Abstract
The transcription factor FliA, also called sigma 28, is a major regulator of bacterial flagellar biosynthesis genes. Growing evidence suggest that in addition to motility, FliA is involved in controlling numerous bacterial behaviors, even though the underlying regulatory mechanism remains unclear. By using a transcriptional fusion to gfp that responds to cyclic (c)-di-GMP, this study revealed a higher c-di-GMP concentration in the fliA deletion mutant of Pseudomonas aeruginosa than in its wild-type strain PAO1. A comparative analysis of transcriptome profiles of P. aeruginosa PAO1 and its fliA deletion mutant revealed an altered expression of several c-di-GMP-modulating enzyme-encoding genes in the fliA deletion mutant. Moreover, the downregulation of PA4367 (bifA), a Glu-Ala-Leu motif-containing phosphodiesterase, in the fliA deletion mutant was confirmed using the β-glucuronidase reporter gene assay. FliA also altered pyocyanin and pyorubin production by modulating the c-di-GMP concentration. Complementing the fliA mutant strain with bifA restored the motility defect and pigment overproduction of the fliA mutant. Our results indicate that in addition to regulating flagellar gene transcription, FliA can modulate the c-di-GMP concentration to regulate the swarming motility and phenazine pigment production in P. aeruginosa.
Introduction
The transcription factor FliA, also called sigma factor 28, is a major transcription factor regulating flagellar biosynthesis [1,2]. The motility of fliA mutants of Escherichia coli, Pseudomonas aeruginosa, and Bacillus subtilis is significantly compromised, whereas the complementation of the fliA gene could rescue the defective motility in fliA mutants [2,3]. The importance of fliA in flagellar biosynthesis and chemotaxis has also been characterized in numerous other bacterial species, including Pseudomonas putida, Vibrio cholerae, Campylobacter jejuni, Edwardsiella tarda, and Yersinia pestis [4–8].
In addition to controlling flagellar biosynthesis at the transcriptional level, bacterial motility is also posttranslationally regulated. A mechanism independent of flagellar biosynthesis involving YhjH and YcgR proteins was observed in E. coli [9]. YhjH, a phosphodiesterase (PDE), governs the downregulation of cyclic (c)-di-GMP (bis-(3ʹ-5ʹ)-c-di-GMP) concentrations. YcgR, a c-di-GMP-binding protein, is a key regulator of flagellar motor control, responding to c-di-GMP and inhibiting motility by interacting with the flagellar switch complex proteins FliG and FliM [9,10]. In E. coli, expression of yhjH is found to be dependent on FliA [11].
C-di-GMP is a universal intracellular second messenger in bacteria [12–14]. It plays crucial roles in complex physiological processes, including virulence, motility, biofilm formation, and cell cycle progression [12,14–17]. Diguanylate cyclases (DGCs) and cyclic nucleotide PDEs cause the synthesis and degradation of c-di-GMP, respectively [18]. DGCs typically contain a conserved Gly-Gly-Asp-Glu-Phe (GGDEF) or Gly-Gly-Glu-Glu-Phe (GGEEF) domain, whereas PDEs typically contain a Glu-Ala-Leu (EAL) domain [19,20].
P. aeruginosa is an opportunistic pathogen possessing a polar flagellum for swimming in a liquid environment and swarming motility on semisolid surfaces [21,22]. P. aeruginosa also requires flagella to adhere to the host epithelium at the initiation of infection [23,24]. As a global regulator, FliA is considered to regulate the expression of genes involving flagellin production, initiation of flagella filament assembly and length control, chemotaxis regulator, motor rotation, and FliA-specific anti-sigma factor in P. aeruginosa [22]. Because there is no E. coli yhjH ortholog present in P. aeruginosa, whether FliA can regulate motility through c-di-GMP in P. aeruginosa is not clear. The present study aimed to determine whether FliA can regulate bacterial motility and pigment production through the second messenger c-di-GMP. Our results indicate that intracellular c-di-GMP concentrations are altered in the fliA deletion mutant and that a PDE, BifA contributes to the FliA-mediated c-di-GMP regulation and further affects the bacterial physiology.
Materials and Methods
Bacterial strains and plasmids
The strains and plasmids of P. aeruginosa used in this study are listed in Table 1. The bacterial strains were propagated in Luria–Bertani (LB) broth supplemented with an appropriate antibiotic, as indicated in Table 1. The growth rates of the strains were determined through spectrometry by measuring the absorbance at 595 nm or by enumerating the colony-forming units after plating the bacterial cells on LB agar and incubating overnight.
Table 1. Bacteria strains and plasmids used in this study.
| Strain/plasmid | Relevant characteristics | Sources |
|---|---|---|
| E. coli | ||
| S17-1 λ pir | Tpr Smr recA, thi, pro, hsdR-M+ RP4: 2-Tc::Mu:Kmr Tn7 λpir | Laboratory stock |
| P. aeruginosa | ||
| PAO1 | Non-mucoid wild type strain | Laboratory stock |
| ΔfliA | fliA deletion mutant of PAO1 | This study |
| PAO1[pMMB66EH] | PAO1, carrying pMMB66EH, Cbr | This study |
| ΔfliA [pMMB66EH] | ΔfliA, carrying pMMB66EH, Cbr | This study |
| ΔfliA[pMMB2133] | ΔfliA, carrying pMMB2133, Cbr | This study |
| ΔfliA[pMMB4367] | ΔfliA, carrying pMMB4367, Cbr | This study |
| Plasmids | ||
| pEX18Tc | Suicide vector; oriT+, sacB+, Tcr | Laboratory stock |
| pMMB66EH | Broad-host-range expression vector, Apr | Laboratory stock |
| pMMBfliA | pMMB66EH containing fliA, Apr | This study |
| pMMB2133 | pMMB66EH containing PA2133, Apr | Laboratory stock |
| pMMB4367 | pMMB66EH containing PA4367, Apr | Laboratory stock |
| pCHUB237 | GUS reporter vector, Gmr | Wen-Ling Deng a |
| pCHUB-PPA4367 | pCHUB237 containing PPA4367, Gmr | This study |
| pCHUB-PPA5017 | pCHUB237 containing PPA5017, Gmr | This study |
| pCdrA-gfpC | c-di-GMP reporter plasmid | Tim Tolker-Nielsen b |
a National Chung Hsing University, Taiwan
b University of Copenhagen, Denmark
Recombinant DNA techniques
Genomic DNA of P. aeruginosa was isolated using the Wizard® Genomic DNA Purification kit (Promega). Plasmid DNA was isolated using the Presto™ Mini Plasmid kit (Geneaid). The DNA quality was determined using agarose gel electrophoresis, and the DNA concentration was identified using a NanoDrop ND-1000 spectrophotometer. Moreover, DNA was amplified using dNTPs (GeneDirex), pfu DNA polymerase (Thermo Scientific), DreamTaq DNA polymerase (Thermo Scientific), an appropriate primer set, and a template DNA. The amplified DNA fragments were then purified using a Gel/PCR DNA fragment extraction kit (Geneaid). Restriction endonucleases were purchased from New England Biolab Inc. and were used according to the manufacturer’s instructions.
