Abstract
Background
Common variable immunodeficiency (CVID) is an antibody deficiency treated with immunoglobulin; however, patients can have noninfectious inflammatory conditions that lead to heightened morbidity and mortality.
Objectives
Modular analyses of RNA transcripts in whole blood previously identified an upregulation of many interferon-responsive genes. In this study we sought the cell populations leading to this signature.
Methods
Lymphoid cells were measured in peripheral blood of 55 patients with CVID (31 with and 24 without inflammatory/autoimmune complications) by using mass cytometry and flow cytometry. Surface markers, cytokines, and transcriptional characteristics of sorted innate lymphoid cells (ILCs) were defined by using quantitative PCR. Gastrointestinal and lung biopsy specimens of subjects with inflammatory disease were stained to seek ILCs in tissues.
Results
The linage-negative, CD127+, CD161+ lymphoid population containing T-box transcription factor, retinoic acid–related orphan receptor (ROR) γt, IFN-γ, IL-17A, and IL-22, all hallmarks of type 3 innate lymphoid cells, were expanded in the blood of patients with CVID with inflammatory conditions (mean, 3.7% of PBMCs). ILCs contained detectable amounts of the transcription factors inhibitor of DNA binding 2, T-box transcription factor, and RORγt and increased mRNA transcripts for IL-23 receptor (IL-23R) and IL-26, demonstrating inflammatory potential. In gastrointestinal and lung biopsy tissues of patients with CVID, numerous IFN-γ+RORγt+CD3− cells were identified, suggesting a role in these mucosal inflammatory states.
Conclusions
An expansion of this highly inflammatory ILC population is a characteristic of patients with CVID with inflammatory disease; ILCs and the interferon signature are markers for the uncontrolled inflammatory state in these patients.
Keywords: Common variable immunodeficiency, inflammatory complications, mucosal disease, innate lymphoid cells
Common variable immunodeficiency (CVID), one of the more prevalent primary immunodeficiency diseases, is characterized by low levels of serum IgG, IgA, and/or IgM and lack of production of specific IgG antibodies.1 Mutations in autosomal genes have been identified in a few patients, but for the majority, genetic or other causes of B-cell failure remain unknown.2 Although replacement immunoglobulin given at frequent intervals reduces the number of infections, it appears to do little to prevent or treat the inflammatory/autoimmune complications that occur in almost 50% of subjects.3–5 These complications include autoimmunity, granulomatous infiltrations, interstitial lung disease, lymphoid hyperplasia, lymphoma, liver disease, and enteropathy. In aggregate, these can lead to an 11-fold increased morbidity and mortality over time in these subjects compared with those seen in patients with CVID who have not had these conditions.5 Although patients with fewer B cells and isotype-switched memory B cells or more impaired T-cell functions are at greater risk for the development of complications,4–6 mRNA transcriptional profiling allowed us to distinguish patients with CVID with and without inflammatory diseases from each other and from control subjects.7 Modular analysis of the differentially expressed RNA transcripts identified a marked upregulation of interferon-related genes, as well as a significant downregulation of both B cell–and T cell–related genes.
In this study we identified an expanded population of innate lymphoid cells (ILCs) with a pronounced IFN-γ signature in both peripheral blood and gastrointestinal and lung tissues of these patients with CVID. ILCs are of lymphoid linage but lack recombination-activating gene–dependent rearranged antigen receptors and myeloid and dendritic cell markers (lineage negative). Because of inflammatory cytokine secretion, transcriptional properties, and location in mucosal tissues, ILCs appear to play important roles in immunity to fungal, bacterial, and viral microbes; wound healing; and inflammation.8 ILC functions are tightly regulated because uncontrolled activation and proliferation can contribute to inflammation and damage in different organs.9 The expansion of IFN-γ+, IL-17A+, and IL-22+ ILCs in peripheral blood in patients with CVID with inflammatory disease (CVIDc) suggests a role for these cells in both inflammation and mucosal damage.
METHODS
Patients and blood and sample processing
Peripheral blood samples were obtained from 22 healthy adult volunteers and 55 patients with CVID (30 female and 25 male patients; age, 14–68 years) by using a protocol and consent approved by the Institutional Review Board of the Icahn School of Medicine at Mount Sinai (IRB#03-1008). One blood sample was used for immediate cell immunophenotyping and another for serum isolation. PBMCs were separated on Ficoll-Histopaque (Pharmacia, Uppsala, Sweden). Thirty-one patients with CVID had inflammatory/autoimmune manifestations (hematologic or organ-specific autoimmunity, biopsy-proved granulomatous disease, interstitial lung disease leading to impaired lung function, lymphoid hyperplasia with splenomegaly, or noninfectious gastrointestinal inflammatory disease), whereas 24 had a history of respiratory or gastrointestinal infections but lacked these conditions (Table I). All subjects were free from concurrent infections and were not taking antibiotics or immune-modifying medications at the time of the study. Blood was taken before interval intravenous immunoglobulin infusions or between subcutaneous immunoglobulin administrations.
TABLE I.
Demographics and clinical parameters for 55 patients with CVID
| Infections only | 24 |
| Subjects with inflammatory/autoimmune complications | 31 |
| Autoimmunity: ITP and/or AIHA | 18 |
| Interstitial lung disease | 18 |
| Splenomegaly/lymphadenopathy | 16 |
| Chronic noninfectious gastrointestinal disease | 9 |
| Granulomatous disease | 7 |
| Splenectomy | 6 |
| Nodular regenerative hyperplasia of the liver | 3 |
| Other autoimmunity (rheumatoid arthritis/nephritis) | 2 |
AIHA, Autoimmune hemolytic anemia; ITP, immune thrombocytopenic purpura.
Immunophenotyping by using flow cytometry
Absolute B-cell, T-cell, and natural killer (NK) cell numbers in peripheral blood were determined by using flow cytometry with CountBright beads (Thermo Fisher, Waltham, Mass) with an Fc-blocking reagent after incubation at 4°C and gating on live CD45+ cells. ILC populations were identified as CD3−CD14−CD11c−CD19−CD117+CD127+ cells (ILCs); CD3−CD14− CD19−CD117−CD127−CD56+ cells were identified as conventional NK cells. For analysis of intracellular transcription factors, PBMCs were permeabilized, fixed with Transcription Factor Buffer Set (eBioscience, San Diego, Calif) and labeled with mAbs to human retinoic acid–related orphan receptor (ROR) γt or T-box transcription factor (T-bet; antibodies are shown Table E1 in this article’s Online Repository at www.jacionline.org). For intracellular cytokine staining, PBMCs were cultured for 4 hours in RPMI medium with GolgiStop (BD PharMingen, San Jose, Calif). Those cells were stained with antibodies to specific surface molecules, fixed, and permeabilized (Cytofix/Cytoperm Kit, BD PharMingen). Cells were examined by using LSR Fortessa (BD) and analyzed with FlowJo software (Tree Star, Ashland, Ore).
Tissues and immunofluorescence
Previous biopsy samples of the lung, gastrointestinal tract, or both from patients with CVID were retrieved with permission and examined with deidentified appropriate control tissues from immunocompetent subjects (Table II). Tissue sections of 5 µm in thickness were stained with primary antibodies (see Table E2 in this article’s Online Repository at www.jacionline.org) and appropriate secondary reagents: Alexa Fluor 488–conjugated anti-rat pAb, Alexa Fluor 546/488–conjugated anti-rabbit pAb, Alexa Fluor 546/647–conjugated anti-mouse pAb, and cyanine 5–conjugated streptavidin (Jackson ImmunoResearch Laboratories, West Grove, Pa). Nuclei were visualized with 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI; Boehringer Mannheim, Indianapolis, Ind). Primary antibodies with irrelevant binding activity and appropriate secondary reagents were used to validate the specificity of tissue staining. Coverslips were applied with FluorSave Reagent (Calbiochem, Nottingham, United Kingdom), and images were acquired with a Zeiss Axioplan 2 microscope (Atto Instruments, Rockville, Md).
