Abstract
Properly functional CNS circuits depend on inhibitory interneurons that in turn rely upon activity-dependent gene expression for morphological development, connectivity and excitatory-inhibitory coordination. Despite its importance, excitation-transcription coupling in inhibitory interneurons is poorly understood. Here, we report that PV+ interneurons employ a novel CaMK-dependent pathway to trigger CREB phosphorylation and gene expression. As in excitatory neurons, voltage-gated Ca2+ influx through CaV1 channels triggers CaM nuclear translocation via local Ca2+ signaling. However, PV+ interneurons are distinct in that nuclear signaling is mediated by γCaMKI, not γCaMKII. CREB phosphorylation also proceeds with slow, sigmoid kinetics, rate-limited by paucity of CaMKIV, protecting against saturation of phospho-CREB in the face of higher firing rates and bigger Ca2+ transients. Our findings support the generality of CaM shuttling to drive nuclear CaMK activity, and are relevant to disease pathophysiology, insofar as dysfunction of PV+ interneurons and molecules underpinning their excitation-transcription coupling both relate to neuropsychiatric disease.
Introduction
Synaptic inhibition by GABAergic interneurons controls information flow and overall stability in neuronal networks (Buzsaki and Chrobak, 1995; Klausberger and Somogyi, 2008). The proper function of inhibitory neurons depends on activity-dependent gene expression, a process critical for regulation of interneuron differentiation (Gu and Spitzer, 1995), maturation (Jiang and Swann, 2005; Patz et al., 2004) and neurotransmitter production (Lau and Murthy, 2012). In neocortical inhibitory neurons, for example, activity-dependent changes in gene expression can influence dendritic morphology and connectivity (De Marco Garcia et al., 2011), GABAergic neurotransmission (Hong et al., 2008; Jiao et al., 2011) and excitatory-inhibitory balance (D’Amour and Froemke, 2015). Modulation of specific activity-regulated genes can strongly influence circuit function, as seen with Npas4 in synaptic plasticity (Spiegel et al., 2014), parvalbumin (PV) in experience-dependent learning (Donato et al., 2013) and GAD67 in inhibitory synaptic homeostasis (Kinney et al., 2006; Lau and Murthy, 2012).
Neuronal firing and changes in gene expression are linked by excitation-transcription (E-T) coupling, of which little is known in interneurons. In excitatory CNS neurons, upregulation of specific genes (e.g., c-Fos, Arc and BDNF) depends on activation of CREB by phosphorylation at Ser133 (pCREB). Signaling to nuclear CREB can be triggered by electrical or chemical activation and is mediated by multiple Ca2+-dependent signaling pathways (Cohen and Greenberg, 2008). A fast Ca2+/calmodulin (CaM)- dependent kinase (CaMK) pathway (operating in tens of seconds) and a slower mitogen-activated protein kinase (MAPK) pathway (requiring many minutes) can trigger CREB phosphorylation and thereby drive gene expression. It has been proposed that interneurons use plasticity pathways distinct from those of principal neurons, on the basis of studies in cortical and hippocampal interneurons reporting low or undetectable levels of αCaMKII (Jones et al., 1994) and calcineurin.
It remains unclear whether findings about E-T coupling in excitatory neurons can be reliably extrapolated to GABAergic interneurons. For example, PV+ interneurons are remarkable for their ‘fast spiking’ phenotype, firing action potentials much faster than excitatory neurons at baseline and with stimulation (Kawaguchi et al., 1987; Royer et al., 2012). The intense activity of PV+ neurons might cause larger and longer elevations of [Ca2+]i that could potentially saturate a replica of the machinery described in excitatory neurons. Accordingly, we hypothesized that the extreme Ca2+ dynamics in PV+ cells might necessitate a distinct E-T coupling pathway, characterized by slower responsiveness and distinct molecular components. Indeed, we uncovered a novel mechanism that differs from that in excitatory neurons in both qualitative and quantitative respects, in using a distinct protein for conveyance of signals to the nucleus, and in restricting levels of the nuclear CREB kinase to slow the dynamics of nuclear CREB phosphorylation.
Results
CREB Phosphorylation Occurs in Response to Sound in PV+ Cells in vivo
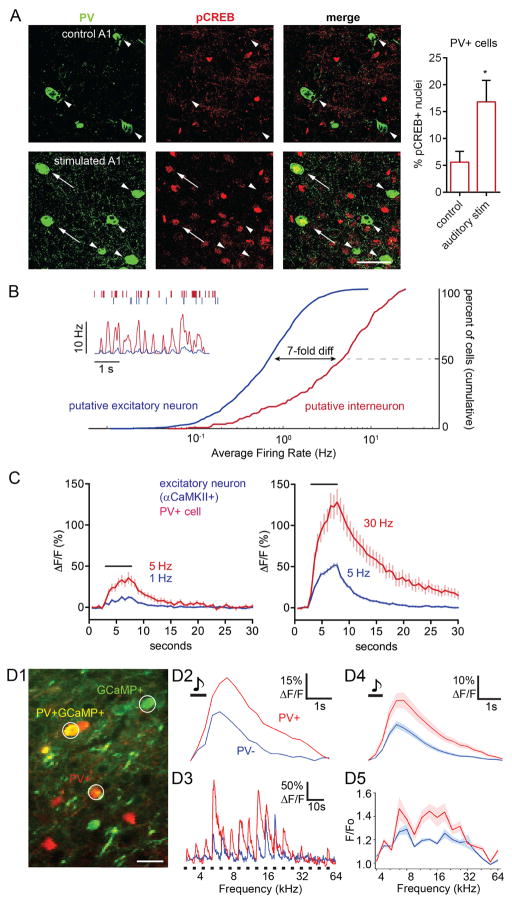
It is well known that activity of excitatory neurons triggers CREB phosphorylation upstream of gene regulation. However, it remains unclear whether CREB phosphorylation also occurs in PV+ inhibitory interneurons in response to physiologic stimuli. To address this in vivo, we subjected anesthetized mice to auditory stimulation, using 70 dB sound pressure level (SPL) broadband noise presented for 20 min. We then co-stained slices from stimulated or non-stimulated (control) mice with antibodies against PV and pCREB, and examined the primary auditory cortex (Figure 1A). We found that the proportion of PV+ cells with pCREB+ nuclei was ~3-fold greater after auditory stimulation than in control. This frames a series of questions about signaling to the nucleus in PV+ neurons.
Figure 1. CREB Phosphorylation Occurs in PV+ Cells in vivo, where PV+ Cells Spike Faster and have Larger Ca2+ increases Compared to Excitatory Neurons.
(A) Left, images from co-staining of PV and pCREB in mouse primary auditory cortex (A1) in stimulated or non-stimulated (control) mice. Stimulation was 70 dB SPL broadband noise for 20 min. Arrows, PV+, pCREB+ cells; arrowheads, PV+, pCREB- cells. Scale bar, 40 μm. Right, proportion of PV+ cells that are pCREB+ in control (461 cells, 2 mice) vs. stimulated (767 cells 2 mice) A1.
(B) in vivo neuronal firing rates. Inset, top, raster plot of exemplar putative PV+ interneuron (red) and putative excitatory neuron (blue). Inset, bottom, smoothed time plot of average instantaneous firing frequency per cell, obtained simultaneously in putative PV+ (n=5) and excitatory (n=24) cells during non-REM sleep. Right, average spike rates in putative PV+ (n=128) and excitatory (n=1031) cells, recorded with silicon probes in rat cortex in vivo across wakefulness and sleep. Note higher average firing rate in putative PV+ interneurons as compared to putative excitatory neurons: frequency at 50th percentile, put. PV+: 4.75 Hz, exc.: 0.69 Hz.
(C) Average Fluo-4 Ca2+ responses (ΔF/F) to field stimulation in cultured rat cortical PV+ neurons (n = 15, red) and in adjacent excitatory neurons (n = 95, blue). Black bar, duration of stimulation (5 s). Left, averaged responses of PV+ neurons to 5 Hz stimulation (red) were superimposed on responses of excitatory neurons at 1 Hz (blue), capturing the >5-fold disparity in firing frequencies in vivo (Figure 1B). Right, similar comparison of 30 Hz PV+ cell responses and 5 Hz excitatory neuron responses.
(D1) Image of mouse A1 with PV− (putative excitatory, green) and PV+ neurons (red/yellow). White circles represent ROIs used for image analysis in (D2) and (D3). Scale bar, 40 μm.
(D2) Example of averaged tone-evoked Ca2+ response (GCaMP6s, ΔF/F) from a PV+ cell and an adjacent PV− neuron. Black bar, duration of stimulation. Note higher amplitude and longer duration of responses in PV+ cell.
(D3) Response of the same neurons in (D2), shown across all individual frequencies (average of 10 repetitions of 17 frequencies at quarter-octave spacing). Note consistently higher amplitude and duration of Ca2+ signals in PV+ cells.
(D4) Average response across all frequencies in 1 mouse of PV+ (n=20, red) and PV− neurons (n=60, blue). Black bar, duration of stimulation.
(D5) Average tuning curve of peak responses, compiled from PV+ (red) and PV− (blue) neurons shown in (D4).
PV+ Cells Fire More Rapidly and Display Larger Ca2+ Increases than Excitatory Neurons
In general, CREB phosphorylation can occur in response to Ca2+ increases triggered by synaptic activity or neuronal depolarization. To quantify differences in neuronal activity between PV+ cells and excitatory neurons, we recorded unitary firing activity from putative interneurons and putative excitatory neurons in vivo using silicon probes implanted in rat frontal cortex. Interneurons (red) exhibited both a markedly higher instantaneous firing frequency, and a ~7-fold faster average firing rate, than excitatory neurons (blue, Figure 1B). Neurons were classified by spike width: a narrow-spiking population was comprised principally of putative PV+ cells (Kawaguchi et al, 1987), while a wide-spiking population consisted mainly of excitatory neurons (Stark et al., 2013). These data quantitatively support the general belief that PV+ cells fire much faster than excitatory neurons in vivo.
To determine how PV+ and excitatory neurons might differ in their Ca2+ dynamics at firing frequencies observed in vivo, cultured rat cortical neurons were driven by direct stimulation over a range of frequencies. Neurons were loaded with the green Ca2+ indicator Fluo-4, and Ca2+ responses to field stimulation were recorded (NBQX/APV present), followed by post-hoc identification of PV+ and excitatory neurons (αCaMKII+) via immunostaining with specific antibodies. Ca2+ rose promptly in response to stimulation at ≥5 Hz (Figures 1C and S1). To capture the difference in firing rates in vivo, we superimposed inhibitory and excitatory responses to stimulus frequencies separated by 5–6 fold. In this context, the bulk Ca2+ increases in PV+ cells were higher and broader than in excitatory neurons. For example, PV+ cells firing at 5 Hz or 30 Hz showed much greater Ca2+ responses, respectively, than those in excitatory neurons firing at 1 Hz or 5 Hz (Figure 1C; Figure S1B compares traces obtained at the same frequencies). The impact of the ~7-fold disparity in firing rates outweighed the ~2-fold lesser sensitivity of PV+ cells to increases in firing rate (Figure S1C). Thus, based on recordings in culture, the relatively high firing rates of cortical PV+ cells in vivo would be expected to give rise to larger and more prolonged Ca2+ elevations than in excitatory neurons.