Construction of an isogenic fliA mutant and a complementary strain
A P. aeruginosa fliA deletion mutant was constructed using homologous recombination. The 5ʹ and 3ʹ regions (approximately 1 kb in length) flanking fliA were cloned into the suicide plasmid pEX18Tc. The resulting plasmid was transformed into E. coli S17-1 λ pir and mobilized into P. aeruginosa PAO1 through conjugation. The first crossover recombinants were selected on LB agar supplemented with 100 μg/ml tetracycline (Sigma), and the second crossover strains were selected on LB agar supplemented with 10% sucrose (J. T. Baker). These strains were further analyzed using PCR for determining fliA deletion. The fliA complementation strains were constructed using pMMB66EH-based plasmids.
Quantification of the phenazine pigment production
P. aeruginosa strains were propagated in a 5-ml Pseudomonas broth (PB) containing 2% (w/v) Bacto Peptone (Difco), 0.14% (w/v) MgCl2 (J. T. Baker), 1% (w/v) K2SO4 (Sigma), and 500 μg/ml carbenicillin (Sigma) and were incubated at 37°C with shaking at 150 rpm for 16 h. The pigments in the bacterial culture were extracted with chloroform (Sigma). After centrifugation, the chloroform layer was mixed with 1 ml of 0.2 N HCl (Sigma), which yielded a pink solution, and the absorbance was measured at OD520. The bacterial cell number was normalized using the cell density (OD595) of the culture to reflect pyocyanin production per cell. Similarly, the OD520 of the aqueous phase was also measured and normalized to indicate pyorubin production per cell.
Transcriptome analysis
The bacterial cells on the periphery (5 mm) of the swarming bacterial mass were collected, and RNA of that was extracted using the Trizol reagent (Invitrogen) and purified using the PureLink™ RNA Mini kit (Ambion). Furthermore, the RNA quality was examined using agarose gel electrophoresis and was quantified using a NanoDrop ND-1000 spectrophotometer. For RNA sequencing, the RNA was reverse transcribed into complementary DNA and then analyzed using HiSeq 2000 (Illumina, Inc.), producing approximately 40.7 million and 44.6 million 100-bp pair-end reads from PAO1 [pMMB66EH] and ΔfliA [pMMB66EH] corresponding to coverage rates of 650.35- and 712.20- fold, respectively.
Bioinformatics analysis
The genomic sequence of P. aeruginosa PAO1 was obtained from the Pseudomonas genome database (www.pseudomonas.com) and Kyoto Encyclopedia of Genes and Genomes (www.genome.jp/kegg/). Nucleotide and protein sequences were compared using BLASTn and BLASTp, respectively; both programs were provided by the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov). Restriction sites and oligonucleotide primer sequences were identified using Vector NTI Advance® software (Invitrogen). Promoter regions were predicted using BPROM software (Softberry, Inc).
Flagellar observation through transmission electron microscopy
Overnight cultures of P. aeruginosa PAO1 [pMMB66EH], ΔfliA [pMMB66EH], and PAO1 [pMMBfliA] were diluted 100 folds with fresh LB broth and were negatively stained with 0.1% (w/v) uranyl acetate (Electron Microscopy Sciences, Hatfield, PA, USA). Flagellar morphologies were observed using a transmission electron microscope (Hitachi HT7700).
Swarming and swimming motility assay
The swarming assay plates contained 0.5% (w/v) glucose (Amersco), 0.5% (w/v) agar (Amersco), and 0.8% (w/v) nutrient broth containing 3.0 g/L beef extract (BD) and 5.0 g/L peptone (BD), supplemented with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG; Amersco) and 500 μg/ml carbenicillin (Sigma). Moreover, 3 μl of overnight liquid cultures of each test strain were inoculated on a swarming plate. After the plate was incubated at 37°C for 24 h, the longest swarming distance of the cells was measured. The swimming assay plates contained 1.0% (w/v) tryptone (Scharlab), 0.3% (w/v) agar (Amersco), and 1.0% (w/v) NaCl (J.T. Baker), supplemented with 500 μg/ml carbenicillin and 0.5 mM IPTG. Moreover, 3 μl of overnight liquid culture of each strain was inoculated on the plate and incubated at 37°C for 24 h; the diameters of the swimming zone were then measured.
Cyclic-di-GMP measurement
The c-di-GMP reporter assay was performed using pCdrA–gfpC, a plasmid carrying the c-di-GMP responsive promoter of the cdrA gene fused with a green fluorescence-encoding gene [25]. The plasmid was introduced into P. aeruginosa wild-type PAO1 and its ΔfliA mutant, followed by growing the transformants under three conditions: in LB broth and on swarming and swimming plates. These pCdrA–gfpC-carrying strains were grown in LB broth supplemented with 60 μg/ml gentamycin (Amersco) at 37°C with overnight shaking at 150 rpm. The cells from 1 ml of overnight culture of each strain were collected by centrifugation at 5000 g for 3 min and were resuspended in sterile phosphate-buffered saline (PBS). The cell density at OD595 was adjusted to 0.3, and 100 μl of the cell suspension was added to each well in a black 96-well microtiter plate. The fluorescence intensity was then measured using excitation and emission wavelengths of 515 nm and 490 nm in a luminometer (Wallac 1420 Victor2). Furthermore, the c-di-GMP reporter assay was performed on plates, and the green fluorescence of the cells was detected using an upright epifluorescence microscope (BX-51; Olympus) equipped with a digital camera. Bright-field and fluorescence images obtained through microscopy were merged and analyzed using SPOT Advanced plus Imaging Software 4.6 (Spot Imaging, Inc.). Similarly, cells at the periphery (approximately 5 mm) of the swarming area were collected and resuspended in PBS, the green fluorescence intensity was measured, and the values were normalized with the cell density.
