Abstract
One of the most crucial steps in the life cycle of a retrovirus is the integration of the viral DNA (vDNA) copy of the RNA genome into the genome of an infected host cell. Integration provides for efficient viral gene expression as well as for the segregation of the viral genomes to daughter cells upon cell division. Some integrated viruses are not well expressed, and cells latently infected with HIV-1 can resist the action of potent antiretroviral drugs and remain dormant for decades. Intensive research has been dedicated to understanding the catalytic mechanism of integration, as well as the viral and cellular determinants that influence integration site distribution throughout the host genome. In this review we summarize the evolution of techniques that have been used to recover and map retroviral integration sites, from the early days that first indicated that integration could occur in multiple cellular DNA locations, to current technologies that map upwards of millions of unique integration sites from single in vitro integration reactions or cell culture infections. We further review important insights gained from the use of such mapping techniques, including the monitoring of cell clonal expansion in patients treated with retrovirus-based gene therapy vectors, or AIDS patients on suppressive antiretroviral therapy (ART). These insights span from integrase (IN) enzyme sequence preferences within target DNA (tDNA) at the sites of integration, to the roles of host cellular proteins in mediating global integration distribution, to the potential relationship between genomic location of vDNA integration site and retroviral latency.
Keywords: Retrovirus, HIV-1, integrase, intasome, integration sites, Illumina, next generation sequencing, gene therapy
Retroviruses are parasites that exploit the replication machinery of their hosts to form stable genetic domains, allowing their consistent replication throughout the lifespan of the infected host cell. Retroviral infection is currently incurable for multiple reasons. First, compounds developed to inhibit specific steps in the viral replication cycle lose potency over time due to the emergence of resistance mutations within the drug target. Second, even under pressure from suppressive ART, a latent reservoir of infected cells persists – the origin of which is not fully recognized. Latently infected cells are capable of persisting primarily because the integrated provirus is not expressed, making it invulnerable to current treatment methodologies. Therefore, the process of integration underlies the current incurability of retroviral infection. Over the last four decades, a wealth of knowledge has been uncovered about the catalytic process of integration, the involvement of host cells in facilitating this process, and the virus genus-specific distributions of integration sites in animal cell genomes. The key to developing this knowledge has been the ability to specifically recover and characterize retroviral integration sites from the bulk of background host genomic DNA (gDNA). Methodologies to accomplish this feat have been exponentially optimized through time. Initial success was gained by detecting proviruses in different locations within infected tissue culture cells, whereas the current state of technology can resolve millions of integration events from a single infection experiment. In this review we summarize the historical improvement of integration site detection techniques, as well as important insights about the mechanics of the integration process. These insights have significantly impacted not only antiretroviral efforts, but also the understanding of cancer gene networks and the development and improvement of gene therapy vectors.
The catalytic process of retroviral integration
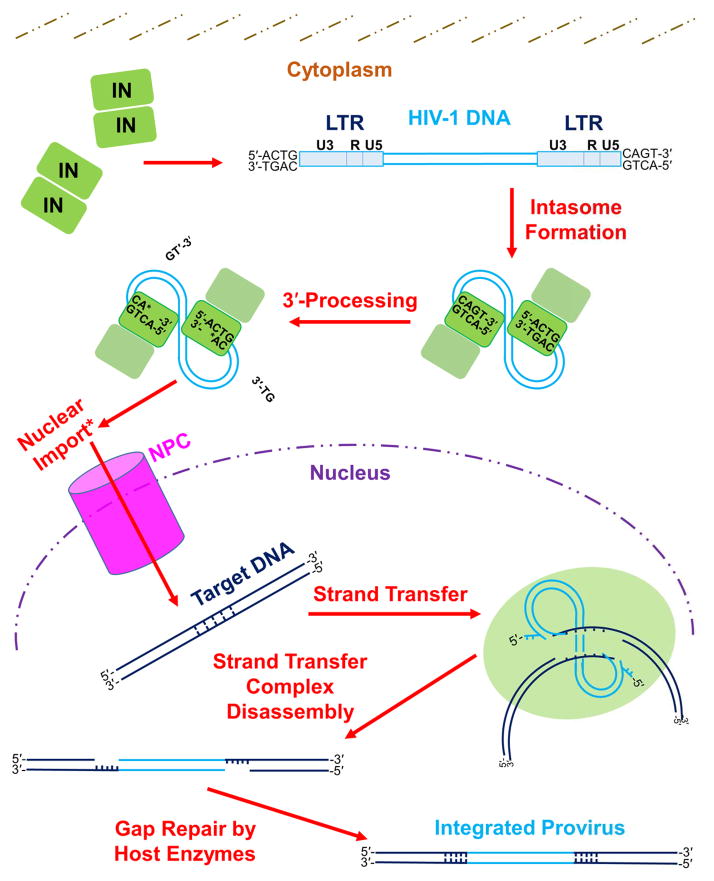
Soon after entering a susceptible target cell, the viral enzyme reverse transcriptase replicates the two plus-sense copies of the viral RNA (vRNA) genome into a linear, double stranded DNA molecule containing a copy of the viral long terminal repeat (LTR) at each end (Hu and Hughes, 2012). The INs catalyze two separate activities, which are known as 3′-processing and DNA strand transfer, to accomplish vDNA integration (Craigie and Bushman, 2012). The catalytic process (depicted for HIV-1 in Figure 1) begins as units of IN within the viral preintegration complex (PIC) multimerize upon the nascent vDNA termini to form the intasome, the IN-vDNA complex from wherein IN catalyzes integration (Chen et al., 1999, Li et al., 2006, Hare et al., 2010, Hare et al., 2012). The X-ray crystal structure of the prototype foamy virus (PFV) intasome has revealed the active configuration of this IN to be a tetramer (Hare et al., 2010, Maertens et al., 2010, Hare et al., 2012), and HIV-1 (Wang et al., 2001, Faure et al., 2005, Li et al., 2006, Bera et al., 2009, Krishnan et al., 2010, Kessl et al., 2011) and α-retroviral avian sarcoma-leukosis virus (ASLV) (Yang et al., 2000, Bao et al., 2003) IN have also been proposed to function as tetramers. IN cleaves the LTR ends of the vDNA adjacent to conserved 5′-CA-3′ dinucleotides during 3′-processing to yield recessed, chemically reactive CAOH-3′ hydroxyl groups (Fujiwara and Mizuuchi, 1988, Roth et al., 1989, Brown et al., 1989, Pauza, 1990, Lee and Coffin, 1991, Hare et al., 2012). Once inside the cell nucleus, the intasome docks with host cellular tDNA to form the pre-catalytic target capture complex (TCC), with the recessed CAOH ends of vDNA poised for chemical attack (Hare et al., 2012, Maertens et al., 2010). During DNA strand transfer, the CAOH-3′ termini are used by IN to cleave the complementary strands of tDNA in a staggered fashion and simultaneously join the vDNA ends to the tDNA 5′-phosphates (Engelman et al., 1991). Due to the nature of the staggered tDNA cleavage, the resulting DNA recombination intermediate contains single stranded tDNA gaps adjacent to the joined vDNA ends (Fujiwara and Mizuuchi, 1988, Brown et al., 1989). These single stranded gaps are repaired by host cell enzymes subsequent to disassembly of the strand transfer complex (STC), which for HIV-1 yields a 5-bp target site duplication (TSD) flanking the stably integrated provirus (Vincent et al., 1990, Vink et al., 1990). The spacing between staggered top and bottom tDNA strand cuts during strand transfer differs depending on the type of retrovirus, and TSDs accordingly range in length from 4-bp for spumaviruses and γ-retroviruses to 6-bp for α-, β-, and δ-retroviruses (Shimotohno et al., 1980, Dhar et al., 1980, Hughes et al., 1981, Majors and Varmus, 1981, Neves et al., 1998, Kim et al., 2010). The size of the TSD that is generated by ε-retroviral integration is currently unknown.
Figure 1. The mechanism of retroviral integration.
The integration process begins in the host cell with formation of the intasome, which consists of multimerized IN bound to the viral LTR ends. The LTR is composed of U3, R, and U5 sections, each of which contains unique elements responsible for mediating viral gene transcription (Pereira et al., 2000). While within the cytoplasm, IN cleaves nucleotides (specifically two nucleotides for the depicted HIV-1 integration pathway) from each 3′ end of the vDNA adjacent to invariant 5′-CA-3′ dinucleotides during 3′-processing. Completion of 3′-processing leaves a recessed and chemically reactive hydroxyl group at each vDNA 3′ end. Lentiviral PICs are actively imported into the cell nucleus (denoted by an asterisk in figure), while γ-retroviruses can only enter the nucleus after dissolution of the nuclear membrane during cell division [reviewed in: (Matreyek and Engelman, 2013)]. Within the nucleus, IN binds tDNA and utilizes the reactive vDNA 3′-hydroxyl groups to simultaneously cleave the tDNA phosphodiester backbone and insert the vDNA molecule, through the process of strand transfer. IN subunits cleave the top and bottom tDNA strands in a staggered fashion. The length of stagger differs among retroviruses; HIV-1 IN cleaves tDNA with the depicted 5-bp stagger. After disassembly of the strand transfer complex, host cell enzymes repair the DNA recombination intermediate to yield the integrated provirus flanked by a host DNA TSD. See main text for descriptions of the roles that other cellular factors play in the process of retroviral DNA integration. Please note that a color version of Figure 1 is available online.
The lentiviruses, which include HIV-1, can efficiently infect non-dividing target cells whereas other types of retroviruses, typified by the γ-retrovirus Moloney murine leukemia virus (Mo-MLV), cannot (Lewis et al., 1992, Roe et al., 1993, Lewis and Emerman, 1994). This biology is reflective of the mechanisms used by different types of retroviruses to access the nuclear environment: whereas HIV-1 PICs are actively transported through the nuclear pore complex (NPC) (Bukrinsky et al., 1992), Mo-MLV PICs require the dissolution of the nuclear membrane in order to access the chromosomal targets for vDNA integration [reviewed in: (Matreyek and Engelman, 2013)]. The ability to usurp the nuclear transport system of the cell is apparently shared by additional types of retroviruses. For examples, the β-retrovirus mouse mammary tumor virus (MMTV) has been reported to transduce growth-arrested cells as efficiently as HIV-1 (Konstantoulas and Indik, 2014), whereas ASLV possesses a phenotype that is intermediary to those of Mo-MLV and HIV-1 (Hatziioannou and Goff, 2001, Katz et al., 2002). HIV-1 integration tends to occur into chromatin that is affiliated with the nuclear periphery, indicating that PIC transport through the NPC and integration may be mechanistically linked (Di Primio et al., 2013, Marini et al., 2015, Lelek et al., 2015).
Early techniques for detecting retroviral integration
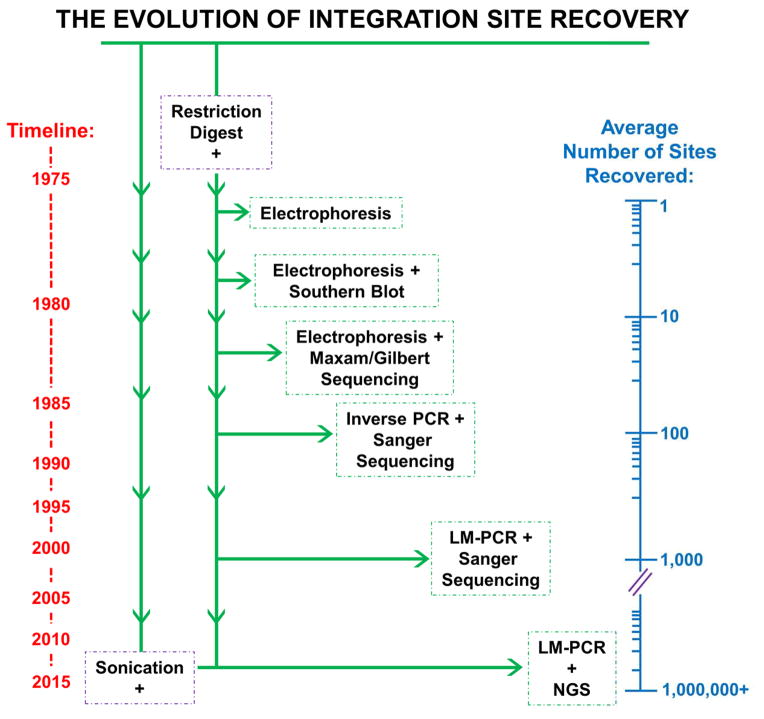
Techniques for retrieving proviral DNA from infected cells and mapping the integration sites have matured greatly over the last four decades, leading to exponential increases in the efficiencies through which sites were recovered (Figure 2). The capacity of cellular gDNA fragments that were made by restriction endonuclease digestion to form infectious virus was established in some of the earliest work (Battula and Temin, 1977, Battula and Temin, 1978). As activity mapped to several gDNA fragments, these studies importantly proved that integration could occur at more than a single site in the host genome. Experimental resolution was incrementally increased in subsequent studies when the digested and electrophoretically separated gDNA was subjected to Southern blotting using vDNA probes (Keshet and Temin, 1978, Steffen and Weinberg, 1978, Bacheler and Fan, 1979, Cohen et al., 1979, Ringold et al., 1979, Peters et al., 1986). The increased sensitivity of these experiments further solidified the understanding that retroviruses can integrate into multiple gDNA locations, but distinguishing between random integration and integration into a large number of preferred sites was not possible at this point. Preliminary insight into integration distribution was gleaned as host gDNA was fractionated based on various parameters such as length or GC content prior to Southern blotting (Franklin et al., 1983, Salinas et al., 1987). This for the first time allowed assignment of integration sites to certain chromosomes or GC-rich gDNA regions.
Figure 2. The evolution of techniques used to determine retroviral integration sites.
Integration site recovery techniques are listed in chronological order from top to bottom, corresponding to a general timeline shown at the left of the figure. The average numbers of integration sites recovered in studies employing these techniques, which exponentially increased over time, are illustrated on a logarithmic axis at the right side of the figure. The initial step of host cell gDNA fragmentation by restriction endonuclease digestion is shared by all of the listed techniques, though sonication has recently become a desirable method of shearing. Please note that a color version of this figure is available online.
The ultimate marriage of next-generation sequencing and ligation-mediated PCR
DNA sequencing technology was obviously critical to obtain integration site sequences. Initial sequences of retroviral integration sites were determined by Maxam Gilbert chemical cleavage (Maxam and Gilbert, 1977) or dideoxy sequencing (Sanger et al., 1977, Maat and Smith, 1978) using fractionated gDNA fragments following restriction endonuclease digestion (Shimotohno et al., 1980, Shimotohno and Temin, 1980, Dhar et al., 1980, Hughes et al., 1981, Fitts and Temin, 1983). These studies laid important groundwork for the classification of different retroviral TSDs, and for the first time highlighted that integration was related to DNA transposition as transposition was known to generate short TSDs flanking these mobile genetic elements (Calos et al., 1978, Johnsrud et al., 1978, Kleckner, 1979). The subsequent development of the polymerase chain reaction (PCR) provided a boon for the burgeoning field of retroviral integration site selection, as the technique greatly simplified the recovery and characterization of specific vDNA-gDNA junctions (Figure 2). Inverse PCR (Ochman et al., 1988, Triglia et al., 1988) was an early application of the technique to integration site sequencing. Specifically, it was used to amplify self-ligated gDNA fragments using outward-facing primers complementary to a known sequence, for example the viral LTR, prior to molecular cloning and dideoxy sequencing (Silver and Keerikatte, 1989, Jin et al., 2002). Using forward and reverse PCR primers that annealed to two different known sequences afforded linear amplification of vDNA-gDNA junctions. An early example of this approach included providing Mo-MLV with a characterized tDNA substrate by co-infecting cells with the SV40 polyomavirus that reaches high copy number as an unintegrated episome in cell nuclei (Pryciak et al., 1992). SV40-specific primers could be used in tandem with Mo-MLV LTR primers to amplify integration sites that occurred into this excess tDNA source. An alternative to viral co-infection involved designing primers to pseudo-randomly prime off of gDNA sequences when integration happened to occur within range (Sorensen et al., 1993, Butler et al., 2001, Gentner et al., 2003). Such work formed the basis of the so-called Alu real-time PCR assays for determining the bulk level of HIV-1 integration in cell culture and in patient samples (Brussel et al., 2005, De Spiegelaere et al., 2014). However, the major breakthrough in recovery efficiency of individual integration sites occurred with the development of ligation-mediated PCR, or LM-PCR (Rosenthal and Jones, 1990, Cavrois et al., 1995). In this method gDNA is digested with restriction enzymes, and then an oligo-cassette (also called linker) containing a compatible “sticky” end is ligated to all digestion fragments. PCR amplification of integration sites is then accomplished using primers complementary to the viral LTR and the linker sequence (Vandegraaff et al., 2001, Schroder et al., 2002, Wu et al., 2003). Along with the completion of the human, mouse, and other host genome reference sequences, this marriage of library construction and DNA sequencing led to a wealth of insights into the integration site distribution patterns of different types of retroviruses (Schroder et al., 2002, Wu et al., 2003, Mitchell et al., 2004).
The combination of LM-PCR amplification with powerful next-generation sequencing (NGS) platforms such as pyrosequencing from 454 Life Sciences (Margulies et al., 2005, Wang et al., 2007) and DNA cluster-based sequencing from Illumina (Gillet et al., 2011) sent the number of recoverable integration sites skyrocketing, from the low hundreds prior to the use of NGS to ultimately hundreds of thousands and millions (LaFave et al., 2014, De Ravin et al., 2014, Maskell et al., 2015) (Figure 2). Substituting sonication for restriction endonuclease digestion as the way to fragment gDNA (Gillet et al., 2011) has additionally afforded the monitoring of identical retroviral integration sites from unique gDNA sources. With NGS, it can be difficult to ascertain if the repeated determination of a unique integration site was due to preferential PCR amplification during library construction or something more interesting, for example the presence of the exact same integration site among different cells in the queried population. Sonication, which cleaves gDNA in a relatively sequence-independent manner, has afforded the identification of unique linker attachment points on multiple sequence reads with the identical integration site (Gillet et al., 2011, Maldarelli et al., 2014, Wagner et al., 2014).
