Abstract
The epigenetic and anti-cancer activities of the nucleoside analog DNA methyltransferase (DNMT) inhibitors decitabine (5-aza-2′-deoxycytidine, DAC), azacitidine, and guadecitabine are thought to require cellular uptake, metabolism to 5-aza-2′-deoxycytidine triphosphate, and incorporation into DNA. This genomic incorporation can then lead to trapping and degradation of DNMT enzymes, and ultimately, passive loss of DNA methylation. To facilitate measurement of critical exposure-response relationships of nucleoside analog DNMT inhibitors, a sensitive and reliable method was developed to simultaneously quantitate 5-aza-2′-deoxycytidine genomic incorporation and genomic 5-methylcytosine content using LC-MS/MS. Genomic DNA was extracted and digested into single nucleosides. Chromatographic separation was achieved with a Thermo Hyperpcarb porous graphite column (100 mm × 2.1 mm, 5μm) and isocratic elution with a 10 mM ammonium acetate:acetonitrile with 0.1% formic acid (70:30, v/v) mobile phase over a 5 minute total analytical run time. An AB Sciex 5500 triple quadrupole mass spectrometer operated in positive electrospray ionization mode was used for the detection of 5-aza-2′-deoxycytidine, 2′-deoxycytidine, and 5-methyl-2′-deoxycytidine. The assay range was 2 – 400 ng/mL for 5-aza-2′-deoxycytidine, 50 – 10,000 ng/mL for 2′-deoxycytidine, and was 5 – 1,000 ng/mL for 5-methyl-2′-deoxycytidine. The assay proved to be accurate (93.0–102.2%) and precise (CV ≤ 6.3%) across all analytes. All analytes exhibited long-term frozen digest matrix stability at −70°C for at least 117 days. The method was applied for the measurement of genomic 5-aza-2′-deoxycytidine and 5-methyl-2′-deoxycytidine content following exposure of in vitro cell culture and in vivo animal models to decitabine.
Keywords: 5-aza-2′-deoxycytidine, decitabine, DNA methyltransferase inhibitor, LC/MS/MS, genomic DNA, global DNA demethylation, 5-methyl-2′-deoxycytidine, 5-methylcytosine
1. Introduction
Epigenetic silencing of genes that encode tumor suppressors, cell cycle regulators, proteins involved in cell-cell interaction and suppression of invasion, and other cancer inhibitory genes, occurs in both hematologic and epithelial malignancies [1]. Such epigenetic silencing can occur by multiple mechanisms influencing chromatin structure including: promoter methylation and histone modifications [2, 3]. The ability to induce gene re-expression by targeting these mechanisms is possible through pharmacological agents that reverse DNA hypermethylation mediated gene silencing by inhibiting DNA methyltransferases (DNMTs) and agents that inhibit “readers”, “writers” and “erasers” of histone modifications [4]. Azacitidine (5-aza-cytidine; 5AC) and decitabine (5-aza-2′-deoxycytidine; DAC) are two nucleoside analog DNMT inhibitors that are FDA approved for use in myelodysplastic syndromes (MDS) [5]. Guadecitabine (GDAC) is an emerging decitabine prodrug (cleaved by phosphodiesterase to produce decitabine [6]) that is currently in phase III clinical trials for treatment of acute myeloid leukemia. Each of these drugs is also being widely tested in multiple hematological and solid organ cancer types, either as monotherapies, or in combination with other epigenetic, targeted, or immunotherapeutic drugs.
With each of these three major nucleoside analog drugs, the mechanism of action for their anti-cancer activity is thought to be via DNMT inhibition, but they can also have other poorly understood cytotoxic effects, particularly at high doses [5, 7, 8]. Their DNMT inhibitory activity requires cellular uptake by nucleoside transporters, convergent metabolism of all three drugs by multiple nucleic acid metabolizing enzymes to DAC triphosphate, and incorporation of into genomic DNA in lieu of 2′-deoxycytidine triphosphate [7, 9, 10]. In the case of azacitidine, approximately 10–20% of the 5AC diphosphate intermediate is converted to DAC diphosphate via ribonucleotide reductase, and the rest is available for further processing and incorporation into RNA [11]. Once incorporated into genomic DNA, DAC can covalently trap DNMT enzymes and trigger their proteosomal degradation [5, 7, 8, 12, 13]. Without DNMTs present to maintain DNA methylation across DNA replication and cell division, passive demethylation of the genome ensues and can reverse DNA hypermethylation and epigenetic gene silencing of cancer protective genes [8, 14, 15]. Given that all of the major nucleoside analog DNMT inhibitors have a presumed mechanism of action for their epigenetic and anti-cancer effects that converges on incorporation of DAC into the genome and DNA demethylation, quantitative measurements of these convergent effects could facilitate a deeper understanding of exposure-response parameters of these drugs. Such an understanding could ultimately allow a greater understanding of their bioavailability, intracellular dynamics, mechanisms of sensitivity and resistance, and informed optimization of dose and schedule of administration for use with different cancer types.
To date, direct measurement of DAC genomic incorporation has likely been impeded by: i) chemical instability and rapid hydrolysis of the parent compounds [16–19] requiring development of efficient pre-analytical processing methods; and ii) the similar molecular weight of DAC and the natural DNA nucleoside 2′-deoxycytidine (2dC) (228.2 vs. 227.2) necessitating robust chromatographic resolution of DAC in the midst of vast excess of 2dC [7, 20]. The closest quantitative method not utilizing radiolabeled drug has been for measurement of 5AC triphosphate [21] and DAC triphosphate [22–24], which were detected in cell lines, peripheral blood mononuclear cell (PBMC) or bone marrow. Incorporation of 3H-DAC ex vivo in leukemic cell lines and patient samples has been assessed as a potential phenotypic probe for the efficacy of therapy [25]. Here, we have overcome these challenges and have developed a robust and reliable method for the quantitative measurement of the incorporation of DAC into DNA combined with a global DNA methylation assessment by quantifying genomic 5-methyl-2′-deoxycytidine (5mC) content, both of which are normalized to 2dC content. The method has been utilized in preclinical experiments to probe the exposure-response properties of DAC.