β-glucuronidase reporter gene assay
The promoter activity assay was performed using pCHUB238, a plasmid carrying a β-glucuronidase (GUS) reporter gene. The GUS reporter enzyme converts the colorless and nonfluorescent substrate 4-methylumbelliferyl-β-D-glucuronide (MUG) to methylumbelliferone, which absorbs light at 365 nm and emits fluorescence at 455 nm [26]. The promoter region of the target gene was cloned at the 5ʹ end of the GUS reporter gene, and the resulting plasmid was transformed into E. coli S17-1 λ pir and mobilized into P. aeruginosa through conjugation. Moreover, 20 μl of overnight liquid culture carrying a reporter vector were inoculated into 2 ml of the LB broth supplemented with 10 μg/ml gentamycin and were allowed to grow for 8 h. Subsequently, 1 ml of the culture was transferred into a clear tube and centrifuged at 12,000 g for 2 min. The supernatant was discarded, and the pellet was mixed with 1 ml GUS buffer (50 mM sodium phosphate, 10 mM β-mercaptoethanol, 10 mM EDTA, 0.1% sodium lauryl sarcosine, and 0.1% Triton X-100, pH 7.0) by vortexing. This solution was maintained on ice for 20 min. The sample was centrifuged at 12,000 g and maintained at 4°C for 3 min, and 25 μl of the supernatant was transferred into each well of a black 96-well microtiter plate containing 50 μl of 3 mM 4-MUG. After incubation at 37°C for 10 min, the reaction was terminated by adding 25 μl of 0.72 M Na2CO3, and the emission intensity at 455 nm under 365 nm excitation was measured using a Wallac 1420 Victor2 luminometer. The protein concentration was determined using the Bradford method for normalizing the GUS results.
Results
FliA affects bacterial motility and phenazine pigment production
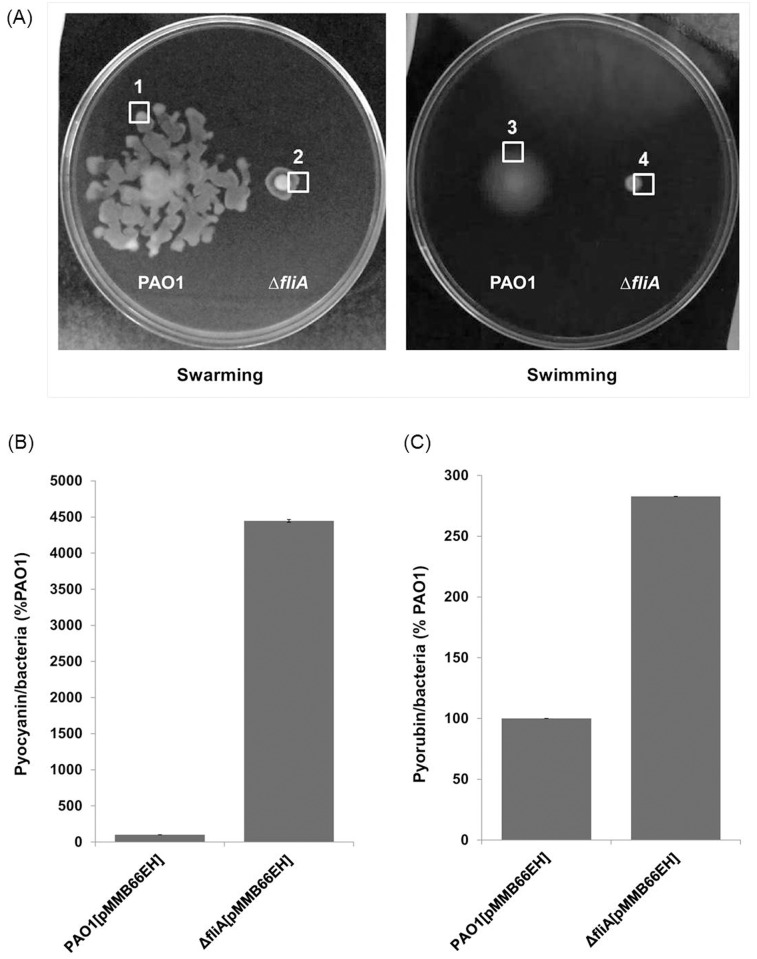
To investigate the physiological role of FliA in P. aeruginosa, we first generated an isogenic fliA deletion mutant in P. aeruginosa PAO1. The motility of the fliA mutant was examined on a swarming agar and compared with that of P. aeruginosa PAO1. The fliA deletion mutant exhibited a significant reduction in swarming motility (Fig 1A), which was consistent with the motility of the expected phenotypes [11]. Among the several other biological properties examined, the production of pyocyanin and pyorubin, which are phenazine pigments associated with oxidative stress-mediated cytotoxicity of mammalian cells [27,28], was altered in the fliA mutant, demonstrating a 40- and 3-fold enhancement, respectively (Fig 1B and 1C) after 36-h growth in the PB broth.
Fig 1. Motility and phenazine pigment production in P. aeruginosa wild-type PAO1 and fliA mutant.
(A) Swarming and swimming motility, (B) Pyocyanin production, and (C) Pyorubin production.