Retroviruses integrate at multiple gDNA locations
As the initial details of retroviral replication were being uncovered (Temin, 1971, Bishop, 1975, Vogt, 1977) it became a point of interest to determine where in the host genome integration occurred and whether the distribution of proviral DNA was random, biased toward certain genomic regions, or perhaps targeted to a limited set of loci. Southern blotting as mentioned provided some insight, as digested gDNA exhibited multiple different restriction patterns when queried with virus-specific probes (Kettmann et al., 1979, Gilmer and Parsons, 1979). The detection of differently sized restriction fragments implied integration into unique regions of the genome, and therefore integration was clearly not always directed to the same place. But did this mean that integration occurred randomly, or rather that specific regions or annotations were favorable? A primer extension-based assay that queried the frequency of integration into a handful of regions of the chicken cell genome indicated that most of that genome was accessible for integration (Withers-Ward et al., 1994), but numerous studies using finer mapping techniques have since determined that distribution is nonrandom by uncovering virus-specific preferences for particular tDNA sequences and chromosomal annotations.
Mechanism of retroviral tDNA base preferences
Retroviruses exhibit reproducible tDNA nucleotide preferences at integration sites (Shih et al., 1988, Chou et al., 1996, Bor et al., 1996, Stevens and Griffith, 1996, Leclercq et al., 2000, Holman and Coffin, 2005, Wu et al., 2005, Valkov et al., 2009, Ballandras-Colas et al., 2013, Serrao et al., 2015). Although these base preferences are relatively weakly conserved, the mechanics behind their selectivity has been resolved for different retroviruses. The groundbreaking crystallization of PFV TCC and STC structures revealed that IN accommodates tDNA with a severe central kink, which facilitates strand transfer by unstacking the base pair at the very center of the targeted stretch of DNA to place scissile phosphodiester bonds at the IN active sites for strand transfer (Maertens et al., 2010). Alignment of PFV integration sites accordingly revealed preference for the most flexible dinucleotides (pyrimidine/purine, or YR) – and selection against the most rigid dinucleotides (purine/pyrimidine, or RY) – at the central position. Target DNA distortion did not involve IN interactions with the flexible, unstacked central base pair, but rather with distal nucleotides at either end of the 4-bp TSD region. PFV IN residues Ala188 and Arg329 mediated side chain-specific contacts with the tDNA, and the sequencing of in vitro integration reaction products that were generated using IN mutant proteins resulted in tDNA base preferences that were altered in ways predicted by the nucleoprotein interactions observed in the crystal structures (Maertens et al., 2010).
HIV-1 IN-tDNA interactions analogous to those illuminated in the PFV intasome crystal structures have been shown to similarly motivate the selection of specific, flexible dinucleotides at the centers of these integration sites (Serrao et al., 2014). As mentioned previously, HIV-1 integration generates 5-bp TSDs, which a dinucleotide step analysis revealed to on average be composed of RYXRY (where X is any base). As was determined for PFV, this specific signature enforces for flexible YR dinucleotides at the two center positions of the 5-bp TSD while selecting against rigid RY dinucleotides at these positions. The structural mechanics of HIV-1 base preferences also resembled those of PFV, as HIV-1 IN residue Ser119 (analogous to PFV IN residue Ala188) was responsible for determining analogous base preferences relative to the points of vDNA insertion (Serrao et al., 2014). This finding was in line with prior (Appa et al., 2001, Harper et al., 2001, Nowak et al., 2009) and subsequent (Demeulemeester et al., 2014) studies that implicated Ser119 in the process of HIV-1 tDNA site selection. The analogous residue in Mo-MLV IN, Pro187, plays the same role as Ala188 in PFV IN and Ser119 in HIV-1 in determining tDNA base selectivity (Aiyer et al., 2015).
A meta-analysis of thousands of integration sites generated by 12 different retroviruses has revealed significant enrichment for flexibility signatures at the central positions of integration sites across the studied viruses (Serrao et al., 2015). The extent of central tDNA flexibility was moreover inversely proportional to TSD length. The examined viruses harbored a neutral, compact amino acid at the position analogous to Ala188 in PFV, and the polarity of the amino acid side chain correlated with the positioning of base preference significance relative to the points of vDNA insertion – a finding that was since confirmed by analyzing the behavior of mutants of the non-polar Pro187 side-chain in Mo-MLV IN (Aiyer et al., 2015). Taken together these studies imply that, though retroviral INs have structurally evolved to target unique nucleotide signatures, the common functional purpose of integration site base preferences may be to generate strand transfer-facilitating central tDNA distortion within the TCC. In this vein, the degenerate nature of tDNA base preference conservation at retroviral integration sites in large part reflects the multitude of nucleotide combinations that on average spawn central (YR) flexibility signatures. Moreover, retroviruses that generate 6-bp TSDs may need to bend tDNA less rigorously to facilitate strand transfer than the viruses that generate 4-bp and 5-bp TSDs (Serrao et al., 2015).
Integration is favored within flexible, nucleosome-bound tDNA
Some of the earliest reports of linkage between integration sites and genomic features involved the association of Mo-MLV and ASLV integration with DNase I hypersensitive sites and actively transcribed regions of the genome (Robinson and Gagnon, 1986, Vijaya et al., 1986, Rohdewohld et al., 1987, Scherdin et al., 1990, Mooslehner et al., 1990, Lewinski et al., 2006, Roth et al., 2011). The level of cellular gene transcriptional activity has since been demonstrated to positively correlate with the integration targeting of additional retroviruses (Zoubak et al., 1994, Jordan et al., 2001, Schroder et al., 2002, Mack et al., 2003, Mitchell et al., 2004, Lewinski et al., 2005, Trobridge et al., 2006, Monse et al., 2006, Kang et al., 2006, Moalic et al., 2006, Hacker et al., 2006). Bent DNA – and specifically DNA bent around nucleosomes – is a highly favored substrate for integration (Milot et al., 1994, Pruss et al., 1994a, Muller and Varmus, 1994, Pruss et al., 1994b, Wang et al., 2007, Roth et al., 2011, Serrao et al., 2015, Naughtin et al., 2015). This may seem counterintuitive, as on the one hand DNase I hypersensitivity and gene transcription are both correlated with open, accessible chromatin (which would be logical for IN to be attracted to), while nucleosomes on the other hand are found with the highest density in heterochromatin [see (Bell et al., 2011) for review]. Also intriguing is that chromatin density in the regions surrounding integration sites can negatively modulate the DNA strand transfer activity of some retroviral INs, while having the opposite effect on other proteins (Taganov et al., 2004, Lesbats et al., 2011, Benleulmi et al., 2015). It nonetheless appears that, regardless of viral preferences for chromatin density in the region surrounding the average integration site, distorted phosphodiester bonds are generally favored by IN at the points of integration (Maertens et al., 2010, Serrao et al., 2014, Serrao et al., 2015, Aiyer et al., 2015), and nucleosomes provide for natural sources of DNA distortion. The mechanism of how nucleosome structure could accommodate the natural propensity to integrate into flexible tDNA sequences was recently revealed through the cryo-electron microscopy structure of the PFV intasome in complex with a bound nucleosome (Maskell et al., 2015). The structure revealed novel interactions between IN amino acids and one H2A-H2B heterodimer, as well as a secondary gyre of wrapped tDNA that was separate from where integration occurred. These interactions afforded a ~7 Å separation of the tDNA sequence from the histone surface, allowing the formation of the severe central bend in tDNA that was observed in the X-ray crystal structure of the TCC with naked tDNA (Maskell et al., 2015).
It is noteworthy that the propensity to integrate into nucleosomal tDNA may not be shared among all retroviruses. In particular, tDNA sequences surrounding the sites of porcine endogenous retrovirus (PERV) and MMTV integration did not exhibit characteristics of nucleosomal DNA, indicating that these two viruses on average select for sites that are not associated with chromatinization (Serrao et al., 2015). While MMTV integration yields 6-bp TSDs, PERV generates 4-bp TSDs. PERV accordingly seems to seek out optimally distorted tDNA without the assistance of nucleosome docking, possibly due to unique IN-tDNA contacts mediated by this intasome.
Retroviruses exhibit unique preferences for genomic annotations
The answer to the question of “why” some retroviruses (Mitchell et al., 2004, Narezkina et al., 2004, Faschinger et al., 2008, de Jong et al., 2014, Konstantoulas and Indik, 2014) favor integration in or nearby genes that exhibit relatively high expression levels is not completely understood, though enhancement of viral transcription is a logical assumption (Jahner and Jaenisch, 1980, Fincham and Wyke, 1991, Zoubak et al., 1994, Jordan et al., 2001, Jordan et al., 2003, Bushman, 2003, Lewinski et al., 2005, Meekings et al., 2008). Despite this, a lot has been learned about the question of “how” integration targeting is accomplished. Cellular cofactors that engage viral PICs can guide integration to distinct areas of the host genome [recently reviewed in (Kvaratskhelia et al., 2014, Craigie and Bushman, 2014, Debyser et al., 2015)]. Dating back to the initial discoveries that HIV-1 and Mo-MLV integration sites cluster within gene bodies (Schroder et al., 2002) and promoters (Wu et al., 2003), respectively, measurements of integrations within RefSeq genes or in proximity to promoter-associated features such as transcription start sites (TSSs) and CpG islands have been used as traditional metrics to quantitatively compare integration site distribution phenotypes among different viruses. Figure 3 provides a summary of the genomic integration site preferences of multiple retroviral genera with respect to the structure of an average transcription unit. Gammaretroviruses such as Mo-MLV exhibit the greatest extent of TSS and CpG targeting of all viral genera (Scherdin et al., 1990, Wu et al., 2003, Mitchell et al., 2004, Moalic et al., 2006, Tsukahara et al., 2006, Dong et al., 2007, Kim et al., 2008, Moalic et al., 2009, De Ravin et al., 2014, LaFave et al., 2014, Serrao et al., 2015), while the lentiviruses target RefSeq transcription units and gene-dense regions of the genome more so than other retroviruses (Schroder et al., 2002, Mack et al., 2003, Mitchell et al., 2004, Crise et al., 2005, Hacker et al., 2006, Kang et al., 2006, Monse et al., 2006, Marshall et al., 2007). The majority of gene-tropic lentiviral integrations occur within introns (Han et al., 2004, Ikeda et al., 2007, Shan et al., 2011, Maldarelli et al., 2014, Wagner et al., 2014, Cohn et al., 2015), presumably due to the fact that introns are generally much larger than exons. The α-, β-, and δ-retroviruses display more random distributions relative to genes and promoter-associated annotations than do either the lenti- or γ-retroviruses (Mitchell et al., 2004, Narezkina et al., 2004, Derse et al., 2007, Faschinger et al., 2008, Konstantoulas and Indik, 2014), while the spumavirus PFV displays the unique phenotype to preferentially avoid gene bodies yet target promoters at a level that is intermediate to the γ- and lentiviruses (Trobridge et al., 2006, Nowrouzi et al., 2006, Maskell et al., 2015). Thus, it is apparent that different cellular forces help to govern the integration site targeting of different classes of retroviruses.
Figure 3. General integration site preferences of different retroviral genera.
Representational integration site patterns with respect to the structure of the average host cell gene are depicted by vertical arrows, with each arrow type corresponding to a different viral genus (or group of genera in the case of α-, β-, and δ-retroviruses). Gammaretroviruses preferentially target active promoters and enhancers (Wu et al., 2003, LaFave et al., 2014, De Ravin et al., 2014) while lentiviruses integrate primarily within the bodies of actively transcribed genes (Schroder et al., 2002). Spumaviruses avoid genes and rather target intergenic regions, while α-, β-, and δ-retroviruses exhibit nearly random distributions. As mentioned in the text, additional features not depicted in the figure can also influence the integration sites of various viruses, such as gene transcription level, proximity to the NPC, and local nucleotide sequence.
The involvement of LEDGF/p75 in tethering lentiviral integration to genes
Integration site targeting by host factors is most thoroughly understood at present for the lentiviruses. A variety of cellular proteins have been characterized for their ability to bind HIV-1 IN, stimulate its catalytic activity in vitro, and to potentially play a role in HIV-1 infection [reviewed in (Van Maele et al., 2006, Engelman, 2007)], but none more so than lens epithelium-derived host factor/p75 (LEDGF/p75). LEDGF/p75 was originally discovered to associate with ectopically expressed HIV-1 IN in HEK293T cells and to stimulate HIV-1 IN catalytic activity in vitro (Cherepanov et al., 2003). Subsequent work revealed that the binding of LEDGF/p75 to IN is lentivirus-specific (Llano et al., 2004, Busschots et al., 2005, Cherepanov, 2007) and that LEDGF/p75 is critical for efficient lentiviral replication (Llano et al., 2006a, Vandekerckhove et al., 2006, Shun et al., 2007, Schrijvers et al., 2012a, Fadel et al., 2014).
LEDGF/p75 functions as a bimodal tether to target the lentiviral intasome to specific regions of chromatin. On one side of the tether is the N-terminal region of LEDGF/p75, which contains a PWWP (Pro-Trp-Trp-Pro) domain – a type of Tudor domain – as well as charged regions and a pair of AT-hook DNA binding motifs that work together to confer constitutive chromatin association (Turlure et al., 2006, Llano et al., 2006b). The PWWP domain in particular recognizes histone 3 tails that are tri-methylated on Lys-36 (H3K36me3) (Pradeepa et al., 2012, Eidahl et al., 2013), an epigenetic mark that positively correlates with transcriptional elongation (Bannister et al., 2005) and sites of HIV-1 integration (Roth et al., 2011). The C-terminal side of the tether contains the IN-binding domain (IBD), which is necessary and sufficient to bind HIV-1 IN in vitro (Cherepanov et al., 2004). Single amino acid substitutions at either end of the tether, for example the W21A change in the PWWP domain or the D366N change in the IBD, are sufficient to effectively break tethering function and render the mutant LEDGF/p75 protein unable to support HIV-1 integration (Cherepanov et al., 2005, Llano et al., 2006a, Shun et al., 2007, Shun et al., 2008, Botbol et al., 2008).
The involvement of LEDGF/p75 in genic integration distribution was validated by depleting the protein from cells prior to lentiviral infection using either RNA interference (Ciuffi et al., 2005) or gene knockout (Shun et al., 2007, Marshall et al., 2007, Schrijvers et al., 2012a). Under these conditions integration into transcription units was reduced, concomitant with increases in integration in the vicinity of normally disfavored promoter-associated features such as TSSs and CpG islands. Although lentiviral infectivity could be considerably reduced by LEDGF/p75 depletion, significant levels of virus infection remained, and the distributions of the residual proviruses were far from random. This implied that although LEDGF/p75 plays an important integration co-factor role, it is not essential for lentiviral infection or integration, and that additional host cell factors likely play supporting roles in directing integration to genic regions. LEDGF/p75 is a member of the hepatoma-derived growth factor (HDGF) related protein (HRP) family, which is defined by similarities amongst N-terminal PWWP domains (Izumoto et al., 1997). One other HRP family member, HRP2, additionally harbors an IBD (Cherepanov et al., 2004), which made it a prime suspect for a role in gene-tropic lentiviral DNA integration. Removal of HRP2 from LEDGF/p75 knockout cells, either by RNA interference or through its own knockout, added to the overall HIV-1 infectivity defect and altered the pattern of integration sites more towards random (Wang et al., 2012, Schrijvers et al., 2012b). However, residual targeting of integration to genes was still apparent, and knockout of HRP2 on its own failed to negatively affect either HIV-1 titer or integration site usage (Wang et al., 2012). The fact that LEDGF/p75 needed to be removed from the system to bring out a subsidiary role for HRP2 indicates that HRP2 normally plays little if any role in determining the sites of HIV-1 integration. At the same time, it became clear that IN is not the only viral determinant of HIV-1 integration site selection, as the interplay among the viral capsid (CA) and CA-binding host proteins has also been shown to play a significant role.
The role of HIV-1 CA-binding host factors in integration targeting
CA forms the protective core that encases retroviral vRNA genomes in association with nucleocapsid protein within the virus particles. Though the core is likely to partially dissolve during the early phase of infection to enable reverse transcription (Hulme et al., 2011, Yang et al., 2013) and trafficking to the nucleus, it is apparent that some fraction of HIV-1 CA remains associated with the PIC as it enters into the cell nucleus (Hulme et al., 2015, Peng et al., 2015). A number of host cell factors that associate with CA, for example cyclophilin A (CypA), the β-karyopherin transportin 3 (TRN-S2 or TNPO3), and nucleoporins (NUPs) 358 and 153, can influence the distribution of HIV-1 integration. Whereas depletion of TNPO3, NUP358, or NUP153 preferentially reduced integration targeting to gene dense regions of chromosomes, disrupting the CA-CypA interaction – either through the addition of a small molecule cyclosporine inhibitor or by genetically altering the CA protein by G89V or P90A mutation – yielded the opposite effect; that is, these manipulations increased the targeting of the virus to gene dense regions (Ocwieja et al., 2011, Schaller et al., 2011, Koh et al., 2013, Di Nunzio et al., 2013). A separate CA mutation, N74D, which rendered HIV-1 resistant to the negative effects of TNPO3, NUP153, or NUP358 depletion (Lee et al., 2010), on its own steered integration to regions of significantly less gene density than the wild-type (WT) control virus (Schaller et al., 2011, Koh et al., 2013). Thus, the N74D mutation in large part phenocopied the effects of depleting TNPO3, NUP358, or NUP153. The N74D virus also mildly resisted the depletion of LEDGF/p75, as it retained relatively efficient infectivity while exhibiting less genic integration targeting than the WT virus (Koh et al., 2013). It has recently been observed that NUP153 and translocated promoter region (TPR), which together comprise the NPC basket (Umlauf et al., 2013), seem to cooperate to maintain chromatin architecture that is favorable for HIV-1 replication by helping to arrange actively transcribed chromatin proximal to the NPC (Marini et al., 2015, Lelek et al., 2015). However, it is currently unclear whether TPR exerts this effect by binding to the PIC through a viral component or perhaps a known cellular binding partner, such as LEDGF/p75. All together, this work underscores the complexity of the interactions that lentiviral PICs utilize to guide integration to favored sites, which are predominantly if not exclusively mediated through interactions of the viral CA and IN proteins with host factors. Additional work that co-depletes CA- and IN-binding cellular factors from cells may reveal whether random or near-random HIV-1 integration distribution is achievable under these circumstances.
