Abstract
Prostate specific antigen (PSA) is currently used as a diagnostic biomarker for prostate cancer. It is a glycoprotein possessing a single glycosylation site at N69. During our previous study of PSA N69 glycosylation, additional glycopeptides were observed in the PSA sample that were not previously reported and did not match glycopeptides of impure glycoproteins existed in the sample. This extra glycosylation site of PSA is associated with mutation in KLK3 genes. Among single nucleotide polymorphisms (SNPs) of KLKs families, the rs61752561 in KLK3 genes is an unusual missense mutation resulting in the conversion of D102 to N in PSA amino acid sequence. Accordingly, a new N-linked glycosylation site is created with an N102MS motif. Here, we report the first qualitative and quantitative glycoproteomic study of PSA N102 glycosylation site by LC-MS/MS. We successfully applied tandem MS to verify the amino acid sequence possessing N102 glycosylation site and associated glycoforms of PSA samples acquired from different suppliers. A total of 21, 7, and 16 glycoforms were detected for LeeBio, Sigma, and EMD PSA samples, respectively. Interestingly, fucosylated glycopeptides were not detected on N102. Among the 3 PSA samples, HexNAc2Hex5 was the predominant glycoform at N102 while HexNAc4Hex5Fuc1NeuAc1 or HexNAc4Hex5Fuc1NeuAc2 were the primary glycoforms at N69.
Keywords: Prostate specific antigen, PSA, missense mutation, rs61752561, D102N, N-linked glycosylation, glycopeptide, glycoproteomics, LC-MS/MS
Graphical Abstract
Introduction
Prostate specific antigen (PSA) is currently used as a diagnostic biomarker for prostate cancer.1 PSA is a member of the kallikrein-related peptidases family.2–4 The kallikrein-related peptidases (KLKs) family consists of 15 members (KLK1~15), which are encoded by the largest cluster of protease-encoding genes (KLK1–15) in the human genome.5, 6 These genes are located on human chromosome 19.5, 6 The KLKs family is a subgroup of secreted trypsin- or chymotrypsin- like serine proteases and PSA is encoded by KLK3 gene.2–4 PSA has been commonly reported as a glycoprotein with a single glycosylation site at N69. An approximately 8% of PSA weight is believed to be due to the microheterogeneity of this glycosylation site.7 Many studies have previously reported the microheterogeneity of PSA on N69. A change in the degree of sialylation of PSA among healthy and malignant origin have also been reported.8–10 It has also been reported that PSA depicts charge heterogeneity due to a different level of sialylation.11–13 PSA N-glycans were also reported to be mostly core fucosylated and possess a minor presence of GalNAc residues with an increasing isoelectric point (pI) of PSA fraction.14–16
Recently, we have reported a comprehensive identification and quantitation of the glycoyslation of two PSA isoforms by LC-MS/MS.17 There were 56 N-glycans associated with PSA while 57 N-glycans were observed in the case of PSA-high pI isoform (PSAH). The high abundance of core-fucosylation and sialylation were noted in PSAH. Also, more GalNAc residue attached to antenna GlcNAc residue and highly branched glycan structures were identified in PSAH. Moreover, these results were compared to the 2012 ABRF Glycoprotein Research Group (gPRG) study18 and a separate study by Behnken et al19. Twenty-six national and international laboratories participated in this 2012 ABRF study. Our laboratory and Behnken et al.19, were among the ABRF participating laboratories. Behnken et al. used top-down approach while we used bottom-up approach. Three studies reported 85 total glycoforms associated with PSA and PSAH including 29 common glycoforms. In regard to the 18 major/intermediate glycoforms determined by the 2012 ABRF study, comparable abundances were observed in both our and Behnken et al. studies.
Analysis of our previously reported data17, 18 revealed the presence of additional glycopeptides that were not originating from impurities related to the samples or commonly known PSA glycosylation site (N69). These glycopeptides were determined to be due to the other glycosylation site originating from the mutation of KLK3 genes. There are several genetic mutations in KLK3 genes.20 Many reports have described the type/level of genetic mutations of KLK3 genes and correlated them with prostate cancer risk and PSA levels in the blood.21–27 They investigated different single nucleotide polymorphisms (SNPs), which may change protein sequence (nonsynonymous SNPs). Rodriguez et al. 26 reported particular SNPs deleting KLK3 genes that lower PSA concentrations in blood, leading to false-negative results in PSA-based diagnostic tests for prostate cancer. Also, Gallagher et al.22 reported two SNPs, rs61752561 in KLK3 and rs2735839 in KLK2–KLK3 intergenic region, which have high correlation with prostate cancer-specific survival. Notably, the rs61752561 in KLK3 genes is a missense mutation that is a type of nonsynonymous SNPs. Missense mutation substitutes a single base pair in a codon promoting the translation of a different amino acid in the protein. This rs61752561 evolves a G to A transition in the 34th codon of KLK3 exon 3 (c.304G>A) resulting in D102 to N conversion (D102N).21 This missense mutation now creates a new N-linked glycosylation site with an N102MS motif. Thus far, the microheterogeneity of this glycosylation site prompted by the missense mutation is not reported.
Here, we report the first qualitative and quantitative glycoproteomic studies of PSA N102 glycosylation site by LC-MS/MS. We successfully applied tandem MS including collision-induced dissociation (CID) and electron transfer dissociation (ETD) to verify the amino acid sequence possessing N102 glycosylation site and the glycoforms of this site. PSA proteins from 3 different vendors were obtained, and the microheterogeneities of their N102 glycosylation site were investigated. Moreover, the levels of glycosylation at N69 and N102 were compared.
Experimental Section
Materials
PSA and PSA-high pI isoform (PSAH) samples were obtained from Lee Biosolutions (St. Louis, MO). Additional PSA samples were purchased from Sigma (St. Louis, MO) and EMD Millipore (Billerica, MA). Sodium chloride and disodium phosphate were obtained from Mallinckrodt Chemicals (Phillipsburg, NJ). DL-dithiothreitol (DTT), iodoacetamide (IAA), and MS-grade formic acid were purchased from Sigma-Aldrich (St. Louis, MO). HPLC-grade solvents, including methanol and isopropanol, were obtained from Fisher Scientific (Pittsburgh, PA). HPLC grade water was obtained from Mallinckrodt (Hazelwood, MO). HPLC grade acetonitrile (ACN) was obtained from J.T. Baker (Phillipsburg, NJ). Mass spectrometry Grade Trypsin Gold was purchased from Promega (Madison, WI). Borane-ammonia complex, dimethyl sulfoxide (DMSO), sodium hydroxide beads, and iodomethane were purchased from Sigma-Aldrich (St. Louis, MO). Empty micro-spine columns were obtained from Harvard Apparatus (Holliston, MA). N-Glycosidase peptide purified from Flavobacterium meningosepticum (PNGase F) was purchased from New England Biolabs Inc. (Ipswich, MA).