2. Experimental
2.1. Chemical and reagents
All analytes purchased had a purity greater than 98%. 5-methyl-2′-deoxycytidine (5mC), 5-azacytidine-15N4 (5AC-15N4), 2′-deoxycytidine-13C15N2 (2dC-13C15N2), and 5-methyl-2′-deoxycytidine-d3 (5mC-d3) were purchased from Toronto Research Chemical (Toronto, ON). DAC and 2′-deoxycytidine (2dC) were purchased from Sigma Aldrich (St. Louis, MO). HPLC grade methanol, acetonitrile, and formic acid (98% v/v, in water) were purchased from EMD Chemical Inc. (Gibbstown, NJ). Deionized water was obtained from Millipore Milli-Q-UF filtration system (Milford, MA). Ammonium acetate was purchased from JT Baker (Phillipburg, NJ). All other chemicals were of molecular biological grade or higher and were obtained from Sigma Aldrich (St. Louis, MO) or Roche Life Science (Indianapolis, IN). DU145, PC3, HOP62 and PC9 cell lines were obtained from American Type Culture Collection (Manassas, VA).
2.2. Chromatography
The LC system was a Waters Acquity with a binary pump and an autosampler (Milford, MA). The autosampler was maintained at 5°C. The analyte separation was achieved using a Thermo Hyperpcarb porous graphite column (PGC), (100 mm × 2.1 mm, 5μm, Pittsburgh, PA) at room temperature. The mobile phase consisted of 10 mM ammonium acetate:acetonitrile with 0.1% formic acid (70:30, v/v) delivered using isocratic elution at a flow rate of 0.3 mL/min for a total runtime of 5 minutes. After each injection, the autosampler needle was washed with 1.6 mL of acetonitrile:water with 0.5% formic acid (60:40, v/v).
2.3. Mass spectrometry
The mass spectrometric detection was carried out using an AB Sciex 5500 triple quadrupole mass spectrometer operating in positive electrospray ionization utilizing multiple reaction monitoring (MRM) mode. The settings for the mass spectrometer were as follows: curtain gas 30 psi, collision gas 7 psi, ion spray voltage 1500 volts, probe temperature 450°C, ion source gas 1 50 psi, ion source gas 2 60 psi, exit potential 13, and collision cell exit potential 10. The declustering potential was 50, 150, 85, 85, 66, and 56 for DAC, 2dC, 5mC, 5AC-15N4, 2dC-13C15N2 and 5mC-d3 respectively. The collision energy was 16, 19, 18, 19, 18, and 16 for DAC, 2dC, 5mC, 5AC-15N4, 2dC-13C15N2 and 5mC-d3 respectively. The MRM m/z transitions were for the following: 228.9 → 113.0 for DAC, 228.0 → 112.0 for 2dC, 242.0 → 126.0 for 5mC, 249.0 → 117.0 for 5AC-15N4 230.8 → 115.0 for 2dC-13C15N2 and 245.8 → 129.0 for 5mC-d3. The LC and the mass spectrometer was controlled by the Analyst software (version 1.6).
2.4. Preparation of calibration standards and quality control (QC) samples
Stock solutions for each analyte were prepared independently. The stock solutions for DAC, 2dC, and 5mC were prepared at a concentration of 1 mg/mL in water. The stock solutions for the internal standards, 5AC-15N4, 2dC-13C15N2, and 5mC-d3 were prepared at concentrations of 0.5 mg/mL in methanol, 1 mg/mL in DMSO, and 1 mg/mL in water, respectively. All stock solutions were stored at −20°C. All working solutions, standards and quality control (QCs) were prepared fresh daily. Working solutions were made in water and spiked into blank digest matrix with no enzymes to make the calibration curve and QC samples. The blank digest matrix contains the following: 0.028 mM deferoxamine mesylate (DFAM), 32.27 mM ammonium acetate, 10.20 mM trizma base, 1.53 mM EDTA and 0.26 mM zinc chloride. The blank matrix was prepared in ~40mL batches and stored at 4°C. The calibration curve consisted of 8 calibrators at the following concentrations: 2, 5, 10, 50, 100, 150, 200 and 400 ng/mL for DAC; 50, 125, 250, 1,250, 2,500, 3,750, 5,000 and 10,000 ng/mL for 2dC; and 5, 12.5, 25, 125, 250, 375, 500 and 1,000 ng/mL for 5mC. The quality control samples were made at 4 different concentration levels for validation: a lower limit of quantitation (LLOQ), low, medium and high QC levels. The QCs were made at the following concentration levels: 2, 3, 80 and 320 ng/mL for DAC; 50, 75, 2,000 and 8,000 ng/mL for 2dC; and 5, 7.5, 200 and 800 ng/mL for 5mC. Blank and zero calibrators were also part of the standard curve and made from blank digest matrix.
2.5. DNA digest and sample preparation
Genomic DNA in 50 μL of HPLC grade water was digested into individual nucleotides by adding 4 Units of Nuclease P1 (Sigma Aldrich, St. Louis, MO) and 100 μL of digest buffer (digest buffer contains: 0.04 mM DFAM, 3.25 mM ammonium acetate pH 5.0 and 0.5 mM zinc chloride) and incubated at 65°C for 10 minutes. Following incubation, the nucleotides were dephosphoralyated by adding 20 μL of 100 mM trizma base, pH 8.5 and 4 Units of alkaline phosphatase (Roche Life Science, Indianapolis, IN) and incubated at 37°C for 1 hour. To stop the digest, 20 μL of 300 mM ammonium acetate, pH 5.0 and 6 μL of a solution containing 0.25 mM DFAM and 50 mM EDTA was added to the sample. To each labelled glass test tube, 100 μL of sample was added and mixed with 20 μL of internal standard (100 ng/mL of 5AC-15N4 and 2dC-13C15N2 and 1,000 ng/mL of 5mC-d3 in water). For blank samples, 20 μL of water was added in lieu of internal standard. Samples were vortex-mixed and transferred to an autosampler vial and 10 μL was injected onto the LC/MS system.
2.6. Method Validation
The validation of this method includes precision and accuracy, sensitivity and specificity, stock and digested sample stability, matrix effects and precision of the DNA digest. The FDA guidelines were followed for all acceptance criteria [26].
2.6.1. Selectivity and specificity
The sensitivity of the assay was determined by the signal to noise ratio of the LLOQ QC from the three precision and accuracy runs. The specificity was performed using blank digest matrix or untreated cell lines assessing for the presence of endogenous or exogenous interfering peaks. The interfering peak area needed to be less than 20% of the peak area of the analytes at the LLOQ in blank digest matrix.
