Abstract
The present study aimed evaluate an on-farm culture system for identification of milk pathogens associated with clinical mastitis in dairy cows using two different gold standard approaches: standard laboratory culture in study 1 and 16S rRNA sequencing in study 2. In study 1, milk from mastitic quarters (i.e. presence of flakes, clots, or serous milk; n = 538) was cultured on-farm using a single plate containing three selective chromogenic media (Accumast—FERA Animal Health LCC, Ithaca, NY) and in a reference laboratory using standard culture methods, which was considered the gold standard. In study 2, mastitic milk was cultured on-farm and analyzed through 16S rRNA sequencing (n = 214). In both studies, plates were cultured aerobically at 37°C for 24 h and read by a single technician masked to gold standard results. Accuracy, sensitivity, specificity, positive (PPV) and negative predictive value (NPV) were calculated based on standard laboratory culture in study 1, and PPV was calculated based on sequencing results in study 2. Overall accuracy of Accumast was 84.9%. Likewise, accuracy for identification of Gram-negative bacteria, Staphylococcus sp., and Streptococcus sp. was 96.4%, 93.8%, and 91.5%, respectively. Sensitivity, specificity, PPV, and NPV were 75.0%, 97.9%, 79.6%, and 97.3% for identification of E. coli, 100.0%, 99.8%, 87.5%, and 100.0% for S. aureus, 70.0%, 95.0%, 45.7%, and 98.1% for other Staphylococcus sp., and 90.0%, 92.9%, 91.8%, and 91.2% for Streptococcus sp. In study 2, Accumast PPV was 96.7% for E. coli, 100.0% for Enterococcus sp., 100.0% for Other Gram-negatives, 88.2% for Staphylococcus sp., and 95.0% for Streptococcus sp., respectively. In conclusion, Accumast is a unique approach for on-farm identification pathogens associated with mastitis, presenting overall sensitivity and specificity of 82.3% and 89.9% respectively.
Introduction
Clinical mastitis remains an important animal health issue and leads to major economic losses to the dairy industry worldwide. From 20% to 30% of dairy cows are diagnosed with clinical mastitis at least once during lactation [1, 2]. Estimated costs per case of clinical mastitis range between $179 and $488 depending upon milk prices, level of production in affected cows, culling policies, and stage of lactation when the disease occurred [3, 4]. In fact, treatment and prevention of mastitis are considered the most common causes of antibiotic use in dairy herds [5, 6]. Although fungi and algae have been observed in the milk of cows diagnosed with clinical mastitis, inflammation of the mammary gland is caused predominantly by bacterial infections. Staphylococcus sp., Streptococcus sp., and coliforms account for approximately 90% of isolates in the milk of mastitic cows [1, 7]. Of particular importance for mastitis control programs, the success of antimicrobial therapy is dependent upon the causal pathogen associated with clinical mastitis. Intramammary antibiotic therapy improves the rate of cure in cows infected with coagulase-negative staphylococci, Staphylococcus sp., and environmental streptococci [8]. On the other hand, the use of an intramammary antibiotic is not recommended for cows with mastitis associated with E. coli [9]. Rapid on-farm identification of milk pathogens is critical for targeted antimicrobial therapy, which helps avoid the indiscriminate use of antibiotics in livestock and reduces the economic burden of clinical mastitis.
Several on-farm culture systems have been developed to characterize milk pathogens and substantiate the decision to treat cows with clinical mastitis. Initial on-farm culture systems were based on blood and MacConkey agar plates, which allowed for categorization of microorganisms into Gram-positive, Gram-negative, or no growth within 24 to 32 h and at relative low cost compared to the use of referral diagnostic laboratories. Previously published studies indicate that the use of selective treatment based on on-farm culture systems results has the potential to reduce antibiotic usage by 50% with no changes in the risk of disease recurrence, bacteriological cure, somatic cell count, milk production, and survival throughout lactation [10, 11]. More sophisticated on-farm culture systems allow for further genus classification of Gram-positive bacteria into Staphylococcus sp. and Streptococcus sp. and evaluation of Staphylococcus aureus presence in milk [12]. Nevertheless, using these other techniques the assessment of colony appearance is required for detailed identification of milk pathogens and may reduce the predictive value of on-farm culture systems when conducted by farm personnel. In fact, the sensitivity of selective culture media to detect S. aureus in milk samples ranged from 43.2% to 59.1% when plates were read by individuals with only limited microbiology training [13].
Chromogenic media have been used for identification of microorganisms in both human and animal specimens [14, 15]. Chromogens incorporated in the culture media are cleaved by specific bacterial enzymes generating chromophores, which can be readily recognized with the naked-eye based on color change. The use of chromogenic culture media for identification of milk pathogens associated with mastitis has not been previously evaluated. However, this technology has the potential to increase the number of bacteria distinguishable using on-farm culture systems without requiring intensive microbiology training by farm personnel. The main hypothesis of this study was that the use of chromogenic culture media allows for identification of milk pathogens associated with clinical mastitis in lactating dairy cows with satisfactory sensitivity and specificity. Therefore, specific objectives were to evaluate the use a of selective chromogenic on-farm culture system designed for identification of specific mastitis pathogens: staphylococci, streptococci, and Gram-negative bacteria, constituted by a single plate containing three selective chromogenic media (Accumast—FERA Animal Health LCC, Ithaca, NY). Predictive values of this on-farm culture system were evaluated based on the results from an official diagnostic laboratory (study 1) and on through molecular identification of cultured pathogens using 16S rRNA sequencing (study 2) as gold standards.
Materials and Methods
In-vitro assessment of colony characteristics of pure bacterial cultures plated onto chromogenic media
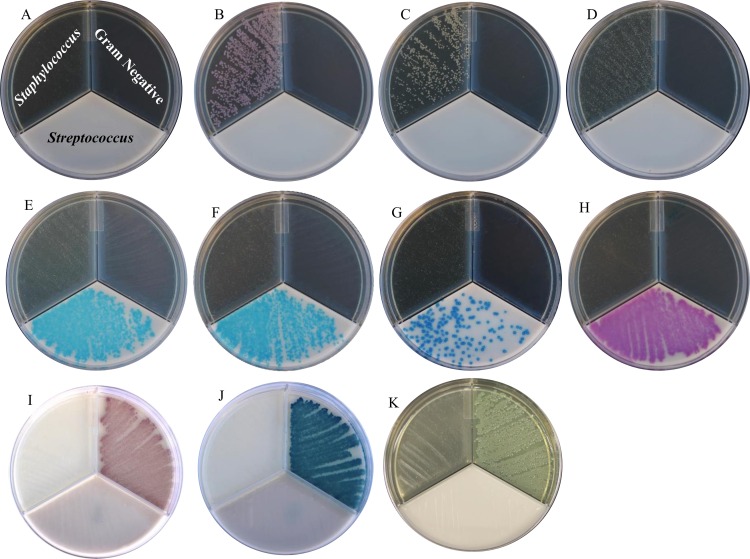
The assessment of growth of pure ATCC strains of pathogens previously described to be associated with bovine mastitis was performed in laboratory using Accumast for the purpose of evaluation of the growth characteristics. The following Gram-positive and Gram-negative ATCC strains (Species, catalog number) were used in the evaluation: (Staphylococcus aureus, 25923; Staphylococcus epidermidis, 12228; Staphylococcus chromogenes, 43764; Streptococcus agalactiae, 27956; Streptococcus dysgalactiae, 43078; Streptococcus uberis, 700407; Enterococcus faecalis, 29212; Escherichia coli, 25922; Klebsiella oxytoca, 49131; Pseudomonas aeruginosa, 15442). Bacteria stocks were activated in unselective tryptic soy agar plates supplemented with 5% sheep blood and 0.1% esculin (BioMerieux, Durhan, NC). Plates were incubated aerobically at 37°C for 24 h to ensure the presence of live bacteria and absence of contamination. For each strain, a single colony was transferred to 5 mL of brain heart infusion broth (Bacto Brain Heart Infusion; Becton, Dickinson and Company, Franklin Lakes, NJ), homogenized, and incubated overnight aerobically at 37°C. Bacterial cultures were diluted to 1:1,000 in sterile PBS solution (Boston BioProducts, Ashland, MA). A sterile cotton swab was used to plate the diluted bacterial culture into each section of Accumast, ensuring that the swab had been saturated in the bacterial sample between plating different sections of the plate. Plates were incubated aerobically at 37°C (Fig 1).
