Summary
The gut microbiota is a dense and diverse microbial community governed by dynamic microbe–microbe and microbe–host interactions, the status of which influences whether enteric pathogens can cause disease. Here we review recent insights into the key roles that nutrients play in bacterial pathogen exploitation of the gut microbial ecosystem. We synthesize recent findings to support a five-stage model describing the transition between a healthy microbiota and one dominated by a pathogen and disease. Within this five-stage model, two stages are critical to the pathogen: (i) an initial expansion phase that must occur in the absence of pathogen-induced inflammation, followed by (ii) pathogen-promoting physiological changes such as inflammation and diarrhoea. We discuss how this emerging paradigm of pathogen life within the lumen of the gut is giving rise to novel therapeutic strategies.
Introduction
The human gut microbiota is an extremely dense microbial community containing 10–100 trillion bacterial cells (Savage, 1977; Turnbaugh and Gordon, 2009). Many of the resident species have likely co-evolved with human hosts for millennia. Although there is high inter-individual variability and extensive diversity at the level of species and strains, the healthy human microbiota is dominated by bacteria in the phyla Bacterioidetes and Firmicutes. Members of the microbiota play important roles in host health and disease, and altered composition, or dysbiosis, has been linked to a number of conditions including obesity, inflammatory bowel disease and cancer (Turnbaugh et al., 2006; Frank et al., 2007; Wang et al., 2012). The presence of a healthy microbiota results in the establishment and maintenance of a robust community generally impervious to enteric pathogens, a property commonly referred to as colonization resistance. In addition to the understanding that a dense microbiota occupies the spatial and nutritional niches within the intestine, a number of specific mechanisms that contribute to colonization resistance have been elucidated, including immune modulation (Mazmanian et al., 2008; Ivanov et al., 2009). Here, we focus on the nutritional and competitive challenges that enteric pathogens must confront within the gut and the strategies they employ during colonization.
Disruption of a healthy microbiota provides an opportunity for several disease-causing bacteria in the gut. Antibiotic use represents one of the most common causes of microbiota disturbance and, not surprisingly, often results in the loss of colonization resistance and increased susceptibility to multiple pathogens. Many pathogens express potent virulence factors that enable them to induce an altered physiological state in the host (e.g. inflammation or diarrhoea), which places non-pathogenic competitors within the ecosystem at a disadvantage. However, one challenge that many pathogens face is achieving sufficient density so that their environment-modulating factors are effective. In the window of time before this density has been reached, pathogens must compete with commensals for nutrients and space to successfully establish a foothold. In this review, we propose a two-step model of enteric pathogen success: an initial expansion/emergence phase largely characterized by metabolic strategies employed by the pathogen to expand and colonize the intestinal ecosystem, followed by an inflammation-dependent stage in which pathogen-encoded virulence factors become crucial for pathogen maintenance and establishment of disease. This two-step model is placed in the context of five stages of microbiota–pathogen interactions (Fig. 1).
Fig. 1.
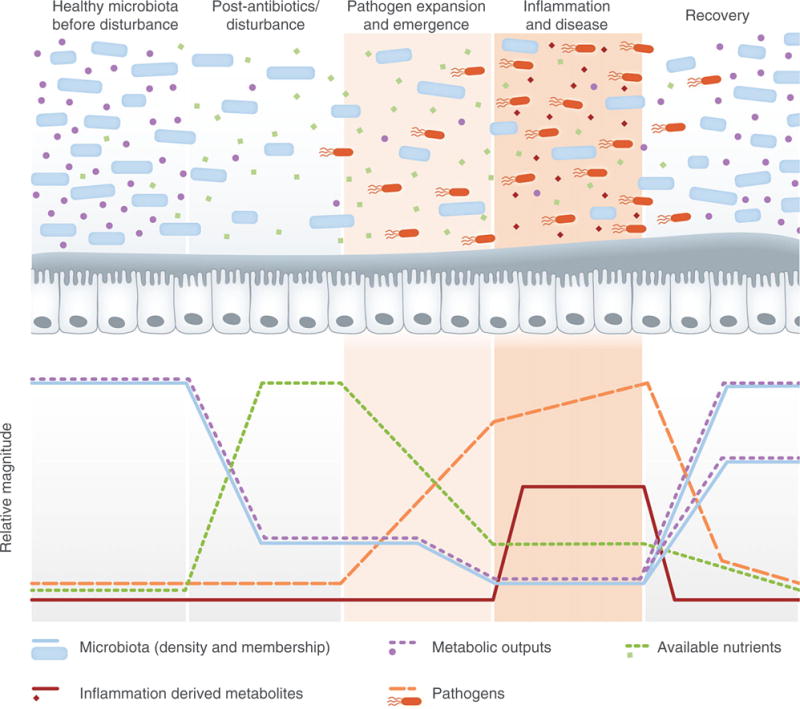
The five stages of microbiota–pathogen interactions. In stage one, a normal, healthy intestinal ecosystem is characterized by a metabolically active microbiota, which consumes available nutrients, produces high levels of metabolic outputs, and keeps pathogens at bay. Upon disturbance (e.g. with antibiotics) in stage two, the density and diversity of the gut commensal microbial community are dramatically decreased, leading to a spike in available nutrients and a decrease in metabolic outputs. In stage three, pathogens capitalize upon the transient increase in available nutrients to expand within the environment. The fourth stage is characterized by pathogen-induced physiological changes, often including inflammation, which continues to shift the balance by further altering the commensal microbial community and the nutrient landscape of the gut. Finally, in the fifth stage, the microbiota begins to recover and expand, consuming available nutrients and increasing metabolic outputs while outcompeting the disease-causing pathogen. Although commensal density will likely increase, the composition and diversity of the microbiota may or may not return to a state identical to the initial healthy state, indicated by the dotted lines at the end of the recovery phase. Light and dark tan shading highlight the enteric two-step, the two distinct stages of pathogen nutritional strategy.
Phase 1: A healthy microbiota
A dynamic network of competitive and cooperative relationships govern interactions within the gut microbiota and between the microbiota and host. Despite compositional differences between healthy individuals, the general functional roles performed by the microbiota appear to be highly conserved (Turnbaugh et al., 2009a). Bacteria that colonize a healthy microbiota have evolved under strong selective pressure to compete efficiently for available nutrients in the gut. Due to the abundance of complex carbohydrates present in host mucins and plant-derived dietary fibres, polysaccharide degradation is a prominent functional attribute found in the human microbiota (Lozupone et al., 2012). Characteristic of a complex microbial ecosystem, much of this nutrient acquisition relies upon key interactions between multiple species. Examples include cross-feeding or synergistic substrate harvest, in which members of the microbiota exhibit interdependent metabolic relationships. It is likely that the inability to isolate several gut resident species in pure monoculture is, in part, due to such highly specialized and intricately linked metabolic niches that are present in the complex gut ecosystem but are difficult to replicate in vitro.