Enzymatic Digestion for Glycoproteomic Study
Since we used PSA data for Lee Biosolutions (LeeBio PSA) from our previous studies,17, 18 the sample preparation and the LC-MS/MS method of PSA samples obtained from Sigma (Sigma PSA) and EMD Millipore (EMD PSA) were the same. Briefly, a 10-μg aliquot of Sigma PSA and EMD PSA were suspended in a 50 mM phosphate buffered saline (PBS, pH 7.5), containing 50 mM disodium phosphate and 150 mM sodium chloride. The samples were reduced by adding by adding a 1.25-μl aliquot of 200 mM DTT prior to incubation at 60° C for 45 min. Those reduced samples were then alkylated with the addition of a 5-μl aliquot of 200mM IAA and incubated at 37.5° C for 45 min in the dark. Excess IAA was consumed through the addition of a second 1.25-μl aliquot of 200 mM DTT. The reaction was allowed to proceed at 37.5° C for 30 min in the dark. The trypsin was added to the samples using the enzyme/substrate ratio of 1:25 w/w and subjected to overnight incubation at 37.5° C for 18 hours. Samples were subjected to microwave digestion at 45° C and 50 W for 30min before adding 0.5-μl aliquot of neat formic acid to the samples to complete enzymatic digestion. The enzymatic digestion was then quenched. Finally, the samples were dried and suspended in 0.1% formic acid prior to LC-MS/MS analysis. The samples were analyzed in technical triplicates.
Permethylation for Glycomic Study
1 μg of LeeBio PSA and PSAH was added into 9μl ammonium bicarbonate buffer (20 mM). The samples were mixed and denatured at 80°C for 1h and cool down to room temperature. 1.2μl of PNGase F (60 unit) was added to each sample and incubated at 37°C for 18h. The released glycans were dried under vacuum. 0.1–0.2 mg of borane-ammonia complex was dissolved in HPLC water to a final concentration of 1μg/μl. Dried samples were resuspended in 10 μl of borane ammonium solution and incubated at 65°C for one hour. Then, the reaction mixtures were dried under vacuum. The remaining borate was removing by adding 300 μl methanol to each sample and dry under vacuum. This process was repeated several times to evaporate all borate salt. The reduced samples were permethylated using the previously published protocols.28–30 Briefly, sodium hydroxide filled spin column was first prepared. The empty column was filled with sodium hydroxide beads and washed with DMSO. The dried samples were resuspended in 1.2 μl water, 30 μl DMSO and 20 μl iodomethane mixture and applied to the sodium hydroxide filled column. The reaction mixtures were kept at room temperature for 25 min. Then, another 20 μl of iodomethane was added to the top of spin column and incubated for another 20 min. The permethylated samples were spun down and collected. 50 μl of ACN was added to the spin column and centrifuged to elute all remaining samples.
LC-MS/MS Analyses of PSA Glycopeptides/Peptides
LC-MS/MS was carried out on Dionex 3000 Ultimate nano-LC system (Dionex, Sunnyvale, CA) interfaced to LTQ Orbitrap Velos mass spectrometer (Thermo Scientific, San Jose, CA) equipped with a nano-ESI source. The PSA and PSAH digests were initially online-purified using a PepMap 100 C18 cartridge (3 μm, 100Å, Dionex). A 2-μg aliquot of Sigma PSA and EMD PSA digests was injected into the trapping cartridges. The purified peptides were then separated using a PepMap 100 C18 capillary column (75 μm id × 150 mm, 2 μm, 100Å, Dionex). The separation was achieved at 350 nl/min flow rate, using the following gradient conditions: 0–10 min 5% solvent B (98% ACN with 0.1% formic acid), 10–40 min ramping of solvent B from 5 to 45%, 40–45 min ramping of solvent B from 45 to 80%, 45–50 min maintaining solvent B at 80%, and 50–51 min reducing solvent B to 5%, and 51–60 min sustaining solvent B at 5%. Solvent A was a 2% ACN aqueous solution containing 0.1 % formic acid. The separation and MS scan time was set to 60 min.
The LTQ Orbitrap Velos mass spectrometer was operated with three scan events. The first scan event was a full MS scan of 500–2000 m/z range with a mass resolution of 15,000. The CID (collision induced dissociation) and HCD the (higher-energy collision dissociation) MS/MS were performed on the 5 most intense ions seen from the first MS scan event. The second scan event was a CID MS/MS of precursor ions selected from the first scan event with an isolation width of 3.0 m/z, a normalized collision energy (CE) of 35%, and an activation Q value of 0.250. The third scan event was set to acquire HCD MS/MS of the parent ions selected from the first scan event. The isolation width of HCD experiment was set to 3.0 m/z while the normalized CE was set to 45% with an activation time of 0.1ms. In a separate LC-MS/MS, ETD was conducted in conjunction with CID and HCD. The first scan event was a full MS scan and 15 scan events were followed alternating between CID, HCD, and ETD. This sequence was performed for the 5 most intense ions observed in the first scan. For ETD, an isolation width was set to 3.0 m/z and a default charge state was set to 4. A reaction time was set to 150 ms with a supplemental activation.
LC-MS/MS Analyses of PSA Permethylated Glycans
The dried samples were re-suspended in 20% ACN and transferred to HPLC vial. Separation was performed using solvent A and solvent B. Solvent A consists of 2% ACN and 98% water with 0.1% formic acid and solvent B consists of 100% ACN and 0.1% formic acid. Samples were first loaded with 100% solvent A with flow rate of 3 μl/min to Acclaim® PepMap100 C18 trap column for online purification.31 At 10 min, the valve switched to the separation column. The separation was started at 20% solvent B. The LC gradient increased from 38% to 45% in 32 min with flow rate of 350 nl/min. Nano-LC system was interfaced to the LTQ Orbitrap Velos mass spectrometer. The mass spectrometer was operated in data-dependent mode. Mass resolution of 15000 was employed for MS full scan (m/z from 500–2000). Eight most abundant ions were selected at ±1.5 Da mass tolerance and subjected to CID and HCD MS/MS. CID MSMS was set with a 0.250 Q-value, 20 ms activation time, and 35% normalized collision energy while HCD was conducted at 7500 resolution with 0.1 ms activation time, 45% normalized collision energy. Dynamic exclusion (peak count was 2, peak duration 30s, exclusion list size 50, exclusion duration 60s) was applied to increase MS/MS spectra for glycan structure interpretation.
Data Processing and Quantitation
The identification of peptides/proteins was confirmed using MASCOT.32 Proteome Discoverer version 1.2 software (Thermo Scientific, San Jose, CA) was used to generate a mascot generic format file (*.mgf) which was subsequently employed for database searching using MASCOT version 2.3.2 (Matrix Science Inc., Boston, MA). Parent ions were selected from a mass range of 300–10000Da with a minimum peak count of 1. The parameters from Mascot Daemon were set to search against UniProtKB (UniProt releases 2014_06). A separate database was created to search mutated PSA amino acid sequence. In original PSA fasta file (Uniprot Accession number: P07288), D102 was modified to N102. Also, other D residues in a potential N-liked glycosylation motif (DXS/T/C) were modified to N residues considering that more missense mutations may occur. Hence, D115, D116, and D182 were reflected as potential glycosylation sites. Oxidation and carbamidomethylation of methionine33 were set as a variable modification while carbamidomethylation of cysteine was set as a fixed modification. Alkylated methionine is formed because of the reaction with iodoacetamide and it creates a neutral loss of 2-(methylthio)acetamide (C3H7NOS, 105.025Da) in CID MS/MS.33 Thus, a neutral loss of 2-(methylthio)acetamide in carbamidomethylation of methionine was set up for searching peptides and scoring ions. Regarding the ETD data, HexNAc2Hex5, HexNAc2Hex6, HexNAc4Hex5NeuAc2, and HexNAc4Hex5Fuc1NeuAc2 were set as additional variable modifications to sequence glycopeptides with glycan intact. Tandem MS ions were searched within 0.8 Da mass tolerances while the peptide sequences were identified within 10ppm. Because trypsin is well-known to produce incomplete cleavage of C-terminus lysine, 2 missed cleavages were allowed to assign tandem MS: only HSLFHPEDTGQVFQVSHSFPHPLYN102MSLLK sequence was scored. The results from MASCOT were imported into Scaffold 3 (Proteome Software, Inc., Portland, OR), where spectral-count quantitation and sequence coverage was checked. The protein identifications were based on ion score cut-off higher than 20 and a minimum number of peptides of 2.
