Abstract
In response to environmental changes, Pseudomonas aeruginosa is able to switch from a planktonic (free swimming) to a sessile (biofilm) lifestyle. The two-component system (TCS) GacS/GacA activates the production of two small non-coding RNAs, RsmY and RsmZ, but four histidine kinases (HKs), RetS, GacS, LadS and PA1611, are instrumental in this process. RetS hybrid HK blocks GacS unorthodox HK autophosphorylation through the formation of a heterodimer. PA1611 hybrid HK, which is structurally related to GacS, interacts with RetS in P. aeruginosa in a very similar manner to GacS. LadS hybrid HK phenotypically antagonizes the function of RetS by a mechanism that has never been investigated. The four sensors are found in most Pseudomonas species but their characteristics and mode of signaling may differ from one species to another. Here, we demonstrated in P. aeruginosa that LadS controls both rsmY and rsmZ gene expression and that this regulation occurs through the GacS/GacA TCS. We additionally evidenced that in contrast to RetS, LadS signals through GacS/GacA without forming heterodimers, either with GacS or with RetS. Instead, we demonstrated that LadS is involved in a genuine phosphorelay, which requires both transmitter and receiver LadS domains. LadS signaling ultimately requires the alternative histidine-phosphotransfer domain of GacS, which is here used as an Hpt relay by the hybrid kinase. LadS HK thus forms, with the GacS/GacA TCS, a multicomponent signal transduction system with an original phosphorelay cascade, i.e. H1LadS→D1LadS→H2GacS→D2GacA. This highlights an original strategy in which a unique output, i.e. the modulation of sRNA levels, is controlled by a complex multi-sensing network to fine-tune an adapted biofilm and virulence response.
Author Summary
P. aeruginosa is able to switch from a planktonic to a sessile lifestyle by regulating the two small RNAs RsmY and RsmZ. The GacS/GacA TCS is the main system involved in their regulation; however, three HKs, RetS, LadS and PA1611, modulate this TCS. Here we elucidate the relationship between the LadS signaling pathway and the GacS/GacA TCS. We indeed show that LadS modulates GacS/GacA through an original phosphorelay mechanism where LadS utilizes its H1 and D1 domains and the H2 domain of the unorthodox GacS HK, thereby forming with GacS HK, a multicomponent signal transduction system. This multicomponent signal transduction system, first evidenced in this work, probably functions in a similar manner in some other pseudomonas species like P. fluorescens but not in all pseumonad genera as illustrated by P. syringae. Multicomponent signaling is an original and pertinent strategy for bacteria, in which a complex multi-sensing network controls a similar output (i.e. the modulation of levels of central regulatory RsmY and RsmZ sRNAs). This mechanism provides P. aeruginosa with the possibility of integrating at least two different signals, one from GacS and the other from LadS, to engage P. aeruginosa in a sessile lifestyle.
Introduction
The ability of bacteria to survive in specific habitats requires coordination of appropriate gene expression in response to encountered environmental changes. It is interesting to note that the complexity of bacterial regulatory networks and the number of regulatory genes of bacterial genomes proportionally increase with the diversity of environments a bacterial species is able to survive [1]. In order to cope with the various environments they encounter, bacteria have evolved several sensing systems, including two-component systems (TCS) that monitor external and internal stimuli (nutrients, ions, temperature, redox states …), and translate these signals into adequate adaptive responses (for a review see, [1]).
A TCS comprises a histidine kinase (HK) protein or sensor mostly inserted into the inner membrane and a cognate partner known as the response regulator (RR). These two proteins function in their simplest version in a two-step phosphorelay mechanism, forming a classical TCS as follows: The detection of the stimulus by the periplasmic or cytoplasmic detection domain of the HK protein triggers autophosphorylation on a conserved histidine (H) residue of the transmitter domain H1. The phosphoryl group is then transferred on a conserved aspartate (D) residue present in the receiver or D domain of the cognate RR [2,3]. In some cases, the phosphorelay mechanism between the HK and the RR requires a four-step phosphorelay. In this case, the HK requires additional domains such as a receiver domain (D1). Although this D1 module could be on a separate protein, it is mostly fused to the HK. Then, an alternative histidine-phosphotransfer domain (Hpt or H2) can either be fused to the HK (H2) or form a third independent component in the cytoplasm called Hpt. The HK carrying both additional D1 and H2 domains are called unorthodox sensors while those carrying only the D1 domain are called hybrid sensors. Autophosphorylation of the first H residue of the H1 domain of hybrid or unorthodox HK initiates a phosphorelay such as HH1→DD1→HH2 or Hpt→DRR. In a few cases, the phosphorelay between the HK and the RR can be more complex and involves another TCS to form a multicomponent signal transduction system. The CblSTR signal transduction pathway of Bukholderia cenocepacia, which controls the expression of the cable pili, is such a system [4]. The cblS and cblT genes encode a hybrid and an unorthodox HK respectively, while the cblR gene encodes the cognate RR. While the first two steps of the phosphorelay require the H1 and D1 domains of CblS, the transphosphorylation of the D domain of CblR by CblS requires the H2 domain of CblT, which serves as a bridge component, increasing the complexity of the transduction pathway.
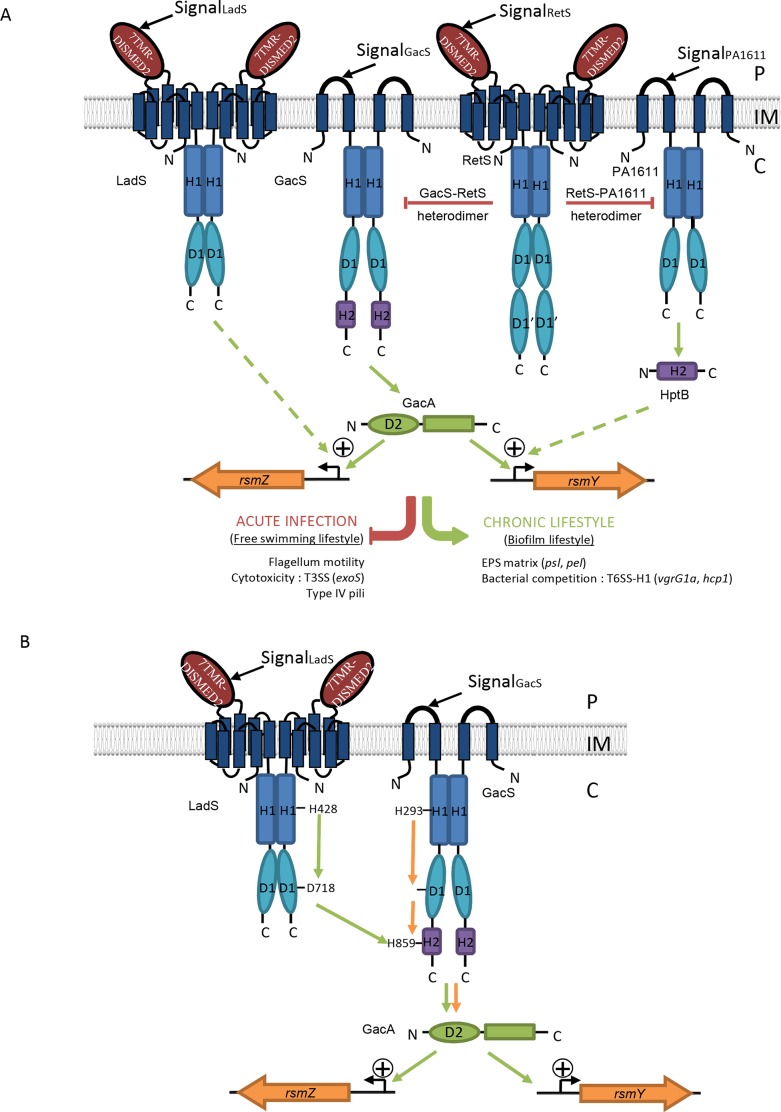
Pseudomonas aeruginosa is a major human pathogen causing severe infections in vulnerable patients such as those with cystic fibrosis or hospitalized with cancer, burns and in intensive care units. It has become a major cause of nosocomial infections. Like other species, P. aeruginosa is able to switch from a planktonic (free swimming) to a sessile (biofilm) lifestyle and several TCSs play a key role in this switch [5–7]. Free swimming cells are characterized by an effective production and injection into host cells of effectors of the Type III secretion system (T3SS). In this free swimming lifestyle, they are thought to represent the vast majority of individuals causing acute infections [8] such as in sepsis, ventilator-associated pneumonia, and infections in postoperative wound and burn patients. In contrast, sessile cells are embedded in a biofilm community sealed by a matrix of exopolysaccharides (EPS) and DNA. In this state, the bacteria concomitantly secrete toxins delivered by the H1-Type VI secretion system (H1-T6SS), which are used for killing and competing with other species in this crowded and enclosed community [8–11]. Cells in biofilms are thought to be in conditions similar to those in chronic infections [12] such as in chronic obstructive pulmonary disease or cystic fibrosis. Several studies have reported an opposing regulation between the expression of molecular determinants involved in acute infection and those involved in chronic infection. Several TCSs have been described as key players controlling this transition, including the central and critical GacS/GacA TCS [13–17] (Fig 1A). GasS is an unorthodox HK with H1/D1/H2 domains. GacA is an RR functioning as a transcriptional regulator, which positively and exclusively controls the expression of two unique target genes encoding two small noncoding RNAs, RsmY and RsmZ [18]. Thus, RsmY and RsmZ have been proposed as key players in controlling the switch between planktonic and biofilm lifestyles [18,19]. These two sRNAs sequester the RNA-binding translational repressor RsmA and thus relieve RsmA binding from its target mRNAs. While bound to target sequences at the site of translational initiation, RsmA exerts a direct translational repression on a limited number of genes grouped in six operons [20], among which are genes encoding the H1-T6SS. Additionally, RsmA has been described as indirectly and positively controlling the expression of a substantial number of genes, including those encoding the T3SS participating in acute infection [20,21]. High expression of rsmY and rsmZ leads to massive biofilm formation due to the production of Pel EPS, and is coupled with H1-T6SS induction and T3SS repression (Fig 1A) [22]. In a gacS mutant, the absence of expression of these sRNAs results in an impaired biofilm formation and induction of T3SS expression [22]. In P. aeruginosa, expression of these two sRNAs is controlled by a complex and sophisticated regulatory network involving the GacS/GacA TCS but also other TCS pathways. The RetS hybrid HK represses expression of both rsmY and rsmZ genes by interfering with the GacS/GacA TCS activity [23]. The PA1611 hybrid HK induces expression of both rsm genes by counteracting the interfering effect of RetS on GacS [24]. The HptB regulatory pathway, which also intersects with the GacS/GacA TCS, only induces rsmY gene expression [22]. The LadS hybrid HK carrying H1/D1 domains has been shown to activate expression of the rsmZ gene. However, it controls target genes in a reciprocal manner as compared to RetS [25], suggesting that it may also control rsmY gene expression although this has not been demonstrated.
Fig 1. The GacSA-RetS-PA1611-LadS signaling network.
(A) Current model for the regulatory elements influencing the expression of two sRNAs, RsmY and RsmZ. See text for details. (B) The multicomponent signal transduction system made of the LadS hybrid HK and the GacS/GacA TCS. In the presented model sustained by results obtained in the present study, this multicomponent signal transduction system made of the LadS hybrid HK and the GacS/GacA TCS forms a multiple-input system probably reflecting the variability of environmental conditions P. aeruginosa is faced with and may result in a range of gradations of chronic infection. IM (Inner Membrane), P (Periplasm), C (Cytoplasm).
While the GacS/GacA TCS is widely distributed throughout the bacterial kingdom, the molecular switch formed by the hybrid LadS, PA1611 and RetS HKs is unique to the Pseudomonas species, though it can function in very different ways in phylogenetically related Pseudomonas species [26–28]. In Pseudomonas fluorescens, it has been proposed that LadS controls rsmX, rsmY and rsmZ expression through GacS, based on the observation that gacS or gacA mutations are epistasic to ladS mutation [29]. In P. syringae, the LadS, GacS and RetS HKs do not control the same targets and this is exemplified for T3SS whose LadS- and RetS-dependent control is GacS-independent in this bacterial species [26,27]. The absence of the D1 domain in P. syringae LadS HK may account for this GacS-independent T3SS regulation [27].
In this network (Fig 1A), the RetS regulatory pathway requires the presence of the GacS/GacA TCS to control rsm gene expression and this occurs via heterodimer formation between RetS HK and GacS HK [23,24,30], impeding GacS autophosphorylation and thereby preventing phosphorylation of its cognate RR GacA [23]. In P. aeruginosa, the PA1611 hybrid HK structurally related to GacS also interacts with RetS in P. aeruginosa in a very similar manner to GacS and RetS and its action is independent of LadS HK [24]. Furthermore, this interaction does not require the conserved phosphorelay residues of PA1611 [24]. Overall, it is still unclear whether in P. aeruginosa, LadS HK triggers rsmZ and possibly rsmY gene expression through GacS or another unorthodox HK or an Hpt protein connected to another TCS [25].
In the present study, we therefore addressed the questions of whether the P. aeruginosa LadS hybrid HK intersects with the GacS/GacA pathway and at which level by using combined genetic, biochemical and phenotypic approaches. We demonstrated that the LadS HK not only controls the expression of rsmZ but also the expression of rsmY, and thus modulates the production of Pel EPS, H1-T6SS and T3SS targets. We further demonstrated that LadS influences its target genes through the GacS/GacA TCS. Specifically, LadS autophosphorylates on its H1 domain, transfers the phosphoryl group on its D1 domain, and then subsequently to the H2 domain of GacS (Fig 1B). These results clearly showed that the LadS HK and the GacS/GacA TCS form a multicomponent signal transduction system, functioning in a mechanism clearly distinct from the one proposed for the RetS or the PA1611 hybrid HKs, the other members of the network.
Results
LadS HK signaling converges on RsmY or RsmZ to control its targets
In P. aeruginosa, the LadS HK had been shown to trigger rsmZ gene expression [25] but no data were available for its action on rsmY gene expression. We thus first addressed the question of whether LadS is able to control rsmY gene expression.
First, to override variable levels of ladS expression and activation (phosphorylation state) due to LadS signal, overexpression of ladS was first undertaken. This approach mimics a constitutive activation of HKs thereby enabling the study of the corresponding signaling pathway. Using chromosomal rsmY–lacZ and rsmZ–lacZ transcriptional fusions in the PAK genetic background (PAKattB::rsmY-lacZ and PAKattB::rsmZ-lacZ), we overexpressed the full-length ladS HK gene using the pBBRladS plasmid [25]. Overexpression of ladS resulted in a significant increase in activity of both rsm fusions (S1A Fig). Maximal rsm gene expression was reached for both fusions at an OD600nm of around 3.8 with an 82-fold and a 42-fold increase for rsmY and rsmZ, respectively, upon ladS overexpression. These results were confirmed by RT-qPCR (S1B Fig).