Fig 1. Visual assessment of Gram-positive and Gram-negative bacterial growth on Accumast plates performed in laboratory.
Pictures were taken onto dark background. Plate without bacteria (panel A), Staphylococcus aureus (panel B), Staphylococcus epidermidis (panel C), Staphylococcus chromogenes (panel D), Streptococcus agalactiae (panel E), Streptococcus dysgalactiae (panel F), Streptococcus uberis (panel G), Enterococcus faecalis (panel H) Escherichia coli (panel I)*, Klebsiella oxytoca (panel J)*, and Pseudomonas aeruginosa (panel K). *Pictures were taken onto light background.
The threshold selected for considering a sample positive for bacterial growth using the Accumast culturing system was the presence of five or more colonies in a single section of the plate. Presence of bacterial growth in each of two different sections of the plate was considered a mixed infection and counted as positive for both types of bacteria. Presence of bacterial growth in each of three sections was considered contamination.
Ethics statement
The present study was carried out in agreement with the recommendations of The Animal Welfare Act of 1966 (P.L. 89–544) and its amendments of 1970 (P.L. 91–579), 1976 (P.L. 94–279), and 1985 (P.L. 99–1998) that regulates the transportation, purchase and treatment of animals used in research. The research protocol was reviewed and approved by the Institutional Animal Care and use Committee of the Cornell University (Protocol number: 2013–0056). Sampling animals that present abnormal milk during forestripping on milking preparation is also routine procedure at the study site.
Conflict of interest statement
The product evaluated in the present study was originally developed in Dr. Bicalho’s research laboratory, in Cornell University, Ithaca, NY. Cornell University requires inventors to assign to the university or its designee all rights and titles of their inventions and related property rights that result from activity conducted in the course of an appointment with the university and/or using university resources, including those provided through an externally funded grant, contract, or other type of award or gift to the university. A U. S. Provisional Patent application (No. 62/212,482) was submitted by Cornell University on August 31, 2015 and listed Dr. Bicalho as the inventor. Dr. Bicalho founded the company FERA Animal Health, LLC and licensed the patent rights from Cornell University on February 25, 2016.
Farm and management
On-farm evaluation of the use of Accumast for identification of pathogens associated with clinical mastitis was performed in a single commercial dairy herd located in Venice Center, NY. During the study, approximately 2,800 cows were milked thrice daily in a double-52 milking parlor and the yearly rolling herd average for milk yield was 13,800 kg, with an average bulk tank somatic cell count of 360.000 cells/mL. Primiparous and multiparous cows were housed separately in free-stall barns equipped with sprinklers, fans, and concrete stalls bedded with manure solids. Cows were fed a total mixed ration to meet or exceed the nutrient requirements of a 650 kg lactating Holstein cow producing 45 kg/d of milk with 3.5% fat and 3.2% true protein when dry matter intake is 25 kg/d [16].
Sample collection
Clinical mastitis was defined as the presence of abnormal milk (i.e. presence of flakes, clots, or serous milk) during forestripping performed at the milking parlor. Milk from affected quarters was sampled aseptically by trained farm personnel following recommendations of the National Mastitis Council. Briefly, teats were cleaned and disinfected using 70% ethanol (vol/vol). The initial three streams were discarded and approximately 5 mL of milk was collected into a sterile plastic tube without preservative (Corning Life Sciences, Tewksbury, MA). Milk samples were kept refrigerated at 4°C in a designated office adjacent to the milking parlor and plated at the farm no longer than 12 h after sample collection.
On-farm culture system
The on-farm culture system evaluated here was created to allow for selective growth and identification of specific mastitis pathogens (i.e. staphylococci, streptococci, and Gram-negative bacteria), using a single plate containing three selective chromogenic media (Accumast, FERA Animal Health LCC, Ithaca, NY).
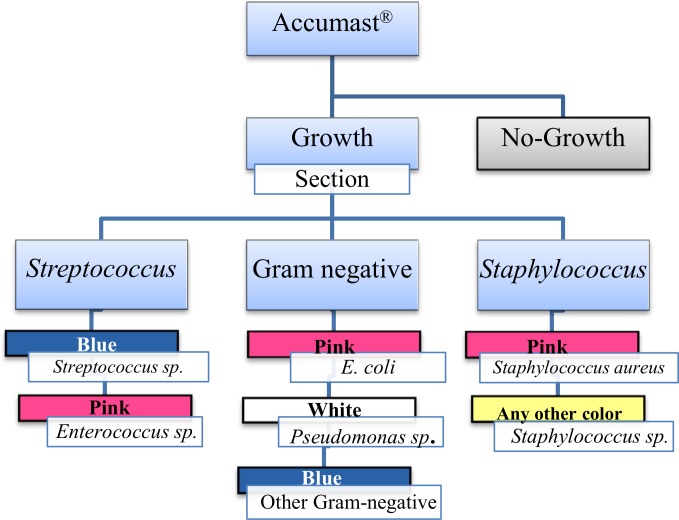
Milk samples were plated onto Accumast using a sterile cotton swab. Before application into each of the three sections of Accumast the swab was immersed in the milk sample. Plates were incubated at 37°C for 24 h and read on-farm by one of the investigators according to the flowchart provided for on-farm diagnosis of mastitis pathogens identifiable by Accumast (Fig 2). Presence of bacterial growth, number of colonies, and color of colonies were recorded. Identification of milk pathogens was performed following instructions of the flowchart developed based on characteristics of growth of American Type Culture Collection (ATCC) strains described above.
Fig 2. Flow-chart for on-farm diagnosis of mastitis pathogens based on Accumast.