FliA affects intracellular c-di-GMP concentrations
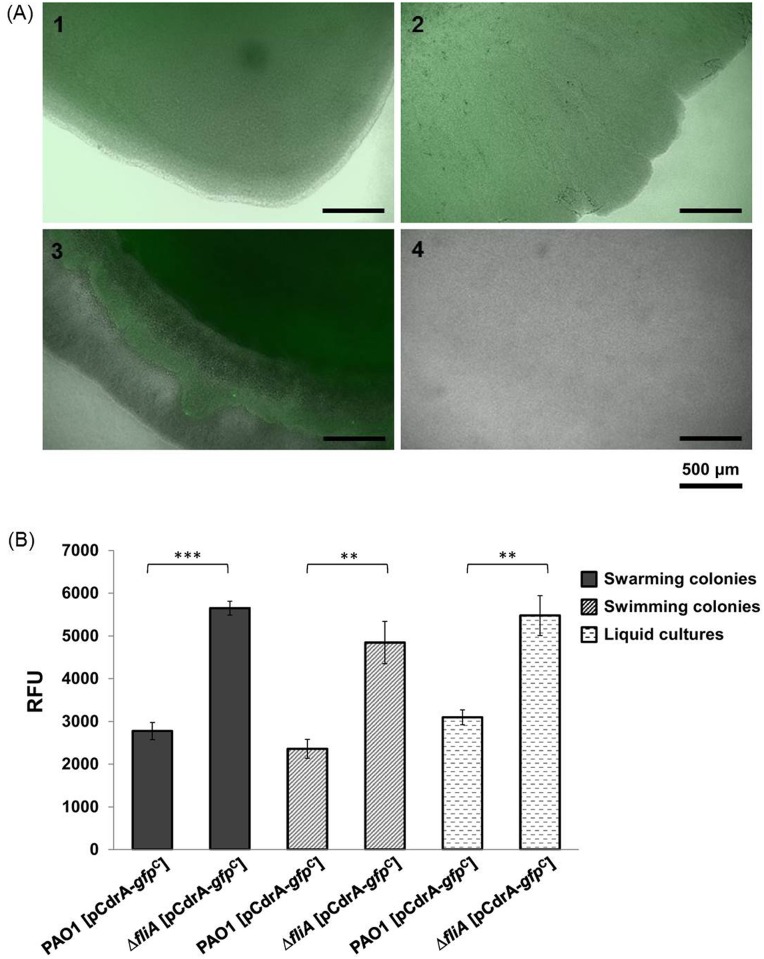
It has been shown previously that c-di-GMP is a regulator of pyocyanin production [29]. The increase in pyocyanin production in the fliA deletion mutant suggested that FliA could affect intracellular c-di-GMP concentrations in P. aeruginosa. This finding, together with a previous report that FliA can modulate expression of a c-di-GMP metabolizing gene yhjH in E. coli [11], prompted us to investigate whether c-di-GMP participates in the FliA regulon in P. aeruginosa. This study first measured the effects of fliA deletion on intracellular c-di-GMP concentrations by using a c-di-GMP reporter system pCdrA–gfpC. The fluorescence intensity of both wild-type and fliA deletion mutant grown overnight under different conditions, namely in LB broth and on swarming and swimming assay plates, were examined through fluorescence microscopy. Fig 2 shows that the fluorescence intensity of the ΔfliA mutant was significantly higher than that of PAO1 particularly in swimming condition, indicating a higher intracellular c-di-GMP concentration in the strain. Further quantitation of the fluorescence intensity using a fluorometer revealed that the fluorescence intensity of PAO1 [pCdrA–gfpC] was significantly lower than that of ΔfliA [pCdrA–gfpC] under the three examined growth conditions, suggesting that FliA modulates intracellular c-di-GMP concentrations.
Fig 2. Intracellular concentration of c-di-GMP in P. aeruginosa wild-type PAO1 and fliA mutant.
(A) Fluorescence intensity of cells on the periphery of swarming (Panels 1 and 2) and swimming (Panels 3 and 4) areas are presented. Panels 1 and 3, PAO1 [pCdrA–gfpC]; 2 and 4, ΔfliA [pCdrA–gfpC]. (B) Relative fluorescence units of PAO1 [pCdrA–gfpC] and ΔfliA [pCdrA–gfpC] under different growth conditions.
Effects of fliA deletion on the expression of c-di-GMP-metabolizing enzyme-encoding genes
This study used RNAseq technology for investigating whether fliA deletion alters the expression of genes that encode c-di-GMP-metabolizing enzymes. Total RNAs isolated from P. aeruginosa strains grown under swarming conditions were reverse transcribed into cDNAs and the transcription profiles of PAO1 and ΔfliA strains were compared (NCBI Accession SRP071734), and several EAL domain-containing PDE-coding genes exhibiting moderate differences in expression levels were identified (Table 2). The PDE genes PA2133, PA3311, PA5017, PA4108, and PA4367 demonstrated lower expression levels in the ΔfliA strain.
Table 2. Expression levels of PDE/DGC-related genes based on the transcriptome analysis of ΔfliA compared with that of PAO1.
| Gene locus | Gene name | Gene product or function | Fold change |
|---|---|---|---|
| PA2133 | - | PDE | -2.21 |
| PA3311 | nbdA | NbdA, PDE | -1.61 |
| PA5017 | dipA | DipA, PDE | -1.56 |
| PA4108 | - | PDE | -1.40 |
| PA4367 | bifA | BifA, PDE | -1.39 |
| PA3825 | - | Catalytically active PDE | -1.23 |
| PA2567 | - | Catalytically active PDE | -1.16 |
| PA4781 | - | PDE | 1.85 |
| PA0861 | rbdA | RbdA, DGC | -1.05 |
| PA1107 | roeA | RoeA, DGC | -1.61 |
| PA0285 | - | Predicted DGC | -2.19 |
| PA5487 | - | DGC | 1.33 |
FliA regulates the PDE-encoding gene PA4367
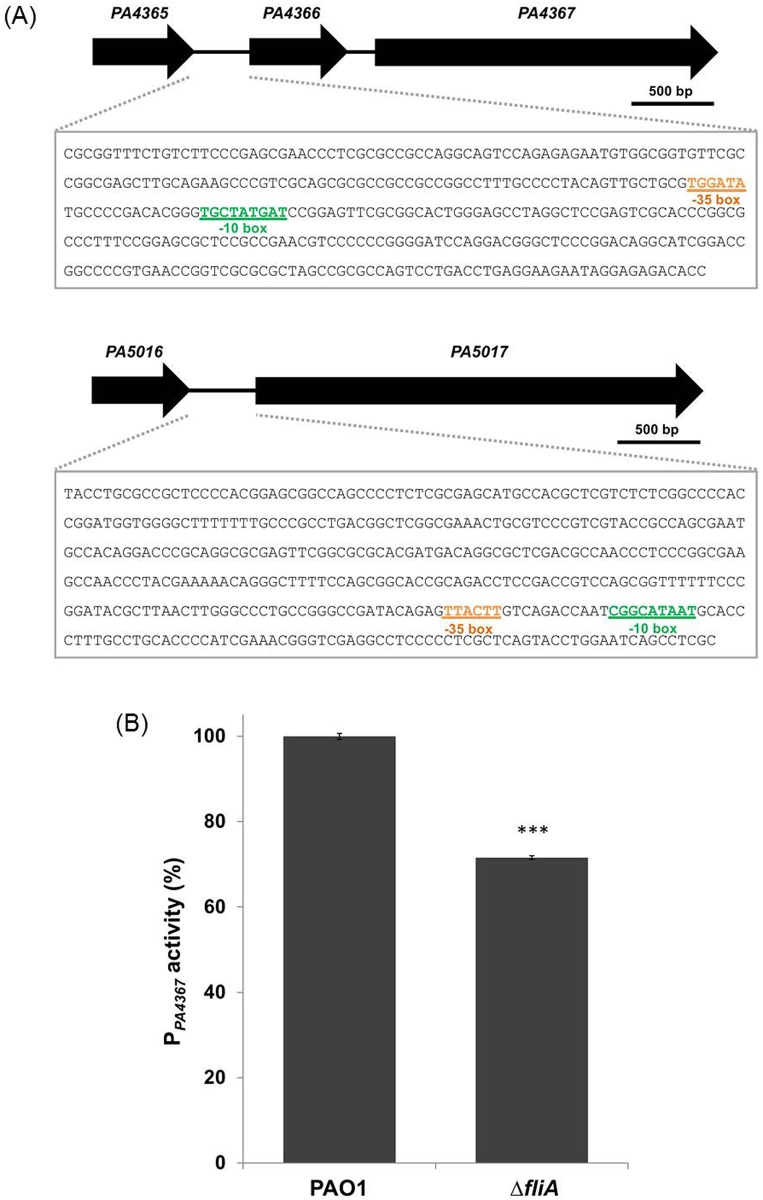