To verify that this conclusion holds for genetically defined PV+ neurons in vivo, we performed two-photon Ca2+ imaging in the auditory cortex of awake, head-fixed mice. PV+ cells were identified by tdTomato expression and GCaMP6s Ca2+ transients were measured (Figure 1D). We recorded Ca2+ transients from PV+ cells and putative excitatory neurons, both spontaneous events and evoked transients in response to sensory input (pure tones). Targeted juxtacellular recordings confirmed that sound-evoked calcium signals were due to neuronal spiking and that GCaMP6s could detect single action potentials in PV+ neurons (Figure S2A). In PV+/tdTomato+ cells, sensory stimulation-evoked Ca2+ transients were broader and ~2-fold higher than in PV−/tdTomato- putative excitatory neurons (Figure 1D4), similar to Figure 1C. The PV+ vs PV− disparity is seen in representative single neuron recordings (Figures 1D2–1D3) and in population data (Figures 1D4–1D5). The larger amplitude and broader waveform were presumably due to PV+ cells firing action potentials at a higher frequency than excitatory neurons, consistent with our in vivo electrical recordings and Ca2+ imaging from cultured neurons. This inference is supported by previous work in auditory cortex showing that PV+ cells fire more rapidly in response to auditory stimuli than excitatory neurons (Li et al., 2014). Spontaneous Ca2+ transients also displayed a similar disparity between PV+ cells and excitatory neurons (Figures S2C–S2D), as if differences in firing rates also held for animals at rest. In sum, these data support the idea that PV+ cells in vivo display particularly large Ca2+ increases due to their rapid firing, with possible implications for downstream Ca2+ signaling.
PV+ Cells Have Distinct Requirements for CREB Phosphorylation
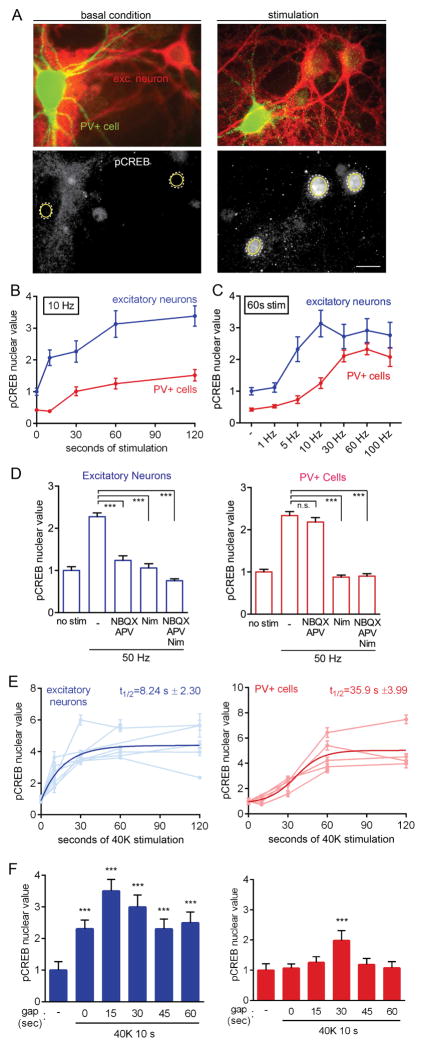
The mechanisms for E-T coupling downstream of Ca2+ transients must ensure that inhibitory neurons stay within their useful dynamic range for signaling to the nucleus. Mindful of the differences in Ca2+ flux between PV+ and excitatory neurons, we set out to describe signaling pathways resulting in CREB phosphorylation and gene expression. To determine the requirements for CREB phosphorylation in PV+ cells, we used immunocytochemistry in rat cortical cultures (Figures 2, S3, S4). Following field stimulation at 10 Hz, pCREB staining in the nucleus increased (average pixel intensity, [nucleus]-[background]). Strikingly, pCREB was seen to increase in the nucleus of excitatory neurons (blue) but not in PV+ cells (red) after 10 s of 10 Hz stimulation (Figure 2B). Kinetic differences were also apparent in analysis of frequency dependence. When action potentials were evoked by field stimulation at various frequencies for 60 s, a saturating level of pCREB in excitatory neurons was triggered by 10 Hz stimulation, while PV+ cells required much higher rates of 30–100 Hz stimulation to reach maximal response (Figure 2C). The amplitude of pCREB response was somewhat lower in PV+ cells, in line with less total CREB staining (Figure S4A). More importantly, the frequency-response relationship for PV+ cells is right-shifted by 6- to 10-fold, reminiscent of the ~7-fold disparity in firing frequency (Figure 1B). This suggests that the sensitivity of PV+ cells to AP firing might be tuned to their high rates of firing and high Ca2+ flux; potentially, these cells might also use significantly different mechanisms of E-T coupling.
Figure 2. PV+ Cells Exhibit Distinct Requirements for and Kinetics of CREB Phosphorylation Compared to Excitatory Neurons.
(A) Image of cultured cortical neurons stained for PV (green) and αCaMKII (excitatory neuron marker, red). Lower panels, pCREB (Ser133) staining of same cells under basal condition (left) and after stimulation (right). Dashed white lines, nuclear outline based on DAPI counterstain; yellow circles, ROIs for calculation of average pixel intensity. Scale bar, 20 μm.
(B) Time course of CREB phosphorylation in PV+ and excitatory neurons in response to 10 Hz field stimulation. Markedly sigmoid kinetics in PV+ cells not seen in excitatory neurons. All values normalized to mean baseline value for excitatory neurons in the same experiment.
(C) pCREB in excitatory neurons and PV+ cells in response to field stimulation at different frequencies. Values normalized as in (B).
(D) pCREB response in excitatory and PV+ neurons following 50 Hz stimulation for 18 s and no stimulation for 42 s prior to fixation. CREB phosphorylation was blocked by either NBQX/APV or nimodipine in excitatory neurons, but only by nimodipine in PV+ cells.
(E) pCREB timecourse for excitatory and PV+ cells in response to 40K. Traces from individual experiments (muted colors) and a fit of pCREB timecourse (bold colors; exponential for excitatory, sigmoid for PV+) reveal slower kinetics of CREB phosphorylation in PV+ cells (t1/2=35.9 s ± 3.99) as compared to excitatory neurons (t1/2=8.24 s ± 2.30).
(F) CREB phosphorylation in excitatory and PV+ cells in response to 40K for 10 s, followed by a gap of variable time prior to fixation. x-axis, length of time gap (s) between end of stimulation and fixation. “-”, basal condition prior to stimulation.
To dissect the relative contribution of various Ca2+ sources towards CREB phosphorylation in PV+ cells, a 50 Hz stimulus was applied with or without pharmacological inhibition of glutamate receptors or CaV1 channels. In PV+ cells, CREB phosphorylation was blocked by specific CaV1 blocker nimodipine (Figure 2D), but not by APV/NBQX. Conversely, pCREB in excitatory neurons was prevented by either APV/NBQX or nimodipine, consistent with previous findings in hippocampal neurons (Deisseroth et al., 1996; Deisseroth et al., 1998). These data suggest that in PV+ cells, CaV1 channels but not glutamate receptors play a privileged role in pCREB formation, in contrast with the joint dependence on CaV1 and glutamate receptors in excitatory neurons. While the role of CaV1 channels in communication with the nucleus has been extensively characterized in excitatory neurons (Cohen et al., 2015a; Ma et al., 2014; Wheeler et al., 2012), it is not well-described in PV+ cells.
To study CaV-dependent signaling in isolation, we depolarized neurons to ~−20 mV with 40 mM K+ (40K) while blocking spiking and neurotransmission with TTX, APV, and NBQX, and monitored the phosphorylation of CREB (Figure 2E). Depolarization with 40K triggered a rapid Ca2+ increase (Figure S4B) as well as formation of pCREB in PV+ cells and excitatory neurons (Figures 2E and S4C). Consistent with pCREB responses to action potential stimulation, CREB phosphorylation in PV+ cells occurred with much slower kinetics than in excitatory neurons (PV+ cells, onset t1/2=35.9 s ± 3.99; excitatory neurons, onset t1/2=8.24 s ± 2.30). This time course seems well matched to the relatively long (tens of seconds) spontaneous Ca2+ transients seen in PV+ cells in vivo (Figure S2C). Slower onset kinetics of pCREB were still apparent in PV+ cells in response to stronger depolarization with 60K or 90K (depolarizing neurons to ~−10 or 0 mV, respectively)(Figure S4D).
It remained possible that a 10 s stimulus was sufficient for nuclear signaling, but that fixation interrupted translocation before nuclear pCREB could become visible. To address this issue, we followed a short depolarization (1 or 10 s) with a variable delay period prior to fixation. Following a 10 s depolarization, pCREB in excitatory neurons continued to rise, peaking 15 s after the stimulus ended (Figure 2F). In contrast, PV+ cells showed only a slight rise in pCREB 30 s after stimulation, with no increase at other time points. Likewise, following a 1 s depolarization, excitatory neurons displayed a prominent rise in pCREB after a 10 s or 60 s time gap; in contrast, a 1 s depolarization failed to trigger any pCREB in PV+ cells (Figure S4E). Thus, the difference in pCREB at early time points cannot be explained simply by slower communication to the nucleus in PV+ cells once the depolarization-induced signal has been initiated. Instead, we hypothesize that PV+ interneurons and excitatory cells differ substantially in their stimulus requirements for cytonuclear communication.