Study 1: Identification of milk pathogens by standard laboratory culture
A total of 538 milk samples obtained from cows affected with clinical mastitis were collected as described above, from April to July, 2014. Immediately after collection and plating onto Accumast, milk samples were refrigerated at 4°C and transported to the Quality Milk Production Services (QMPS) laboratory (Cornell University, Ithaca, NY) on ice. Milk samples were cultured following standard procedures for mastitis associated pathogens identification [17] and standard laboratory culture results were used as gold standard for evaluation of on-farm culture system. In summary, milk samples were plated onto trypticase soy agar plates supplemented with 5% of sheep blood and 0.1% esculin using a sterile cotton swab. Plates were incubated aerobically at 35°C to 38°C for at least 24 h but no longer than 48 h. Culture characteristics evaluated included size, color, hemolytic pattern, and odor. Ancillary tests for further bacterial classification included Gram stain and wet mount microscopic evaluations. Biochemical tests comprised evaluation of the presence of catalase using 3% hydrogen peroxide, coagulase using EDTA rabbit plasma tubes, indole using SpotTest (Hardy Diagnostics), KOH string test using 3% potassium hydroxide, oxidase, lactose, sorbitol fermentation, and CAMP tests. Additionally, surface carbohydrates group typing (BactiStaph and PathoDx, Thermo Scientific) and selective differential agars such as MacConkey, Edwards, and bile esculin were used when needed. Samples were considered mixed infections when two clearly distinct bacterial types in a well distributed growth pattern were detected, and both pathogens were reported. Identification of more than two distinct colony types and no contagious pathogens such as S. aureus or S. agalactiae present was considered contamination of the sample. Samples were considered negative when no aerobic bacterial growth was observed in the first 48 h of incubation following guidelines for accredited diagnostic laboratories.
Study 2: Identification of milk pathogens by 16S rRNA sequencing
To further evaluate the accuracy of Accumast for on-farm identification of milk pathogens, a second study utilized bacterial 16S rRNA sequencing from pathogens isolated from mastitic milk samples cultured on-farm between October to December of 2014 using Accumast plates as described above (n = 214) utilizing the Illumina platform.
On-farm identification of milk pathogens was performed as previously described. Plates were read at the farm and those with bacterial growth were transported at room temperature to the laboratory for bacterial isolation, DNA extraction, and sequencing. For each sample, one colony was selected from Accumast, collected using a sterile inoculating loop, and plated onto unselective tryptic soy agar plates supplemented with 5% sheep blood and 0.1% esculin (BioMerieux, Durhan, NC). Plates were incubated aerobically at 37°C for 24 h. This procedure was repeated twice using blood agar plates (Northeast Laboratory Services, Winslow, ME) to ensure that the colony isolated from Accumast was free of contamination.
Genomic DNA was isolated using InstaGene matrix (Bio-Rad laboratories, Hercules, CA) according to manufacturer’s recommendations. Concentration and purity of extracted DNA were evaluated based on optical density using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Rockland, DE). Amplification of the V4 hypervariable region of the bacterial 16S rRNA gene was performed from genomic DNA by PCR utilizing the primers 515F (AATGATACGGCGACCACCGAGATCTACACTATGGTAATTGTGTGCCAGCMGCCGCGGTAA) and barcoded 806R (CAAGCAGAAGACGGCATACGAGATXXXXXXXXXXXX AGTCAGTCAGCCGGACTACHVGGGTWTCTAAT) which have been optimized for the Illumina MiSeq platform [18]. To allow for multiplex sample sequencing, each sample was tagged with a unique 12-bp error-correcting Golay barcode for the 16S rRNA PCR selected using the earth microbiome project (http://www.earthmicrobiome.org/) as formerly described [18, 19]. Barcoded amplicons were generated in triplicate using 3 μL DNA template, 1X EconoTaq Plus Green Master Mix (Lucigen, Middleton, WI), and 5 μM of each primer. Thermocycler conditions were as follows: an initial denaturing step at 94°C for 3 min, 35 cycles of 94°C for 45 s, 50°C for 1 min, and 72°C for 90 s, and a final elongation step of 72°C for 10 min. Blank controls were incorporated in each reaction batch to ensure the absence of bacterial DNA contamination. Replicate amplicons were pooled and and visualized by electrophoresis through 1.2% (wt/vol) agarose gels stained with 0.5 mg/mL ethidium bromide. Purification of amplicons was performed using Gel PCR DNA Fragment Extraction kit (IBI Scientific, Peosta, IA). The NanoDrop ND-1000 spectrophotometer was used for quantification.
Aliquots of 16S rRNA amplicons were standardized to the same concentration (i.e. 16 ng/μL) and sequentially diluted to 20 pM following DNA denaturation. Because bacterial DNA was harvested from a single purified colony, amplicons were combined with 70% of PhiX (Illumina Inc., San Diego, CA). Amplicons were pooled into a single run and final equimolar library was sequenced using the MiSeq reagent kit V2 Nano for 300 cycles on the MiSeq platform (Illumina Inc., San Diego, CA). The 16S rRNA gene sequences were processed using the MiSeq Reporter analysis software version 2.5. Indexed reads were demultiplexed for generation of individual FASTQ files and reads were aligned to the Illumina-curated version of Greengenes database for genus-level classification of milk pathogens.
Statistical analyses
The predictive values of Accumast for the identification of pathogens associated with mastitis were evaluated based on comparing Accumast results with those from standard laboratory culture and results from 16S rRNA sequencing, which were considered the gold standards for comparison in study 1 and 2, respectively. Samples considered contaminated either by the reference laboratory (n = 3) or Accumast (n = 6) were not included in the analysis. Results are presented as parameter estimates and 95% confidence intervals. Confidence intervals were calculated based on the standard error obtained from a binomial distribution following the formulas: SE = and CI = estimate ± 1.96 × SE.
Study 1: Standard laboratory culture as gold standard
Sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) were calculated based on true positives, true negatives, false positives and false negatives as stated by [20] comparing results from on-farm culture of milk samples collected between April and July 2014 and reference laboratory results using the same milk samples. In addition, accuracy was calculated by dividing the number of true positives and true negatives by the total number of tests. The simple Cohen's kappa coefficient (κ) was calculated using the FREQ procedure of SAS version 9.3 (SAS/STAT, SAS Institute Inc., Cary, NC). This parameter assumes that the two response variables (on-farm culture system and gold standard) are independent ratings, and the coefficient equals 1 when there is complete agreement between the two tests. The null hypothesis for this test is that if agreement happens due to chance the Kappa coefficient is equal to zero. Under this null hypothesis, P-values associated with this test equal or smaller than 0.05 were considered significant.
Study 2: 16S rRNA sequencing as gold standard
Because only positive results from Accumast plates were analyzed by 16S rRNA sequencing, the PPV and the simple Cohen's kappa coefficient between sequencing and Accumast were assessed as previously described. Interpretation of Cohen's kappa coefficient was applied following description in [20]. A Kappa coefficient between 0.81 and 1.0 corresponded to almost perfect agreement, 0.61 to 0.80 represented substantial agreement, an estimate between 0.41 and 0.60 was considered moderate agreement and a value of 0.21 to 0.40 denoted fair agreements.