Dietary and host glycan utilization
The influx of dietary and host mucin glycans into the colonic lumen accounts for a major proportion of nutrients available to the microbiota (Koropatkin et al., 2012). The dietary fibres that reach the large intestine include resistant starch and plant cell wall polysaccharides, and are composed of complex sugar molecules containing a high diversity of complex glycosidic linkages. The human genome encodes an estimated 17 glycoside hydrolases and no polysaccharide lyases that participate in dietary carbohydrate degradation. However, an individual’s microbiota encodes thousands of glycoside hydrolases and polysaccharide lyases, which greatly expands the polysaccharide degradation capabilities and illustrates a key ‘profession’ within the gut microbiota (El Kaoutari et al., 2013).
While dietary glycans can change dramatically between meals, a more stable, albeit limited source of nutrients is derived from the host. Host mucus functions as a protective barrier, separating the epithelium from the bacterial community, but also functions as the main host-derived nutrient source for the microbiota. Some commensals have evolved to preferentially consume dietary plant glycans over mucin glycans when both are present, capitalizing on the potentially transient availability of the dietary glycans (Sonnenburg et al., 2010; Lynch and Sonnenburg, 2012; Rogers et al., 2013).
Gut commensals can also shape the landscape of host mucin glycan production in order to provide themselves with exclusive nutritional substrates. For instance, the human gut symbiont Bacteroides thetaiotaomicron induces fucosylation of host mucins in gnotobiotic mice, but loses this ability when its fucose catabolism has been genetically ablated (Hooper et al., 1999). In addition to specific co-evolved ‘gardening’ host–commensal relationships, host mucus secretion is generally responsive to microbial cues from pathogens and commensals (Wrzosek et al., 2013). Alteration of mucin production and glycosylation patterns are likely to have cascading effects across the ecosystem, by promoting the growth of mucin utilizers which may remodel the network of syntrophic interactions.
Fermentation and microbiota metabolic output
As members of the microbiota catabolize carbohydrates they produce end-products including heat, gases (e.g. CO2, CH4 and H2), and organic acids such as the short-chain fatty acids (SCFAs). The SCFAs acetate, propionate and butyrate are the most abundant microbiota fermentation end-products (Wong et al., 2006). Their relative concentrations are affected by several factors including microbial composition, gut transit time and dietary substrates. While commonly viewed as metabolic end-products, SCFAs serve as important metabolic intermediates for nutrient exchange between specific gut bacteria and between the microbiota and the host (Macfarlane and Macfarlane, 2003). For instance, commensal-derived acetate is consumed by several important butyrate producers, such as Faecalibacterium prausnitzii, Roseburia intestinalis and Eubacterium rectale (Duncan et al., 2004; Mahowald et al., 2009).
Consistent with their ubiquitous production by the microbiota, SCFAs are involved in many important processes within the colon. Humans obtain an estimated 10% of their total calories from SCFAs, and butyrate, which is the preferred energy source for the colonic epithelium, has been shown to prevent colorectal cancer and colitis (Bergman, 1990; Wong et al., 2006). The numerous roles that SCFAs play in microbe–microbe and microbe–host interactions make them likely candidates to impact pathogen resistance.
Phase 2: Disturbance
Antibiotic models of infection
Different antibiotic treatments shift the density and diversity of the gut microbiota in nuanced ways. While some antibiotics induce only short-term changes, others impart long-lasting alterations to the microbiota composition and function (Antonopoulos et al., 2009; Buffie et al., 2012). Both the personalized and dynamic properties of the microbiota complicate the predictability of how a single individual’s microbial community will react to antibiotics (Dethlefsen and Relman, 2011). An important area of investigation is aimed at understanding how changes in the microbiota composition during and after antibiotic use influence the metabolic networks between species, the production of small molecule metabolites, and changes in host responses.
Metabolomic studies are providing a new broad view of changes in the spectrum of small molecules made by the gut microbiota and how this profile varies between conditions (Wikoff et al., 2009; Marcobal et al., 2013). Metabolomic analysis of faecal samples from mice treated with a single dose of streptomycin revealed dramatic changes in catabolism, as well as the synthesis of eicosanoid and steroid hormones and the production of bile acids (Antunes et al., 2011). Additionally, SCFA concentrations decrease following antibiotic treatment, and antibiotic-associated diarrhoea has been correlated with reduced levels of SCFAs (Clausen et al., 1991). The loss of SCFAs presumably results from a decrease in the density of the microbiota and a loss of microbes performing fermentation reactions. However, SCFA irrigation as a treatment for inflammatory disorders has led to inconsistent results (Wong et al., 2006). It still remains unclear how altered concentrations of SCFAs and other metabolites impact pathogenesis, but with potential effects on the metabolism of the microbiota, pathogens and host (Smith et al., 2013), the answers that address these relationships are likely to be complex.
Antibiotics are frequently accompanied by antibiotic-associated diarrhoea (AAD) in humans, and in hamster and mouse models antibiotics are often required for pathogens to colonize and cause disease within the gut. A single dose of clindamycin is sufficient to predispose mice to C. difficile infection for as long as ten days post-antibiotic treatment (Buffie et al., 2012). The majority (94%) of hospital-acquired cases of C. difficile-associated disease (CDAD) occur after recent antibiotic treatment (Khanna et al., 2012). Studies with germ-free mice which lack commensal bacteria have demonstrated the importance of the gut microbial community in keeping C. difficile at bay (Onderdonk et al., 1980). Together, these and many other studies support the healthy commensal gut microbial community as a key in preventing pathogen-induced disease within the intestine.
What qualifies as a disturbance?
Epidemiological studies have indicated that while the majority of cases of C. difficile-associated disease were correlated with antibiotic use, 22% of community-acquired C. difficile cases had no history of antibiotic exposure within 90 days (Khanna et al., 2012). These data suggest that there are other susceptibility factors that remain to be understood. Considering that antibiotic use by humans has only become a widespread treatment option over the last several decades, there are likely other factors or ecological disturbances that contribute to the establishment of opportunistic pathogens within the gut.
