Abstract
Low-grade neuroepithelial tumors (LGNTs) are diverse CNS tumors presenting in children and young adults, often with a history of epilepsy. While the genetic profiles of common LGNTs, such as the pilocytic astrocytoma and ‘adult-type’ diffuse gliomas, are largely established, those of uncommon LGNTs remain to be defined.
In this study, we have used massively parallel sequencing and various targeted molecular genetic approaches to study alterations in 91 LGNTs, mostly from children but including young adult patients. These tumors comprise dysembryoplastic neuroepithelial tumors (DNETs; n=22), diffuse oligodendroglial tumors (d-OTs; n=20), diffuse astrocytomas (DAs; n=17), angiocentric gliomas (n=15), and gangliogliomas (n=17).
Most LGNTs (84%) analyzed by whole-genome sequencing (WGS) were characterized by a single driver genetic alteration. Alterations of FGFR1 occurred frequently in LGNTs composed of oligodendrocyte-like cells, being present in 82% of DNETs and 40% of d-OTs. In contrast, a MYB-QKI fusion characterized almost all angiocentric gliomas (87%), and MYB fusion genes were the most common genetic alteration in DAs (41%). A BRAF:p.V600E mutation was present in 35% of gangliogliomas and 18% of DAs. Pathogenic alterations in FGFR1/2/3, BRAF, or MYB/MYBL1 occurred in 78% of the series. Adult-type d-OTs with an IDH1/2 mutation occurred in four adolescents, the youngest aged 15 years at biopsy. Despite a detailed analysis, novel genetic alterations were limited to two fusion genes, EWSR1-PATZ1 and SLMAP-NTRK2, both in gangliogliomas.
Alterations in BRAF, FGFR1, or MYB account for most pathogenic alterations in LGNTs, including pilocytic astrocytomas, and alignment of these genetic alterations and cytologic features across LGNTs has diagnostic implications. Additionally, therapeutic options based upon targeting the effects of these alterations are already in clinical trials.
Keywords: Glioma, glioneuronal, RNA-seq, FGFR1, MYB, BRAF
Introduction
Low-grade gliomas (LGGs) and low-grade glioneuronal tumors, together designated ‘low-grade neuroepithelial tumors’ (LGNTs), represent an important group of central nervous system (CNS) neoplasms in children and young adults [34]. LGNTs include the pilocytic astrocytoma, the commonest pediatric brain tumor, but encompass a large variety of uncommon CNS tumors, some of which – ganglioglioma, dysembryoplastic neuroepithelial tumor (DNET), angiocentric glioma – are associated with a history of chronic epilepsy and are referred to as ‘long-term epilepsy-associated tumors’ (LEATs) [7,59].
Overall survival among the broad category of LGNTs is good, largely because many pilocytic astrocytomas are circumscribed non-infiltrative tumors and present at anatomic sites that make them amenable to surgical resection, such as the cerebellar hemisphere [4,17]. However, a significant proportion of less common LGNTs diffusely infiltrates the CNS or presents at a site where surgical resection is not feasible without significantly damaging the patient. Such tumors cause premature death, following a prolonged period of increasing morbidity that exacts a significant economic and emotional toll on families and health care providers [57].
Over the past decade, our understanding of genetic alterations in pediatric LGNTs has increased significantly, beyond recognition of the association between neurofibromatosis type 1 (NF1) and optic pathway glioma [2,24,45]. The NF1 gene product, neurofibromin 1, is a negative regulator of the MAPK pathway, and recurrent genetic alterations in other members of this pathway have been discovered in multiple types of LGNT [25,67]. However, genetic alterations in various uncommon types of LGNT, particularly some LEATs, have not been studied in a systematic manner.
We report a study of genetic alterations in a series of 91 uncommon LGNTs from children and young adults, which consists of diffuse astrocytomas (DAs), dysembryoplastic neuroepithelial tumors (DNETs), oligodendrogliomas, oligoastrocytomas, angiocentric gliomas (AGs), and gangliogliomas. Our analysis demonstrates a limited repertoire of recurrent genetic alterations that align strongly with morphology.
Methods
Sample series and processing
The study cohort comprised 91 primary cerebral LGNTs (Supp. Table S1), presenting in 48 (53%) male and 43 (47%) female patients, with a median age at surgery of 10 years (range 1 – 41 years). Three broad categories of LGNT were represented: (i) tumors with a dominant oligodendroglial phenotype – dysembryoplastic neuroepithelial tumors (DNETs, n=22) and the diffuse oligodendroglial tumors (d-OTs, n=20), oligodendrogliomas and oligoastrocytomas, (ii) tumors with a dominant astrocytic phenotype – angiocentric gliomas (AGs, n=15) and diffuse astrocytomas (DAs, n=17), and (iii) gangliogliomas (n=17). The histopathological diagnosis was reviewed in each case (DWE). Fifty-nine samples were snap-frozen at the time of first resection. Thirty-two samples were available only as archived formalin-fixed paraffin wax-embedded (FFPE) material. Tumor and germline blood samples were processed according to starting material and as previously described (see Supplemental Methods & Supp. Fig. S1) [22,67].
Whole genome, whole exome and transcriptome sequencing and data analysis
Whole genome, whole exome, and transcriptome sequencing (WGS, WES, RNA-seq) were undertaken using derivatives from frozen samples as previously described [65,66]. WGS and WES mapping, coverage and quality assessment, single nucleotide variation (SNV) and insertion/deletion (indel) detection, tier annotation for sequence mutations, prediction of the deleterious effects of missense mutations, structural variant (SV) detection, and identification of loss of heterozygosity (LOH) have been described previously [65,66]. The reference human genome assembly NCBI Build 37 was used to map all samples. Mapping statistics and coverage for each sample on different sequencing platforms are summarized in the supplemental methods/analysis and Supp. Table S2.
Mutation hotspot analysis by Sanger sequencing
Mutational hotspots in BRAF, KRAS, IDH1, IDH2 and H3F3A were sequenced in gDNA from the entire series of tumors using previously published primers [22,58,62]. Point mutations within FGFR1 and FGFR3 were sequenced using primer pairs listed in Supp. Table S3. Amplicons were generated using GoTaq™ DNA Polymerase (Promega, Madison, WI), and PCR products were sequenced using BigDye version 3.1 chemistry and a 3730XL DNA analyzer (Applied Biosystems, Foster City, CA). Output data were analyzed using CLC Main Workbench sequence analysis software version 6.0.2 (CLC bio, Cambridge, MA).
Genome-wide copy number analysis and clustering analysis by DNA methylation profiling
Genome-wide DNA methylation profiles were generated for 89 tumor samples, using the Illumina Infinium HumanMethylation450 BeadChip array. Frozen and FFPE-derived genomic DNA (500ng) was bisulfite-treated using the Zymo EZ DNA Methylation Kit according to the following thermocycling conditions (16 cycles: 95°C for 30sec, 50°C for 1hr). Following bisulfite treatment, DNA samples were desulphonated, column purified, then eluted using 12μl of elution buffer (Zymo Research). FFPE-derived DNA samples were then processed using the Illumina Infinium HD FFPE Restore kit according to the manufacturer’s protocol. All bisulfite-converted DNA (frozen) and bisulfite-converted and restored DNA (FFPE) samples were processed using the Illumina Infinium Methylation Assay, including hybridization to HumanMethylation450 BeadChips, single base extension assay, staining and scanning using the Illumina HiScan system, according to manufacturer’s recommendations. Data were preprocessed using the SWAN normalization method in R package minfi to normalize Type I and Type II probe bias. The combined intensity values of the methylation probes were then analyzed using R package conumee to generate genome-wide DNA copy number profiles. For unsupervised hierarchical clustering analysis, the following additional filters were applied: (i) removal of probes targeting X and Y chromosomes, (ii) removal of probes with CpG-sites overlapping a single nucleotide polymorphism (dbSNP138), and (iii) removal of probes with detection p-value>0.05 in more than 5% of the samples. Beta values were calculated, and the top 5% most variable methylated CpG probes as measured by median absolute deviation were selected. Tumor samples were clustered by applying a “ward” method on euclidean distance.