TABLE II.
Pathology samples from patients with CVID and control subjects
| Source | Tissue | Pathologic diagnosis | Fixation |
|---|---|---|---|
| CVID, 33-year-old man | Terminal ileum | Patchy, mildly active nonspecific colitis with marked depletion of plasma cells |
FFPE |
| CVID, 47-year-old woman | Small bowel, jejunum | Small intestine with moderate intraepithelial lymphocytosis, mild villous atrophy, moderate lymphocytic and eosinophilic infiltration, with focal aggregation of the lymphocytes; marked decrease of plasma cells; no granuloma or parasites |
FFPE |
| Control, 38-year-old man | Terminal ileum | Obstructed, not inflamed | FFPE |
| Control, 27-year-old woman | Terminal ileum | Crohn disease, mildly inflamed | FFPE |
| Control, 39-year-old man | Right colon | Active Crohn enteritis with transmural chronic inflammation | FFPE |
| CVID, 49-year-old mam | Lung | Interstitial lung disease, lymphoid hyperplasia, organizing pneumonia, and occasional poorly formed granulomas |
FFPE |
| CVID, 49-year-old woman | Lung | Interstitial lung disease, lymphoid hyperplasia, organizing pneumonia, and occasional poorly formed granulomas |
FFPE |
| CVID, 46-year-old man | Lung | Reactive lymphoid hyperplasia, interstitial lung disease and few poorly formed nonnecrotizing granulomas |
FFPE |
| Control, 47-year-old woman | Lung | Lung nodule: nodular lymphoid hyperplasia | FFPE |
| 18-year-old man | Spleen | Resection after trauma | Fresh |
| 34-year-old man | Spleen | Resection after trauma | Fresh |
FFPE, Formalin fixed and paraffin embedded.
Mass cytometric analyses
PBMCs from 4 patients with CVID with interstitial lung disease, chronic enteropathy, or both or 4 healthy sex-matched adult donors were stained immediately or after a 5-hour incubation in monensin (for intracellular staining) and examined by means of mass cytometry (CyTOF).10 In brief, 3 million cells were washed with PBS containing 0.1% BSA and incubated with antibodies against selected surface markers for 30 minutes on ice. Antibodies were preconjugated to metal tags or conjugated in house by using MaxPar X8 conjugation kits (Fluidigm, South San Francisco, Calif). Cells were incubated in cisplatin (Fluidigm) to label dead cells, washed, fixed, and permeabilized with a commercial kit (FoxP3/Transcription Buffer Staining Kit, eBioscience) and stained with antibodies against intracellular cytokines and transcription factors (see Table E2). The samples were then incubated overnight in PBS containing 1.6% formaldehyde and 1:3000 dilution of the nucleic acid Intercalator-Ir (Fluidigm), washed with PBS and diH20, and resuspended in diH20 with a 1:10 dilution of EQ 4 Element Calibration beads and cells acquired on a CyTOF2 Mass Cytometer (Fluidigm). Data files were concatenated and normalized by using a bead-based normalization algorithm (CyTOF software) and uploaded to Cytobank. The gated populations were clustered by using spanning-tree progression analysis of density-normalized events,10 and cell populations were annotated based on expression of key canonical markers while preserving visualization of novel populations within the data set.
Sorting and culturing of ILCs
ILCs were sorted from fresh PBMCs as CD117+CD127+CD56+ cells, as described elsewhere.11 Separately, CD3−CD14−CD19−CD117−CD127− CD56+ NK cells and CD3+ T cells were also sorted. For comparison with ILCs from patients with CVID and control subjects, ILCs were similarly sorted from splenocytes of fresh spleens from healthy subjects removed because of trauma.11 Cells were stained with appropriate mixtures of fluorochrome-labeled antibodies (see Table E1) and sorted with a FACSAria II (BD Biosciences) after exclusion of dead cells by using the LIVE/DEAD Fixable Violet Cell Stain Kit (Invitrogen, Carlsbad, Calif). The purity of sorted cells was consistently greater than 97%. To further examine sorted circulating ILCs, cells (5 × 104/well) were plated in 96-well U-bottom plates and then cultured for 3 to 5 days in complete RPMI medium with 10% FBS, penicillin, and streptomycin (10 U/mL), with or without 50 ng/mL IL-7, 50 ng/mL IL-1β, or both (PeproTech, Rocky Hills, NJ), as previously described.11 The survival of the sorted ILC population after culture was measured with the Annexin V Apoptosis Detection Kit I (BD PharMingen). Cells were acquired with an LSR Fortessa (BD) and analyzed by using FlowJo software (Tree Star), with comparisons with isotype-matched antibody controls.
RNA extraction and real-time PCR
Total RNA was extracted from cell populations by using the RNAqueous-4 PCR Isolation Kit (Ambion, Foster City, Calif), followed by cDNA synthesis with the qScript cDNA Synthesis kit (Quanta Biosciences, Gaithersburg, Md). The relative abundance of transcripts was measured by using quantitative RT-PCR with primers (see Table E3 in this article’s Online Repository at www.jacionline.org) with PerfeCTa SYBR Green Super Mix (Quanta Biosciences). Results for ILCs were normalized to Actb (β-actin) mRNA and presented as relative expression (or abundance) compared with that of total PBMCs.
ELISAs
Sera collected from patients with CVID and control subjects were analyzed with BD OptEIA human INF-γ ELISA set (BD Biosciences), the ELISA MAX set for human IL-17A (BioLegend, San Diego, Calif), and ELISA Ready-SET-Go! for human IL-22 (eBioscience). Cytokines were measured in 1:10 diluted serum in picograms per milliliter, according to the manufacturer’s instructions, and by recording absorbance at 450 nm.
Statistical analysis
Values were expressed as means ± SEMs or means ± SDs. Statistical significance was assessed with the 2-tailed Student t test and 1-way ANOVA, unless otherwise specified. A Mann-Whitney U test and the Kruskal-Wallis test with the Dunn multiple comparison test for matched pairs were used for analysis of nonparametric data. Correlations between data pairs were examined by using the Spearman rank order coefficient. Results were analyzed with GraphPad Prism software (version 5; GraphPad Software, La Jolla, Calif), and P values of less than .05 were considered significant.
RESULTS
Microarray analysis of interferon-related genes
RNA microarray studies previously showed that whole blood of 47 patients with CVID with inflammatory features contained a significant upregulation of numerous interferon-responsive genes not found in 44 subjects without these conditions, control subjects, or patients with X-linked agammaglobulinemia.7 Examining IFN-γ and α1, α2, and β1 gene transcripts in microarray data of these 91 subjects (http://www.ncbi.nlm.nih.gov/geo) demonstrated upregulation of only IFN-γ transcripts, which were significantly increased in blood of patients with CVIDc (P < .01). As activated, peripheral blood T cells of these subjects produced little IFN-γ.7 We hypothesized that other populations might be responsible for the interferon signature.