Among the PDE genes with a lower expression level in the ΔfliA, we further analyzed PA4367 [30] and PA5017 [31], the two most effectively characterized genes of the group. To investigate whether these two genes were directly regulated by FliA, we analyzed the promoter activities of both genes in the ΔfliA and wild-type PAO1 by using the GUS gene reporter system. We employed the bioinformatics tool BPROM to identify the possible promoter regions of PA4367 and PA5017 (Fig 3A). We then cloned the DNA fragment comprising the predicted promoters of PA4367 and PA5017 into pCHUB238, a plasmid carrying a GUS reporter gene. The resulting plasmids were individually introduced into wild-type PAO1 and the ΔfliA mutant, and the GUS activity of the transconjugants was determined. The results revealed that the GUS activity of PPA4367, but not that of PPA5017 (data not shown), significantly decreased in the ΔfliA mutant (Fig 3B). These findings strongly suggest that PA4367 is positively regulated by FliA. Nevertheless, the putative FliA binding sequence (TAAAGTTT-N11-GCCGATAA) in Salmonella typhimurium [32] could not be found in the 500-bp upstream region of PA4367. The lack of this canonical FliA promoter sequence suggests that this regulation may be indirect.
Fig 3. Promoter activity of PDE-coding genes in P. aeruginosa wild-type PAO1 and fliA mutant strains.
(A) Promoters of PA4367 and PA5017 were predicted using the BPROM program. The DNA sequence of the predicted promoter regions, which were used in the GUS reporter assay, are shown in the block. The possible sigma factor binding sites are also indicated. (B) The promoter activity of PA4367 in wild-type PAO1 and ΔfliA is presented. All experiments were conducted in triplicate, and data are represented as mean ± standard error (n = 8; ***, p < 0.001).
PA4367 overexpression in the fliA mutant rescues the defect in swarming motility and restores phenazine pigment production to normal levels
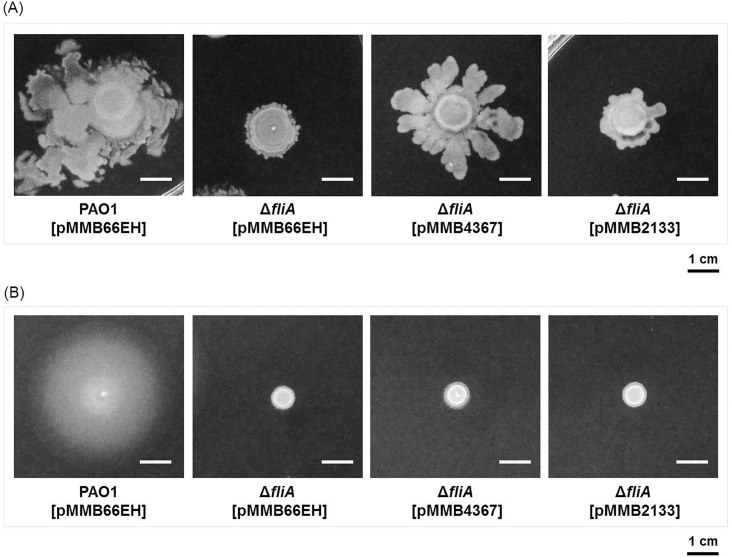
To investigate whether the differences in the swarming motility and phenazine pigment production in the ΔfliA mutant were mediated by increases in intracellular c-di-GMP concentrations, we complemented the mutant with a PDE-encoding gene and examined the phenotypes of the complement strain. The deletion of the BifA-coding gene PA4367 in P. aeruginosa resulted in a phenotype defective in swarming motility [30], whereas PA2133 overexpression in P. aeruginosa reduced the c-di-GMP concentration and impaired biofilm formation [33]. On the basis of these findings, we complemented PA4367 and PA2133 in the ΔfliA strain and examined their motility. The results revealed that PA2133 overexpression partly rescued the swarming motility of the ΔfliA mutant, whereas PA4367 complementation restored the swarming motility nearly to the degree of the wild-type strain (Fig 4A). The swimming motility of the ΔfliA mutant was not rescued by PA2133 or PA4367 overexpression (Fig 4B). The finding further suggests that fliA influences the swarming motility but not the swimming motility by modulating the c-di-GMP concentration.
Fig 4. Effects of the c-di-GMP PDE-coding genes PA4367 and PA2133 on the swarming motility of the fliA mutant.
Bacterial culture of each strain were inoculated on the swarming and swimming plates and incubated at 37°C for 24 h. Images of the swarming (A) and swimming (B) pattern of the bacterial strains are presented.