Interphase fluorescence in situ hybridizaton (iFISH)
Dual-color iFISH was performed on 4μm FFPE tissue sections, as previously described [20]. BAC clones (BACPAC Resources, Oakland, CA) used to develop both break-apart and copy number iFISH probe sets are detailed in Supp. Table S4.
URLs
NCBI RefSeq, http://www.ncbi.nlm.nih.gov/refseq/; NCBI reference human genome, http://www.ncbi.nlm.nih.gov/genome/51; National Heart, Lung, and Blood Institute GO Exome Sequencing Project (ESP), http://evs.gs.washington.edu/EVS/.
Accession codes
Our entire genomic data set, including whole-genome sequencing, whole-exome sequencing and RNA-seq has been deposited at the European Bioinformatics Institute (EBI) under accession number EGAS00001000255.
Results
NGS overview
A total of 1,047 somatic sequence mutations, including only 28 non-silent coding mutations (Supp. Table S5), and 47 SVs (Supp. Table S6) were identified by WGS of 19 LGNTs (Supp. Fig. 2). In addition, 72 somatic mutations (38 non-silent) were found by WES (Supp. Table S7). Transcriptome sequencing captured 77 SVs (Supp. Table S8). The background mutation rate derived from SNVs in non-repetitive regions in tumors analyzed by WGS ranged from 8.05×10−10 to 2.66×10−7 mutations per base, with an average mutation rate of 4.39×10−8 mutations per base (Supp. Table S9). Overall, LGNTs showed a median of 1 non-silent somatic mutation per case (range 0–7); and the cohort showed a median of 1 SV per case (range 0–14). Despite the paucity of genetic alterations overall, we found multiple recurrent abnormalities in different LGNT subtypes.
Eighty-nine percent (55/62) NGS cases had at least 1 recurrent abnormality involving FGFR1/FGFR3, MYB/MYBL1, BRAF, PDGFRA, NF1, and NTRK2 (Supp. Table S10). Sixty one percent (38/62) of NGS cases contained an SV, consistent with our previous observation that SVs are frequent driver events in LGNTs [67]. Four cases were found to contain multiple driver abnormalities: LGNT57 contained mutations within FGFR1 and BRAF, and LGNT58 contained mutations within FGFR1, KRAS and NF1, and LGNT69 and LGNT63 contained both a MYB-QKI gene fusion and a BRAF:p.V600E mutation (Supp. Table S10).
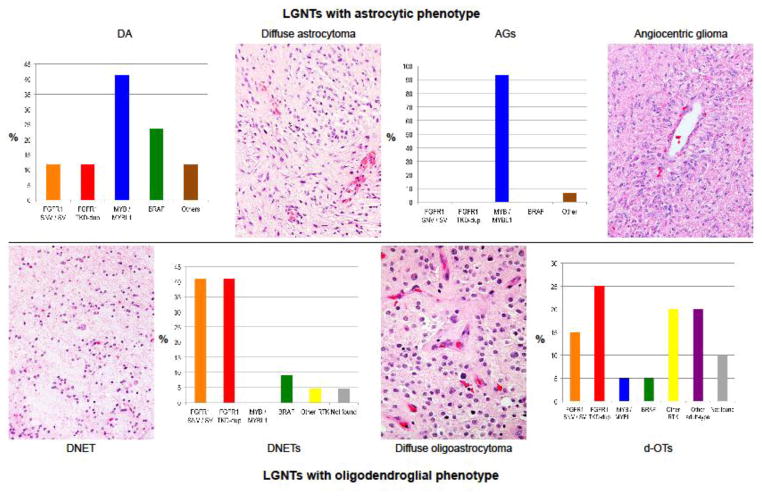
Different sequencing strategies revealed recurrent genetic alterations in all types of LGNT. These alterations were dominated by SNVs, SVs, or tyrosine kinase domain (TKD) duplications in FGFR1 (33% of tumors), SVs involving MYB (22%), and BRAF fusions or mutations (18%), mainly BRAF:p.V600E. Each of these principal genetic alterations was associated with a morphological phenotype. FGFR1 abnormalities were greatly enriched in LGNTs with an oligodendroglial phenotype – dysembryoplastic neuroepithelial tumors (DNETs) and the diffuse oligodendroglial tumors (d-OTs), while practically all MYB abnormalities were detected in angiocentric gliomas and diffuse astrocytomas, and BRAF alterations were most frequent among gangliogliomas.
FGFR1 alterations in cerebral LGNTs with oligodendroglial phenotype
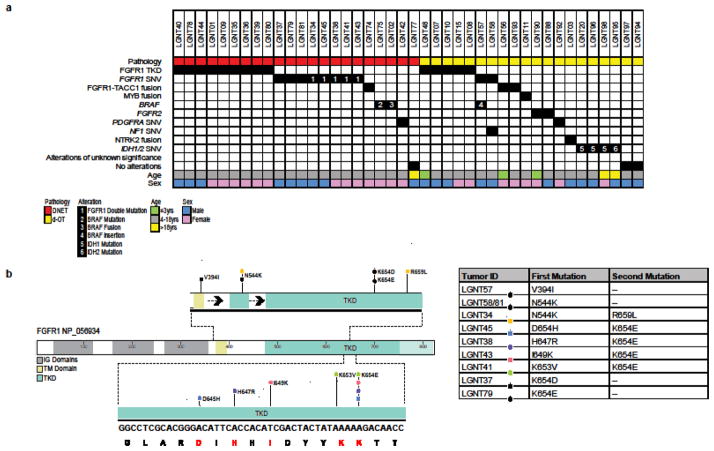
Genetic alterations were detected in 39/42 (93%) of LGNTs with an oligodendroglial phenotype – DNETs and d-OTs (Fig. 1). FGFR1 alterations were detected in 26/42 (62%) and comprised SNVs, TKD duplications, and fusions. Pathogenic FGFR1 SNVs, which were single or ‘doublet’ missense mutations in the TKD, occurred only in DNETs or d-OTs across the LGNT series. Doublet missense mutations occurred on the same allele within 1–9 codons of each other (Fig. 1, Supp. Fig. 3). In LGNT34, two FGFR1 mutations (N544K & R659L) were identified on different alleles. Most (14/17; 82%) FGFR1 TKD duplications were detected among DNETs or d-OTs; only two were in diffuse astrocytomas and one occurred in a ganglioglioma (Supp. Fig. 4). Three of five FGFR1 fusions, all FGFR1-TACC1, were detected in DNETs or d-OTs; two others, one FGFR1-TACC1 and one FGFR3-TACC3, occurred in diffuse astrocytomas.
Figure 1.
Genetic alterations among 42 LGNTs with an oligodendroglial phenotype (a). FGFR1 SNVs represented on the protein structure and tabulated, including four tumors with ‘doublet’ missense mutations on the same allele – lower part of diagram – and one tumor with two mutations, each on a different allele – upper part of diagram, with single mutations (b).
Other pathogenic alterations occurred at low frequency in DNETs and d-OTs: two CNAs in FGFR2 (5%), two PDGFRA mutations (5%), one BRAF:p.V600E (2%), one BRAF:p.G503>GVLR (2%), one BRAF-RNF130 (2%), one NAV1-NTRK2 (2%), and one MYB-MAML2 (2%). Four dOTs harbored an IDH1/2 mutation and other genetic alterations characteristic of ‘adult-type’ diffuse grade II oligodendrogliomas. These tumors occurred in teenagers and young adults aged 15–22 years at diagnosis. No alteration was found in three LGNTs with an oligodendroglial phenotype.
All FGFR1 SNVs occurred in DNETs, with one exception, an oligoastrocytoma that had an FGFR1:p.N544K mutation and concurrent NF1:p.T653fs and KRAS:p.G12D mutations. Tumors with an FGFR1 SNV presented at an older mean age, 14.2 years, than those with an FGFR1 TKD duplication, 7.4 years (p=0.0002), though this partly reflects a higher mean age at presentation among patients with DNET, 12.4 years, than those with d-OTs, 9.2 (p=0.036). No association between any genetic alteration and tumor location was evident.