Shifts in diet have been shown to affect the composition of the microbiota and certainly are met with transient perturbation as the community adjusts to accommodate the new diet (Turnbaugh et al., 2009b; McNulty et al., 2013). Some pathogens, as well as other opportunistic commensals, may have evolved mechanisms to capitalize upon acute ecosystem alterations, such as those that occur meal-to-meal (Lozupone et al., 2013). Changes in diet can affect motility and SCFAs, and are likely to affect spatial proximity of species to one another and the host (Kashyap et al., 2013). How such ecosystem-wide changes influence the favourability of conditions for pathogens is currently not well understood. One study showed that changing mice from a low-fibre to a high-fibre diet results not only in compositional and metabolite changes within the microbiota, but also increased expression of the GP3 receptor for Shiga toxin by the colonic epithelium, leading to an increase in colonization by Shiga-toxin-producing Escherichia coli (Zumbrun et al., 2013). Consumption of live microbes (i.e. probiotics) also has the capacity to influence microbiota functional status and can directly impact pathogen viability in vivo (Corr et al., 2007; McNulty et al., 2011). Fermented foods have been part of our diet for a large part of human evolution, and much suggestive data exists for their potentially diverse roles in human health. The low quality Western diet consumed by much of the modern world, and the corresponding loss of beneficial food-borne microbes is a marked departure from the ancestral human’s high-fibre diet rich in microbes, and the consequences for enteric pathogen susceptibility remain to be determined.
Phase 3: Pathogen expansion
Colonization resistance and the emergence of pathogens
One of the defining features of a ‘healthy’ microbiota is colonization resistance to enteric pathogens. The pathogens Shigella flexneri, Salmonella typhimurium and Vibrio cholerae are unable to colonize mice to high densities without pre-treatment with antibiotics (Freter, 1956; Barthel et al., 2003; Lawley et al., 2008). This requirement of antibiotics to facilitate mouse infection models is consistent with the association between antibiotic usage and susceptibility to enteric pathogens such as Salmonella and C. difficile observed in humans (Pavia et al., 1990; Doorduyn et al., 2006; Khanna et al., 2012). Many data suggest that commensal occupation of niches that would otherwise be exploited by the pathogen contributes to colonization resistance. In anaerobic continuous flow cultures, growth of C. difficile in the presence of a conventional microbiota was observed only when monosaccharides were added, indicating that efficient nutrient depletion represents a key mechanism used by the microbiota to outcompete C. difficile (Wilson and Perini, 1988). It is essential for pathogens to quickly benefit from niches that open up after antibiotic treatment, a transient window of opportunity given the highly efficient resource partitioning and metabolic networks in a healthy, resistant microbiota. For instance, avirulent S. typhimurium rapidly expands in the mouse gut, but without the ability to induce inflammation, recedes as the microbiota recovers (Stecher et al., 2007). Pre-colonization with commensal strains of E. coli and C. difficile can effectively prevent colonization of their pathogenic counterparts, suggesting that careful dissection of nutritional profiles and targeted occupation of nutritional niches with commensal relatives is an effective strategy for preventing infection (Leatham et al., 2009; Maltby et al., 2013; Nagaro et al., 2013).
The metabolic robustness and composition of a stable microbiota in the absence of external perturbations is an important predictor of pathogen invasion. S. typhimurium susceptibility increases in gnotobiotic mice colonized with the low complexity altered Schaedler flora, as well as conventionalized mice which harbour more Enterobacteriaceae (Stecher et al., 2010). Therefore, pathogens may benefit when a microbiota has low diversity. Additionally, the presence of closely related commensals may provide a measure of overall hospitability to colonization by pathogens with similar growth requirements and selective pressures.
One important question is what happens during the transition between a healthy microbiota and one dominated by a pathogen that is causing inflammation and disruption of the gut ecosystem. Inflammation-independent (or pre-inflammatory) mechanisms allow pathogens such as S. typhimurium to grow to high densities before widespread inflammation is triggered. Mutants in the Salmonella pathogenicity islands (SPI) 1 and 2 are unable to cause inflammation, and yet on days 1 and 2 post-antibiotics are able to reach the same intestinal levels as wild-type S. typhimurium (Hapfelmeier et al., 2004; Stecher et al., 2007). Thus, there appears to be a two-step process of successful enteric pathogen colonization (Fig. 1). First, an ‘expansion’ phase, when bacteria must use specific nutritional strategies to compete for nutrients and multiply to high densities, followed by an ‘inflammatory’ phase, when these bacteria attain sufficient density to induce the inflammation and disease from which they benefit, and other strategies for survival become important.
Pathogen nutrient sources during emergence
Competition for commensal-liberated mucosal sugars is key for the expansion of many enteric pathogens. E. coli and S. typhimurium are poor mucin-degraders (i.e. possess few mucus-degrading glycoside hydrolases) and are primarily capable of metabolizing monosaccharides. Therefore, these pathogens appear to depend upon the activity of commensal anaerobes such as Ruminococcus and Bacteroides species that encode wide arsenals of glycoside hydrolases (Xu et al., 2003; Pultz et al., 2006). Although antibiotic treatment kills many mucin-degrading commensals, it is feasible that in addition to secreted extracellular hydrolases, membrane-associated hydrolases would continue to function after cell lysis, leading to an accumulation of monosaccharides in the absence of commensal catabolism (Shipman et al., 1999). Numerous pathogens appear to capitalize upon the short-term increased nutrient availability after antibiotic treatment. A single dose of streptomycin treatment leads to a transient increase in mucus-derived sialic acids in the caecum. The distantly related pathogens S. typhimurium and C. difficile utilize this sugar during the expansion phase of infection, and mutants in sialic acid utilization are at a disadvantage relative to wild-type bacteria during infection (Ng et al., 2013). Other host-derived sugars such as fucose also appear to be important during pathogen expansion (Deatherage Kaiser et al., 2013; Ng et al., 2013). Fucosylated glycans also appear to transiently spike in abundance after antibiotic treatment, suggesting that there is an infection-independent accumulation of fucose that Salmonella may capitalize on in the early stages of expansion (Deatherage Kaiser et al., 2013).
Pathogen-specific metabolic strategies during expansion
Many pathogenic strains appear to exhibit distinct nutrient utilization strategies relative to their commensal counterparts. In a streptomycin mouse model of E. coli colonization, pathogens simultaneously metabolize multiple sugars and display different carbohydrate preferences compared to commensals in vivo, but not in vitro (Chang et al., 2004; Fabich et al., 2008). This differential preference and cometabolism of carbohydrates may allow the pathogen to capitalize upon niches unoccupied by its commensal relative, as well to exploit shared food sources as rapidly as possible.