CaV1-mediated Ca2+ Influx, Local Ca2+ and CaM Kinases Are Critical for pCREB and Gene Expression
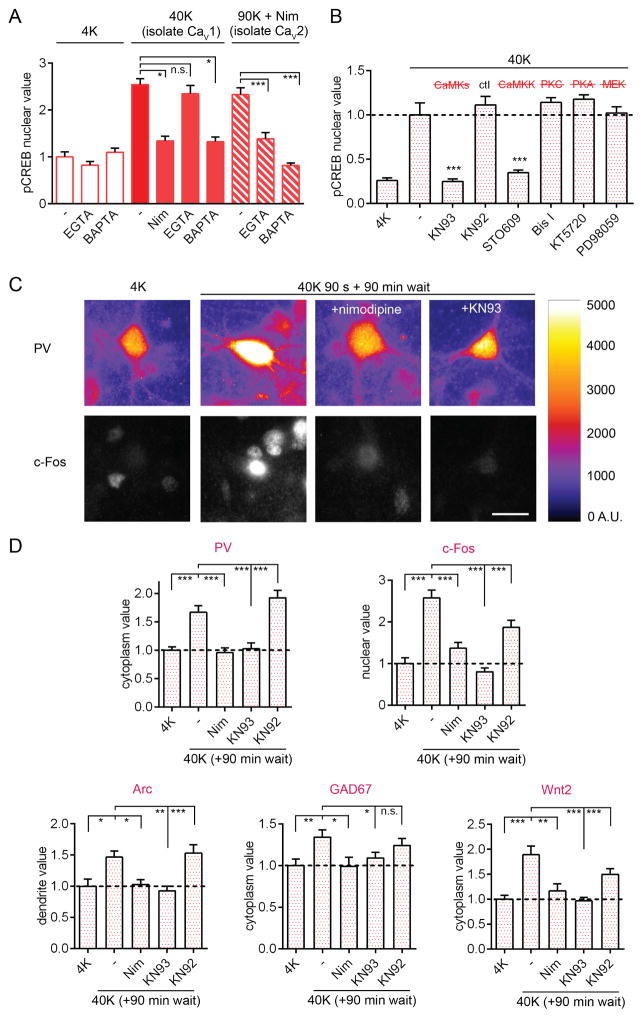
In sympathetic and excitatory neurons, Ca2+ is known to act locally within nanodomains near the CaV1 channel pore to activate a CaMK cascade that triggers translocation of Ca2+/CaM to the nucleus(Deisseroth et al., 1998; Li, 2016; Ma et al., 2014; Wheeler et al., 2012). We first checked that depolarization-triggered pCREB in PV+ cells required CaV1-mediated Ca2+ influx. Indeed, nimodipine prevented pCREB induction upon 40K depolarization (Figure 3A). We next asked whether the target of Ca2+ action in PV+ cells was also localized at the CaV1 channel or whether more distant increases in cytoplasmic Ca2+ were necessary. We compared effects of EGTA, a slow Ca2+ chelator, which buffers global Ca2+ but allows local Ca2+ increases within 1 μm of a channel pore, and of the fast Ca2+ chelator BAPTA, which buffers both global and local Ca2+ (Adler et al., 1991; Deisseroth et al., 1996). CREB phosphorylation was blocked by BAPTA to the same extent as with nimodipine, but not by EGTA (Figure 3A). Thus, a local Ca2+ rise at CaV1 is necessary and sufficient to trigger pCREB. In contrast, following selective activation of CaV2 channels (90K + nimodipine), the increase in pCREB was prevented by EGTA as well as BAPTA. This indicates that Ca2+ does not act locally near CaV2 to initiate signaling to the nucleus; rather, Ca2+ entering the cell via CaV2 must act over a distance, possibly at CaV1 channels (Wheeler et al., 2012).
Figure 3. Depolarization Triggers CaMK-dependent CREB Phosphorylation and Gene Expression in PV+ Interneurons.
(A) pCREB response in PV+ cells to 40K for 1 min (largely isolates CaV1; Wheeler et al., 2008), or 90K + nimodipine (isolates CaV2) +/− nimodipine, BAPTA or EGTA.
(B) pCREB response to 40K for 30 s was blocked by KN93 or STO609, but not by KN92 (inactive congener of KN93), Bis I (inhibits PKC), KT5720 (inhibits PKA), or PD98059 (inhibits MAP2K1/MEK1). Values normalized to 40K mean value.
(C) Images of PV/c-Fos co-staining. Cells were depolarized with 40K for 90 s or mock-stimulated in 4K (+/− nimodipine or KN93), and then incubated for 90 min without stimulation before fixation. Right, heatmap of PV staining (A.U., artificial units). Scale bar, 20 μm.
(D) 40K for 90 s triggered increases in PV, c-Fos, Arc, GAD67, and Wnt2 assessed 90 min later. Nimodipine or KN93 prevented activity-dependent increases of all five genes.
In excitatory neurons, local Ca2+ is important for activating a CaMK-mediated pathway (Wheeler et al., 2012). To test whether CaMKs are important for nuclear signaling in PV+ cells, we applied KN93, a blocker of Ca2+/CaM binding to CaMKs, before 40K stimulation. Indeed, CREB phosphorylation was completely abolished by KN93 but not its inactive congener KN92. The specific CaMKK inhibitor STO609 also prevented pCREB. In contrast, pCREB was not prevented by inhibitors of PKC (bisindolylmaleimide I), PKA (KT5720), or MEK1 (PD98059)(Figures 3B and S5A). Clearly, in the context of a relatively brief stimulation, CaMKs mediate signaling to pCREB.
CREB phosphorylation regulates transcription of many target genes (Impey et al., 2004). To assay activity-dependent gene expression in PV+ cells, we depolarized neurons with 40K (or mock-stimulated in 4K) for 90 s, then waited 90 min before fixation and staining. 40K triggered significant increases in the immunostaining intensity of protein products of c-Fos, Arc, Gad1 (encoding GAD67), Pvalb (encoding PV) and Wnt2 (Figures 3C, 3D, S5B, and S5C), all of which have a consensus CRE upstream of the cognate gene. The importance of CREB for regulation of PV and GAD67, a new finding, was verified by expression of a dominant-negative CREB (Figure S5D). Thus, depolarization triggered pCREB in tens of seconds, boosting levels of CREB target genes.
To determine the role of CaV1 and CaMKs in mediating upregulation of these genes, we treated cells with nimodipine, KN93, or KN92 and assayed gene expression (Figures 3C, 3D, and S5C). Increases in expression of c-Fos, Arc, Gad1, Pvalb, and Wnt2 were all prevented by nimodipine or KN93. The control drug KN92 did not affect the increase in Arc, Pvalb, Gad1, or Wnt2 expression, while increases in c-Fos were partially reduced, possibly because of the inhibitory action of KN92 on CaV1 (Ca2+ flux reduced by ~55% at 4 μM Gao et al., 2006). MEK1 inhibitor PD98059 did not significantly affect increases in expression of Arc, Gad1, Pvalb, or Wnt2, and partially reduced but partly spared the increases in c-Fos (Figure S5E). Overall, activation of CaV1 channels and CaMK signaling is critical in PV+ neurons for a wide range of activity-dependent gene expression, likely acting through Ser133 phosphorylation of CREB.
Signaling to the Nucleus Is Mediated by CaM Translocation
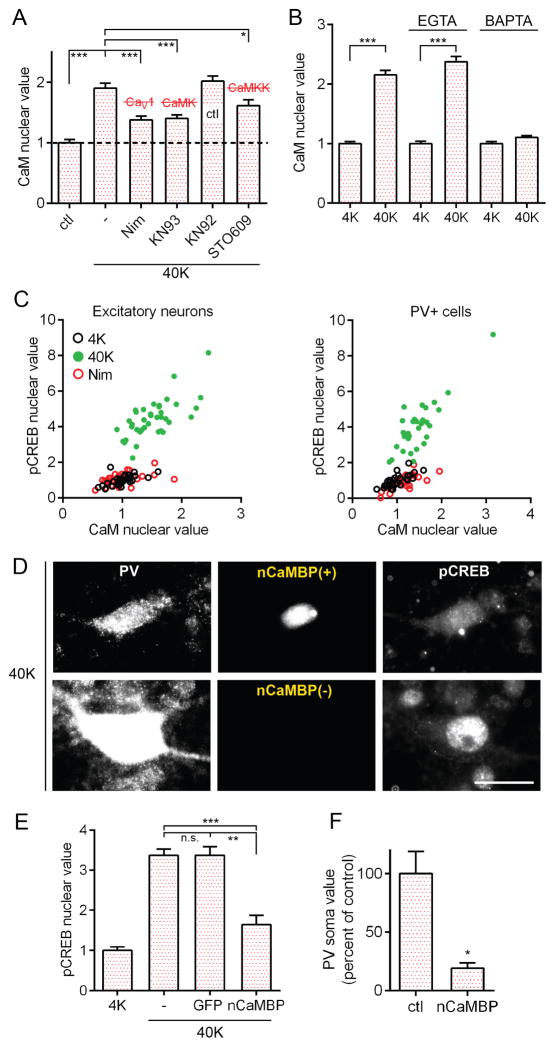
How is a local Ca2+ signal propagated to the nucleus? We tested the possibility that, as in excitatory neurons (Deisseroth et al., 1998), CaM translocation underlies signaling to the nucleus in in PV+ neurons. Consistent with this idea, increases in nuclear CaM in PV+ neurons were triggered by 40K (Figures 4A and S6A) or 50 Hz stimulation (Figure S6B) and blocked by nimodipine (Figures 4A and S6B). Furthermore, 40K-triggered increases were blocked by BAPTA but spared by EGTA (Figure 4B). CaM translocation was also blocked by KN93 or STO609, but not by KN92 (Figure 4A). These findings support the hypothesis that CaM translocation depends on local Ca2+ near the CaV1 channel pore and is regulated by a CaM kinase, rather than by free diffusion alone.
Figure 4. The Mechanism of Cytonuclear Communication in PV+ Neurons Involves CaM and CaMKs.
(A) Following 1 min of 40K, nuclear CaM increased in PV+ cells; effect inhibited by nimodipine, KN93, or STO609, but not by KN92.
(B) Increased nuclear CaM (40K, 1 min) ; prevented by BAPTA but spared by EGTA.
(C) In excitatory and PV+ cells, CaM increased in the nucleus with 40K for 1 min, correlated with increases in pCREB. Increases prevented by nimodipine. Pearson correlation for 40K condition: excitatory neurons R2: 0.4825, p<0.001; PV+ cells R2: 0.6937, p<0.001.
(D) Image of a PV+ cell expressing nuclear CaM-Binding Protein (nCaMBP, top row) and a wild-type PV+ cell (bottom row). Both pCREB and PV staining intensity reduced in the cell expressing nCaMBP. Scale bar, 20 μm.
(E) nCaMBP expression inhibited pCREB formation as compared to GFP-expressing or non-transfected control PV+ cells.
(F) nCaMBP expression resulted in reduced somatic PV staining in PV+ cells.
Is CaM translocation required for CREB phosphorylation in PV cells? This idea was supported by correlation between increases in nuclear CaM and nuclear pCREB (Figure 4C; R2: 0.69). Once Ca2+/CaM arrives in the nucleus, it could in principle activate a potent nuclear CaMK cascade that depends steeply on [Ca2+/CaM] (Racioppi and Means, 2008; Wayman et al., 2008). We tested the necessity of nuclear CaM for CREB phosphorylation, using a “nuclear CaM Binding Protein” (nCaMBP; (Wang et al., 1995), that can intercept CaM in the nucleus. In PV+ cells, nCaMBP largely prevented pCREB in response to 40K (Figures 4D–4E). In addition, nCaMBP expression attenuated PV staining intensity in PV+ cells (Figure 4F), supporting the idea that CaM signaling acts through CREB to regulate PV expression. In the wider population of GAD67+ neurons, nCaMBP also prevented pCREB (Figures S6C–S6D) and reduced GAD67 expression (Figure S6E). Thus, nuclear translocation of CaM may participate in activating CREB-dependent gene expression across the broader category of GABAergic interneurons.