MYB alterations in cerebral LGNTs with astrocytic phenotype
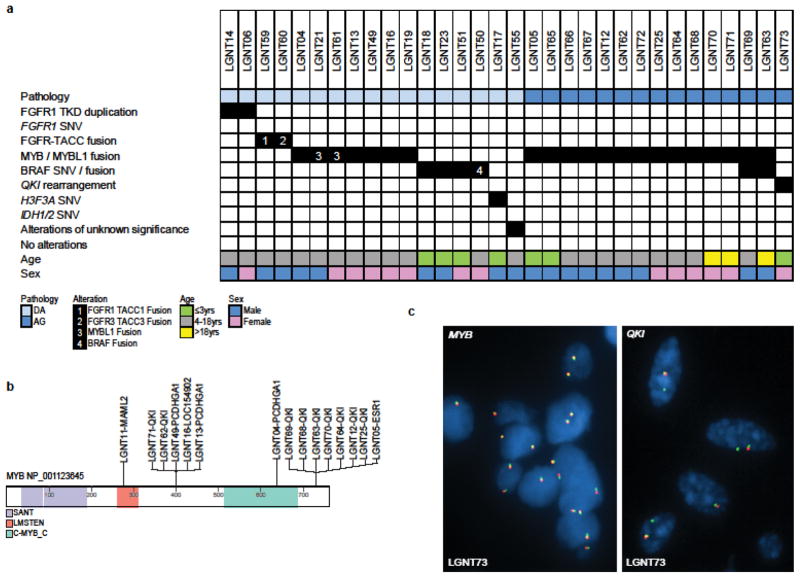
Pathogenic alterations were detected in 31/32 (97%) of LGNTs with an astrocytic morphology – diffuse astrocytomas and angiocentric gliomas (Fig. 2), and the dominant lesions were MYB fusions (21/32; 66%). All angiocentric gliomas harbored a MYB-QKI fusion (13/15; 87%), MYB-ESR1 fusion (1/15), or QKI rearrangement (1/15) (Supp. Figs. 5, 6). The last of these had no apparent involvement of MYB or MYBL1, showing only split signals with iFISH break-apart probes across QKI. We did not have sufficient derivatives from this tumor for FFPE RNA-seq, so were unable to determine whether a cryptic MYB-QKI aberration was present in this case, or whether QKI was fused to another gene partner. Most MYB-QKI fusions (78%) were between MYB exon 15 and QKI exon 5. Fusions were also identified between MYB exon 9 and QKI exon 5 (Supp. Fig. S5). Two tumors with a MYB-QKI fusion also harbored a BRAF:p.V600E mutation (Supp. Fig. S7).
Figure 2.
Genetic alterations among 32 LGNTs with an astrocytic phenotype (a). Location of MYB fusion genes represented on protein structure (b). LGNT73 - iFISH profiles using MYB and QKI ‘break-apart’ probe sets and demonstrating rearrangement/deletion of QKI, but not MYB.
Diffuse astrocytomas were more heterogeneous. MYB or MYBL1 rearrangements were detected in 7/17 tumors (41%), while BRAF or FGFR1/3 alterations were present in 4 tumors each (Fig. 2, Supp. Fig. S7). One tumor had an H3F3A:p.K27M mutation, which we have reported before [67]. Another tumor harbored only an ELAC2 duplication of uncertain significance, although variants in this gene have been implicated in susceptibility to hereditary prostatic cancer [1].
Only one other tumor across the series, an oligodendroglioma with a MYB-MAML2 fusion, harbored a MYB or MYBL1 alteration. Among diffuse astrocytomas, there was no obvious difference in radiological characteristics, including anatomic site, or outcome between those tumors with MYB, those with MYBL1 alterations, and those with other genetic alterations (data not shown).
BRAF alterations in gangliogliomas
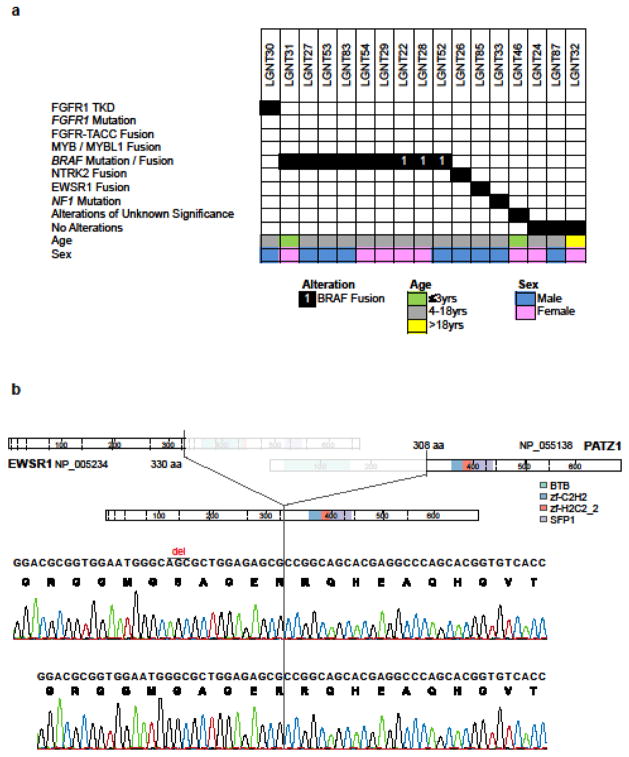
WGS, WES, or RNA-seq was undertaken on 15/17 (88%) gangliogliomas, but in two of these fifteen (13%) no significant alteration was detected (Supp. Fig. S7). Otherwise, abnormalities in gangliogliomas most frequently involved BRAF (9/17; 53%), specifically a BRAF:p.V600E mutation in 6/17 tumors (35%), and a MACF1-BRAF, AGK-BRAF, or GNAI1-BRAF fusion in each of three others (Fig. 3). All of these alterations are predicted to activate the downstream effects of BRAF in a constitutive manner. Individual gangliogliomas contained single miscellaneous pathogenic alterations: NF1 SNV, FGFR1 TKD duplication, and an EWSR1-PATZ1 or SLMAP-NTRK2 fusion (Fig. 3). No particular clinicopathological associations were found for these diverse genetic abnormalities.
Figure 3.
Genetic alterations among 17 gangliogliomas (a). WGS, WES, or RNA-seq data were available for 15/17 tumors; LGNT83 and LGNT87 were analyzed by targeted approaches but not by NGS. Of nine gangliogliomas with BRAF alterations, six had a BRAF:p.V600E mutation. EWSR1-PATZ1 fusion (b). Two clones, differing only with respect to a 3-bp deletion (‘del’ over sequence trace), were independently validated. In the rearrangement, PATZ1 loses its transcriptional repressor domain (BTB/POZ domain) at the N-terminus.
LGNT methylation profiling
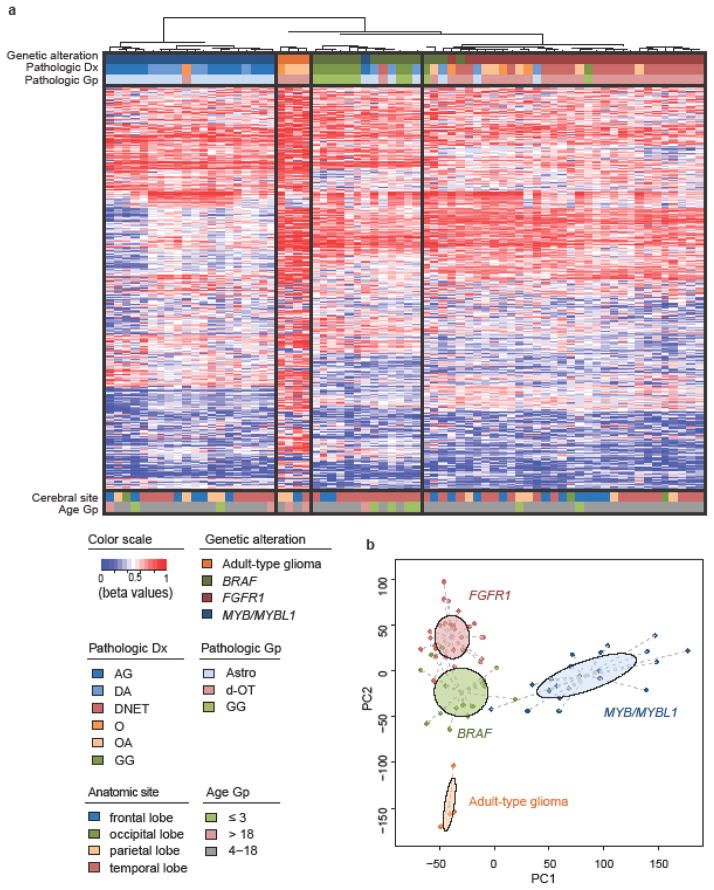
Hierarchical clustering analysis of the DNA methylation profiles of 71 LGNTs with FGFR1, MYB/MYBL1, BRAF, or IDH1/2 alterations segregated tumors primarily by genetic alteration (Fig. 4), although the strong association between genetic alteration and morphology was also apparent. Principal component analysis of these data demonstrated clear separation of the four genetic subgroups, consistent with the results from hierarchical clustering (Fig. 4b). A similar analysis including the entire study cohort showed a similar result and allowed review of the assignment of those tumors for which no genetic alteration was discovered (Supp. Fig. S8). Of six such tumors, two gangliogliomas segregated with most other gangliogliomas in a genetic group characterized mainly by BRAF alterations, while a DNET and oligodendroglioma were placed among tumors with FGFR1 alterations.