BET proteins influence γ-retroviral integration target site selection
As discussed above, lentiviruses are not alone in utilizing interactions with host cell proteins to target integration to specific gDNA features. Additional examples include the retrotransposons Ty5 and Tf1 from yeast, which are directed by host proteins to target heterochromatin (Xie et al., 2001, Zhu et al., 2003) and promoters (Leem et al., 2008), respectively. Also, the catalytic activities of γ-retroviral INs are stimulated through the binding of bromodomain and extraterminal (BET) proteins, with BET-mediated bimodal tethering underlying the characteristic predilection of these viruses to integrate adjacent to promoter-associated annotations (Sharma et al., 2013, Gupta et al., 2013, De Rijck et al., 2013, Larue et al., 2014, Aiyer et al., 2014, El Ashkar et al., 2014, Serrao et al., 2015). Unlike the LEDGF/p75 bimodal tether and lentiviral DNA integration, small molecules exist that compete for BET protein binding to chromatin, and these compounds have been shown to diminish Mo-MLV infectivity and integration targeting without preventing the BET-IN interaction (Sharma et al., 2013, Gupta et al., 2013, De Rijck et al., 2013). This observation has potential implications for the future of ART, as small molecules that reduce infectivity by binding to and modulating the activity of cellular factors at positions that are distinct from the locations of cell-virus factor interaction sites would be expected to remain potent in the face of viral mutations. Readers are directed to several comprehensive reviews that have been recently published on the role of host proteins in directing lentiviral and γ-retroviral integration for additional details (Kvaratskhelia et al., 2014, Craigie and Bushman, 2014, Debyser et al., 2015).
Towards safer vector integration profiles
Understanding the mechanisms that govern the targeting of retroviral integration to specific regions of gDNA has far-reaching implications for the development and improvement of gene therapy technologies. Gene therapy has faced several hurdles over the years, including the integration of Mo-MLV-based vectors into unintended genomic sites that led to oncogene activation in an unacceptable number of cases (Marshall, 2002, Marshall, 2003, Hacein-Bey-Abina et al., 2003, Howe et al., 2008, Hacein-Bey-Abina et al., 2008). Gammaretrovirus- and lentivirus-based vectors display the integration site profiles described above for the parental viruses (Hematti et al., 2004, Laufs et al., 2006, Deichmann et al., 2007, Schwarzwaelder et al., 2007, Cattoglio et al., 2007, Mantovani et al., 2009, Wang et al., 2010), so customizing vector integration profiles to target “safe” regions of the genome is a current topic of interest. The design of completely safe vectors is not within reach at present, but knowledge of retrotransposon, lentivirus, and γ-retrovirus tethering mechanisms has been exploited to create prototype vectors with redirected, potentially less dangerous insertion profiles. For example, Mo-MLV vectors with lowered propensity to target promoter-proximal regions have been generated by mutating the C-terminal tail of IN to prevent the BET protein interaction (Aiyer et al., 2014, El Ashkar et al., 2014), and the mutant IN viruses generated similar distribution profiles as those observed using small molecules that block the BET-chromatin association. However, the distributions of the IN mutant viruses were still significantly different from random, with residual promoter proximal targeting evident.
A different approach to potentially safer retroviral-based gene therapy includes the development of self-inactivating (SIN) vectors, whereby the LTR U3 promoter and enhancer region is deleted (Schambach et al., 2006, Thornhill et al., 2008). A recent clinical trial employing Mo-MLV-based SIN vectors to correct X-linked severe combined immunodeficiency revealed that, though the genomic distribution of the vector integration sites was indistinguishable from that of enhancer-containing Mo-MLV-based vectors, integrations adjacent to proto-oncogenes and genes associated with adverse events in earlier trials was significantly reduced (Hacein-Bey-Abina et al., 2014). It is not fully understood why a SIN vector would exhibit selective reductions in integration near proto-oncogenes without a noticeable global change in integration site distribution, but a potential explanation is that the Mo-MLV LTR enhancer sequence contributed to the in vivo selection of engrafted cells containing such integrations, possibly through stimulating gene expression and ultimately cell growth rate. As the above-mentioned trial was comprised of only eight subjects, it will be informative to study the utility of SIN vectors to correct human genetic ailments using larger sample sizes moving forward.
Retargeting integration with customized chromatin tethers
The development of custom-targeted vectors is another option to avoid integration in genomic areas that promote unwanted side effects. Such customization research studies were originally conducted in vitro, employing HIV-1 or ASLV IN fused to DNA binding proteins such as λ-repressor (Bushman, 1994) or bacterial LexA (Goulaouic and Chow, 1996, Katz et al., 1996). The catalytic activity of these fusions resembled that of the WT INs, and integration into λ-repressor- and LexA-binding sequences was observed, thus providing optimism that IN protein hybrids could eventually be useful in vivo. Such experiments did in large part prove successful in the context of tissue culture infection, as vectors containing IN fused to zinc finger proteins (Bushman and Miller, 1997, Tan et al., 2006) displayed evidence for the expected altered integration distribution profile. However, while fusions of IN to large proteins or protein domains can exhibit WT-like levels of IN catalysis in vitro, the infectivity of vectors carrying such fusions is significantly diminished in cell culture (Bushman and Miller, 1997, Tan et al., 2006), thus limiting their potential clinical utility. An alternative approach to redirect retroviral integration targeting is to alter the chromatin binding activity of the IN-binding chromosomal DNA tether rather than the IN protein itself. Such approaches have been successfully developed for both LEDGF/p75 (Ciuffi et al., 2006, Ferris et al., 2010, Gijsbers et al., 2010, Silvers et al., 2010) and BET (De Rijck et al., 2013) fusion proteins. The fusion protein construct in these cases was expressed in target cells rather than packaging within the cargo-size-limited virion, thus permitting efficient redirection of integration into altered gDNA regions. Despite these promising results, the clinical utility of these constructs is complicated by the fact that these hybrids would have to compete with ubiquitously expressed endogenous LEDGF/p75 or BET proteins in target cells. An approach to circumvent this limitation has been developed by altering IN and LEDGF/p75 amino acid side chains that mediate the protein-protein interaction. The interaction between the IBD and the N-terminal domain of lentivirus IN is in particular mediated through salt-bridge contacts (Hare et al., 2009), and reversing the charge of the side chains (from acidic to basic for IN, and vice-versa for the complementary interacting LEDGF/p75 mutant) afforded selective IN mutant viral infection and integration in cells expressing the reverse-charge LEDGF/p75 variant (Hare et al., 2009, Wang et al., 2014). Although intriguing, the methodology has the disadvantage of requiring expression of the customized LEDGF/p75 partner in target cells.
Given the slow pace at which potential translational applications have evolved, interest in developing directed integration targeting vectors through modifying either IN or host cofactors may very well wane due to the development of other more promising gene editing technologies. The basis of the prokaryotic adaptive immune system that recognizes and degrades exogenous genetic elements (the CRISPR/Cas system) has been manipulated and applied toward customized editing in a variety of species including human embryos and non-human primates (Jinek et al., 2012, Niu et al., 2014, Liang et al., 2015, Chen et al., 2015, Wan et al., 2015). The details of this system involve a customized guide RNA directing a CRISPR-associated (Cas) nuclease to genomic DNA in a sequence-specific fashion, leading to a double-stranded DNA cut at virtually any desired location in the genome (Cong et al., 2013). Although the CRISPR/Cas system is particularly promising for eventual clinical applications, similar to the zinc finger protein and transcription activator-like effector nuclease/TALEN (Miller et al., 2011) technologies that preceded it, the threat of low-frequency off-target events remains a potential hurdle (Pattanayak et al., 2011, Mussolino et al., 2011, Cho et al., 2014, Fu et al., 2014). Lentiviral vectors have notably been designed to introduce CRISPR/Cas system components into target cells (Kabadi et al., 2014, Albers et al., 2015).
Retroviruses as tools to identify cancer genes through insertional mutagenesis
Although retroviruses preferentially target particular tDNA nucleotides and genomic annotations for integration, the bulk of the nucleotides and genes within the host genome are available as insertion targets (Carteau et al., 1998). The tendency of retroviruses to integrate at innumerable gDNA locations has been harnessed to identify cancer-related genes through insertional mutagenesis [reviewed in (Mikkers and Berns, 2003, Carlson and Largaespada, 2005, Ranzani et al., 2013)]. Initial observations that tumors from multiple different birds contained evidence of ASLV integration adjacent to a specific cellular gene (c-myc) suggested that insertion of the viral promoter adjacent to this gene resulted in its enhanced expression, thereby leading to neoplastic transformation and positive selection for the provirus-harboring cells (Neel et al., 1981, Hayward et al., 1981, Fung et al., 1981, Noori-Daloii et al., 1981, Robinson and Gagnon, 1986). It was subsequently shown that retrovirus-mediated activation of additional genes could also stimulate tumor formation (Nusse and Varmus, 1982, Peters et al., 1983, Seiki et al., 1984, Sorensen et al., 1996), which led to the realization that retroviruses could be used as tools to probe entire genomes for theoretically all regions relevant to cancer. With completion of the Mouse Genome Project and development of powerful integration site sequencing methodologies, it became possible to conduct in vivo genetic screens using insertional mutagenesis, which is also referred to as retroviral tagging (Li et al., 1999, Hansen et al., 2000, Lund et al., 2002, Mikkers et al., 2002, Suzuki et al., 2002). These original high-throughput screens recovered hundreds of recurrent integration sites in different tumor samples (Akagi et al., 2004), thus leading to the classification of oncogenesis-relevant signaling pathways. The ability to focus on large networks of proteins as potentially being associated with cancer allowed for comprehensive follow-up biochemical studies, thus representing a major strength of the technique. One procedural limitation, however, has been the underrepresentation of identified tumor suppressor genes (TSGs), which are arguably as important for study as proto-oncogenes. This drawback results from the fact that powerful enhancer features in retroviral promoters cause most integrations to be activating rather than inactivating. Furthermore, whereas insertional mutagenesis proceeds via integration nearby one copy of a proto-oncogene, both copies of the TSG in general need to be inactivated to promote tumorigenesis (Knudson, 1971).
Surveillance of repopulating stem cell expansion during gene therapy
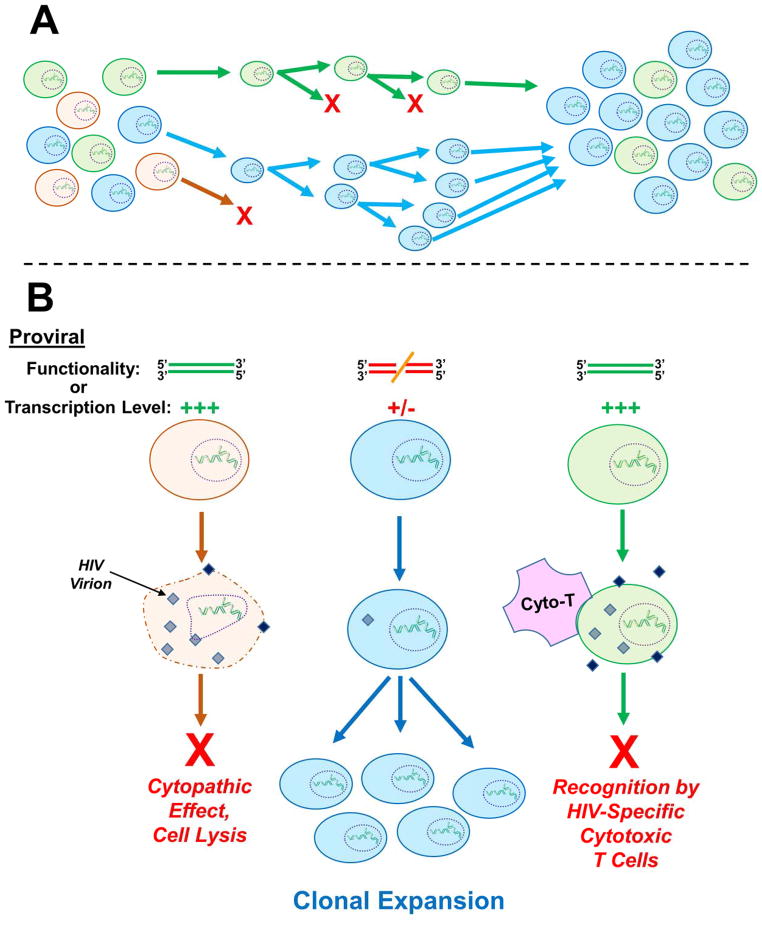
Aside from putative oncogene identification, retroviral tagging has been used to follow the repopulation dynamics of hematopoietic cells (Williams et al., 1984, Dick et al., 1985, Lemischka et al., 1986). It was ultimately discovered that individual retrovirally-transduced stem cell clones exhibited extensive variability in terms of lifespan and proliferative capacity (Guenechea et al., 2001). The clonal dominance of certain cellular subsets (depicted in Figure 4A) may be due to increased fitness caused specifically by integration-mediated transcriptional dysregulation of gene(s) involved in cell survival or self-renewal. Initial evidence described that integrations clustering within the transcriptional regulator gene Evi1 in mice and nonhuman primates were associated with long-term clonal persistence without signs of dominance or abnormal proliferation (Barjesteh van Waalwijk van Doorn-Khosrovani et al., 2003, Kustikova et al., 2005, Calmels et al., 2005), despite the fact that Evi1 expression had been correlated with leukemic disease progression (Barjesteh van Waalwijk van Doorn-Khosrovani et al., 2003, Buonamici et al., 2004). Surveillance of transduced cell engraftment in subsequent human gene therapy trials clarified that not only were integrations within EVI1 and other DNA replication- and cancer-related genes like PRDM16, SETBP1, CCND2, and HMGA2 recurrently present in recipient patients, but cell clones harboring these integrations had extensively expanded to dominate over time (Li et al., 2006, Deichmann et al., 2007, Cavazzana-Calvo et al., 2010). Interestingly, a β-thalassemia gene correction study in mice (Ocwieja et al., 2011), which employed the same vector utilized in the above-cited human trial (Cavazzana-Calvo et al., 2010), failed to find evidence for clonal expansion of cells harboring vectors integrated near genes involved in growth control. Therefore, taken together, it seems possible that species-specific factors play tangible roles in determining whether retroviral vector integrations influence host cell survival, engraftment, or proliferation. It is noteworthy that recent gene therapy trials utilizing optimized lentiviral vectors to correct metachromatic leukodystrophy and Wiskott-Aldrich syndrome have achieved efficient gene correction in multiple human patients without any evidence for clonal expansion among the tens of thousands of integration sites analyzed (Aiuti et al., 2013).
Figure 4. Clonal expansion of provirus-harboring cells.
A) General schematic of cellular clonal expansion. From an initially diverse population of cells, certain clones can exhibit relatively increased viability or growth rate, thus leading to persistence and ultimate enrichment. In this example the orange cell clone is dividing slowly or not at all, while the green cell clone exhibits an intermediate rate of expansion, and the blue cell clone expands most quickly. Thus in the final cell population, the blue clone predominates. B) Host cells harboring a particular integration site(s) can also become enriched over time within infected patients. The fact that clonally expanded, provirus-harboring cells can persist in the face of suppressive ART suggests that additional factors beyond integration site-mediated stimulation of cell growth rate contribute to clonal persistence. Two suggestions have been that the resident proviruses within the expanded cell clones are 1) nonfunctional due to non-tolerable insertions or deletions or 2) are molecularly intact but are expressed at a relatively low level (or not at all). In either case, the clones would escape viral replication-associated cytopathic effects and cell lysis (orange clone), as well as host cell recognition and purging by HIV-specific cytotoxic T-cells (green clone), leading to the enrichment of particular integration sites over time (blue clone). Neither of these possibilities, though, has thus far been definitively proven. A color version of this figure is available online.
Following integrated proviruses to classify host cell clonal expansion
Interestingly, cellular clonal expansion has recently been observed with provirus-harboring cells in HIV-1-infected patients undergoing ART, and the most highly expanded clones correspondingly exhibited strong integration clustering in cancer-related genes such as BACH2 (Ikeda et al., 2007, Maldarelli et al., 2014, Wagner et al., 2014). The presumption is that enhancement of cell cycle-regulating gene transcription by HIV-1 proviruses allows certain cells to persist in the face of ART for long periods of time and expand at a higher rate than cells that harbor less growth-promoting integration events. Accordingly, it has been suggested that the clonally expanded cells form a significant component of the latent viral reservoir (Maldarelli et al., 2014, Wagner et al., 2014), though a subsequent report failed to find supporting evidence for this model (Cohn et al., 2015). Consistent with the suggestion that clonally expanded cells can contribute to residual viremia is the fact that during suppressive ART the residual HIV-1 virion population shifts from diverse sets of variants to one or a small number of persisting clusters with genetic uniformity (Bailey et al., 2006, Josefsson et al., 2013, Kearney et al., 2014).
The means by which clonally expanded cells persist in the face of ART while other cells die is currently unknown, though there are two likely possibilities (Figure 4B). First, in addition to proliferating at a relatively increased rate, these clones could express the virus at an extremely low level. In this case the provirus would be duplicated continuously while remaining relatively dormant and thereby escaping not only drug treatment but also the added dangers of host cell recognition by HIV-specific cytotoxic T lymphocytes and viral protein-induced cytopathic effects. Though not pertinent to clonally expanded cells specifically, attempts have been made to quantify the relationship between integration site and latent provirus expression, and the results between systems have been inconsistent. In some cell culture models “latent” clones selectively harbor proviruses in heterochromatin (Jordan et al., 2001, Jordan et al., 2003), suggesting that low proviral transcription is a function of the surrounding genomic environment. Alternatively, replication-competent non-transcribed proviruses from treated patients have been found with high frequency in active transcription units (Han et al., 2004, Ikeda et al., 2007, Ho et al., 2013), while still other cell culture models failed to detect any chromatin feature that served as better latency predictors than pure chance (Sherrill-Mix et al., 2013). Therefore, the extent to which the chromatin environment of any given latent provirus contributes to viral transcriptional activity, host cell clonal expansion, and persistence is currently unresolved.