Because overexpression of genes encoding HKs can have adverse effects [31], we further assessed by RT-qPCR whether LadS produced at the chromosomal level could have the same impact on rsmY and rsmZ expression in the PAK and its isogenic mutant PAKΔladS strains. Levels of rsmY and rsmZ expression were, respectively, reduced by a 17-fold and an 11-fold factor in the ladS mutant as compared to the wild-type strain (S1B Fig). This suggests that the ladS gene is indeed expressed and that LadS signal, although unknown, is present in our testing conditions. This further proved that LadS controls rsmY and rsmZ gene expression at the basal level. The basal level of ladS expression in the wild-type strain PAK was further determined by RT-qPCR. An absolute number of 4,600 copies of ladS gene mRNA copies per μg of total RNA retrotranscribed was monitored in the wild-type PAK strain bearing or not the mild copy empty vector pBBRMCS4 and no copy was detectable in its counterpart ladS mutant. This number increased to 130,000 copies when ladS gene was expressed from the pBBRMCS4 vector (S1C Fig). From these results, overexpression of ladS with a mild copy vector appeared to be a suitable way to investigate the LadS signaling pathway: it can reproduce LadS signaling without adverse effect since the increase in rsm gene expression observed in ladS overexpression conditions was consistent with the one obtained under ladS basal expression (S1A and S1B Fig).
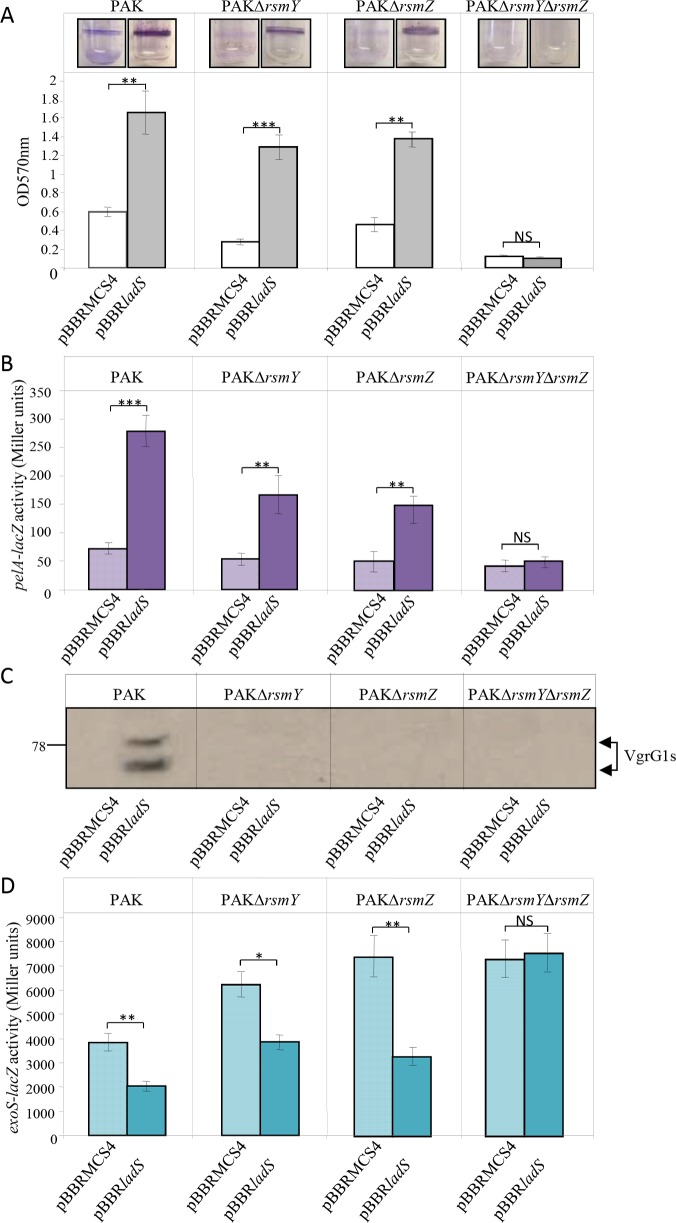
LadS was shown to antagonistically control several of the RetS targets such as Pel, T6SS and T3SS [25]. We next investigated whether it exclusively occurs through the two sRNAs RsmY and RsmZ. The pBBRladS and pBBRMCS4 plasmids were separately conjugated in the PAK, PAKΔrsmY, PAKΔrsmZ and PAKΔrsmYΔrsmZ strains. Overexpression of the ladS HK gene resulted in a 2.65-, 4.3- and 2.8-fold increase in biofilm formation in PAK and in the single rsmY and rsmZ mutants, respectively, while the double mutation abrogated biofilm formation (Fig 2A). Since in the PAK background the biofilm built-in response to the activation of the rsm genes is mostly dependent on pel gene expression [10,22], we further investigated whether the ladS-dependent biofilm formation could rely on the transcriptional activity of the pel locus. The activity of the chromosomal pelA–lacZ transcriptional fusion was therefore assessed in the PAK, PAKΔrsmY, PAKΔrsmZ and PAKΔrsmYΔrsmZ strains transformed with either the pBBRladS or the pBBRMCS4 plasmids. Upon ladS overexpression, the β-galactosidase activity of the pelA transcriptional fusion measured after 4 hours of growth (OD600nm≈3.5) was significantly induced in the wild-type strain, intermediately induced in the rsmY or rsmZ single mutants and abolished in the double rsmYrsmZ mutant (Fig 2B). Another known LadS target is the H1-T6SS, whose production was further checked by immunodetection of the VgrG1 proteins. While ladS HK overexpression induced production of VgrG1 proteins in the PAK strain, these T6SS proteins were undetectable in the PAKΔrsmY, PAKΔrsmZ and PAKΔrsmYΔrsmZ strains (Fig 2C). Finally, LadS control on T3SS was checked using a chromosomal exoS–lacZ transcriptional fusion in the PAK, PAKΔrsmY, PAKΔrsmZ and PAKΔrsmYΔrsmZ strains, which had received the pBBRladS plasmid or the corresponding empty vector pBBRMCS4. Cells were grown in the presence of EDTA, a Ca2+ chelator, a condition known to activate T3SS expression. Upon ladS overexpression, the β-galactosidase activity of the exoS fusion was reduced 2-fold in the wild-type strain, 1.65-fold in the rsmY mutant and 1.9-fold in the rsmZ mutant but had no effect in the double rsmYrsmZ mutant (Fig 2D). These ladS-overexpression effects on pelA, vgrG1b and exoS genes were confirmed by RT-qPCR (S1B Fig). The LadS signaling pathway was found to exert the same control on these targets (Pel, T6SS and ExoS) for ladS expression at the basal chromosomal level (S1B Fig).
Fig 2. Biofilm production, Pel EPS expression, H1-T6SS production and T3SS expression in the LadS signaling pathway.
The pBBRladS plasmid containing the ladS HK gene (dark bars) and the pBBRMCS4 corresponding empty cloning vector (light bars) were conjugated in the PAK, PAKΔrsmY, PAKΔrsmZ or PAKΔrsmYΔrsmZ strains. (A) Biofilm production in glass tubes was illustrated (upper panel) and quantified after Crystal Violet-staining (lower panel). Corresponding levels of biofilm production represent mean values and standard deviations obtained from three independent experiments. (B) Activity of the pelA–lacZ transcriptional chromosomal fusion was monitored in the same strains with the pBBRladS plasmid containing the ladS HK gene (dark violet bars) and the pBBRMCS4 corresponding empty cloning vector (light violet bars) after 4 hours of growth (OD600nm≈3.5). Corresponding β-galactosidase activities are expressed in Miller units and correspond to mean values (with error bars) obtained from three independent experiments. (C) Production of the H1-T6SS VgrG1s proteins was detected in whole cell extracts using western blot with an anti-VgrG1 polyclonal antibody. Numbers on the left side correspond to molecular weight standards (kDa). (D) Activity of the exoS–lacZ transcriptional chromosomal fusion was monitored in the same strains with the pBBRladS plasmid containing the ladS HK gene (dark royal blue bars) and the pBBRMCS4 corresponding empty cloning vector (light royal blue bars) after 6 hours of growth (OD600nm≈4). Corresponding β-galactosidase activities are expressed in Miller units and correspond to mean values (with error bars) obtained from three independent experiments. Wilcoxon-Mann-Whitney tests were performed and *, **, *** and ns referred to p<0.05, p<0.01 and p<0.001 and nonsignificant difference, respectively.
Taken together, these results demonstrated that like RetS, LadS HK controls the expression of both rsmY and rsmZ genes and impacts biofilm formation and Pel production, H1-T6SS and T3SS. Since these sRNAs are the exclusive targets of the GacA RR, our results strongly suggest that such control could occur through the GacS/GacA pathway.
LadS signaling pathway requires GacS and GacA
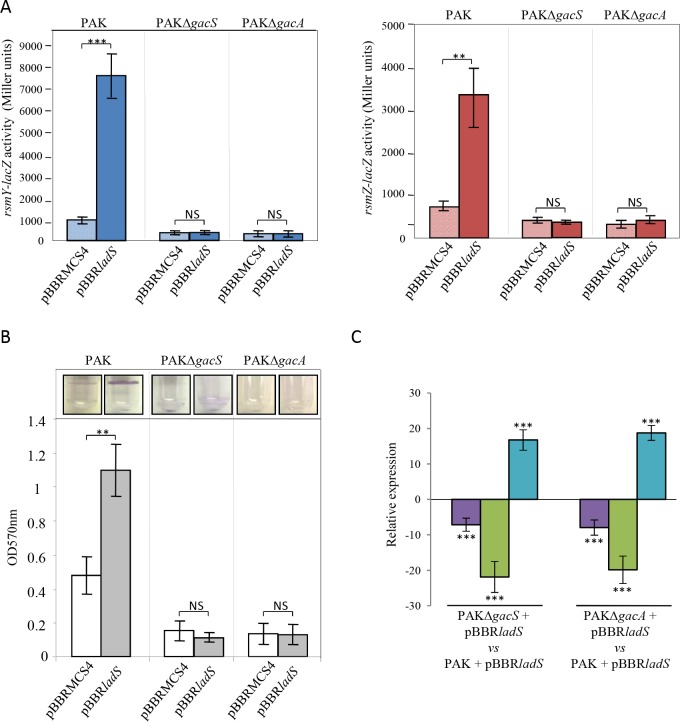
We next investigated whether in P. aeruginosa, LadS signaling converges on the GacS/GacA TCS, as previously demonstrated for RetS [16,23] and HptB [22] signaling pathways. We first examined whether ladS HK overexpression promotes rsm gene expression in gacA and gacS mutants by monitoring the activity of chromosomal rsmY-lacZ and rsmZ-lacZ fusions in both mutants. Overexpression of ladS was no longer able to induce rsmY or rsmZ expression in both mutants (Fig 3A), indicating that LadS control of rsm gene expression occurs through the GacS/GacA TCS. This was confirmed by examining phenotypes highly dependent on GacS/GacA and on the sRNAs RsmY and RsmZ. As illustrated in Fig 3B, the ability of LadS to promote biofilm in the wild-type strain was fully abolished in both gacS and gacA mutants. Similarly, LadS was unable to control the expression of pel, vgrG1b and exoS genes in gac mutants (Fig 3C). These results confirmed that in P. aeruginosa, LadS-dependent induction of rsmY and rsmZ gene expression and of their targets is strictly dependent on the GacS/GacA TCS.
Fig 3. Role of the GacS/GacA TCS in the LadS signaling pathway.
The pBBRladS plasmid containing the ladS HK gene (dark bars) and the pBBRMCS4 corresponding empty cloning vector (light bars) were conjugated in the PAK, PAKΔgacS or PAKΔgacA strains. (A) Activities of the rsmY–lacZ (left panel, blue bars) and rsmZ–lacZ (right panel, brick-red-colored bars) transcriptional chromosomal fusions were monitored after 6 hours of growth (OD600nm≈4) and corresponding β-galactosidase activities are expressed in Miller units and correspond to mean values (with error bars) obtained from three independent experiments. (B) Biofilm production in glass tubes was illustrated (upper panel) and quantified after crystal violet-staining (lower panel). Corresponding levels of biofilm production represent mean values and standard deviations obtained from three independent experiments. Wilcoxon-Mann-Whitney tests were performed and *, **, *** and ns referred to p<0.05, p<0.01 and p<0.001 and nonsignificant difference, respectively. (C) Transcript levels of PelA (violet bars), VgrG1b (T6SS) (green bars) and ExoS (T3SS) (royal blue bars) were monitored by RT-qPCR in PAK, PAKΔgacS and PAKΔgacA strains with the pBBRladS plasmid containing the ladS HK gene and the pBBRMCS4 corresponding empty cloning vector and fold induction was presented for the two mutant strains as compared to the PAK strain. Moderated t-tests were performed; *, ** and *** referred respectively to p<0.05, p<0.01 and p<0.001.
H1 domain of the LadS HK does not heterodimerize with H1 domains of GacS or RetS HK
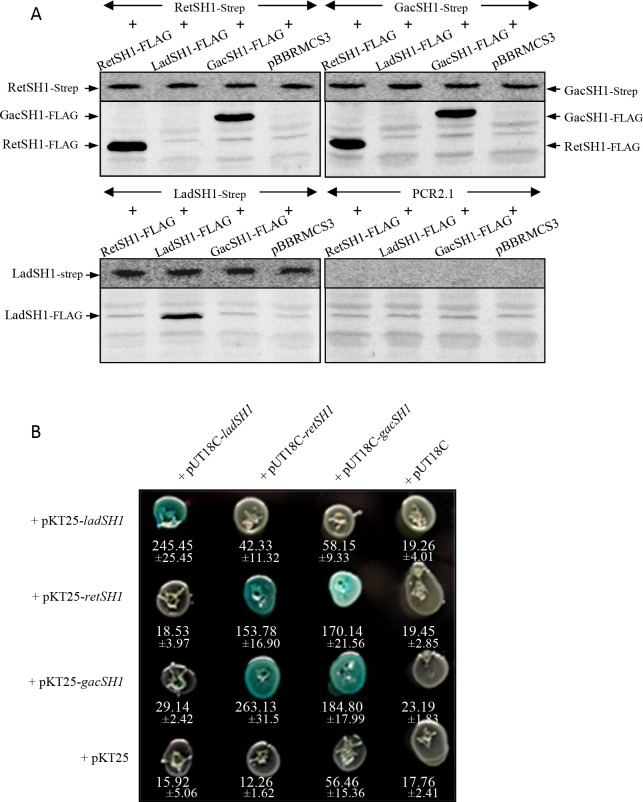
It was shown earlier that RetS forms a heterodimer via its H1 domain, with the H1 domain of GacS, and that such heterodimerization prevents GacS autophosphorylation independently of any phosphorelay residue of RetS HK [23]. Because of the antagonism between RetS and LadS HK, we next examined whether LadS HK could form an active heterodimer with GacS HK or an inactive heterodimer with RetS HK counteracting the inhibitory effect of RetS HK on GacS HK by using pull-down and two-hybrid experiments.