2.6.2. Matrix Effects
Matrix effects were assessed to determine suppression/enhancement for DAC from the digest matrix and from the DNA input prior to digestion. To determine the matrix effects from the digest matrix, the low, medium and high QCs were evaluated in neat solution and in the digest matrix. The second matrix effect experiment was to assess the effects on DNA input on DAC. Genomic DNA that was not treated with DAC was analyzed using the following DNA input: 0 μg, 1 μg, 2.5 μg, 5 μg and 10 μg. All samples were spiked with DAC at 3 QC concentration levels (low, medium and high). Each sample for both experiments was prepared and processed in triplicate.
2.6.3. Precision and Accuracy
The precision and accuracy validation runs were performed on three consecutive days. This included a calibration curve processed in duplicate, and QC samples at four different concentrations (LLOQ, low, medium and high QC) in quintuplicate; a single blank and zero-level standard (blank with internal standard). Calibration curves for DAC, 2dC and 5mC were computed using the ratio of the peak area of analyte to the internal standard (5AC-15N4, 2dC-13C15N2 and 5mC-d3) by using a least-squares quadratic regression analysis with 1/x2 weight for all analytes. The parameters of each calibration curve were used to back-calculate concentrations and to obtain values for the QC samples and unknown samples by interpolation. Estimates of the inter-day precision, intra-day precision and accuracy were obtained.
2.6.4. Precision of the DNA Digest Procedure
To assess the precision of the digest procedures, three different lab technicians, digested genomic DNA from four different cell lines (DU145, PC3, HOP62 and PC9) using 5 μg of DNA input in triplicate. These four different cell lines were seeded in T75 flasks overnight at 25% confluence; and then treated with a single-dose of 250 nM DAC for 48 hours. Following cell harvest, genomic DNA samples were prepared using Blood and Cell Culture DNA Mini Kit (Qiagen, Germantown, MD). Samples were then analyzed to determine the concentration of DAC, 2dC and 5mC in each sample. Inter- and intra-individual variability was assessed.
2.6.5. Stability
The long-term stock stability was evaluated at −20°C and determined by injecting the test stock solution and compared to freshly prepared stock solution. Each stock was injected five times. Long term stability was evaluated at approximately 1 and 2 months. Short-term bench top stability of the stock solution was evaluated at room temperature over 6 hours. Each time point was analyzed in triplicate and the area counts were compared to time zero. Long term stability was determined by comparing fresh prepared low and high QCs in digest matrix and QCs made in digest matrix that was stored at −70°C. Each sample was prepared in triplicate. Long term stability was evaluated monthly for 3 months. The processed sample stability was determined by injecting freshly prepared low and high QCs and storing at 5°C (autosampler temperature) for ~5 day and reinjecting the sample. The concentration of the fresh samples was compared to the reinjected samples using the original fresh injected standard curve.
2.7. Assay application
The method was applied to both in vitro cell culture work and an in vivo mouse model after DAC treatment.
2.7.1. Cell culture and genomic DNA preparation
All cell lines were maintained in a humidified incubator at 5% CO2 and 37°C. DAC was freshly dissolved in DMSO at a concentration of 10mM and stored in −20°C for up to 1 month. Human prostate cancer DU145 cells were seeded in T75 flasks overnight at 25% confluence, and then treated with a single dose of DAC at various concentrations. Cells were allowed to grow for 48 hours prior to cell harvest and genomic DNA extraction using Blood and Cell Culture DNA Mini Kit (Qiagen, Germantown, MD) according to the manufacturer’s recommended protocols, except that DNA was eluted in pure molecular biology grade water.
2.7.2. In vivo DAC treatment
Eight to ten week old athymic nude mice (Harlan, Indianapolis, IN) were randomly assigned to four treatment groups of 3 mice each to receive vehicle control or one of three dose levels of DAC (1, 2, 5 mg/kg) DAC was dissolved in normal saline and administered by intraperitoneal injection. Doses were administered for two 1-week cycles consisting of every day administration for 5 days followed by two days of rest. Upon completion of two cycles of treatment, mice were euthanized with CO2 inhalation; colon tissue and whole bone marrow were collected and stored in liquid nitrogen for genomic DNA preparation as described previously [27]. Briefly, mouse tissue samples (up to 100 mg wet weight) were sliced into small pieces and grinded with liquid nitrogen followed by overnight incubation at 50°C in 1000 μL DNA extraction buffer (10 mM NaCl, 20 mM Tris-HCl, pH 8.0, 1 mM EDTA) supplemented with 20 μL of 10 mg/mL proteinase K (Life Technologies, Carlsbad, CA) and 50 μL 10% SDS. Digested samples were treated with RNase A (Life Technologies, Carlsbad, CA) at 100 μg/mL for 1 hour at 37°C. After triple extractions with 1 volume of phenol:chloroform:isoamyl alcohol (25:24:1), DNA was precipitated with 0.7 volume of prechilled isopropanol. Pelleted DNA samples were washed with 70% ethanol, air-dried and dissolved in pure molecular biology grade water.
3.0. Results and Discussion
3.1. Development of the LC/ESI MS/MS conditions for the simultaneous quantitation of DAC, 2dC, and 5mC
The LC/MS/MS method was developed and validated to determine DAC, 2dC and 5mC in digested genomic DNA. The biggest challenge that was overcome was the detection of DAC and 2dC. The two analytes are one dalton different in molecular weight for both the parent and the daughter ions creating crosstalk between the two mass spectrometer MRM channels. Since there was inadequate mass spectrometric resolution, chromatographic resolution was essential to adequately quantify DAC and 2dC. Many different column chemistries were assessed such as: multiple manufacture of C18’s, Agilent Zorbax phenyl-hexyl, Phenomenix Luna CN, Waters BEH HILIC, Waters’ HSS PFP, and Waters’ HSS T3. These columns could not retain the analytes, completely separate and resolve DAC and 2dC, or provide sufficient sensitivity for detection of DAC. The Thermo Hypercarb porous graphite column was the only analytical column that was able to routinely and robustly provide the chromatographic resolution necessary to separate DAC and 2dC.