A second, equally plausible explanation for clonal cell resilience during ART could be that the proviruses that reside within these cells are defective due to inactivating insertions, deletions, or other mutations. In this case cells could continue to proliferate during ART without virion production while intact, integrated LTRs would still be sensed by integration site sequencing methodology. These integration sites would appear to be longitudinally enriched, since the bulk of other integrated proviruses would be eliminated over the course of time on treatment. Both sides of the coin have landed face-up in this research realm as well. Some studies have observed that up to 12% of clonally expanded proviruses are fully functional (Ho et al., 2013), while others have reported that every provirus that was analyzed contained an inactivating mutation of some sort (Josefsson et al., 2013, Imamichi et al., 2014, Cohn et al., 2015). Only a relatively low number of full-length proviruses have been sequenced when all of these studies are merged, mainly due to the laborious procedures required to obtain full HIV-1 genome (~10 kb) sequencing reads. A consensus in this realm may be thus reached in the near future when sample sizes are ultimately increased. This is becoming a possibility due to promising new technologies such as Single Molecule, Real-Time (SMRT®) Sequencing from Pacific Biosciences, which allows long and continuous DNA sequencing reads in a multiplexed format (Dilernia et al., 2015). The application of this particular technology to high-throughput HIV-1 full-genome sequencing is currently being optimized, and it holds the promise of becoming an invaluable new tool for following HIV-1 population dynamics over time in infected patients.
Perspectives
All retroviruses replicate in a similar fashion, reverse transcribing their RNA genome, integrating the vDNA product into host cell chromatin, and ultimately budding out of the host cell to infect a new target. However, different patterns and preferences evident at many steps of the infection process highlight close similarities and major differences among viral genera. These patterns are particularly evident at stages surrounding the integration step of the life cycle. As one example, γ-retroviruses preferentially target gene promoter and active enhancer regions, while viruses like MMTV integrate in a largely random fashion throughout the genome. Such differences not only provide an important springboard for understanding the dynamics of viral infection, but also for the gradual development of clinically applicable gene therapy vectors. Alternatively, the use of retroviruses as tagging agents has led to huge leaps forward in both the recognition of complex gene networks involved in cancer, as well as in the understanding of infused stem cell repopulation dynamics. And most recently the ability to distinguish clonally expanded proviruses from PCR-amplified sequence duplicates has begun to shed light on the elusive components of the latent HIV-1 reservoir. This important basic science and translational research has been made possible by extremely powerful, specific, and high-throughput integration site sequencing methodologies combined with thoroughly annotated reference genomes. These techniques have progressed from electrophoretic resolution of a handful of differently sized provirus-containing restriction fragments to the precise mapping of millions of unique tDNA sequences. To say that the methods have come a long way would be an understatement, but there is still ample room for further improvement. One weakness of the current state of integration site sequencing technology is that only about 30% of all LTRs within a DNA library can be expected to be amplified and recovered in a single sequencing run, even from the best of library preparations (unpublished observations). This potential limitation can be addressed by repeatedly sequencing the same library preparation and/or by sequencing multiple parallel libraries prepared from the same gDNA source. Further improvements in assay sensitivity should allow for more translational questions to be asked and answered as to the relationship between retroviral integration site and host disease state.
Acknowledgments
We would like to sincerely thank Drs. Peter Cherepanov, Stephen Hughes, Xiaolin Wu, Henry Levin, and Parmit Singh for helpful discussions regarding next generation sequencing technologies and their application to retroviral integration site sequencing.
Footnotes
Declaration of interest
The authors declare no conflict of financial interest. This work was supported by US National Institutes of Health grants AI052014 and AI039394 (to A.N.E.) as well as AI007386 (to E.S.).
References
- AIUTI A, BIASCO L, SCARAMUZZA S, FERRUA F, CICALESE MP, BARICORDI C, DIONISIO F, CALABRIA A, GIANNELLI S, CASTIELLO MC, BOSTICARDO M, EVANGELIO C, ASSANELLI A, CASIRAGHI M, DI NUNZIO S, CALLEGARO L, BENATI C, RIZZARDI P, PELLIN D, DI SERIO C, SCHMIDT M, VON KALLE C, GARDNER J, MEHTA N, NEDUVA V, DOW DJ, GALY A, MINIERO R, FINOCCHI A, METIN A, BANERJEE PP, ORANGE JS, GALIMBERTI S, VALSECCHI MG, BIFFI A, MONTINI E, VILLA A, CICERI F, RONCAROLO MG, NALDINI L. Lentiviral hematopoietic stem cell gene therapy in patients with Wiskott-Aldrich syndrome. Science. 2013;341:1233151. doi: 10.1126/science.1233151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AIYER S, ROSSI P, MALANI N, SCHNEIDER WM, CHANDAR A, BUSHMAN FD, MONTELIONE GT, ROTH MJ. Structural and sequencing analysis of local target DNA recognition by MLV integrase. Nucleic Acids Res. 2015;43:5647–5663. doi: 10.1093/nar/gkv410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AIYER S, SWAPNA GVT, MALANI N, ARAMINI JM, SCHNEIDER WM, PLUMB MR, GHANEM M, LARUE RC, SHARMA A, STUDAMIRE B, KVARATSKHELIA M, BUSHMAN FD, MONTELIONE GT, ROTH MJ. Altering murine leukemia virus integration through disruption of the integrase and BET protein family interaction. Nucleic Acids Res. 2014;42:5917–5928. doi: 10.1093/nar/gku175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AKAGI K, SUZUKI T, STEPHENS RM, JENKINS NA, COPELAND NG. RTCGD: retroviral tagged cancer gene database. Nucleic Acids Res. 2004;32:D523–D527. doi: 10.1093/nar/gkh013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ALBERS J, DANZER C, RECHSTEINER M, LEHMANN H, BRANDT LP, HEJHAL T, CATALANO A, BUSENHART P, GONÇALVES AF, BRANDT S, BODE PK, BODE-LESNIEWSKA B, WILD PJ, FREW IJ. A versatile modular vector system for rapid combinatorial mammalian genetics. J Clin Invest. 2015;125:1603–1619. doi: 10.1172/JCI79743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- APPA RS, SHIN CG, LEE P, CHOW SA. Role of the nonspecific DNA-binding region and alpha helices within the core domain of retroviral integrase in selecting target DNA sites for integration. J Biol Chem. 2001;276:45848–45855. doi: 10.1074/jbc.M107365200. [DOI] [PubMed] [Google Scholar]
- BACHELER LT, FAN H. Multiple integration sites for Moloney murine leukemia virus in productively infected mouse fibroblasts. J Virol. 1979;30:657–667. doi: 10.1128/jvi.30.3.657-667.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BAILEY JR, SEDAGHAT AR, KIEFFER T, BRENNAN T, LEE PK, WIND-ROTOLO M, HAGGERTY CM, KAMIREDDI AR, LIU Y, LEE J, PERSAUD D, GALLANT JE, COFRANCESCO J, JR, QUINN TC, WILKE CO, RAY SC, SILICIANO JD, NETTLES RE, SILICIANO RF. Residual human immunodeficiency virus type 1 viremia in some patients on antiretroviral therapy is dominated by a small number of invariant clones rarely found in circulating CD4+ T cells. J Virol. 2006;80:6441–6457. doi: 10.1128/JVI.00591-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BALLANDRAS-COLAS A, NARAHARISETTY H, LI X, SERRAO E, ENGELMAN A. Biochemical characterization of novel retroviral integrase proteins. PLoS One. 2013;8:e76638. doi: 10.1371/journal.pone.0076638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BANNISTER AJ, SCHNEIDER R, MYERS FA, THORNE AW, CRANE-ROBINSON C, KOUZARIDES T. Spatial distribution of di- and tri-methyl lysine 36 of histone H3 at active genes. J Biol Chem. 2005;280:17732–17736. doi: 10.1074/jbc.M500796200. [DOI] [PubMed] [Google Scholar]
- BAO KK, WANG H, MILLER JK, ERIE DA, SKALKA AM, WONG I. Functional oligomeric state of avian sarcoma virus integrase. J Biol Chem. 2003;278:1323–1327. doi: 10.1074/jbc.C200550200. [DOI] [PubMed] [Google Scholar]
- BARJESTEH VAN WAALWIJK VAN DOORN-KHOSROVANI S, ERPELINCK C, VAN PUTTEN WL, VALK PJ, VAN DER POEL-VAN DE LUYTGAARDE S, HACK R, SLATER R, SMIT EM, BEVERLOO HB, VERHOEF G, VERDONCK LF, OSSENKOPPELE GJ, SONNEVELD P, DE GREEF GE, LOWENBERG B, DELWEL R. High EVI1 expression predicts poor survival in acute myeloid leukemia: a study of 319 de novo AML patients. Blood. 2003;101:837–845. doi: 10.1182/blood-2002-05-1459. [DOI] [PubMed] [Google Scholar]
- BATTULA N, TEMIN HM. Infectious DNA of spleen necrosis virus is integrated at a single site in the DNA of chronically infected chicken fibroblasts. Proc Natl Acad Sci USA. 1977;74:281–285. doi: 10.1073/pnas.74.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BATTULA N, TEMIN HM. Sites of integration of infectious DNA of avian reticuloendotheliosis viruses in different avian cellular DNAs. Cell. 1978;13:387–398. doi: 10.1016/0092-8674(78)90207-6. [DOI] [PubMed] [Google Scholar]
- BELL O, TIWARI VK, THOMÄ NH, SCHÜBELER D. Determinants and dynamics of genome accessibility. Nat Rev Genet. 2011;12:554–564. doi: 10.1038/nrg3017. [DOI] [PubMed] [Google Scholar]
- BENLEULMI MS, MATYSIAK J, HENRIQUEZ DR, VAILLANT C, LESBATS P, CALMELS C, NAUGHTIN M, LEON O, SKALKA AM, RUFF M, LAVIGNE M, ANDREOLA ML, PARISSI V. Intasome architecture and chromatin density modulate retroviral integration into nucleosome. Retrovirology. 2015;12:13. doi: 10.1186/s12977-015-0145-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BERA S, PANDEY KK, VORA AC, GRANDGENETT DP. Molecular Interactions between HIV-1 integrase and the two viral DNA ends within the synaptic complex that mediates concerted integration. J Mol Biol. 2009;389:183–198. doi: 10.1016/j.jmb.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BISHOP JM, VARMUS HE. The Molecular Biology of RNA Tumor Viruses. In: BECKER FF, editor. Cancer, a Comprehensive Treatise. New York: Plenum Press; 1975. [Google Scholar]
- BOR YC, MILLER MD, BUSHMAN FD, ORGEL LE. Target-sequence preferences of HIV-1 integration complexes in vitro. Virology. 1996;222:283–288. doi: 10.1006/viro.1996.0422. [DOI] [PubMed] [Google Scholar]
- BOTBOL Y, RAGHAVENDRA NK, RAHMAN S, ENGELMAN A, LAVIGNE M. Chromatinized templates reveal the requirement for the LEDGF/p75 PWWP domain during HIV-1 integration in vitro. Nucleic Acids Res. 2008;36:1237–1246. doi: 10.1093/nar/gkm1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BROWN PO, BOWERMAN B, VARMUS HE, BISHOP JM. Retroviral integration: structure of the initial covalent product and its precursor, and a role for the viral IN protein. Proc Natl Acad Sci USA. 1989;86:2525–2529. doi: 10.1073/pnas.86.8.2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BRUSSEL A, DELELIS O, SONIGO P. Alu-LTR real-time nested PCR assay for quantifying integrated HIV-1 DNA. Methods Mol Biol. 2005;304:139–154. doi: 10.1385/1-59259-907-9:139. [DOI] [PubMed] [Google Scholar]
- BUKRINSKY MI, SHAROVA N, DEMPSEY MP, STANWICK TL, BUKRINSKAYA AG, HAGGERTY S, STEVENSON M. Active nuclear import of human immunodeficiency virus type 1 preintegration complexes. Proc Natl Acad Sci USA. 1992;89:6580–6584. doi: 10.1073/pnas.89.14.6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BUONAMICI S, LI D, CHI Y, ZHAO R, WANG X, BRACE L, NI H, SAUNTHARARAJAH Y, NUCIFORA G. EVI1 induces myelodysplastic syndrome in mice. J Clin Invest. 2004;114:713–719. doi: 10.1172/JCI21716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BUSHMAN FD. Tethering human immunodeficiency virus 1 integrase to a DNA site directs integration to nearby sequences. Proc Natl Acad Sci USA. 1994;91:9233–9237. doi: 10.1073/pnas.91.20.9233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BUSHMAN FD. Targeting survival: integration site selection by retroviruses and LTR-retrotransposons. Cell. 2003;115:135–138. doi: 10.1016/s0092-8674(03)00760-8. [DOI] [PubMed] [Google Scholar]
- BUSHMAN FD, MILLER MD. Tethering human immunodeficiency virus type 1 preintegration complexes to target DNA promotes integration at nearby sites. J Virol. 1997;71:458–464. doi: 10.1128/jvi.71.1.458-464.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BUSSCHOTS K, VERCAMMEN J, EMILIANI S, BENAROUS R, ENGELBORGHS Y, CHRIST F, DEBYSER Z. The interaction of LEDGF/p75 with integrase Is lentivirus-specific and promotes DNA binding. J Biol Chem. 2005;280:17841–17847. doi: 10.1074/jbc.M411681200. [DOI] [PubMed] [Google Scholar]
- BUTLER SL, HANSEN MS, BUSHMAN FD. A quantitative assay for HIV DNA integration in vivo. Nat Med. 2001;7:631–634. doi: 10.1038/87979. [DOI] [PubMed] [Google Scholar]
- CALMELS B, FERGUSON C, LAUKKANEN MO, ADLER R, FAULHABER M, KIM HJ, SELLERS S, HEMATTI P, SCHMIDT M, VON KALLE C, AKAGI K, DONAHUE RE, DUNBAR CE. Recurrent retroviral vector integration at the Mds1/Evi1 locus in nonhuman primate hematopoietic cells. Blood. 2005;106:2530–2533. doi: 10.1182/blood-2005-03-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CALOS MP, JOHNSRUD L, MILLER JH. DNA sequence at the integration sites of the insertion element IS1. Cell. 1978;13:411–418. doi: 10.1016/0092-8674(78)90315-x. [DOI] [PubMed] [Google Scholar]
- CARLSON CM, LARGAESPADA DA. Insertional mutagenesis in mice: new perspectives and tools. Nature Rev Genet. 2005;6:568–580. doi: 10.1038/nrg1638. [DOI] [PubMed] [Google Scholar]
- CARTEAU S, HOFFMANN C, BUSHMAN F. Chromosome structure and human immunodeficiency virus type 1 cDNA integration: Centromeric alphoid repeats are a disfavored target. J Virol. 1998;72:4005–4014. doi: 10.1128/jvi.72.5.4005-4014.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CATTOGLIO C, FACCHINI G, SARTORI D, ANTONELLI A, MICCIO A, CASSANI B, SCHMIDT M, VON KALLE C, HOWE S, THRASHER AJ, AIUTI A, FERRARI G, RECCHIA A, MAVILIO F. Hot spots of retroviral integration in human CD34+ hematopoietic cells. Blood. 2007;110:1770–1778. doi: 10.1182/blood-2007-01-068759. [DOI] [PubMed] [Google Scholar]
- CAVAZZANA-CALVO M, PAYEN E, NEGRE O, WANG G, HEHIR K, FUSIL F, DOWN J, DENARO M, BRADY T, WESTERMAN K, CAVALLESCO R, GILLET-LEGRAND B, CACCAVELLI L, SGARRA R, MAOUCHE-CHRETIEN L, BERNAUDIN F, GIROT R, DORAZIO R, MULDER GJ, POLACK A, BANK A, SOULIER J, LARGHERO J, KABBARA N, DALLE B, GOURMEL B, SOCIE G, CHRETIEN S, CARTIER N, AUBOURG P, FISCHER A, CORNETTA K, GALACTEROS F, BEUZARD Y, GLUCKMAN E, BUSHMAN F, HACEIN-BEY-ABINA S, LEBOULCH P. Transfusion independence and HMGA2 activation after gene therapy of human beta-thalassaemia. Nature. 2010;467:318–322. doi: 10.1038/nature09328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CAVROIS M, WAIN-HOBSON S, WATTEL E. Stochastic events in the amplification of HTLV-I integration sites by linker-mediated PCR. Res Virol. 1995;146:179–184. doi: 10.1016/0923-2516(96)80578-4. [DOI] [PubMed] [Google Scholar]
- CHEN H, WEI SQ, ENGELMAN A. Multiple integrase functions are required to form the native structure of the human immunodeficiency virus type I intasome. J Biol Chem. 1999;274:17358–17364. doi: 10.1074/jbc.274.24.17358. [DOI] [PubMed] [Google Scholar]
- CHEN Y, ZHENG Y, KANG Y, YANG W, NIU Y, GUO X, TU Z, SI C, WANG H, XING R, PU X, YANG SH, LI S, JI W, LI XJ. Functional disruption of the dystrophin gene in rhesus monkey using CRISPR/Cas9. Hum Mol Genet. 2015;24:3764–3774. doi: 10.1093/hmg/ddv120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHEREPANOV P. LEDGF/p75 interacts with divergent lentiviral integrases and modulates their enzymatic activity in vitro. Nucleic Acids Res. 2007;35:113–124. doi: 10.1093/nar/gkl885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHEREPANOV P, DEVROE E, SILVER PA, ENGELMAN A. Identification of an evolutionarily conserved domain in human lens epithelium-derived growth factor/transcriptional co-activator p75 (LEDGF/p75) that binds HIV-1 integrase. J Biol Chem. 2004;279:48883–48892. doi: 10.1074/jbc.M406307200. [DOI] [PubMed] [Google Scholar]