The different N-terminal-tagged (FLAG or Strep) versions of H1 domains of GacS, LadS and RetS HKs were efficiently co-produced in E. coli (S2 Fig). Pull-down experiments were performed using anti-Strep antibody-coupled beads. The LadSH1-FLAG protein was only pulled down in LadSH1-Strep-producing cells (Fig 4A, lower left panel) while the RetSH1-FLAG and GacSH1-FLAG proteins were pulled down in both RetSH1-Strep- and GacSH1-Strep-producing cells (Fig 4A, upper left and right panel), confirming the capacity of the H1 RetS and the H1 GacS domains to form homo- and heterodimers [23,24,32]. The absence of heterodimer formation involving the H1 domain of LadS HK was further confirmed by using two-hybrid experiments. As shown in Fig 4B, the H1 domain of LadS HK was unable to interact with either the H1 domain of GacS HK or the H1 domain of RetS HK. The H1 domain of each HK, LadS, RetS and GacS was able to homodimerize and interaction was also observed between H1 domains of RetS HK and GacS HK reflecting RetS/GacS heterodimerization as previously reported [23] (Fig 4B shows strains having received both vectors on X-gal-containing plates as well as corresponding levels of measured ß-galactosidase activities). Altogether, these results demonstrate that the H1 domain of LadS HK does not form heterodimers with H1 domains of RetS and of GacS, while H1 domains of RetS and GacS HKs form homo- and heterodimers.
Fig 4. Interactions between H1 domains of the LadS hybrid HK, the GacS unorthodox HK and the RetS hybrid HK using pull-down and two-hybrid experiments.
(A) Pull-down experiments. N-terminal FLAG or Strep versions of the H1 domain of GacS, LadS and RetS HKs were constructed in pBBRMCS3 and pCR2.1 vectors, respectively, and expressed in E. coli. Cell lysates were immunoprecipitated using anti-Strep antibody-coupled beads, and FLAG and Strep derivatives were further detected using StrepTactin Alkaline Phosphatase conjugate (upper panel) and anti-FLAG antibody detection (lower panel). (B) In two-hybrid experiments, the ladSH1, retSH1 and gacSH1 DNA regions were cloned into the two-hybrid pUT18C or pKT25 vectors and corresponding vectors were co-transformed in BTH101 cells that were further streaked on LB plates containing X-gal. A blue color of colonies reflects interaction between chimeric proteins, while white color attests to the absence of interaction. The interactions were further quantified by measuring the corresponding ß-galactosidase levels expressed in Miller units (values and standard deviations of 3 independent clones below corresponding colonies).
LadS signaling pathway requires functional H1 and D1 domains
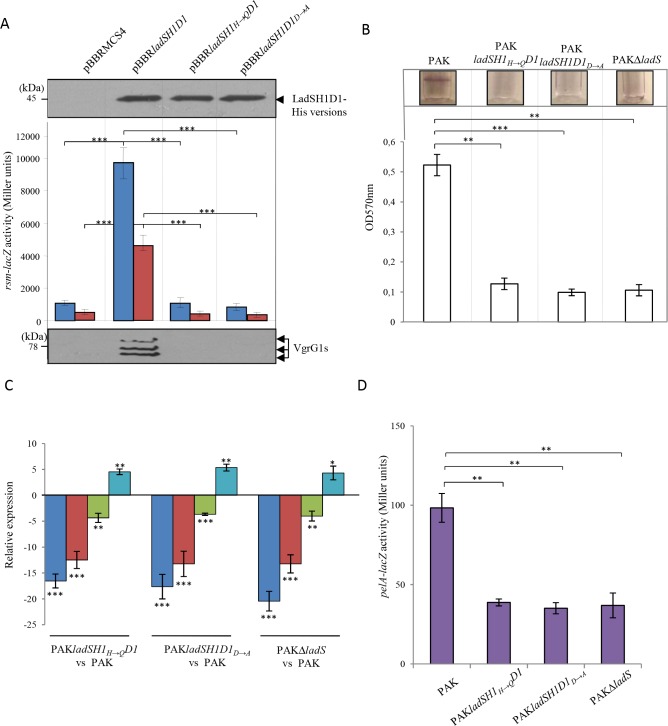
We next tested whether functional H1 and D1 domains of the LadS hybrid HK are required for the LadS-dependent signaling pathway. For that purpose, we engineered a truncated His-tagged version of the whole LadS cytoplasmic part of the HK including the H1 and D1 domains thus referred to as LadSH1D1 yielding to pBBRladSH1D1 (S3 Fig). We then performed site-directed mutagenesis in the corresponding wild-type LadS protein, to generate LadSH1H→QD1 and LadSH1D1D→A versions in which histidine residue in position 428 of the H1 domain and aspartate residue in position 718 of the D1 domain were substituted with a glutamine and an alanine, respectively yielding pBBRladSH1H→QD1 and pBBRladSH1D1D→A (S3 Fig). All truncated versions, although lacking the transmembrane domains anchoring them in the inner membrane, were effectively and equivalently produced in the cytoplasm as detected by western blot (Fig 5A, upper panel). The LadSH1D1 version was able to promote expression of rsmY and rsmZ gene (Fig 5A, middle panel) to levels comparable with those obtained with the full-length version (compare rsmZ-lacZ activities with those presented in Fig 3A) while the LadSH1H→QD1 and LadSH1D1D→A versions were unable to do so (Fig 5A, middle panel). Moreover, the wild-type LadSH1D1 version was able to induce the production of VgrG1 proteins while the LadSH1H→QD1 and LadSH1D1D→A versions were not (Fig 5A, lower panel). To confirm these results, we directly engineered the same point mutations in the ladS gene of the PAK strain, leading to the PAKladSH1H→QD1 and PAKladSH1D1D→A strains. Equivalent results were obtained with point chromosomal ladS mutants of the full-length ladS gene and ladS mutant for biofilm (Fig 5B), expression of rsmY, rsmZ, T6SS-H1 and T3SS genes (Fig 5C) and EPS production (Fig 5D). Taken together, these results demonstrated that the LadS signaling pathway requires both the histidine residue of its H1 domain and the aspartate residue of its D1 domain to activate rsm gene expression and further target genes.
Fig 5. H1 and D1 domain involvement of the LadS hybrid HK in the LadS signaling pathway.
(A) The pBBRladSH1D1 plasmid containing the ladSH1D1 cytoplasmic DNA region of the LadS hybrid HK fused to a C-terminal His-tag, the pBBRladSH1H→QD1 and pBBRladSH1D1D→A variant plasmids and the pBBRMCS4 corresponding empty cloning vector were conjugated in the PAK strain. Production of the corresponding cytoplasmic versions of LadS was checked in whole cell extracts using western blot and a monoclonal anti-His antibody. Numbers on the left side are molecular weight standards (kDa) (upper panel). Activity of the rsmY–lacZ (blue bars) and rsmY–lacZ (brick-red-colored bars) transcriptional chromosomal fusions were monitored after 6 hours of growth (OD600nm≈4) and corresponding β-galactosidase activities are expressed in Miller units and correspond to mean values (with error bars) obtained from three independent experiments. Wilcoxon-Mann-Whitney tests were performed and *, **, *** and ns referred to p<0.05, p<0.01 and p<0.001 and nonsignificant difference, respectively (middle panel). Production of the H1-T6SS VgrG1 protein was detected in whole cell extracts using western blot with an anti-VgrG1 polyclonal antibody. Numbers on the left side are molecular weight standards (kDa) (lower panel). (B) Biofilm production in glass tubes of PAK, PAKΔladS and of point chromosomal mutants PAKladSH1H→QD1 and PAKladSH1D1D→A was presented (upper panel) and quantified after crystal violet-staining and extraction (lower panel). Corresponding levels of biofilm production represented by mean values and standard deviations were obtained from three independent experiments. Wilcoxon-Mann-Whitney tests were performed and *, **, *** and ns referred to p<0.05, p<0.01 and p<0.001 and nonsignificant difference, respectively. (C) Transcript levels of RsmY (blue bars), RsmZ (brick-red-colored bars), VgrG1b (T6SS) (green bars) and ExoS (T3SS) (royal blue bars) were monitored in PAK, PAKΔladS and in point chromosomal mutants PAKladSH1H→QD1 and PAKladSH1D1D→A strains using RT-qPCR. Fold induction was presented for the three mutant strains as compared to the PAK strain. Moderated t-tests were performed; *, ** and *** referred respectively to p<0.05, p<0.01 and p<0.001. (D) Activity of the pelA–lacZ (violet bars) transcriptional chromosomal fusion was monitored after 6 hours of growth (OD600nm≈5) and corresponding β-galactosidase activities are expressed in Miller units and correspond to mean values (with error bars) obtained from three independent experiments. Statistical tests were performed and ** referred to p<0.01.
LadS requires only the H2 domain of GacS to control its target genes
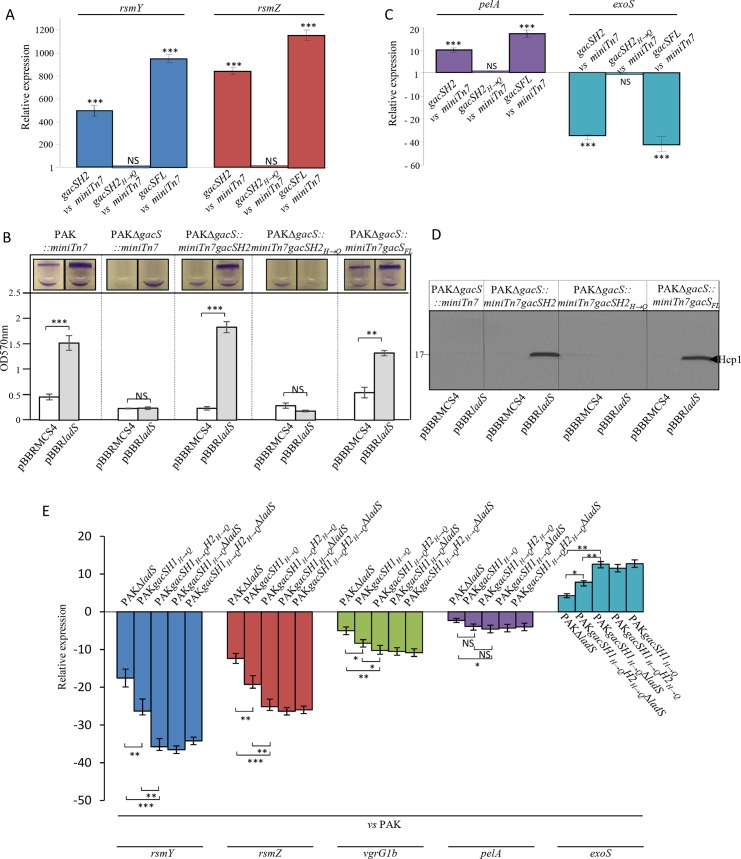
Since we evidenced that LadS requires its phosphorelay residues for signaling, we next investigated whether such phosphorelay requires the H2 domain of GacS. To test this, we engineered a truncated version of the GacS HK formed from its H2 domain (GacSH2) and its counterpart version GacSH2H→Q in which the histidine residue in position 859 was mutated into a glutamine residue (S3 Fig), the corresponding H863 residue being crucial for GacS H2 domain activity in P. fluorescens CHA0 [33]. The corresponding gacSH2 and gacSH2H→Q gene versions as well as the gacS full-length gene (gacSFL) were introduced into the chromosome of a gacS mutant at the miniTn7 site in which we further introduced the pBBRladS or the corresponding empty vector. GacSH2 was able to trigger a ≈ 600- and a 800-fold induction of rsmY and rsmZ transcript levels in cells overexpressing ladS as compared to cells carrying the corresponding empty vector, respectively. In contrast, GacSH2H→Q could not transduce LadS signaling (Fig 6A). Upon ladS overexpression, the production of GacSH2 was sufficient to induce biofilm formation (Fig 6B), pel expression assessed by RT-qPCR (Fig 6C), T3SS gene repression (Fig 6C) and production of T6SS proteins (Fig 6D) while the GacSH2H→Q had no ability to affect the rsm-dependent phenotypes tested here. Interestingly, when the gacSFL gene was introduced in the gacS mutant, the complementation level of each rsm-dependent phenotype under ladS overexpression was similar to that obtained with the gacSH2 gene version. As these results were obtained upon LadS overproduction, we further addressed the question of whether the LadS phosphotransfer to GacS occurs at the natural levels of expression of LadS and GacS HKs. We thus engineered the mutations gacSH1H→QH2 and gacSH1H→QH2H→Q in the wild-type and ladS mutant strains to disable autophosphorylation of the GacSH1 domain and the functionality of the GacsH2 domain, respectively. These strains were evaluated for their capacity to control the expression of rsmY, rsmZ, T6SS, pelA and T3SS genes by RT-qPCR (Fig 6E). GacSH1 domain autophosphorylation disabled in the strain PAKgacSH1H→Q led to a reduction of rsmY, rsmZ, pelA and T6SS gene expression and an induction of T3SS gene expression as compared to the PAK strain. In this genetic context, further alteration of GacSH2 domain functionality (PAKgacSH1H→QH2H→Q, strain) resulted in a higher impact on rsmY, rsmZ, T6SS, pelA and T3SS gene expression. We further engineered deletion of the ladS gene in these two strains yielding respectively the PAKgacSH1H→QΔladS and PAKgacSH1H→QH2H→QΔladS strains. The ladS gene deletion in the PAKgacSH1H→Q strain impacted rsmY, rsmZ, T6SS, pelA and T3SS gene expression to a similar level as observed in the PAKgacSH1H→QH2H→Q strain and ladS gene deletion in the PAKgacSH1H→QH2H→Q strain did not further impact the expression of these target genes. Taken together, all of these results demonstrated that the LadS pathway solely utilizes the H2 domain of GacS HK for signaling and that the histidine residue in position 859 of the GacSH2 is the cross point between the LadS and GacS signaling pathways.
