the antimicrobial activity in a variety of biological settings.
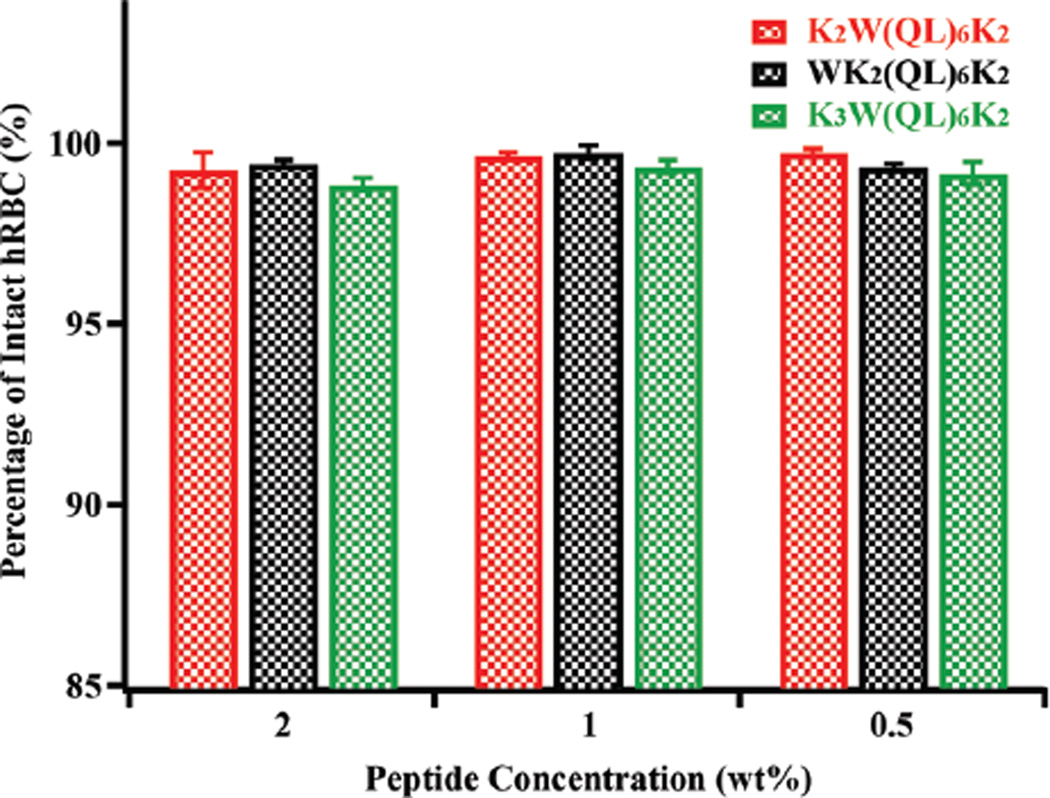
Finally, hemolytic activity is an important parameter to be evaluated for antimicrobial therapy development. A hemolysis assay was conducted by incubating human red blood cells (hRBC) with different MDP hydrogels at 37 °C for 1 h. The leakage of hemoglobin was quantified by UV spectroscopy for MDP-treated samples and the relative percentage of hemolysis was plotted with respect to the positive control of Triton X-100 treated hRBCs. Fig. 6 shows the results of the hemolytic activity of various MDP hydrogels at 2 wt%, 1 wt% and 0.5 wt%. More than 98% of hRBCs are intact upon incubation with MDP hydrogels suggesting minimum cytotoxicity to hRBC. The difference in membrane composition between bacteria and human red blood cells (HRBCs) may account for bacterial cell selectivity as mammalian cells lack anionic lipids to interact with the cationic hydrogels. The exceptional cell selectivity could also be due to the formation of cationic supramolecular nanofibers which bacteria are susceptible to. Instead, HRBCs were not significantly affected presumably due to the unique RBC membrane structure and rheology. Related study on comprehensive understanding of the interaction of MDP hydrogels with HRBCs and their hemocompatibility has been initiated in our lab and the results will be reported separately.
Fig. 6.
Hemolytic activity of peptide hydrogels at three different peptide concentrations. The data was plotted by comparing the UV absorbance at 405 nm of hemoglobin from each peptide treated sample with that from Triton X-100 treated hRBCs.
Conclusions
In summary, by using de novo designed cationic multidomain peptides, hydrogels with tunable supramolecular nanostructure, rheological properties and inherent antimicrobial activity were produced. Present studies suggest a distinct bactericidal mechanism displayed by supramolecular peptides reflecting different assembly dimensions. In terms of antimicrobial hydrogel design, tuning the surface chemistry of the supramolecular nanofibers and the rheological properties of the hydrogels can be highly effective for improving the antimicrobial activity. The structure–property–activity relationship developed through this study will provide important guidelines for designing biocompatible peptide hydrogels with built-in antimicrobial activity for various biomedical applications.
Acknowledgments
Clarkson University is acknowledged for the support of this work. We thank the Clarkson-Trudeau Partnership for providing seed fund to support the project. We would like to thank Prof. Paul Goulet and Lina Bian for providing kind support for FT-IR analysis. We acknowledge the confocal imaging core facility at St. Lawrence University for excellent technical support.