To gain further insight into the basis of CaM translocation, we turned to pharmacology. In agreement with previous results, inhibition of protein phosphatase 1 and 2A (PP1 and PP2A) with 2 μM okadaic acid (OA) increased CREB phosphorylation (Figure S6F). PP1 is likely acting as a CREB phosphatase (Bito et al., 1996). Interestingly, in PV+ cells, as in sympathetic neurons (Ma et al., 2014), inhibition of PP2A with 20 nM OA had the opposite effect: a significant reduction of pCREB (Figures S6F and S6G). Thus, we conclude that an interplay between CaM kinases and S/T phosphatases helps control CREB phosphorylation in PV+ cells.
CaMKII Is Not a CaM Shuttle in PV+ cells
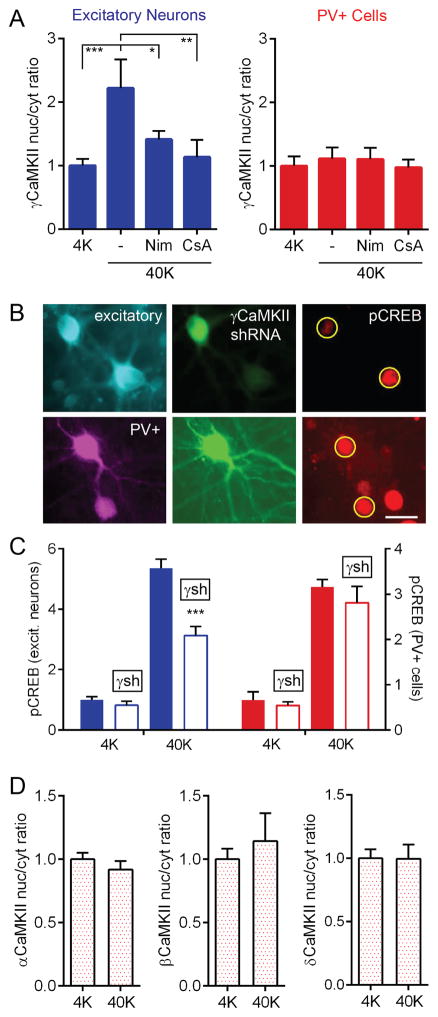
We recently addressed the conundrum of surface-nucleus communication in excitatory cells by demonstrating that γCaMKII supports a CaM shuttle that mediates cytonuclear signaling (Ma et al., 2014). To test whether PV+ cells employ the same shuttle mechanism, we compared the nucleus/cytoplasm (nuc/cyt) ratio of γCaMKII before and after 40K stimulation (Figure 5A). In cortical excitatory neurons, this ratio sharply increased with depolarization, prevented by nimodipine. Additionally, cyclosporine A (CsA), prevented γCaMKII translocation, consistent with calcineurin-mediated unmasking of a nuclear localization signal (NLS). In contrast, in PV+ cells, there was no detectable γCaMKII translocation. To test further for γCaMKII involvement in CREB phosphorylation, we knocked down γCaMKII protein levels via shRNA (Figures 5B–5C) and observed reduced pCREB in excitatory neurons, but not in PV+ cells. Consistent with data in excitatory neurons (Deisseroth et al., 1998), no other CaMKII isoform underwent nuclear translocation in PV+ cells (Figures 5D and S7A). Furthermore, we found that shRNA knockdown of βCaMKII, a critical component of the γCaMKII shuttling pathway (Ma et al., 2014), reduced pCREB in cortical excitatory neurons but not in PV+ cells (Figure S7B). Taken together, these results unveil a key difference in E-T coupling between excitatory and PV+ neurons, and raise the question of what mediates CaM nuclear translocation.
Figure 5. Unlike Excitatory Neurons, PV+ cells Do Not Use γCaMKII as a CaM Shuttle.
(A) γCaMKII translocated to the nucleus following 40K for 1 min in excitatory neurons (prevented by nimodipine or CsA), but not PV+ cells.
(B) Images of pCREB staining (red, right panel) in excitatory (top) and PV+ cells (bottom), some transfected with γCaMKII shRNA (green). Transfected excitatory neurons had reduced pCREB compared to control, but transfected PV+ neurons were unaffected. Yellow circles, ROIs used to calculate nuclear pixel intensity. Scale bar, 20 μm.
(C) γCaMKII shRNA knockdown reduced pCREB in excitatory neurons (blue, left y-axis) but not in PV+ cells (red, right y-axis).
(D) 40K did not induce a redistribution of α, β, or δCaMKII within PV+ cells.
CaMKI Translocates to the Nucleus in PV+ Cells
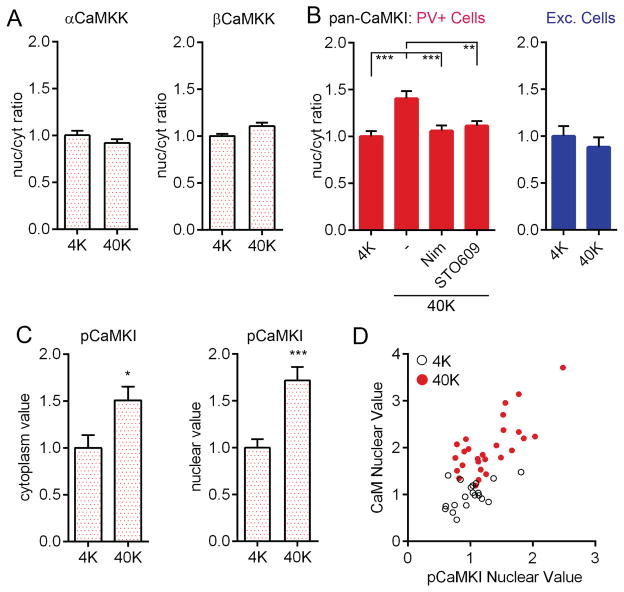
We asked whether CaMKK or its substrate CaMKI mediate CaM translocation in PV+ cells. Antibodies against α or βCaMKK showed no change in distribution following depolarization (Figure 6A). However, we found a clear increase in CaMKI nuc/cyt ratio (antibody against α/β/γ/δ isoforms) with 40K in PV+ cells (Figure 6B) but not excitatory neurons. This increase was prevented by nimodipine or STO609, implicating both CaV1-mediated Ca2+ flux and CaMKK-dependent phosphorylation in CaMKI translocation. Following depolarization, pCaMKI (Thr177/178) increased in both the cytoplasm and in the nucleus of PV+ cells (Figure 6C)(likewise for excitatory cells, Figure S7C). This result does not distinguish between a scheme where CaMKI, residing in both nucleus and cytoplasm, is phosphorylated upon Ca2+ influx, or a scenario wherein phosphorylation of CaMKI triggers CaMKI nuclear translocation and possibly co-transport of CaM (Schild et al., 2014; Yu et al., 2014). To explore the latter scenario, we performed a cell-by-cell analysis of nuclear pCaMKI and nuclear CaM in PV+ cells and observed a striking correlation (Figure 6D; R2=0.52). This result is consistent with the notion that some species of phosphorylated CaMKI helps support the rise in nuclear CaM, and thereby promotes CREB phosphorylation.
Figure 6. Depolarization Triggers Redistribution of CaMKI to the Nucleus in PV+ Cells.
(A) Nuc/cyt ratio of α or βCaMKK in PV+ cells did not change following 40K for 2 min.
(B) Nuc/cyt ratio of pan-CaMKI staining increased in PV+ but not excitatory neurons, with 40K for 1 min, and was prevented by nimodipine or by STO609.
(C) pCaMKI immunostaining increased in both cytoplasm and nucleus of PV+ cells following 3 min of 40K.
(D) Following 40K for 1 min, nuclear CaM values were correlated with nuclear pCaMKI values in PV+ cells (Pearson correlation, R2 = 0.52, p<0.001).
γCaMKI Translocates to the Nucleus, Supports CREB Phosphorylation, and Regulates Dendritic Branching
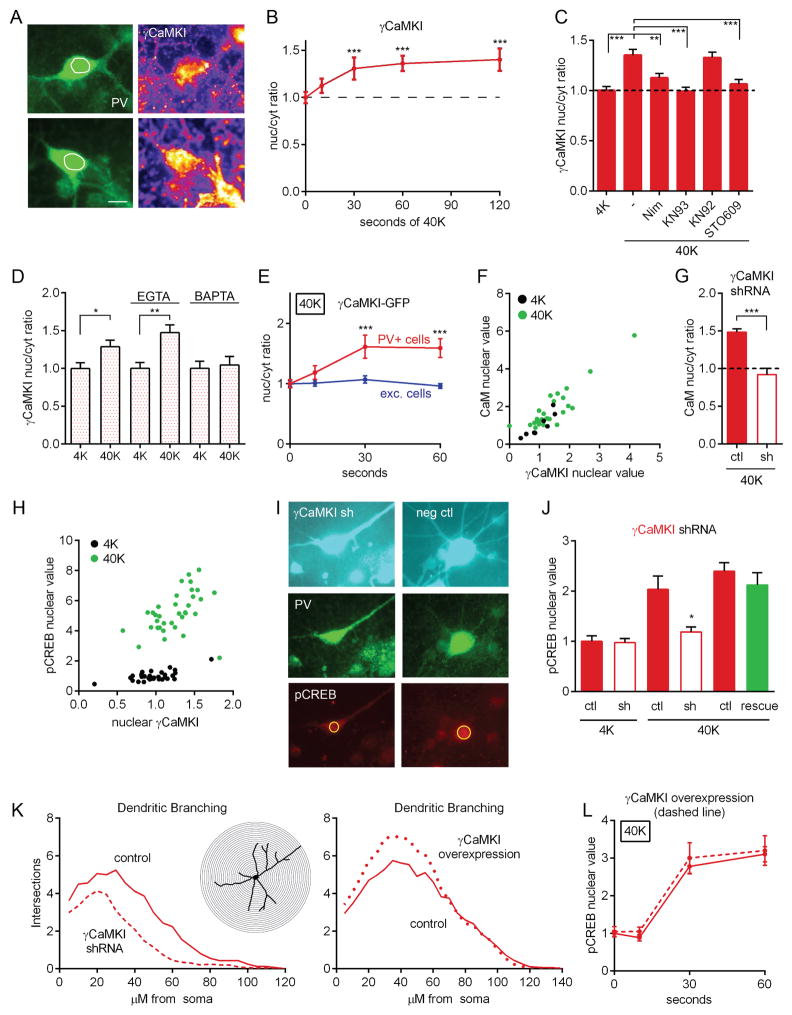
We used specific antibodies to test if a particular isoform of CaMKI translocated to the nucleus. 40K triggered a progressive increase in γCaMKI nuc/cyt ratio: the nucleus filled in (Figure 7A) and the elevated ratio leveled off after ~60 s (Figures 7B, S8A, and S8B). In contrast, neither α, β, nor δCaMKI displayed an altered distribution after 40K (Figure S8C). The increase in γCaMKI nuc/cyt ratio was prevented by nimodipine, KN93, or STO609, but not by KN92 (Figure 7C), and was also blocked by BAPTA, but spared by EGTA (Figure 7D); this suggests that local Ca2+ flux through CaV1 channels is necessary to trigger γCaMKI translocation. Consistent with immunostaining results, 40K triggered an increase in γCaMKI-GFP nuc/cyt ratio in PV+ cells (Figure 7E), but not in excitatory neurons. These results demonstrate that in PV+ cells, neuronal depolarization initiates γCaMKI nuclear translocation, which requires CaMKK activity.