Another tactic that appears to be used by intestinal pathogens is rapid growth on nutrients with the trade-off of decreased metabolic efficiency. This divergence of metabolic strategies has been observed in growth of different E. coli strains in chemostats and in bovine intestinal contents (Maharjan et al., 2007; Bertin et al., 2013). The slower but more efficient growth of commensals on mucosal sugars may translate into better adaptation to the nutrient-poor, highly competitive environment of the healthy gut and may explain the long-term nature of commensal colonization. Rapidly expanding pathogens in the antibiotic-disrupted intestine benefit from potentially inefficient metabolic strategies, as swift utilization of nutrients deprives commensal counterparts of substrates while facilitating sufficient growth to enable remodelling the environment via expression of virulence factors.
Phase 4: Pathogen-induced inflammation and disease
The ecological opportunity offered to pathogens after disturbance of the intestinal ecosystem appears to be transient. Therefore, many pathogens change the playing field, inducing high levels of inflammation and causing other physiological changes, such as diarrhoea, thus shaping the intestinal environment to promote their own growth. Since other reviews cover this phase of infection thoroughly (Stecher and Hardt, 2011; Winter et al., 2013a), we will focus on a few of the key mechanisms that illustrate the importance of inflammation-derived metabolites which pathogens capitalize upon to maintain colonization.
Remodelling the gut community
After the emergence phase, some pathogens such as C. rodentium and S. typhimurium trigger a positive feedback loop that promotes a disturbed environment advantageous for their growth (Lupp et al., 2007; Sekirov et al., 2008; Lawley et al., 2009). The defect in colonization maintenance by SPI-1 and SPI-2 effector mutants can be rescued by co-infection with a wild-type strain, suggesting that generation of the inflammatory environment is crucial for Salmonella’s long-term survival in the gut (Stecher et al., 2007). However, the functional changes that result from pathogen-induced alterations in microbiota composition and the role altered communities play in pathogen persistence remain intriguing topics for future investigation.
Inflammation: Generation of electron acceptors and nutrients
Inflammation profoundly alters intestinal nutritional resource availability, and pathogens appear to use the products of this disruption to their advantage. During inflammation, S. typhimurium localizes closer to the mucosa to capitalize on increased mucus secretion (Stecher et al., 2008). The immune response to many pathogens involves the production of reactive oxygen and nitrogen species, which undergo a variety of transformations including interacting with lumenal metabolites to form a number of compounds, including nitrate, amine oxides and sulfur oxides. Many members of the Enterobacteriaceae are able to utilize these compounds as electron acceptors to perform anaerobic respiration. Some non-typhoidal Salmonella enterica strains express an effector protein, sopE, which induces caecal nitric oxide synthase 2 (Nos2) expression, providing Salmonella with increased nitrate and promoting growth to higher densities in the inflamed intestine (Lopez et al., 2012). However, E. coli, another member of the Enterobacteriaceae, is also able to profit within the gut from nitrate-based anaerobic respiration (Winter et al., 2013b). Thus, although nitrate is the preferred terminal electron acceptor for Salmonella, it must compete with resident Enterobacteriaceae for this substrate.
Respiration of a different electron acceptor has been demonstrated to promote S. typhimurium densities in the inflamed intestine, potentially a strategy employed to sidestep the competition from commensals for nitrate. In the presence of inflammation, tetrathionate is formed by the oxidation of thiosulfate, a product of host detoxification of the microbiota metabolite hydrogen sulfide. The inability of E. coli to utilize tetrathionate provides an exclusive substrate for Salmonella’s use, and the presence of tetrathionate allows Salmonella to gain energy through anaerobic respiration of compounds that it cannot ferment, such as ethanolamine (Price-Carter et al., 2001; Thiennimitr et al., 2011). Thus, it appears that after expansion in the gut, Salmonella induces inflammation and harnesses its unique capability to utilize nutrients that most commensals cannot use.
Phase 5: Recovery
After a pathogenic insult, the microbiota and the host face the challenge of rebounding from a disturbed and inflammatory state to a normal healthy state. Host innate and adaptive immune responses often play a large role in pathogen clearance, and microbiota recovery may be aided by therapeutic intervention (e.g. antibiotics or faecal transplantation) (Bry and Brenner, 2004; Lawley et al., 2009). During this recovery phase, the microbiota begins to regain its density and diversity, consuming available resources, increasing its metabolic outputs, and outcompeting the invading pathogen (Fig. 1). The new healthy state that excludes the pathogen may or may not resemble the original ecological configuration and microbial species composition (Costello et al., 2012; Lee et al., 2013). The factors that determine the resilience of different disturbed microbial communities to resist transition back to a normal state will be an interesting frontier for future research to explore and may reveal new treatment strategies (Dethlefsen and Relman, 2011).
Probiotics
Probiotics, or intentionally consumed microbial species that exert a positive health impact, have been touted for protection or alleviation from pathogenic insults. Currently, the molecular mechanisms underlying the impact that probiotics exert on the host, pathogens, and the microbiota are largely lacking, with some notable exceptions (Corr et al., 2007; Yan et al., 2013). Probiotic studies vary widely in design, which has led to difficult to interpret and sometimes conflicting results (McFarland, 2006; Hempel et al., 2012). Although several studies have found probiotic consumption to be useful in treating diarrhoea and antibiotic-associated diarrhoea (Siitonen et al., 1990; Pochapin, 2000), the mode of activity of probiotic species, which often do not reach substantial densities in vivo, remains a mystery. Whether individual-specific aspects of host microbiome or genome may influence the effectiveness of a probiotic remain to be understood, and whether probiotics are effective in combination with other therapies or only for certain types of disease is also of great interest. Soon, we may be able to select novel probiotics based on their capabilities to fill certain niches that are exposed after antibiotic treatment to preventatively exclude potential invading pathogens.
Diet as a treatment
A variety of studies have shown that dietary non-digestible oligosaccharides, known as prebiotics, present a useful approach for promoting a healthy indigenous microbiota. Prebiotic interventions have shown a variety of effects in human studies, including altering abundance of metabolites such as phenols, ammonia and SCFAs (Cummings et al., 1996; Muir et al., 2004). Altering host diet has also been shown to promote recovery from pathogen-induced disease. Consumption of cooked green bananas has been shown to effectively treat severe Shigella-induced dysentery in children in two independent studies (Alvarez-Acosta et al., 2009; Rabbani et al., 2009); the increased SCFA production suggests that providing resistant starch and pectin to the microbiota may provide a path from a pathogen-dominated state back to a healthy microbiota. Starch-based solutions have been effective in treating cholera and diarrhoea, presumably by promoting recuperation of the commensal microbiota, although the mechanism is unknown and may differ between studies (Ramakrishna et al., 2000; Alam et al., 2005).