The identification of glycopeptides was attained by manual confirmation and MASCOT database searching. Since a maximum number of modifications that can be set in MASCOT database searching are 7, a limited number of glycan structures were imported into MASCOT software. Therefore, the theoretical m/z values of glycopeptides were created based on theoretical peptide backbone sequence using Peptide Mass tool from ExPASy website. These m/z values were used to create extracted ion chromatograms (EICs) using Xcalibur Qual Broswer 2.1 (Thermo Scientific, San Jose, CA). Also, the glycopeptides were searched through GlycoSeq software (available as open source software at http://sourceforge.net/projects/glycoseq/, manuscript in preparation). A default glycan library contains 413 glycan compositions with combinations of GlcNAc, Man, Fuc, and NeuAc. GlycoSeq was used to determine glycan sequences (i.e. topology or cartoon-graph representation) using a de-novo sequencing algorithm. A mass accuracy of 7ppm or better was then applied to confirm ions prior to manual evaluation of tandem MS data. As a result, 3.51ppm, 2.47ppm, and 2.96ppm were obtained as an averaged mass accuracy from LeeBio, Sigma, and EMD PSA samples, respectively.
For quantitation, peak areas were acquired using Xcalibur Qual Browser. The software was used to generate EICs. Mass range was set to full FTMS scan with 7-point smoothing enabled and mass tolerance of 10ppm allowed. The obtained peak areas were normalized by a total abundance of glycopeptides. Then, the normalized relative abundances of identified glycopeptides were compared between N69 and N102 glycosylation sites as well as among 3 PSA samples. Heatmap was created using GENE-E (version 3.0.230, http://www.broadinstitute.org/cancer/software/GENE-E).
LC/MS data for glycomic study was processed using Xcarlibur Qual Browser. Extracted ion chromatograms were generated using isotopic mass with 10 ppm tolerance. A 7 points boxcar smoothing was utilized to produce smooth peak. Peak areas were used to represent the abundance of glycan structure.
Results and Discussion
Confirmation and Identification of Glycopeptides in D102N Glycosylation Site
PSA was identified with sequence coverages of 81%, 82%, 82%, and 83% for LeeBio PSA, LeeBio PSAH, Sigma PSA, and EMD PSA samples, respectively. The identification of PSA sequence is illustrated in Supplementary Figure 1. The quantitative values and percentage of PSA proteins based on spectral counts are listed in Supplementary Table 1. According to spectral count data generated by Scaffold software, spectral counts of PSA accounted for 84%, 78%, 93%, and 72% of the total spectral count data of LeeBio PSA, LeeBio PSAH, Sigma, and EMD samples, respectively. Other proteins were detected such as triosephosphate isomerase, prolactin-inducible protein, or prostaglandin-H2 D-isomerase. The Y1 ions of these proteins do not match any of the Y1 ions originating from PSA samples. For example, prostaglandin-H2 D-isomerase has 2 potential N glycosylation sites. The tryptic peptides containing these glycosylation sites are WFSAGLASNSSWLR (potential Y1+2= 892m/z) and SVVAPATDGGLNLTSTFLR (potential Y1+2=1061 m/z). The Y1 ions of these two peptide backbones do not match either of the two glycopeptides originating from PSA, namely NKSVILLGR (Y1+2=601) and HSLFHPEDTGQVFQVSHSFPHPLYN102MSLLK (Y1+4=925). LeeBio PSA samples are two isolates purified at different pI that were used in our previous publication.17, 18 Thus, quantitative values of identified glycopeptides for LeeBio PSA and PSAH samples were combined to compare those with other PSA samples.
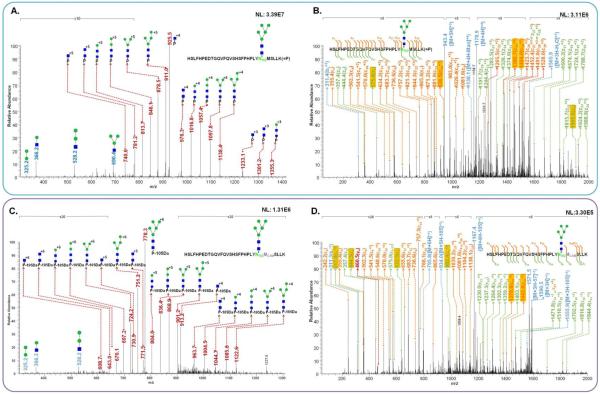
The glycosylation of HSLFHPEDTGQVFQVSHSFPHPLYN102MSLLK peptide backbone were detected, and confirmed by tandem MS as depicted in Figure 1. The glycan fragments of HexNAc2Hex5 on HSLFHPEDTGQVFQVSHSFPHPLYN102MSLLK backbone from LeeBio PSA samples were assigned to diagnostic ions originating from this glycan residue as shown in the CID MS/MS (Figure 1A). Three different charge states of Y1 ions were detected at m/z values of 740.6 (+5), 925.5 (+4), and 1233.1 (+3). The HCD spectrum was very comparable to CID MS/MS (data not shown). The fragments of HSLFHPEDTGQVFQVSHSFPHPLYN102MSLLK backbone were confirmed with HexNAc2Hex5 intact through ETD MS/MS (Figure 1B). This mutated sequence in D102N is confirmed by detecting 19 c ions and 15 z ions. Moreover, the presence of fragment ions at m/z 575.4 (z5) and 1905.0 (z6) affirms the presence of HexNAc2Hex5 on the N102 glycosylation site. Also, other ions including m/z 936.5 (c24+3), 1380.7 (c25+3), and 1404.0 (c24+2) supports the N102 glycosylation. Accordingly, the glycosylation site at N102MS created by one of the PSA missense mutation is successfully confirmed by tandem MS in this study.
Figure 1.
Examples of identified glycoforms and N102 glycosylation were confirmed by CID MS/MS and ETD MS/MS. The assignment of fragments of HexNAc2Hex5 on HSLFHPEDTGQVFQVSHSFPHPLYN102MSLLK with m/z value of 942.6390 in CID MS/MS(A) and ETD MS/MS (B). The assignment of fragments of HexNAc2Hex5 on HSLFHPEDTGQVFQVSHSFPHPLYN102MCAMSLLK with m/z value of 795.2017 in CID MS/MS(C) and in ETD MS/MS (D). The fragments indicating N102 glycosylation and the carbamidomethylaion of methionine was highlighted in yellow. In Figure (C), P* indicates HSLFHPEDTGQVFQVSHSFPHPLYN102MCAMSLLK with a loss of 2-(methylthio)acetamidyl group corresponding to 105Da.