Figure 4.
DNA methylation-based data from LGNTs with BRAF, FGFR1, or MYB/MYBL1 pathogenic alterations used in unsupervised hierarchical clustering (a), or principal components analysis (b). AG, angiocentric glioma; DA, diffuse astrocytoma; DNET, dysembryoplastic neuroepithelial tumor; O, oligodendroglioma; OA, oligoastrocytoma; GG, ganglioglioma; Astro, astrocytic; d-OT, diffuse oligodendroglial tumor.
Copy number alterations among LGNTs
Very few CNAs were discovered among the series of LGNTs using the Illumina methylation array or Affymetrix SNP 6.0 platforms (Supp. Fig. S9). Recurrent changes included 1p/19q codeletion among the four adult-type d-OTs, and gains of chromosomes 5, 7, and 12 were found among disparate LGNTs with a various genetic alterations. Focal deletions at 6q23, involving the 3′ terminal region of MYB and the 5′ region of QKI were seen in 4 angiocentric gliomas, as previously reported [58]. However, the majority of angiocentric gliomas with confirmed MYB-QKI fusions did not display CNAs at these loci. No other recurrent focal CNA was identified.
Discussion
The present study extends our earlier WGS analysis of low-grade gliomas by focusing on uncommon LGNTs [67], including several LEATs – DNETs, angiocentric gliomas, and gangliogliomas. In addition, we have increased the number of ‘pediatric-type’ WHO grade II diffuse LGGs analyzed on NGS platforms, both astrocytomas and d-OTs. WGS has consistently shown that pediatric LGGs contain only one pathogenic alteration, and this principle extends to most of the uncommon LGNTs; 84% of the present series analyzed by WGS had just a single SNV or SV in a known oncogene. Combinations of common genetic alterations did occur in a few tumors from the series, including an oligoastrocytoma with pathogenic FGFR1, KRAS, and NF1 mutations and two angiocentric gliomas with a MYB-QKI fusion and BRAF:p.V600E mutation. Very few novel genetic alterations were discovered in our study. Instead, FGFR1, MYB, and BRAF alterations dominated our series of LGNTs and showed clear alignment with tumor morphology.
LGNTs with an oligodendroglial phenotype and composed primarily of oligodendrocyte-like cells (OLCs) were characterized by recurrent FGFR1 alterations. OLCs dominate the histopathology of DNETs and the diffuse oligodendroglial tumors (d-OTs), oligodendroglioma and oligoastrocytoma. While d-OTs are diffuse tumors and DNETs are generally nodular tumors, overlap between their architectural, as well as cytologic, features can occur; DNETs sometimes demonstrate a diffuse element, in which OLCs invade the cortex and subcortical white matter to resemble an oligodendroglioma [14,6]. FGFR1 alterations in this axis of tumors encompassed TKD duplications, fusion genes, and mutations within the TKD, all of which have been reported before in diverse tumor types of varying malignancy, including LGGs, malignant melanoma, and glioblastoma [5,31,48,67], though FGFR1 amplification is a more common alteration in several types of carcinoma, including lung, breast, and bladder [41,49,50]. Such alterations result in constitutively activated signaling pathways downstream of FGFR1 [55,67]. Published studies of genetic alterations in DNETs are limited, focusing on BRAF, rather then FGFR1 [10,43,51]. These find BRAF alterations at a higher frequency than in our dataset (Table 1). Aside from those in our earlier publication [67], there are no data on pediatric d-OTs.
Table 1.
Summary of published genetic alterations by histopathology
| Study | Tumor | Number of cases | FGFR1 alteration (%) | MYB/MYBL1 alteration (%) | BRAF alteration (%) |
|---|---|---|---|---|---|
| Zhang et al., Nat Genet 2013 [67] | AG | 2 | 0 | 100 | 0 |
| DA | 23 | 17 | 26 | 26 | |
| DNET | 1 | 100 | 0 | 0 | |
| d-OT | 13 | 69 | 8 | 8 | |
| GG | 9 | 0 | 0 | 56 | |
|
| |||||
| Roth et al., Cancer Genet, 2014 [51] | AG | 1 | nd | 0 | 0 |
| DA | 6 | nd | 0 | 17 | |
| DNET | 2 | nd | 0 | 50 | |
| GG | 2 | nd | 0 | 50 | |
|
| |||||
| Ramkissoon et al., PNAS, 2013 [46] | AG | 3 | 0 | 66 | 0 |
| DA | 18 | 0 | 28 | 22 | |
| GG | 9 | 0 | 0 | 56 | |
|
| |||||
| Prabowo et al., Brain Pathol, 2013 [43] | DNET | 77 | nd | nd | 30 |
| GG | 93 | nd | nd | 41 | |
|
| |||||
| Chappé et al., Brain Pathol 2013 [10] | DNET | 20 | nd | nd | 30 |
| GG | 31 | nd | nd | 39 | |
|
| |||||
| Cruz et al., J Neurooncol 2014 [12] | DA | 17 | nd | nd | 29 |
AG, angiocentric glioma; DA, diffuse astrocytoma; DNET, dysembryoplastic neuroepithelial tumor; d-OT, diffuse oligodendroglial tumor; GG, ganglioglioma nd, not determined
References in brackets
With the exception of a single oligoastrocytoma, FGFR1 SNVs occurred only in DNETs, and pathogenic FGFR1 alterations, including TKD duplications, dominated these tumors, occurring in 82%. In contrast, d-OTs mainly harbored FGFR1 TKD duplications, rather than SNVs, and were more heterogeneous, with FGFR2, PDGFRA, BRAF, and single NTRK and MYB alterations. These genetic alterations would be predicted to have similar effects on the MAPK and PI3K pathways as the recurrent FGFR1 alterations [30,67]. In 4/8 DNETs with FGFR1 SNVs, ‘doublet’ missense FGFR1 mutations were located 1–9 codons apart on the same allele (Fig. 1). Similar ‘doublet’ mutations have been recorded in other genes, such as KRAS in pilocytic astrocytoma or EGFR in lung carcinoma [16,25], but the mechanistic basis of any selective advantage that ‘doublet’ FGFR1 mutations might have is unknown. No FGFR2/3 activating mutations were identified in this study cohort; FGFR2 CNAs were seen, but were rare findings in just 2% of cases.
Our series of 15 angiocentric gliomas largely comprised tumors for which only FFPE-derived material was available, so it was not possible to gain a comprehensive insight into the genomic landscape of these tumors. However, our data show that nearly all angiocentric gliomas contain a MYB-QKI fusion. Other partner genes for MYB appear to be rare in this tumor. In contrast, the histologically related DAs appear more heterogeneous; some tumors harbor MYB or MYBL1 fusions or CNAs, while others show mutations or SVs of BRAF or TKD duplication or SVs of FGFR1. These findings for two types of LGNT with a predominantly astrocytic phenotype mirror our findings for LGNTs with an oligodendroglial phenotype – on each axis, diffuse WHO grade II gliomas are more heterogeneous than the more circumscribed WHO grade I tumors (Fig. 5).
Figure 5.
Proportions of key genetic alterations in four types of LGNT divided into two axes according to histogenesis – astrocytic and oligodendroglial.