FliA affects the production of phenazine pigments by modulating the intracellular concentration of c-di-GMP
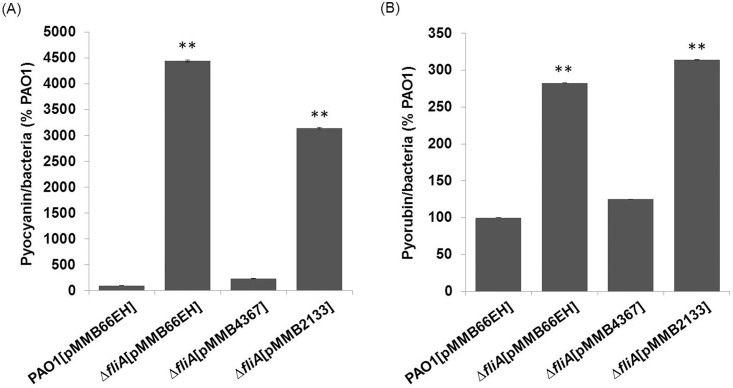
We further investigated whether c-di-GMP is involved in the production of phenazine pigments in the ΔfliA mutant. In this study, both PA4367- and PA2133-overexpressing strains were examined for their production of phenazine pigments of P. aeruginosa. Introducing functional PA4367, but not PA2133, into the ΔfliA mutant restored the overproduction of pyocyanin and pyorubin phenotype to a concentration comparable with that of the wild-type strain (Fig 5). This finding suggests that fliA regulates pyocyanin and pyorubin production through a c-di-GMP-dependent pathway.
Fig 5. Effects of fliA and the c-di-GMP PDE-coding genes PA4367 and PA2133 on phenazine pigment production.
Pyocyanin (A) and pyorubin (B) produced by different P. aeruginosa strains were quantified after the cells were grown in a PB medium for 36 h. All experiments were conducted in triplicate, and data are represented as mean ± standard error (n = 3; **, p < 0.01).
FliA affects swarming motility not only by controlling flagellar biosynthesis but also by modulating the c-di-GMP concentration
Similar to the findings of the swarming assay, the ΔfliA mutant exhibited a defective phenotype in the swimming assay. However, in contrast to the results of the swarming assay, complementing the ΔfliA mutant with PA4367 or PA2133 failed to restore the defective swimming motility to normal (Fig 4). To elucidate the possible mechanisms underlying the difference in complementation, we examined the flagellar morphology of ΔfliA [pMMB66EH], ΔfliA [pMMB4367], and ΔfliA [pMMB2133] through transmission electron microscopy (TEM). The results revealed that the flagella were not only absent in ΔfliA [pMMB66EH] but also in ΔfliA [pMMB4367] and ΔfliA [pMMB2133] (Fig 6), suggesting that fliA regulates swarming motility through PDE-coding genes and is independent of flagellar biosynthesis.
Fig 6. Effects of the c-di-GMP PDE coding genes PA4367 and PA2133 on flagellar synthesis in the P. aeruginosa fliA mutant.
Cells from overnight cultures grown in LB broth supplemented with 500 μg/ml carbenicillin and 0.5 mM IPTG were examined through TEM.
Discussion
C-di-GMP has been reported to be a regulator of bacterial motility [34–36]. Whether the secondary messenger crosstalks to the major motility transcription factor FliA is not clear. It has been shown that cellular c-di-GMP levels can affect expression of Clostridium difficile sigD, a fliA ortholog, and hence regulate toxin production in the bacterium [37]. On the other hand, FliA may regulate the expression of c-di-GMP metabolizing genes, as in the case of E. coli yhjH [11]. However, because of the presence of numerous c-di-GMP-metabolizing genes in a bacterium, less information exists on genes responsible for regulating the intracellular c-di-GMP concentration and hence the motility. This study demonstrated that FliA, in addition to transcriptionally controlling flagellar biosynthesis, regulated P. aeruginosa motility by modulating the c-di-GMP concentration. We identified the c-di-GMP-metabolizing genes responsible for regulating the c-di-GMP concentration in bacterial motility, and the result suggests that the PA4367 gene is a target of FliA. Overall, these findings establish a basis for elucidating the mechanism underlying the FliA-mediated regulation of bacterial motility.
By using the RNAseq technology, we demonstrated that FliA regulates the expression of several PDE-encoding genes, including PA4367. However, moderate fold changes were observed in expression levels of the PDE-encoding genes between the wild-type and mutant ΔfliA. The GUS reporter gene assay revealed that PA4367 is regulated by FliA. However, the involvement of additional PDEs in the FliA-mediated regulation of motility could not be eliminated. The presence of more than one PDE gene, with each contributing to a part of the c-di-GMP metabolism required in the bacterial motility regulation, may explain the slight fold changes in the expression of most PDE genes in the ΔfliA mutant.
TEM revealed that PA4367 overexpression in the ΔfliA mutant did not restore the flagella (Fig 6); however, the defective swarming motility, and not the swimming motility, of the mutant could be rescued (Fig 4). The swimming motility of cells primarily depends on the flagella, whereas additional factors are involved in swarming motility [38,39]. Thus, under environmental conditions suitable for swimming, FliA may regulate bacterial motility through flagellar biosynthesis. When P. aeruginosa is cultivated on a solid surface, FliA may affect PA4367 expression, which subsequently affects the c-di-GMP concentration and thus regulates swarming-associated factors, such as biosurfactants and type IV pili. Biosurfactants as wetting agents can reduce the surface tension required for swarming [39,40]. The hypothesis that PA4367 overexpression results in increased pili production, which is associated with swarming [41], could not be confirmed through TEM, possibly because type IV pili are commonly present in a dynamic state and may not always be detected through TEM. Future studies examining the presence of type IV pili through Western blotting or fluorescence microscopy [42] may be required for verifying this hypothesis.
Complementation of the fliA mutant with pMMBfliA, a low copy number plasmid carrying a functional fliA gene, was conducted. The production of pyorubin and flagella biogenesis (examined under electron microscopy) could be restored partially in the complementation strain, indicating that fliA is responsible for these two phenotypes. However, the swimming and swarming phenotype of the fliA mutant could not be restored by complementation. The finding is not surprising because that FliA is known to involve in complex regulation of flagellar biogenesis in P. aeruginosa [43,22,44]. One possible explanation is that excessive FliA leads to feedback inhibition through activating expression of flgM which encodes an anti-FliA activity. Therefore an optimal quantity of FliA is critical to the bacterial motility.