The goal for sample preparation was to maximize the amount of DAC recovered from genomic DNA, due to the expected low levels in the sample. In order to remove any of the salts that are not mass spectrometer friendly, solid-phase extraction (SPE) was tried first. Water’s Oasis HLB was unable to retain all of the analytes. From previous experience, it was known that Water’s Oasis mixed cation exchange utilized for azacitidine in plasma resulted in very poor recovery [28]. Since clean-up methods resulted in inconsistent DNA yields, the samples were injected with no clean-up. The mass spectrometer was continually monitored for any salt build up to ensure the LLOQ was detectable. The mass spectrometer was routinely cleaned after approximately 500 injections.
3.2. Chromatographic separation and detection
The assay was validated with a Thermo Hypercarb porous graphite column with a 5 minute run time. The column was able to retain all 3 analytes and have baseline separation between DAC and 2dC. DAC, 2dC, and 5mC eluted at approximately 1.2, 1.6, and 2.5 minutes, respectively. The column eluent was diverted to waste for the first 0.4 minutes of each injection.
3.3. Method Validation
3.3.1. Sensitivity and Selectivity
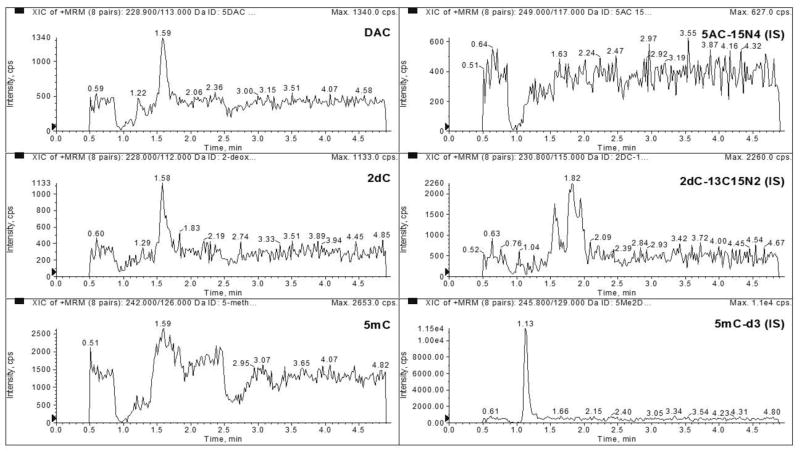
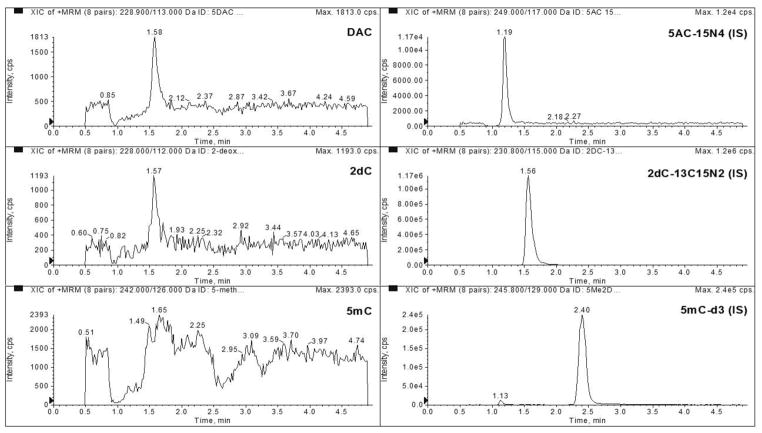
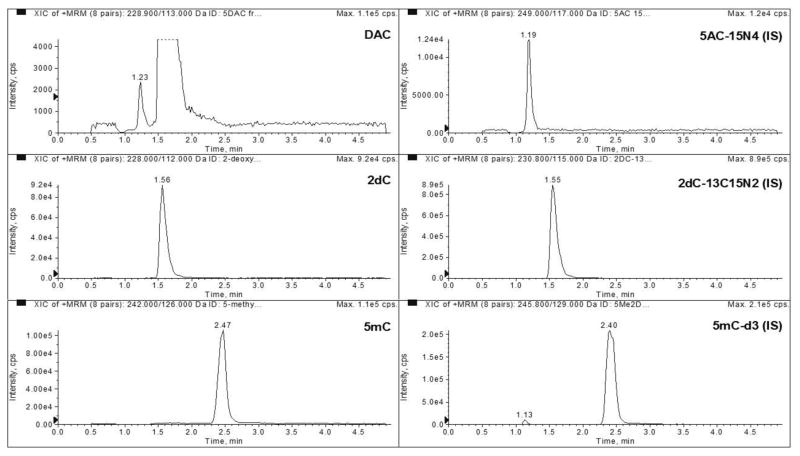
The LLOQ was determined to be 2 ng/mL for DAC, 50 ng/mL for 2dC, and 5 ng/mL for 5mC in 3 precision and accuracy runs (Table 1). No major interferences were seen at the same retention time as our analytes (Figures 1 and 2). The average signal to noise ratio of the LLOQ was 79, 1151, and 857 for DAC, 2dC, and 5mC, respectively (Figure 3).
Table 1.
Validation Characteristics for DAC, 2dC, and 5mC
| Analyte | Lower Limit of Quantitation | Low QC | Medium QC | High QC |
|---|---|---|---|---|
| DAC | 2 ng/mL | 3 ng/mL | 80 ng/mL | 320 ng/mL |
| Average ± St. Dev | 1.9 ± 0.1 | 2.9 ± 0.1 | 79.6 ± 2.1 | 315.4 ± 11.2 |
| Intra-Day Accuracy (%) | 90.6 – 95.6 | 94.3 – 96.5 | 98.0 – 100.8 | 97.2 – 99.6 |
| Intra-Day Precision (%) | 4.7 – 8.1 | 0.8 – 4.7 | 2.3 – 2.9 | 2.4 – 4.6 |
| Inter-Day Accuracy (%) | 93.0 | 95.4 | 99.4 | 98.6 |
| Inter-Day Precision (%) | 6.3 | 3.3 | 2.7 | 3.5 |
|
| ||||
| 2dC | 50 ng/mL | 75 ng/mL | 2000 ng/mL | 8000 ng/mL |
| Average ± St. Dev | 49.2 ± 1.0 | 73.6 ± 1.1 | 2044.5 ± 88.7 | 7853.2 ± 482.4 |
| Intra-Day Accuracy (%) | 97.4 – 99.8 | 97.8 – 98.7 | 100.3 – 103.4 | 96.1 – 100.6 |
| Intra-Day Precision (%) | 1.2 – 2.6 | 1.3 – 1.9 | 3.8 – 4.9 | 3.7 – 8.3 |
| Inter-Day Accuracy (%) | 98.5 | 98.2 | 102.2 | 98.2 |
| Inter-Day Precision (%) | 2.0 | 1.6 | 4.3 | 6.1 |
|
| ||||
| 5mC | 5 ng/mL | 7.5 ng/mL | 200 ng/mL | 800 ng/mL |
| Average ± St. Dev | 4.9 ± 0.1 | 7.4 ± 0.1 | 204.4 ± 3.9 | 801.7 ± 13.1 |
| Intra-Day Accuracy (%) | 96.2 – 99.8 | 96.2 – 99.9 | 101.8 – 102.9 | 98.8 – 101.9 |
| Intra-Day Precision (%) | 0.7 – 1.2 | 0.7 – 2.1 | 1.0 – 3.0 | 0.6 – 1.2 |
| Inter-Day Accuracy (%) | 98.1 | 98.0 | 102.2 | 100.2 |
| Inter-Day Precision (%) | 1.8 | 2.1 | 1.9 | 1.6 |
Figure 1.