CyTOF analyses
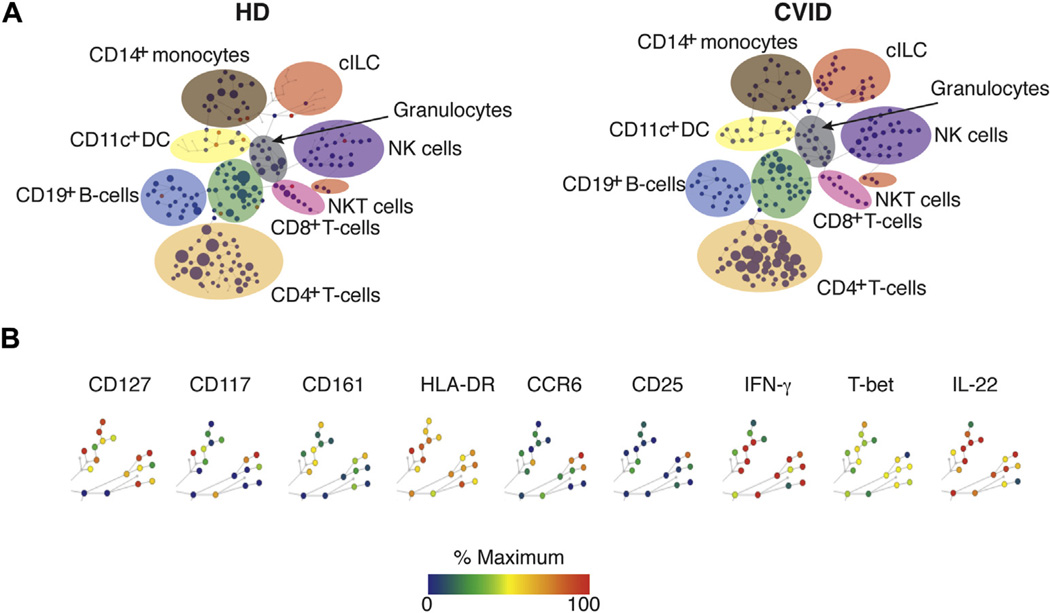
As an unbiased approach, we used CyTOF to seek IFN-γ+ lymphoid cells in the peripheral blood of patients with CVID with chronic inflammatory disease. For this, neighboring cells are grouped by using unsupervised hierarchical clustering with spanning-tree progression analysis of density-normalized events algorithm in which nodes are linked by a minimum-spanning tree. These studies revealed a population of cells bearing surface markers associated with ILCs, including CD127 (IL-7 receptor) and the pan-ILC marker CD161, but lacking the lineage markers CD8, CD4, CD11c, CD14, and CD19 and having low levels of CD56 (Fig 1). These cell populations expressed various amounts of surface CD25 and CD117 (c-kit) and, as expected, were positive for intracellular IFN-γ, T-bet, RORγt, and IL-22, suggesting that ILCs were present in blood of patients with CVID. Other markers examined by using CyTOF are shown in Fig E1 in this article’s Online Repository at www.jacionline.org.
FIG 1.
CyTOF analyses. A, CyTOF was used to compare cell lineages in the blood of 4 patients with CVIDc compared with control subjects (HD). B, Lin− cells positive for CD127 and CD161 were observed in samples from patients with CVID. Other markers tested are shown here. ILC populations are highlighted. Node color is scaled to the median intensity of marker expression. One representative experiment is displayed.
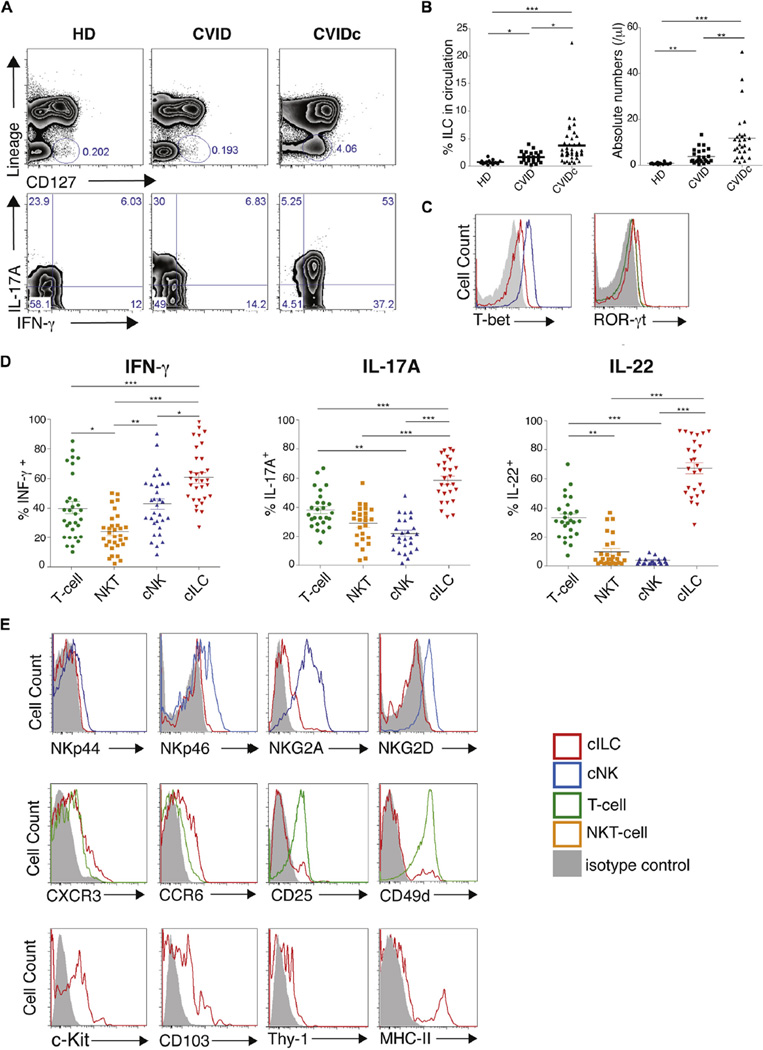
Phenotypic characteristics of circulating ILCs
ILCs are further categorized by both cytokine capability and transcriptional features,12 and thus we tested the cell populations of 55 patients with CVID with panels of antibodies using fluorescence-activated cell sorting. The upper panels of Fig 2, A, show the expanded, lineage-negative, CD127+ ILC population in a representative patient with CVIDc compared with a patient with CVID and a healthy control subject. This cell population was both IFN-γ+ and IL-17A+, as shown (Fig 2, A, bottom panel). Overall, ILC counts were found to be significantly increased in the blood of these 55 patients with CVID (0.62% of CD45+ lymphocytes) compared with those in 22 healthy donors (0.03%). However, these cells were significantly increased in the blood of 31 patients with CVIDc (mean, 3.7%) compared with a mean of 1.6% in the 24 patients with CVID without complications (P < .001, 1-way ANOVA; Fig 2, B). ILCs are categorized based on both cytokine capability and transcriptional features.8 The ILCs identified in the circulation of patients with CVID contained both T-bet and RORγt, correlating with a proinflammatory type 3 innate lymphoid cell (ILC3) population (Fig 2, C).13,14 These cells also contained large amounts of intracellular IFN-γ, IL-17, and especially IL-22 compared with levels in T cells, NK T cells, or conventional NK cells of the same patients with CVID (Fig 2, D). Comparing ILC3s from patients with CVID with NK cells from the same 55 patients, ILCs from patients with CVID showed less expression of the natural cytotoxicity receptors NKp44 and NKp46 and the activating NK receptor NKG2D (Fig 2, E, upper row).13,15 Comparing the features of ILC3s from patients with CVID with those of T cells from the same subjects,11,16,17 ILCs showed detectable levels of the chemokine receptors CXCR3 and CCR6,18,19 with some expression of CD25 and the integrin CD49d (Fig 2, E, middle row). As expected, circulating ILCs, as in previous studies, showed significant levels of c-Kit9,17 and other markers of innate cells, such as the integrin αE (CD103) and Thy-1,20 and demonstrated the capacity for antigen presentation (MHC-II; Fig 2, E, bottom row).9
FIG 2.