Additionally, the glycosylation of HSLFHPEDTGQVFQVSHSFPHPLYN102MCAMSLLK peptide was identified, where MCAM represents carbamidomethylaion of methionine residue. This structure results from a side chain reaction of methionine during alkylation of cysteine.33 A tandem MS feature of this structure is the presence of a dominant neutral loss of 105 Da (2-(methylthio)acetamide).33 In Figure 1C, a neutral loss of 105 Da from the peptide sequence is observed. Glycan fragments of HexNAcHex5-HSLFHPEDTGQVFQVSHSFPHPLYN102MCAMSLLK are also detected in the spectrum (Figure 1C). The highest peak in the tandem MS is observed at m/z 778.3, corresponding to a neutral loss of 105Da from the precursor ion representing HexNAc2Hex5-HSLFHPEDTGQVFQVSHSFPHPLYN102MCAMSLLK glycopeptide. Accordingly, the mass of Y1 ions detected in the spectra is accounting for this neutral loss. In Figure 1C, 3 different charge states of Y1 ions are detected at m/z values of 608.7 (+6), 730.9 (+5), and 913.2 (+4), which accounts for the abovementioned neutral loss. Also, an additional permanent proton is created after the loss of 2-(methylthio)acetamide) on methionine.33 This positively influences the ionization efficiency of this glycopeptide. Accordingly, this modified glycopeptide is observed carrying 6 or 5 protons (data not shown). This observation is accounted for in quantitation. A charge state of 6 is employed for the quantitation of the glycopeptides with MCAM modification while a charge state of 5 is used for the quantitation of the original glycopeptides.
The methionine alkylation and glycosylation of HSLFHPEDTGQVFQVSHSFPHPLYN102MCAMSLLK backbone were confirmed by ETD MS/MS as shown in Figure 1D. The HSLFHPEDTGQVFQVSHSFPHPLYN102MCAMSLLK with HexNAc2Hex5 glycopeptide is affirmed with assigning 15 c ions and 14 z ions. The fragment ions at m/z 317.0 (z5+2) and 982.4 (z6+2) confirm the presence of HexNAc2Hex5 on N102 glycosylation site. In c-series ions, m/z 1379.8 (c25+3) is observed to support the presence of HexNAc2Hex5 on N102 glycosylation. The occurrence of methionine alkylation is confirmed by the fragments at m/z values of 444.5 (z4), 632.5 (z5), 1379.8 (c25+3), and 1442.6 (c26+3). Also, a neutral loss of 105Da was observed in ETD MS/MS. The ions at m/z values of 934.0 (+5), 1167.4 (+4), and 1555.9 (+3) represent a loss of 105Da from the precursor ion with different charge states. The annotations of CID MS/MS of some N102 glycopeptides are illustrated in Supplementary Figure 2.
PSA has 5 protein isoforms with identifier P07288-1 through P07288-5 from UniProt KB (http://www.uniprot.org/uniprot/P07288). The tryptic peptide HSLFHPEDTGQVFQVSHSFPHPLYD102MSLLK are present in P07288-1 (canonical sequence), P07288-2,34 and P07288-5.35 Both protein sequences of P07288-2 and P07288-5 have different amino acid sequence from 211 positions compared to P07288-1. Therefore, it is possible to observe HSLFHPEDTGQVFQVSHSFPHPLYN102MSLLK glycosylation associated with these protein isoforms. In the previous 2012 ABRF study,18 some laboratories reported the detection of additional glycopeptides in PSA samples. For example, a laboratory observed SVILLGRHSLFHPEDTGQVFQVSHSFPHPLYNMSLLK with HexNAc2Hex5 glycoform. They noted that it may originate from KLK2 since the sequence homology between KLK2 and KLK3 is 77%. However, SVILLGRHSLFHPEDTGQVFQVSHSFPHPLYNMSLLK is different from NSQVWLGRHNLFEPEDTGQRVPVSHSFPHPLYNMSLLK originating from KLK2. Moreover, KLK2 was not detected in the proteomics data of the 2012 ABRF study and our proteomics data (data not shown).
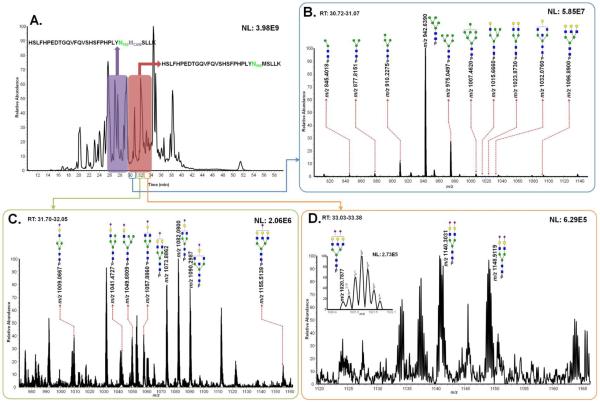
The retention time of HSLFHPEDTGQVFQVSHSFPHPLYN102MSLLK glycoforms is 30~34min while that of HSLFHPEDTGQVFQVSHSFPHPLYN102MCAMSLLK glycoforms is 26~30min as shown in Figure 2A. There are 21 glycoforms associated with N102 glycosylation site observed from LeeBio PSA samples as summarized in Table 1. All glycoforms were identified with HSLFHPEDTGQVFQVSHSFPHPLYN102MSLLK sequence while 17 structures were seen with the methionine alkylated sequence. For the same glycoforms identified from both sequences, the abundances were added. Figures 2B, 2C, and 2D illustrate averaged full MS (5 scans) with observed N-glycans on HSLFHPEDTGQVFQVSHSFPHPLYN102MSLLK peptide backbone. There were 10 neutral N-glycans observed at 30.7~31.1min (Figure 2B), 8 monosialylated N-glycans were observed at 31.7~32.1min (Figure 2C), and 3 disialylated N-glycans were detected at 33.0~33.4min (Figure 2D). One of the disialylated glycan structures was only detected in LeeBio PSAH sample as shown in the inset of Figure 2D. All the identified glycopeptides at N102 glycosylation site for Sigma PSA and EMD PSA samples are listed in Supplementary Table 2 and 3, respectively.
Figure 2.
Total ion chromatogram of LeeBio PSA digests with different retention times of two peptide backbones possessing its glycosylation site of N102 (A). Assigned glycoforms in MS are presented with HSLFHPEDTGQVFQVSHSFPHPLYN102MSLLK backbone. Different retention times correspond to the detection of neutral glycoforms (B), monosialylated glycoforms (C), and disialylated glycoforms (D).
Table 1.
Summary of identified glycoforms of HSLFHPEDTGQVFQVSHSFPHPLYN102MSLLK peptide sequence in LeeBio PSA (#) and PSAH (≠) samples.