Representative chromatograms of blank matrix monitoring DAC, 2dC, 5mC, 5AC-15N4 (I.S.), 2dC-13C15N2 (I.S.), and 5mC-d3 (I.S.).
Figure 2.
Representative chromatograms of blank matrix spiked with internal standard monitoring DAC, 2dC, 5mC, 5AC-15N4 (I.S.), 2dC-13C15N2 (I.S.), and 5mC-d3 (I.S.).
Figure 3.
Representative chromatograms of the lowest calibration standard spiked with 2 ng/mL DAC, 5 ng/mL 5mC and 50 ng/mL 2dC monitoring DAC, 2dC, 5mC, 5AC-15N4 (I.S.), 2dC-13C15N2 (I.S.), and 5mC-d3 (I.S.).
3.3.2. Matrix Effects
An experiment was set up to evaluate matrix effects from the DNA digest buffer in 3 QC concentration levels for DAC. The matrix effect for DAC was found to have 70% – 81% ion suppression from the blank matrix (Table 2). The next experiment was set up to evaluate the effects of DNA input on DAC. Using different amounts of DNA input, DAC was spiked into the samples with different amounts of DNA input at 3 QC levels (low, medium and high). Samples were analyzed and compared to neat solutions at the same concentration levels. The different amounts of DNA input show there is not a significant difference with DNA input and that the ion suppression is from the matrix (Table 2).
Table 2.
Matrix Effects on DAC from DNA Input
| DNA Input | Neat Solution | Blank Matrix (0 μg) | 1 μg | 2.5 μg | 5 μg | 10 μg | |
|---|---|---|---|---|---|---|---|
| QC Low | Average ± St. Dev | 85900 ± 3236 | 16433 ± 153 | 17433 ± 808 | 19533 ± 503 | 17000 ± 346 | 16533 ± 945 |
| % CV | 3.77 | 0.93 | 4.64 | 2.58 | 2.04 | 5.72 | |
| Matrix Suppression (%) | −80.9 | −79.7 | −77.3 | −80.2 | −80.8 | ||
| QC Mid | Average ± St. Dev | 1683333 ± 5774 | 444667 ± 10214 | 510667 ± 1155 | 497667 ± 3214 | 485333 ± 12503 | 455333 ± 19088 |
| % CV | 0.34 | 2.3 | 0.23 | 0.65 | 2.58 | 4.19 | |
| Matrix Suppression (%) | −73.6 | −69.7 | −70.4 | −71.2 | −73 | ||
| QC High | Average ± St. Dev | 6053333 ± 311823 | 1836667 ± 55076 | 1976667 ± 40415 | 1986667 ± 23094 | 1966667 ± 5774 | 1836667 ± 25166 |
| % CV | 5.15 | 3 | 2.04 | 1.16 | 0.29 | 1.37 | |
| Matrix Suppression (%) | −69.7 | −67.4 | −67.2 | −67.5 | −69.7 |
Calculations are based on area counts of DAC
3.3.3. Precision and Accuracy
The calibration curve for DAC, 2dC and 5mC was constructed from the peak area ratio of the analyte to the peak area of its internal standard (5AC-15N4, 2dC-13C15N2 and 5mC-d3) by using the least-squares quadratic regression analysis with 1/x2 weight. Validation experiments revealed excellent goodness of fit, with the r2 ≥ 0.99 with a calibration range of 2 to 500 ng/mL for DAC, 50 to 10,000 ng/mL for 2dC and 5 to 1,000 ng/mL for 5mC. The accuracy, intra-day precision and inter-day precision at the LLOQ was: 93.0%, 2.7% and 6.3% respectively for DAC (at LLOQ of 2ng/mL); 98.5%, 1.2% and 2.0% respectively for 2dC (at LLOQ of 50 ng/mL); and 98.1%, 1.8% and 1.8% respectively for 5mC (at LLOQ of 5 ng/mL). The accuracy, intra-day precision and the inter-day precision for the QCs ranged from 95.4% to 99.1%, 1.1% to 1.4%, and 2.7% to 3.5% respectively for DAC; 98.2% to 102.2%, 0.5% to 2.4% and 1.6% to 6.1% respectively for 2dC; and 98.0% to 102.2%, 0.6% to 3.2% and 1.6% to 2.1% respectively for 5mC (Table 1).
3.3.4. Precision of the DNA Digest Process
To determine the reproducibility of the DNA digest, three lab technicians digested four different cell lines independently in triplicate. The inter- and intra-individual precision was determined for all 3 analytes. After initial determinations, it was found that 2 samples for the DAC analyte only were considered outliers by Grubbs outlier test. After removing the outliers all %CV were well within 15% (Table 3).
Table 3.