Results
Study 1: Prevalence of milk pathogens associated with clinical mastitis
The prevalence reported herein was calculated based on results from standard laboratory culture which was considered the gold standard and is known as the true prevalence. The most prevalent pathogens in the milk of cows diagnosed with clinical mastitis according to results from QMPS standard laboratory culture were S. uberis, Streptococcus sp., and E. coli (Table 1). Only a small proportion of quarters were diagnosed with mixed infections and 31.2% of milk samples did not result in growth of significant organisms. Three samples were characterized as contaminated by the reference laboratory and 6 samples were characterized as contaminated based on Accumast and were excluded from subsequent analyses. Detailed information is provided in Tables A and B in S1 File.
Table 1. Prevalence of pathogens associated with clinical mastitis.
| Bacteria identified by laboratory culture1 | Number | Prevalence, % |
|---|---|---|
| Streptococcus uberis | 134 | 24.9 |
| Streptococcus sp. | 56 | 10.4 |
| Escherichia coli | 49 | 9.1 |
| Streptococcus dysgalactiae | 40 | 7.8 |
| Staphylococcus sp. | 28 | 5.2 |
| Klebsiella sp. | 16 | 3.0 |
| Mixed infection | 14 | 2.2 |
| Trueperella pyogenes | 10 | 1.9 |
| Staphylococcus aureus | 7 | 1.3 |
| Enterococcus sp. | 7 | 1.3 |
| Gram-negative bacilli | 5 | 0.9 |
| Pseudomonas sp. | 1 | 0.2 |
| No growth | 168 | 31.2 |
| Contamination | 3 | 0.6 |
| Total | 538 | 100 |
1 Results from standard laboratory culture performed by the Quality Milk Production Services laboratory at Cornell University (Ithaca, NY).
Study 1: Test characteristics of Accumast plates for identification of milk pathogens compared to standard laboratory culture
The overall sensitivity, specificity, PPV, NPV, accuracy, and κ coefficient of Accumast for identification of mastitis associated pathogens are presented on (Table 2). Among the Gram-negative bacteria observed in milk samples, the sensitivity and PPV were smaller for the detection of other Gram-negatives compared with E. coli and Pseudomonas sp. (Table 3). The overall sensitivity, specificity, and accuracy for detection of Staphylococcus sp. were 78.4%, 94.9%, and 93.8%, respectively (Table 4). Nevertheless, the accuracy for the identification of S. aureus was greater than for other Staphylococcus sp. Additionally, overall sensitivity, specificity and accuracy for identification of bacteria belonging to the streptococci group was high and the Cohen’s kappa coefficient among Accumast and standard laboratory culture for this bacterial group was denoted substantial.
Table 2. Overall test characteristics of selective chromogenic culture plates to identify bacteria associated with clinical mastitis determined by standard laboratory culture.
| Parameter | Accumast | 95% Confidence Interval |
|---|---|---|
| Number of Tests | 529 | |
| True Prevalence, % (n/n) | 66.2 (350/529) | (66.0–66.3) |
| Sensitivity, % | 82.3 | (82.1–82.5) |
| Specificity, % | 89.9 | (89.6–90.3) |
| PPV1, % | 94.1 | (94.0–94.3) |
| NPV2, % | 72.2 | (71.8–72.6) |
| Accuracy, % | 84.9 | (84.7–85.0) |
| κ 3, % | 0.68 | (0.61–0.74) |
| κ P-value | <0.0001 |
Each milk sample was cultured for identification of bacteria associated with clinical mastitis and culture results from the reference laboratory were considered the gold standard. Each plate was capable of identifying Gram-negative bacteria (E. coli, Pseudomonas sp., and other Gram-negatives), Staphylococcus sp. (S. aureus and Staphylococcus sp.), and Streptococcus sp. Only sections with more than five colonies were considered positive. Samples considered contaminated in either standard culture (n = 3) or on-farm culture (n = 6) were not included in the analysis (n = 9). Plates with no bacterial growth (n = 168) were considered in all calculations.
1 Positive predictive value.
2 Negative predictive value.
3 Cohen’s kappa coefficient. κ ≤ 0 denotes poor agreement; 0.01 to 0.20 denotes slight agreement; 0.21 to 0.40 denotes fair agreement; 0.41 to 0.60 denotes moderate agreement; 0.61 to 0.80 denotes substantial agreement and 0.81 to 1.00 denotes almost perfect agreement.
Table 3. Test characteristics of Accumast plates to identify Gram-negative bacteria associated with clinical mastitis determined by standard laboratory culture.
| Plate results | ||||
|---|---|---|---|---|
| Parameter | Overall Gram-negative | E. coli | Pseudomonas sp. | Other Gram-negatives |
| True Prevalence, % (CI1) n/n | 14.4 (14.2–14.5) 76/529 | 9.8 (9.7–9.9) 52/529 | 0.2 (0.2–0.2) 1/529 | 4.3 (4.3–4.4) 23/529 |
| Sensitivity, % (CI) | 81.6 (80.6–82.6) | 75.0 (73.4–76.6) | 100.0 (100.0–100.0) | 52.2 (47.9–56.4) |
| Specificity, % (CI) | 98.9 (98.9–98.9) | 97.9 (97.8–98.0) | 99.8 (99.8–99.8) | 99.2 (99.2–99.2) |
| PPV2, % (CI) | 92.5 (91.8–93.3) | 79.6 (78.0–81.2) | 50.0 (1.0–99.0) | 75.0 (69.7–80.3) |
| NPV3, % (CI) | 97.0 (96.9–97.0) | 97.3 (97.2–97.4) | 100.0 (100.0–100.0) | 97.9 (97.8–97.9) |
| Accuracy, % (CI) | 96.4 (96.3–96.5) | 95.7 (95.6–95.7) | 99.8 (99.8–99.8) | 97.2 (97.1–97.2) |
| κ4, % (CI) | 0.84 (0.77–0.91) | 0.74 (0.65–0.84) | 0.66 (0.04–1.0) | 0.60 (0.41–0.78) |
| κ P-value | <0.0001 | <0.0001 | <0.0001 | <0.0001 |
Each milk sample was cultured for identification of bacteria associated with clinical mastitis and culture results from the reference laboratory were considered the gold standard. Only sections with more than five colonies were considered positive. Samples considered contaminated in either standard culture (n = 3) or on-farm culture (n = 6) were not included in the analysis (n = 9). Plates with no bacterial growth (n = 168) were considered in all columns. Pure and mixed cultures with the species of interest were combined. Growth on Gram-negative section regardless of color of colonies was considered positive for overall calculations. Only correct identification of bacterial group based on color was considered positive for within group calculations.
1 95% confidence interval.