| Glycan | Molecular mass of glycopeptide | Theoretical m/z | Observed m/z | ppm | Normalized relative abundance (×10−2) | STD (×10−2) |
|---|---|---|---|---|---|---|
| HexNAc2Hex2# | 4221.9728 | 845.4018 | 845.4018 | 0.01 | 0.66 | 0.07 |
| HexNAc2Hex3# | 4384.0256 | 877.8124 | 877.8151 | −3.11 | 0.97 | 0.02 |
| HexNAc2Hex4#≠ | 4546.0784 | 910.2229 | 910.2267 | −4.14 | 11.03 | 2.57 |
| 4603.0998* | 768.1906 | 768.1906 | −0.07 | |||
| HexNAc2Hex5#≠ | 4708.1312 | 942.6335 | 942.6390 | −5.85 | 51.58 | 3.02 |
| 4765.1526* | 795.1994 | 795.2017 | −2.96 | |||
| HexNAc2Hex6#≠ | 4870.1840 | 975.0441 | 975.0483 | −4.36 | 13.51 | 0.57 |
| 4927.2054* | 822.2082 | 822.2100 | −2.25 | |||
| HexNAc2Hex7# | 5032.2368 | 1007.4546 | 1007.4620 | −7.24 | 0.98 | 0.09 |
| 5089.2582* | 849.2170 | 849.2200 | −3.59 | |||
| HexNAc3Hex6#≠ | 5073.2634 | 1015.6599 | 1015.6650 | −4.70 | 0.66 | 0.10 |
| 5130.2848* | 856.0547 | 856.0559 | −1.38 | |||
| HexNAc4Hex5#≠ | 5114.2900 | 1023.8653 | 1023.8690 | −3.37 | 0.80 | 0.10 |
| 5171.3114* | 862.8925 | 862.8962 | −4.31 | |||
| HexNAc5Hex4#≠ | 5155.3166 | 1032.0706 | 1032.0760 | −5.46 | 0.52 | 0.05 |
| 5212.3380* | 869.7303 | 869.7246 | 6.50 | |||
| HexNAc5Hex6# | 5479.4222 | 1096.8917 | 1096.8900 | 2.00 | 0.03 | 0.01 |
| HexNAc3Hex4NeuAc1#≠ | 5040.2532 | 1009.0579 | 1009.0670 | −8.73 | 0.93 | 0.08 |
| 5097.2746* | 850.5530 | 850.5558 | −3.27 | |||
| HexNAc3Hex5NeuAc1# | 5202.3060 | 1041.4685 | 1041.4730 | −4.08 | 0.35 | 0.04 |
| 5259.3274* | 877.5618 | 877.5616 | 0.25 | |||
| HexNAc3Hex6NeuAc1#≠ | 5364.3588 | 1073.8790 | 1073.8860 | −6.70 | 7.32 | 1.41 |
| 5421.3802* | 904.5706 | 904.5758 | −5.73 | |||
| HexNAc4Hex4NeuAc1# | 5243.3326 | 1049.6738 | 1049.6810 | −6.79 | 0.72 | 0.08 |
| 5300.3540* | 884.3996 | 884.3989 | 0.77 | |||
| HexNAc4Hex5NeuAc1#≠ | 5405.3854 | 1082.0843 | 1082.0850 | −0.62 | 2.95 | 0.43 |
| 5462.4068* | 911.4084 | 911.4080 | 0.42 | |||
| HexNAc5Hex3NeuAc1# | 5284.3592 | 1057.8791 | 1057.8860 | −6.53 | 0.55 | 0.07 |
| 5341.3806* | 891.2374 | 891.2421 | −5.33 | |||
| HexNAc5Hex4NeuAc1#≠ | 5446.4120 | 1090.2897 | 1090.2890 | 0.96 | 2.62 | 0.60 |
| 5503.4334* | 918.2462 | 918.2463 | −0.16 | |||
| HexNAc5Hex6NeuAc1#≠ | 5770.5176 | 1155.1108 | 1155.1170 | −4.96 | 0.51 | 0.15 |
| 5827.5390* | 972.2638 | 972.2653 | −1.59 | |||
| HexNAc4Hex5NeuAc2#≠ | 5696.4808 | 1140.3034 | 1140.3030 | 0.27 | 1.79 | 0.32 |
| 5753.5022* | 959.9243 | 959.9213 | 3.11 | |||
| HexNAc5Hex4NeuAc2#≠ | 5737.5074 | 1148.5087 | 1148.5090 | −0.24 | 1.36 | 0.08 |
| 5794.5288* | 966.7621 | 966.7584 | 3.78 | |||
| HexNAc5Hex6NeuAc2≠ | 6118.6344 | 1020.7797 | 1020.7880 | −7.89 | 0.17 | 0.04 |
The molecular mass of glycopeptide is the modified mass accounting for carbamidomethylation of methionine.
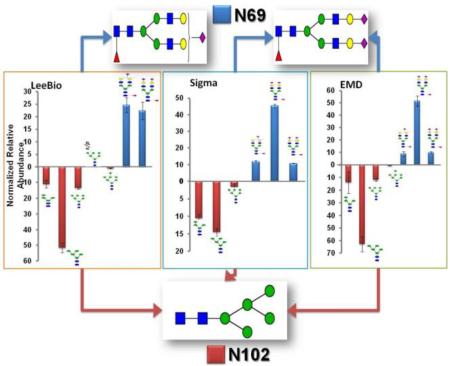
In total, 7 glycan structures were observed for Sigma PSA (Supplementary Table 2) while 16 glycan structures were detected for EMD PSA (Supplementary Table 3). As mentioned above, 21 glycan structures were identified for LeeBio PSA samples (Table 1). Out of 23 glycan structures, 7 glycan structures were typical in the 3 PSA samples: HexNAc2Hex4, HexNAc2Hex5, HexNAc2Hex6, HexNAc4Hex5NeuAc1, HexNAc5Hex4NeuAc1, HexNAc4Hex5NeuAc2, and HexNAc5Hex4NeuAc2. These are all the glycan structures identified in the case of Sigma PSA (Supplementary Table 2). Between LeeBio PSA and EMD PSA, more structures are observed, including HexNAc2Hex7 and 7 other monosialylated glycopeptides (Table 1 and Supplementary Table 3).
Interestingly, no fucosylated glycopeptides were detected on N102 glycosylation site for all 3 samples. The glycosylation at N69 of PSA is known to be highly fucosylated.17–19 From our previous study, a total of 68 glycan structures were identified for PSA N69 glycosylation site.17 37 of these structures have fucoses ranging from 1 to 3 on core or antenna of glycan structures. However, this is not true for N120 glycosylation site. This could be explained by a limited accessibility of specific glycosyltransferases/exoglycosidase to this new glycosylation site. Less glycosyltransferases/exoglycosidases, especially related to fucosylation, might be involved in trimming or attachment of glycan residues at N102 glycosylation site compared to N69 glycosylation during the biosynthesis of N-glycans. This is because N102 glycosylation site is substituted from D102 by a mutation on nucleotide sequence of the KLK3 genes. Thus, the glycosylation at N102 is not the same as at N69. Moreover, D102 is a starting amino acid of “kallikrein loop”, which is thought to be important for controlling the activity, substrate and inhibitor specificity, and function in autolytic regulation of PSA.3, 4, 36 The D102 is nearby one of the zinc binding sites H98, which influences the PSA activity and the binding of substrate.3, 4, 36 Also, the D102 is close to the catalytic triad consisting of H65, D120, and S213 residues, which are three amino acid residues involved in catalysis.3, 4, 36–38 Since the H-D-S triad should function together at the center of the active sites during the catalysis, the missense mutation of D102N and its glycosylation might affect many physiological function of PSA.
Quantitation of Glycopeptides at D102N Glycosylation Site and Comparisons with N69 Glycosylation
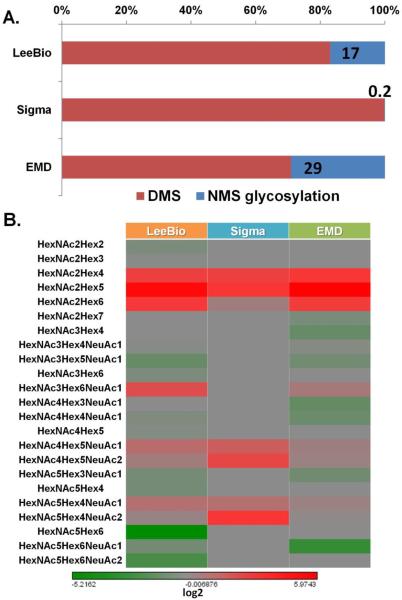
The D102N missense mutation led to the novel glycosylation of PSA as shown in the three PSA samples. Similar to N69 glycosylation site, N102 glycosylation site is fully occupied with glycans if the mutation occurs. This finding was supported by the fact that non-glycosylated peptide possessing N102 was not observed in the LC-MS/MS analysis. Thus, it suggests that glycosylation evolves if the mutation at D102 occurs. As shown in Figure 3A, the different extent of glycosylation at N102 evolves for 3 PSA samples. The intensity of non-mutated sequence HSLFHPEDTGQVFQVSHSFPHPLYD102MSLLK (namely DMS) is shown as a red bar while the percentage of glycosylation is shown as blue bar. Notably, the mutation/glycosylation occurs very less in Sigma PSA sample indicating that D102 is highly conserved. Accordingly, 17%, 0.2%, and 29% of N102 glycosylation is observed for LeeBio PSA, Sigma PSA, and EMD PSA samples, respectively. The relative abundances of all identified glycopeptides are illustrated in a heat map shown in Figure 3B. A distinct glycosylation was observed with different number of identified glycoforms among the 3 PSA samples. The most intense glycan structure at N102 was HexNAc2Hex5.