DNA Digest Protocol/Lab Technician Reproducibility
| Cell Line | DAC | 2dC | 5mC | |||
|---|---|---|---|---|---|---|
| Average ± St. Dev | % CV | Average ± St. Dev | % CV | Average ± St. Dev | % CV | |
| DU145 | ||||||
| Tech 1 | 10.8 ± 0.30 | 2.8 | 1233.33 ± 30.55 | 2.5 | 27.93 ± 0.84 | 3.0 |
| Tech 2 | 9.38 ± 0.40 | 4.2 | 1180 ± 30.00 | 2.5 | 25.23 ± 0.15 | 0.6 |
| Tech 3 | 10.23 ± 0.06 | 0.6 | 1203.33 ± 37.86 | 3.2 | 25.5 ± 0.44 | 1.7 |
| Overall | 10.14 ± 0.67 | 6.6 | 1205.56 ± 36.78 | 3.1 | 26.22 ± 1.37 | 5.2 |
|
| ||||||
| PC3 | ||||||
| Tech 1 | 8.02 ± 0.72 | 9.0 | 1523.33 ± 32.15 | 2.1 | 24.43 ± 1.05 | 4.3 |
| Tech 2 | 8.00 ± 0.14 | 1.8 | 1520 ± 17.32 | 1.1 | 23.13 ± 0.15 | 0.7 |
| Tech 3 | 8.36 ± 0.10* | 1.2* | 1516.67 ± 75.06 | 5.0 | 22.8 ± 0.85 | 3.8 |
| Overall | 8.10 ± 0.43* | 5.3 | 1520 ± 41.83 | 2.8 | 23.46 ± 1.01 | 4.3 |
|
| ||||||
| HOP62 | ||||||
| Tech 1 | 17.73 ± 2.62 | 14.8 | 1426.67 ± 41.63 | 2.9 | 16.83 ± 0.21 | 1.2 |
| Tech 2 | 15.53 ± 0.4 | 2.6 | 1380 ± 65.57 | 4.8 | 15.6 ± 0.44 | 2.8 |
| Tech 3 | 17.4 ± 0.46 | 2.6 | 1386.67 ± 15.28 | 1.1 | 16.13 ± 0.29 | 1.8 |
| Overall | 16.89 ± 1.69 | 10.0 | 1397.78 ± 45.22 | 3.2 | 16.19 ± 0.61 | 3.7 |
|
| ||||||
| PC9 | ||||||
| Tech 1 | 8.70 ± 0.26* | 3.0* | 1106.67 ± 15.28 | 1.4 | 16.3 ± 0.2 | 1.2 |
| Tech 2 | 9.59 ± 0.48 | 5.0 | 1153.33 ± 5.77 | 0.5 | 16.43 ± 0.67 | 4.1 |
| Tech 3 | 9.56 ± 0.10 | 1.0 | 1110 ± 10 | 0.9 | 15.97 ± 0.15 | 1.0 |
| Overall | 9.36 ± 0.49* | 5.3* | 1123.33 ± 24.49 | 2.2 | 16.23 ± 0.41 | 2.5 |
Samples have one statistical outlier that was not used in the calculations
3.3.5. Stability
The master stock stability was tested at 27 and 77 days. It was determined that DAC, 2dC and 5mC are stable to 77 days in 100% water at −20°C. Bench top stability for the stock solution was tested up to 6 hours. All 3 analytes were found to be stable at room temperature for up to 6 hours in water. Stability was determined in the DNA digest matrix at the low and high QC level stored at −70°C at 35, 76 and 117 days. It was determined that DAC, 2dC and 5mC were stable up to 117 days (Table 4).
Table 4.
Stability of DAC, 2dC, and 5mC*
| Analyte | Low QC | High QC | Stock Solution in Water |
|---|---|---|---|
| DAC | 3 ng/mL | 320 ng/mL | 1 mg/mL |
| Digest Buffers Stability (−70 C) (117 Days) (%) | 89.4 | 101.2 | |
| Reinjection Stability (5 Days) (5 C) (%) | 88.6 | 92.5 | |
| Stock Stability (−20 C) (77 Days) (%) | 103.9 | ||
| Benchtop Stability (6 hours) | 107.1 | ||
|
| |||
| 2dC | 75 ng/mL | 8000 ng/mL | 1 mg/mL |
| Digest Buffers Stability (−70 C) (117 Days) (%) | 108.8 | 112.3 | |
| Reinjection Stability (5 Days) (5 C) (%) | 108.0 | 106.0 | |
| Stock Stability (77 Days) (%) | 105.0 | ||
| Benchtop Stability (6 hours) | 101.9 | ||
|
| |||
| 5mC | 75 ng/mL | 800 ng/mL | 1 mg/mL |
| Digest Buffers Stability (−70 C) (117 Days) (%) | 94.2 | 97.7 | |
| Reinjection Stability (5 Days) (5 C) (%) | 100.8 | 100.4 | |
| Stock Stability (77 Days) (%) | 100.8 | ||
| Benchtop Stability (6 hours) | 102.3 | ||
Stability experiments are continuing for the digest buffer and stock solution.
3.4. Application of the method
When DU145 human prostate cancer cells were treated with increasing doses of DAC, the amount of DAC per thousand 2dC showed a strong linear correlation with the initial treatment dose (Figure 4).
Figure 4.
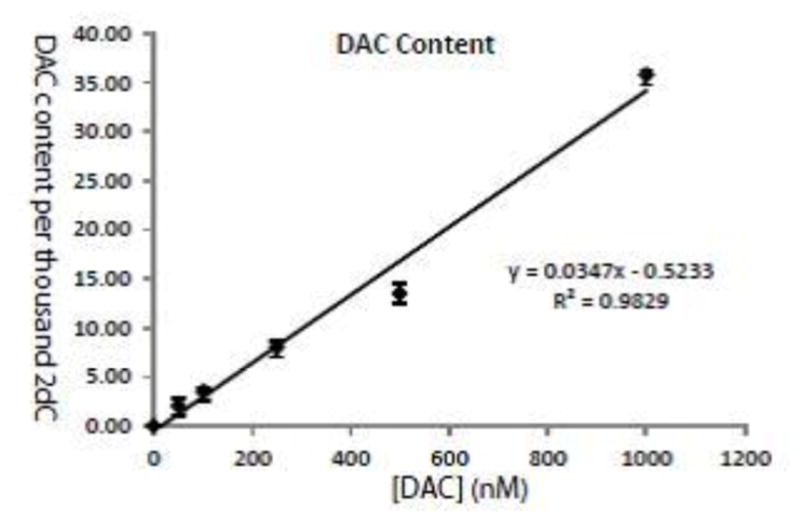
Incorporation of DAC into genomic DNA follows a linear correlation with dose of DAC exposure. DU145 human prostate cancer cells were treated with increasing doses of DAC for 48 hours. The amount of DAC per thousand 2dC in the genomic DNA showed a strong linear correlation with the initial treatment dose. Shown are the mean ± standard error of the mean (SEM) of three replicate measurements.