2 Positive predictive value.
3 Negative predictive value.
4 Cohen’s kappa coefficient. κ ≤ 0 denotes poor agreement; 0.01 to 0.20 denotes slight agreement; 0.21 to 0.40 denotes fair agreement; 0.41 to 0.60 denotes moderate agreement; 0.61 to 0.80 denotes substantial agreement and 0.81 to 1.00 denotes almost perfect agreement.
Table 4. Test characteristics of Accumast to identify Gram-positive bacteria associated with clinical mastitis determined by standard laboratory culture.
| Plate results | ||||
|---|---|---|---|---|
| Parameter | Overall Gram-positive Streptococcus | Overall Gram-positive Staphylococcus | S. aureus | Staphylococcus sp. |
| True Prevalence, % (CI1) n/n | 47.1 (46.9–47.3) 249/529 | 7.0 (6.9–7.1) 37/529 | 1.3 (1.3–1.4) 7/529 | 5.7 (5.6–5.8) 30/529 |
| Sensitivity, % (CI) | 90.0 (89.7–92.7) | 78.4 (76.2–80.6) | 100.0 (100.0–100.0) | 70.0 (67.0–73.0) |
| Specificity, % (CI) | 92.9 (92.7–93.0) | 94.9 (94.8–95.0) | 99.8 (99.8–99.8) | 95.0 (94.9–95.1) |
| PPV2, % (CI) | 91.8 (91.6–92.0) | 53.7 (51.9–55.5) | 87.5 (79.4–95.6) | 45.7 (43.5–47.8) |
| NPV3, % (CI) | 91.2 (91.0–91.4) | 98.3 (98.3–98.4) | 100.0 (100.0–100.0) | 98.1 (98.1–98.2) |
| Accuracy, % (CI) | 91.5 (91.4–91.6) | 93.8 (93.7–93.9) | 99.8 (99.8–99.8) | 93.6 (93.5–93.7) |
| κ4, % (CI) | 0.82 (0.78–0.87) | 0.60 (0.48–0.72) | 0.93 (0.80–1.00) | 0.52 (0.37–0.66) |
| κ P-value | <0.0001 | <0.0001 | <0.0001 | <0.0001 |
Each milk sample was cultured for identification of bacteria associated with clinical mastitis and culture results from the reference laboratory were considered the gold standard. Only sections with more than five colonies were considered positive. Samples considered contaminated in either standard culture (n = 3) or on-farm culture (n = 6) were not included in the analysis (n = 9). Plates with no bacterial growth (n = 168) were considered in all columns. Pure and mixed cultures with the species of interest were combined. Growth on Gram-positive section regardless of color of colonies was considered positive for overall calculations. Only correct identification of bacterial group based on color was considered positive for within group calculations.
1 95% confidence interval.
2 Positive predictive value.
3 Negative predictive value.
4 Cohen’s kappa coefficient. κ ≤ 0 denotes poor agreement; 0.01 to 0.20 denotes slight agreement; 0.21 to 0.40 denotes fair agreement; 0.41 to 0.60 denotes moderate agreement; 0.61 to 0.80 denotes substantial agreement and 0.81 to 1.00 denotes almost perfect agreement.
Study 2: Test characteristics of Accumast plates for identification of milk pathogens compared to 16S rRNA gene sequencing
Results from 16S rRNA gene sequencing confirmed the precision of Accumast plates for identification of milk pathogens associated with clinical mastitis in dairy cows (Table 5). Cohen’s kappa coefficient was above 85% for all bacterial groups evaluated. Likewise, PPV was above 88% across all groups.
Table 5. Test characteristics Accumast plates to identify bacteria associated with clinical mastitis determined by 16S rRNA sequencing.
| Parameter | ||||
|---|---|---|---|---|
| Bacterial group | Tests, (n/n) | PPV1, % (CI2) | κ 3, % | κ P-value |
| Overall | 214 | 95.3 (95.1–95.5) | 0.89 (0.83–0.95) | <0.0001 |
| Escherichia | 30/214 | 96.7 (95.5–97.8) | 0.85 (0.76–0.95) | <0.0001 |
| Enterococcus | 3/214 | 100.0 (100.0–100.0) | 1.000 (1.0–1.00) | <0.0001 |
| Other Gram-negatives | 23/214 | 100.0 (100.0–100.0) | 0.95 (0.88–1.00) | <0.0001 |
| Staphylococcus | 17/214 | 88.2 (84.5–91.9) | 0.93 (0.83–1.00) | <0.0001 |
| Streptococcus | 141/214 | 95.0 (94.7–95.3) | 0.91 (0.86–0.97) | <0.0001 |
Isolates from cases of clinical mastitis cultured using Accumast plates from October 2014 to December 2014 were subjected to 16S rRNA gene sequencing for genus level determination of bacterial growth. Only positive results were available for comparison, therefore only PPV and the Cohen’s kappa coefficient between 16S rRNA sequencing and Accumast were calculated.
1 Positive predictive value.
2 95% confidence interval.
3 Cohen’s kappa coefficient. κ ≤ 0 denotes poor agreement; 0.01 to 0.20 denotes slight agreement; 0.21 to 0.40 denotes fair agreement; 0.41 to 0.60 denotes moderate agreement; 0.61 to 0.80 denotes substantial agreement and 0.81 to 1.00 denotes almost perfect agreement.
Discussion
Knowing the etiology of mammary infections is extremely valuable for the development of strategies to control mastitis [21]. Selective treatment of cows diagnosed with clinical mastitis present major advantages to dairy herds including smaller costs associated with antimicrobials, reduction in the number of animals managed in the hospital pen, less discarded milk, and a potential reduction in the rate of development of antibiotic resistance in livestock [10, 22]. Targeted therapy relies on rapid and accurate identification of milk pathogens [2, 21, 23, 24]. The on-farm culture system evaluated in this study was designed to enable farm personnel to identify major bacteria associated with clinical mastitis in dairy cows in a straightforward manner. The spectrum of microorganisms identifiable using Accumast encompasses both Gram-positive (i.e. streptococci, staphylococci, and S. aureus) and Gram-negative pathogens (i.e. E. coli, Pseudomonas sp. and other Gram-negative pathogens). These bacteria have been reported to represent 80% and 100% of all milk pathogens isolated from mastitic cows in the states of New York [25] and Wisconsin [26]. When compared to a referral laboratory, the overall accuracy of Accumast to identify milk pathogens was 84.9%; with individual accuracies of 96.4%, 93.8%, and 91.5% for Gram-negative bacteria, staphylococci, and streptococci, respectively. Additionally, when compared to 16S rRNA sequencing results, the overall positive predictive value of Accumast was 95.3% and the Cohen's kappa coefficient was 0.89, which according to [20], is considered almost perfect agreement. The present results support the use of this culture system for on-farm identification of pathogens associated with clinical mastitis, for decision making in targeted treatment protocols, and for pathogen prevalence surveillance. It is important to acknowledge that certain pathogens were present in very low prevalence in our study (e. g. S. aureus) and further research should be conducted to validate current findings.