Immunophenotyping of circulating ILC3s. A, Fluorescence-activated cell sorting analyses show Lin−IFN-γ+ and IL-17A+CD127+ cells in peripheral blood in patients with CVIDc. B, ILC3s were significantly increased in these patients with CVID. C, Intracellular T-bet and RORγt of ILC3 populations (red), NK cells (blue), NK T cells (green), or T cells (orange). D, Percentages of IFN-γ+, IL17-A+, and IL-22+ immune cells in these patients with CVID. E, Fluorescence-activated cell sorting comparison of ILC3s from patients with CVID with NK cells (NKp44, NKp46, and NKG2D). Middle row, CXCR3, CCR6, CD25, and CD49d levels for ILC3s from patients with CVID to T cells. Bottom row, ILC3s from patients with CVID are c-Kit+, CD103+, and Thy-1+. Representative plots from 22 healthy control subjects, 31 patients with CVID with complications, and 24 patients with CVID without complications are shown in Fig 2, A, C, and E. For Fig 2, B and D: *P < .05, **P < .01, and ***P < .001 (1-way ANOVA). HD, Healthy donors.
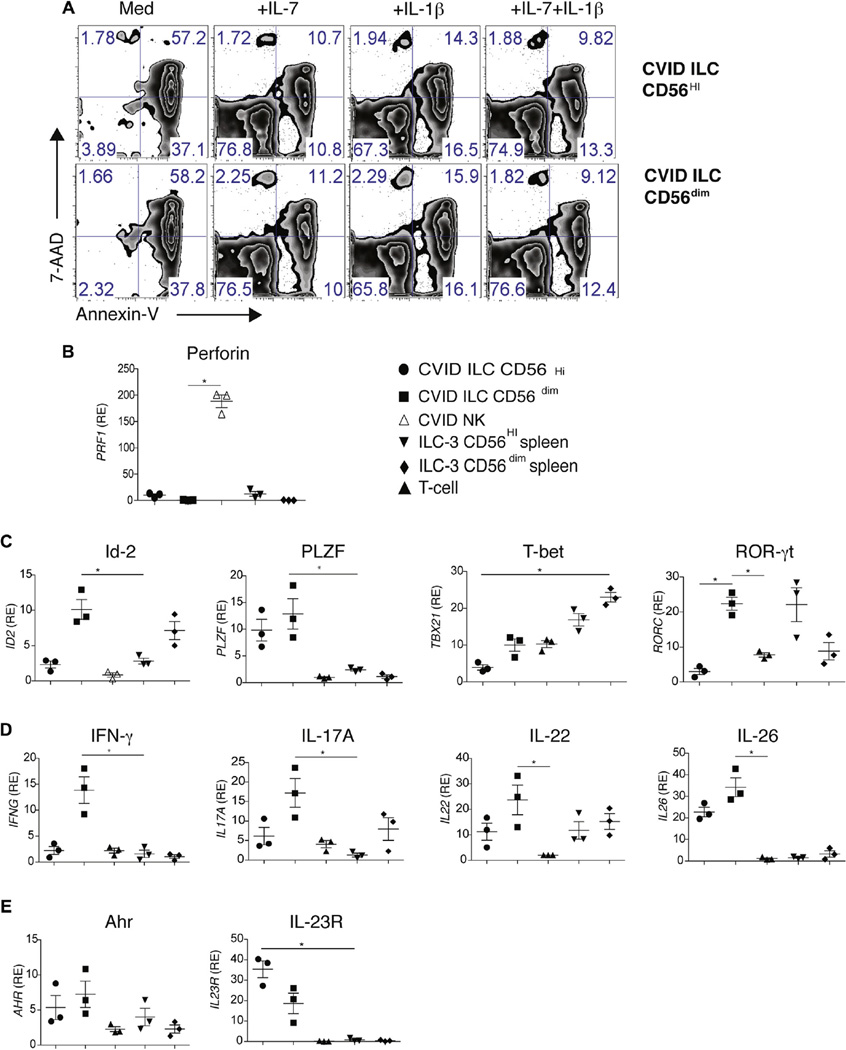
Ex vivo culture and transcriptional features of isolated ILCs
ILCs from peripheral blood of 4 patients with inflammatory complications were sorted and cultured ex vivo into CD56+ and CD5dim populations to compare them with ILC3s described in human spleens.11 As for these cells, CD56+ ILC subsets from patients with CVID were capable of surviving for 5 days in culture when supplemented with IL-7, IL-1β, or both (Fig 3, A). We also compared the transcriptional features of freshly sorted ILCs from patients with CVID with similarly sorted CD56+ and CD5dim ILCs isolated from spleens of immunocompetent donors.11 Although ILC3s were CD56+, perforin-1 mRNA was detected solely in the sorted CD127−CD56+ NK cells but not in the CD127+ ILC3 cells from blood or normal spleen tissue of patients with CVID, differentiating these ILC3s from cytotoxic NK cells (Fig 3, B).
FIG 3.
CD56 expression and transcriptional features. A, Viability of sorted CD56hi and CD56lo ILC3s from patients with CVIDc cultured with medium alone or IL-7, IL-1β, or both × 5 days, as assessed by using fluorescence-activated cell sorting. B, RT-PCR for perforin (PRF1) mRNA expression of sorted cILC3s or NK cells from spleens of patients with CVID or control subjects. C, Quantitative RT-PCR analysis of mRNA for Id-2 (ID2), PLZF, T-bet (TBX21), and RORγt (RORC) in sorted ILC3s, NK cells, or CD3+ T cells. D, Quantitative RT-PCR mRNA encoding for IFN-γ, IL-17A, IL-22, and IL-26. Results were normalized and expressed as above (Fig 3, C). E, Quantitative RT-PCR mRNA for aryl hydrocarbon receptor and the cytokine receptor IL-23R. All RT-PCR results were normalized to β-actin mRNA expressed as relative expression (RE) to PBMCs. Data are from 3 experiments. For Fig 3, B–E: *P < .05, 1-way ANOVA. Error bars = SEMs.