Figure 7. γCaMKI Translocates to the Nucleus and Is Important for CREB Phosphorylation in PV+ Cells.
(A) Image of γCaMKI staining in PV+ cells before and after 40K. Note how nucleus is more uniformly filled with γCaMKI following stimulation. White circle represents nuclear outline, traced from DAPI staining. Scale bar, 10 μm.
(B) Time course of nuc/cyt ratio of immunostaining against γCaMKI.
(C) Nuc/cyt ratio of staining against γCaMKI increased with 5 min of 40K; increase prevented by nimodipine, KN93, or STO609 but not by KN92.
(D) γCaMKI nuc/cyt ratio increased in PV+ cells following 40K; increase prevented by BAPTA but spared by EGTA.
(E) Nuc/cyt ratio of γCaMKI-GFP increased following 40K in PV+ but not excitatory neurons.
(F) Following 40K for 5 min, nuclear CaM was correlated with nuclear γCaMKI. (Pearson correlation, R2 = 0.80, p<0.001).
(G) Increased CaM nuc/cyt ratio following 40K for 5 min was prevented by shRNA knockdown of γCaMKI. 40K value for shRNA-expressing cells was not different from baseline (4K) value for control cells (p>0.3). All values normalized to baseline value for control, non-transfected cells.
(H) Following 40K for 5 min, pCREB was correlated with nuclear γCaMKI. (Pearson correlation, R2 = 0.18, p<0.05).
(I) Image of neurons transfected with γCaMKI shRNA (left, cyan) or with non-silencing control (right, cyan). Cells treated with 40K for 30 s and co-stained for PV (middle row, green) and pCREB (bottom row, red). Note that γCaMKI knockdown reduced CREB phosphorylation and dendritic branching.
(J) pCREB response to 40K for 30 s was reduced by shRNA knockdown of γCaMKI and rescued by shRNA-resistant construct (green bar). Third/fifth bar from the left, non-transfected cells on coverslips transfected with shRNA/shRNA+rescue. Data shown are representative of 4 shRNA experiments.
(K) Two-dimensional Sholl analysis of dendritic arbor complexity 7 days after transfection with γCaMKI shRNA (left) or γCaMKI-GFP overexpression construct (right). Number of intersections between branches and successive concentric circles (inset, 5 μm increments). Left, area under curve reduced by 42% in neurons expressing γCaMKI shRNA (dashed line) compared to control (solid line). Right, area under curve increased by 15% in neurons overexexpressing γCaMKI-GFP (dotted line) compared to control (solid line).
(L) Overexpression of γCaMKI-GFP (dashed line) did not affect the kinetics of pCREB.
To test further the role of γCaMKI in signaling to the nucleus, we analyzed the relationship between γCaMKI, CaM, and CREB. The level of nuclear γCaMKI was strongly correlated with nuclear CaM, both at baseline and upon stimulation (Figure 7F; R2=0.80). To test causality, we assayed CaM translocation following γCaMKI shRNA knockdown. Indeed, γCaMKI knockdown prevented the depolarization-triggered increase in CaM nuc/cyt ratio (Figures 7G and S8D), suggesting that CaM movement to the nucleus depends on γCaMKI. Following 40K, nuclear γCaMKI was significantly correlated with nuclear pCREB (Figure 7H, R2=0.18).
Neuronal cultures were then transfected with either the γCaMKI shRNA construct or a nonsilencing control, and co-stained for PV and pCREB. γCaMKI shRNA resulted in a sharp reduction in pCREB (Figures 7I–7J). This effect was rescued by co-transfection of an shRNA-resistant γCaMKI overexpression plasmid, verifying the specificity of the shRNA effect; transfection with either a control shRNA or knockdown of α or βCaMKI did not reduce pCREB (Figure S8E). These results suggest that γCaMKI is indeed active upstream of pCREB in PV+ cells, acting as a molecular shuttle for CaM or as a CREB kinase itself (Figure S8F).
We further examined the impact of manipulation of γCaMKI levels on dendritic organization (Figure 7K). In accord with previous work (Takemoto-Kimura et al., 2007; Wayman et al., 2006), shRNA knockdown of γCaMKI reduced dendritic branching in cultured PV+ neurons (representative image, Figure 7I, top panel; Sholl analysis of branching patterns, Figure 7K, left panel). Conversely, we found that overexpression of γCaMKI resulted in increased dendritic branching in PV+ cells (Figure 7K, right panel), also in line with previous findings in cultured excitatory neurons (Takemoto-Kimura et al., 2007). Taken together, the data on CREB phosphorylation and dendritic arborization align with the hypothesis that γCaMKI signaling affects gene expression and thereby regulates dendritic branching in PV+ cells.
CaMKIV Is a Nuclear CREB Kinase Responsible for Limiting the Rate of CREB Phosphorylation in PV+ cells
Could use of a distinct shuttle protein, γCaMKI, be responsible for the slower dynamics of CREB phosphorylation in PV+ cells? If γCaMKI translocation were rate-limiting, its overexpression should accelerate CREB phosphorylation. On the contrary, γCaMKI-GFP overexpression did not increase the rate of pCREB at any timepoint, including 10 s (Figure 7L), when its nuclear level had already started to rise (Figures 7B and 7E). The biological efficacy of this construct was reflected by its effect on dendritic branching (Figure 7K, right). Taken together, γCaMKI translocation is likely not the rate-limiting step in CREB phosphorylation in PV+ cells.
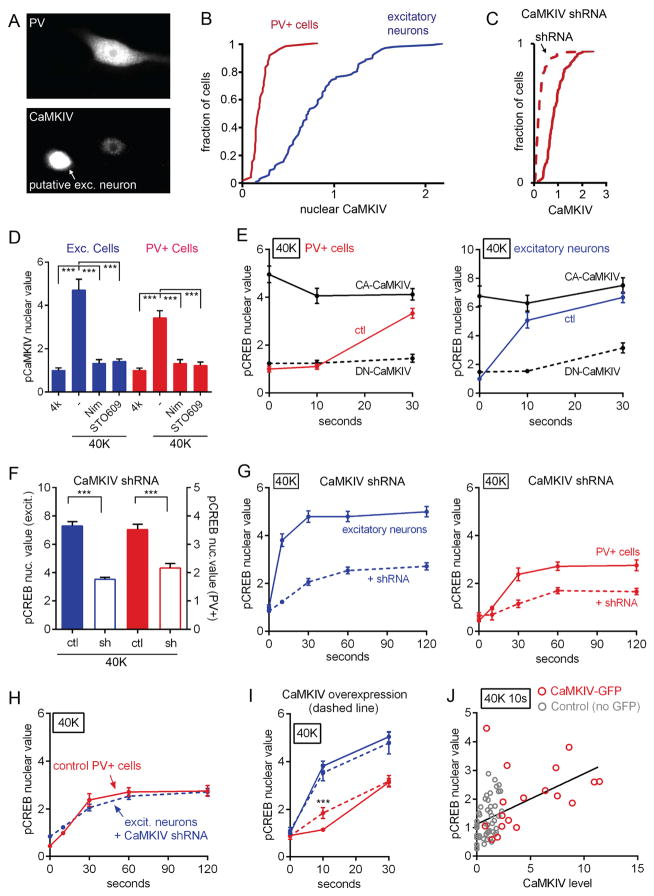
Accordingly, we asked whether the sigmoid kinetics of CREB phosphorylation might depend on on CaMKIV, the dominant CREB kinase in CaMK signaling in excitatory neurons (Bito et al., 1996; Wayman et al., 2008). CaMKIV is not known to be phosphorylated by CaMKI, but it could be activated by nuclear delivery of CaM. We found that CaMKIV was present at very low levels in PV+ cells, at ~15% of the staining intensity observed in excitatory neurons (Figures 8A and 8B). To exclude the possibility that the observed CaMKIV immunoreactivity was due to nonspecific staining, we knocked down CaMKIV levels with shRNA and verified that CaMKIV immunostaining was indeed reduced (Figure 8C). Thus, CaMKIV is demonstrably present in PV+ cells. Using an antibody against pCaMKIV (Thr196), we observed that CaMKIV was phosphorylated following 40K in PV+ cells as well as excitatory neurons; this increase was prevented in both cell types by nimodipine or STO609 (Figure 8D), consistent with a role for pCaMKIV as a critical player in the overall signal transduction pathway.
Figure 8. Nuclear CaMKIV is a CREB Kinase in PV+ cells and Acts as the Rate-Limiting Factor Slowing CREB Phosphorylation.
(A) CaMKIV present at very low levels in PV+ cells. Bottom, CaMKIV staining in a PV+ cell (right) and in an adjacent putative excitatory neuron (at left).
(B) PV+ cells expressed CaMKIV at levels much lower than excitatory neurons (K-S test, p<0.001). CaMKIV value plotted in arbitrary units.
(C) CaMKIV nuclear staining in PV+ cells was further decreased by shRNA knockdown of CaMKIV (dashed line)(K-S test, p<0.001). Data normalized to mean control CaMKIV value.
(D) phospho-CaMKIV increased in excitatory and PV+ neurons following 40K for 1 min; increase prevented in both cell types by nimodipine or STO609.
(E) Dominant negative CaMKIV (DN-CaMKIV, dashed line) prevented pCREB at 10 s and 30 s following 40K in PV+ and excitatory neurons. Constitutively active CaMKIV (CA-CaMKIV, solid black line) induced maximal pCREB formation at baseline, 10 s and 30 s following 40K in PV+ and excitatory neurons. Red (PV+) and blue (excitatory) lines, non-transfected control.
(F) CaMKIV shRNA knockdown resulted in reduction in pCREB in excitatory (blue, left y-axis) and PV+ cells (red, right y-axis). All values normalized to control at basal condition for each cell type.
(G) CaMKIV knockdown (dashed line) slowed kinetics and reduced amplitude of pCREB response to 40K in both excitatory and PV+ neurons. All values normalized to mean 4K value for excitatory neurons.
(H) Rate and amplitude of pCREB in excitatory neurons with CaMKIV knockdown (blue, dashed line) closely resembled that of wild type PV+ cells (red, solid line). By ANOVA, no significant difference at any time point.
(I) CaMKIV-GFP overexpression speeded rise of pCREB in PV+ (red) but not excitatory neurons (blue).
(J) Increased pCREB following 10 s of 40K correlated with higher CaMKIV in PV+ cells. Gray circles, non-transfected; red circles, CaMKIV-GFP overexpression plasmid. Black line, linear regression of pCREB vs. CaMKIV value in all cells (Pearson Correlation, R2=0.26, p<0.001).