- CHEREPANOV P, MAERTENS G, PROOST P, DEVREESE B, VAN BEEUMEN J, ENGELBORGHS Y, DE CLERCQ E, DEBYSER Z. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J Biol Chem. 2003;278:372–381. doi: 10.1074/jbc.M209278200. [DOI] [PubMed] [Google Scholar]
- CHEREPANOV P, SUN ZY, RAHMAN S, MAERTENS G, WAGNER G, ENGELMAN A. Solution structure of the HIV-1 integrase-binding domain in LEDGF/p75. Nat Struct Mol Biol. 2005;12:526–532. doi: 10.1038/nsmb937. [DOI] [PubMed] [Google Scholar]
- CHO SW, KIM S, KIM Y, KWEON J, KIM HS, BAE S, KIM JS. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 2014;24:132–141. doi: 10.1101/gr.162339.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHOU KS, OKAYAMA A, SU IJ, LEE TH, ESSEX M. Preferred nucleotide sequence at the integration target site of human T-cell leukemia virus type I from patients with adult T-cell leukemia. Int J Cancer. 1996;65:20–24. doi: 10.1002/(SICI)1097-0215(19960103)65:1<20::AID-IJC4>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- CIUFFI A, DIAMOND TL, HWANG Y, MARSHALL HM, BUSHMAN FD. Modulating target site selection during human immunodeficiency virus DNA integration in vitro with an engineered tethering factor. Hum Gene Ther. 2006;17:960–967. doi: 10.1089/hum.2006.17.960. [DOI] [PubMed] [Google Scholar]
- CIUFFI A, LLANO M, POESCHLA E, HOFFMANN C, LEIPZIG J, SHINN P, ECKER JR, BUSHMAN F. A role for LEDGF/p75 in targeting HIV DNA integration. Nat Med. 2005;11:1287–1289. doi: 10.1038/nm1329. [DOI] [PubMed] [Google Scholar]
- COHEN JC, SHANK PR, MORRIS VL, CARDIFF R, VARMUS HE. Integration of the DNA of mouse mammary tumor virus in virus-infected normal and neoplastic tissue of the mouse. Cell. 1979;16:333–345. doi: 10.1016/0092-8674(79)90010-2. [DOI] [PubMed] [Google Scholar]
- COHN LB, SILVA IT, OLIVEIRA TY, ROSALES RA, PARRISH EH, LEARN GH, HAHN BH, CZARTOSKI JL, MCELRATH MJ, LEHMANN C, KLEIN F, CASKEY M, WALKER BD, SILICIANO JD, SILICIANO RF, JANKOVIC M, NUSSENZWEIG MC. HIV-1 integration landscape during latent and active infection. Cell. 2015;160:420–432. doi: 10.1016/j.cell.2015.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CONG L, RAN FA, COX D, LIN S, BARRETTO R, HABIB N, HSU PD, WU X, JIANG W, MARRAFFINI LA, ZHANG F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CRAIGIE R, BUSHMAN FD. HIV DNA integration. Cold Spring Harb Perspect Med. 2012;2:a006890. doi: 10.1101/cshperspect.a006890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CRAIGIE R, BUSHMAN FD. Host factors in retroviral integration and the selection of integration target sites. Microbiol Spectr. 2014;2 doi: 10.1128/microbiolspec.MDNA3-0026-2014. MDNA3-0026-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CRISE B, LI Y, YUAN C, MORCOCK DR, WHITBY D, MUNROE DJ, ARTHUR LO, WU X. Simian immunodeficiency virus integration preference is similar to that of human immunodeficiency virus type 1. J Virol. 2005;79:12199–12204. doi: 10.1128/JVI.79.19.12199-12204.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DE JONG J, AKHTAR W, BADHAI J, RUST AG, RAD R, HILKENS J, BERNS A, VAN LOHUIZEN M, WESSELS LF, DE RIDDER J. Chromatin landscapes of retroviral and transposon integration profiles. PLoS Genet. 2014;10:e1004250. doi: 10.1371/journal.pgen.1004250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DE RAVIN SS, SU L, THEOBALD N, CHOI U, MACPHERSON JL, POIDINGER M, SYMONDS G, POND SM, FERRIS AL, HUGHES SH, MALECH HL, WU X. Enhancers are major targets for murine leukemia virus vector integration. J Virol. 2014;88:4504–4513. doi: 10.1128/JVI.00011-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DE RIJCK J, DE KOGEL C, DEMEULEMEESTER J, VETS S, EL ASHKAR S, MALANI N, BUSHMAN FD, LANDUYT B, HUSSON SJ, BUSSCHOTS K, GIJSBERS R, DEBYSER Z. The BET family of proteins targets moloney murine leukemia virus integration near transcription start sites. Cell Rep. 2013;5:886–894. doi: 10.1016/j.celrep.2013.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DE SPIEGELAERE W, MALATINKOVA E, LYNCH L, VAN NIEUWERBURGH F, MESSIAEN P, O’DOHERTY U, VANDEKERCKHOVE L. Quantification of integrated HIV DNA by repetitive-sampling Alu-HIV PCR on the basis of poisson statistics. Clin Chem. 2014;60:886–895. doi: 10.1373/clinchem.2013.219378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DEBYSER Z, CHRIST F, DE RIJCK J, GIJSBERS R. Host factors for retroviral integration site selection. Trends Biochem Sci. 2015;40:108–116. doi: 10.1016/j.tibs.2014.12.001. [DOI] [PubMed] [Google Scholar]
- DEICHMANN A, HACEIN-BEY-ABINA S, SCHMIDT M, GARRIGUE A, BRUGMAN MH, HU J, GLIMM H, GYAPAY G, PRUM B, FRASER CC, FISCHER N, SCHWARZWAELDER K, SIEGLER ML, DE RIDDER D, PIKE-OVERZET K, HOWE SJ, THRASHER AJ, WAGEMAKER G, ABEL U, STAAL FJ, DELABESSE E, VILLEVAL JL, ARONOW B, HUE C, PRINZ C, WISSLER M, KLANKE C, WEISSENBACH J, ALEXANDER I, FISCHER A, VON KALLE C, CAVAZZANA-CALVO M. Vector integration is nonrandom and clustered and influences the fate of lymphopoiesis in SCID-X1 gene therapy. J Clin Invest. 2007;117:2225–2232. doi: 10.1172/JCI31659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DEMEULEMEESTER J, VETS S, SCHRIJVERS R, MADLALA P, DE MAEYER M, DE RIJCK J, NDUNG’U T, DEBYSER Z, GIJSBERS R. HIV-1 integrase variants retarget viral integration and are associated with disease progression in a chronic infection cohort. Cell Host Microbe. 2014;16:651–662. doi: 10.1016/j.chom.2014.09.016. [DOI] [PubMed] [Google Scholar]
- DERSE D, CRISE B, LI Y, PRINCLER G, LUM N, STEWART C, MCGRATH CF, HUGHES SH, MUNROE DJ, WU X. Human T-cell leukemia virus type 1 integration target sites in the human genome: comparison with those of other retroviruses. J Virol. 2007;81:6731–6741. doi: 10.1128/JVI.02752-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DHAR R, MCCLEMENTS WL, ENQUIST LW, VANDE WOUDE GF. Nucleotide sequences of integrated Moloney sarcoma provirus long terminal repeats and their host and viral junctions. Proc Natl Acad Sci USA. 1980;77:3937–3941. doi: 10.1073/pnas.77.7.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DI NUNZIO F, FRICKE T, MICCIO A, VALLE-CASUSO JC, PEREZ P, SOUQUE P, RIZZI E, SEVERGNINI M, MAVILIO F, CHARNEAU P, DIAZ-GRIFFERO F. Nup153 and Nup98 bind the HIV-1 core and contribute to the early steps of HIV-1 replication. Virology. 2013;440:8–18. doi: 10.1016/j.virol.2013.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DI PRIMIO C, QUERCIOLI V, ALLOUCH A, GIJSBERS R, CHRIST F, DEBYSER Z, AROSIO D, CERESETO A. Single-cell imaging of HIV-1 provirus (SCIP) Proc Natl Acad Sci USA. 2013;110:5636–5641. doi: 10.1073/pnas.1216254110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DICK JE, MAGLI MC, HUSZAR D, PHILLIPS RA, BERNSTEIN A. Introduction of a selectable gene into primitive stem cells capable of long-term reconstitution of the hemopoietic system of W/Wv mice. Cell. 1985;42:71–79. doi: 10.1016/s0092-8674(85)80102-1. [DOI] [PubMed] [Google Scholar]
- DILERNIA DA, CHIEN JT, MONACO DC, BROWN MP, ENDE Z, DEYMIER MJ, YUE L, PAXINOS EE, ALLEN S, TIRADO-RAMOS A, HUNTER E. Multiplexed highly-accurate DNA sequencing of closely-related HIV-1 variants using continuous long reads from single molecule, real-time sequencing. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv630. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DONG B, KIM S, HONG S, DAS GUPTA J, MALATHI K, KLEIN EA, GANEM D, DERISI JL, CHOW SA, SILVERMAN RH. An infectious retrovirus susceptible to an IFN antiviral pathway from human prostate tumors. Proc Natl Acad Sci USA. 2007;104:1655–1660. doi: 10.1073/pnas.0610291104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EIDAHL JO, CROWE BL, NORTH JA, MCKEE CJ, SHKRIABAI N, FENG L, PLUMB M, GRAHAM RL, GORELICK RJ, HESS S, POIRIER MG, FOSTER MP, KVARATSKHELIA M. Structural basis for high-affinity binding of LEDGF PWWP to mononucleosomes. Nucleic Acids Res. 2013;41:3924–3936. doi: 10.1093/nar/gkt074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EL ASHKAR S, DE RIJCK J, DEMEULEMEESTER J, VETS S, MADLALA P, CERMAKOVA K, DEBYSER Z, GIJSBERS R. BET-independent MLV-based vectors target away from promoters and regulatory elements. Mol Ther Nucleic Acids. 2014;3:e179. doi: 10.1038/mtna.2014.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENGELMAN A. Host cell factors and HIV-1 integration. Future HIV Ther. 2007;1:415–426. [Google Scholar]
- ENGELMAN A, MIZUUCHI K, CRAIGIE R. HIV-1 DNA integration: mechanism of viral DNA cleavage and DNA strand transfer. Cell. 1991;67:1211–1221. doi: 10.1016/0092-8674(91)90297-c. [DOI] [PubMed] [Google Scholar]
- FADEL HJ, MORRISON JH, SAENZ DT, FUCHS JR, KVARATSKHELIA M, EKKER SC, POESCHLA EM. TALEN Knockout of the PSIP1 Gene in Human Cells: Analyses of HIV-1 Replication and Allosteric Integrase Inhibitor Mechanism. J Virol. 2014;88:9704–9717. doi: 10.1128/JVI.01397-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FASCHINGER A, ROUAULT F, SOLLNER J, LUKAS A, SALMONS B, GUNZBURG WH, INDIK S. Mouse mammary tumor virus integration site selection in human and mouse genomes. J Virol. 2008;82:1360–1367. doi: 10.1128/JVI.02098-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAURE A, CALMELS C, DESJOBERT C, CASTROVIEJO M, CAUMONT-SARCOS A, TARRAGO-LITVAK L, LITVAK S, PARISSI V. HIV-1 integrase crosslinked oligomers are active in vitro. Nucleic Acids Res. 2005;33:977–986. doi: 10.1093/nar/gki241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FERRIS AL, WU X, HUGHES CM, STEWART C, SMITH SJ, MILNE TA, WANG GG, SHUN MC, ALLIS CD, ENGELMAN A, HUGHES SH. Lens epithelium-derived growth factor fusion proteins redirect HIV-1 DNA integration. Proc Natl Acad Sci USA. 2010;107:3135–3140. doi: 10.1073/pnas.0914142107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FINCHAM VJ, WYKE JA. Differences between cellular integration sites of transcribed and nontranscribed Rous sarcoma proviruses. J Virol. 1991;65:461–463. doi: 10.1128/jvi.65.1.461-463.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FITTS R, TEMIN HM. Cellular DNA surrounding integration sites of an avian retrovirus. J Gen Virol. 1983;64(Pt 2):267–274. doi: 10.1099/0022-1317-64-2-267. [DOI] [PubMed] [Google Scholar]
- FRANKLIN JP, BOSE HR, JR, KANG CY. Reticuloendotheliosis virus-transformed cells contain infectious and noninfectious proviral sequences in different chromosomes. Can J Microbiol. 1983;29:354–363. doi: 10.1139/m83-059. [DOI] [PubMed] [Google Scholar]
- FU Y, SANDER JD, REYON D, CASCIO VM, JOUNG JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol. 2014;32:279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FUJIWARA T, MIZUUCHI K. Retroviral DNA integration: structure of an integration intermediate. Cell. 1988;54:497–504. doi: 10.1016/0092-8674(88)90071-2. [DOI] [PubMed] [Google Scholar]
- FUNG YK, FADLY AM, CRITTENDEN LB, KUNG HJ. On the mechanism of retrovirus-induced avian lymphoid leukosis: deletion and integration of the proviruses. Proc Natl Acad Sci USA. 1981;78:3418–3422. doi: 10.1073/pnas.78.6.3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GENTNER B, LAUFS S, NAGY KZ, ZELLER WJ, FRUEHAUF S. Rapid detection of retroviral vector integration sites in colony-forming human peripheral blood progenitor cells using PCR with arbitrary primers. Gene Ther. 2003;10:789–794. doi: 10.1038/sj.gt.3301935. [DOI] [PubMed] [Google Scholar]
- GIJSBERS R, RONEN K, VETS S, MALANI N, DE RIJCK J, MCNEELY M, BUSHMAN FD, DEBYSER Z. LEDGF hybrids efficiently retarget lentiviral integration into heterochromatin. Mol Ther. 2010;18:552–560. doi: 10.1038/mt.2010.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GILLET NA, MALANI N, MELAMED A, GORMLEY N, CARTER R, BENTLEY D, BERRY C, BUSHMAN FD, TAYLOR GP, BANGHAM CR. The host genomic environment of the provirus determines the abundance of HTLV-1-infected T-cell clones. Blood. 2011;117:3113–3122. doi: 10.1182/blood-2010-10-312926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GILMER TM, PARSONS JT. Analysis of cellular integration sites in avian sarcoma virus infected duck embryo cells. J Virol. 1979;32:762–769. doi: 10.1128/jvi.32.3.762-769.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GOULAOUIC H, CHOW SA. Directed integration of viral DNA mediated by fusion proteins consisting of human immunodeficiency virus type 1 integrase and Escherichia coli LexA protein. J Virol. 1996;70:37–46. doi: 10.1128/jvi.70.1.37-46.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GUENECHEA G, GAN OI, DORRELL C, DICK JE. Distinct classes of human stem cells that differ in proliferative and self-renewal potential. Nat Immunol. 2001;2:75–82. doi: 10.1038/83199. [DOI] [PubMed] [Google Scholar]
- GUPTA SS, MAETZIG T, MAERTENS GN, SHARIF A, ROTHE M, WEIDNER-GLUNDE M, GALLA M, SCHAMBACH A, CHEREPANOV P, SCHULZ TF. Bromo- and extraterminal domain chromatin regulators serve as cofactors for murine leukemia virus integration. J Virol. 2013;87:12721–12736. doi: 10.1128/JVI.01942-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HACEIN-BEY-ABINA S, GARRIGUE A, WANG GP, SOULIER J, LIM A, MORILLON E, CLAPPIER E, CACCAVELLI L, DELABESSE E, BELDJORD K, ASNAFI V, MACINTYRE E, DAL CORTIVO L, RADFORD I, BROUSSE N, SIGAUX F, MOSHOUS D, HAUER J, BORKHARDT A, BELOHRADSKY BH, WINTERGERST U, VELEZ MC, LEIVA L, SORENSEN R, WULFFRAAT N, BLANCHE S, BUSHMAN FD, FISCHER A, CAVAZZANA-CALVO M. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J Clin Invest. 2008;118:3132–3142. doi: 10.1172/JCI35700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HACEIN-BEY-ABINA S, PAI SY, GASPAR HB, ARMANT M, BERRY CC, BLANCHE S, BLEESING J, BLONDEAU J, DE BOER H, BUCKLAND KF, CACCAVELLI L, CROS G, DE OLIVEIRA S, FERNANDEZ KS, GUO D, HARRIS CE, HOPKINS G, LEHMANN LE, LIM A, LONDON WB, VAN DER LOO JC, MALANI N, MALE F, MALIK P, MARINOVIC MA, MCNICOL AM, MOSHOUS D, NEVEN B, OLEASTRO M, PICARD C, RITZ J, RIVAT C, SCHAMBACH A, SHAW KL, SHERMAN EA, SILBERSTEIN LE, SIX E, TOUZOT F, TSYTSYKOVA A, XU-BAYFORD J, BAUM C, BUSHMAN FD, FISCHER A, KOHN DB, FILIPOVICH AH, NOTARANGELO LD, CAVAZZANA M, WILLIAMS DA, THRASHER AJ. A modified gamma-retrovirus vector for X-linked severe combined immunodeficiency. N Engl J Med. 2014;371:1407–1417. doi: 10.1056/NEJMoa1404588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HACEIN-BEY-ABINA S, VON KALLE C, SCHMIDT M, MCCORMACK MP, WULFFRAAT N, LEBOULCH P, LIM A, OSBORNE CS, PAWLIUK R, MORILLON E, SORENSEN R, FORSTER A, FRASER P, COHEN JI, DE SAINT BASILE G, ALEXANDER I, WINTERGERST U, FREBOURG T, AURIAS A, STOPPA-LYONNET D, ROMANA S, RADFORD-WEISS I, GROSS F, VALENSI F, DELABESSE E, MACINTYRE E, SIGAUX F, SOULIER J, LEIVA LE, WISSLER M, PRINZ C, RABBITTS TH, LE DEIST F, FISCHER A, CAVAZZANA-CALVO M. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- HACKER CV, VINK CA, WARDELL TW, LEE S, TREASURE P, KINGSMAN SM, MITROPHANOUS KA, MISKIN JE. The integration profile of EIAV-based vectors. Mol Ther. 2006;14:536–545. doi: 10.1016/j.ymthe.2006.06.006. [DOI] [PubMed] [Google Scholar]