In E. coli, bacterial motility could be regulated by PDE YhjH and YcgR. The mechanism underlying the regulated expression of yhjH in E.coli remains unclear. The YcgR ortholog could be unambiguously identified in P. aeruginosa PAO1. We previously constructed an isogenic mutant of YcgR in P. aeruginosa PAO1, and its phenotype was consistent with that reported for E. coli YcgR.[9] By contrast, the corresponding gene of E. coli yhjH in P. aeruginosa PAO1 remains unclear. Restoring the defective swarming motility of P. aeruginosa ΔfliA by complementing it with PA4367 (Fig 4) suggests that this gene plays a functional role equivalent to that of yhjH in E. coli [9,10]. Finally, both RNAseq and GUS reporter assay confirmed that PA4367 is a downstream target of FliA in P. aeruginosa PAO1 (Table 2). This finding is also consistent with that of a previous study stating that FliA regulates yhjH in E. coli [26].
Acknowledgments
We thank Wen-Ling Deng (National Chung Hsing University, Taiwan) for plasmid pCHUB237 which is used for promoter activity assay; Tim Tolker-Nielsen (University of Copenhagen, Denmark) for plasmid pCdrA-gfpC which is used for intracellular c-di-GMP measurement; Ming-Der Peng (National Tsing Hua University, Taiwan) for assistance in technical support of TEM.
Data Availability
All relevant data within the manuscript expect trasncriptome data; all transcriptome raw data files are available from the NCBI database (accession number SRP071734).
Funding Statement
The study was supported by the Ministry of Science and Technology, Taiwan ROC (www.most.gov.tw), the grant number of that is 103-2314-B-182A-089-MY3, received by H.Y. Chang. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Jagannathan A, Constantinidou C, Penn CW. Roles of rpoN, fliA, and flgR in expression of flagella in Campylobacter jejuni. J Bacteriol. 2001;183(9):2937–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Starnbach MN, Lory S. The fliA (rpoF) gene of Pseudomonas aeruginosa encodes an alternative sigma factor required for flagellin synthesis. Mol Microbiol. 1992;6(4):459–69. [DOI] [PubMed] [Google Scholar]
- 3.Chen YF, Helmann JD. Restoration of motility to an Escherichia coli fliA flagellar mutant by a Bacillus subtilis sigma factor. Proc Natl Acad Sci U S A. 1992;89(11):5123–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Syed KA, Beyhan S, Correa N, Queen J, Liu J, Peng F, et al. The Vibrio cholerae flagellar regulatory hierarchy controls expression of virulence factors. J Bacteriol. 2009;191(21):6555–70. 10.1128/JB.00949-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rodriguez-Herva JJ, Duque E, Molina-Henares MA, Navarro-Aviles G, Van Dillewijn P, De La Torre J, et al. Physiological and transcriptomic characterization of a fliA mutant of Pseudomonas putida KT2440. Environ Microbiol Rep. 2010;2(3):373–80. 10.1111/j.1758-2229.2009.00084.x [DOI] [PubMed] [Google Scholar]
- 6.Boll JM, Hendrixson DR. A regulatory checkpoint during flagellar biogenesis in Campylobacter jejuni initiates signal transduction to activate transcription of flagellar genes. mBio. 2013;4(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kapatral V, Olson JW, Pepe JC, Miller VL, Minnich SA. Temperature-dependent regulation of Yersinia enterocolitica Class III flagellar genes. Mol Microbiol. 1996;19(5):1061–71. [DOI] [PubMed] [Google Scholar]
- 8.Xu T, Su Y, Xu Y, He Y, Wang B, Dong X, et al. Mutations of flagellar genes fliC12, fliA and flhDC of Edwardsiella tarda attenuated bacterial motility, biofilm formation and virulence to fish. J Appl Microbiol. 2014;116(2):236–44. 10.1111/jam.12357 [DOI] [PubMed] [Google Scholar]
- 9.Paul K, Nieto V, Carlquist WC, Blair DF, Harshey RM. The c-di-GMP binding protein YcgR controls flagellar motor direction and speed to affect chemotaxis by a “Backstop Brake” mechanism. Molecular Cell. 2010;38(1):128–39. 10.1016/j.molcel.2010.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fang X, Gomelsky M. A post-translational, c-di-GMP-dependent mechanism regulating flagellar motility. Mol Microbiol. 2010;76(5):1295–305. 10.1111/j.1365-2958.2010.07179.x [DOI] [PubMed] [Google Scholar]
- 11.Claret L, Miquel S, Vieille N, Ryjenkov DA, Gomelsky M, Darfeuille-Michaud A. The flagellar sigma factor FliA regulates adhesion and invasion of Crohn disease-associated Escherichia coli via a cyclic dimeric GMP-dependent pathway. J Biol Chem. 2007;282(46):33275–83. [DOI] [PubMed] [Google Scholar]
- 12.Römling U, Gomelsky M, Galperin MY. C-di-GMP: the dawning of a novel bacterial signalling system. Mol Microbiol. 2005;57(3):629–39. [DOI] [PubMed] [Google Scholar]
- 13.Mills E, Pultz IS, Kulasekara HD, Miller SI. The bacterial second messenger c-di-GMP: mechanisms of signalling. Cell Microbiol. 2011;13(8):1122–9. 10.1111/j.1462-5822.2011.01619.x [DOI] [PubMed] [Google Scholar]
- 14.Tamayo R, Pratt JT, Camilli A. Roles of cyclic diguanylate in the regulation of bacterial pathogenesis. Annu Rev Microbiol. 2007;61(1):131–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jenal U. Cyclic di-guanosine-monophosphate comes of age: a novel secondary messenger involved in modulating cell surface structures in bacteria? Curr Opin Microbiol. 2004;7(2):185–91. [DOI] [PubMed] [Google Scholar]
- 16.Jenal U, Malone J. Mechanisms of cyclic-di-GMP signaling in bacteria. Annu Rev Genet. 2006;40(1):385–407. [DOI] [PubMed] [Google Scholar]
- 17.Römling U, Amikam D. Cyclic di-GMP as a second messenger. Curr Opin Microbiol. 2006;9(2):218–28. [DOI] [PubMed] [Google Scholar]
- 18.Tal R, Wong HC, Calhoon R, Gelfand D, Fear AL, Volman G, et al. Three cdg operons control cellular turnover of cyclic di-GMP in Acetobacter xylinum: Genetic organization and occurrence of conserved domains in isoenzymes. J Bacteriol. 1998;180(17):4416–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stock AM. Diguanylate cyclase activation: It takes two. Structure. 2007;15(8):887–8. [DOI] [PubMed] [Google Scholar]