To test the contribution of CaMKIV as a CREB kinase, we transfected neuronal cultures with dominant negative (DN) or constitutively active (CA) CaMKIV constructs (Figure 8E). DN-CaMKIV expression prevented pCREB at 30 s, suggesting that CaMKIV is a necessary CREB kinase in PV+ cells. Expression of CA-CaMKIV resulted in maximal pCREB staining at baseline, demonstrating the sufficiency of CaMKIV for inducing CREB phosphorylation. To assay the importance of endogenous CaMKIV, we stained for pCREB in PV+ cells transfected with the CaMKIV shRNA construct (Figure 8F). The increase in pCREB upon 40K stimulation was reduced by CaMKIV knockdown. Taken together, these results demonstrate that CaMKIV acts as a CREB kinase in the nuclei of PV+ cells.
We then asked whether the low levels of CaMKIV in PV+ cells were responsible for the slow, sigmoid kinetics of CREB phosphorylation. We knocked down CaMKIV levels with shRNA to ~8% of baseline protein levels (Figure S8G), and assayed the dynamics of pCREB following depolarization. In both excitatory and PV+ cells, CaMKIV knockdown slowed the kinetics and reduced the amplitude of CREB phosphorylation (Figure 8G). Strikingly, the kinetics and amplitude of pCREB in excitatory neurons with CaMKIV knockdown nearly matched those seen in control PV+ cells (Figure 8H). This suggests that high CaMKIV levels are necessary for kinetically rapid CREB phosphorylation. To test for sufficiency, we transfected cells with a CaMKIV-GFP overexpression plasmid and stained for pCREB after 10 s of 40K depolarization (Figures 8I–8J). When CaMKIV was overexpressed, no difference was seen in pCREB kinetics in excitatory neurons, as if their basal levels of CaMKIV were not rate-limiting. In contrast, overexpression of CaMKIV accelerated the kinetics of pCREB in PV+ cells, eliminating the sigmoid onset by driving a significant rise in pCREB even following a 10 s depolarization (Figure 8I). Indeed, CaMKIV levels were positively correlated with pCREB levels in both wild type and CaMKIV-overexpressing cells (Figure 8J; R2=0.26). These results indicate that the level of CaMKIV is critical to the kinetics of CREB phosphorylation. At its low endogenous level, CaMKIV imposes a kinetic constraint, rendering the time course of CREB phosphorylation sigmoid, even though the redistribution of γCaMKI (and by extension, CaM) proceeds linearly (Figures 7B and 7E); elevation of CaMKIV eliminates the sigmoid rise. By rate-limiting CREB phosphorylation, CaMKIV controls the dynamics of CREB phosphorylation in a manner appropriate to the firing pattern and the Ca2+ regulation in PV+ cells.
Discussion
CaM kinases have been largely ignored in inhibitory neurons, possibly because they are deficient in αCaMKII, the best-known CaMK family member. Here we have identified a CaMK-dependent pathway of nuclear signaling in PV+ interneurons. Our experiments demonstrate that in PV+ cells, γCaMKI translocates to the nucleus and thereby mediates CaM nuclear translocation, CREB phosphorylation, gene transcription, and dendritic branching. Furthermore, PV+ cells express relatively low levels of the CREB kinase CaMKIV, which dampens the kinetics of nuclear pCREB signaling, in accordance with rapid firing of PV+ neurons in vivo.
CaMK Translocation as an Evolutionarily Conserved Strategy in Surface-to-Nucleus Communication
Surface-to-nucleus communication is important for cellular functioning, but relevant proteins in interneurons have remained elusive. CRTC1 and Jacob, two recently identified synapse-to-nucleus trafficking proteins in excitatory neurons, do not appear to translocate in interneurons (Ch’ng et al., 2012; Mikhaylova et al., 2014). In addition, γCaMKII shuttles CaM to the nucleus in excitatory neurons but not in PV+ cells (Figure 5A). Thus, our finding that γCaMKI redistributes to the nucleus reveals an activity-dependent nuclear translocation event in interneurons. Interestingly, CMK-1 (the ortholog of CaMKI) translocates to the nucleus in sensory neurons of C. elegans to regulate thermosensory plasticity (Schild et al., 2014; Yu et al., 2014). Thus, CaMKI translocation may be an evolutionarily conserved mechanism to regulate gene expression and long-term cellular plasticity.
Our evidence for the importance of γCaMKI brings fresh perspective to previous anatomical data. At embryonic day 13, γCaMKI is preferentially expressed in the medial ganglionic eminence, which is composed of PV+ and SST+ cell precursors (Kamata et al., 2007). In the adult brain, γCaMKI is expressed at particularly high levels in non-pyramidal cell layers of the hippocampus (Allen Brain Atlas) and in the central amygdala (Takemoto-Kimura et al., 2003), which is principally composed of GABAergic cells. High expression in these brain regions is consistent with a privileged role of γCaMKI for intracellular signaling in inhibitory neurons.
Our experiments establish that γCaMKI translocation is important for CREB phosphorylation in PV+ neurons, but leave important mechanistic questions unsettled. γCaMKI does not possess a classic NLS, but it does contain a putative nuclear export signal (NES, 315VVHHMRKLHM324, (Stedman et al., 2004). Regulation of γCaMKI cytonuclear trafficking could occur by way of exclusion from the nucleus which is removed by an activity-dependent masking of the NES, possibly by phosphorylation at Ser328, a consensus site. Work in lung epithelial cells (Agassandian et al., 2012) suggests another possible mechanism of CaMKI nuclear translocation: CaMKI phosphorylates CCTα, triggering formation of a putative CaMKI-CCTα complex whose Ca2+-induced nuclear entry is dependent on a binding partner, the protein 14-3-3ζ. γCaMKI nuclear translocation may, in a similar fashion, be facilitated by an interaction with these or other binding partners. This scenario fits with our finding that STO609 prevented the translocation of γCaMKI (Figure 7C) and CaM (Figure 4A). STO609 also blocked phosphorylation of CaMKIV (Figure 8D) and CREB (Figure 3B), consistent with the idea that catalytic activity of CaMKK is necessary to drive cytonuclear communication, either by triggering γCaMKI translocation as just mentioned, or by directly phosphorylating nuclear CaMKIV, or both.
CaMKs Support Regulation of Gene Expression in PV cells
In the context of the depolarizing stimuli studied here, upregulation of CREB target genes depends critically on the CaV1-CaMK-CaM pathway (Figure 3D) and not the MAPK pathway (Figure S5E). The importance of such regulation extends from neuronal excitability and survival (c-Fos, Zhang et al., 2002) to synaptic plasticity (Arc, Plath et al., 2006 and PV, Donato et al., 2013).
Our results are also consistent with previous studies demonstrating that γCaMKI regulates dendritic branching in excitatory neurons in vitro. One mechanism involves γCaMKI-mediated signaling to MAPKs and CREB, leading to upregulation of Wnt2 (Wayman et al., 2006). This pathway requires Ca2+ influx, CaMKK-mediated activation of γCaMKI, and gene transcription. A second, distinct pathway involves local regulation and is independent of transcription (Takemoto-Kimura et al., 2007). Our data in PV+ neurons suggest that Ca2+ influx via CaV1 activates a γCaMKI-mediated pathway to CREB, Wnt2 transcription and dendritic branching. These findings are largely consistent with the first proposed mechanism, while not excluding a role for γCaMKI in local dendritic regulation.
Mechanism of Cytonuclear Signaling in PV+ cells Is Tailored to their Physiology
We found in vivo that PV+ cells undergo CREB phosphorylation in response to sensory stimulation (Figure 1A), and that the dynamics of CREB phosphorylation in PV+ cells are distinct from those in excitatory neurons. The contrast in kinetics is consistent with fundamental differences in their physiological properties. On average, putative PV+ cells fire at higher rates than excitatory neurons (Figure 1B), leading to longer and larger increases in intracellular Ca2+, as verified by our Ca2+ imaging in vitro (Figure 1C) and in vivo (Figure 1D). High frequency firing, and the large Ca2+ influxes that it triggers, could easily overwhelm the fast and sensitive E-T coupling mechanisms in excitatory neurons, were they duplicated in PV+ neurons. Our data suggest that PV+ neurons differ in two substantive ways. First, transient rises in Ca2+ in PV+ cells are ~2-fold less sensitive to increasing stimulation frequency (Figure S1C), possibly because of their narrower spikes or distinct Ca2+ buffering properties (Aponte et al., 2008). Second, the frequency-dependence of pCREB in PV+ neurons is roughly 6–10-fold less sensitive to elevation of stimulus frequency (Figure 2C). Furthermore, the kinetics of CREB phosphorylation are slower and more obviously sigmoid in PV+ neurons than in pyramidal cells (Figure 2E). Each of these differences tunes PV+ cells to the patterns of activity that they are likely to encounter in situ (Figures 1D and S2). In turn, these disparities suggest PV+ neurons must employ a different strategy, downstream of Ca2+ elevation, to couple excitation to transcription. Indeed, we find that PV+ cells express CaMKIV at significantly lower levels than in excitatory neurons (Figures 8A–8B), contributing to slow, sigmoid kinetics of pCREB (Figures 8G–8I). Experiments with CA- and DN-CaMKIV establish the dominance of CaMKIV in CREB phosphorylation upon depolarization for seconds to minutes. Taken together, our results leave little doubt about the importance of CaMKIV: as a crucial enzyme, present at rate-limiting levels, it effectively dampens the kinetics of CREB phosphorylation in PV+ cells, in accord with their high frequency firing. However, it remains possible that in other contexts CREB phosphorylation may depend on such kinases as Rsk1+Rsk2, Msk1+Msk2, p38, PKG, AKT/PKB, or PI3K (Johannessen et al., 2004).
Significance for Learning and Memory and Neuropsychiatric Disease
Given that the activity of interneurons is critical for morphological development (De Marco Garcia et al., 2011) and synaptic plasticity (D’Amour and Froemke, 2015), a critical question is how their firing is linked to cellular and behavioral phenotypes. Here we show that activity-dependent transcriptional regulation depends on a pathway of CaMK-dependent signaling to the nucleus. Such activity-triggered changes in PV and GAD67 expression were shown recently to be important for excitatory-to-inhibitory synaptic-density ratios and structural synaptic plasticity (Donato et al., 2013). Environmental enrichment or fear conditioning affected levels of PV and GAD67 expression in vivo, with intriguing implications for memory consolidation and retrieval. By elucidating a biochemical mechanism that connects PV+ cell activation to expression of these key genes, we provide a missing link between neuronal activity and memory consolidation.