Major advances in the control of contagious pathogens implicated in mastitis have been accomplished through improvement of milking hygiene and management practices [27–29]. However, mammary infections with S. aureus remain a concern in dairy herds and require constant surveillance, aggressive antibiotic therapy, and segregation or culling of infected cows [30, 31]. On-farm culture systems have been used successfully to characterize bacteria present in the milk of mastitic cows as Gram-positive, Gram-negative, or no growth after 24 h to 32 h of incubation. Nonetheless, inconsistent results were observed when classification at the genus and/or species level was attempted by readers lacking extensive microbiology training. For instance, the sensitivity and specificity of University of Minnesota Triplate for identification of S. aureus by four readers with limited microbiology training ranged from 43.2% to 59.1% and 93.8% to 95.9%, respectively [13]. In another study in which two readers without extensive microbiology training used the same system to identify S. aureus, sensitivity ranged from 52% to 78% and specificity from 92% to 98% [12]. Likewise, other investigators used a different triplate for identification of Gram-negative bacteria, staphylococci, and other Gram-positive bacteria and achieved sensitivity and specificity for identification of S. aureus of 65% and 94% [32]. In the present study, the use of Accumast resulted in sensitivity and specificity of 100% and 99.8% for identification of S. aureus under field conditions when compared to standard laboratory culture. These results were confirmed by our in vitro studies, in which ATCC strains of S. aureus, S. chromogenes, and S. epidermidis were plated in all sections of the Accumast plate and the growth of S. aureus colonies were of pink coloration and markedly different from the other two species of staphylococci. The high predictive value of this system compared with other on-farm culture systems can be partially explained by the clear difference in color between S. aureus and other staphylococci in Accumast, as opposed to the need of identification of more subtle differences in colony characteristics and β-hemolysis in other methods [13]. In fact, hemolysin production by S. aureus has been shown to be variable [33]. It is important to acknowledge that this particular species comprised only 1.3% of the pathogen prevalence in the study sample and further research is needed to confirm the findings presented here. Additionally, when compared to 16S rRNA, Accumast presented PPV and a Cohen's kappa coefficient of 0.89, considered almost perfect agreement [20] for all bacterial groups evaluated. Unfortunately the use of this technique as the gold standard does not allow for calculation of NPV, Sensitivity and Specificity, since only positive results from the test being evaluated are available for comparison. However, as mentioned before, bacterial characteristics often used for species identification such as hemolysin production have been shown to be variable. This inconsistency is not an issue when targeted sequencing of the 16S rRNA gene is performed, once this gene has been proved to be highly conserved among different phenotypes of the same bacterial species.
The specificity and NPV of Accumast for identification of Staphylococcus sp. were above 95%; however, its sensitivity and PPV were much smaller compared to that of S. aureus, which can be a limitation of the on-farm culture system evaluated here. Similarly, previous reports also observed a reduced sensitivity for discrimination between S. aureus and other staphylococci [12, 13]. Although not ideal, the reduced capacity of on-farm culture system to identify other Staphylococcus sp. has a smaller impact on its applicability in dairy herds when compared to the capacity of correctly identifying S. aureus. Coagulase-negative staphylococci (CNS) are considered pathogens of minor importance compared with other bacteria while S. aureus remain a major concern because of its contagious behavior [34–36]. In fact, CNS has been associated with subclinical or moderate clinical mastitis and with high spontaneous cure rates [37–40]. Other studies argue that CNS are the main species responsible for mammary gland infection in ruminants, causing significant changes in milk metabolites that play an important role in the quality of dairy products [41, 42]. Regardless of the effect of CNS in mammary infections, milk yield, and downstream milk quality, we acknowledge that improvement on the capability of correctly diagnosing Staphylococcus sp. would be advantageous for the on-farm culture system presented here.
Environmental streptococci were the most prevalent bacteria isolated in the present study, which is in agreement with previous studies [7, 13, 26, 43]. Among environmental pathogens, S. uberis plays an important role in intramammary infections because of its invasive comportment and association with recurring infections [23, 29, 44]. The ability to identify cows infected with Streptococcus sp. is critical for health management in dairy herds as these infections respond well to commercially available intramammary antimicrobials [8]. The use of Accumast resulted in high overall sensitivity and specificity for identification of environmental streptococci independent of the species characterized by standard laboratory culture. The sensitivity and overall accuracy of Accumast plates for identification of Streptococcus sp. were comparable to the ones reported for the methods evaluated by McCarron et al. [43] and Royster et al. [12]. Although the differentiation among Streptococcus species using Accumast was not attempted, visual inspection of ATCC cultures indicate a lighter blue associated with S. agalactiae and S. dysgalactiae compared with that of S. uberis. Similar patterns were also observed during the field study; however, such nuances in tonality were not recorded and further research is necessary to evaluate the predictive values of Accumast for differentiation of Streptococcus sp. in the species level.
Bovine mastitis associated with E. coli has been reported to have high self-cure rates. In an elegant review, Suojala et al., (2013) compiled data from studies that evaluated the treatment of E. coli caused bovine mastitis and concluded that intramammary antibiotic therapy should not be recommended in mild and moderate cases [9]. For this reason, identifying mastitic cows infected primarily with E. coli is a critical step towards reducing the use of antibiotics in dairy herds. On the other hand, results reported by Schukken et al., (2011) support the use of intramammary antimicrobials for treatment of mild and moderate cases of Gram-negative mastitis. In that study, a randomized clinical trial was conducted and revealed a significant increase in the odds of clinical and bacteriological cure in treated animals when compared to non-treated controls [45]. The accuracy of Accumast to identify E. coli was 95.7% compared with standard laboratory culture, with sensitivity and specificity of 75.0% and 97.9%, respectively which are greater than the results from Viora et al. [32] that reported a sensitivity and specificity of 67% and 92% for identification of E. coli in the milk of mastitic cows using a triplate containing selective culture media. Nevertheless, the use of systemic antimicrobial therapy combined with support therapy and anti-inflammatory drugs are recommended in severe cases of E. coli mastitis due to the high indices of bacteremia experienced in cows undergoing this presentation of the disease [46, 47].
Conclusions
The on-farm culture system evaluated in the present study is suitable for use under field conditions and presented substantial overall accuracy for detection of common mastitis pathogens, which was confirmed by 16S rRNA gene sequencing. Accumast provides a unique approach for on-farm identification of mastitis associated pathogens, mostly through its straightforward color-based classification of bacteria. Identification of bacteria based on color allows for easy interpretation by individuals with limited microbiological training; thus, providing the basis for selective antimicrobial therapy of mastitic cows based on causal microorganisms. Further research is warranted to evaluate test characteristics of Accumast between multiple study sites with distinct mastitis pathogens prevalence profiles and among readers without microbiology experience.
Supporting Information
Table A represents True Positives, True Negatives, False Positives and False Negatives according to standard laboratory culture. Table B contains the distribution of results between on-farm culture system and standard laboratory culture.