Faecal microbiota transplantation
Faecal microbiota transplantation (FMT), or the exogenous administration of a healthy human donor’s faeces into a sick patient, is the most successful treatment for recurrent C. difficile infections in humans and is far more effective than the standard vancomycin treatment (van Nood et al., 2013). The success of FMT supports the notion that the pathogen creates a self-potentiating dysbiotic inflammatory state. Studies on FMT in humans and mice demonstrate that pathogen eradication is accompanied by restoration of a ‘healthy’ microbiota resembling the composition of the donor microbiota (Khoruts et al., 2010). Although the complexity of the donor microbiota required to re-boot a diseased microbiota is not well-defined, administration of a community of six bacterial species has recently been shown to be sufficient to displace C. difficile in a mouse model of persistent C. difficile disease. Notably, several other six species cocktails were ineffective, and colonizations with individual species from the ‘magic’ combination of six species were not able to outcompete C. difficile (Lawley et al., 2012).
Prospectus
Research in microbiota–pathogen nutritional interactions has progressed significantly over the past 5 years. With numerous -omics technologies and increasingly cheap sequencing technology, researchers are able to probe new aspects of complex ecosystems at unprecedented depth (Medini et al., 2008; Lichtman et al., 2013; Marcobal et al., 2013). Germ-free mice colonized with defined or complete human gut communities will continue to provide valuable tools in the mechanistic understanding of microbiota–pathogen dynamics. In the coming age of personalized medicine, it is likely that microbiome information will help to identify resident opportunistic pathogenic strains or other predictors of disease risk and inform individualized treatments, such as custom diets or defined cocktails of microbiota-derived bacterial strains (i.e. ‘second generation’ probiotics).
As the process of FMT becomes more refined, it still remains to be seen whether certain individuals are endowed with more pathogen-resistant or sensitive microbiotas and would serve as better donor (or recipient) candidates for FMT, and whether FMT is appropriate for use in conditions other than C. difficile recurrence. Alternatively, faecal transplants may even be simplified to a defined community of bacteria. Reaching this stage will require much more data to understand the specific species, molecular determinants and important ecological principles which are key to the success of FMT, as well as a better assessment of the still uncertain potential risks.
A major problem faced by patients undergoing antibiotic treatment is the unintended indiscriminate ablation of the commensal microbes. Antibiotics with increased precision in targeting specific pathogens may help to solve the problem of microbiota disruption (Tannock et al., 2010). Application of principles from synthetic biology to create genetically engineered gut resident microbes that secrete specific effectors, complement metabolic pathways, block pathogen adherence or neutralize harmful toxins will likely be among the arsenal of new tools. For instance, bacteriocins have shown promise in directly killing pathogens in vivo with exceptional species specificity (Corr et al., 2007). Additionally, ecological engineering approaches to strengthen the existing microbial community may sprout from increased insight into the rules that govern microbial ecosystem stability and resilience. Gut microbiota manipulations could be designed based on co-occurrence principles or syntrophic relationships between different commensal microbes, relationships whose characterization is well underway.
As this review highlights, new therapeutic strategies aimed to undercut the nutritional and functional niches used by pathogens are a new frontier. There may be an opportunity to leverage new drugs to target recently elucidated nutritional pathways exploited by antibiotic-associated pathogens, such as administration of a sialidase inhibitor to block sialic acid liberation by commensal microbes after antibiotic treatment (Ng et al., 2013). By considering the dynamics of nutrient use and metabolite production by the microbiota, we may discover novel treatments or improve existing treatments for various opportunistic and antibiotic-associated pathogens.
Acknowledgments
This research was supported by R01-DK085025 (to J.L.S.), NSF graduate fellowships (to K.M.N. and J.A.F.) and a Stanford Graduate Fellowship (to K.M.N.). Justin L. Sonnenburg holds an Investigators in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund.
Footnotes
The authors declare no conflicts of interest.
References
- Alam NH, Meier R, Sarker SA, Bardhan PK, Schneider H, Gyr N. Partially hydrolysed guar gum supplemented comminuted chicken diet in persistent diarrhoea: a randomised controlled trial. Arch Dis Child. 2005;90:195–199. doi: 10.1136/adc.2003.040089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Acosta T, Leon C, Acosta-Gonzalez S, Parra-Soto H, Cluet-Rodriguez I, Rossell MR, Colina-Chourio JA. Beneficial role of green plantain [Musa paradisiaca] in the management of persistent diarrhea: a prospective randomized trial. J Am Coll Nutr. 2009;28:169–176. doi: 10.1080/07315724.2009.10719768. [DOI] [PubMed] [Google Scholar]
- Antonopoulos DA, Huse SM, Morrison HG, Schmidt TM, Sogin ML, Young VB. Reproducible community dynamics of the gastrointestinal microbiota following antibiotic perturbation. Infect Immun. 2009;77:2367–2375. doi: 10.1128/IAI.01520-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antunes LCM, Han J, Ferreira RBR, Lolić P, Borchers CH, Finlay BB. Effect of antibiotic treatment on the intestinal metabolome. Antimicrob Agents Chemother. 2011;55:1494–1503. doi: 10.1128/AAC.01664-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthel M, Hapfelmeier S, Quintanilla-Martínez L, Kremer M, Rohde M, Hogardt M, et al. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect Immun. 2003;71:2839–2858. doi: 10.1128/IAI.71.5.2839-2858.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman EN. Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol Rev. 1990;70:567–590. doi: 10.1152/physrev.1990.70.2.567. [DOI] [PubMed] [Google Scholar]