The relevance of E-T coupling in PV+ interneurons for neuropsychiatric disease therefore comes as no surprise. There is mounting evidence suggesting that schizophrenia may result from dysfunction of GABAergic neurons in general (Benes and Berretta, 2001) and PV+ interneurons in particular (Cohen et al., 2015b; Lewis et al., 2012). In the schizophrenic brain, PV+ cells have reduced levels of GAD67 and PV mRNA (Beasley et al., 2002; Hashimoto et al., 2003). Further, PV+ cell dysfunction may underlie the disruption of gamma oscillations in schizophrenia (Carlen et al., 2012). Multiple genes linked to schizophrenia or autism participate in nuclear signaling as described here for cortical PV+ neurons: they encode the CaV1 channel (Bhat et al., 2012), βCaMKK (Luo et al., 2014), CaM (Broadbelt and Jones, 2008), CaMKIV (Waltes et al., 2014) and CREB (Kawanishi et al., 1999). Thus, our findings raise the possibility that genetic defects in such signaling proteins disturb E-T coupling in PV+ interneurons and thus contribute to their specific dysfunction in brain disorders.
Experimental Procedures
Additional information regarding DNA constructs, image analysis, antibodies, and buffers is provided in the Extended Experimental Procedures.
In vivo Auditory Stimulation
Mice were subjected to a 100 ms-long, 70 dB white noise stimulus played at 0.5 Hz for 20 min, followed by PFA perfusion 10 min later (see Extended Experimental Procedures).
In vivo Electrical Recordings
Measurement of spike rates were performed in 11 rats implanted with 64-site silicon probe electrodes. Spikes were clustered using KlustaKwik, manually reviewed, and a separatrix was created (Stark et al., 2013) (see Extended Experimental Procedures).
In vivo Ca2+ Imaging
PV-tdTomato mice were stimulated with pure tones (70 dB SPL, 4–64 kHz) and GCaMP6s signals were acquired and juxtacellular recordings were performed in A1 (see Extended Experimental Procedures).
Cortical Cultures
Cortical neurons were cultured as previously described (Ma et al., 2014), with minor modifications (see Extended Experimental Procedures).
In vitro Ca2+ Imaging
Cultured rat cortical neurons were loaded with Fluo-4 and subjected to field stimulation in Tyrode’s solution (see Extended Experimental Procedures).
Drug Treatments and Stimulation
Cortical neurons were stimulated with the indicated KCl solutions (with TTX, APV, NBQX) or field stimulation (with TTX) at 37°C for the indica ted times, with drugs present as noted (see Extended Experimental Procedures).
Transfection
Cortical neurons were transfected 6 days after plating using a high efficiency Ca2+-phosphate transfection method (Jiang and Chen, 2006).
Immuno(cyto/histo)chemistry
16 μm-thick cryostat sections or fixed cultured neurons were blocked, and incubated with primary and secondary antibodies, in donkey serum and Triton X-100 (see Extended Experimental Procedures).
Image Analysis
Immunostaining quantification was performed as previously described (Ma et al., 2014) with modifications (see Extended Experimental Procedures).
Statistical Analysis
All data were normalized to corresponding basal condition unless otherwise noted. All data are shown as mean ± standard error of the mean (SEM), unless otherwise noted. ≥ 24 neurons were scored from 2–3 separate experiments, unless otherwise noted (see Extended Experimental Procedures). Statistical analysis with one-way ANOVA followed by t-test, unless otherwise noted. *, p ≤ 0.05, **, p ≤ 0.01, ***, p ≤ 0.001.
Supplementary Material
Highlights.
Voltage-gated Ca2+ influx triggers nuclear translocation of CaM in PV+ interneurons.
CaMK signaling promotes CREB phosphorylation and activates key genes in PV+ cells.
γCaMKI, not γCaMKII, operates to shuttle CaM to the nucleus in PV+ cells.
Low CaMKIV levels rate-limit CREB phosphorylation in PV+ cells. eTOC blurb
Acknowledgments
We thank S. Takemoto-Kimura and H. Bito for the generous gifts of γCaMKI-GFP, CaMKIV-GFP, and CaMKIV, α and γCaMKI shRNA constructs, and G. Wayman for the generous gift of a βCaMKI shRNA construct. We thank N. Chenouard for help with math and programming, S. Sanchez for cultures, and R. Priya, G. Fishell, B. Li, J.B. Cohen, and all Tsien lab members for useful advice and technical assistance. This work was supported by research grants to R.W.T. from the IGMS (GM058234), NINDS (NS24067), the NIMH (MH071739), and Simons, Mathers, and Burnett Family Foundations. S.M.C. is supported by a Medical Scientist Research Service Award (T32GM007308).
Footnotes
Author Contributions
S.M.C. and K.V.K. performed auditory cortex experiments and analyzed data. B.O.W. performed silicon probe recordings and analyzed data. S.M.C, H.M., and R.W.T conceived project. S.M.C. performed experiments with cultured neurons and analyzed data. S.M.C. and R.W.T. wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adler EM, Augustine GJ, Duffy SN, Charlton MP. Alien intracellular calcium chelators attenuate neurotransmitter release at the squid giant synapse. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1991;11:1496–1507. doi: 10.1523/JNEUROSCI.11-06-01496.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agassandian M, Chen BB, Pulijala R, Kaercher L, Glasser JR, Mallampalli RK. Calcium-calmodulin kinase I cooperatively regulates nucleocytoplasmic shuttling of CCTalpha by accessing a nuclear export signal. Molecular biology of the cell. 2012;23:2755–2769. doi: 10.1091/mbc.E11-10-0863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aponte Y, Bischofberger J, Jonas P. Efficient Ca2+ buffering in fast-spiking basket cells of rat hippocampus. The Journal of physiology. 2008;586:2061–2075. doi: 10.1113/jphysiol.2007.147298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beasley CL, Zhang ZJ, Patten I, Reynolds GP. Selective deficits in prefrontal cortical GABAergic neurons in schizophrenia defined by the presence of calcium-binding proteins. Biological psychiatry. 2002;52:708–715. doi: 10.1016/s0006-3223(02)01360-4. [DOI] [PubMed] [Google Scholar]
- Benes FM, Berretta S. GABAergic interneurons: implications for understanding schizophrenia and bipolar disorder. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 2001;25:1–27. doi: 10.1016/S0893-133X(01)00225-1. [DOI] [PubMed] [Google Scholar]
- Bhat S, Dao DT, Terrillion CE, Arad M, Smith RJ, Soldatov NM, Gould TD. CACNA1C (Cav1.2) in the pathophysiology of psychiatric disease. Progress in neurobiology. 2012;99:1–14. doi: 10.1016/j.pneurobio.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bito H, Deisseroth K, Tsien RW. CREB phosphorylation and dephosphorylation: a Ca(2+)- and stimulus duration-dependent switch for hippocampal gene expression. Cell. 1996;87:1203–1214. doi: 10.1016/s0092-8674(00)81816-4. [DOI] [PubMed] [Google Scholar]
- Broadbelt K, Jones LB. Evidence of altered calmodulin immunoreactivity in areas 9 and 32 of schizophrenic prefrontal cortex. Journal of psychiatric research. 2008;42:612–621. doi: 10.1016/j.jpsychires.2007.07.006. [DOI] [PubMed] [Google Scholar]
- Buzsaki G, Chrobak JJ. Temporal structure in spatially organized neuronal ensembles: a role for interneuronal networks. Curr Opin Neurobiol. 1995;5:504–510. doi: 10.1016/0959-4388(95)80012-3. [DOI] [PubMed] [Google Scholar]
- Carlen M, Meletis K, Siegle JH, Cardin JA, Futai K, Vierling-Claassen D, Ruhlmann C, Jones SR, Deisseroth K, Sheng M, et al. A critical role for NMDA receptors in parvalbumin interneurons for gamma rhythm induction and behavior. Molecular psychiatry. 2012;17:537–548. doi: 10.1038/mp.2011.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ch’ng TH, Uzgil B, Lin P, Avliyakulov NK, O’Dell TJ, Martin KC. Activity-dependent transport of the transcriptional coactivator CRTC1 from synapse to nucleus. Cell. 2012;150:207–221. doi: 10.1016/j.cell.2012.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S, Greenberg ME. Communication between the synapse and the nucleus in neuronal development, plasticity, and disease. Annual review of cell and developmental biology. 2008;24:183–209. doi: 10.1146/annurev.cellbio.24.110707.175235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen SM, Li B, Tsien RW, Ma H. Evolutionary and functional perspectives on signaling from neuronal surface to nucleus. Biochem Biophys Res Commun. 2015a;460:88–99. doi: 10.1016/j.bbrc.2015.02.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen SM, Tsien RW, Goff DC, Halassa MM. The impact of NMDA receptor hypofunction on GABAergic neurons in the pathophysiology of schizophrenia. Schizophrenia research. 2015b doi: 10.1016/j.schres.2014.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Amour JA, Froemke RC. Inhibitory and excitatory spike-timing-dependent plasticity in the auditory cortex. Neuron. 2015;86:514–528. doi: 10.1016/j.neuron.2015.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Marco Garcia NV, Karayannis T, Fishell G. Neuronal activity is required for the development of specific cortical interneuron subtypes. Nature. 2011;472:351–355. doi: 10.1038/nature09865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deisseroth K, Bito H, Tsien RW. Signaling from synapse to nucleus: postsynaptic CREB phosphorylation during multiple forms of hippocampal synaptic plasticity. Neuron. 1996;16:89–101. doi: 10.1016/s0896-6273(00)80026-4. [DOI] [PubMed] [Google Scholar]
- Deisseroth K, Heist EK, Tsien RW. Translocation of calmodulin to the nucleus supports CREB phosphorylation in hippocampal neurons. Nature. 1998;392:198–202. doi: 10.1038/32448. [DOI] [PubMed] [Google Scholar]
- Donato F, Rompani SB, Caroni P. Parvalbumin-expressing basket-cell network plasticity induced by experience regulates adult learning. Nature. 2013;504:272–276. doi: 10.1038/nature12866. [DOI] [PubMed] [Google Scholar]
- Gao L, Blair LA, Marshall J. CaMKII-independent effects of KN93 and its inactive analog KN92: reversible inhibition of L-type calcium channels. Biochem Biophys Res Commun. 2006;345:1606–1610. doi: 10.1016/j.bbrc.2006.05.066. [DOI] [PubMed] [Google Scholar]
- Gu X, Spitzer NC. Distinct aspects of neuronal differentiation encoded by frequency of spontaneous Ca2+ transients. Nature. 1995;375:784–787. doi: 10.1038/375784a0. [DOI] [PubMed] [Google Scholar]