Further comparing transcriptional features of ILCs from patients with CVID with sorted splenic ILC3 populations, CD56dim ILC3s from patients with CVID were found to have higher amounts of the transcriptional regulator inhibitor of DNA binding 2 (Id-2) and comparable levels of RORγt (Fig 3, C).15,17,21 In contrast, ILC3s from patients with CVID had higher amounts of promyelocytic leukemia zinc finger (PLZF) when compared with splenic ILC3s, indicating a different developmental stage (Fig 3, C).22 Although splenic ILC3s contained greater amounts of T-bet, both subsets of ILCs from patients with CVID contained this transcription factor (Fig 3, C). Displaying their unique signature and in concert with intracellular staining, both CD56+ and CD56dim subsets of ILCs from patients with CVID contained higher levels of mRNA transcripts for the proinflammatory cytokines IFN-γ, IL-17A, IL-22, and IL-26 when compared with peripheral T cells or NK cells of the same subjects or splenic ILC3s (Fig 3, D). CD56+ and CD56dim ILC3s from patients with CVID also contained higher levels of aryl hydrocarbon receptor (important in IL-22 production) than splenic ILC3, and only ILC3s from patients with CVID were positive for IL-23 receptor (IL-23R), a part of the inflammatory IL-17/IL-23 axis (Fig 3, E).23,24
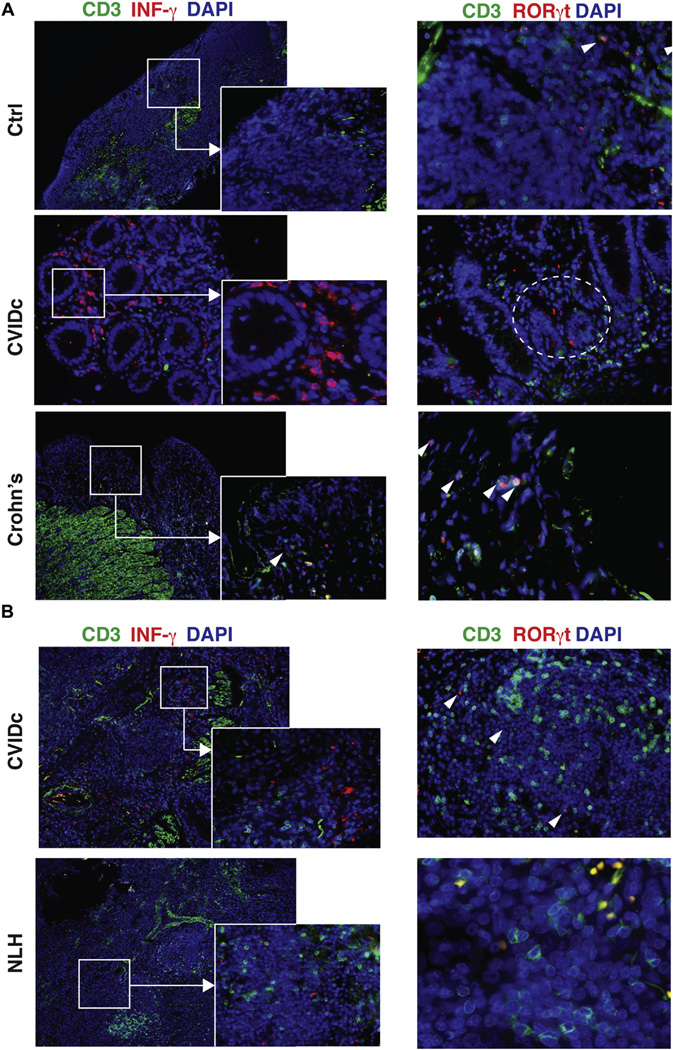
ILCs are found in gastrointestinal and pulmonary tissues
In healthy human subjects about 0.01% to 0.1% of circulating lymphocytes are CD127+ cells, and these are mostly nonactivated type 2 ILCs.9 In contrast, ILC3s, with a few exceptions,25,26 are very rare in the circulation and are generally considered to be restricted to the mucosa, where they appear to play important roles in immunity to fungal, bacterial, and viral microbes, as well as pathologic states of inflammation.9,11,27 Thus we examined tissues from mucosal sites, where patients with CVID have significant tissue damage. Immunofluorescence analysis of small bowel biopsy specimens of patients with enteropathy revealed a distinct population of CD3−RORγt+IFN-γ+ cells, which are not present in noninflamed gastrointestinal tissues of control subjects undergoing unrelated surgery or mucosa of patients with active Crohn disease, where RORγt+ cells were CD3+ (Fig 4, A). CD3−IFN-γ+ populations were detected in other portions of the small bowel in other patients with CVID who had enteropathy (see Fig E2 in this article’s Online Repository at www.jacionline.org).
FIG 4.
ILC3s are detected in mucosal tissues. A, Ileal biopsy specimens of a patient with CVID with enteropathy were compared with tissue from a nonimmunodeficient donor (top) and a patient with Crohn disease (bottom). Tissue was stained for CD3+ T cells (green), IFN-γ (left panels), and RORc (right panels; red) and counterstained with 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI) for nuclei (blue). Solid squares, Area of magnification; dashed circles, IFN-γ+ and RORc+ CD3− cells. CD3+RORc+ double-positive T cells (arrowheads) in noninflamed control ileum (upper rightmost panel) and T cells in patients with Crohn disease. Magnification ×10; inset magnification ×40. Magnification ×20 and ×40 for Crohn disease. B, Lung biopsy specimen of a patient with CVID with lymphoid hyperplasia (CVIDc). The control subject was a nonimmunodeficient patient with nodular lymphoid hyperplasia. Tissue was stained as in Fig 4, A. Magnification ×10; inset magnification ×40. Right panel magnification ×20 and ×40 for nodular lymphoid hyperplasia. Data are from one 4 experiments.
Examining lung biopsy samples of patients with CVID given a diagnosis of lymphocytic interstitial lung disease also showed CD3−IFN-γ+RORγt+ cells, which are suggestive of inflammatory ILC3s. Cells of this sort were not noted in the lungs of an immunocompetent subject given a diagnosis of nodular lymphoid hyperplasia,28 examined here as a pertinent control subject (Fig 4, B, and see Fig E3 in this article’s Online Repository at www.jacionline.org).
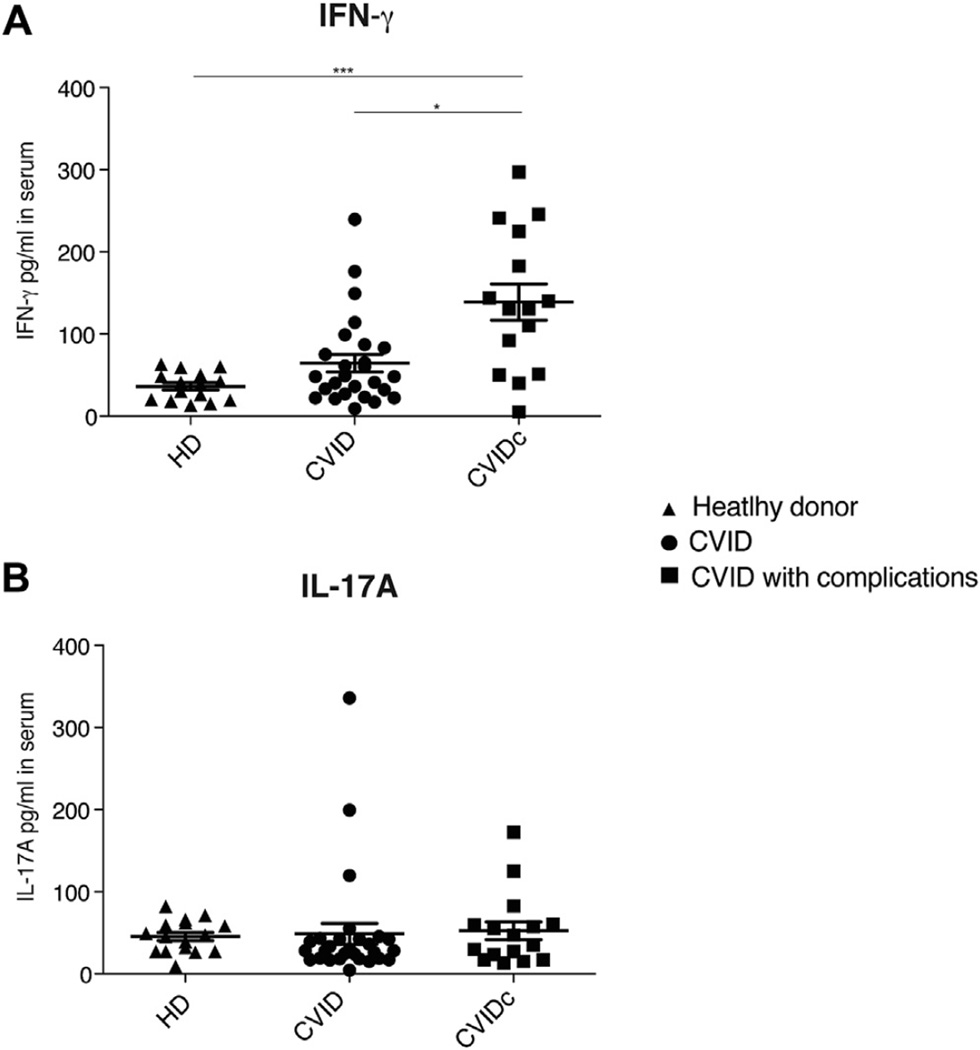
Detection of IFN-γ in serum of patients with CVID
With the significant mRNA IFN-γ signature in blood and expanded IFN-γ ILC3s in the circulation, we then tested sera of patients with CVID for levels of IFN-γ, IL-17A, and IL-22. Sera from patients with CVIDc had significantly increased amounts of IFN-γ (P <.001, 1-way ANOVA; Fig 5, A). Although IL-17A was detected in sera of a few subjects, no significant differences were noted between groups (Fig 5, B). IL-22, which was also measured, was not detectable in these sera (data not shown).