Fig 6. Involvement of the H2 domain of the GacS unorthodox HK in the LadS signaling pathway.
The pBBRladS plasmid containing the ladS HK gene and the pBBRMCS4 corresponding empty cloning vector were conjugated in the PAKΔgacS strain in which the gacSH2 or gacSH2H→Q gene versions or the corresponding suicide vector were chromosomally integrated at the Tn7 site. (A) RsmY (blue bars) and RsmZ (brick-red-colored bars) transcript levels were monitored using RT-qPCR and fold induction was presented in the strains PAKΔgacS::miniTn7gacSH2 (gacSH2) and PAKΔgacS::miniTn7gacSH2H→Q (gacSH2H→Q) as compared to the PAKΔgacS::miniTn7 strain (miniTn7). (B) Biofilm production in glass tubes was illustrated (upper panel) and quantified after crystal violet-staining (lower panel). Corresponding levels of biofilm production represent mean values and standard deviations obtained from three independent experiments. Wilcoxon-Mann-Whitney tests were performed; ** and ns referred to p<0.01 and nonsignificant difference. C. PelA (violet bars) and ExoS (royal blue bars) transcript levels were monitored using RT-qPCR and fold induction was presented in the strains PAKΔgacS::miniTn7gacSH2 (gacSH2) and PAKΔgacS::miniTn7gacSH2H→Q (gacSH2H→Q) as compared to the PAKΔgacS::miniTn7 strain (miniTn7). (D) Production of the H1-T6SS Hcp1 proteins was detected in whole cell extracts using western blot with an anti-Hcp1 polyclonal antibody. Numbers on the left side are molecular weight standards (kDa). Moderated t-tests were performed and *, **, *** and ns referred to p<0.05, p<0.01 and p<0.001 and nonsignificant difference, respectively. (E) Transcript levels of RsmY (blue bars), RsmZ (brick-red-colored bars), VgrG1 (green bars), PelA (violet bars) and ExoS (royal blue bars) were monitored using RT-qPCR. Fold induction was presented in the strains PAK, PAKΔladS, PAKgacSH1H→Q, PAKgacSH1H→QΔladS, PAKgacSH1H→QH2H→Q and PAKgacSH1H→QH2H→QΔladS in order to disable autophosphorylation of the GacSH1 domain and the functionality of the GacsH2 domain, respectively. Moderated t-tests were performed and *, **, *** and ns referred to p<0.05, p<0.01 and p<0.001 and nonsignificant difference, respectively.
In vitro transphosphorylation of GacS H2 domain by LadS HK
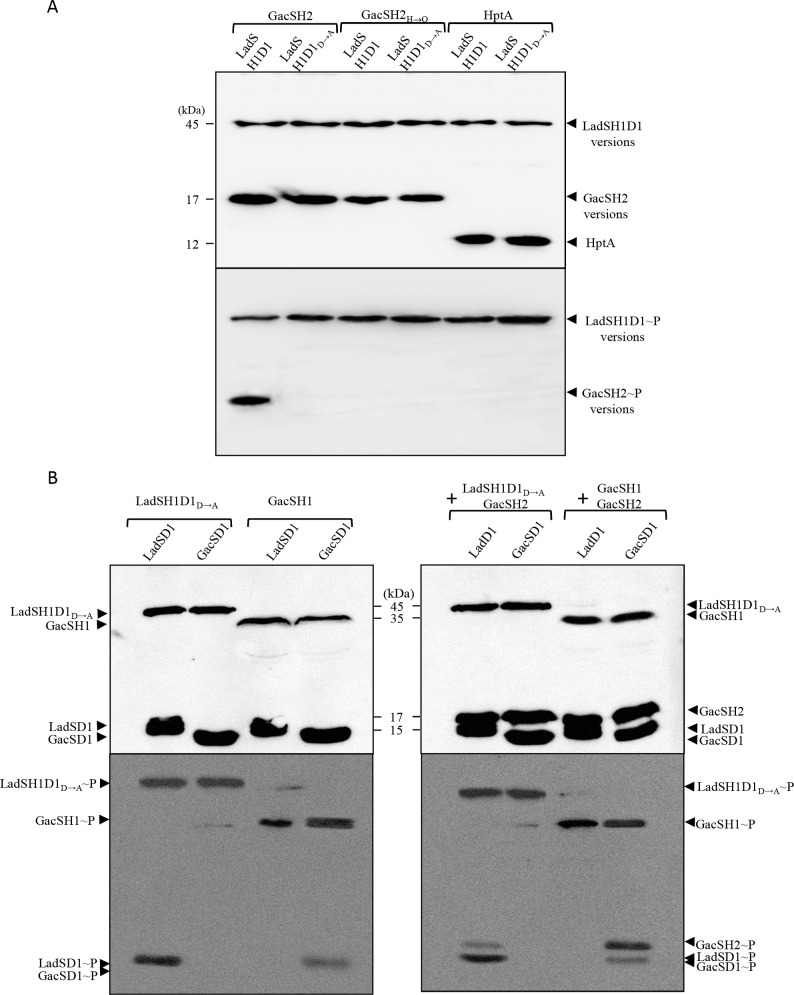
The results presented above strongly suggest that LadS HK could use the H2 domain of GacS HK to activate rsm gene transcription, probably via a transphosphorylation mechanism. To further confirm this, in vitro phosphorylation experiments were performed. The C-terminal His-tag forms of LadSH1D1, LadSH1D1D→A, GacSD1, LadSD1, GacSH1D1, GacSH2, GacSH2H→Q and HptA proteins were produced in E. coli and purified close to homogeneity from soluble fractions with nickel affinity columns and subjected to autophosphorylation assays by using [γ-32P]ATP (S4A and S4B Fig) (see Materials and Methods). LadSH1D1 and LadSH1D1D→A proteins were found in an autophosphorylated form, while GacSH2, GacSH2H→Q and HptA were not (S4C Fig), confirming that LadSH1D1 and LadSH1D1D→A are functional for their kinase activity. When mixed with LadSH1D1, GacSH2 was phosphorylated whereas GacSH2H→Q and HptA were not (Fig 7A), demonstrating that LadS HK transphosphorylates the H2 domain of GacS HK on its H859 residue. LadSH1D1D→A protein, while capable of autophosphorylation (S4C Fig), was unable to transphosphorylate the GacS H2 domain, demonstrating that D718 residue on D1 domain of LadS HK is fully required for the transphosphorylation process and that direct transphosphorylation cannot directly occur between the H1 domain of LadS HK and the H2 domain of GacS HK (Fig 7A).
Fig 7. In vitro transphosphorylation assays.
(A) For transphosphorylation assay between LadS or GacS variants and GacSH2 variants or HptA protein, 2 mM of LadSH1D1 or LadSH1D1D→A recombinant proteins were incubated with [γ-32P] ATP and GacSH2 (lanes 1 and 2), GacSH2H→Q (lanes 3 and 4) or HptA (lanes 5 and 6) at room temperature for 20 min (see Materials and Methods) then separated in an SDS-polyacrylamide gel in duplicate. (B) Transphosphorylation assay between the LadSH1D1D→A or GacSH1 and LadSD1 or GacsD1 domains with or without the GacSH2 domain. Two mM of LadSD1 or GacSD1 recombinant proteins were incubated with [γ-32P] ATP and LadSH1D1D→A or GacSH1 (left panel) together with the GacSH2 domain (right panel) at room temperature for 20 min. In both experiments mixtures of proteins were separated in an SDS-polyacrylamide gel in duplicate. Numbers on the left side are molecular weight standards (kDa). Locations of the recombinant proteins are indicated by arrowheads. For each experiment presented in panels A and B, one gel was detected by western blot using an anti-penta-His antibody (upper panel) while the other was autoradiographied (lower panel).
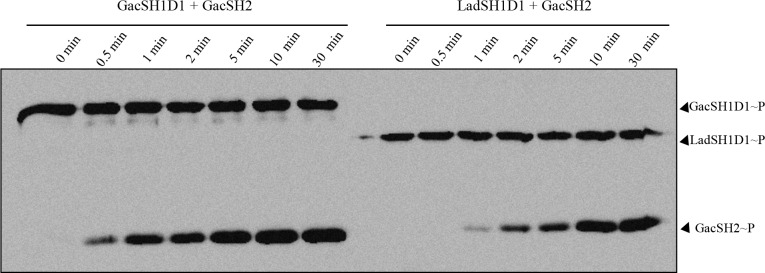
To further exclude the crosstalk between LadSH1 and GacSD1 or between GacSH1 and LadSD1 domains, we produced the corresponding isolated domains (GacSH1, GacSD1, LadSD1) (S4B Fig). No phosphotransfer could be observed from the H1 domain of LadS to the D1 domain of GacS or from the H1 domain of GacS to the D1 domain of LadS. Meanwhile, phosphotransfer occurred between the H1 domain of LadS and the D1 domain of LadS and between the H1 domain of GacS and the D1 domain of GacS (Fig 7B, left panel). When GacSH2 was further added, we observed that the GacSH2 domain can only receive phosphate for the LadSH1D1Q→A/LadSD1 domain and for the GacSH1/GacSD1 domain combinations (Fig 7B, right panel), thereby finally proving that LadS signaling is H1LadS→D1LadS→H2GacS→D2GacA. From this last experiment, it clearly appeared that GacSH2 phosphorylation was less effective through LadS HK than through GacS HK. As the sampling was done at the same time in both experiments, this strongly suggests that the GacS signaling is faster than the LadS signaling. To confirm this observation, we performed kinetic experiments and clearly observed that phosphotransfer to the GacSH2 domain occurred 0.5–1 min earlier with the GacSH1D1 domain compared to the LadSH1D1 domain as a phosphodonor. This result suggests that the LadS hybrid HK forms a multicomponent signal transduction pathway with the GacS/GacA TCA and adds a supplementary level of regulation triggering chronic infection (Fig 8).
Fig 8. Transphosphorylation kinetic between the LadSH1D1 or GacSH1D1 and GacSH2 domains.
Two mM of LadSH1D1 or GacSH1D1 and GacSH2 recombinant proteins were incubated with [γ-32P] ATP at room temperature. The reaction was stopped at different time points (see Materials and Methods) and the samples were separated in an SDS-polyacrylamide gel and autoradiographied.
Discussion
In the present study, we demonstrated that in P. aeruginosa the LadS hybrid HK forms a multicomponent signal transduction system with the GacS/GacA TCS. This multicomponent signal transduction system involves an H1LadS→D1LadS→H2GacS→D2GacA signaling pathway, first through a phosphorelay between the H1 and D1 domains of LadS HK, and thus a transphosphorylation of the H2 domain of GacS HK. This LadS signaling pathway triggers both rsmY and rsmZ gene expression, the two sole direct targets controlled by the RR GacA [18] and further targets such as biofilm formation, H1-T6SS and T3SS expression.
As suggested or observed in other very closely phylogenetically related species to P. aeruginosa such as P. fluorescens [29] but never demonstrated in P. aeruginosa [7,25], our results showed that LadS controls both rsmY and rsmZ gene expression. These results also highlight the fact that LadS signaling occurs through the GacS/GacA TCS, reinforcing the notion that LadS and RetS HK reciprocally regulate the virulence factors under the rsm gene dependency [23,25]. This reciprocal regulation of LadS and RetS converging on GacS seems to be specific to some pseudomonas species such as P. aeruginosa or P. fluorescens but not generalizable to all, since in P. syringae the LadS, GacS and RetS HK do not control the same targets. This is exemplified for T3SS whose LadS- and RetS-dependent control is GacS-independent in this bacterial species [26,27]. In P. syringae, the absence of D1 domain in LadS HK may account for this GacS-independent T3SS regulation [27], while the P. aeruginosa LadS version without its D1 domain is nonfunctional (S5 Fig). Thus, in P. aeruginosa, LadS HK may control T3SS through its D1 domain and GacS HK. Another example of the crucial involvement of the H1D1 subdomain of P. aeruginosa LadS HK in LadS signaling is provided by the observed specific and natural ladS mutation in the PA14 strain, a 49 nucleotide duplication that leads to possible frameshift and results in a truncated and nonfunctional LadS protein lacking H1 and D1 domains [34]. In PA14, this nonfunctional mutation of the ladS gene can be reversed by trans-complementation with the PAK ladS gene.
The RetS hybrid HK [16,35,36] control of GacS involves a heterodimer formation between their H1 domains, which leads to suppression of GacS autophosphorylation in a phosphorelay-independent manner [23]. RetS conserved phosphorelay residues have been found dispensable [23] or not fully required [30] to control GacS, probably depending on the genetic background used. Conversely, we demonstrated that LadS control of GacS does not involve such a heterodimer mechanism in P. aeruginosa. In P. aeruginosa, LadS and GacS do not form heterodimers through their H1 domains as proven here by: i) pull-down and two-hybrid experiments and ii) the simple requirement of the H2 domain of the GacS HK for LadS signaling through GacS. Thus, the interaction between LadS H1 and GacS H1 domains as a mechanism of signaling between LadS HK and GacS HK can be ruled out. The functional bridge (phosphorelay) between the D1 domain of LadS and the H2 domain of GacS requires a transient interaction that has been potentially evidenced in P. fluorescens with the full-length proteins [32] and that we weakly observed between the LadS D1 domain and the GacS H2 domain using a two-hybrid approach (S6A Fig). Undoubtedly, as demonstrated in the present study, LadS signaling involves a phosphorelay mechanism along GacS, ruling out GacS HK titration by LadS, as RetS does. Although RetS suppresses GacS autophosphorylation, we demonstrated that LadS transphosphorylates the GacS HK by a phosphorelay involving the H428 of H1LadS domain → the D718 of the D1LadS domain → the H859 of the H2GacS domain. Thus, LadS HK and the GacS/GacA TCS certainly form a multicomponent signal transduction system as with the CblS and CblT/CblR in B. cenocepacia [4]. The LadS signaling through the GacS H2 domain was GacS H2 domain-specific since i) the LadS hybrid HK did not use any of the three Hpt proteins (HptA, HptB, HptC) or any of the H2 domains of the four other unorthodox HK encoded in the P. aeruginosa genome (RocS1, RocS2, PA4112, PA4982) as demonstrated for the control of rsmZ gene expression (S6B Fig), ii) no interaction was observed between the LadS D1 domain and any of the three Hpt proteins or the H2 domains of the four other unorthodox HK (RocS1, RocS2, PA4112, PA4982) encoded in the P. aeruginosa genome (S6A Fig), and iii) a weak but significant interaction was observed between the LadS D1 and GacS H2 domains (S6A Fig). This confirms that the LadS signaling pathway exclusively use the Hpt module (the H2 domain) of the GacS unorthodox HK to control its targets.