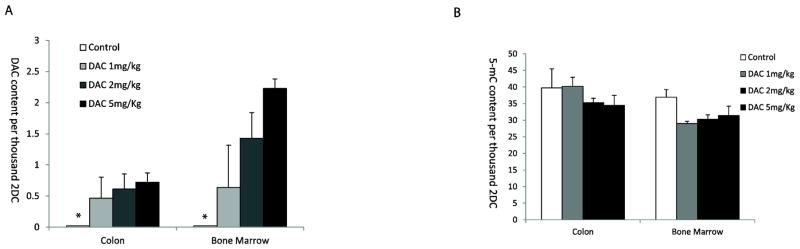
We next assessed the degree of DAC incorporation in tissue samples from mice treated with DAC. Mice were treated with a dose response of DAC in normal saline at 1, 2, or 5 mg/kg by intraperitoneal injection for a total of 10 administrations. Our results for DAC and 5mC content per thousand 2-DC show a dose responsive increase in incorporation in colon and bone marrow cells with different efficiencies (Figure 5). Interestingly, 5mC content was inversely correlated with DAC incorporation in colon cells, but not in bone marrow cells, suggesting that 5mC measurement alone is a poor surrogate for understanding DAC incorporation due to complex cell and tissue dynamics.
Figure 5.
DAC and 5mC content in genomic DNA of colon and bone marrow cells from mice treated with a dose response of DAC. Mice were treated with a dose response of DAC in normal saline at various doses for two 1-week cycles consisting of every day administration for 5 days followed by two days of rest. Results for DAC and 5mC content per thousand 2dC are reported. The * indicates that there is no detectable DAC. Shown are the mean values from 3 mice in each drug treated group and the mean value of four replicate measurements for Control. Error bars represent standard deviations.
4. Conclusion
An analytical assay method was developed and validated to quantify DAC, 2dC and 5mC levels in digested DNA. The analytical assay and the DNA digest was shown to be robust and reproducible. The method has been applied to show DAC incorporation into genomic DNA in cell lines and in tissue samples collected from mice treated with DAC. Future experiments will be aimed at interrogating nucleoside analog DNMT inhibitor mechanisms of action and exposure-response relationships when using these drugs alone or in combination modalities for both current and emerging indications. These measurements may also allow a better understanding of mechanisms of resistance and sensitivity to these agents.
Highlights.
First quantitative method for incorporation of 5-aza-2′-deoxycytidine in genomic DNA by UHPLC MS/MS.
Method also simultaneously assesses global demethylation by measuring genomic content of 5-methyl-2′-deoxycytidine and 2′-deoxycytidine.
This method can be utilized to probe exposure-response relationships of nucleoside analog DNA methyltransferase inhibitors.
Acknowledgments
We would like to thank Linping Xu for her quality assurance review of the data. The project described was supported by the Flight Attendants Medical Research Institution (FAMRI) Center for Excellence at the Johns Hopkins University School of Medicine (M.A.R.), the Prostate Cancer Foundation Challenge Grant (to S.Y. and W.G.N.), NIH/NCI grant P50CA058236, and the Commonwealth Foundation, and the Analytical Pharmacology Core of the Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins [NIH grants P30CA006973 and UL1TR001079, and the Shared Instrument Grant (S10RR026824)]. The project described was also supported by grant number UL1 TR001079 from the National Center for Advancing Translational Sciences (NCATS), a component of the National Institutes of Health (NIH), and the NIH Roadmap for Medical Research. Its contents are solely the responsibility of the authors and do not necessarily represent the official view of the NCATS or NIH.
Footnotes
Disclosure: Dr. Rudek receives research funding from Celgene Corporation via Johns Hopkins University.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. The New England journal of medicine. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 2.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gilbert J, Gore SD, Herman JG, Carducci MA. The clinical application of targeting cancer through histone acetylation and hypomethylation. Clin Cancer Res. 2004;10:4589–4596. doi: 10.1158/1078-0432.CCR-03-0297. [DOI] [PubMed] [Google Scholar]
- 4.Gardner KE, Allis CD, Strahl BD. Operating on chromatin, a colorful language where context matters. Journal of molecular biology. 2011;409:36–46. doi: 10.1016/j.jmb.2011.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Derissen EJ, Beijnen JH, Schellens JH. Concise drug review: azacitidine and decitabine. The oncologist. 2013;18:619–624. doi: 10.1634/theoncologist.2012-0465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Issa JP, Roboz G, Rizzieri D, Jabbour E, Stock W, O’Connell C, Yee K, Tibes R, Griffiths EA, Walsh K, Daver N, Chung W, Naim S, Taverna P, Oganesian A, Hao Y, Lowder JN, Azab M, Kantarjian H. Safety and tolerability of guadecitabine (SGI-110) in patients with myelodysplastic syndrome and acute myeloid leukaemia: a multicentre, randomised, dose-escalation phase 1 study. The Lancet Oncology. 2015;16:1099–1110. doi: 10.1016/S1470-2045(15)00038-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stresemann C, Lyko F. Modes of action of the DNA methyltransferase inhibitors azacytidine and decitabine. International journal of cancer Journal international du cancer. 2008;123:8–13. doi: 10.1002/ijc.23607. [DOI] [PubMed] [Google Scholar]
- 8.Oki Y, Aoki E, Issa JP. Decitabine--bedside to bench. Critical reviews in oncology/hematology. 2007;61:140–152. doi: 10.1016/j.critrevonc.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 9.Momparler RL, Derse D. Kinetics of phosphorylation of 5-aza-2′-deoxyycytidine by deoxycytidine kinase. Biochem Pharmacol. 1979;28:1443–1444. doi: 10.1016/0006-2952(79)90454-4. [DOI] [PubMed] [Google Scholar]