Footnotes
Electronic supplementary information (ESI) available. See DOI: 10.1039/c5nr05233e
References
- 1.Yang C, Mariner PD, Nahreini JN, Anseth KS. J. Controlled Release. 2012;162:612–618. doi: 10.1016/j.jconrel.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tokuda EY, Leight JL, Anseth KS. Biomaterials. 2014;35:4310–4318. doi: 10.1016/j.biomaterials.2014.01.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mariner PD, Wudel JM, Miller DE, Genova EE, Streubel SO, Anseth KS. J. Orthop. Res. 2013;31:401–406. doi: 10.1002/jor.22243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mabry KM, Lawrence RL, Anseth KS. Biomaterials. 2015;49:47–56. doi: 10.1016/j.biomaterials.2015.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galler KM, Hartgerink JD, Cavender AC, Schmalz G, D’Souza RN. Tissue Eng., Part A. 2012;18:176–184. doi: 10.1089/ten.tea.2011.0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ng VW, Chan JM, Sardon H, Ono RJ, Garcia JM, Yang YY, Hedrick JL. Adv. Drug Delivery Rev. 2014;78:46–62. doi: 10.1016/j.addr.2014.10.028. [DOI] [PubMed] [Google Scholar]
- 7.Ghobril C, Grinstaff MW. Chem. Soc. Rev. 2015;44:1820–1835. doi: 10.1039/c4cs00332b. [DOI] [PubMed] [Google Scholar]
- 8.Kharkar PM, Kiick KL, Kloxin AM. Chem. Soc. Rev. 2013;42:7335–7372. doi: 10.1039/c3cs60040h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tian YF, Devgun JM, Collier JH. Soft Matter. 2011;7:6005–6011. doi: 10.1039/C1SM05504F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peppas NA, Hilt JZ, Khademhosseini A, Langer R. Adv. Mater. 2006;18:1345–1360. [Google Scholar]
- 11.Collier JH, Rudra JS, Gasiorowski JZ, Jung JP. Chem. Soc. Rev. 2010;39:3413–3424. doi: 10.1039/b914337h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu J. Soft Matter. 2011;7:6757–6767. [Google Scholar]
- 13.Kisiday J, Jin M, Kurz B, Hung H, Semino C, Zhang S, Grodzinsky AJ. Proc. Natl. Acad. Sci. U. S. A. 2002;99:9996–10001. doi: 10.1073/pnas.142309999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu J, Song H, Zhang L, Xu H, Zhao X. Macromol. Biosci. 2010;10:1164–1170. doi: 10.1002/mabi.200900450. [DOI] [PubMed] [Google Scholar]
- 15.Gao Y, Zhao F, Wang Q, Zhang Y, Xu B. Chem. Soc. Rev. 2010;39:3425–3433. doi: 10.1039/b919450a. [DOI] [PubMed] [Google Scholar]
- 16.Liang G, Yang Z, Zhang R, Li L, Fan Y, Kuang Y, Gao Y, Wang T, Lu WW, Xu B. Langmuir. 2009;25:8419–8422. doi: 10.1021/la804271d. [DOI] [PubMed] [Google Scholar]
- 17.Zhang L, Cao Z, Bai T, Carr L, Ella-Menye JR, Irvin C, Ratner BD, Jiang S. Nat. Biotechnol. 2013;31:553–556. doi: 10.1038/nbt.2580. [DOI] [PubMed] [Google Scholar]
- 18.Malmsten M. Soft Matter. 2011;7:8725–8736. [Google Scholar]
- 19.Veiga AS, Schneider JP. Biopolymers. 2013;100:637–644. doi: 10.1002/bip.22412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li P, Poon YF, Li W, Zhu HY, Yeap SH, Cao Y, Qi X, Zhou C, Lamrani M, Beuerman RW, Kang ET, Mu Y, Li CM, Chang MW, Leong SS, Chan-Park MB. Nat. Mater. 2011;10:149–156. doi: 10.1038/nmat2915. [DOI] [PubMed] [Google Scholar]
- 21.Tsao CT, Chang CH, Lin YY, Wu MF, Wang JL, Han JL, Hsieh KH. Carbohydr. Res. 2010;345:1774–1780. doi: 10.1016/j.carres.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 22.Liu SQ, Yang C, Huang Y, Ding X, Li Y, Fan WM, Hedrick JL, Yang YY. Adv. Mater. 2012;24:6484–6489. doi: 10.1002/adma.201202225. [DOI] [PubMed] [Google Scholar]
- 23.Andrews MA, Figuly GD, Chapman JS, Hunt TW, Glunt CD, Rivenbark JA, Chenault HK. J. Appl. Polym. Sci. 2011;119:3244–3252. [Google Scholar]
- 24.Veiga AS, Sinthuvanich C, Gaspar D, Franquelim HG, Castanho MA, Schneider JP. Biomaterials. 2012;33:8907–8916. doi: 10.1016/j.biomaterials.2012.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Madsen J, Armes SP, Bertal K, Lomas H, MacNeil S, Lewis AL. Biomacromolecules. 2008;9:2265–2275. doi: 10.1021/bm8005006. [DOI] [PubMed] [Google Scholar]