- Bertin Y, Chaucheyras-Durand F, Robbe-Masselot C, Durand A, de la Foye A, Harel J, et al. Carbohydrate utilization by enterohaemorrhagic Escherichia coli O157:H7 in bovine intestinal content. Environ Microbiol. 2013;15:610–622. doi: 10.1111/1462-2920.12019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bry L, Brenner MB. Critical role of T cell-dependent serum antibody, but not the gut-associated lymphoid tissue, for surviving acute mucosal infection with Citrobacter rodentium, an attaching and effacing pathogen. J Immunol. 2004;172:433–441. doi: 10.4049/jimmunol.172.1.433. [DOI] [PubMed] [Google Scholar]
- Buffie CG, Jarchum I, Equinda M, Lipuma L, Gobourne A, Viale A, et al. Profound alterations of intestinal microbiota following a single dose of clindamycin results in sustained susceptibility to Clostridium difficile-induced colitis. Infect Immun. 2012;80:62–73. doi: 10.1128/IAI.05496-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang DE, Smalley DJ, Tucker DL, Leatham MP, Norris WE, Stevenson SJ, et al. Carbon nutrition of Escherichia coli in the mouse intestine. Proc Natl Acad Sci USA. 2004;101:7427–7432. doi: 10.1073/pnas.0307888101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen MR, Bonnen H, Tvede M, Mortensen PB. Colonic fermentation to short-chain fatty acids is decreased in antibiotic-associated diarrhea. Gastroenterology. 1991;101:1497–1504. doi: 10.1016/0016-5085(91)90384-w. [DOI] [PubMed] [Google Scholar]
- Corr SC, Li Y, Riedel CU, O’Toole PW, Hill C, Gahan CG. Bacteriocin production as a mechanism for the antiinfective activity of Lactobacillus salivarius UCC118. Proc Natl Acad Sci USA. 2007;104:7617–7621. doi: 10.1073/pnas.0700440104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello EK, Stagaman K, Dethlefsen L, Bohannan BJ, Relman DA. The application of ecological theory toward an understanding of the human microbiome. Science. 2012;336:1255–1262. doi: 10.1126/science.1224203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings JH, Beatty ER, Kingman SM, Bingham SA, Englyst HN. Digestion and physiological properties of resistant starch in the human large bowel. Br J Nutr. 1996;75:733–747. doi: 10.1079/bjn19960177. [DOI] [PubMed] [Google Scholar]
- Deatherage Kaiser BL, Li J, Sanford JA, Kim YM, Kronewitter SR, Jones MB, et al. A multi-omic view of host-pathogen-commensal interplay in Salmonella-mediated intestinal infection. PLoS ONE. 2013;8:e67155. doi: 10.1371/journal.pone.0067155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci USA. 2011;108(Suppl. 1):4554–4561. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doorduyn Y, Van Den Brandhof WE, Van Duynhoven YTHP, Wannet WJB, Van Pelt W. Risk factors for Salmonella enteritidis and Typhimurium (DT104 and non-DT104) infections in The Netherlands: predominant roles for raw eggs in Enteritidis and sandboxes in Typhimurium infections. Epidemiol Infect. 2006;134:617–626. doi: 10.1017/S0950268805005406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan SH, Holtrop G, Lobley GE, Calder AG, Stewart CS, Flint HJ. Contribution of acetate to butyrate formation by human faecal bacteria. Br J Nutr. 2004;91:915–923. doi: 10.1079/BJN20041150. [DOI] [PubMed] [Google Scholar]
- El Kaoutari A, Armougom F, Gordon JI, Raoult D, Henrissat B. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat Rev Microbiol. 2013;11:497–504. doi: 10.1038/nrmicro3050. [DOI] [PubMed] [Google Scholar]
- Fabich AJ, Jones SA, Chowdhury FZ, Cernosek A, Anderson A, Smalley D, et al. Comparison of carbon nutrition for pathogenic and commensal Escherichia coli strains in the mouse intestine. Infect Immun. 2008;76:1143–1152. doi: 10.1128/IAI.01386-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freter R. Experimental enteric Shigella and Vibrio infections in mice and guinea pigs. J Exp Med. 1956;104:411–418. doi: 10.1084/jem.104.3.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hapfelmeier S, Ehrbar K, Stecher B, Barthel M, Kremer M, Hardt WD. Role of the Salmonella pathogenicity island 1 effector proteins SipA, SopB, SopE, and SopE2 in Salmonella enterica subspecies 1 serovar Typhimurium colitis in streptomycin-pretreated mice. Infect Immun. 2004;72:795–809. doi: 10.1128/IAI.72.2.795-809.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hempel S, Newberry SJ, Maher AR, Wang Z, Miles JN, Shanman R, et al. Probiotics for the prevention and treatment of antibiotic-associated diarrhea: a systematic review and meta-analysis. JAMA. 2012;307:1959–1969. doi: 10.1001/jama.2012.3507. [DOI] [PubMed] [Google Scholar]
- Hooper LV, Xu J, Falk PG, Midtvedt T, Gordon JI. A molecular sensor that allows a gut commensal to control its nutrient foundation in a competitive ecosystem. Proc Natl Acad Sci USA. 1999;96:9833–9838. doi: 10.1073/pnas.96.17.9833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashyap PC, Marcobal A, Ursell LK, Larauche M, Duboc H, Earle KA, et al. Complex interactions among diet, gastrointestinal transit, and gut microbiota in humanized mice. Gastroenterology. 2013;144:967–977. doi: 10.1053/j.gastro.2013.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna S, Pardi DS, Aronson SL, Kammer PP, Orenstein R, St Sauver JL, et al. The epidemiology of community-acquired Clostridium difficile infection: a population-based study. Am J Gastroenterol. 2012;107:89–95. doi: 10.1038/ajg.2011.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoruts A, Dicksved J, Jansson JK, Sadowsky MJ. Changes in the composition of the human fecal microbiome after bacteriotherapy for recurrent Clostridium difficile-associated diarrhea. J Clin Gastroenterol. 2010;44:354–360. doi: 10.1097/MCG.0b013e3181c87e02. [DOI] [PubMed] [Google Scholar]
- Koropatkin NM, Cameron EA, Martens EC. How glycan metabolism shapes the human gut microbiota. Nat Rev Microbiol. 2012;10:323–335. doi: 10.1038/nrmicro2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawley TD, Bouley DM, Hoy YE, Gerke C, Relman DA, Monack DM. Host transmission of Salmonella enterica serovar Typhimurium is controlled by virulence factors and indigenous intestinal microbiota. Infect Immun. 2008;76:403–416. doi: 10.1128/IAI.01189-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawley TD, Clare S, Walker AW, Goulding D, Stabler RA, Croucher N, et al. Antibiotic treatment of Clostridium difficile carrier mice triggers a supershedder state, spore-mediated transmission, and severe disease in immunocompromised hosts. Infect Immun. 2009;77:3661–3669. doi: 10.1128/IAI.00558-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawley TD, Clare S, Walker AW, Stares MD, Connor TR, Raisen C, et al. Targeted restoration of the intestinal microbiota with a simple, defined bacteriotherapy resolves relapsing Clostridium difficile disease in mice. PLoS Pathog. 