(DOCX)
Acknowledgments
The authors thank the owners and staff of the collaborating dairy farm for the use of their samples and facilities, as well as for their assistance during experimental procedures. The authors also thank all personnel from the QMPS laboratory for the help with this study.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
These authors have no support or funding to report.
References
- 1.Sargeant JM, Scott HM, Leslie KE, Ireland MJ, Bashiri A. Clinical mastitis in dairy cattle in Ontario: frequency of occurrence and bacteriological isolates. The Canadian veterinary journal La revue veterinaire canadienne. 1998;39(1):33–8. [PMC free article] [PubMed] [Google Scholar]
- 2.Hertl JA, Schukken YH, Welcome FL, Tauer LW, Grohn YT. Effects of pathogen-specific clinical mastitis on probability of conception in Holstein dairy cows. Journal of dairy science. 2014;97(11):6942–54. 10.3168/jds.2014-8203 [DOI] [PubMed] [Google Scholar]
- 3.Bar D, Tauer LW, Bennett G, Gonzalez RN, Hertl JA, Schukken YH, et al. The cost of generic clinical mastitis in dairy cows as estimated by using dynamic programming. Journal of dairy science. 2008;91(6):2205–14. 10.3168/jds.2007-0573 [DOI] [PubMed] [Google Scholar]
- 4.Hagnestam-Nielsen C, Ostergaard S. Economic impact of clinical mastitis in a dairy herd assessed by stochastic simulation using different methods to model yield losses. Animal. 2009;3:315–28. 10.1017/S1751731108003352 [DOI] [PubMed] [Google Scholar]
- 5.Pol M, Ruegg PL. Treatment practices and quantification of antimicrobial drug usage in conventional and organic dairy farms in Wisconsin. Journal of dairy science. 2007;90(1):249–61. [DOI] [PubMed] [Google Scholar]
- 6.Erskine RJ, Wagner SA, DeGraves FJ. Mastitis therapy and pharmacology. Vet Clin North Am Food Anim Pract. 2003;19:109–38. [DOI] [PubMed] [Google Scholar]
- 7.Olde Riekerink RG, Barkema HW, Kelton DF, Scholl DT. Incidence rate of clinical mastitis on Canadian dairy farms. Journal of dairy science. 2008;91(4):1366–77. 10.3168/jds.2007-0757 [DOI] [PubMed] [Google Scholar]
- 8.Roberson JR. Treatment of clinical mastitis. The Veterinary clinics of North AmericaFood animal practice. 2012;28(2):271–88. [DOI] [PubMed] [Google Scholar]
- 9.Suojala L, Kaartinen L, Pyorala S. Treatment for bovine Escherichia coli mastitis—an evidence-based approach. Journal of veterinary pharmacology and therapeutics. 2013;36(6):521–31. 10.1111/jvp.12057 [DOI] [PubMed] [Google Scholar]
- 10.Lago A, Godden SM, Bey R, Ruegg PL, Leslie K. The selective treatment of clinical mastitis based on on-farm culture results: I. Effects on antibiotic use, milk withholding time, and short-term clinical and bacteriological outcomes. Journal of dairy science. 2011;94(9):4441–56. 10.3168/jds.2010-4046 [DOI] [PubMed] [Google Scholar]
- 11.Lago A, Godden SM, Bey R, Ruegg PL, Leslie K. The selective treatment of clinical mastitis based on on-farm culture results: II. Effects on lactation performance, including clinical mastitis recurrence, somatic cell count, milk production, and cow survival. J Dairy Sci. 2011;94(9):4457–67. 10.3168/jds.2010-4047 [DOI] [PubMed] [Google Scholar]
- 12.Royster E, Godden S, Goulart D, Dahlke A, Rapnicki P, Timmerman J. Evaluation of the Minnesota Easy Culture System II Bi-Plate and Tri-Plate for identification of common mastitis pathogens in milk. Journal of dairy science. 2014;97(6):3648–59. 10.3168/jds.2013-7748 [DOI] [PubMed] [Google Scholar]
- 13.McCarron JL, Keefe GP, McKenna SL, Dohoo IR, Poole DE. Evaluation of the University of Minnesota Tri-plate and 3M Petrifilm for the isolation of Staphylococcus aureus and Streptococcus species from clinically mastitic milk samples. Journal of dairy science. 2009;92(10):5326–33. 10.3168/jds.2009-2333 [DOI] [PubMed] [Google Scholar]
- 14.Perry JD, Freydiere AM. The application of chromogenic media in clinical microbiology. Journal of applied microbiology. 2007;103(6):2046–55. [DOI] [PubMed] [Google Scholar]
- 15.Kalchayanand N, Arthur TM, Bosilevac JM, Wells JE, Wheeler TL. Chromogenic agar medium for detection and isolation of Escherichia coli serogroups O26, O45, O103, O111, O121, and O145 from fresh beef and cattle feces. Journal of food protection. 2013;76(2):192–9. 10.4315/0362-028X.JFP-12-182 [DOI] [PubMed] [Google Scholar]
- 16.NRC. Nutrient Requirements of Dairy Cattle Natl Acad Press; 2001;7th rev. ed. [Google Scholar]
- 17.NMC. Laboratory Handbook on Bovine Mastitis. National Mastitis Council. 1999;(Rev. ed.), Madison, WI.