- 26.Li Y, Fukushima K, Coady DJ, Engler AC, Liu S, Huang Y, Cho JS, Guo Y, Miller LS, Tan JP, Ee PL, Fan W, Yang YY, Hedrick JL. Angew. Chem., Int. Ed. 2013;52:674–678. doi: 10.1002/anie.201206053. [DOI] [PubMed] [Google Scholar]
- 27.Salick DA, Pochan DJ, Schneider JP. Adv. Mater. 2009;21:4120–4123. [Google Scholar]
- 28.Debnath S, Shome A, Das D, Das PK. J. Phys. Chem. B. 2010;114:4407–4415. doi: 10.1021/jp909520w. [DOI] [PubMed] [Google Scholar]
- 29.Colak S, Nelson CF, Nusslein K, Tew GN. Biomacromolecules. 2009;10:353–359. doi: 10.1021/bm801129y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salick DA, Kretsinger JK, Pochan DJ, Schneider JP. J. Am. Ceram. Soc. 2007;129:14793–14799. doi: 10.1021/ja076300z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Y, Yang Y, Wang C, Zhao X. Nanoscale. 2013;5:6413–6421. doi: 10.1039/c3nr00225j. [DOI] [PubMed] [Google Scholar]
- 32.Cao B, Tang Q, Li L, Humble J, Wu H, Liu L, Cheng G. Adv. Healthcare Mater. 2013;2:1096–1102. doi: 10.1002/adhm.201200359. [DOI] [PubMed] [Google Scholar]
- 33.Hughes M, Debnath S, Knapp CW, Ulijn RV. Biomater. Sci. 2013;1:1138–1142. doi: 10.1039/c3bm60135h. [DOI] [PubMed] [Google Scholar]
- 34.Kretsinger JK, Haines LA, Ozbas B, Pochan DJ, Schneider JP. Biomaterials. 2005;26:5177–5186. doi: 10.1016/j.biomaterials.2005.01.029. [DOI] [PubMed] [Google Scholar]
- 35.Xu D, Jiang L, Singh A, Dustin D, Yang M, Liu L, Lund R, Sellati TJ, Dong H. Chem. Commun. 2015;51:1289–1292. doi: 10.1039/c4cc08808e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Galler KM, Aulisa L, Regan KR, D’Souza RN, Hartgerink JD. J. Am. Chem. Soc. 2010;132:3217–3223. doi: 10.1021/ja910481t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kang MK, Colombo JS, D’Souza RN, Hartgerink JD. Biomacromolecules. 2014;15:2004–2011. doi: 10.1021/bm500075r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shai Y. Biochim. Biophys. Acta. 1999;1462:55–70. doi: 10.1016/s0005-2736(99)00200-x. [DOI] [PubMed] [Google Scholar]
- 39.Bakota EL, Aulisa L, Galler KM, Hartgerink JD. Biomacromolecules. 2011;12:82–87. doi: 10.1021/bm1010195. [DOI] [PubMed] [Google Scholar]
- 40.Bakota EL, Wang Y, Danesh FR, Hartgerink JD. Biomacromolecules. 2011;12:1651–1657. doi: 10.1021/bm200035r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wickremasinghe NC, Kumar VA, Hartgerink JD. Biomacromolecules. 2014;15:3587–3595. doi: 10.1021/bm500856c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bakota EL, Sensoy O, Ozgur B, Sayar M, Hartgerink JD. Biomacromolecules. 2013;14:1370–1378. doi: 10.1021/bm4000019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dong H, Paramonov SE, Aulisa L, Bakota EL, Hartgerink JD. J. Am. Chem. Soc. 2007;129:12468–12472. doi: 10.1021/ja072536r. [DOI] [PubMed] [Google Scholar]
- 44.Aulisa L, Dong H, Hartgerink JD. Biomacromolecules. 2009;10:2694–2698. doi: 10.1021/bm900634x. [DOI] [PubMed] [Google Scholar]
- 45.Long MM, Urry DW, Stoeckenius W. Biochem. Biophys. Res. Commun. 1977;75:725–731. doi: 10.1016/0006-291x(77)91532-7. [DOI] [PubMed] [Google Scholar]
- 46.Pandya MJ, Spooner GM, Sunde M, Thorpe JR, Rodger A, Woolfson DN. Biochemistry. 2000;39:8728–8734. doi: 10.1021/bi000246g. [DOI] [PubMed] [Google Scholar]
- 47.Newcomb CJ, Sur S, Ortony JH, Lee OS, Matson JB, Boekhoven J, Yu JM, Schatz GC, Stupp SI. Nat. Commun. 2014;5:1–10. doi: 10.1038/ncomms4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chu-Kung AF, Nguyen R, Bozzelli KN, Tirrell M. J. Colloid Interface Sci. 2010;345:160–167. doi: 10.1016/j.jcis.2009.11.057. [DOI] [PubMed] [Google Scholar]