- Hashimoto T, Volk DW, Eggan SM, Mirnics K, Pierri JN, Sun Z, Sampson AR, Lewis DA. Gene expression deficits in a subclass of GABA neurons in the prefrontal cortex of subjects with schizophrenia. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2003;23:6315–6326. doi: 10.1523/JNEUROSCI.23-15-06315.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong EJ, McCord AE, Greenberg ME. A biological function for the neuronal activity-dependent component of Bdnf transcription in the development of cortical inhibition. Neuron. 2008;60:610–624. doi: 10.1016/j.neuron.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Impey S, McCorkle SR, Cha-Molstad H, Dwyer JM, Yochum GS, Boss JM, McWeeney S, Dunn JJ, Mandel G, Goodman RH. Defining the CREB regulon: a genome-wide analysis of transcription factor regulatory regions. Cell. 2004;119:1041–1054. doi: 10.1016/j.cell.2004.10.032. [DOI] [PubMed] [Google Scholar]
- Jiang M, Chen G. High Ca2+-phosphate transfection efficiency in low-density neuronal cultures. Nature protocols. 2006;1:695–700. doi: 10.1038/nprot.2006.86. [DOI] [PubMed] [Google Scholar]
- Jiang M, Swann JW. A role for L-type calcium channels in the maturation of parvalbumin-containing hippocampal interneurons. Neuroscience. 2005;135:839–850. doi: 10.1016/j.neuroscience.2005.06.073. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Zhang Z, Zhang C, Wang X, Sakata K, Lu B, Sun QQ. A key mechanism underlying sensory experience-dependent maturation of neocortical GABAergic circuits in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:12131–12136. doi: 10.1073/pnas.1105296108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannessen M, Delghandi MP, Moens U. What turns CREB on? Cell Signal. 2004;16:1211–1227. doi: 10.1016/j.cellsig.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Jones EG, Huntley GW, Benson DL. Alpha calcium/calmodulin-dependent protein kinase II selectively expressed in a subpopulation of excitatory neurons in monkey sensory-motor cortex: comparison with GAD-67 expression. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1994;14:611–629. doi: 10.1523/JNEUROSCI.14-02-00611.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamata A, Sakagami H, Tokumitsu H, Owada Y, Fukunaga K, Kondo H. Spatiotemporal expression of four isoforms of Ca2+/calmodulin-dependent protein kinase I in brain and its possible roles in hippocampal dendritic growth. Neuroscience research. 2007;57:86–97. doi: 10.1016/j.neures.2006.09.013. [DOI] [PubMed] [Google Scholar]
- Kawaguchi Y, Katsumaru H, Kosaka T, Heizmann CW, Hama K. Fast spiking cells in rat hippocampus (CA1 region) contain the calcium-binding protein parvalbumin. Brain Res. 1987;416:369–374. doi: 10.1016/0006-8993(87)90921-8. [DOI] [PubMed] [Google Scholar]
- Kawanishi Y, Harada S, Tachikawa H, Okubo T, Shiraishi H. Novel variants in the promoter region of the CREB gene in schizophrenic patients. Journal of human genetics. 1999;44:428–430. doi: 10.1007/s100380050196. [DOI] [PubMed] [Google Scholar]
- Kinney JW, Davis CN, Tabarean I, Conti B, Bartfai T, Behrens MM. A specific role for NR2A-containing NMDA receptors in the maintenance of parvalbumin and GAD67 immunoreactivity in cultured interneurons. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2006;26:1604–1615. doi: 10.1523/JNEUROSCI.4722-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klausberger T, Somogyi P. Neuronal diversity and temporal dynamics: the unity of hippocampal circuit operations. Science (New York, NY) 2008;321:53–57. doi: 10.1126/science.1149381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau CG, Murthy VN. Activity-dependent regulation of inhibition via GAD67. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2012;32:8521–8531. doi: 10.1523/JNEUROSCI.1245-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis DA, Curley AA, Glausier JR, Volk DW. Cortical parvalbumin interneurons and cognitive dysfunction in schizophrenia. Trends Neurosci. 2012;35:57–67. doi: 10.1016/j.tins.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li BT, MR, Tsien RW. A slow coincidence detector that requires Ca2+ channel flux and conformational signals. Science (New York, NY) 2016 in press. [Google Scholar]
- Li LY, Xiong XR, Ibrahim LA, Yuan W, Tao HW, Zhang LI. Differential Receptive Field Properties of Parvalbumin and Somatostatin Inhibitory Neurons in Mouse Auditory Cortex. Cerebral cortex (New York, NY : 1991) 2014 doi: 10.1093/cercor/bht417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo XJ, Li M, Huang L, Steinberg S, Mattheisen M, Liang G, Donohoe G, Shi Y, Chen C, Yue W, et al. Convergent lines of evidence support CAMKK2 as a schizophrenia susceptibility gene. Molecular psychiatry. 2014;19:774–783. doi: 10.1038/mp.2013.103. [DOI] [PubMed] [Google Scholar]
- Ma H, Groth RD, Cohen SM, Emery JF, Li B, Hoedt E, Zhang G, Neubert TA, Tsien RW. gammaCaMKII Shuttles Ca(2+)/CaM to the Nucleus to Trigger CREB Phosphorylation and Gene Expression. Cell. 2014;159:281–294. doi: 10.1016/j.cell.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikhaylova M, Karpova A, Bar J, Bethge P, YuanXiang P, Chen Y, Zuschratter W, Behnisch T, Kreutz MR. Cellular distribution of the NMDA-receptor activated synapto-nuclear messenger Jacob in the rat brain. Brain structure & function. 2014;219:843–860. doi: 10.1007/s00429-013-0539-1. [DOI] [PubMed] [Google Scholar]
- Patz S, Grabert J, Gorba T, Wirth MJ, Wahle P. Parvalbumin expression in visual cortical interneurons depends on neuronal activity and TrkB ligands during an Early period of postnatal development. Cerebral cortex (New York, NY : 1991) 2004;14:342–351. doi: 10.1093/cercor/bhg132. [DOI] [PubMed] [Google Scholar]
- Plath N, Ohana O, Dammermann B, Errington ML, Schmitz D, Gross C, Mao X, Engelsberg A, Mahlke C, Welzl H, et al. Arc/Arg3.1 is essential for the consolidation of synaptic plasticity and memories. Neuron. 2006;52:437–444. doi: 10.1016/j.neuron.2006.08.024. [DOI] [PubMed] [Google Scholar]
- Racioppi L, Means AR. Calcium/calmodulin-dependent kinase IV in immune and inflammatory responses: novel routes for an ancient traveller. Trends in immunology. 2008;29:600–607. doi: 10.1016/j.it.2008.08.005. [DOI] [PubMed] [Google Scholar]
- Royer S, Zemelman BV, Losonczy A, Kim J, Chance F, Magee JC, Buzsaki G. Control of timing, rate and bursts of hippocampal place cells by dendritic and somatic inhibition. Nat Neurosci. 2012;15:769–775. doi: 10.1038/nn.3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schild LC, Zbinden L, Bell HW, Yu YV, Sengupta P, Goodman MB, Glauser DA. The Balance between Cytoplasmic and Nuclear CaM Kinase-1 Signaling Controls the Operating Range of Noxious Heat Avoidance. Neuron. 2014;84:983–996. doi: 10.1016/j.neuron.2014.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegel I, Mardinly AR, Gabel HW, Bazinet JE, Couch CH, Tzeng CP, Harmin DA, Greenberg ME. Npas4 regulates excitatory-inhibitory balance within neural circuits through cell-type-specific gene programs. Cell. 2014;157:1216–1229. doi: 10.1016/j.cell.2014.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark E, Eichler R, Roux L, Fujisawa S, Rotstein HG, Buzsaki G. Inhibition-induced theta resonance in cortical circuits. Neuron. 2013;80:1263–1276. doi: 10.1016/j.neuron.2013.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stedman DR, Uboha NV, Stedman TT, Nairn AC, Picciotto MR. Cytoplasmic localization of calcium/calmodulin-dependent protein kinase I-alpha depends on a nuclear export signal in its regulatory domain. FEBS letters. 2004;566:275–280. doi: 10.1016/j.febslet.2004.04.042. [DOI] [PubMed] [Google Scholar]
- Takemoto-Kimura S, Ageta-Ishihara N, Nonaka M, Adachi-Morishima A, Mano T, Okamura M, Fujii H, Fuse T, Hoshino M, Suzuki S, et al. Regulation of dendritogenesis via a lipid-raft-associated Ca2+/calmodulin-dependent protein kinase CLICK-III/CaMKIgamma. Neuron. 2007;54:755–770. doi: 10.1016/j.neuron.2007.05.021. [DOI] [PubMed] [Google Scholar]
- Takemoto-Kimura S, Terai H, Takamoto M, Ohmae S, Kikumura S, Segi E, Arakawa Y, Furuyashiki T, Narumiya S, Bito H. Molecular cloning and characterization of CLICK-III/CaMKIgamma, a novel membrane-anchored neuronal Ca2+/calmodulin-dependent protein kinase (CaMK) The Journal of biological chemistry. 2003;278:18597–18605. doi: 10.1074/jbc.M300578200. [DOI] [PubMed] [Google Scholar]
- Waltes R, Duketis E, Knapp M, Anney RJ, Huguet G, Schlitt S, Jarczok TA, Sachse M, Kampfer LM, Kleinbock T, et al. Common variants in genes of the postsynaptic FMRP signalling pathway are risk factors for autism spectrum disorders. Human genetics. 2014;133:781–792. doi: 10.1007/s00439-013-1416-y. [DOI] [PubMed] [Google Scholar]
- Wang J, Campos B, Jamieson GA, Jr, Kaetzel MA, Dedman JR. Functional elimination of calmodulin within the nucleus by targeted expression of an inhibitor peptide. The Journal of biological chemistry. 1995;270:30245–30248. doi: 10.1074/jbc.270.51.30245. [DOI] [PubMed] [Google Scholar]
- Wayman GA, Impey S, Marks D, Saneyoshi T, Grant WF, Derkach V, Soderling TR. Activity-dependent dendritic arborization mediated by CaM-kinase I activation and enhanced CREB-dependent transcription of Wnt-2. Neuron. 2006;50:897–909. doi: 10.1016/j.neuron.2006.05.008. [DOI] [PubMed] [Google Scholar]
- Wayman GA, Lee YS, Tokumitsu H, Silva AJ, Soderling TR. Calmodulin-kinases: modulators of neuronal development and plasticity. Neuron. 2008;59:914–931. doi: 10.1016/j.neuron.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler DG, Barrett CF, Groth RD, Safa P, Tsien RW. CaMKII locally encodes L-type channel activity to signal to nuclear CREB in excitation-transcription coupling. The Journal of cell biology. 2008;183:849–863. doi: 10.1083/jcb.200805048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler DG, Groth RD, Ma H, Barrett CF, Owen SF, Safa P, Tsien RW. Ca(V)1 and Ca(V)2 channels engage distinct modes of Ca(2+) signaling to control CREB-dependent gene expression. Cell. 2012;149:1112–1124. doi: 10.1016/j.cell.2012.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu YV, Bell HW, Glauser DA, Van Hooser SD, Goodman MB, Sengupta P. CaMKI-Dependent Regulation of Sensory Gene Expression Mediates Experience-Dependent Plasticity in the Operating Range of a Thermosensory Neuron. Neuron. 2014;84:919–926. doi: 10.1016/j.neuron.2014.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Zhang D, McQuade JS, Behbehani M, Tsien JZ, Xu M. c-fos regulates neuronal excitability and survival. Nature genetics. 2002;30:416–420. doi: 10.1038/ng859. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.