FIG 5.
Cytokine profile of patients with CVID. IFN-γ (A) and IL-17A (B) levels in serum, as measured by using ELISA. *P < .05 and ***P < .001, 1-way ANOVA, followed by the Dunn multiple comparison post hoc test). Data for 15 healthy subjects (HD), 26 patients with CVID without inflammatory complications, and 16 patients with CVID with such complications were pooled for Fig 5, B. Bars denote means with SEMs.
DISCUSSION
One of the more puzzling features of patients with CVID is clinical heterogeneity, with a significant number of subjects whose symptoms clinically stable for years receiving replacement immunoglobulin with very little need for additional therapy. In contrast, up to 50% of subjects have a series of often interlinked complications with the hallmarks of immune dysregulation.4,5,29–31 These complications require treatment with immunosuppressants, immunomodulators, or, in the worst cases, stem cell transplantation.32–34 Seeking to understand the underlying differences in CVID groups, investigators have examined immunologic profiles, clinical phenotypes, cell populations and functions,4,6,35 molecular aspects,36 and cytokines, as previously reviewed.37 Our recent work used a modular whole blood RNA approach to probe the inflammatory signature in patients with CVID.7 We found that the most overactive genes were those in interferon pathways and showed that this signature identified subjects with inflammatory complications. Examination of interferon gene transcripts showed that of these cytokines, only levels of IFN-γ were upregulated and significantly increased in patients with CVIDc. Although cytokine dysregulation has been a recurring theme in patients with CVID, many studies have shown that activated T cells from patients with CVID might produce less IFN-γ than control cells.37 Nevertheless, IFNG mRNA was previously found to be enriched in both gastrointestinal and liver tissues of patients with CVID with enteropathy or nodular regenerative hyperplasia.38–40 Here we investigated the possibly that an expanded IFN-γ+ ILC population might be responsible for the IFN-γ signature observed.
ILCs have emerged as a family of developmentally related hematopoietic effector cells involved in innate immunity, tissue development, and remodeling; however, these cells appear to be essential players in inflammatory diseases and capable of rapid and potent cytokine production. ILCs are divided based on phenotypic and functional characteristics: group 1 contains typical NK cells and other noncytotoxic ILCs, which secrete IFN-γ and express T-bet; group 2 cells depend on GATA-3 and produce IL-5 and IL-13; and group 3 ILCs produce IL-17 and IL-22 and contain the transcription factor RORγt, and depending of the cytokine environment, might produce IFN-γ.8,9 Perhaps reflecting their defensive/inflammatory role, ILCs have been described in the oral mucosa, respiratory and gastrointestinal tracts, and spleen.17,23,26,41–43 In health ILCs secrete cytokines needed for neutrophil recruitment and promote an equilibrium between mucosal immunity and microbial colonization. However, in disease states, such as eczema, psoriasis, Crohn disease, asthma, or multiple sclerosis, these cytokines can foster mucosal damage, alteration of microbial flora, or even tumorigenesis.27
In contrast to a previous study that suggested reduced numbers of peripheral blood IL-17+CD127+Lin− cells in patients with CVID,44 in peripheral blood and tissues of patients with CVIDc, we identified a markedly expanded population of Lin− IL-7 receptor–positive cells that contain IFN-γ, IL-17A, and IL-22, characteristics of ILC3s described in spleen and other lymphoid tissues, skin, brain, and the gastrointestinal and respiratory mucosa.8,9,11,45 Peripheral blood ILCs in patients with CVID, being CD127+ and containing intracellular IFN-γ, IL-17A, IL-22, and the transcription factors T-bet and RORγt, are similar to the proinflammatory “double-negative” ILC3 population previously described.13
Although ILCs have been most extensively investigated at mucosal sites, very rare ILC populations have been detected in peripheral blood. In healthy human subjects about 0.01% to 0.1% of lymphocytes are CD127+ cells, mostly classified as nonactivated type 2 ILCs.9 However, ILC3s in the circulation in patients with CVID were much more numerous, and subjects with inflammatory complications had significantly more of these cells (mean, 3.7%) than subjects without these complications. On sorting ILCs and culture with appropriate cytokines, these cells displayed the distinctive transcriptional features associated with ILCs: Id-2 and PLZF, as well as IL-26, aryl hydrocarbon receptor, and IL-23R.9,13,22 We also found numerous CD3− IFN-γ+ and RORγt+ cells in mucosal biopsy specimens of patients with CVID with enteropathy and interstitial lung disease. Because biopsy specimens were obtained from patients with chronic mucosal disease, we suspect that these are likely to play a role in ongoing mucosal inflammation.
We conclude that inflammatory conditions in patients with CVID are characterized by an expansion of activated mucosal and circulating ILC3s, which might contribute to the interferon mRNA gene signature and cytokine levels in whole blood. Recombination-activating gene 1–deficient mice, lacking T and B cells, also have increased ILC numbers and increased IL-22 secretion, suggesting that adaptive immunity exerts controls on innate cell populations, which proliferate when these are defective.46 On this basis, we predict that an expansion of ILCs will be a feature of other primary immune deficiency states in which there are inflammatory complications. Potentially, the loss of mucosal immunity leading to infections and/or alteration of the microbiome, or bacterial translocation,47 could drive recruitment of ILC3s to these sites. Once recruited, the inflammatory nature of these cells is likely to contribute to further tissue damage.9 Although immunoglobulin treatment reversed the CD4+ T-cell defects ascribed to absorbed bacterial endotoxins in patients with CVID,47 we and others have not found that immunoglobulin therapy alters the clinical course of the noninfectious inflammatory complications in patients with CVID.5,34,40 We also did not find that immunoglobulin therapy prevented the appearance of the whole blood interferon signature in these subjects.7 As the inflammatory potential of ILCs has been increasingly recognized, therapeutic means to control these cells and re-exert homeostasis have not emerged. Further studies to understand the emergence of the ILC population and therapeutic measures to better address these inflammatory complications are required.
Supplementary Material
Clinical implications.
The IFN-γ signature in patients with CVIDc is correlated with a highly proinflammatory ILC population in circulation and mucosal tissues.
Acknowledgments
Supported by the National Institutes of Health (AI 101093, AI-086037, AI-48693, and T32-GM007280), the Jeffrey Modell Foundation, and the David S. Gottesman Immunology Chair.