AG, angiocentric glioma; DA, diffuse astrocytoma; DNET, dysembryoplastic neuroepithelial tumor; d-OTs, diffuse oligodendroglial tumors.
MYB has a long history as a proto-oncogene, but has so far been implicated in the oncogenesis of few tumors [47,56]. MYB duplications and balanced reciprocal t(6;7)(q23;q34) translocations between the regulatory sequence for the T-cell receptor-beta gene and MYB have been demonstrated in some patients with T-ALL, and high-level amplifications have been found in one third of hereditary (BRAC1) breast cancers [27,11,29]. A translocation between MYB and the nuclear transcription factor NFIB has been detected in approximately 30% of adenoid cystic carcinomas from the breast or salivary glands [42]. The rearrangement removes the final exon and 3′UTR of MYB. The 3′UTR contains binding sites for several microRNAs (miR-15a, miR-16-1, and miR-150) that negatively regulate MYB transcription; MYB overexpression at the RNA and protein levels results from the loss of this regulatory site. MYB-QKI rearrangements in AGs that fuse exon 15 of MYB with exon 5 of QKI also remove this 3′UTR regulatory site, while the fusion between MYB exon 9 and QKI exon 5 in LGNT62 and LGNT71 also removes the entire inhibitory C-terminal domain of MYB (Supp. Fig. S5). Other mechanisms that increase expression of MYB, which we have previously demonstrated at the protein level in AGs and diffuse LGGs, are amplification and fusions with other gene partners [58,67].
QKI is a member of the highly conserved signal transduction and activator of RNA (STAR) family of RNA-binding proteins and has a role in myelination through the regulation of differentiation in oligodendrocytes and Schwann cells [21]. Nuclear QKI levels decline throughout CNS maturation, while intracytoplasmic forms rise during myelination. In AGs, MYB fuses to exon 5 of QKI, which means that the KH domain responsible for RNA-binding is lost. However, it is possible that the C-terminal domain of QKI influences the location and activity of MYB, enhancing its oncogenic activities.
Angiocentric gliomas have an astrocytic morphology, but also demonstrate features of ependymal differentiation, which manifests as a distinctive punctate intracytoplasmic immunoreactivity for EMA and extracellular cilia, microvilli-containing lumens, and zonulae adherentes at the ultrastructural level [44,61]. However, our data demonstrate that angiocentric gliomas harbor alterations that are found in pediatric-type low-grade astrocytomas, rather than ependymomas. Most pediatric supratentorial ependymomas harbor a characteristic C11orf95-RELA fusion [38], but this SV was not found in any angiocentric glioma from the present series. In addition, we have analyzed two large cerebral ependymomas with an infiltrative subcortical component that resembled the phenotype of angiocentric glioma (Suppl Fig. S10). Both of these tumors, which were not included among LGNTs in the present series, demonstrated an iFISH profile consistent with the presence of a C11orf95-RELA fusion, but no alteration in MYB or QKI.
The ganglioglioma is a low-grade glioneuronal tumor composed of large dystrophic neuronal cells and a glial element that generally resembles a pilocytic astrocytoma or diffuse astrocytoma. The latter may infiltrate surrounding CNS parenchyma, although most gangliogliomas are relatively circumscribed tumors in imaging studies. Alterations in BRAF are recognized as the most frequent genetic alteration in gangliogliomas, particularly BRAF:p.V600E mutation, which has been reported in up to 50% of these tumors [10,15,43,46,53]. In the present series, 53% of gangliogliomas harbored BRAF alterations, both BRAF:p.V600E mutation and fusions. Other alterations were found in NF1, NTRK2, and EWSR1, but no other alteration was found at high frequency in these tumors.
A novel SLMAP-NTRK2 gene fusion was identified by RNAseq in one parietal ganglioglioma (Supp. Fig. S11). This fusion employs a similar mechanism to the NAV1-NTRK2 fusion previously reported by our group in an oligoastrocytoma [67], retaining the tyrosine kinase domain of NTRK2. QKI-NTRK2 and NACC2-NTRK2 fusions have previously been reported in pilocytic astrocytoma, in pediatric high-grade glioma and adult glioblastoma [25,63,23]. NTRK receptors modulate developmental signals within the nervous system, influencing induction of neurite outgrowth and neuronal differentiation, survival, and death. They activate the MAPK pathway following neurotrophin binding [36].
A novel EWSR1-PATZ1 fusion was present in one ganglioglioma. A similar fusion involving these two genes has been described once before in an undifferentiated sarcoma that presented in the chest wall of a teenager [33]. PATZ1 is a BTB-ZF transcription factor that inhibits the ability of p53 to bind to its response elements and is involved in keeping embryonic stem cells in an undifferentiated state [28,37]. Rearrangement of EWSR1 is found in intracranial Ewing sarcomas that involve the inner table of the skull and extraosseous meningeal ‘peripheral’ primitive neuroectodermal tumors [35,52]. Additionally, exceptional examples of other intracranial neoplasms with an EWSR1 rearrangement and variable malignancy have been reported [13,18]. However, this is the first report of such a fusion gene in a primary neuroepithelial tumor.
Platelet-derived growth factor receptor α (PDGFRA) amplification and mutations have previously been reported in pediatric high-grade glioma and adult glioblastoma [40,9]. Dinucleotide PDGFRA somatic mutations were identified in 2 LGNTs; 1 DNET (LGNT42) and 1 oligoastrocytoma (LGNT92) (Supp. Fig. S12). Neither tumor was associated with an aggressive clinical course. LGNT42 had dinucleotide mutations on the same allele at chr4:55136831 and chr4:55136832 (AA>TT), while LGNT92 had dinucleotide mutations on the same allele at chr4:55136832 and cht4:55136833 (AG>TC). These changes introduced K385L and K385I mutations in LGNT42 and LGNT92, respectively. Notably, both mutations changed K385 from a basic amino acid to a hydrophobic amino acid. A dinucleotide PDGFRA mutation at K385 (AG>TA) introducing K385I has previously been reported in one pediatric high-grade glioma [54]. Additional PDGFRA K385M mutations have also been reported in one adult LGG and one adult HGG [67,54]. BRAF:p.V600E mutation was the most prevalent SNV (12/91) identified in the cohort, followed by SNVs in FGFR1 (9/91) and IDH1 (3/91). No additional recurrent SNVs or indels were identified in the study cohort. Genomic aberrations could not be determined for 6/91 (7%) cases; 3 gangliogliomas, 2 d-OTs and 1 DNET.
Recent genetic studies have helped to delineate pediatric-type and adult-type diffuse LGGs, which have very similar histopathological features. Adult-type disease demonstrates a high frequency of IDH1/2 mutation [8,19,39,64], while recurrent genetic alterations in MYB, BRAF, or FGFR1 have been found in small series of pediatric tumors [12,15,46,67]. Adult-type disease can present in adolescence as shown in the present data and other studies, and its distinction from pediatric-type disease is important, because anaplastic progression occurs in most adult-type grade II gliomas but rarely in histopathologically comparable pediatric-type tumors [26,60]. In this way, the molecular genetic characterization of LGGs has important implications for how they are classified in the WHO classification of nervous system tumors [32].
Aside from supporting the important distinction between pediatric-type and adult-type diffuse LGGs, our findings have further diagnostic implications. Many CNS neuroepithelial tumors can contain OLCs or appear similar to diffuse astrocytomas, and in histopathologic preparations of small CNS biopsies distinguishing between such tumors can be difficult. Genetic analysis may aid diagnosis in this situation, by demonstrating one of the key alterations that would align a tumor to either the DNET/d-OT or AG/DA axis. Such analysis need not involve NGS; the key alterations described in our study can all be determined by conventional molecular pathology or cytogenetic methods on FFPE tissue preparations.
Additionally, targeted therapies are currently being tested for the key LGNT genetic alterations. Clinical trials investigating BRAF:p.V600E inhibitors are under way in pediatric patients, and a study of the mTOR inhibitor, Everolimus, in progressive pediatric LGG has recently been completed. Dovitinib (TKI258), an FGFR1/2/3 inhibitor, has shown anti-tumor activity in breast cancer patients with FGFR-amplified breast cancer [3]. On the basis of our findings, these developments would be predicted to benefit children and young adults with LGNTs.