- HAN Y, LASSEN K, MONIE D, SEDAGHAT AR, SHIMOJI S, LIU X, PIERSON TC, MARGOLICK JB, SILICIANO RF, SILICIANO JD. Resting CD4+ T cells from human immunodeficiency virus type 1 (HIV-1)-infected individuals carry integrated HIV-1 genomes within actively transcribed host genes. J Virol. 2004;78:6122–6133. doi: 10.1128/JVI.78.12.6122-6133.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HANSEN GM, SKAPURA D, JUSTICE MJ. Genetic profile of insertion mutations in mouse leukemias and lymphomas. Genome Res. 2000;10:237–243. doi: 10.1101/gr.10.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HARE S, GUPTA SS, VALKOV E, ENGELMAN A, CHEREPANOV P. Retroviral intasome assembly and inhibition of DNA strand transfer. Nature. 2010;464:232–236. doi: 10.1038/nature08784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HARE S, MAERTENS GN, CHEREPANOV P. 3′-processing and strand transfer catalysed by retroviral integrase in crystallo. EMBO J. 2012;31:3020–3028. doi: 10.1038/emboj.2012.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HARE S, SHUN MC, GUPTA SS, VALKOV E, ENGELMAN A, CHEREPANOV P. A novel co-crystal structure affords the design of gain-of-function lentiviral integrase mutants in the presence of modified PSIP1/LEDGF/p75. PLoS Pathog. 2009;5:e1000259. doi: 10.1371/journal.ppat.1000259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HARPER AL, SKINNER LM, SUDOL M, KATZMAN M. Use of patient-derived human immunodeficiency virus type 1 integrases to identify a protein residue that affects target site selection. J Virol. 2001;75:7756–7762. doi: 10.1128/JVI.75.16.7756-7762.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HATZIIOANNOU T, GOFF SP. Infection of nondividing cells by Rous sarcoma virus. J Virol. 2001;75:9526–9531. doi: 10.1128/JVI.75.19.9526-9531.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HAYWARD WS, NEEL BG, ASTRIN SM. Activation of a cellular onc gene by promoter insertion in ALV-induced lymphoid leukosis. Nature. 1981;290:475–480. doi: 10.1038/290475a0. [DOI] [PubMed] [Google Scholar]
- HEMATTI P, HONG BK, FERGUSON C, ADLER R, HANAWA H, SELLERS S, HOLT IE, ECKFELDT CE, SHARMA Y, SCHMIDT M, VON KALLE C, PERSONS DA, BILLINGS EM, VERFAILLIE CM, NIENHUIS AW, WOLFSBERG TG, DUNBAR CE, CALMELS B. Distinct genomic integration of MLV and SIV vectors in primate hematopoietic stem and progenitor cells. PLoS Biol. 2004;2:e423. doi: 10.1371/journal.pbio.0020423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HO YC, SHAN L, HOSMANE NN, WANG J, LASKEY SB, ROSENBLOOM DI, LAI J, BLANKSON JN, SILICIANO JD, SILICIANO RF. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell. 2013;155:540–551. doi: 10.1016/j.cell.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOLMAN AG, COFFIN JM. Symmetrical base preferences surrounding HIV-1, avian sarcoma/leukosis virus, and murine leukemia virus integration sites. Proc Natl Acad Sci USA. 2005;102:6103–6107. doi: 10.1073/pnas.0501646102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOWE SJ, MANSOUR MR, SCHWARZWAELDER K, BARTHOLOMAE C, HUBANK M, KEMPSKI H, BRUGMAN MH, PIKE-OVERZET K, CHATTERS SJ, DE RIDDER D, GILMOUR KC, ADAMS S, THORNHILL SI, PARSLEY KL, STAAL FJ, GALE RE, LINCH DC, BAYFORD J, BROWN L, QUAYE M, KINNON C, ANCLIFF P, WEBB DK, SCHMIDT M, VON KALLE C, GASPAR HB, THRASHER AJ. Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. J Clin Invest. 2008;118:3143–3150. doi: 10.1172/JCI35798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HU WS, HUGHES SH. HIV-1 reverse transcription. Cold Spring Harb Perspect Med. 2012;2:a006882. doi: 10.1101/cshperspect.a006882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUGHES SH, MUTSCHLER A, BISHOP JM, VARMUS HE. A Rous sarcoma virus provirus is flanked by short direct repeats of a cellular DNA sequence present in only one copy prior to integration. Proc Natl Acad Sci USA. 1981;78:4299–4303. doi: 10.1073/pnas.78.7.4299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HULME AE, KELLEY Z, FOLEY D, HOPE TJ. Complementary assays reveal a low level of CA associated with viral complexes in the nuclei of HIV-1-infected cells. J Virol. 2015;89:5350–5361. doi: 10.1128/JVI.00476-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HULME AE, PEREZ O, HOPE TJ. Complementary assays reveal a relationship between HIV-1 uncoating and reverse transcription. Proc Natl Acad Sci USA. 2011;108:9975–9980. doi: 10.1073/pnas.1014522108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IKEDA T, SHIBATA J, YOSHIMURA K, KOITO A, MATSUSHITA S. Recurrent HIV-1 integration at the BACH2 locus in resting CD4+ T cell populations during effective highly active antiretroviral therapy. J Infect Dis. 2007;195:716–725. doi: 10.1086/510915. [DOI] [PubMed] [Google Scholar]
- IMAMICHI H, NATARAJAN V, ADELSBERGER JW, REHM CA, LEMPICKI RA, DAS B, HAZEN A, IMAMICHI T, LANE HC. Lifespan of effector memory CD4+ T cells determined by replication-incompetent integrated HIV-1 provirus. AIDS. 2014;28:1091–1099. doi: 10.1097/QAD.0000000000000223. [DOI] [PubMed] [Google Scholar]
- IZUMOTO Y, KURODA T, HARADA H, KISHIMOTO T, NAKAMURA H. Hepatoma-derived growth factor belongs to a gene family in mice showing significant homology in the amino terminus. Biochem Biophys Res Commun. 1997;238:26–32. doi: 10.1006/bbrc.1997.7233. [DOI] [PubMed] [Google Scholar]
- JAHNER D, JAENISCH R. Integration of Moloney leukaemia virus into the germ line of mice: correlation between site of integration and virus activation. Nature. 1980;287:456–458. doi: 10.1038/287456a0. [DOI] [PubMed] [Google Scholar]
- JIN YF, ISHIBASHI T, NOMOTO A, MASUDA M. Isolation and analysis of retroviral integration targets by solo long terminal repeat inverse PCR. J Virol. 2002;76:5540–5547. doi: 10.1128/JVI.76.11.5540-5547.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JINEK M, CHYLINSKI K, FONFARA I, HAUER M, DOUDNA JA, CHARPENTIER E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JOHNSRUD L, CALOS MP, MILLER JH. The transposon Tn9 generates a 9 bp repeated sequence during integration. Cell. 1978;15:1209–1219. doi: 10.1016/0092-8674(78)90047-8. [DOI] [PubMed] [Google Scholar]
- JORDAN A, BISGROVE D, VERDIN E. HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. EMBO J. 2003;22:1868–1877. doi: 10.1093/emboj/cdg188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JORDAN A, DEFECHEREUX P, VERDIN E. The site of HIV-1 integration in the human genome determines basal transcriptional activity and response to Tat transactivation. EMBO J. 2001;20:1726–1738. doi: 10.1093/emboj/20.7.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JOSEFSSON L, VON STOCKENSTROM S, FARIA NR, SINCLAIR E, BACCHETTI P, KILLIAN M, EPLING L, TAN A, HO T, LEMEY P, SHAO W, HUNT PW, SOMSOUK M, WYLIE W, DOUEK DC, LOEB L, CUSTER J, HOH R, POOLE L, DEEKS SG, HECHT F, PALMER S. The HIV-1 reservoir in eight patients on long-term suppressive antiretroviral therapy is stable with few genetic changes over time. Proc Natl Acad Sci USA. 2013;110:E4987–E4996. doi: 10.1073/pnas.1308313110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KABADI AM, OUSTEROUT DG, HILTON IB, GERSBACH CA. Multiplex CRISPR/Cas9-based genome engineering from a single lentiviral vector. Nucleic Acids Res. 2014;42:e147. doi: 10.1093/nar/gku749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KANG Y, MORESSI CJ, SCHEETZ TE, XIE L, TRAN DT, CASAVANT TL, AK P, BENHAM CJ, DAVIDSON BL, MCCRAY PB., JR Integration site choice of a feline immunodeficiency virus vector. J Virol. 2006;80:8820–8823. doi: 10.1128/JVI.00719-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KATZ RA, GREGER JG, DARBY K, BOIMEL P, RALL GF, SKALKA AM. Transduction of interphase cells by avian sarcoma virus. J Virol. 2002;76:5422–5434. doi: 10.1128/JVI.76.11.5422-5434.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KATZ RA, MERKEL G, SKALKA AM. Targeting of retroviral integrase by fusion to a heterologous DNA binding domain: in vitro activities and incorporation of a fusion protein into viral particles. Virology. 1996;217:178–190. doi: 10.1006/viro.1996.0105. [DOI] [PubMed] [Google Scholar]
- KEARNEY MF, SPINDLER J, SHAO W, YU S, ANDERSON EM, O’SHEA A, REHM C, POETHKE C, KOVACS N, MELLORS JW, COFFIN JM, MALDARELLI F. Lack of detectable HIV-1 molecular evolution during suppressive antiretroviral therapy. PLoS Pathog. 2014;10:e1004010. doi: 10.1371/journal.ppat.1004010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KESHET E, TEMIN HM. Sites of integration of reticuloendotheliosis virus DNA in chicken DNA. Proc Natl Acad Sci USA. 1978;75:3372–3376. doi: 10.1073/pnas.75.7.3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KESSL JJ, LI M, IGNATOV M, SHKRIABAI N, EIDAHL JO, FENG L, MUSIER-FORSYTH K, CRAIGIE R, KVARATSKHELIA M. FRET analysis reveals distinct conformations of IN tetramers in the presence of viral DNA or LEDGF/p75. Nucleic Acids Res. 2011;39:9009–9022. doi: 10.1093/nar/gkr581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KETTMANN R, MEUNIER-ROTIVAL M, CORTADAS J, CUNY G, GHYSDAEL J, MAMMERICKX M, BURNY A, BERNARDI G. Integration of bovine leukemia virus DNA in the bovine genome. Proc Natl Acad Sci USA. 1979;76:4822–4826. doi: 10.1073/pnas.76.10.4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KIM S, KIM N, DONG B, BOREN D, LEE SA, DAS GUPTA J, GAUGHAN C, KLEIN EA, LEE C, SILVERMAN RH, CHOW SA. Integration site preference of xenotropic murine leukemia virus-related virus, a new human retrovirus associated with prostate cancer. J Virol. 2008;82:9964–9977. doi: 10.1128/JVI.01299-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KIM S, RUSMEVICHIENTONG A, DONG B, REMENYI R, SILVERMAN RH, CHOW SA. Fidelity of target site duplication and sequence preference during integration of xenotropic murine leukemia virus-related virus. PLoS One. 2010;5:e10255. doi: 10.1371/journal.pone.0010255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KLECKNER N. DNA sequence analysis of Tn10 insertions: origin and role of 9 bp flanking repetitions during Tn10 translocation. Cell. 1979;16:711–720. doi: 10.1016/0092-8674(79)90087-4. [DOI] [PubMed] [Google Scholar]
- KNUDSON AG., JR Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci USA. 1971;68:820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KOH Y, WU X, FERRIS AL, MATREYEK KA, SMITH SJ, LEE K, KEWALRAMANI VN, HUGHES SH, ENGELMAN A. Differential effects of human immunodeficiency virus type 1 capsid and cellular factors nucleoporin 153 and LEDGF/p75 on the efficiency and specificity of viral DNA integration. J Virol. 2013;87:648–658. doi: 10.1128/JVI.01148-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KONSTANTOULAS CJ, INDIK S. Mouse mammary tumor virus-based vector transduces non-dividing cells, enters the nucleus via a TNPO3-independent pathway and integrates in a less biased fashion than other retroviruses. Retrovirology. 2014;11:34. doi: 10.1186/1742-4690-11-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KRISHNAN L, LI X, NARAHARISETTY HL, HARE S, CHEREPANOV P, ENGELMAN A. Structure-based modeling of the functional HIV-1 intasome and its inhibition. Proc Natl Acad Sci USA. 2010;107:15910–15915. doi: 10.1073/pnas.1002346107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KUSTIKOVA O, FEHSE B, MODLICH U, YANG M, DULLMANN J, KAMINO K, VON NEUHOFF N, SCHLEGELBERGER B, LI Z, BAUM C. Clonal dominance of hematopoietic stem cells triggered by retroviral gene marking. Science. 2005;308:1171–1174. doi: 10.1126/science.1105063. [DOI] [PubMed] [Google Scholar]
- KVARATSKHELIA M, SHARMA A, LARUE RC, SERRAO E, ENGELMAN A. Molecular mechanisms of retroviral integration site selection. Nucleic Acids Res. 2014;42:10209–10225. doi: 10.1093/nar/gku769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LAFAVE MC, VARSHNEY GK, GILDEA DE, WOLFSBERG TG, BAXEVANIS AD, BURGESS SM. MLV integration site selection is driven by strong enhancers and active promoters. Nucleic Acids Res. 2014;42:4257–4269. doi: 10.1093/nar/gkt1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LARUE RC, PLUMB MR, CROWE BL, SHKRIABAI N, SHARMA A, DIFIORE J, MALANI N, AIYER SS, ROTH MJ, BUSHMAN FD, FOSTER MP, KVARATSKHELIA M. Bimodal high-affinity association of Brd4 with murine leukemia virus integrase and mononucleosomes. Nucleic Acids Res. 2014;42:4868–4881. doi: 10.1093/nar/gku135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LAUFS S, GUENECHEA G, GONZALEZ-MURILLO A, ZSUZSANNA NAGY K, LUZ LOZANO M, DEL VAL C, JONNAKUTY S, HOTZ-WAGENBLATT A, JENS ZELLER W, BUEREN JA, FRUEHAUF S. Lentiviral vector integration sites in human NOD/SCID repopulating cells. J Gene Med. 2006;8:1197–1207. doi: 10.1002/jgm.958. [DOI] [PubMed] [Google Scholar]
- LECLERCQ I, MORTREUX F, CAVROIS M, LEROY A, GESSAIN A, WAIN-HOBSON S, WATTEL E. Host sequences flanking the human T-cell leukemia virus type 1 provirus in vivo. J Virol. 2000;74:2305–2312. doi: 10.1128/jvi.74.5.2305-2312.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEE K, AMBROSE Z, MARTIN TD, OZTOP I, MULKY A, JULIAS JG, VANDEGRAAFF N, BAUMANN JG, WANG R, YUEN W, TAKEMURA T, SHELTON K, TANIUCHI I, LI Y, SODROSKI J, LITTMAN DR, COFFIN JM, HUGHES SH, UNUTMAZ D, ENGELMAN A, KEWALRAMANI VN. Flexible use of nuclear import pathways by HIV-1. Cell Host Microbe. 2010;7:221–233. doi: 10.1016/j.chom.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEE YM, COFFIN JM. Relationship of avian retrovirus DNA synthesis to integration in vitro. Mol Cell Biol. 1991;11:1419–1430. doi: 10.1128/mcb.11.3.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEEM YE, RIPMASTER TL, KELLY FD, EBINA H, HEINCELMAN ME, ZHANG K, GREWAL SI, HOFFMAN CS, LEVIN HL. Retrotransposon Tf1 is targeted to Pol II promoters by transcription activators. Mol Cell. 2008;30:98–107. doi: 10.1016/j.molcel.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LELEK M, CASARTELLI N, PELLIN D, RIZZI E, SOUQUE P, SEVERGNINI M, DI SERIO C, FRICKE T, DIAZ-GRIFFERO F, ZIMMER C, CHARNEAU P, DI NUNZIO F. Chromatin organization at the nuclear pore favours HIV replication. Nature Commun. 2015;6:6483. doi: 10.1038/ncomms7483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEMISCHKA IR, RAULET DH, MULLIGAN RC. Developmental potential and dynamic behavior of hematopoietic stem cells. Cell. 1986;45:917–927. doi: 10.1016/0092-8674(86)90566-0. [DOI] [PubMed] [Google Scholar]
- LESBATS P, BOTBOL Y, CHEVEREAU G, VAILLANT C, CALMELS C, ARNEODO A, ANDREOLA ML, LAVIGNE M, PARISSI V. Functional coupling between HIV-1 integrase and the SWI/SNF chromatin remodeling complex for efficient in vitro integration into stable nucleosomes. PLoS Pathog. 2011;7:e1001280. doi: 10.1371/journal.ppat.1001280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEWINSKI MK, BISGROVE D, SHINN P, CHEN H, HOFFMANN C, HANNENHALLI S, VERDIN E, BERRY CC, ECKER JR, BUSHMAN FD. Genome-wide analysis of chromosomal features repressing human immunodeficiency virus transcription. J Virol. 2005;79:6610–6619. doi: 10.1128/JVI.79.11.6610-6619.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEWINSKI MK, YAMASHITA M, EMERMAN M, CIUFFI A, MARSHALL H, CRAWFORD G, COLLINS F, SHINN P, LEIPZIG J, HANNENHALLI S, BERRY CC, ECKER JR, BUSHMAN FD. Retroviral DNA integration: viral and cellular determinants of target-site selection. PLoS Pathog. 2006;2:e60. doi: 10.1371/journal.ppat.0020060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEWIS P, HENSEL M, EMERMAN M. Human immunodeficiency virus infection of cells arrested in the cell cycle. EMBO J. 1992;11:3053–3058. doi: 10.1002/j.1460-2075.1992.tb05376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEWIS PF, EMERMAN M. Passage through mitosis is required for oncoretroviruses but not for the human immunodeficiency virus. J Virol. 1994;68:510–516. doi: 10.1128/jvi.68.1.510-516.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LI J, SHEN H, HIMMEL KL, DUPUY AJ, LARGAESPADA DA, NAKAMURA T, SHAUGHNESSY JD, JR, JENKINS NA, COPELAND NG. Leukaemia disease genes: large-scale cloning and pathway predictions. Nat Genet. 1999;23:348–353. doi: 10.1038/15531. [DOI] [PubMed] [Google Scholar]
- LI M, MIZUUCHI M, BURKE TR, JR, CRAIGIE R. Retroviral DNA integration: reaction pathway and critical intermediates. EMBO J. 2006;25:1295–1304. doi: 10.1038/sj.emboj.7601005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIANG P, XU Y, ZHANG X, DING C, HUANG R, ZHANG Z, LV J, XIE X, CHEN Y, LI Y, SUN Y, BAI Y, SONGYANG Z, MA W, ZHOU C, HUANG J. CRISPR/Cas9-mediated gene editing in human tripronuclear zygotes. Protein Cell. 2015;6:363–372. doi: 10.1007/s13238-015-0153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LLANO M, SAENZ DT, MEEHAN A, WONGTHIDA P, PERETZ M, WALKER WH, TEO W, POESCHLA EM. An essential role for LEDGF/p75 in HIV integration. Science. 2006a;314:461–464. doi: 10.1126/science.1132319. [DOI] [PubMed] [Google Scholar]