2012;8:e1002995. doi: 10.1371/journal.ppat.1002995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leatham MP, Banerjee S, Autieri SM, Mercado-Lubo R, Conway T, Cohen PS. Precolonized human commensal Escherichia coli strains serve as a barrier to E. coli O157:H7 growth in the streptomycin-treated mouse intestine. Infect Immun. 2009;77:2876–2886. doi: 10.1128/IAI.00059-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SM, Donaldson GP, Mikulski Z, Boyajian S, Ley K, Mazmanian SK. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature. 2013;501:426–429. doi: 10.1038/nature12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtman JS, Marcobal A, Sonnenburg JL, Elias JE. Host-centric proteomics of stool: a novel strategy focused on intestinal responses to the gut microbiota. Mol Cell Proteomics. 2013;12:3310–3318. doi: 10.1074/mcp.M113.029967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez CA, Winter SE, Rivera-Chávez F, Xavier MN, Poon V, Nuccio SP, et al. Phage-mediated acquisition of a type III secreted effector protein boosts growth of salmonella by nitrate respiration. MBio. 2012;3:e00143–12. doi: 10.1128/mBio.00143-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone CA, Stombaugh J, Gonzalez A, Ackermann G, Wendel D, Vazquez-Baeza Y, et al. Meta-analyses of studies of the human microbiota. Genome Res. 2013;23:1704–1714. doi: 10.1101/gr.151803.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupp C, Robertson ML, Wickham ME, Sekirov I, Champion OL, Gaynor EC, Finlay BB. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe. 2007;2:119–129. doi: 10.1016/j.chom.2007.06.010. [DOI] [PubMed] [Google Scholar]
- Lynch JB, Sonnenburg JL. Prioritization of a plant polysaccharide over a mucus carbohydrate is enforced by a Bacteroides hybrid two-component system. Mol Microbiol. 2012;85:478–491. doi: 10.1111/j.1365-2958.2012.08123.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlane S, Macfarlane GT. Regulation of short-chain fatty acid production. Proc Nutr Soc. 2003;62:67–72. doi: 10.1079/PNS2002207. [DOI] [PubMed] [Google Scholar]
- McFarland LV. Meta-analysis of probiotics for the prevention of antibiotic associated diarrhea and the treatment of Clostridium difficile disease. Am J Gastroenterol. 2006;101:812–822. doi: 10.1111/j.1572-0241.2006.00465.x. [DOI] [PubMed] [Google Scholar]
- McNulty NP, Wu M, Erickson AR, Pan C, Erickson BK, Martens EC, et al. Effects of diet on resource utilization by a model human gut microbiota containing Bacteroides cellulosilyticus WH2, a symbiont with an extensive glycobiome. PLoS Biol. 2013;11:e1001637. doi: 10.1371/journal.pbio.1001637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNulty NP, Yatsunenko T, Hsiao A, Faith JJ, Muegge BD, Goodman AL, et al. The impact of a consortium of fermented milk strains on the gut microbiome of gnotobiotic mice and monozygotic twins. Science translational medicine. 2011;3:106ra106. doi: 10.1126/scitranslmed.3002701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maharjan RP, Seeto S, Ferenci T. Divergence and redundancy of transport and metabolic rate-yield strategies in a single Escherichia coli population. J Bacteriol. 2007;189:2350–2358. doi: 10.1128/JB.01414-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahowald MA, Rey FE, Seedorf H, Turnbaugh PJ, Fulton RS, Wollam A, et al. Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla. Proc Natl Acad Sci USA. 2009;106:5859–5864. doi: 10.1073/pnas.0901529106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maltby R, Leatham-Jensen MP, Gibson T, Cohen PS, Conway T. Nutritional basis for colonization resistance by human commensal Escherichia coli strains HS and Nissle 1917 against E. coli O157:H7 in the mouse intestine. PLoS ONE. 2013;8:e53957. doi: 10.1371/journal.pone.0053957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcobal A, Southwick AM, Earle KA, Sonnenburg JL. A refined palate: bacterial consumption of host glycans in the gut. Glycobiology. 2013;23:1038–1046. doi: 10.1093/glycob/cwt040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- Medini D, Serruto D, Parkhill J, Relman DA, Donati C, Moxon R, et al. Microbiology in the post-genomic era. Nat Rev Microbiol. 2008;6:419–430. doi: 10.1038/nrmicro1901. [DOI] [PubMed] [Google Scholar]
- Muir JG, Yeow EG, Keogh J, Pizzey C, Bird AR, Sharpe K, et al. Combining wheat bran with resistant starch has more beneficial effects on fecal indexes than does wheat bran alone. Am J Clin Nutr. 2004;79:1020–1028. doi: 10.1093/ajcn/79.6.1020. [DOI] [PubMed] [Google Scholar]
- Nagaro KJ, Phillips ST, Cheknis AK, Sambol SP, Zukowski WE, Johnson S, Gerding DN. Nontoxigenic Clostridium difficile protects hamsters against challenge with historic and epidemic strains of toxigenic BI/NAP1/027 C. difficile. Antimicrob Agents Chemother. 2013;57:5266–5270. doi: 10.1128/AAC.00580-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng KM, Ferreyra JA, Higginbottom SK, Lynch JB, Kashyap PC, Gopinath S, et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature. 2013;502:96–99. doi: 10.1038/nature12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM, et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med. 2013;368:407–415. doi: 10.1056/NEJMoa1205037. [DOI] [PubMed] [Google Scholar]
- Onderdonk AB, Cisneros RL, Bartlett JG. Clostridium difficile in gnotobiotic mice. Infect Immun. 1980;28:277–282. doi: 10.1128/iai.28.1.277-282.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavia AT, Shipman LD, Wells JG, Puhr ND, Smith JD, McKinley TW, Tauxe RV. Epidemiologic evidence that prior antimicrobial exposure decreases resistance to infection by antimicrobial-sensitive Salmonella. J Infect Dis. 1990;161:255–260. doi: 10.1093/infdis/161.2.255. [DOI] [PubMed] [Google Scholar]
- Pochapin M. The effect of probiotics on Clostridium difficile diarrhea. Am J Gastroenterol. 2000;95:S11–S13. doi: 10.1016/s0002-9270(99)00809-6. [DOI] [PubMed] [Google Scholar]
- Price-Carter M, Tingey J, Bobik TA, Roth JR. The alternative electron acceptor tetrathionate supports B12-dependent anaerobic growth of Salmonella enterica serovar typhimurium on ethanolamine or 1,2-propanediol. J Bacteriol. 2001;183:2463–2475. doi: 10.1128/JB.183.8.2463-2475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pultz NJ, Hoskins LC, Donskey CJ. Vancomycin-resistant Enterococci may obtain nutritional support by scavenging carbohydrate fragments generated during mucin degradation by the anaerobic microbiota of the colon. Microb Drug Resist. 2006;12:63–67. doi: 10.1089/mdr.2006.12.63. [DOI] [PubMed] [Google Scholar]