Supplementary Material
Acknowledgments
We are grateful for support from Anatomic Pathology and the Hartwell Center of Biotechnology and Bioinformatics at St. Jude Children’s Research Hospital. We acknowledge the St. Jude Children’s Research Hospital Biorepository, from which tissue samples were obtained in accordance with institutional review board approval for the Pediatric Cancer Genome Project. This work was supported by the St. Jude Children’s Research Hospital–Washington University Pediatric Cancer Genome Project and the American Lebanese Syrian Associated Charities (ALSAC) of St. Jude Children’s Research Hospital.
References
- 1.Alvarez-Cubero MJ, Saiz M, Martinez-Gonzalez LJ, Alvarez JC, Lorente JA, Cozar JM. Genetic analysis of the principal genes related to prostate cancer: a review. Urol Oncol. 2013;31:1419–1429. doi: 10.1016/j.urolonc.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 2.Anderson JL, Gutmann DH. Neurofibromatosis type 1. Handb Clin Neurol. 2015;132:75–86. doi: 10.1016/B978-0-444-62702-5.00004-4. [DOI] [PubMed] [Google Scholar]
- 3.Andre F, Bachelot T, Campone M, Dalenc F, Perez-Garcia JM, Hurvitz SA, Turner N, Rugo H, Smith JW, Deudon S, Shi M, Zhang Y, Kay A, Porta DG, Yovine A, Baselga J. Targeting FGFR with dovitinib (TKI258): preclinical and clinical data in breast cancer. Clin Cancer Res. 2013;19:3693–3702. doi: 10.1158/1078-0432.CCR-13-0190. [DOI] [PubMed] [Google Scholar]
- 4.Bandopadhayay P, Bergthold G, London WB, Goumnerova LC, Morales La Madrid A, Marcus KJ, Guo D, Ullrich NJ, Robison NJ, Chi SN, Beroukhim R, Kieran MW, Manley PE. Long-term outcome of 4,040 children diagnosed with pediatric low-grade gliomas: an analysis of the Surveillance Epidemiology and End Results (SEER) database. Pediatr Blood Cancer. 2014;61:1173–1179. doi: 10.1002/pbc.24958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Becker AP, Scapulatempo-Neto C, Carloni AC, Paulino A, Sheren J, Aisner DL, Musselwhite E, Clara C, Machado HR, Oliveira RS, Neder L, Varella-Garcia M, Reis RM. KIAA1549: BRAF Gene Fusion and FGFR1 Hotspot Mutations Are Prognostic Factors in Pilocytic Astrocytomas. J Neuropathol Exp Neurol. 2015;74:743–754. doi: 10.1097/NEN.0000000000000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bird-Lieberman G, Sethi K, Childs AM, Chumas P, Crimmins D, Ismail A, Livingston J. Diffuse hemispheric dysembryoplastic neuroepithelial tumor: a new radiological variant associated with early-onset severe epilepsy. Journal of neurosurgery Pediatrics. 2011;7:416–420. doi: 10.3171/2011.1.PEDS10258. [DOI] [PubMed] [Google Scholar]
- 7.Blumcke I, Aronica E, Urbach H, Alexopoulos A, Gonzalez-Martinez JA. A neuropathology-based approach to epilepsy surgery in brain tumors and proposal for a new terminology use for long-term epilepsy-associated brain tumors. Acta Neuropathol. 2014;128:39–54. doi: 10.1007/s00401-014-1288-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brat DJ, Verhaak RG, Aldape KD, Yung WK, Salama SR, Cooper LA, Rheinbay E, Miller CR, Vitucci M, Morozova O, Robertson AG, Noushmehr H, Laird PW, Cherniack AD, Akbani R, Huse JT, Ciriello G, Poisson LM, Barnholtz-Sloan JS, Berger MS, Brennan C, Colen RR, Colman H, Flanders AE, Giannini C, Grifford M, Iavarone A, Jain R, Joseph I, Kim J, Kasaian K, Mikkelsen T, Murray BA, O’Neill BP, Pachter L, Parsons DW, Sougnez C, Sulman EP, Vandenberg SR, Van Meir EG, von Deimling A, Zhang H, Crain D, Lau K, Mallery D, Morris S, Paulauskis J, Penny R, Shelton T, Sherman M, Yena P, Black A, Bowen J, Dicostanzo K, Gastier-Foster J, Leraas KM, Lichtenberg TM, Pierson CR, Ramirez NC, Taylor C, Weaver S, Wise L, Zmuda E, Davidsen T, Demchok JA, Eley G, Ferguson ML, Hutter CM, Mills Shaw KR, Ozenberger BA, Sheth M, Sofia HJ, Tarnuzzer R, Wang Z, Yang L, Zenklusen JC, Ayala B, Baboud J, Chudamani S, Jensen MA, Liu J, Pihl T, Raman R, Wan Y, Wu Y, Ally A, Auman JT, Balasundaram M, Balu S, Baylin SB, Beroukhim R, Bootwalla MS, Bowlby R, Bristow CA, Brooks D, Butterfield Y, Carlsen R, Carter S, Chin L, Chu A, Chuah E, Cibulskis K, Clarke A, Coetzee SG, Dhalla N, Fennell T, Fisher S, Gabriel S, Getz G, Gibbs R, Guin R, Hadjipanayis A, Hayes DN, Hinoue T, Hoadley K, Holt RA, Hoyle AP, Jefferys SR, Jones S, Jones CD, Kucherlapati R, Lai PH, Lander E, Lee S, Lichtenstein L, Ma Y, Maglinte DT, Mahadeshwar HS, Marra MA, Mayo M, Meng S, Meyerson ML, Mieczkowski PA, Moore RA, Mose LE, Mungall AJ, Pantazi A, Parfenov M, Park PJ, Parker JS, Perou CM, Protopopov A, Ren X, Roach J, Sabedot TS, Schein J, Schumacher SE, Seidman JG, Seth S, Shen H, Simons JV, Sipahimalani P, Soloway MG, Song X, Sun H, Tabak B, Tam A, Tan D, Tang J, Thiessen N, Triche T, Jr, Van Den Berg DJ, Veluvolu U, Waring S, Weisenberger DJ, Wilkerson MD, Wong T, Wu J, Xi L, Xu AW, Yang L, Zack TI, Zhang J, Aksoy BA, Arachchi H, Benz C, Bernard B, Carlin D, Cho J, DiCara D, Frazer S, Fuller GN, Gao J, Gehlenborg N, Haussler D, Heiman DI, Iype L, Jacobsen A, Ju Z, Katzman S, Kim H, Knijnenburg T, Kreisberg RB, Lawrence MS, Lee W, Leinonen K, Lin P, Ling S, Liu W, Liu Y, Liu Y, Lu Y, Mills G, Ng S, Noble MS, Paull E, Rao A, Reynolds S, Saksena G, Sanborn Z, Sander C, Schultz N, Senbabaoglu Y, Shen R, Shmulevich I, Sinha R, Stuart J, Sumer SO, Sun Y, Tasman N, Taylor BS, Voet D, Weinhold N, Weinstein JN, Yang D, Yoshihara K, Zheng S, Zhang W, Zou L, Abel T, Sadeghi S, Cohen ML, Eschbacher J, Hattab EM, Raghunathan A, Schniederjan MJ, Aziz D, Barnett G, Barrett W, Bigner DD, Boice L, Brewer C, Calatozzolo C, Campos B, Carlotti CG, Jr, Chan TA, Cuppini L, Curley E, Cuzzubbo S, Devine K, DiMeco F, Duell R, Elder JB, Fehrenbach A, Finocchiaro G, Friedman W, Fulop J, Gardner J, Hermes B, Herold-Mende C, Jungk C, Kendler A, Lehman NL, Lipp E, Liu O, Mandt R, McGraw M, McLendon R, McPherson C, Neder L, Nguyen P, Noss A, Nunziata R, Ostrom QT, Palmer C, Perin A, Pollo B, Potapov A, Potapova O, Rathmell WK, Rotin D, Scarpace L, Schilero C, Senecal K, Shimmel K, Shurkhay V, Sifri S, Singh R, Sloan AE, Smolenski K, Staugaitis SM, Steele R, Thorne L, Tirapelli DP, Unterberg A, Vallurupalli M, Wang Y, Warnick R, Williams F, Wolinsky Y, Bell S, Rosenberg M, Stewart C, Huang F, Grimsby JL, Radenbaugh AJ, Zhang J. Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. N Engl J Med. 2015;372:2481–2498. doi: 10.1056/NEJMoa1402121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, Zheng S, Chakravarty D, Sanborn JZ, Berman SH, Beroukhim R, Bernard B, Wu CJ, Genovese G, Shmulevich I, Barnholtz-Sloan J, Zou L, Vegesna R, Shukla SA, Ciriello G, Yung WK, Zhang W, Sougnez C, Mikkelsen T, Aldape K, Bigner DD, Van Meir EG, Prados M, Sloan A, Black KL, Eschbacher J, Finocchiaro G, Friedman W, Andrews DW, Guha A, Iacocca M, O’Neill BP, Foltz G, Myers J, Weisenberger DJ, Penny R, Kucherlapati R, Perou CM, Hayes DN, Gibbs R, Marra M, Mills GB, Lander E, Spellman P, Wilson R, Sander C, Weinstein J, Meyerson M, Gabriel S, Laird PW, Haussler D, Getz G, Chin L, Network TR. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chappé C, Padovani L, Scavarda D, Forest F, Nanni-Metellus I, Loundou A, Mercurio S, Fina F, Lena G, Colin C, Figarella-Branger D. Dysembryoplastic neuroepithelial tumors share with pleomorphic xanthoastrocytomas and gangliogliomas BRAF(V600E) mutation and expression. Brain Pathol. 2013;23:574–583. doi: 10.1111/bpa.12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clappier E, Cuccuini W, Kalota A, Crinquette A, Cayuela JM, Dik WA, Langerak AW, Montpellier B, Nadel B, Walrafen P, Delattre O, Aurias A, Leblanc T, Dombret H, Gewirtz AM, Baruchel A, Sigaux F, Soulier J. The C-MYB locus is involved in chromosomal translocation and genomic duplications in human T-cell acute leukemia (T-ALL), the translocation defining a new T-ALL subtype in very young children. Blood. 2007;110:1251–1261. doi: 10.1182/blood-2006-12-064683. [DOI] [PubMed] [Google Scholar]
- 12.Cruz GR, Dias Oliveira I, Moraes L, Del Giudice Paniago M, de Seixas Alves MT, Capellano AM, Saba-Silva N, Cavalheiro S, Cerutti JM, Toledo SR. Analysis of KIAA1549-BRAF fusion gene expression and IDH1/IDH2 mutations in low grade pediatric astrocytomas. Journal of neuro-oncology. 2014;117:235–242. doi: 10.1007/s11060-014-1398-1. [DOI] [PubMed] [Google Scholar]
- 13.Cummings TJ, Bridge JA, Fukushima T. Extraskeletal myxoid chondrosarcoma of the jugular foramen. Clinical neuropathology. 2004;23:232–237. [PubMed] [Google Scholar]
- 14.Daumas-Duport C. Dysembryoplastic neuroepithelial tumours. Brain Pathol. 1993;3:283–295. doi: 10.1111/j.1750-3639.1993.tb00755.x. [DOI] [PubMed] [Google Scholar]
- 15.Dougherty MJ, Santi M, Brose MS, Ma C, Resnick AC, Sievert AJ, Storm PB, Biegel JA. Activating mutations in BRAF characterize a spectrum of pediatric low-grade gliomas. Neuro Oncol. 2010;12:621–630. doi: 10.1093/neuonc/noq007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Driver BR, Portier BP, Mody DR, Deavers M, Bernicker EH, Kim MP, Teh BS, Santacruz JF, Kopas L, Munden RF, Cagle PT. Next-Generation Sequencing of a Cohort of Pulmonary Large Cell Carcinomas Reclassified by World Health Organization 2015 Criteria. Archives of pathology & laboratory medicine. 2015 doi: 10.5858/arpa.2015-0361-OA. [DOI] [PubMed] [Google Scholar]
- 17.Dudley RW, Torok MR, Gallegos DR, Mulcahy-Levy JM, Hoffman LM, Liu AK, Handler MH, Hankinson TC. Pediatric low-grade ganglioglioma: epidemiology, treatments, and outcome analysis on 348 children from the surveillance, epidemiology, and end results database. Neurosurgery. 2015;76:313–319. doi: 10.1227/NEU.0000000000000619. discussion 319; quiz 319–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dunham C, Hussong J, Seiff M, Pfeifer J, Perry A. Primary intracerebral angiomatoid fibrous histiocytoma: report of a case with a t(12;22)(q13;q12) causing type 1 fusion of the EWS and ATF-1 genes. Am J Surg Pathol. 2008;32:478–484. doi: 10.1097/PAS.0b013e3181453451. [DOI] [PubMed] [Google Scholar]
- 19.Eckel-Passow JE, Lachance DH, Molinaro AM, Walsh KM, Decker PA, Sicotte H, Pekmezci M, Rice T, Kosel ML, Smirnov IV, Sarkar G, Caron AA, Kollmeyer TM, Praska CE, Chada AR, Halder C, Hansen HM, McCoy LS, Bracci PM, Marshall R, Zheng S, Reis GF, Pico AR, O’Neill BP, Buckner JC, Giannini C, Huse JT, Perry A, Tihan T, Berger MS, Chang SM, Prados MD, Wiemels J, Wiencke JK, Wrensch MR, Jenkins RB. Glioma Groups Based on 1p/19q, IDH, and TERT Promoter Mutations in Tumors. N Engl J Med. 2015;372:2499–2508. doi: 10.1056/NEJMoa1407279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ellison DW, Kocak M, Dalton J, Megahed H, Lusher ME, Ryan SL, Zhao W, Nicholson SL, Taylor RE, Bailey S, Clifford SC. Definition of disease-risk stratification groups in childhood medulloblastoma using combined clinical, pathologic, and molecular variables. J Clin Oncol. 2011;29:1400–1407. doi: 10.1200/JCO.2010.30.2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feng Y, Bankston A. The star family member QKI and cell signaling. Adv Exp Med Biol. 2010;693:25–36. [PubMed] [Google Scholar]
- 22.Forshew T, Tatevossian RG, Lawson AR, Ma J, Neale G, Ogunkolade BW, Jones TA, Aarum J, Dalton J, Bailey S, Chaplin T, Carter RL, Gajjar A, Broniscer A, Young BD, Ellison DW, Sheer D. Activation of the ERK/MAPK pathway: a signature genetic defect in posterior fossa pilocytic astrocytomas. J Pathol. 2009;218:172–181. doi: 10.1002/path.2558. [DOI] [PubMed] [Google Scholar]
- 23.Frattini V, Trifonov V, Chan JM, Castano A, Lia M, Abate F, Keir ST, Ji AX, Zoppoli P, Niola F, Danussi C, Dolgalev I, Porrati P, Pellegatta S, Heguy A, Gupta G, Pisapia DJ, Canoll P, Bruce JN, McLendon RE, Yan H, Aldape K, Finocchiaro G, Mikkelsen T, Prive GG, Bigner DD, Lasorella A, Rabadan R, Iavarone A. The integrated landscape of driver genomic alterations in glioblastoma. Nat Genet. 2013;45:1141–1149. doi: 10.1038/ng.2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huse JT, Rosenblum MK. The Emerging Molecular Foundations of Pediatric Brain Tumors. J Child Neurol. 2015;30:1838–1850. doi: 10.1177/0883073815579709. [DOI] [PubMed] [Google Scholar]