Once the GacS/GacA signaling pathway is activated upon a signal that remains to be identified, P. aeruginosa is engaged in a chronic infection lifestyle characterized by biofilm and H1-T6SS production and the shutting down of the T3SS. The multicomponent signal transduction system made of the LadS hybrid HK and the GacS/GacA TCS (Fig 1B) is therefore able to integrate at least two different signals, one from GacS and the other from LadS, which also remains to be identified. LadS contains a putative 7-transmembrane (7TMR) region anchoring the HK into the inner membrane and a periplasmic sensor domain (diverse intracellular signaling module extracellular 2, DISMED2), whose predicted fold exhibits a putative binding site, highly conserved in carbohydrate-binding modules (CBMs) [37]. The activity of the multicomponent transduction system described here and the subsequent output response were also certainly modulated by the ratio of kinase to phosphatase activity [38,39]. Phosphatase-like activity leading to catalyzed dephosphorylation of phospho-response regulators described for unorthodox and hybrid sensors by reverse phosphotransfer [40–42] can be embodied in the cognate sensor kinase itself [42], carried out by the response regulator itself [40] or by another partner protein [43]. Thus, whether phosphatase-like activity leading to GacA dephosphorylation is assumed by GacS, LadS, any other HK of the network, by itself or by a partner protein requires extensive additional studies.
Thus, P. aeruginosa probably shares with P. fluorescens but not with P. syringae a unique molecular switch controlling Rsm sRNA-dependent virulence made of a central unorthodox HK and three hybrid HKs among which LadS HK makes a unique multicomponent system with the TCS GacS-GacA. This multiple input system probably reflects the variability of the environmental conditions P. aeruginosa faces and may result in a range of gradations of chronic infection through the integration of variable environmental signals.
Materials and Methods
Bacterial strains, growth conditions and media
The bacterial strains and plasmids used in this study are described in Tables 1 and 2 and the oligonucleotides used in S1 Table. Strains were grown aerobically in Luria–Bertani (LB) broth or on LB agar at 37°C or 30°C. To visualize bacterial two-hybrid interactions on solid medium, LB agar plates supplemented with the chromogenic substrate X-gal (5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside, 40 μg/mL), isopropyl β-D-1-thiogalactopyranoside (IPTG) (100 μM), ampicillin (Ap, 100 μg/mL) and kanamycin (Km, 50 μg/mL) were used. Plasmids were introduced into P. aeruginosa by electroporation or by triparental mating using the conjugative properties of pRK2013. The transformants were selected on Pseudomonas isolation agar. Antibiotics were used at the following concentrations for Escherichia coli: 50 μg/mL ampicillin, 50 μg/mL streptomycin and 15 μg/mL tetracycline. For P. aeruginosa, 500 μg/mL carbenicillin, 2,000 μg/mL streptomycin and 200 μg/mL tetracycline were used.
Table 1. Strains used in this study.
| Strains | Relevant characteristics* | Source |
|---|---|---|
| E. coli | ||
| TG1 | K-12, Δ(lac-pro) supE thi hsdD5/F' traD36 proA+B+ lacIq lacZΔM15 | Lab collection |
| DH5α | endA1 hsdR17 supE44 thi-1 recA1 gyrA relA1 Δ(lacZYA-argF)U169 deoR (phi 80lacZ Δ M15) | Lab collection |
| BTH101 | F- cya-99 araD139 galE15 galK16 rpsL1 (Smr) hsdR2 mcrA1 mcrB1 | [45] |
| Top10F’ | F’ (lacIq Tn10 (TetR)) mrcA Δ(mrr-hsdRMS-mcrBC) Φ80 lacZΔM15 ΔlacX74 recA1 | Invitrogen |
| CC118(λpir) | Host strain for pKNG101 replication, Δ(ara-leu) araD ΔlacX74 galE galK phoA20 thi-1 rpsE rpoB argE(Am) recA1 RfR (λpir) | Lab collection |
| P. aeruginosa | ||
| PAK | Wild-type | [48] |
| PAKΔrsmY | PAK deletion mutant for rsmY gene | [24] |
| PAKΔrsmZ | PAK deletion mutant for rsmZ gene | [22] |
| PAKΔrsmYΔrsmZ | PAK deletion mutant for rsmY and rsmZ genes | [22] |
| PAKΔgacS | PAK deletion mutant for gacS gene | [22] |
| PAKΔgacA | PAK deletion mutant for gacA gene | [22] |
| PAKΔhptA | PAK deletion mutant for hptA gene | [22] |
| PAKΔhptB | PAK deletion mutant for hptB gene | [22] |
| PAKΔhptC | PAK deletion mutant for hptC gene | [22] |
| PAKΔhptAΔhptBΔhptC | PAK deletion mutant for hptA, hptB and hptC genes | This study |
| PAKΔrocS1 | PAK deletion mutant for rocS1 gene | [49] |
| PAKΔrocS2 | PAK deletion mutant for rocS2 gene | [49] |
| PAKΔPA4112 | PAK deletion mutant for PA4112 gene | This study |
| PAKΔPA4982 | PAK deletion mutant for PA4982 gene | This study |
| PAKattB::pelA-lacZ | PAK strain with pelA-lacZ inserted at attB sites | This study |
| PAKattB::exoS-lacZ | PAK strain with exoS-lacZ inserted at attB sites | This study |
| PAKattB::rsmY-lacZ | PAK strain with rsmY-lacZ inserted at attB sites | This study |
| PAKattB::rsmZ-lacZ | PAK strain with rsmZ-lacZ inserted at attB sites | This study |
| PAKΔrsmYattB::pelA-lacZ | PAKΔrsmY strain with pelA-lacZ inserted at attB sites | This study |
| PAKΔrsmYattB::exoS-lacZ | PAKΔrsmY strain with exoS-lacZ inserted at attB sites | This study |
| PAKΔrsmZattB::pelA-lacZ | PAKΔrsmZ strain with pelA-lacZ inserted at attB sites | This study |
| PAKΔrsmZattB::exoS-lacZ | PAKΔrsmZ strain with exoS-lacZ inserted at attB sites | This study |
| PAKΔrsmYΔrsmZattB::pelA-lacZ | PAKΔrsmYΔrsmZ strain with pelA-lacZ inserted at attB sites | This study |
| PAKΔrsmYΔrsmZattB::exoS-lacZ | PAKΔrsmYΔrsmZ strain with exoS-lacZ inserted at attB sites | This study |
| PAKΔgacSattB::rsmY-lacZ | PAKΔgacS strain with rsmY-lacZ inserted at attB sites | This study |
| PAKΔgacSattB::rsmZ-lacZ | PAKΔgacS strain with rsmZ-lacZ inserted at attB sites | This study |
| PAKΔgacAattB::rsmY-lacZ | PAKΔgacA strain with rsmY-lacZ inserted at attB sites | This study |
| PAKΔgacAattB::rsmZ-lacZ | PAKΔgacA strain with rsmZ-lacZ inserted at attB sites | This study |
| PAKΔhptAattB::rsmZ-lacZ | PAKΔhptA strain with rsmZ-lacZ inserted at attB sites | This study |
| PAKΔhptBattB::rsmZ-lacZ | PAKΔhptB strain with rsmZ-lacZ inserted at attB sites | This study |
| PAKΔhptCattB::rsmZ-lacZ | PAKΔhptC strain with rsmZ-lacZ inserted at attB sites | This study |
| PAKΔhptAΔhptBΔhptCattB::rsmZ-lacZ | PAKΔhptAΔhptBΔhptC strain with rsmZ-lacZ inserted at attB sites | This study |
| PAK::miniTn7 | PAKstrain with empty mini Tn7 construct; GmR | This study |
| PAKΔgacS::miniTn7 | PAKΔgacS strain with empty mini Tn7 construct; GmR | This study |
| PAKΔgacS::miniTn7gacSFL | PAKΔgacS strain with gacS in a mini Tn7 construct; GmR | This study |
| PAKΔgacS::miniTn7gacSH2 | PAKΔgacS strain with gacSH2WT in a mini Tn7 construct; GmR | This study |
| PAKΔgacS::miniTn7gacSH2H→Q | PAKΔgacS strain with gacSH2 H→Q in a mini Tn7 construct; GmR | This study |
| PAKΔladS | PAK deletion mutant for ladS gene | [25] |
| PAKΔladSD1 | PAK deletion mutant for D1 domain of ladS gene | This study |
| PAKladSH1H→QD1 | Punctual chromosomal mutant H428Q of ladS gene in PAK | This study |
| PAKladSH1D1D→A | Punctual chromosomal mutant D718A of ladS gene in PAK | This study |
| PAKgacSH1H→Q | Punctual chromosomal mutant H293Q of gacS gene in PAK | This study |
| PAKgacSH2H→Q | Punctual chromosomal mutant H859Q of gacS gene in PAK | This study |
| PAKgacSH1 H→Q,H2H→Q | Punctual chromosomal mutant H293Q and H859Q of gacS gene in PAK | This study |
| PAKgacSH1H→QΔladS | Punctual chromosomal mutant H293Q of gacS gene and deletion mutant for ladS gene. | This study |
| PAKgacSH1 H→Q,H2H→QΔladS | Punctual chromosomal mutant H293Q and H859Q of gacS gene and deletion mutant for ladS gene. | This study |
* SmR, streptomycin resistance, GmR gentamicin resistance
Table 2. Plasmids used in this study.
| Plasmids | Relevant characteristics* | Source |
|---|---|---|
| pLic03 | Vector containing T7 promoter with pBR322 origin of replication, KmR | [50] |
| pLic03_ LadSD1 | pLic03 carrying the Nt-6his tagged LadSD1 subdomain DNA region | This study |
| pLic03_ GacSH1D1 | pLic03 carrying the Nt-6his tagged GacSH1D1 subdomain DNA region | This study |
| pLic03_ GacSH1 | pLic03 carrying the Nt-6his tagged GacSH1 subdomain DNA region | This study |
| pLic03_ GacSD1 | pLic03 carrying the Nt-6his tagged GacSD1 subdomain DNA region | This study |
| pJF_hptA | pJF119EH carrying the 6his tagged hptA DNA region | This study |
| pBBRMCS3 | Broad host range plasmid, TcR | [51] |
| pBBRMCS4 | Broad host range plasmid, ApR | [51] |
| pBBRladS | pBBRMCS4 carrying the ladS gene | [25] |
| pBBRladSH1D1 | pBBRMCS4 carrying the ladSH1D1 subdomain DNA region | This study |
| pBBRladSH1H→QD1 | pBBRMCS4 carrying the ladSH1H→QD1 subdomain DNA region | This study |
| pBBRladSH1D1D→A | pBBRMCS4 carrying the ladSH1D1D→A subdomain DNA region | This study |
| pBBR_FLAG-ladSH1 | pBBRMCS3 carrying the FLAG tagged ladSH1 subdomain DNA region | This study |
| pBBR_FLAG-gacSH1 | pBBRMCS3 carrying the FLAG tagged gacSH1 subdomain DNA region | This study |
| pBBR_FLAG-retSH1 | pBBRMCS3 carrying the FLAG tagged retSH1 subdomain DNA region | This study |
| PCR2.1 | TA cloning vector for PCR products, lacZα ColE1 f1 ori, ApR, KmR | Invitrogen |
| pCR2.1_Strep-ladSH1 | pCR2.1 carrying the Strep tagged ladSH1 subdomain DNA region | This study |
| pCR2.1_Strep-gacSH1 | pCR2.1 carrying the Strep tagged gacSH1 subdomain DNA region | This study |
| pCR2.1_Strep-retSH1 | pCR2.1 carrying the Strep tagged retSH1 subdomain DNA region | This study |
| pCR2.1_FLAG-ladSH1 | pCR2.1 carrying the FLAG tagged ladSH1 subdomain DNA region | This study |
| pCR2.1_FLAG-gacSH1 | pCR2.1 carrying the FLAG tagged gacSH1 subdomain DNA region | This study |
| pCR2.1_FLAG-retSH1 | pCR2.1 carrying the FLAG tagged retSH1 subdomain DNA region | This study |
| pCR2.1ladSH1D1 | pCR2.1 carrying the carrying the ladSH1D1 subdomain DNA region | This study |
| pCR2.1H1H→QD1 | pCR2.1 carrying the ladSH1H→QD1 subdomain DNA region | This study |
| pCR2.1H1D1D→A | pCR2.1 carrying the ladSH1D1D→A subdomain DNA region | This study |
| pCR2.1gacSH2 | pCR2.1 carrying the gacSH2 subdomain DNA region | This study |
| pCR2.1intladSH1H→Q | pCR2.1 carrying the internal fragment ladSH1H→Q | This study |
| pCR2.1intladSD1D→A | pCR2.1 carrying the internal fragment ladSD1D→A | This study |
| pUC18-miniTn7 | mini-Tn7 base vector with MCS; for cloning of suitable selection markers or other functional and selectable elements GmR | [52] |
| pUC18-miniTn7-gacSH2 | pUC18-miniTn7 carrying the gacSH2 subdomain DNA region | This study |
| pUC18-miniTn7-gacSH2H→Q | pUC18-miniTn7 carrying the gacSH2H→Q subdomain DNA region | This study |
| pRK2013 | Tra+ Mob+ KmR | Lab collection |
| pKNG101ΔPA4112 | Mutator plasmid for PA4112 deletion SmR | This study |
| pKNG101ΔPA4982 | Mutator plasmid for PA4982 deletion SmR | This study |
| pKNG101ΔhptA | Mutator plasmid for hptA deletion SmR | [22] |
| pKNG101ΔhptB | Mutator plasmid for hptB deletion SmR | [22] |
| pKNG101ΔhptC | Mutator plasmid for hptC deletion SmR | [22] |
| pKNGladSH1H→QD1 | Mutator plasmid for point mutation H428Q in ladS gene SmR | This study |
| pKNGladSH1D1D→A | Mutator plasmid for point mutation D718A in ladS gene SmR | This study |
| pKNGgacSH1H→Q | Mutator plasmid for point mutation H293Q in gacS gene SmR | This study |
| pKNGgacSH2H→Q | Mutator plasmid for point mutation H859Q in gacS gene SmR | This study |
| pKΔS | Mutator plasmid for ladS deletion SmR | [25] |
| miniCTX-lacZ | Tcr lacZ+; self-proficient integration vector with tet, V-FRT-attPMCS, ori, int, and oriT | [51] |
| miniCTX-rsmY-lacZ | Promoter region of rsmY gene inserted into miniCTX-lacZ, TcR | This study |
| miniCTX-rsmZ-lacZ | Promoter region of rsmZ gene inserted into miniCTX-lacZ, TcR | This study |
| miniCTX-pelA-lacZ | Promoter region of pelA gene inserted into miniCTX-lacZ, TcR | This study |
| miniCTX-exoS-lacZ | Promoter region of exoS gene inserted into miniCTX-lacZ, TcR | This study |
| pUT18C | Two-hybrid plasmid, cyaAT18 fusion, ApR | [53] |
| pUT18C-ladSH1 | Two-hybrid plasmid containing cyaAT18-ladS H1 domain fusion | This study |
| pUT18C-retSH1 | Two-hybrid plasmid containing cyaAT18-retS H1 domain fusion | [23] |
| pUT18C-gacSH1 | Two-hybrid plasmid containing cyaAT18-gacS H1 domain fusion | [23] |
| pUT18C-ladSD1 | Two-hybrid plasmid containing cyaAT18-ladS D1 domain fusion | This study |
| pKT25 | Two-hybrid plasmid, cyaAT25 fusion, KmR | [53] |
| pKT25-ladSH1 | Two-hybrid plasmid containing cyaAT25–ladS H1 domain fusion | This study |
| pKT25-retSH1 | Two-hybrid plasmid containing cyaAT25–retS H1 domain fusion | [23] |
| pKT25-gacSH1 | Two-hybrid plasmid containing cyaAT25–gacS H1 domain fusion | [23] |
| pKT25-gacSH2 | Two-hybrid plasmid containing cyaAT25–gacS H2 domain fusion | [49] |
| pKT25-rocS1H2 | Two-hybrid plasmid containing cyaAT25–rocS1 H2 domain fusion | [49] |
| pKT25-rocS2H2 | Two-hybrid plasmid containing cyaAT25–rocS2 H2 domain fusion | [49] |
| pKT25-hptA | Two-hybrid plasmid containing cyaAT25–hptA gene fusion | This study |
| pKT25-hptB | Two-hybrid plasmid containing cyaAT25–hptB gene fusion | This study |
| pKT25-hptC | Two-hybrid plasmid containing cyaAT25–hptC gene fusion | This study |
| pKT25-PA4112H2 | Two-hybrid plasmid containing cyaAT25–PA4112 H2 domain fusion | This study |
| pKT25-PA4982H2 | Two-hybrid plasmid containing cyaAT25–PA4982 H2 domain fusion | This study |
* SmR, streptomycin resistance, GmR gentamicin resistance, TCR tetracyclin resistance, ApR ampicillin, KmR Kanamycin
Transcriptional fusions
The rsmY, rsmZ, pelA and exoS promoter regions were amplified by PCR with PAK genomic DNA using appropriate oligonucleotide pairs (S1 Table) and corresponding PCR products were cloned into pCR2.1 vector (Invitrogen) by TA cloning. After DNA sequencing, each promoter region cloned into pCR2.1 vector was excised by HindIII/BamHI and inserted into the linearized miniCTX-lacZ vector [44], thereby generating miniCTX-rsmY-lacZ, miniCTX-rsmZ-lacZ, miniCTX-pelA-lacZ and miniCTX-exoS-lacZ constructs. These plasmids were introduced in the different P. aeruginosa strains and site-specific recombination at the attB site, generating chromosomal rsmY-lacZ, rsmZ-lacZ, pelA-lacZ and exoS-lacZ fusions. The FRT cassette-excision step was performed, resulting in the generation of strains without tetracycline resistance.