- LLANO M, VANEGAS M, FREGOSO O, SAENZ D, CHUNG S, PERETZ M, POESCHLA EM. LEDGF/p75 determines cellular trafficking of diverse lentiviral but not murine oncoretroviral integrase proteins and is a component of functional lentiviral preintegration complexes. J Virol. 2004;78:9524–9537. doi: 10.1128/JVI.78.17.9524-9537.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LLANO M, VANEGAS M, HUTCHINS N, THOMPSON D, DELGADO S, POESCHLA EM. Identification and characterization of the chromatin-binding domains of the HIV-1 integrase interactor LEDGF/p75. J Mol Biol. 2006b;360:760–773. doi: 10.1016/j.jmb.2006.04.073. [DOI] [PubMed] [Google Scholar]
- LUND AH, TURNER G, TRUBETSKOY A, VERHOEVEN E, WIENTJENS E, HULSMAN D, RUSSELL R, DEPINHO RA, LENZ J, VAN LOHUIZEN M. Genome-wide retroviral insertional tagging of genes involved in cancer in Cdkn2a-deficient mice. Nat Genet. 2002;32:160–165. doi: 10.1038/ng956. [DOI] [PubMed] [Google Scholar]
- MAAT J, SMITH AJ. A method for sequencing restriction fragments with dideoxynucleoside triphosphates. Nucleic Acids Res. 1978;5:4537–4545. doi: 10.1093/nar/5.12.4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MACK KD, JIN X, YU S, WEI R, KAPP L, GREEN C, HERNDIER B, ABBEY NW, ELBAGGARI A, LIU Y, MCGRATH MS. HIV insertions within and proximal to host cell genes are a common finding in tissues containing high levels of HIV DNA and macrophage-associated p24 antigen expression. J Acquir Immune Defic Syndr. 2003;33:308–320. doi: 10.1097/00126334-200307010-00004. [DOI] [PubMed] [Google Scholar]
- MAERTENS GN, HARE S, CHEREPANOV P. The mechanism of retroviral integration from X-ray structures of its key intermediates. Nature. 2010;468:326–329. doi: 10.1038/nature09517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MAJORS JE, VARMUS HE. Nucleotide sequences at host-proviral junctions for mouse mammary tumour virus. Nature. 1981;289:253–258. doi: 10.1038/289253a0. [DOI] [PubMed] [Google Scholar]
- MALDARELLI F, WU X, SU L, SIMONETTI FR, SHAO W, HILL S, SPINDLER J, FERRIS AL, MELLORS JW, KEARNEY MF, COFFIN JM, HUGHES SH. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science. 2014;345:179–183. doi: 10.1126/science.1254194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MANTOVANI J, CHARRIER S, ECKENBERG R, SAURIN W, DANOS O, PEREA J, GALY A. Diverse genomic integration of a lentiviral vector developed for the treatment of Wiskott-Aldrich syndrome. J Gene Med. 2009;11:645–654. doi: 10.1002/jgm.1346. [DOI] [PubMed] [Google Scholar]
- MARGULIES M, EGHOLM M, ALTMAN WE, ATTIYA S, BADER JS, BEMBEN LA, BERKA J, BRAVERMAN MS, CHEN YJ, CHEN Z, DEWELL SB, DU L, FIERRO JM, GOMES XV, GODWIN BC, HE W, HELGESEN S, HO CH, IRZYK GP, JANDO SC, ALENQUER ML, JARVIE TP, JIRAGE KB, KIM JB, KNIGHT JR, LANZA JR, LEAMON JH, LEFKOWITZ SM, LEI M, LI J, LOHMAN KL, LU H, MAKHIJANI VB, MCDADE KE, MCKENNA MP, MYERS EW, NICKERSON E, NOBILE JR, PLANT R, PUC BP, RONAN MT, ROTH GT, SARKIS GJ, SIMONS JF, SIMPSON JW, SRINIVASAN M, TARTARO KR, TOMASZ A, VOGT KA, VOLKMER GA, WANG SH, WANG Y, WEINER MP, YU P, BEGLEY RF, ROTHBERG JM. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARINI B, KERTESZ-FARKAS A, ALI H, LUCIC B, LISEK K, MANGANARO L, PONGOR S, LUZZATI R, RECCHIA A, MAVILIO F, GIACCA M, LUSIC M. Nuclear architecture dictates HIV-1 integration site selection. Nature. 2015;521:227–231. doi: 10.1038/nature14226. [DOI] [PubMed] [Google Scholar]
- MARSHALL E. Clinical research. Gene therapy a suspect in leukemia-like disease. Science. 2002;298:34–35. doi: 10.1126/science.298.5591.34. [DOI] [PubMed] [Google Scholar]
- MARSHALL E. Gene therapy. Second child in French trial is found to have leukemia. Science. 2003;299:320. doi: 10.1126/science.299.5605.320. [DOI] [PubMed] [Google Scholar]
- MARSHALL HM, RONEN K, BERRY C, LLANO M, SUTHERLAND H, SAENZ D, BICKMORE W, POESCHLA E, BUSHMAN FD. Role of PSIP1/LEDGF/p75 in lentiviral infectivity and integration targeting. PLoS One. 2007;2:e1340. doi: 10.1371/journal.pone.0001340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MASKELL DP, RENAULT L, SERRAO E, LESBATS P, MATADEEN R, HARE S, LINDEMANN D, ENGELMAN AN, COSTA A, CHEREPANOV P. Structural basis for retroviral integration into nucleosomes. Nature. 2015;523:366–369. doi: 10.1038/nature14495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MATREYEK KA, ENGELMAN A. Viral and cellular requirements for the nuclear entry of retroviral preintegration nucleoprotein complexes. Viruses. 2013;5:2483–2511. doi: 10.3390/v5102483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MAXAM AM, GILBERT W. A new method for sequencing DNA. Proc Natl Acad Sci USA. 1977;74:560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MEEKINGS KN, LEIPZIG J, BUSHMAN FD, TAYLOR GP, BANGHAM CR. HTLV-1 integration into transcriptionally active genomic regions is associated with proviral expression and with HAM/TSP. PLoS Pathog. 2008;4:e1000027. doi: 10.1371/journal.ppat.1000027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MIKKERS H, ALLEN J, KNIPSCHEER P, ROMEIJN L, HART A, VINK E, BERNS A. High-throughput retroviral tagging to identify components of specific signaling pathways in cancer. Nat Genet. 2002;32:153–159. doi: 10.1038/ng950. [DOI] [PubMed] [Google Scholar]
- MIKKERS H, BERNS A. Retroviral insertional mutagenesis: tagging cancer pathways. Adv Cancer Res. 2003;88:53–99. doi: 10.1016/s0065-230x(03)88304-5. [DOI] [PubMed] [Google Scholar]
- MILLER JC, TAN S, QIAO G, BARLOW KA, WANG J, XIA DF, MENG X, PASCHON DE, LEUNG E, HINKLEY SJ, DULAY GP, HUA KL, ANKOUDINOVA I, COST GJ, URNOV FD, ZHANG HS, HOLMES MC, ZHANG L, GREGORY PD, REBAR EJ. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- MILOT E, BELMAAZA A, RASSART E, CHARTRAND P. Association of a host DNA structure with retroviral integration sites in chromosomal DNA. Virology. 1994;201:408–412. doi: 10.1006/viro.1994.1310. [DOI] [PubMed] [Google Scholar]
- MITCHELL RS, BEITZEL BF, SCHRODER AR, SHINN P, CHEN H, BERRY CC, ECKER JR, BUSHMAN FD. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2004;2:E234. doi: 10.1371/journal.pbio.0020234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MOALIC Y, BLANCHARD Y, FELIX H, JESTIN A. Porcine endogenous retrovirus integration sites in the human genome: features in common with those of murine leukemia virus. J Virol. 2006;80:10980–10988. doi: 10.1128/JVI.00904-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MOALIC Y, FELIX H, TAKEUCHI Y, JESTIN A, BLANCHARD Y. Genome areas with high gene density and CpG island neighborhood strongly attract porcine endogenous retrovirus for integration and favor the formation of hot spots. J Virol. 2009;83:1920–1929. doi: 10.1128/JVI.00856-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MONSE H, LAUFS S, KUATE S, ZELLER WJ, FRUEHAUF S, UBERLA K. Viral determinants of integration site preferences of simian immunodeficiency virus-based vectors. J Virol. 2006;80:8145–8150. doi: 10.1128/JVI.00373-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MOOSLEHNER K, KARLS U, HARBERS K. Retroviral integration sites in transgenic Mov mice frequently map in the vicinity of transcribed DNA regions. J Virol. 1990;64:3056–3058. doi: 10.1128/jvi.64.6.3056-3058.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MULLER HP, VARMUS HE. DNA bending creates favored sites for retroviral integration: an explanation for preferred insertion sites in nucleosomes. EMBO J. 1994;13:4704–4714. doi: 10.1002/j.1460-2075.1994.tb06794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MUSSOLINO C, MORBITZER R, LUTGE F, DANNEMANN N, LAHAYE T, CATHOMEN T. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res. 2011;39:9283–9293. doi: 10.1093/nar/gkr597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NAREZKINA A, TAGANOV KD, LITWIN S, STOYANOVA R, HAYASHI J, SEEGER C, SKALKA AM, KATZ RA. Genome-wide analyses of avian sarcoma virus integration sites. J Virol. 2004;78:11656–11663. doi: 10.1128/JVI.78.21.11656-11663.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NAUGHTIN M, HAFTEK-TERREAU Z, XAVIER J, MEYER S, SILVAIN M, JASZCZYSZYN Y, LEVY N, MIELE V, BENLEULMI MS, RUFF M, PARISSI V, VAILLANT C, LAVIGNE M. DNA physical properties and nucleosome positions are major determinants of HIV-1 integrase selectivity. PLoS One. 2015;10:e0129427. doi: 10.1371/journal.pone.0129427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NEEL BG, HAYWARD WS, ROBINSON HL, FANG J, ASTRIN SM. Avian leukosis virus-induced tumors have common proviral integration sites and synthesize discrete new RNAs: oncogenesis by promoter insertion. Cell. 1981;23:323–334. doi: 10.1016/0092-8674(81)90128-8. [DOI] [PubMed] [Google Scholar]
- NEVES M, PERIES J, SAIB A. Study of human foamy virus proviral integration in chronically infected murine cells. Res Virol. 1998;149:393–401. doi: 10.1016/s0923-2516(99)80007-7. [DOI] [PubMed] [Google Scholar]
- NIU Y, SHEN B, CUI Y, CHEN Y, WANG J, WANG L, KANG Y, ZHAO X, SI W, LI W, XIANG AP, ZHOU J, GUO X, BI Y, SI C, HU B, DONG G, WANG H, ZHOU Z, LI T, TAN T, PU X, WANG F, JI S, ZHOU Q, HUANG X, JI W, SHA J. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell. 2014;156:836–843. doi: 10.1016/j.cell.2014.01.027. [DOI] [PubMed] [Google Scholar]
- NOORI-DALOII MR, SWIFT RA, KUNG HJ, CRITTENDEN LB, WITTER RL. Specific integration of REV proviruses in avian bursal lymphomas. Nature. 1981;294:574–576. doi: 10.1038/294574a0. [DOI] [PubMed] [Google Scholar]
- NOWAK MG, SUDOL M, LEE NE, KONSAVAGE WMJ, KATZMAN M. Identifying amino acid residues that contribute to the cellular-DNA binding site on retroviral integrase. Virology. 2009;389:141–148. doi: 10.1016/j.virol.2009.04.014. [DOI] [PubMed] [Google Scholar]
- NOWROUZI A, DITTRICH M, KLANKE C, HEINKELEIN M, RAMMLING M, DANDEKAR T, VON KALLE C, RETHWILM A. Genome-wide mapping of foamy virus vector integrations into a human cell line. J Gen Virol. 2006;87:1339–1347. doi: 10.1099/vir.0.81554-0. [DOI] [PubMed] [Google Scholar]
- NUSSE R, VARMUS HE. Many tumors induced by the mouse mammary tumor virus contain a provirus integrated in the same region of the host genome. Cell. 1982;31:99–109. doi: 10.1016/0092-8674(82)90409-3. [DOI] [PubMed] [Google Scholar]
- OCHMAN H, GERBER AS, HARTL DL. Genetic applications of an inverse polymerase chain reaction. Genetics. 1988;120:621–623. doi: 10.1093/genetics/120.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OCWIEJA KE, BRADY TL, RONEN K, HUEGEL A, ROTH SL, SCHALLER T, JAMES LC, TOWERS GJ, YOUNG JA, CHANDA SK, KONIG R, MALANI N, BERRY CC, BUSHMAN FD. HIV integration targeting: a pathway involving Transportin-3 and the nuclear pore protein RanBP2. PLoS Pathog. 2011;7:e1001313. doi: 10.1371/journal.ppat.1001313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PATTANAYAK V, RAMIREZ CL, JOUNG JK, LIU DR. Revealing off-target cleavage specificities of zinc-finger nucleases by in vitro selection. Nat Methods. 2011;8:765–770. doi: 10.1038/nmeth.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PAUZA CD. Two bases are deleted from the termini of HIV-1 linear DNA during integrative recombination. Virology. 1990;179:886–889. doi: 10.1016/0042-6822(90)90161-j. [DOI] [PubMed] [Google Scholar]
- PENG K, MURANYI W, GLASS B, LAKETA V, YANT SR, TSAI L, CIHLAR T, MÜLLER B, KRÄUSSLICH HG. Quantitative microscopy of functional HIV post-entry complexes reveals association of replication with the viral capsid. Elife. 2015;3:e04114. doi: 10.7554/eLife.04114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PEREIRA LA, BENTLEY K, PEETERS A, CHURCHILL MJ, DEACON NJ. A compilation of cellular transcription factor interactions with the HIV-1 LTR promoter. Nucleic Acids Res. 2000;28:663–668. doi: 10.1093/nar/28.3.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PETERS G, BROOKES S, SMITH R, DICKSON C. Tumorigenesis by mouse mammary tumor virus: evidence for a common region for provirus integration in mammary tumors. Cell. 1983;33:369–377. doi: 10.1016/0092-8674(83)90418-x. [DOI] [PubMed] [Google Scholar]
- PETERS G, PLACZEK M, BROOKES S, KOZAK C, SMITH R, DICKSON C. Characterization, chromosome assignment, and segregation analysis of endogenous proviral units of mouse mammary tumor virus. J Virol. 1986;59:535–544. doi: 10.1128/jvi.59.3.535-544.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PRADEEPA MM, SUTHERLAND HG, ULE J, GRIMES GR, BICKMORE WA. Psip1/Ledgf p52 binds methylated histone H3K36 and splicing factors and contributes to the regulation of alternative splicing. PLoS Genet. 2012;8:e1002717. doi: 10.1371/journal.pgen.1002717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PRUSS D, BUSHMAN FD, WOLFFE AP. Human immunodeficiency virus integrase directs integration to sites of severe DNA distortion within the nucleosome core. Proc Natl Acad Sci USA. 1994a;91:5913–5917. doi: 10.1073/pnas.91.13.5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PRUSS D, REEVES R, BUSHMAN FD, WOLFFE AP. The influence of DNA and nucleosome structure on integration events directed by HIV integrase. J Biol Chem. 1994b;269:25031–25041. [PubMed] [Google Scholar]
- PRYCIAK PM, MULLER HP, VARMUS HE. Simian virus 40 minichromosomes as targets for retroviral integration in vivo. Proc Natl Acad Sci USA. 1992;89:9237–9241. doi: 10.1073/pnas.89.19.9237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RANZANI M, ANNUNZIATO S, ADAMS DJ, MONTINI E. Cancer gene discovery: exploiting insertional mutagenesis. Mol Cancer Res. 2013;11:1141–1158. doi: 10.1158/1541-7786.MCR-13-0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RINGOLD GM, SHANK PR, VARMUS HE, RING J, YAMAMOTO KR. Integration and transcription of mouse mammary tumor virus DNA in rat hepatoma cells. Proc Natl Acad Sci USA. 1979;76:665–669. doi: 10.1073/pnas.76.2.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROBINSON HL, GAGNON GC. Patterns of proviral insertion and deletion in avian leukosis virus-induced lymphomas. J Virol. 1986;57:28–36. doi: 10.1128/jvi.57.1.28-36.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROE T, REYNOLDS TC, YU G, BROWN PO. Integration of murine leukemia virus DNA depends on mitosis. EMBO J. 1993;12:2099–3108. doi: 10.1002/j.1460-2075.1993.tb05858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROHDEWOHLD H, WEIHER H, REIK W, JAENISCH R, BREINDL M. Retrovirus integration and chromatin structure: Moloney murine leukemia proviral integration sites map near DNase I-hypersensitive sites. J Virol. 1987;61:336–343. doi: 10.1128/jvi.61.2.336-343.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROSENTHAL A, JONES DS. Genomic walking and sequencing by oligo-cassette mediated polymerase chain reaction. Nucleic Acids Res. 1990;18:3095–3096. doi: 10.1093/nar/18.10.3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROTH MJ, SCHWARTZBERG PL, GOFF SP. Structure of the termini of DNA intermediates in the integration of retroviral DNA: dependence on IN function and terminal DNA sequence. Cell. 1989;58:47–54. doi: 10.1016/0092-8674(89)90401-7. [DOI] [PubMed] [Google Scholar]