P. J. Maglione receives research support from the Primary Immune Deficiency Treatment Consortium and the Jeffrey Model Foundation. C. Cunningham-Rundles receives research funding from the National Institute of Health.
We thank the Mount Sinai Microscopy CORE facility at the Icahn School of Medicine. We thank Dr Anthony Bonito for supplying the CD1d tetramer (originally from National Institutes of Health Tetramer Core Facility). We thank Dr Ruiji of the Mount Sinai Core Facility Biorepository, who aided us in obtaining tissues.
Abbreviations used
- CVID
Common variable immunodeficiency
- CVIDc
CVID with inflammatory disease
- CyTOF
Mass cytometry
- Id-2
Inhibitor of DNA binding 2
- ILC
Innate lymphoid cell
- ILC3
Type 3 innate lymphoid cell
- IL-23R
IL-23 receptor
- PLZF
Promyelocytic leukemia zinc finger
- ROR
Retinoic acid–related orphan receptor γt
- T-bet
T-box transcription factor
Footnotes
Disclosure of potential conflict of interest: The rest of the authors declare that they have no relevant conflicts of interest.
REFERENCES
- 1.Al-Herz W, Bousfiha A, Casanova JL, Chatila T, Conley ME, Cunningham-Rundles C, et al. Primary immunodeficiency diseases: an update on the classification from the international union of immunological societies expert committee for primary immunodeficiency. Front Immunol. 2014;5:162. doi: 10.3389/fimmu.2014.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Warnatz K, Voll RE. Pathogenesis of autoimmunity in common variable immunodeficiency. Front Immunol. 2012;3:210. doi: 10.3389/fimmu.2012.00210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Quinti I, Soresina A, Spadaro G, Martino S, Donnanno S, Agostini C, et al. Long-term follow-up and outcome of a large cohort of patients with common variable immunodeficiency. J Clin Immunol. 2007;27:308–316. doi: 10.1007/s10875-007-9075-1. [DOI] [PubMed] [Google Scholar]
- 4.Wehr C, Kivioja T, Schmitt C, Ferry B, Witte T, Eren E, et al. The EUROclass trial: defining subgroups in common variable immunodeficiency. Blood. 2008;111:77–85. doi: 10.1182/blood-2007-06-091744. [DOI] [PubMed] [Google Scholar]
- 5.Resnick ES, Moshier EL, Godbold JH, Cunningham-Rundles C. Morbidity and mortality in common variable immune deficiency over 4 decades. Blood. 2012;119:1650–1657. doi: 10.1182/blood-2011-09-377945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giovannetti A, Pierdominici M, Mazzetta F, Marziali M, Renzi C, Mileo AM, et al. Unravelling the complexity of T cell abnormalities in common variable immunodeficiency. J Immunol. 2007;178:3932–3943. doi: 10.4049/jimmunol.178.6.3932. [DOI] [PubMed] [Google Scholar]
- 7.Park J, Munagala I, Xu H, Blankenship D, Maffucci P, Chaussabel D, et al. Interferon signature in the blood in inflammatory common variable immune deficiency. PLoS One. 2013;8:e74893. doi: 10.1371/journal.pone.0074893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spits H, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, et al. Innate lymphoid cells—a proposal for uniform nomenclature. Nat Rev Immunol. 2013;13:145–149. doi: 10.1038/nri3365. [DOI] [PubMed] [Google Scholar]
- 9.Hazenberg MD, Spits H. Human innate lymphoid cells. Blood. 2014;124:700–709. doi: 10.1182/blood-2013-11-427781. [DOI] [PubMed] [Google Scholar]
- 10.Bendall SC, Simonds EF, Qiu P, Amir el AD, Krutzik PO, Finck R, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332:687–696. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Magri G, Miyajima M, Bascones S, Mortha A, Puga I, Cassis L, et al. Innate lymphoid cells integrate stromal and immunological signals to enhance antibody production by splenic marginal zone B cells. Nat Immunol. 2014;15:354–364. doi: 10.1038/ni.2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Artis D, Spits H. The biology of innate lymphoid cells. Nature. 2015;517:293–301. doi: 10.1038/nature14189. [DOI] [PubMed] [Google Scholar]
- 13.Cording S, Medvedovic J, Cherrier M, Eberl G. Development and regulation of RORgammat innate lymphoid cells. FEBS Lett. 2014;588:4176–4181. doi: 10.1016/j.febslet.2014.03.034. [DOI] [PubMed] [Google Scholar]
- 14.Crellin NK, Trifari S, Kaplan CD, Cupedo T, Spits H. Human NKp44 + IL-22+ cells and LTi-like cells constitute a stable RORC+ lineage distinct from conventional natural killer cells. J Exp Med. 2010;207:281–290. doi: 10.1084/jem.20091509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klose CS, Flach M, Mohle L, Rogell L, Hoyler T, Ebert K, et al. Differentiation of type 1 ILCs from a common progenitor to all helper-like innate lymphoid cell lineages. Cell. 2014;157:340–356. doi: 10.1016/j.cell.2014.03.030. [DOI] [PubMed] [Google Scholar]
- 16.Cella M, Fuchs A, Vermi W, Facchetti F, Otero K, Lennerz JK, et al. A human natural killer cell subset provides an innate source of IL-22 for mucosal immunity. Nature. 2009;457:722–725. doi: 10.1038/nature07537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bernink JH, Peters CP, Munneke M, te Velde AA, Meijer SL, Weijer K, et al. Human type 1 innate lymphoid cells accumulate in inflamed mucosal tissues. Nat Immunol. 2013;14:221–229. doi: 10.1038/ni.2534. [DOI] [PubMed] [Google Scholar]
- 18.Kunkel EJ, Boisvert J, Murphy K, Vierra MA, Genovese MC, Wardlaw AJ, et al. Expression of the chemokine receptors CCR4, CCR5, and CXCR3 by human tissue-infiltrating lymphocytes. Am J Pathol. 2002;160:347–355. doi: 10.1016/S0002-9440(10)64378-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rivino L, Gruarin P, Haringer B, Steinfelder S, Lozza L, Steckel B, et al. CCR6 is expressed on an IL-10-producing, autoreactive memory T cell population with context-dependent regulatory function. J Exp Med. 2010;207:565–577. doi: 10.1084/jem.20091021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buonocore S, Ahern PP, Uhlig HH, Ivanov II, Littman DR, Maloy KJ, et al. Innate lymphoid cells drive interleukin-23-dependent innate intestinal pathology. Nature. 2010;464:1371–1375. doi: 10.1038/nature08949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mjosberg J, Bernink J, Peters C, Spits H. Transcriptional control of innate lymphoid cells. Eur J Immunol. 2012;42:1916–1923. doi: 10.1002/eji.201242639. [DOI] [PubMed] [Google Scholar]
- 22.Constantinides MG, McDonald BD, Verhoef PA, Bendelac A. A committed precursor to innate lymphoid cells. Nature. 2014;508:397–401. doi: 10.1038/nature13047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Geremia A, Arancibia-Carcamo CV, Fleming MP, Rust N, Singh B, Mortensen NJ, et al. IL-23-responsive innate lymphoid cells are increased in inflammatory bowel disease. J Exp Med. 2011;208:1127–1133. doi: 10.1084/jem.20101712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee JS, Cella M, McDonald KG, Garlanda C, Kennedy GD, Nukaya M, et al. AHR drives the development of gut ILC22 cells and postnatal lymphoid tissues via pathways dependent on and independent of Notch. Nat Immunol. 2012;13:144–151. doi: 10.1038/ni.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ciccia F, Guggino G, Rizzo A, Saieva L, Peralta S, Giardina A, et al. Type 3 innate lymphoid cells producing IL-17 and IL-22 are expanded in the gut, in the peripheral blood, synovial fluid and bone marrow of patients with ankylosing spondylitis. Ann Rheum Dis. 2015;74:1739–1747. doi: 10.1136/annrheumdis-2014-206323. [DOI] [PubMed] [Google Scholar]