- Rabbani GH, Ahmed S, Hossain I, Islam R, Marni F, Akhtar M, Majid N. Green banana reduces clinical severity of childhood shigellosis: a double-blind, randomized, controlled clinical trial. Pediatr Infect Dis J. 2009;28:420–425. doi: 10.1097/INF.0b013e31819510b5. [DOI] [PubMed] [Google Scholar]
- Ramakrishna BS, Venkataraman S, Srinivasan P, Dash P, Young GP, Binder HJ. Amylase-resistant starch plus oral rehydration solution for cholera. N Engl J Med. 2000;342:308–313. doi: 10.1056/NEJM200002033420502. [DOI] [PubMed] [Google Scholar]
- Rogers TE, Pudlo NA, Koropatkin NM, Bell JS, Moya Balasch M, Jasker K, Martens EC. Dynamic responses of Bacteroides thetaiotaomicron during growth on glycan mixtures. Mol Microbiol. 2013;88:876–890. doi: 10.1111/mmi.12228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage DC. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol. 1977;31:107–133. doi: 10.1146/annurev.mi.31.100177.000543. [DOI] [PubMed] [Google Scholar]
- Sekirov I, Tam NM, Jogova M, Robertson ML, Li Y, Lupp C, Finlay BB. Antibiotic-induced perturbations of the intestinal microbiota alter host susceptibility to enteric infection. Infect Immun. 2008;76:4726–4736. doi: 10.1128/IAI.00319-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shipman JA, Cho KH, Siegel HA, Salyers AA. Physiological characterization of SusG, an outer membrane protein essential for starch utilization by Bacteroides thetaiotaomicron. J Bacteriol. 1999;181:7206–7211. doi: 10.1128/jb.181.23.7206-7211.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siitonen S, Vapaatalo H, Salminen S, Gordin A, Saxelin M, Wikberg R, Kirkkola AL. Effect of Lactobacillus GG yoghurt in prevention of antibiotic associated diarrhoea. Ann Med. 1990;22:57–59. doi: 10.3109/07853899009147243. [DOI] [PubMed] [Google Scholar]
- Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg ED, Zheng H, Joglekar P, Higginbottom SK, Firbank SJ, Bolam DN, Sonnenburg JL. Specificity of polysaccharide use in intestinal bacteroides species determines diet-induced microbiota alterations. Cell. 2010;141:1241–1252. doi: 10.1016/j.cell.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecher B, Hardt WD. Mechanisms controlling pathogen colonization of the gut. Curr Opin Microbiol. 2011;14:82–91. doi: 10.1016/j.mib.2010.10.003. [DOI] [PubMed] [Google Scholar]
- Stecher B, Robbiani R, Walker AW, Westendorf AM, Barthel M, Kremer M, et al. Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 2007;5:2177–2189. doi: 10.1371/journal.pbio.0050244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecher B, Barthel M, Schlumberger MC, Haberli L, Rabsch W, Kremer M, Hardt WD. Motility allows S. Typhimurium to benefit from the mucosal defence. Cell Microbiol. 2008;10:1166–1180. doi: 10.1111/j.1462-5822.2008.01118.x. [DOI] [PubMed] [Google Scholar]
- Stecher B, Chaffron S, Käppeli R, Hapfelmeier S, Freedrich S, Weber TC, et al. Like will to like: abundances of closely related species can predict susceptibility to intestinal colonization by pathogenic and commensal bacteria. PLoS Pathog. 2010;6:e1000711. doi: 10.1371/journal.ppat.1000711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannock GW, Munro K, Taylor C, Lawley B, Young W, Byrne B, et al. A new macrocyclic antibiotic, fidaxomicin (OPT-80), causes less alteration to the bowel microbiota of Clostridium difficile-infected patients than does vancomycin. Microbiology. 2010;156:3354–3359. doi: 10.1099/mic.0.042010-0. [DOI] [PubMed] [Google Scholar]
- Thiennimitr P, Winter SE, Winter MG, Xavier MN, Tolstikov V, Huseby DL, et al. Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc Natl Acad Sci USA. 2011;108:17480–17485. doi: 10.1073/pnas.1107857108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Gordon JI. The core gut microbiome, energy balance and obesity. J Physiol. 2009;587:4153–4158. doi: 10.1113/jphysiol.2009.174136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, et al. A core gut microbiome in obese and lean twins. Nature. 2009a;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009b;1:6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Cai G, Qiu Y, Fei N, Zhang M, Pang X, et al. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. 2012;6:320–329. doi: 10.1038/ismej.2011.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci USA. 2009;106:3698–3703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson KH, Perini F. Role of competition for nutrients in suppression of Clostridium difficile by the colonic microflora. Infect Immun. 1988;56:2610–2614. doi: 10.1128/iai.56.10.2610-2614.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter SE, Lopez CA, Bäumler AJ. The dynamics of gut-associated microbial communities during inflammation. EMBO Rep. 2013a;14:319–327. doi: 10.1038/embor.2013.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter SE, Winter MG, Xavier MN, Thiennimitr P, Poon V, Keestra AM, et al. Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science. 2013b;339:708–711. doi: 10.1126/science.1232467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JM, de Souza R, Kendall CW, Emam A, Jenkins DJ. Colonic health: fermentation and short chain fatty acids. J Clin Gastroenterol. 2006;40:235–243. doi: 10.1097/00004836-200603000-00015. [DOI] [PubMed] [Google Scholar]
- Wrzosek L, Miquel S, Noordine ML, Bouet S, Joncquel Chevalier-Curt M, Robert V, et al. Bacteroides thetaiotaomicron and Faecalibacterium prausnitzii influence the production of mucus glycans and the development of goblet cells in the colonic epithelium of a gnotobiotic model rodent. BMC Biol. 2013;11:61. doi: 10.1186/1741-7007-11-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Bjursell MK, Himrod J, Deng S, Carmichael LK, Chiang HC, et al. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science. 2003;299:2074–2076. doi: 10.1126/science.1080029. [DOI] [PubMed] [Google Scholar]
- Yan F, Liu L, Dempsey PJ, Tsai YH, Raines EW, Wilson CL, et al. A Lactobacillus rhamnosus GG-derived Soluble protein, p40, stimulates ligand release from intestinal epithelial cells to transactivate epidermal growth factor receptor. J Biol Chem. 2013;288:30742–30751. doi: 10.1074/jbc.M113.492397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumbrun SD, Melton-Celsa AR, Smith MA, Gilbreath JJ, Merrell DS, O’Brien AD. Dietary choice affects Shiga toxin-producing Escherichia coli (STEC) O157:H7 colonization and disease. Proc Natl Acad Sci USA. 2013;110:E2126–E2133. doi: 10.1073/pnas.1222014110. [DOI] [PMC free article] [PubMed] [Google Scholar]