- ROTH SL, MALANI N, BUSHMAN FD. Gammaretroviral integration into nucleosomal target DNA in vivo. J Virol. 2011;85:7393–7401. doi: 10.1128/JVI.00635-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SALINAS J, ZERIAL M, FILIPSKI J, CREPIN M, BERNARDI G. Nonrandom distribution of MMTV proviral sequences in the mouse genome. Nucleic Acids Res. 1987;15:3009–3022. doi: 10.1093/nar/15.7.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SANGER F, NICKLEN S, COULSON AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHALLER T, OCWIEJA KE, RASAIYAAH J, PRICE AJ, BRADY TL, ROTH SL, HUE S, FLETCHER AJ, LEE K, KEWALRAMANI VN, NOURSADEGHI M, JENNER RG, JAMES LC, BUSHMAN FD, TOWERS GJ. HIV-1 capsid-cyclophilin interactions determine nuclear import pathway, integration targeting and replication efficiency. PLoS Pathog. 2011;7:e1002439. doi: 10.1371/journal.ppat.1002439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHAMBACH A, MUELLER D, GALLA M, VERSTEGEN MM, WAGEMAKER G, LOEW R, BAUM C, BOHNE J. Overcoming promoter competition in packaging cells improves production of self-inactivating retroviral vectors. Gene Ther. 2006;13:1524–1533. doi: 10.1038/sj.gt.3302807. [DOI] [PubMed] [Google Scholar]
- SCHERDIN U, RHODES K, BREINDL M. Transcriptionally active genome regions are preferred targets for retrovirus integration. J Virol. 1990;64:907–912. doi: 10.1128/jvi.64.2.907-912.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHRIJVERS R, DE RIJCK J, DEMEULEMEESTER J, ADACHI N, VETS S, RONEN K, CHRIST F, BUSHMAN FD, DEBYSER Z, GIJSBERS R. LEDGF/p75-independent HIV-1 replication demonstrates a role for HRP-2 and remains sensitive to inhibition by LEDGINs. PLoS Pathog. 2012a;8:e1002558. doi: 10.1371/journal.ppat.1002558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHRIJVERS R, VETS S, DE RIJCK J, MALANI N, BUSHMAN FD, DEBYSER Z, GIJSBERS R. HRP-2 determines HIV-1 integration site selection in LEDGF/p75 depleted cells. Retrovirology. 2012b;9:84. doi: 10.1186/1742-4690-9-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHRODER AR, SHINN P, CHEN H, BERRY C, ECKER JR, BUSHMAN F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- SCHWARZWAELDER K, HOWE SJ, SCHMIDT M, BRUGMAN MH, DEICHMANN A, GLIMM H, SCHMIDT S, PRINZ C, WISSLER M, KING DJ, ZHANG F, PARSLEY KL, GILMOUR KC, SINCLAIR J, BAYFORD J, PERAJ R, PIKE-OVERZET K, STAAL FJ, DE RIDDER D, KINNON C, ABEL U, WAGEMAKER G, GASPAR HB, THRASHER AJ, VON KALLE C. Gammaretrovirus-mediated correction of SCID-X1 is associated with skewed vector integration site distribution in vivo. J Clin Invest. 2007;117:2241–2249. doi: 10.1172/JCI31661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SEIKI M, EDDY R, SHOWS TB, YOSHIDA M. Nonspecific integration of the HTLV provirus genome into adult T-cell leukaemia cells. Nature. 1984;309:640–642. doi: 10.1038/309640a0. [DOI] [PubMed] [Google Scholar]
- SERRAO E, BALLANDRAS-COLAS A, CHEREPANOV P, MAERTENS GN, ENGELMAN AN. Key determinants of target DNA recognition by retroviral intasomes. Retrovirology. 2015;12:39. doi: 10.1186/s12977-015-0167-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SERRAO E, KRISHNAN L, SHUN MC, LI X, CHEREPANOV P, ENGELMAN A, MAERTENS GN. Integrase residues that determine nucleotide preferences at sites of HIV-1 integration: implications for the mechanism of target DNA binding. Nucleic Acids Res. 2014;42:5164–5176. doi: 10.1093/nar/gku136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHAN L, YANG HC, RABI SA, BRAVO HC, SHROFF NS, IRIZARRY RA, ZHANG H, MARGOLICK JB, SILICIANO JD, SILICIANO RF. Influence of host gene transcription level and orientation on HIV-1 latency in a primary-cell model. J Virol. 2011;85:5384–5393. doi: 10.1128/JVI.02536-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHARMA A, LARUE RC, PLUMB MR, MALANI N, MALE F, SLAUGHTER A, KESSL JJ, SHKRIABAI N, COWARD E, AIYER SS, GREEN PL, WU L, ROTH MJ, BUSHMAN FD, KVARATSKHELIA M. BET proteins promote efficient murine leukemia virus integration at transcription start sites. Proc Natl Acad Sci USA. 2013;110:12036–12041. doi: 10.1073/pnas.1307157110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHERRILL-MIX S, LEWINSKI MK, FAMIGLIETTI M, BOSQUE A, MALANI N, OCWIEJA KE, BERRY CC, LOONEY D, SHAN L, AGOSTO LM, PACE MJ, SILICIANO RF, O’DOHERTY U, GUATELLI J, PLANELLES V, BUSHMAN FD. HIV latency and integration site placement in five cell-based models. Retrovirology. 2013;10:90. doi: 10.1186/1742-4690-10-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHIH CC, STOYE JP, COFFIN JM. Highly preferred targets for retrovirus integration. Cell. 1988;53:531–537. doi: 10.1016/0092-8674(88)90569-7. [DOI] [PubMed] [Google Scholar]
- SHIMOTOHNO K, MIZUTANI S, TEMIN HM. Sequence of retrovirus provirus resembles that of bacterial transposable elements. Nature. 1980;285:550–554. doi: 10.1038/285550a0. [DOI] [PubMed] [Google Scholar]
- SHIMOTOHNO K, TEMIN HM. No apparent nucleotide sequence specificity in cellular DNA juxtaposed to retrovirus proviruses. Proc Natl Acad Sci USA. 1980;77:7357–7361. doi: 10.1073/pnas.77.12.7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHUN MC, BOTBOL Y, LI X, DI NUNZIO F, DAIGLE JE, YAN N, LIEBERMAN J, LAVIGNE M, ENGELMAN A. Identification and Characterization of PWWP Domain Residues Critical for LEDGF/p75 Chromatin Binding and Human Immunodeficiency Virus Type 1 Infectivity. J Virol. 2008;82:11555–11567. doi: 10.1128/JVI.01561-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHUN MC, RAGHAVENDRA NK, VANDEGRAAFF N, DAIGLE JE, HUGHES S, KELLAM P, CHEREPANOV P, ENGELMAN A. LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes Dev. 2007;21:1767–1778. doi: 10.1101/gad.1565107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SILVER J, KEERIKATTE V. Novel use of polymerase chain reaction to amplify cellular DNA adjacent to an integrated provirus. J Virol. 1989;63:1924–1928. doi: 10.1128/jvi.63.5.1924-1928.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SILVERS RM, SMITH JA, SCHOWALTER M, LITWIN S, LIANG Z, GEARY K, DANIEL R. Modification of integration site preferences of an HIV-1-based vector by expression of a novel synthetic protein. Hum Gene Ther. 2010;21:337–349. doi: 10.1089/hum.2009.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SORENSEN AB, DUCH M, AMTOFT HW, JORGENSEN P, PEDERSEN FS. Sequence tags of provirus integration sites in DNAs of tumors induced by the murine retrovirus SL3-3. J Virol. 1996;70:4063–4070. doi: 10.1128/jvi.70.6.4063-4070.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SORENSEN AB, DUCH M, JORGENSEN P, PEDERSEN FS. Amplification and sequence analysis of DNA flanking integrated proviruses by a simple two-step polymerase chain reaction method. J Virol. 1993;67:7118–7124. doi: 10.1128/jvi.67.12.7118-7124.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STEFFEN D, WEINBERG RA. The integrated genome of murine leukemia virus. Cell. 1978;15:1003–1010. doi: 10.1016/0092-8674(78)90284-2. [DOI] [PubMed] [Google Scholar]
- STEVENS SW, GRIFFITH JD. Sequence analysis of the human DNA flanking sites of human immunodeficiency virus type 1 integration. J Virol. 1996;70:6459–6462. doi: 10.1128/jvi.70.9.6459-6462.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SUZUKI T, SHEN H, AKAGI K, MORSE HC, MALLEY JD, NAIMAN DQ, JENKINS NA, COPELAND NG. New genes involved in cancer identified by retroviral tagging. Nat Genet. 2002;32:166–174. doi: 10.1038/ng949. [DOI] [PubMed] [Google Scholar]
- TAGANOV KD, CUESTA I, DANIEL R, CIRILLO LA, KATZ RA, ZARET KS, SKALKA AM. Integrase-specific enhancement and suppression of retroviral DNA integration by compacted chromatin structure in vitro. J Virol. 2004;78:5848–5855. doi: 10.1128/JVI.78.11.5848-5855.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TAN W, DONG Z, WILKINSON TA, BARBAS CF, 3RD, CHOW SA. Human immunodeficiency virus type 1 incorporated with fusion proteins consisting of integrase and the designed polydactyl zinc finger protein E2C can bias integration of viral DNA into a predetermined chromosomal region in human cells. J Virol. 2006;80:1939–1948. doi: 10.1128/JVI.80.4.1939-1948.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TEMIN HM. Mechanism of cell transformation by RNA tumor viruses. Ann Rev Microbiol. 1971;25:609–648. doi: 10.1146/annurev.mi.25.100171.003141. [DOI] [PubMed] [Google Scholar]
- THORNHILL SI, SCHAMBACH A, HOWE SJ, ULAGANATHAN M, GRASSMAN E, WILLIAMS D, SCHIEDLMEIER B, SEBIRE NJ, GASPAR HB, KINNON C, BAUM C, THRASHER AJ. Self-inactivating gammaretroviral vectors for gene therapy of X-linked severe combined immunodeficiency. Mol Ther. 2008;16:590–598. doi: 10.1038/sj.mt.6300393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TRIGLIA T, PETERSON MG, KEMP DJ. A procedure for in vitro amplification of DNA segments that lie outside the boundaries of known sequences. Nucleic Acids Res. 1988;16:8186. doi: 10.1093/nar/16.16.8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TROBRIDGE GD, MILLER DG, JACOBS MA, ALLEN JM, KIEM HP, KAUL R, RUSSELL DW. Foamy virus vector integration sites in normal human cells. Proc Natl Acad Sci USA. 2006;103:1498–1503. doi: 10.1073/pnas.0510046103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TSUKAHARA T, AGAWA H, MATSUMOTO S, MATSUDA M, UENO S, YAMASHITA Y, YAMADA K, TANAKA N, KOJIMA K, TAKESHITA T. Murine leukemia virus vector integration favors promoter regions and regional hot spots in a human T-cell line. Biochem Biophys Res Commun. 2006;345:1099–1107. doi: 10.1016/j.bbrc.2006.05.007. [DOI] [PubMed] [Google Scholar]
- TURLURE F, MAERTENS G, RAHMAN S, CHEREPANOV P, ENGELMAN A. A tripartite DNA-binding element, comprised of the nuclear localization signal and two AT-hook motifs, mediates the association of LEDGF/p75 with chromatin in vivo. Nucleic Acids Res. 2006;34:1653–1665. doi: 10.1093/nar/gkl052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UMLAUF D, BONNET J, WAHARTE F, FOURNIER M, STIERLE M, FISCHER B, BRINO L, DEVYS D, TORA L. The human TREX-2 complex is stably associated with the nuclear pore basket. J Cell Sci. 2013;126:2656–2667. doi: 10.1242/jcs.118000. [DOI] [PubMed] [Google Scholar]
- VALKOV E, GUPTA SS, HARE S, HELANDER A, ROVERSI P, MCCLURE M, CHEREPANOV P. Functional and structural characterization of the integrase from the prototype foamy virus. Nucleic Acids Res. 2009;37:243–255. doi: 10.1093/nar/gkn938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VAN MAELE B, BUSSCHOTS K, VANDEKERCKHOVE L, CHRIST F, DEBYSER Z. Cellular co-factors of HIV-1 integration. Trends Biochem Sci. 2006;31:98–105. doi: 10.1016/j.tibs.2005.12.002. [DOI] [PubMed] [Google Scholar]
- VANDEGRAAFF N, KUMAR R, BURRELL CJ, LI P. Kinetics of human immunodeficiency virus type 1 (HIV) DNA integration in acutely infected cells as determined using a novel assay for detection of integrated HIV DNA. J Virol. 2001;75:11253–11260. doi: 10.1128/JVI.75.22.11253-11260.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VANDEKERCKHOVE L, CHRIST F, VAN MAELE B, DE RIJCK J, GIJSBERS R, VAN DEN HAUTE C, WITVROUW M, DEBYSER Z. Transient and stable knockdown of the integrase cofactor LEDGF/p75 reveals its role in the replication cycle of human immunodeficiency virus. J Virol. 2006;80:1886–1896. doi: 10.1128/JVI.80.4.1886-1896.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VIJAYA S, STEFFEN DL, ROBINSON HL. Acceptor sites for retroviral integrations map near DNase I-hypersensitive sites in chromatin. J Virol. 1986;60:683–692. doi: 10.1128/jvi.60.2.683-692.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VINCENT KA, YORK-HIGGINS D, QUIROGA M, BROWN PO. Host sequences flanking the HIV provirus. Nucleic Acids Res. 1990;18:6045–6047. doi: 10.1093/nar/18.20.6045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VINK C, GROENINK M, ELGERSMA Y, FOUCHIER RA, TERSMETTE M, PLASTERK RH. Analysis of the junctions between human immunodeficiency virus type 1 proviral DNA and human DNA. J Virol. 1990;64:5626–5627. doi: 10.1128/jvi.64.11.5626-5627.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VOGT PK. The Genetics of RNA Tumor Viruses. In: FRAENKEL-CONRAT HRW, editor. Comprehensive Virology. New York: Plenum Press; 1977. [Google Scholar]
- WAGNER TA, MCLAUGHLIN S, GARG K, CHEUNG CY, LARSEN BB, STYRCHAK S, HUANG HC, EDLEFSEN PT, MULLINS JI, FRENKEL LM. Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science. 2014;345:570–573. doi: 10.1126/science.1256304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WAN H, FENG C, TENG F, YANG S, HU B, NIU Y, XIANG AP, FANG W, JI W, LI W, ZHAO X, ZHOU Q. One-step generation of p53 gene biallelic mutant Cynomolgus monkey via the CRISPR/Cas system. Cell Res. 2015;25:258–261. doi: 10.1038/cr.2014.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WANG GP, BERRY CC, MALANI N, LEBOULCH P, FISCHER A, HACEIN-BEY-ABINA S, CAVAZZANA-CALVO M, BUSHMAN FD. Dynamics of gene-modified progenitor cells analyzed by tracking retroviral integration sites in a human SCID-X1 gene therapy trial. Blood. 2010;115:4356–4366. doi: 10.1182/blood-2009-12-257352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WANG GP, CIUFFI A, LEIPZIG J, BERRY CC, BUSHMAN FD. HIV integration site selection: analysis by massively parallel pyrosequencing reveals association with epigenetic modifications. Genome Res. 2007;17:1186–1194. doi: 10.1101/gr.6286907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WANG H, JURADO KA, WU X, SHUN MC, LI X, FERRIS AL, SMITH SJ, PATEL PA, FUCHS JR, CHEREPANOV P, KVARATSKHELIA M, HUGHES SH, ENGELMAN A. HRP2 determines the efficiency and specificity of HIV-1 integration in LEDGF/p75 knockout cells but does not contribute to the antiviral activity of a potent LEDGF/p75-binding site integrase inhibitor. Nucleic Acids Res. 2012;40:11518–11530. doi: 10.1093/nar/gks913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WANG H, SHUN MC, LI X, DI NUNZIO F, HARE S, CHEREPANOV P, ENGELMAN A. Efficient transduction of LEDGF/p75 mutant cells by gain-of-function HIV-1 integrase mutant viruses. Mol Ther Methods Clin Dev. 2014;1 doi: 10.1038/mtm.2013.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WANG JY, LING H, YANG W, CRAIGIE R. Structure of a two-domain fragment of HIV-1 integrase: implications for domain organization in the intact protein. EMBO J. 2001;20:7333–7343. doi: 10.1093/emboj/20.24.7333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WILLIAMS DA, LEMISCHKA IR, NATHAN DG, MULLIGAN RC. Introduction of new genetic material into pluripotent haematopoietic stem cells of the mouse. Nature. 1984;310:476–480. doi: 10.1038/310476a0. [DOI] [PubMed] [Google Scholar]
- WITHERS-WARD ES, KITAMURA Y, BARNES JP, COFFIN JM. Distribution of targets for avian retrovirus DNA integration in vivo. Genes Dev. 1994;8:1473–1487. doi: 10.1101/gad.8.12.1473. [DOI] [PubMed] [Google Scholar]
- WU X, LI Y, CRISE B, BURGESS SM. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- WU X, LI Y, CRISE B, BURGESS SM, MUNROE DJ. Weak palindromic consensus sequences are a common feature found at the integration target sites of many retroviruses. J Virol. 2005;79:5211–5214. doi: 10.1128/JVI.79.8.5211-5214.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- XIE W, GAI X, ZHU Y, ZAPPULLA DC, STERNGLANZ R, VOYTAS DF. Targeting of the yeast Ty5 retrotransposon to silent chromatin is mediated by interactions between integrase and Sir4p. Mol Cell Biol. 2001;21:6606–6614. doi: 10.1128/MCB.21.19.6606-6614.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YANG Y, FRICKE T, DIAZ-GRIFFERO F. Inhibition of reverse transcriptase activity increases stability of the HIV-1 core. J Virol. 2013;87:683–687. doi: 10.1128/JVI.01228-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YANG ZN, MUESER TC, BUSHMAN FD, HYDE CC. Crystal structure of an active two-domain derivative of Rous sarcoma virus integrase. J Mol Biol. 2000;296:535–548. doi: 10.1006/jmbi.1999.3463. [DOI] [PubMed] [Google Scholar]
- ZHU Y, DAI J, FUERST PG, VOYTAS DF. Controlling integration specificity of a yeast retrotransposon. Proc Natl Acad Sci USA. 2003;100:5891–5895. doi: 10.1073/pnas.1036705100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZOUBAK S, RICHARDSON JH, RYNDITCH A, HOLLSBERG P, HAFLER DA, BOERI E, LEVER AM, BERNARDI G. Regional specificity of HTLV-I proviral integration in the human genome. Gene. 1994;143:155–163. doi: 10.1016/0378-1119(94)90091-4. [DOI] [PubMed] [Google Scholar]