Truncated versions of LadS and GacS HK
DNA fragments corresponding to the cytoplasmic LadS1D1 part of the LadS hybrid HK (H1 and D1 subdomains), the LadSD1 domain, the cytoplasmic GacSH1D1 part of the GacS unorthodox HK (H1 and D1 subdomains), the GacSH1, the GacSD1 and the GacSH2 domains all fused to a His tag were amplified by PCR using appropriate oligonucleotide pairs (S1 Table). The DNA fragments corresponding to LadSH1D1 and GacSH2 were cloned into pCR2.1 vector yielding respectively pCR2.1ladSH1D1 and pCR2.1gacSH2 plasmids while LadSD1, GacSH1D1, GacSH1 and GacSD1 were cloned into pLic03 vector yielding pLic03_ladSD1, pLic03_gacSH1D1, pLic03_gacSH1 and pLic03_gacSD1, respectively. After DNA sequencing, digestion was performed using EcoRI/BamHI for LadSH1D1 and BamHI/HindIII for GacSH2 for subcloning into pBBRMCS4 and pUC18-miniTn7 vectors, respectively, yielding pBBRLadSH1D1, referred to as LadSH1D1, and pUC18-miniTn7-gacSH2, referred to as GacSH2. Site-directed mutations in the DNA sequence of LadSH1D1 and GacSH2 were introduced respectively into pCR2.1ladSH1D1 and pCR2.1gacSH2 plasmids by Quick exchange site-directed mutagenesis method. Briefly, the conserved histidine residue at position 428 and the conserved aspartate residue at position 718 of LadS HK were changed into glutamine and alanine residues, respectively generating LadSH1H→QD1 and LadSH1D1D→A variants. The conserved histidine residue at position 859 of GacS HK was changed into glutamine, leading to the GacSH2H→Q variant. This was done by using pCR2.1ladSH1D1 and pCR2.1gacSH2 vectors as matrices and by PCR using pfu turbo DNA polymerase (Stratagene) and 39-mer primers that incorporated appropriate mismatches to introduce the expected mutations (S1 Table). The resulting PCR products were digested with DpnI for 1 hour. After DNA sequencing, each DNA sequence corresponding to LadSH1H→QD1 and LadSH1D1D→A or GacSH2H→Q variants cloned into pCR2.1 were released by EcoRI/BamHI or BamHI/HindIII digestion, respectively, and inserted into pBBRMCS4 or pUC18-miniTn7, respectively, to generate pBBRladSH1H→QD1 and pBBRladSH1D1D→A and pUC18-miniTn7-gacSH2H→Q.
Construction of deletion mutants
PCR was used to generate a 500 bp DNA fragment upstream (Up) and a 500 bp DNA fragment downstream (Dn) of the PA4112 and PA4982 and of the LadSD1 domain of the ladS gene using the appropriate pairs of primers (S1 Table). Each PCR upstream and downstream product was linked together by overlapping PCR and products were cloned into pCR2.1. The linked DNA fragment was digested with XbaI and SpeI and cloned in the suicide vector pKNG101, yielding pKNG101ΔPA4112 and pKNG101ΔPA4982, respectively. The suicide plasmids were introduced into P. aeruginosa via a three-partner procedure and the deletion mutants were obtained by double selection on LB agar supplemented with Irgasan (25 μg/mL) and streptomycin (1000 μg/mL) at 37°C and NaCl-free LB agar containing 6% sucrose at 30°C. The PAKΔhpAtΔhptBΔhptC triple mutant was constructed as follows: the hptA mutator cloned into the suicide pKNG101 vector [22] was introduced by mating into the PAKΔhptB leading to PAKΔhptAΔhptB. The hptC mutator cloned into the suicide pKNG101 vector was further introduced by mating into the PAKΔhptAΔhptB strain leading to the PAKΔhptAΔhptBΔhptC strain. The PAKgacSH1H→QΔladS and PAKgacSH1H→QH2H→QΔladS strains were constructed by introducing the pKNG101ΔladS vector by mating into the PAKgacSH1H→Q and the PAKgacSH1H→QH2H→Q strains, respectively.
Construction of chromosomal punctual mutants
For engineering strains: 1) PAKladSH1H→QD1 and PAKladSH1D1D→A harboring in their chromosomal copy of ladS gene a point mutation of the conserved histidine residue at position 428 or of the conserved aspartate residue at position 718, respectively, and 2) PAKgacSH1H→Q and PAKgacSH1 H→Q H2H→Q harboring in their chromosomal copy of gacS gene a point mutation of the conserved histidine at position 293 or of both conserved histidine residues at positions 293 and 859, respectively, the upstream and downstream sequences (approximately 500 bp) were amplified from PAK genomic DNA using the appropriate pairs of primers (S1 Table). Each PCR upstream and downstream product was linked together by overlapping PCR. For the ladS and the gacSH2H→Q variants, the PCR products were cloned into pCR2.1. Then, each linked DNA fragment was digested with XbaI and SpeI and cloned into the suicide vector pKNG101, yielding pKNGladSH1H→QD1, pKNGladSH1D1D→A and pKNGgacSH2H→Q, respectively. For the gacSH1H→Q variant, the PCR product was digested with ApaI and SpeI and cloned into the suicide vector pKNG101, yielding pKNGgacSH1H→Q. The suicide plasmids were introduced into the PAK strain and a double recombination event was selected using NaCl-free LB plates supplemented with 6% sucrose. The presence of mutations was checked by sequencing. For the gacSH1H→QH2H→Q variant, the pKNGgacSH1H→Q suicide vector was further introduced by mating into the PAKgacSH2H→Q strain to further obtain after the double recombination event the PAKgacSH1H→QH2H→Q strain.
Biofilm assay
The P. aeruginosa adherence assay was performed in individual glass tubes containing 1 mL of medium as described previously [22]. After 5 hours of incubation at 30°C, the cultures were incubated with 1% Crystal Violet for 10 min and washed twice. Staining was extracted by treatment with 400 μL 95% ethanol. Subsequently, 600 μL of water was added and OD570nm was measured. All quantification assays were performed at least in triplicate.
Bacterial two-hybrid experiments
The DNA regions encoding the HptA, HptB and HptC proteins, the H1 and D1 domains of LadS HK and the H2 domains of PA4112 and PA4982 HKs were PCR amplified by using PAK genomic DNA with appropriate oligonucleotide pairs (S1 Table). PCR product of HptA was digested by PstI/KpnI, while PCR products of HptB, HptC and H2 domains and of PA4112 and PA4982 HKs were digested by KpnI/XbaI and cloned into pKT25, yielding pKT25-hptA, pKT25-hptB, pKT25-hptC, pKT25-PA4112H2 and pKT25-PA4982H2, respectively. PCR product of the D1 domain of LadS HK was digested by PstI/ClaI and cloned into pUT18C, yielding pUT18C-ladSD1. The DNA region encoding the H1 domain of LadS HK was digested by XbaI/SacI and cloned into pKT25 or pUT18C vectors, yielding pKT25-ladSH1 and pUT18C-ladSH1, respectively. The adenylate cyclase-deficient E. coli strain BTH101 was used to screen for positive interactions [45,46]. BTH101 competent cells were transformed simultaneously with pKT25 and pUT18C derivatives and transformants were selected on agar plates supplemented with ampicillin (100 μg/mL) and kanamycin (50 μg/mL). Single colonies were spotted on solid medium, LB-agar plates supplemented with the chromogenic substrate X-gal (5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside, 40 μg/mL), isopropyl β-D-1-thiogalactopyranoside (IPTG) (100 μM), ampicillin (100 μg/mL) and kanamycin (50 μg/mL). Positive interactions were identified as blue colonies after 24 hours’ incubation at 30°C. The co-transformants of interest were grown in liquid LB medium at 37°C at 250 rpm for 16 hours. Five hundred μL of each culture was pelleted and mixed with 900 μL of Z buffer (10.7 g l−1 Na2HPO4 2H2O, 5.5 g l−1 NaH2PO4, 0.75 g l−1 KCl, 0.246 g l−1 MgSO4.7H2O, 2.7 mL l− 1β-mercaptoethanol, pH 7) before addition of 20 μL of 0.1(w/v) SDS and 100 μL of CHCl3 for permeabilization. A 40 μL volume of orthonitrophenyl-β-galactoside (ONPG) solution (4 mg/mL in Z buffer without β-mercaptoethanol) was added to 20 μL of permeabilized cells diluted in 180 μL of Z buffer, and β-galactosidase activity was then calculated and expressed in Miller units.
Pull-down experiments
A DNA fragment corresponding to H1 domains of GacS HK, RetS HK and LadS HK was amplified by PCR with an N-terminal FLAG or Strep tag (see S1 Table) and cloned in pCR2.1. The sequences of each construction were checked by sequencing and the DNA fragment of each H1 domain with FLAG tag was further digested with XbaI/SacI (LadS HK, RetS HK) or PstI/SacI (GacS HK) for subcloning into pBBRMCS3 vector. Production of each H1 FLAG- or Strep-tagged domain was checked in E. coli TG1 cells. The 16 following combinations were further examined: pBBR_FLAG-retSH1/pCR2.1_Strep-retSH1, pBBR_FLAG-ladSH1/pCR2.1_Strep-retSH1, pBBR_FLAG-gacSH1/pCR2.1_Strep-retSH1, pBBRMCS3/pCR2.1_Strep-retSH1, pBBR_FLAG-retSH1/pCR2.1_Strep-ladSH1, pBBR_FLAG-ladSH1/pCR2.1_Strep-ladSH1, pBBR_FLAG-gacSH1/pCR2.1_Strep-ladSH1, pBBRMCS3/pCR2.1_Strep-ladSH1, pBBR_FLAG-retSH1/pCR2.1_Strep-gacSH1, pBBR_FLAG-ladSH1/pCR2.1_Strep-gacSH1, pBBR_FLAG-gacSH1/pCR2.1_Strep-gacSH1, pBBRMCS3/pCR2.1_Strep-gacSH1, pBBR_FLAG-retSH1/pCR2.1, pBBR_FLAG-ladSH1/pCR2.1, pBBR_FLAG-gacSH1/pCR2.1 and pBBRMCS3/pCR2.1. Thirty milliliters of cell cultures at OD600nm 0.6 were cultured with 1 mM IPTG for 3 hours. Forty OD units of cells were harvested and resuspended in 2mL of 10 mM Tris buffer pH 8.0 supplemented with cOmplete, EDTA-free Protease Inhibitor Cocktail (Roche) and 100 mM NaCl. Ten μg/mL of DNase and RNase were added and cells were lysed by sonication. Total lysates were mixed with 50 μL of agarose beads coupled with antibody against Strep peptide (Streptactin Superflow IBA) and incubated on a rotating wheel for 1 hour at 4°C. The unbound fraction was collected by centrifugation for 2 min at 2000 rpm. Beads were washed three times with 1 mL of 10 mM Tris 50 mM NaCl, and were collected by centrifugation, resuspended in loading buffer and heated for 10 min at 95°C before analysis by SDS-PAGE and immunoblotting.