- 20.Ryjenkov DA, Tarutina M, Moskvin OV, Gomelsky M. Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: Insights into biochemistry of the GGDEF protein domain. J Bacteriol. 2005;187(5):1792–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murray TS, Ledizet M, Kazmierczak BI. Swarming motility, secretion of type 3 effectors and biofilm formation phenotypes exhibited within a large cohort of Pseudomonas aeruginosa clinical isolates. J Med Microbiol. 2010;59(Pt 5):511–20. 10.1099/jmm.0.017715-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dasgupta N, Wolfgang MC, Goodman AL, Arora SK, Jyot J, Lory S, et al. A four-tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa. Mol Microbiol. 2003;50(3):809–24. [DOI] [PubMed] [Google Scholar]
- 23.Haiko J, Westerlund-Wikstrom B. The role of the bacterial flagellum in adhesion and virulence. Biology. 2013;2(4):1242–67. 10.3390/biology2041242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feldman M, Bryan R, Rajan S, Scheffler L, Brunnert S, Tang H, et al. Role of flagella in pathogenesis of Pseudomonas aeruginosa pulmonary infection. Infect Immun. 1998;66(1):43–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rybtke MT, Borlee BR, Murakami K, Irie Y, Hentzer M, Nielsen TE, et al. Fluorescence-based reporter for gauging cyclic di-GMP levels in Pseudomonas aeruginosa. Appl Environ Microbiol. 2012;78(15):5060–9. 10.1128/AEM.00414-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jefferson RA, Burgess SM, Hirsh D. beta-Glucuronidase from Escherichia coli as a gene-fusion marker. Proc Natl Acad Sci U S A. 1986;83(22):8447–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Britigan BE, Roeder TL, Rasmussen GT, Shasby DM, McCormick ML, Cox CD. Interaction of the Pseudomonas aeruginosa secretory products pyocyanin and pyochelin generates hydroxyl radical and causes synergistic damage to endothelial cells. Implications for Pseudomonas-associated tissue injury. J Clin Invest. 1992;90(6):2187–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilson R, Pitt T, Taylor G, Watson D, MacDermot J, Sykes D, et al. Pyocyanin and 1-hydroxyphenazine produced by Pseudomonas aeruginosa inhibit the beating of human respiratory cilia in vitro. J Clin Invest. 1987;79(1):221–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ryan RP, Lucey J, O'Donovan K, McCarthy Y, Yang L, Tolker-Nielsen T, Dow JM. HD-GYP domain proteins regulate biofilm formation and virulence in Pseudomonas aeruginosa. Environ Microbiol. 2009; 11(5): 1126–36. 10.1111/j.1462-2920.2008.01842.x [DOI] [PubMed] [Google Scholar]
- 30.Kuchma SL, Brothers KM, Merritt JH, Liberati NT, Ausubel FM, O'Toole GA. BifA, a cyclic-di-GMP phosphodiesterase, inversely regulates biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J Bacteriol. 2007;189(22):8165–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roy AB, Petrova OE, Sauer K. The phosphodiesterase DipA (PA5017) is essential for Pseudomonas aeruginosa biofilm dispersion. J Bacteriol. 2012;194(11):2904–15. 10.1128/JB.05346-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kutsukake K, Ohya Y, Iino T. Transcriptional analysis of the flagellar regulon of Salmonella typhimurium. J. Bacteriol. 1990; 172(2):741–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cole SJ, Lee VT. Cyclic-di-GMP signaling contributes to Pseudomonas aeruginosa mediated catheter-associated urinary tract infection. J Bacteriol. 2015; 10.1128/JB.00410-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hengge R. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol. 2009;7(4):263–73. 10.1038/nrmicro2109 [DOI] [PubMed] [Google Scholar]
- 35.Zorraquino V, García B, Latasa C, Echeverz M, Toledo-Arana A, Valle J, et al. Coordinated cyclic-di-GMP repression of Salmonella motility through YcgR and cellulose. J Bacteriol. 2013;195(3):417–28. 10.1128/JB.01789-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Römling U, Galperin MY, Gomelsky M. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev. 2013;77(1):1–52. 10.1128/MMBR.00043-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McKee RW, Mangalea MR, Purcell EB, Borchardt EK, Tamayo R. The second messenger cyclic di-GMP regulates Clostridium difficile toxin production by controlling expression of sigD. J Bacteriol. 2013;195(22):5174–85. 10.1128/JB.00501-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O'May C, Tufenkji N. The swarming motility of Pseudomonas aeruginosa is blocked by cranberry proanthocyanidins and other tannin-containing materials. Appl Environ Microbiol. 2011;77(9):3061–7. 10.1128/AEM.02677-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kearns DB. A field guide to bacterial swarming motility. Nat Rev Microbiol. 2010;8(9):634–44. 10.1038/nrmicro2405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Déziel E, Lépine F, Milot S, Villemur R. rhlA is required for the production of a novel biosurfactant promoting swarming motility in Pseudomonas aeruginosa: 3-(3-hydroxyalkanoyloxy)alkanoic acids (HAAs), the precursors of rhamnolipids. Microbiology. 2003;149(8):2005–13. [DOI] [PubMed] [Google Scholar]
- 41.Köhler T, Curty LK, Barja F, van Delden C, Pechère JC. Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pili. J Bacteriol. 2000;182(21):5990–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barocchi MA, Ries J, Zogaj X, Hemsley C, Albiger B, Kanth A, et al. A pneumococcal pilus influences virulence and host inflammatory responses. Proc Natl Acad Sci U S A. 2006;103(8):2857–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frisk A, Jyot J, Arora SK, Ramphal R. Identification and functional characterization of flgM, a gene encoding the anti-sigma 28 factor in Pseudomonas aeruginosa. J Bacteriol. 2002;184(6):1514–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jyot J, Sonawane A, Wu W, Ramphal R. Genetic mechanisms involved in the repression of flagellar assembly by Pseudomonas aeruginosa in human mucus. Mol Microbiol. 2007;63(4):1026–38. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data within the manuscript expect trasncriptome data; all transcriptome raw data files are available from the NCBI database (accession number SRP071734).