Figure 3.
The comparisons of the level of glycosylation among LeeBio, Sigma, and EMD PSA samples. The intensity of non-mutated sequence possessing D102 and the level of glycosylation is illustrated for 3 PSA samples (A). The overall relative abundance of identified glycopeptides at N102 is represented in the heat map (B). In common, HexNAc2Hex5 is the most abundant glycoform among 3 samples.
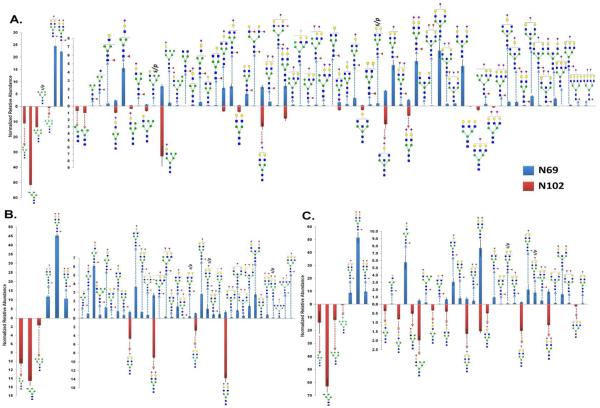
The quantitative values of glycopeptides for the N102 glycosylation site was compared to that at N69 glycosylation site as shown in Figure 4. The upper panel represents the relative abundances of glycopeptides at N69 glycosylation site while the lower panel represents that at N102 glycosylation site. The quantitative values are listed in Table 1 (LeeBio PSA), Supplementary Table 2 (Sigma PSA), and 3 (EMD PSA). The three PSA samples appear to be slightly dissimilar in regard to the number and the extent of identified glycopeptides. For LeeBio PSA samples (Figure 4A), 68 total glycoforms were observed at N69 glycosylation while 21 total glycoforms were identified at N102 glycosylation site. The most abundant glycoforms at N69 glycosylation site were HexNAc4Hex5Fuc1NeuAc1 and HexNAc4Hex5Fuc1NeuAc2 while that at N102 glycosylation site was HexNAc2Hex5. This glycan occupied N102 glycosylation site at 51.58%. The next abundant glycoforms are HexNAc2Hex4 and HexNAc2Hex6, which corresponded to 11.03% and 13.51% occupancies. In the case of the N69 glycosylation site, two glycoforms of HexNAc4Hex5Fuc1NeuAc1 and HexNAc4Hex5Fuc1NeuAc2 occupied 24.56% and 22.32%, respectively. Accordingly, the fucosylated sialylated glycopeptides (46.88%) are dominant for N69 glycosylation site while the high-mannosylated glycopeptides (76.10%) are dominant for N102 glycosylation site. This might explain the discrepancy of data between glycomics and glycoproteomics of PSA. In Supplementary Figure 3, the relative abundances of identified glycans from LeeBio PSA and PSAH samples are illustrated. One of the interesting findings is that the intensity of HexNAc2Hex5 is as high as that of HexNAc4Hex5Fuc1NeuAc1 and HexNAc4Hex5Fuc1NeuAc2. It suggests that the glycans are released from both N69 and N102 glycosylation sites. Accordingly, glycomic studies of PSA shows the entire microheterogeneity of PSA, including both glycosylation sites.
Figure 4.
Quantitative values (relative abundance) of N-glycans at N102 glycosylation site. A total 68 N-glycans at N69 were quantitatively compared to 21 glycoforms at N102 for LeeBio PSA (A), a total 39 N-glycans at N69 were quantitatively compared to 7 glycoforms at N102 for Sigma PSA (B), and a total 27 N-glycans at N69 were quantitatively compared to 16 glycoforms at N102 for EMD PSA (C).
Figure 4B and 4C depict the quantitation of glycopeptides observed in Sigma and EMD PSA samples, respectively. As shown in Figure 4B, 39 glycoforms were seen for N69 glycosylation site while 7 glycoforms were identified for N102 glycosylation site. The major glycoforms for N69 glycosylation site are HexNAc4Hex5Fuc1NeuAc2 (45.27%), HexNAc4Hex5Fuc1NeuAc1 (11.76%), and HexNAc5Hex4Fuc1NeuAc2 (10.64%). On the other hand, the dominant glycoforms for N102 glycosylation site are HexNAc2Hex5 (14.54%), HexNAc5Hex4NeuAc2 (13.74%), HexNAc2Hex4 (10.47%), and HexNAc4Hex5NeuAc2 (9.01%). In the case of the N102 glycosylation site of Sigma PSA, the abundance of two sialylated glycoforms appeared to be comparable to that of high mannose glycoforms. In the case of EMD PSA (Figure 4C), the prevailing glycoforms of N69 glycosylation site are HexNAc4Hex5Fuc1NeuAc2 (51.58%), HexNAc5Hex4Fuc1NeuAc2 (9.74%), and HexNAc4Hex5Fuc1NeuAc1 (8.87%). On the other hand, HexNAc2Hex5 predominantly occupied N102 glycosylation site with 62.87% relative abundance. Two high mannose glycoforms HexNAc2Hex4 (13.97%) and HexNAc2Hex6 (11.89%) were the next abundance glycoforms at N102. Overall, HexNAc2Hex5 was the primary glycoform of N102 glycosylation site among the 3 PSA samples.
Conclusion
The identification and quantitation of a mutant glycosylation site of PSA were successfully achieved using LC-MS/MS. This study is the first work dedicated to deciphering the glycosylation of PSA N102 site created by a missense mutation. The mutated peptide sequence possessing N102 was successfully verified using CID and ETD tandem MS. PSA proteins from 3 different vendors were obtained and their various microheterogeneities of N102 glycosylation site were investigated. A total of 21, 7, and 16 glycoforms were detected for LeeBio, Sigma, and EMD PSA samples. Notably, only high-mannose and sialylated glycopeptides were observed at N102, which is remarkably different from N69 glycosylation. Among 3 PSA samples, HexNAc2Hex5 is the predominant glycoform at N102 while HexNAc4Hex5Fuc1NeuAc1 or HexNAc4Hex5Fuc1NeuAc2 is the primary glycoforms at N69.
Supplementary Material
Acknowledgments
This work was supported by NIH (1R01GM093322-05 and 1R01GM112490-01).
Footnotes
Supporting Information Available: This material is available free of charge via the Internet at http://pubs.acs.org.