Overexpression and purification of proteins
Recombinant His-tagged LadSH1D1, LadSH1D1D→A, LadSD1, GacSH1D1, GacSH1, GacSD1, GacSH2, GacSH2H→Q and HptA proteins were purified from soluble extracts of the TG1 strain containing either pJF_ladSH1D1, pJF_ladSH1D1D→A, pJF_gacSH2, pLic03_ gacSH1D1, pLic03_ gacSH1, pLic03_gacSD1, pLic03_ladSD1, pJF_gacSH2H→Q or pJF_hptA. Cultures were grown aerobically at 37°C until OD600nm 0.6 and induced for 3 hours with 1 mM IPTG for recombinant proteins produced by the pJF119EH vector or 250 μM IPTG for recombinant proteins produced by the pLic03 vector. A one-step purification via affinity chromatography was facilitated by the presence of a His_Tag at the C-terminal extremity of LadSH1D1, LadSH1D1D→A, GacSH2, GacSH2H→Q and HptA and at the N-terminal extremity of LadSD1, GacSH1D1, GacSH1 and GacSD1, using nickel columns (HiTrap HP chelating column) as described by the manufacturer (GE healthcare). Proteins were eluted in an imidazole gradient buffer (20 mM to 500 mM) and analyzed by SDS-PAGE.
In vitro phosphorylation assay
Evidence of a phosphotransfer between LadS variants (LadSH1D1 and LadSH1D1D→A) or GacS variants (GacSH1D1 and GacSH1) and GacSH2 variants (GacSH2 and GacSH2H→Q), GacSD1 domain, LadSD1 domain or HptA protein was tested by in vitro phosphorylation assays. These assays were carried out in 10 μL of reaction buffer (50 mM Tris-HCl [pH 7.6], 50 mM KCl, 5 mM MgCl2, 1 mM dithiothreitol containing 0.1 mM [γ-32P] ATP) with 2 mM of purified proteins, and the mixture was incubated at room temperature for 20 min. The reaction was stopped by adding 5 μl of loading buffer (120 mM Tris–HCl (pH 8.8), 3.5 mM EDTA, 0.6 M sucrose, 0.06% (w/v) bromophenol blue, 6% (w/v) SDS, 0.1 M DTT and 1.6% (v/v) β-mercaptoethanol). All samples were analyzed by SDS-PAGE and radioactivity was revealed 12 hours after exposition by using a PhosphorImager screen (Molecular Dynamics).
Phosphotransfer kinetic experiments
The phosphotransfer kinetic between the LadSH1D1 or GacSH1D1 and GacS H2 domains was further followed in vitro. These assays were conducted as follows: 2 mM of purified LadSH1D1 or GacSH1D1 proteins was incubated at room temperature for 20 min in 10 μL of reaction buffer (50 mM Tris-HCl [pH 7.6], 50 mM KCl, 5 mM MgCl2, 1 mM dithiothreitol containing 0.1 mM [γ-32P] ATP). Then 2 mM of purified GacSH2 protein was added and the transphosphorylation reaction was stopped after 0, 0.5, 1, 2, 5, 10 or 30 min by adding 5 μl of loading buffer as described above. All samples were analyzed by SDS-PAGE and radioactivity was revealed 10 hours after exposition by using a PhosphorImager screen (Molecular Dynamics).
Western blot
Bacterial cell pellets were resuspended in loading buffer (Tris HCl 0.1M pH 8.8; EDTA 3mM; Saccharose 0.6M; SDS 6%; DTT 0.1 M; β-mercaptoethanol 1.5% bromophenol blue 0.03%). The samples were boiled and separated in SDS gels containing 20% acrylamide and blotted onto nitrocellulose membranes. After overnight saturation at 4°C in phosphate-buffered saline (PBS), 0.1% Tween 20 and 10% skimmed milk, the membrane was incubated for 1 hour in PBS 0.1% Tween 20, and 10% skimmed milk with appropriated antibodies (1:5,000 for pentaHis; 1:2,000 anti-VgrG1a and 1:2,500 for anti-Hcp1), washed three times with PBS 0.1% Tween 20, incubated for 1 hour in PBS 0.1% Tween 20, and 10% skimmed milk with anti-mouse conjugate HRP antibody (Sigma) (1:5,000), washed three times with PBS 0.1% Tween 20 and then revealed with a Super Signal Chemiluminescence system (Pierce).
Measurements of β-galactosidase activity
Strains carrying the lacZ transcriptional fusions were grown in LB under agitation at 37°C. The bacterial cells were collected by centrifugation at different growth times. The β-galactosidase activity was measured using the method of Miller. Experiments with strains carrying the exoS-lacZ fusion were performed similarly except that EGTA (5 mM) and MgCl2 (20 mM) were added in the growth medium.
RT-qPCR
The PAKΔgacS::miniTn7gacSH2 and PAKΔgacS::miniTn7gacSH2H→Q strains that had received the pBBRladS plasmid or the corresponding pBBRMCS4 empty vector and the PAK, PAKΔladS, PAKladSH1H→QD1 and PAKladSH1D1D→A strains were grown in the presence of EDTA at 37°C under agitation until OD600nm reached 4. Total cellular RNA from 10 mL of cultures was isolated, using the PureYield RNA Midiprep System (Promega), cleaned up and concentrated using the RNeasy kit (Qiagen). The yield, purity and integrity of RNA were further evaluated on Nanodrop and Experion devices. Reverse transcription was performed on 2 μg of RNA by using the SuperScript III first-strand synthesis system (Invitrogen). Real-time PCR runs were carried out on a CFX96 Real-Time System (Bio-Rad). Cycling parameters of the real-time PCR were 98°C for 2 min, followed by 45 cycles of 98°C for 5 s and 60°C for 10 s, ending with a melting curve from 65°C to 95°C to determine the specificity of the amplification. To determine the amplification kinetics of each product, the fluorescence derived from the incorporation of EvaGreen into the double-stranded PCR products was measured at the end of each cycle using a SsoFast EvaGreen Supermix 2X Kit (Bio-Rad). The results were analyzed using Bio-Rad CFX Manager Software 3.0 (Bio-Rad). The uvrD gene was used as a reference for normalization, in particular because transcription of uvrD is fairly stable in bacteria exposed to antibiotics even at relatively high concentrations [47].
Supporting Information
(A) Activities of the rsmY–lacZ (blue circles) and rsmZ–lacZ (brick-red-colored triangles) transcriptional chromosomal fusions were monitored at different growth stages in the PAK strain, which had received the pBBRMCS4 (empty symbols) or the pBBRladS (filled symbols) vectors. Corresponding β-galactosidase activities are expressed in Miller units and correspond to mean values (with error bars) obtained from three independent experiments. (B) RsmY (blue bars), RsmZ (brick-red-colored bars), VgrG1 (green bars), PelA (violet bars) and ExoS (royal blue bars) transcript levels were monitored using RT-qPCR and fold induction was presented in the strains PAK, PAKΔladS, PAKpBBRMCS4 and PAKpBBRladS. Moderated t-tests were performed and *, **, *** and ns referred to p<0.05, p<0.01 and p<0.001 and nonsignificant difference, respectively. (C) Number of LadS mRNA copies were expressed per μg of total RNA retrotranscribed in PAK, PAKΔladS, PAK + pBBRMCS4 and PAK + pBBRladS vectors. Mean values (with error bars) were obtained from three independent experiments. Wilcoxon-Mann-Whitney tests were performed and *** referred to p<0.001.
(TIF)
Production of each FLAG- or Strep-tagged proteins was detected in whole cell extracts using western blot using StrepTactin Alkaline Phosphatase conjugate (upper panel) and anti-FLAG antibody detection (lower panel).
(TIF)
(TIF)
Purified His-tagged forms of LadSH1D1, LadSH1D1D→A, GacSH2, GacSH2H→Q, HptA (A), LadSD1, GacSH1D1, GacSD1 and GacSH1 (B) proteins separated in an SDS-polyacrylamide gel stained with coomassie blue (upper panel) or detected by western blot using an anti-penta-His antibody (lower panel). Numbers on the left side are molecular weight standards (kDa) and locations of the recombinant proteins are indicated by arrowheads. In vitro phosphorylation assays of LadSH1D1, LadSH1D1D→A, GacSH2, GacSH2H→Q, HptA (C), LadSD1, GacSH1D1, GacSD1 and GacSH1 (D) proteins. Each protein was incubated with [γ-32P] ATP at room temperature for 20 min (see materials and methods) then resolved by an SDS-polyacrylamide gel and autoradiographied. Locations of the recombinant proteins are indicated by arrowheads.
(TIF)
Transcript levels of RsmY (blue bars) and RsmZ (brick-red-colored bars) were monitored in the PAK, PAKΔladS and PAKΔladSD1 strains using RT-qPCR and fold induction was presented for the two mutant strains as compared to the PAK strain. Moderated t-tests were performed; *, ** and *** referred respectively to p<0.05, p<0.01 and p<0.001.
(TIF)
(A) The hptA, hptB, hptC, rocS1H2, rocS2H2, PA4112H2, PA4982H2 and gacSH2 DNA regions were cloned into the two-hybrid pKT25 and the ladS-D1 DNA region was cloned into pUT18C. All of the pKT25 construction as well as the empty vector were co-transformed in BTH101 cells with pUT18C vector containing or not ladS-D1 DNA regions and β-galactosidase activities were measured after 16 hours of growth. All experiments were carried out in at least triplicate, and error bars represent standard deviation. (B) The pBBRladS plasmid containing the ladS HK gene (dark bars) and the pBBRMCS4 corresponding empty cloning vector (light bars) were conjugated in the PAK, PAKΔhptA, PAKΔhptB, PAKΔhptC, PAKΔhptAΔhptBΔhptC, PAKΔrocS1, PAKΔrocS2, PAKΔPA4112, PAKΔPA4982 and PAKΔgacS strains. Activity of the rsmZ–lacZ (brick-red-colored) transcriptional chromosomal fusion was monitored after 6 hours of growth (OD600nm≈4) and corresponding β-galactosidase activities are expressed in Miller units and correspond to mean values (with error bars) obtained from three independent experiments.
(TIF)
(DOCX)
Acknowledgments
We thank Yann Denis for technical assistance at the transcriptome platform of the Institut de Microbiologie de la Méditerranée and Ina Attrée-Delic for the gift of the Hcp1 antibody. We thank also Florence Vincent and Yves Bourne for their helpful discussion.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
SdB and CB are supported by the French Cystic Fibrosis Foundation (VLM), GDR3171, ANR grants: TWOCOMPNET ANR-05-MIIM-040-01, REGALAD ANR-14-CE09-0005-02 and ERA-NET ADHRES 27481. GC was supported by Aix-Marseille University. LR was supported by the French Cystic Fibrosis Foundation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Jung K, Fried L, Behr S, Heermann R. Histidine kinases and response regulators in networks. Curr Opin Microbiol. 2012;15: 118–24. 10.1016/j.mib.2011.11.009 [DOI] [PubMed] [Google Scholar]
- 2.Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69: 183–215. [DOI] [PubMed] [Google Scholar]
- 3.Buckler DR, Anand GS, Stock AM. Response-regulator phosphorylation and activation: a two-way street? Trends Microbiol. 2000;8: 153–6. Available: http://www.ncbi.nlm.nih.gov/pubmed/10754569 [DOI] [PubMed] [Google Scholar]
- 4.Tomich M, Mohr CD. Genetic characterization of a multicomponent signal transduction system controlling the expression of cable pili in Burkholderia cenocepacia. J Bacteriol. 2004;186: 3826–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gooderham WJ, Hancock REW. Regulation of virulence and antibiotic resistance by two-component regulatory systems in Pseudomonas aeruginosa. FEMS Microbiol Rev. 2009;33: 279–94. 10.1111/j.1574-6976.2008.00135.x [DOI] [PubMed] [Google Scholar]
- 6.Bordi C, de Bentzmann S. Hacking into bacterial biofilms: a new therapeutic challenge. Ann Intensive Care. 2011;1: 19 10.1186/2110-5820-1-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balasubramanian D, Schneper L, Kumari H, Mathee K. A dynamic and intricate regulatory network determines Pseudomonas aeruginosa virulence. Nucleic Acids Res. 2013;41: 1–20. 10.1093/nar/gks1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hood RD, Singh P, Hsu F, Güvener T, Carl MA, Trinidad RRS, et al. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe. 2010;7: 25–37. 10.1016/j.chom.2009.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mougous JD, Cuff ME, Raunser S, Shen A, Zhou M, Gifford CA, et al. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science. 2006;312: 1526–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moscoso JA, Mikkelsen H, Heeb S, Williams P, Filloux A. The Pseudomonas aeruginosa sensor RetS switches type III and type VI secretion via c-di-GMP signalling. Environ Microbiol. 2011;13: 3128–38. 10.1111/j.1462-2920.2011.02595.x [DOI] [PubMed] [Google Scholar]
- 11.Russell AB, Hood RD, Bui NK, LeRoux M, Vollmer W, Mougous JD. Type VI secretion delivers bacteriolytic effectors to target cells. Nature. 2011;475: 343–7. 10.1038/nature10244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bielecki P, Komor U, Bielecka A, Müsken M, Puchałka J, Pletz MW, et al. Ex vivo transcriptional profiling reveals a common set of genes important for the adaptation of Pseudomonas aeruginosa to chronically infected host sites. Environ Microbiol. 2013;15: 570–87. 10.1111/1462-2920.12024 [DOI] [PubMed] [Google Scholar]
- 13.Kitten T, Kinscherf TG, McEvoy JL, Willis DK. A newly identified regulator is required for virulence and toxin production in Pseudomonas syringae. Mol Microbiol. 1998;28: 917–29. Available: http://www.ncbi.nlm.nih.gov/pubmed/9663679 [DOI] [PubMed] [Google Scholar]
- 14.Rahme LG, Ausubel FM, Cao H, Drenkard E, Goumnerov BC, Lau GW, et al. Plants and animals share functionally common bacterial virulence factors. Proc Natl Acad Sci U S A. 2000;97: 8815–21. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=34017&tool=pmcentrez&rendertype=abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coleman FT, Mueschenborn S, Meluleni G, Ray C, Carey VJ, Vargas SO, et al. Hypersusceptibility of cystic fibrosis mice to chronic Pseudomonas aeruginosa oropharyngeal colonization and lung infection. Proc Natl Acad Sci U S A. 2003;100: 1949–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goodman AL, Kulasekara B, Rietsch A, Boyd D, Smith RS, Lory S. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev Cell. 2004;7: 745–54. [DOI] [PubMed] [Google Scholar]