- 10.Hubeek I, Stam RW, Peters GJ, Broekhuizen R, Meijerink JP, van Wering ER, Gibson BE, Creutzig U, Zwaan CM, Cloos J, Kuik DJ, Pieters R, Kaspers GJ. The human equilibrative nucleoside transporter 1 mediates in vitro cytarabine sensitivity in childhood acute myeloid leukaemia. Br J Cancer. 2005;93:1388–1394. doi: 10.1038/sj.bjc.6602881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li LH, Olin EJ, Buskirk HH, Reineke LM. Cytotoxicity and mode of action of 5-azacytidine on L1210 leukemia. Cancer research. 1970;30:2760–2769. [PubMed] [Google Scholar]
- 12.Christman JK. 5-Azacytidine and 5-aza-2′-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene. 2002;21:5483–5495. doi: 10.1038/sj.onc.1205699. [DOI] [PubMed] [Google Scholar]
- 13.Ghoshal K, Datta J, Majumder S, Bai S, Kutay H, Motiwala T, Jacob ST. 5-Aza-deoxycytidine induces selective degradation of DNA methyltransferase 1 by a proteasomal pathway that requires the KEN box, bromo-adjacent homology domain, and nuclear localization signal. Molecular and cellular biology. 2005;25:4727–4741. doi: 10.1128/MCB.25.11.4727-4741.2005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Baylin SB, Herman JG, Graff JR, Vertino PM, Issa JP. Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv Cancer Res. 1998;72:141–196. [PubMed] [Google Scholar]
- 15.Baylin SB. Mechanisms underlying epigenetically mediated gene silencing in cancer. Semin Cancer Biol. 2002;12:331–337. doi: 10.1016/s1044-579x(02)00053-6. [DOI] [PubMed] [Google Scholar]
- 16.Kissinger LD, Stemm NL. Determination of the antileukemia agents cytarabine and azacitidine and their respective degradation products by high-performance liquid chromatography. J Chromatogr. 1986;353:309–318. doi: 10.1016/s0021-9673(01)87101-6. [DOI] [PubMed] [Google Scholar]
- 17.Notari RE, DeYoung JL. Kinetics and mechanisms of degradation of the antileukemic agent 5-azacytidine in aqueous solutions. J Pharm Sci. 1975;64:1148–1157. doi: 10.1002/jps.2600640704. [DOI] [PubMed] [Google Scholar]
- 18.Rogstad DK, Herring JL, Theruvathu JA, Burdzy A, Perry CC, Neidigh JW, Sowers LC. Chemical decomposition of 5-aza-2′-deoxycytidine (Decitabine): kinetic analyses and identification of products by NMR, HPLC, and mass spectrometry. Chemical research in toxicology. 2009;22:1194–1204. doi: 10.1021/tx900131u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Z, Marcucci G, Byrd JC, Grever M, Xiao J, Chan KK. Characterization of decomposition products and preclinical and low dose clinical pharmacokinetics of decitabine (5-aza-2′-deoxycytidine) by a new liquid chromatography/tandem mass spectrometry quantification method. Rapid communications in mass spectrometry : RCM. 2006;20:1117–1126. doi: 10.1002/rcm.2423. [DOI] [PubMed] [Google Scholar]
- 20.Rivard GE, Momparler RL, Demers J, Benoit P, Raymond R, Lin K, Momparler LF. Phase I study on 5-aza-2′-deoxycytidine in children with acute leukemia. Leuk Res. 1981;5:453–462. doi: 10.1016/0145-2126(81)90116-8. [DOI] [PubMed] [Google Scholar]
- 21.Derissen EJ, Hillebrand MJ, Rosing H, Otten HM, Laille E, Schellens JH, Beijnen JH. Quantitative determination of azacitidine triphosphate in peripheral blood mononuclear cells using liquid chromatography coupled with high-resolution mass spectrometry. Journal of pharmaceutical and biomedical analysis. 2014;90:7–14. doi: 10.1016/j.jpba.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 22.Jansen RS, Rosing H, Wijermans PW, Keizer RJ, Schellens JH, Beijnen JH. Decitabine triphosphate levels in peripheral blood mononuclear cells from patients receiving prolonged low-dose decitabine administration: a pilot study. Cancer Chemother Pharmacol. 2012;69:1457–1466. doi: 10.1007/s00280-012-1850-x. [DOI] [PubMed] [Google Scholar]
- 23.Wang H, Chen P, Wang J, Santhanam R, Aimiuwu J, Saradhi UV, Liu Z, Schwind S, Mims A, Byrd JC, Grever MR, Villalona-Calero MA, Klisovic R, Walker A, Garzon R, Blum W, Chan KK, Marcucci G. In vivo quantification of active decitabine-triphosphate metabolite: a novel pharmacoanalytical endpoint for optimization of hypomethylating therapy in acute myeloid leukemia. AAPS J. 2013;15:242–249. doi: 10.1208/s12248-012-9427-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aimiuwu J, Wang H, Chen P, Xie Z, Wang J, Liu S, Klisovic R, Mims A, Blum W, Marcucci G, Chan KK. RNA-dependent inhibition of ribonucleotide reductase is a major pathway for 5-azacytidine activity in acute myeloid leukemia. Blood. 2012;119:5229–5238. doi: 10.1182/blood-2011-11-382226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oz S, Raddatz G, Rius M, Blagitko-Dorfs N, Lubbert M, Maercker C, Lyko F. Quantitative determination of decitabine incorporation into DNA and its effect on mutation rates in human cancer cells. Nucleic Acids Res. 2014;42:e152. doi: 10.1093/nar/gku775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.U.S. Department of Health and Human Services, Food and Drug Administration, Center for Drug Evaluation and Research (CDER), and Center for Veterinary Medicine (CVM) Guidance for industry bioanalytical method validation. May, 2001. [Google Scholar]
- 27.Haffner MC, Aryee MJ, Toubaji A, Esopi DM, Albadine R, Gurel B, Isaacs WB, Bova GS, Liu W, Xu J, Meeker AK, Netto G, De Marzo AM, Nelson WG, Yegnasubramanian S. Androgen-induced TOP2B-mediated double-strand breaks and prostate cancer gene rearrangements. Nature genetics. 2010;42:668–675. doi: 10.1038/ng.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anders NM, Wanjiku TM, He P, Azad NS, Rudek MA. A robust and rapid liquid chromatography tandem mass spectrometric method for the quantitative analysis of 5-azacytidine. Biomedical chromatography : BMC. 2015 doi: 10.1002/bmc.3562. [DOI] [PMC free article] [PubMed] [Google Scholar]