- 18.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. The ISME journal. 2012;6(8):1621–4. 10.1038/ismej.2012.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gilbert JA, Meyer F, Antonopoulos D, Balaji P, Brown CT, Brown CT, et al. Meeting report: the terabase metagenomics workshop and the vision of an Earth microbiome project. Standards in genomic sciences. 2010;3(3):243–8. 10.4056/sigs.1433550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dohoo I, Martin W, and Stryhn H. Screening and diagnostic tests In: Veterinary Epidemiologic Research. Charlottetown: VER Inc; 2009. pp. 91–134. [Google Scholar]
- 21.Cha E, Kristensen AR, Hertl JA, Schukken YH, Tauer LW, Welcome FL, et al. Optimal insemination and replacement decisions to minimize the cost of pathogen-specific clinical mastitis in dairy cows. Journal of dairy science. 2014;97(4):2101–17. 10.3168/jds.2013-7067 [DOI] [PubMed] [Google Scholar]
- 22.Oliver SP, Murinda SE. Antimicrobial resistance of mastitis pathogens. The Veterinary clinics of North AmericaFood animal practice. 2012;28(2):165–85. [DOI] [PubMed] [Google Scholar]
- 23.McDougall S, Arthur DG, Bryan MA, Vermunt JJ, Weir AM. Clinical and bacteriological response to treatment of clinical mastitis with one of three intramammary antibiotics. New Zealand veterinary journal. 2007;55(4):161–70. [DOI] [PubMed] [Google Scholar]
- 24.Royster E, Wagner S. Treatment of mastitis in cattle. The Veterinary clinics of North AmericaFood animal practice. 2015;31(1):17–46, v. [DOI] [PubMed] [Google Scholar]
- 25.Hertl JA, Schukken YH, Welcome FL, Tauer LW, Grohn YT. Pathogen-specific effects on milk yield in repeated clinical mastitis episodes in Holstein dairy cows. Journal of dairy science. 2014;97(3):1465–80. 10.3168/jds.2013-7266 [DOI] [PubMed] [Google Scholar]
- 26.Oliveira L, Ruegg PL. Treatments of clinical mastitis occurring in cows on 51 large dairy herds in Wisconsin. Journal of dairy science. 2014;97(9):5426–36. 10.3168/jds.2013-7756 [DOI] [PubMed] [Google Scholar]
- 27.Bushnell RB. The importance of hygienic procedures in controlling mastitis. The Veterinary clinics of North AmericaLarge animal practice. 1984;6(2):361–70. [DOI] [PubMed] [Google Scholar]
- 28.Hogan JS, Smith KL, Hoblet KH, Schoenberger PS, Todhunter DA, Hueston WD, et al. Field survey of clinical mastitis in low somatic cell count herds. J Dairy Sci. 1989;72:1547–56. [DOI] [PubMed] [Google Scholar]
- 29.Hillerton JE, Berry EA. The management and treatment of environmental streptococcal mastitis. The Veterinary clinics of North AmericaFood animal practice. 2003;19(1):157–69. [DOI] [PubMed] [Google Scholar]
- 30.Zecconi A, Binda E, Borromeo V, Piccinini R. Relationship between some Staphylococcus aureus pathogenic factors and growth rates and somatic cell counts. The Journal of dairy research. 2005;72(2):203–8. [DOI] [PubMed] [Google Scholar]
- 31.Boss R, Naskova J, Steiner A, Graber HU. Mastitis diagnostics: quantitative PCR for Staphylococcus aureus genotype B in bulk tank milk. Journal of dairy science. 2011;94(1):128–37. 10.3168/jds.2010-3251 [DOI] [PubMed] [Google Scholar]
- 32.Viora L, Graham EM, Mellor DJ, Reynolds K, Simoes PB, Geraghty TE. Evaluation of a culture-based pathogen identification kit for bacterial causes of bovine mastitis. The Veterinary record. 2014;175(4):89 10.1136/vr.102499 [DOI] [PubMed] [Google Scholar]
- 33.Boerlin P, Kuhnert P, Hussy D, Schaellibaum M. Methods for identification of Staphylococcus aureus isolates in cases of bovine mastitis. Journal of clinical microbiology. 2003;41(2):767–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pyorala S, Taponen S. Coagulase-negative staphylococci—emerging mastitis pathogens. Vet Microbiol. 2009;134(1–2):3–8. 10.1016/j.vetmic.2008.09.015 [DOI] [PubMed] [Google Scholar]
- 35.Boss R, Cosandey A, Luini M, Artursson K, Bardiau M, Breitenwieser F, et al. Bovine Staphylococcus aureus: Subtyping, evolution, and zoonotic transfer. J Dairy Sci. 2016;99(1):515–28. 10.3168/jds.2015-9589 [DOI] [PubMed] [Google Scholar]
- 36.da Costa LB, Rajala-Schultz PJ, Schuenemann GM. Management practices associated with presence of Staphylococcus aureus in bulk tank milk from Ohio dairy herds. J Dairy Sci. 2016;99(2):1364–73. 10.3168/jds.2015-9870 [DOI] [PubMed] [Google Scholar]
- 37.McDougall S. Efficacy of two antibiotic treatments in curing clinical and subclinical mastitis in lactating dairy cows. New Zealand veterinary journal. 1998;46(6):226–32. [DOI] [PubMed] [Google Scholar]
- 38.Wilson DJ, Gonzalez RN, Case KL, Garrison LL, Grohn YT. Comparison of seven antibiotic treatments with no treatment for bacteriological efficacy against bovine mastitis pathogens. Journal of dairy science. 1999;82(8):1664–70. [DOI] [PubMed] [Google Scholar]
- 39.Taponen S, Simojoki H, Haveri M, Larsen HD, Pyorala S. Clinical characteristics and persistence of bovine mastitis caused by different species of coagulase-negative staphylococci identified with API or AFLP. Veterinary microbiology. 2006;115(1–3):199–207. [DOI] [PubMed] [Google Scholar]
- 40.Tomazi T, Goncalves JL, Barreiro JR, Arcari MA, dos Santos MV. Bovine subclinical intramammary infection caused by coagulase-negative staphylococci increases somatic cell count but has no effect on milk yield or composition. Journal of dairy science. 2015;98(5):3071–8. 10.3168/jds.2014-8466 [DOI] [PubMed] [Google Scholar]
- 41.Silanikove N, Merin U, Shapiro F, Leitner G. Milk metabolites as indicators of mammary gland functions and milk quality. The Journal of dairy research. 2014;81(3):358–63. 10.1017/S0022029914000260 [DOI] [PubMed] [Google Scholar]
- 42.Silanikove N, Merin U, Shapiro F, Leitner G. Subclinical mastitis in goats is associated with upregulation of nitric oxide-derived oxidative stress that causes reduction of milk antioxidative properties and impairment of its quality. Journal of dairy science. 2014;97(6):3449–55. 10.3168/jds.2013-7334 [DOI] [PubMed] [Google Scholar]
- 43.McCarron JL, Keefe GP, McKenna SL, Dohoo IR, Poole DE. Laboratory evaluation of 3M Petrifilms and University of Minnesota Bi-plates as potential on-farm tests for clinical mastitis. Journal of dairy science. 2009;92(5):2297–305. 10.3168/jds.2008-1661 [DOI] [PubMed] [Google Scholar]
- 44.Abureema S, Smooker P, Malmo J, Deighton M. Molecular epidemiology of recurrent clinical mastitis due to Streptococcus uberis: evidence of both an environmental source and recurring infection with the same strain. Journal of dairy science. 2014;97(1):285–90. 10.3168/jds.2013-7074 [DOI] [PubMed] [Google Scholar]
- 45.Schukken YH, Bennett GJ, Zurakowski MJ, Sharkey HL, Rauch BJ, Thomas MJ, et al. Randomized clinical trial to evaluate the efficacy of a 5-day ceftiofur hydrochloride intramammary treatment on nonsevere gram-negative clinical mastitis. Journal of dairy science. 2011;94(12):6203–15. 10.3168/jds.2011-4290 [DOI] [PubMed] [Google Scholar]
- 46.Wenz JR, Barrington GM, Garry FB, McSweeney KD, Dinsmore RP, Goodell G, et al. Bacteremia associated with naturally occuring acute coliform mastitis in dairy cows. J Am Vet Med Assoc. 2001;219(7):976–81. [DOI] [PubMed] [Google Scholar]
- 47.Erskine RJ, Bartlett PC, VanLente JL, Phipps CR. Efficacy of systemic ceftiofur as a therapy for severe clinical mastitis in dairy cattle. J Dairy Sci. 2002;85(10):2571–5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table A represents True Positives, True Negatives, False Positives and False Negatives according to standard laboratory culture. Table B contains the distribution of results between on-farm culture system and standard laboratory culture.
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.