- 17.Sall KM, Casabona MG, Bordi C, Huber P, de Bentzmann S, Attrée I, et al. A gacS Deletion in Pseudomonas aeruginosa Cystic Fibrosis Isolate CHA Shapes Its Virulence. PLoS One. 2014;9: e95936 10.1371/journal.pone.0095936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brencic A, McFarland KA, McManus HR, Castang S, Mogno I, Dove SL, et al. The GacS/GacA signal transduction system of Pseudomonas aeruginosa acts exclusively through its control over the transcription of the RsmY and RsmZ regulatory small RNAs. Mol Microbiol. 2009;73: 434–45. 10.1111/j.1365-2958.2009.06782.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lapouge K, Schubert M, Allain FH- T, Haas D. Gac/Rsm signal transduction pathway of gamma-proteobacteria: from RNA recognition to regulation of social behaviour. Mol Microbiol. 2008;67: 241–53. [DOI] [PubMed] [Google Scholar]
- 20.Brencic A, Lory S. Determination of the regulon and identification of novel mRNA targets of Pseudomonas aeruginosa RsmA. Mol Microbiol. 2009;72: 612–32. 10.1111/j.1365-2958.2009.06670.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burrowes E, Baysse C, Adams C, O’Gara F. Influence of the regulatory protein RsmA on cellular functions in Pseudomonas aeruginosa PAO1, as revealed by transcriptome analysis. Microbiology. 2006;152: 405–18. [DOI] [PubMed] [Google Scholar]
- 22.Bordi C, Lamy M- C, Ventre I, Termine E, Hachani A, Fillet S, et al. Regulatory RNAs and the HptB/RetS signalling pathways fine-tune Pseudomonas aeruginosa pathogenesis. Mol Microbiol. 2010;76: 1427–1443. 10.1111/j.1365-2958.2010.07146.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goodman AL, Merighi M, Hyodo M, Ventre I, Filloux A, Lory S. Direct interaction between sensor kinase proteins mediates acute and chronic disease phenotypes in a bacterial pathogen. Genes Dev. 2009;23: 249–259. 10.1101/gad.1739009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kong W, Chen L, Zhao J, Shen T, Surette MG, Shen L, et al. Hybrid sensor kinase PA1611 in Pseudomonas aeruginosa regulates transitions between acute and chronic infection through direct interaction with RetS. Mol Microbiol. 2013;88: 784–97. 10.1111/mmi.12223 [DOI] [PubMed] [Google Scholar]
- 25.Ventre I, Goodman AL, Vallet-Gely I, Vasseur P, Soscia C, Molin S, et al. Multiple sensors control reciprocal expression of Pseudomonas aeruginosa regulatory RNA and virulence genes. Proc Natl Acad Sci U S A. 2006;103: 171–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chatterjee A, Cui Y, Yang H, Collmer A, Alfano JR, Chatterjee AK. GacA, the response regulator of a two-component system, acts as a master regulator in Pseudomonas syringae pv. tomato DC3000 by controlling regulatory RNA, transcriptional activators, and alternate sigma factors. Mol Plant Microbe Interact. 2003;16: 1106–17. [DOI] [PubMed] [Google Scholar]
- 27.Records AR, Gross DC. Sensor kinases RetS and LadS regulate Pseudomonas syringae type VI secretion and virulence factors. J Bacteriol. 2010;192: 3584–96. 10.1128/JB.00114-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wei X, Huang X, Tang L, Wu D, Xu Y. Global control of GacA in secondary metabolism, primary metabolism, secretion systems, and motility in the rhizobacterium Pseudomonas aeruginosa M18. J Bacteriol. 2013;195: 3387–400. 10.1128/JB.00214-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Humair B, González N, Mossialos D, Reimmann C, Haas D. Temperature-responsive sensing regulates biocontrol factor expression in Pseudomonas fluorescens CHA0. ISME J. 2009;3: 955–65. 10.1038/ismej.2009.42 [DOI] [PubMed] [Google Scholar]
- 30.Laskowski MA, Kazmierczak BI. Mutational analysis of RetS, an unusual sensor kinase-response regulator hybrid required for Pseudomonas aeruginosa virulence. Infect Immun. 2006;74: 4462–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wise AA, Fang F, Lin Y- H, He F, Lynn DG, Binns AN. The receiver domain of hybrid histidine kinase VirA: an enhancing factor for vir gene expression in Agrobacterium tumefaciens. J Bacteriol. 2010;192: 1534–42. 10.1128/JB.01007-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Workentine ML, Chang L, Ceri H, Turner RJ. The GacS-GacA two-component regulatory system of Pseudomonas fluorescens: a bacterial two-hybrid analysis. FEMS Microbiol Lett. 2009;292: 50–6. 10.1111/j.1574-6968.2008.01445.x [DOI] [PubMed] [Google Scholar]
- 33.Zuber S, Carruthers F, Keel C, Mattart A, Blumer C, Pessi G, et al. GacS sensor domains pertinent to the regulation of exoproduct formation and to the biocontrol potential of Pseudomonas fluorescens CHA0. Mol Plant Microbe Interact. The American Phytopathological Society; 2003;16: 634–44. [DOI] [PubMed] [Google Scholar]
- 34.Mikkelsen H, McMullan R, Filloux A. The Pseudomonas aeruginosa reference strain PA14 displays increased virulence due to a mutation in ladS. PLoS One. 2011;6: e29113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laskowski MA, Osborn E, Kazmierczak BI. A novel sensor kinase-response regulator hybrid regulates type III secretion and is required for virulence in Pseudomonas aeruginosa. Mol Microbiol. 2004;54: 1090–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zolfaghar I, Angus AA, Kang PJ, To A, Evans DJ, Fleiszig SMJ. Mutation of retS, encoding a putative hybrid two-component regulatory protein in Pseudomonas aeruginosa, attenuates multiple virulence mechanisms. Microbes Infect. 2005;7: 1305–16. [DOI] [PubMed] [Google Scholar]
- 37.Vincent F, Round A, Reynaud A, Bordi C, Filloux A, Bourne Y. Distinct oligomeric forms of the Pseudomonas aeruginosa RetS sensor domain modulate accessibility to the ligand binding site. Environ Microbiol. 2010;12: 1775–1786. 10.1111/j.1462-2920.2010.02264.x [DOI] [PubMed] [Google Scholar]
- 38.Kenney LJ. How important is the phosphatase activity of sensor kinases? Curr Opin Microbiol. 2010;13: 168–76. 10.1016/j.mib.2010.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Capra EJ, Laub MT. Evolution of two-component signal transduction systems. Annu Rev Microbiol. 2012;66: 325–47. 10.1146/annurev-micro-092611-150039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Freeman JA, Lilley BN, Bassler BL. A genetic analysis of the functions of LuxN: a two-component hybrid sensor kinase that regulates quorum sensing in Vibrio harveyi. Mol Microbiol. 2000;35: 139–49. Available: http://www.ncbi.nlm.nih.gov/pubmed/10632884 [DOI] [PubMed] [Google Scholar]
- 41.Ansaldi M, Jourlin-Castelli C, Lepelletier M, Théraulaz L, Méjean V. Rapid dephosphorylation of the TorR response regulator by the TorS unorthodox sensor in Escherichia coli. J Bacteriol. 2001;183: 2691–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Georgellis D, Kwon O, De Wulf P, Lin EC. Signal decay through a reverse phosphorelay in the Arc two-component signal transduction system. J Biol Chem. 1998;273: 32864–9. Available: http://www.ncbi.nlm.nih.gov/pubmed/9830034 [DOI] [PubMed] [Google Scholar]
- 43.Ninfa AJ, Magasanik B. Covalent modification of the glnG product, NRI, by the glnL product, NRII, regulates the transcription of the glnALG operon in Escherichia coli. Proc Natl Acad Sci U S A. 1986;83: 5909–13. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=386406&tool=pmcentrez&rendertype=abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Becher A, Schweizer HP. Integration-proficient Pseudomonas aeruginosa vectors for isolation of single-copy chromosomal lacZ and lux gene fusions. Biotechniques. 2000;29: 948–50, 952 Available: http://www.ncbi.nlm.nih.gov/pubmed/11084852 [DOI] [PubMed] [Google Scholar]
- 45.Karimova G, Pidoux J, Ullmann A, Ladant D. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci U S A. 1998;95: 5752–6. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=20451&tool=pmcentrez&rendertype=abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Houot L, Fanni A, de Bentzmann S, Bordi C. A bacterial two-hybrid genome fragment library for deciphering regulatory networks of the opportunistic pathogen Pseudomonas aeruginosa. Microbiology. 2012. pp. 1964–1971. 10.1099/mic.0.057059-0 [DOI] [PubMed] [Google Scholar]
- 47.Jo JTH, Brinkman FSL, Hancock REW. Aminoglycoside efflux in Pseudomonas aeruginosa: involvement of novel outer membrane proteins. Antimicrob Agents Chemother. 2003;47: 1101–11. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=149301&tool=pmcentrez&rendertype=abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sastry PA, Pearlstone JR, Smillie LB, Paranchych W. Amino acid sequence of pilin isolated from pseudomonas aeruginosa PAK. FEBS Lett. 1983;151: 253–6. Available: http://www.ncbi.nlm.nih.gov/pubmed/6131838 [DOI] [PubMed] [Google Scholar]
- 49.Sivaneson M, Mikkelsen H, Ventre I, Bordi C, Filloux A. Two-component regulatory systems in Pseudomonas aeruginosa: an intricate network mediating fimbrial and efflux pump gene expression. Mol Microbiol. 2011;79: 1353–1366. 10.1111/j.1365-2958.2010.07527.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stathopulos J, Cambillau C, Cascales E, Roussel A, Leone P. Crystallization and preliminary X-ray analysis of the C-terminal fragment of PorM, a subunit of the Porphyromonas gingivalis type IX secretion system. Acta Crystallogr Sect F, Struct Biol Commun. 2015;71: 71–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hoang TT, Kutchma AJ, Becher A, Schweizer HP. Integration-proficient plasmids for Pseudomonas aeruginosa: site-specific integration and use for engineering of reporter and expression strains. Plasmid. 2000;43: 59–72. [DOI] [PubMed] [Google Scholar]
- 52.Choi K- H, Schweizer HP. mini-Tn7 insertion in bacteria with single attTn7 sites: example Pseudomonas aeruginosa. Nat Protoc. 2006;1: 153–61. [DOI] [PubMed] [Google Scholar]
- 53.Karimova G, Ullmann A, Ladant D. A bacterial two-hybrid system that exploits a cAMP signaling cascade in Escherichia coli. Methods Enzymol. 2000;328: 59–73. Available: http://www.ncbi.nlm.nih.gov/pubmed/11075338 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Activities of the rsmY–lacZ (blue circles) and rsmZ–lacZ (brick-red-colored triangles) transcriptional chromosomal fusions were monitored at different growth stages in the PAK strain, which had received the pBBRMCS4 (empty symbols) or the pBBRladS (filled symbols) vectors. Corresponding β-galactosidase activities are expressed in Miller units and correspond to mean values (with error bars) obtained from three independent experiments. (B) RsmY (blue bars), RsmZ (brick-red-colored bars), VgrG1 (green bars), PelA (violet bars) and ExoS (royal blue bars) transcript levels were monitored using RT-qPCR and fold induction was presented in the strains PAK, PAKΔladS, PAKpBBRMCS4 and PAKpBBRladS. Moderated t-tests were performed and *, **, *** and ns referred to p<0.05, p<0.01 and p<0.001 and nonsignificant difference, respectively. (C) Number of LadS mRNA copies were expressed per μg of total RNA retrotranscribed in PAK, PAKΔladS, PAK + pBBRMCS4 and PAK + pBBRladS vectors. Mean values (with error bars) were obtained from three independent experiments. Wilcoxon-Mann-Whitney tests were performed and *** referred to p<0.001.
(TIF)
Production of each FLAG- or Strep-tagged proteins was detected in whole cell extracts using western blot using StrepTactin Alkaline Phosphatase conjugate (upper panel) and anti-FLAG antibody detection (lower panel).
(TIF)
(TIF)
Purified His-tagged forms of LadSH1D1, LadSH1D1D→A, GacSH2, GacSH2H→Q, HptA (A), LadSD1, GacSH1D1, GacSD1 and GacSH1 (B) proteins separated in an SDS-polyacrylamide gel stained with coomassie blue (upper panel) or detected by western blot using an anti-penta-His antibody (lower panel). Numbers on the left side are molecular weight standards (kDa) and locations of the recombinant proteins are indicated by arrowheads. In vitro phosphorylation assays of LadSH1D1, LadSH1D1D→A, GacSH2, GacSH2H→Q, HptA (C), LadSD1, GacSH1D1, GacSD1 and GacSH1 (D) proteins. Each protein was incubated with [γ-32P] ATP at room temperature for 20 min (see materials and methods) then resolved by an SDS-polyacrylamide gel and autoradiographied. Locations of the recombinant proteins are indicated by arrowheads.
(TIF)
Transcript levels of RsmY (blue bars) and RsmZ (brick-red-colored bars) were monitored in the PAK, PAKΔladS and PAKΔladSD1 strains using RT-qPCR and fold induction was presented for the two mutant strains as compared to the PAK strain. Moderated t-tests were performed; *, ** and *** referred respectively to p<0.05, p<0.01 and p<0.001.
(TIF)
(A) The hptA, hptB, hptC, rocS1H2, rocS2H2, PA4112H2, PA4982H2 and gacSH2 DNA regions were cloned into the two-hybrid pKT25 and the ladS-D1 DNA region was cloned into pUT18C. All of the pKT25 construction as well as the empty vector were co-transformed in BTH101 cells with pUT18C vector containing or not ladS-D1 DNA regions and β-galactosidase activities were measured after 16 hours of growth. All experiments were carried out in at least triplicate, and error bars represent standard deviation. (B) The pBBRladS plasmid containing the ladS HK gene (dark bars) and the pBBRMCS4 corresponding empty cloning vector (light bars) were conjugated in the PAK, PAKΔhptA, PAKΔhptB, PAKΔhptC, PAKΔhptAΔhptBΔhptC, PAKΔrocS1, PAKΔrocS2, PAKΔPA4112, PAKΔPA4982 and PAKΔgacS strains. Activity of the rsmZ–lacZ (brick-red-colored) transcriptional chromosomal fusion was monitored after 6 hours of growth (OD600nm≈4) and corresponding β-galactosidase activities are expressed in Miller units and correspond to mean values (with error bars) obtained from three independent experiments.
(TIF)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.