References
- (1).Diamandis EP. Prostate-specific Antigen: Its Usefulness in Clinical Medicine. Trends Endocrinol Metab. 1998;9(8):310–6. doi: 10.1016/s1043-2760(98)00082-4. [DOI] [PubMed] [Google Scholar]
- (2).Watt KW, Lee PJ, M'Timkulu T, Chan WP, Loor R. Human prostate-specific antigen: structural and functional similarity with serine proteases. Proc Natl Acad Sci U S A. 1986;83(10):3166–70. doi: 10.1073/pnas.83.10.3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Borgoño CA, Michael IP, Diamandis EP. Human Tissue Kallikreins: Physiologic Roles and Applications in Cancer. Molecular Cancer Research. 2004;2(5):257–280. [PubMed] [Google Scholar]
- (4).Emami N, Diamandis EP. New insights into the functional mechanisms and clinical applications of the kallikrein-related peptidase family. Molecular Oncology. 2007;1(3):269–287. doi: 10.1016/j.molonc.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Yousef GM, Diamandis EP. The new human tissue kallikrein gene family: structure, function, and association to disease. Endocr Rev. 2001;22(2):184–204. doi: 10.1210/edrv.22.2.0424. [DOI] [PubMed] [Google Scholar]
- (6).Grimwood J, Gordon LA, Olsen A, Terry A, Schmutz J, Lamerdin J, Hellsten U, Goodstein D, Couronne O, Tran-Gyamfi M, Aerts A, Altherr M, Ashworth L, Bajorek E, Black S, Branscomb E, Caenepeel S, Carrano A, Caoile C, Chan YM, Christensen M, Cleland CA, Copeland A, Dalin E, Dehal P, Denys M, Detter JC, Escobar J, Flowers D, Fotopulos D, Garcia C, Georgescu AM, Glavina T, Gomez M, Gonzales E, Groza M, Hammon N, Hawkins T, Haydu L, Ho I, Huang W, Israni S, Jett J, Kadner K, Kimball H, Kobayashi A, Larionov V, Leem SH, Lopez F, Lou Y, Lowry S, Malfatti S, Martinez D, McCready P, Medina C, Morgan J, Nelson K, Nolan M, Ovcharenko I, Pitluck S, Pollard M, Popkie AP, Predki P, Quan G, Ramirez L, Rash S, Retterer J, Rodriguez A, Rogers S, Salamov A, Salazar A, She X, Smith D, Slezak T, Solovyev V, Thayer N, Tice H, Tsai M, Ustaszewska A, Vo N, Wagner M, Wheeler J, Wu K, Xie G, Yang J, Dubchak I, Furey TS, DeJong P, Dickson M, Gordon D, Eichler EE, Pennacchio LA, Richardson P, Stubbs L, Rokhsar DS, Myers RM, Rubin EM, Lucas SM. The DNA sequence and biology of human chromosome 19. Nature. 2004;428(6982):529–35. doi: 10.1038/nature02399. [DOI] [PubMed] [Google Scholar]
- (7).Armbruster DA. Prostate-specific antigen: biochemistry, analytical methods, and clinical application. Clin Chem. 1993;39(2):181–95. [PubMed] [Google Scholar]
- (8).Huber PR, Schmid HP, Mattarelli G, Strittmatter B, van Steenbrugge GJ, Maurer A. Serum free prostate specific antigen: isoenzymes in benign hyperplasia and cancer of the prostate. Prostate. 1995;27(4):212–9. doi: 10.1002/pros.2990270406. [DOI] [PubMed] [Google Scholar]
- (9).Ohyama C, Hosono M, Nitta K, Oh-eda M, Yoshikawa K, Habuchi T, Arai Y, Fukuda M. Carbohydrate structure and differential binding of prostate specific antigen to Maackia amurensis lectin between prostate cancer and benign prostate hypertrophy. Glycobiology. 2004;14(8):671–9. doi: 10.1093/glycob/cwh071. [DOI] [PubMed] [Google Scholar]
- (10).Tabares G, Radcliffe CM, Barrabes S, Ramirez M, Aleixandre RN, Hoesel W, Dwek RA, Rudd PM, Peracaula R, de Llorens R. Different glycan structures in prostate-specific antigen from prostate cancer sera in relation to seminal plasma PSA. Glycobiology. 2006;16(2):132–45. doi: 10.1093/glycob/cwj042. [DOI] [PubMed] [Google Scholar]
- (11).Zhang WM, Leinonen J, Kalkkinen N, Dowell B, Stenman UH. Purification and characterization of different molecular forms of prostate-specific antigen in human seminal fluid. Clin Chem. 1995;41(11):1567–73. [PubMed] [Google Scholar]
- (12).Charrier JP, Tournel C, Michel S, Dalbon P, Jolivet M. Two-dimensional electrophoresis of prostate-specific antigen in sera of men with prostate cancer or benign prostate hyperplasia. Electrophoresis. 1999;20(4–5):1075–81. doi: 10.1002/(SICI)1522-2683(19990101)20:4/5<1075::AID-ELPS1075>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- (13).Sarrats A, Saldova R, Comet J, O'Donoghue N, de Llorens R, Rudd PM, Peracaula R. Glycan characterization of PSA 2-DE subforms from serum and seminal plasma. OMICS. 2010;14(4):465–74. doi: 10.1089/omi.2010.0050. [DOI] [PubMed] [Google Scholar]
- (14).Okada T, Sato Y, Kobayashi N, Sumida K, Satomura S, Matsuura S, Takasaki M, Endo T. Structural characteristics of the N-glycans of two isoforms of prostate-specific antigens purified from human seminal fluid. Biochim Biophys Acta. 2001;1525(1–2):149–60. doi: 10.1016/s0304-4165(00)00182-3. [DOI] [PubMed] [Google Scholar]
- (15).Peracaula R, Tabares G, Royle L, Harvey DJ, Dwek RA, Rudd PM, de Llorens R. Altered glycosylation pattern allows the distinction between prostate-specific antigen (PSA) from normal and tumor origins. Glycobiology. 2003;13(6):457–70. doi: 10.1093/glycob/cwg041. [DOI] [PubMed] [Google Scholar]
- (16).Tajiri M, Ohyama C, Wada Y. Oligosaccharide profiles of the prostate specific antigen in free and complexed forms from the prostate cancer patient serum and in seminal plasma: a glycopeptide approach. Glycobiology. 2008;18(1):2–8. doi: 10.1093/glycob/cwm117. [DOI] [PubMed] [Google Scholar]
- (17).Song E, Mayampurath A, Yu CY, Tang H, Mechref Y. Glycoproteomics: Identifying the Glycosylation of Prostate Specific Antigen at Normal and High Isoelectric Points by LC-MS/MS. J Proteome Res. 2014;13(12):5570–80. doi: 10.1021/pr500575r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Leymarie N, Griffin PJ, Jonscher K, Kolarich D, Orlando R, McComb M, Zaia J, Aguilan J, Alley WR, Altmann F, Ball LE, Basumallick L, Bazemore-Walker CR, Behnken H, Blank MA, Brown KJ, Bunz SC, Cairo CW, Cipollo JF, Daneshfar R, Desaire H, Drake RR, Go EP, Goldman R, Gruber C, Halim A, Hathout Y, Hensbergen PJ, Horn DM, Hurum D, Jabs W, Larson G, Ly M, Mann BF, Marx K, Mechref Y, Meyer B, Moginger U, Neususs C, Nilsson J, Novotny MV, Nyalwidhe JO, Packer NH, Pompach P, Reiz B, Resemann A, Rohrer JS, Ruthenbeck A, Sanda M, Schulz JM, Schweiger-Hufnagel U, Sihlbom C, Song E, Staples GO, Suckau D, Tang H, Thaysen-Andersen M, Viner RI, An Y, Valmu L, Wada Y, Watson M, Windwarder M, Whittal R, Wuhrer M, Zhu Y, Zou C. Interlaboratory Study on Differential Analysis of Protein Glycosylation by Mass Spectrometry: the ABRF Glycoprotein Research Multi-Institutional Study 2012. Mol Cell Proteomics. 2013 doi: 10.1074/mcp.M113.030643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Behnken HN, Ruthenbeck A, Schulz JM, Meyer B. Glycan analysis of Prostate Specific Antigen (PSA) directly from the intact glycoprotein by HR-ESI/TOF-MS. J Proteome Res. 2014;13(2):997–1001. doi: 10.1021/pr400999y. [DOI] [PubMed] [Google Scholar]