- 26.Teunissen MB, Munneke JM, Bernink JH, Spuls PI, Res PC, Te Velde A, et al. Composition of innate lymphoid cell subsets in the human skin: enrichment of NCR(+) ILC3 in lesional skin and blood of psoriasis patients. J Invest Dermatol. 2014;134:2351–2360. doi: 10.1038/jid.2014.146. [DOI] [PubMed] [Google Scholar]
- 27.McKenzie AN, Spits H, Eberl G. Innate lymphoid cells in inflammation and immunity. Immunity. 2014;41:366–374. doi: 10.1016/j.immuni.2014.09.006. [DOI] [PubMed] [Google Scholar]
- 28.Abbondanzo SL, Rush W, Bijwaard KE, Koss MN. Nodular lymphoid hyperplasia of the lung: a clinicopathologic study of 14 cases. Am J Surg Pathol. 2000;24:587–597. doi: 10.1097/00000478-200004000-00015. [DOI] [PubMed] [Google Scholar]
- 29.Cunningham-Rundles C. How I treat common variable immune deficiency. Blood. 2010;116:7–15. doi: 10.1182/blood-2010-01-254417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boursiquot JN, Gerard L, Malphettes M, Fieschi C, Galicier L, Boutboul D, et al. Granulomatous disease in CVID: retrospective analysis of clinical characteristics and treatment efficacy in a cohort of 59 patients. J Clin Immunol. 2013;33:84–95. doi: 10.1007/s10875-012-9778-9. [DOI] [PubMed] [Google Scholar]
- 31.Gathmann B, Mahlaoui N, Gerard L, Oksenhendler E, Warnatz K, Warnatz K, et al. Clinical picture and treatment of 2212 patients with common variable immunodeficiency. J Allergy Clin Immunol. 2014;134:116–126. doi: 10.1016/j.jaci.2013.12.1077. [DOI] [PubMed] [Google Scholar]
- 32.Wehr C, Gennery AR, Lindemans C, Schulz A, Hoenig M, Marks R, et al. Multicenter experience in hematopoietic stem cell transplantation for serious complications of common variable immunodeficiency. J Allergy Clin Immunol. 2015;135:988.e6–997.e6. doi: 10.1016/j.jaci.2014.11.029. [DOI] [PubMed] [Google Scholar]
- 33.Gobert D, Bussel JB, Cunningham-Rundles C, Galicier L, Dechartres A, Berezne A, et al. Efficacy and safety of rituximab in common variable immunodeficiency-associated immune cytopenias: a retrospective multicentre study on 33 patients. Br J Haematol. 2011;155:498–508. doi: 10.1111/j.1365-2141.2011.08880.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chase NM, Verbsky JW, Hintermeyer MK, Waukau JK, Tomita-Mitchell A, Casper JT, et al. Use of combination chemotherapy for treatment of granulomatous and lymphocytic interstitial lung disease (GLILD) in patients with common variable immunodeficiency (CVID) J Clin Immunol. 2013;33:30–39. doi: 10.1007/s10875-012-9755-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ochtrop ML, Goldacker S, May AM, Rizzi M, Draeger R, Hauschke D, et al. T and B lymphocyte abnormalities in bone marrow biopsies of common variable immunodeficiency. Blood. 2011;118:309–318. doi: 10.1182/blood-2010-11-321695. [DOI] [PubMed] [Google Scholar]
- 36.Orange JS, Glessner JT, Resnick E, Sullivan KE, Lucas M, Ferry B, et al. Genome-wide association identifies diverse causes of common variable immunodeficiency. J Allergy Clin Immunol. 2011;127:1360.e6–1367.e6. doi: 10.1016/j.jaci.2011.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Varzaneh FN, Keller B, Unger S, Aghamohammadi A, Warnatz K, Rezaei N. Cytokines in common variable immunodeficiency as signs of immune dysregulation and potential therapeutic targets—a review of the current knowledge. J Clin Immunol. 2014;34:524–543. doi: 10.1007/s10875-014-0053-0. [DOI] [PubMed] [Google Scholar]
- 38.Mannon PJ, Fuss IJ, Dill S, Friend J, Groden C, Hornung R, et al. Excess IL-12 but not IL-23 accompanies the inflammatory bowel disease associated with common variable immunodeficiency. Gastroenterology. 2006;131:748–756. doi: 10.1053/j.gastro.2006.06.022. [DOI] [PubMed] [Google Scholar]
- 39.Agarwal S, Mayer L. Pathogenesis and treatment of gastrointestinal disease in antibody deficiency syndromes. J Allergy Clin Immunol. 2009;124:658–664. doi: 10.1016/j.jaci.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fuss IJ, Friend J, Yang Z, He JP, Hooda L, Boyer J, et al. Nodular regenerative hyperplasia in common variable immunodeficiency. J Clin Immunol. 2013;33:748–758. doi: 10.1007/s10875-013-9873-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perry JS, Han S, Xu Q, Herman ML, Kennedy LB, Csako G, et al. Inhibition of LTi cell development by CD25 blockade is associated with decreased intrathecal inflammation in multiple sclerosis. Sci Transl Med. 2012;4:145ra06. doi: 10.1126/scitranslmed.3004140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salimi M, Barlow JL, Saunders SP, Xue L, Gutowska-Owsiak D, Wang X, et al. A role for IL-25 and IL-33-driven type-2 innate lymphoid cells in atopic dermatitis. J Exp Med. 2013;210:2939–2950. doi: 10.1084/jem.20130351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Villanova F, Flutter B, Tosi I, Grys K, Sreeneebus H, Perera GK, et al. Characterization of innate lymphoid cells in human skin and blood demonstrates increase of NKp44+ ILC3 in psoriasis. J Invest Dermatol. 2014;134:984–991. doi: 10.1038/jid.2013.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ganjalikhani-Hakemi M, Yazdani R, Sherkat R, Homayouni V, Masjedi M, Hosseini M. Evaluation of the T helper 17 cell specific genes and the innate lymphoid cells counts in the peripheral blood of patients with the common variable immunodeficiency. J Res Med Sci. 2014;19:30–35. [PMC free article] [PubMed] [Google Scholar]
- 45.Cella M, Otero K, Colonna M. Expansion of human NK-22 cells with IL-7, IL-2, and IL-1beta reveals intrinsic functional plasticity. Proc Natl Acad Sci U S A. 2010;107:10961–10966. doi: 10.1073/pnas.1005641107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sawa S, Lochner M, Satoh-Takayama N, Dulauroy S, Berard M, Kleinschek M, et al. RORgammat+ innate lymphoid cells regulate intestinal homeostasis by integrating negative signals from the symbiotic microbiota. Nat Immunol. 2011;12:320–326. doi: 10.1038/ni.2002. [DOI] [PubMed] [Google Scholar]
- 47.Perreau M, Vigano S, Bellanger F, Pellaton C, Buss G, Comte D, et al. Exhaustion of bacteria-specific CD4 T cells and microbial translocation in common variable immunodeficiency disorders. J Exp Med. 2014;211:2033–2045. doi: 10.1084/jem.20140039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.