- (20).Couzin-Frankel J. Genetic Analysis Points the Way to Individualized PSA Tests. Science. 2010;330(6011):1602. doi: 10.1126/science.330.6011.1602. [DOI] [PubMed] [Google Scholar]
- (21).Mononen N, Seppala EH, Duggal P, Autio V, Ikonen T, Ellonen P, Saharinen J, Saarela J, Vihinen M, Tammela TL, Kallioniemi O, Bailey-Wilson JE, Schleutker J. Profiling genetic variation along the androgen biosynthesis and metabolism pathways implicates several single nucleotide polymorphisms and their combinations as prostate cancer risk factors. Cancer Res. 2006;66(2):743–7. doi: 10.1158/0008-5472.CAN-05-1723. [DOI] [PubMed] [Google Scholar]
- (22).Gallagher DJ, Vijai J, Cronin AM, Bhatia J, Vickers AJ, Gaudet MM, Fine S, Reuter V, Scher HI, Hallden C, Dutra-Clarke A, Klein RJ, Scardino PT, Eastham JA, Lilja H, Kirchhoff T, Offit K. Susceptibility loci associated with prostate cancer progression and mortality. Clin Cancer Res. 2010;16(10):2819–32. doi: 10.1158/1078-0432.CCR-10-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Parikh H, Deng Z, Yeager M, Boland J, Matthews C, Jia J, Collins I, White A, Burdett L, Hutchinson A, Qi L, Bacior J, Lonsberry V, Rodesch M, Jeddeloh J, Albert T, Halvensleben H, Harkins T, Ahn J, Berndt S, Chatterjee N, Hoover R, Thomas G, Hunter D, Hayes R, Chanock S, Amundadottir L. A comprehensive resequence analysis of the KLK15-KLK3-KLK2 locus on chromosome 19q13.33. Human Genetics. 2010;127(1):91–99. doi: 10.1007/s00439-009-0751-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Klein RJ, Hallden C, Cronin AM, Ploner A, Wiklund F, Bjartell AS, Stattin P, Xu J, Scardino PT, Offit K, Vickers AJ, Gronberg H, Lilja H. Blood biomarker levels to aid discovery of cancer-related single-nucleotide polymorphisms: kallikreins and prostate cancer. Cancer Prev Res (Phila) 2010;3(5):611–9. doi: 10.1158/1940-6207.CAPR-09-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Penney KL, Schumacher FR, Kraft P, Mucci LA, Sesso HD, Ma J, Niu Y, Cheong JK, Hunter DJ, Stampfer MJ, Hsu SI. Association of KLK3 (PSA) genetic variants with prostate cancer risk and PSA levels. Carcinogenesis. 2011;32(6):853–9. doi: 10.1093/carcin/bgr050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Rodriguez S, Al-Ghamdi OA, Burrows K, Guthrie PAI, Lane JA, Davis M, Marsden G, Alharbi KK, Cox A, Hamdy FC, Neal DE, Donovan JL, Day INM. Very Low PSA Concentrations and Deletions of the KLK3 Gene. Clin Chem. 2013;59(1):234–244. doi: 10.1373/clinchem.2012.192815. [DOI] [PubMed] [Google Scholar]
- (27).Knipe DW, Evans DM, Kemp JP, Eeles R, Easton DF, Kote-Jarai Z, Al Olama AA, Benlloch S, Donovan JL, Hamdy FC, Neal DE, Davey Smith G, Lathrop M, Martin RM. Genetic variation in prostate-specific antigen-detected prostate cancer and the effect of control selection on genetic association studies. Cancer Epidemiol Biomarkers Prev. 2014;23(7):1356–65. doi: 10.1158/1055-9965.EPI-13-0889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Mechref Y, Kang P, Novotny MV. Solid-phase permethylation for glycomic analysis. Methods Mol Biol. 2009;534:53–64. doi: 10.1007/978-1-59745-022-5_4. [DOI] [PubMed] [Google Scholar]
- (29).Kang P, Mechref Y, Novotny MV. High-throughput solid-phase permethylation of glycans prior to mass spectrometry. Rapid Commun Mass Spectrom. 2008;22(5):721–34. doi: 10.1002/rcm.3395. [DOI] [PubMed] [Google Scholar]
- (30).Kang P, Mechref Y, Klouckova I, Novotny MV. Solid-phase permethylation of glycans for mass spectrometric analysis. Rapid Communications in Mass Spectrometry. 2005;19(23):3421–3428. doi: 10.1002/rcm.2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Desantos-Garcia JL, Khalil SI, Hussein A, Hu Y, Mechref Y. Enhanced sensitivity of LC-MS analysis of permethylated N-glycans through online purification. Electrophoresis. 2011;32(24):3516–3525. doi: 10.1002/elps.201100378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Brosch M, Yu L, Hubbard T, Choudhary J. Accurate and sensitive peptide identification with Mascot Percolator. J Proteome Res. 2009;8(6):3176–81. doi: 10.1021/pr800982s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Kruger R, Hung CW, Edelson-Averbukh M, Lehmann WD. Iodoacetamidealkylated methionine can mimic neutral loss of phosphoric acid from phosphopeptides as exemplified by nano-electrospray ionization quadrupole time-of-flight parent ion scanning. Rapid Commun Mass Spectrom. 2005;19(12):1709–16. doi: 10.1002/rcm.1976. [DOI] [PubMed] [Google Scholar]
- (34).Lundwall Å, Lilja H. Molecular cloning of human prostate specific antigen cDNA. FEBS Letters. 1987;214(2):317–322. doi: 10.1016/0014-5793(87)80078-9. [DOI] [PubMed] [Google Scholar]
- (35).Heuze-Vourc'h N, Leblond V, Courty Y. Complex alternative splicing of the hKLK3 gene coding for the tumor marker PSA (prostate-specific-antigen) Eur J Biochem. 2003;270(4):706–14. doi: 10.1046/j.1432-1033.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- (36).Ménez R, Michel S, Muller BH, Bossus M, Ducancel F, Jolivet-Reynaud C, Stura EA. Crystal Structure of a Ternary Complex between Human Prostate-specific Antigen, Its Substrate Acyl Intermediate and an Activating Antibody. Journal of Molecular Biology. 2008;376(4):1021–1033. doi: 10.1016/j.jmb.2007.11.052. [DOI] [PubMed] [Google Scholar]
- (37).Villoutreix BO, Getzoff ED, Griffin JH. A structural model for the prostate disease marker, human prostate-specific antigen. Protein Sci. 1994;3(11):2033–44. doi: 10.1002/pro.5560031116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Coombs GS, Bergstrom RC, Pellequer J-L, Baker SI, Navre M, Smith MM, Tainer JA, Madison EL, Corey DR. Substrate specificity of prostate-specific antigen (PSA) Chemistry & Biology. 1998;5(9):475–488. doi: 10.1016/s1074-5521(98)90004-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.