- 25.Jones DT, Hutter B, Jager N, Korshunov A, Kool M, Warnatz HJ, Zichner T, Lambert SR, Ryzhova M, Quang DA, Fontebasso AM, Stutz AM, Hutter S, Zuckermann M, Sturm D, Gronych J, Lasitschka B, Schmidt S, Seker-Cin H, Witt H, Sultan M, Ralser M, Northcott PA, Hovestadt V, Bender S, Pfaff E, Stark S, Faury D, Schwartzentruber J, Majewski J, Weber UD, Zapatka M, Raeder B, Schlesner M, Worth CL, Bartholomae CC, von Kalle C, Imbusch CD, Radomski S, Lawerenz C, van Sluis P, Koster J, Volckmann R, Versteeg R, Lehrach H, Monoranu C, Winkler B, Unterberg A, Herold-Mende C, Milde T, Kulozik AE, Ebinger M, Schuhmann MU, Cho YJ, Pomeroy SL, von Deimling A, Witt O, Taylor MD, Wolf S, Karajannis MA, Eberhart CG, Scheurlen W, Hasselblatt M, Ligon KL, Kieran MW, Korbel JO, Yaspo ML, Brors B, Felsberg J, Reifenberger G, Collins VP, Jabado N, Eils R, Lichter P, Pfister SM International Cancer Genome Consortium PedBrain Tumor P. Recurrent somatic alterations of FGFR1 and NTRK2 in pilocytic astrocytoma. Nat Genet. 2013;45:927–932. doi: 10.1038/ng.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones DT, Mulholland SA, Pearson DM, Malley DS, Openshaw SW, Lambert SR, Liu L, Backlund LM, Ichimura K, Collins VP. Adult grade II diffuse astrocytomas are genetically distinct from and more aggressive than their paediatric counterparts. Acta Neuropathol. 2011;121:753–761. doi: 10.1007/s00401-011-0810-6. [DOI] [PubMed] [Google Scholar]
- 27.Kauraniemi P, Hedenfalk I, Persson K, Duggan DJ, Tanner M, Johannsson O, Olsson H, Trent JM, Isola J, Borg A. MYB oncogene amplification in hereditary BRCA1 breast cancer. Cancer Res. 2000;60:5323–5328. [PubMed] [Google Scholar]
- 28.Keskin N, Deniz E, Eryilmaz J, Un M, Batur T, Ersahin T, Cetin Atalay R, Sakaguchi S, Ellmeier W, Erman B. PATZ1 Is a DNA Damage-Responsive Transcription Factor That Inhibits p53 Function. Molecular and cellular biology. 2015;35:1741–1753. doi: 10.1128/MCB.01475-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lahortiga I, De Keersmaecker K, Van Vlierberghe P, Graux C, Cauwelier B, Lambert F, Mentens N, Beverloo HB, Pieters R, Speleman F, Odero MD, Bauters M, Froyen G, Marynen P, Vandenberghe P, Wlodarska I, Meijerink JP, Cools J. Duplication of the MYB oncogene in T cell acute lymphoblastic leukemia. Nat Genet. 2007;39:593–595. doi: 10.1038/ng2025. [DOI] [PubMed] [Google Scholar]
- 30.Lannon CL, Martin MJ, Tognon CE, Jin W, Kim SJ, Sorensen PH. A highly conserved NTRK3 C-terminal sequence in the ETV6-NTRK3 oncoprotein binds the phosphotyrosine binding domain of insulin receptor substrate-1: an essential interaction for transformation. J Biol Chem. 2004;279:6225–6234. doi: 10.1074/jbc.M307388200. [DOI] [PubMed] [Google Scholar]
- 31.Lin WM, Baker AC, Beroukhim R, Winckler W, Feng W, Marmion JM, Laine E, Greulich H, Tseng H, Gates C, Hodi FS, Dranoff G, Sellers WR, Thomas RK, Meyerson M, Golub TR, Dummer R, Herlyn M, Getz G, Garraway LA. Modeling genomic diversity and tumor dependency in malignant melanoma. Cancer research. 2008;68:664–673. doi: 10.1158/0008-5472.CAN-07-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Louis DN, Perry A, Burger P, Ellison DW, Reifenberger G, von Deimling A, Aldape K, Brat D, Collins VP, Eberhart C, Figarella-Branger D, Fuller GN, Giangaspero F, Giannini C, Hawkins C, Kleihues P, Korshunov A, Kros JM, Beatriz Lopes M, Ng HK, Ohgaki H, Paulus W, Pietsch T, Rosenblum M, Rushing E, Soylemezoglu F, Wiestler O, Wesseling P. International Society Of Neuropathology--Haarlem consensus guidelines for nervous system tumor classification and grading. Brain Pathol. 2014;24:429–435. doi: 10.1111/bpa.12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mastrangelo T, Modena P, Tornielli S, Bullrich F, Testi MA, Mezzelani A, Radice P, Azzarelli A, Pilotti S, Croce CM, Pierotti MA, Sozzi G. A novel zinc finger gene is fused to EWS in small round cell tumor. Oncogene. 2000;19:3799–3804. doi: 10.1038/sj.onc.1203762. [DOI] [PubMed] [Google Scholar]
- 34.McKean-Cowdin R, Razavi P, Barrington-Trimis J, Baldwin RT, Asgharzadeh S, Cockburn M, Tihan T, Preston-Martin S. Trends in childhood brain tumor incidence, 1973–2009. Journal of neuro-oncology. 2013;115:153–160. doi: 10.1007/s11060-013-1212-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mobley BC, Roulston D, Shah GV, Bijwaard KE, McKeever PE. Peripheral primitive neuroectodermal tumor/Ewing’s sarcoma of the craniospinal vault: case reports and review. Human pathology. 2006;37:845–853. doi: 10.1016/j.humpath.2006.02.011. [DOI] [PubMed] [Google Scholar]
- 36.Nguyen N, Lee SB, Lee YS, Lee KH, Ahn JY. Neuroprotection by NGF and BDNF against neurotoxin-exerted apoptotic death in neural stem cells are mediated through Trk receptors, activating PI3-kinase and MAPK pathways. Neurochem Res. 2009;34:942–951. doi: 10.1007/s11064-008-9848-9. [DOI] [PubMed] [Google Scholar]
- 37.Ow JR, Ma H, Jean A, Goh Z, Lee YH, Chong YM, Soong R, Fu XY, Yang H, Wu Q. Patz1 regulates embryonic stem cell identity. Stem Cells Dev. 2014;23:1062–1073. doi: 10.1089/scd.2013.0430. [DOI] [PubMed] [Google Scholar]
- 38.Parker M, Mohankumar KM, Punchihewa C, Weinlich R, Dalton JD, Li Y, Lee R, Tatevossian RG, Phoenix TN, Thiruvenkatam R, White E, Tang B, Orisme W, Gupta K, Rusch M, Chen X, Li Y, Nagahawhatte P, Hedlund E, Finkelstein D, Wu G, Shurtleff S, Easton J, Boggs K, Yergeau D, Vadodaria B, Mulder HL, Becksfort J, Gupta P, Huether R, Ma J, Song G, Gajjar A, Merchant T, Boop F, Smith AA, Ding L, Lu C, Ochoa K, Zhao D, Fulton RS, Fulton LL, Mardis ER, Wilson RK, Downing JR, Green DR, Zhang J, Ellison DW, Gilbertson RJ. C11orf95-RELA fusions drive oncogenic NF-kappaB signalling in ependymoma. Nature. 2014;506:451–455. doi: 10.1038/nature13109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, Olivi A, McLendon R, Rasheed BA, Keir S, Nikolskaya T, Nikolsky Y, Busam DA, Tekleab H, Diaz LA, Jr, Hartigan J, Smith DR, Strausberg RL, Marie SK, Shinjo SM, Yan H, Riggins GJ, Bigner DD, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. 1164382 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paugh BS, Zhu X, Qu C, Endersby R, Diaz AK, Zhang J, Bax DA, Carvalho D, Reis RM, Onar-Thomas A, Broniscer A, Wetmore C, Zhang J, Jones C, Ellison DW, Baker SJ. Novel oncogenic PDGFRA mutations in pediatric high-grade gliomas. Cancer Res. 2013;73:6219–6229. doi: 10.1158/0008-5472.CAN-13-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peifer M, Fernandez-Cuesta L, Sos ML, George J, Seidel D, Kasper LH, Plenker D, Leenders F, Sun R, Zander T, Menon R, Koker M, Dahmen I, Muller C, Di Cerbo V, Schildhaus HU, Altmuller J, Baessmann I, Becker C, de Wilde B, Vandesompele J, Bohm D, Ansen S, Gabler F, Wilkening I, Heynck S, Heuckmann JM, Lu X, Carter SL, Cibulskis K, Banerji S, Getz G, Park KS, Rauh D, Grutter C, Fischer M, Pasqualucci L, Wright G, Wainer Z, Russell P, Petersen I, Chen Y, Stoelben E, Ludwig C, Schnabel P, Hoffmann H, Muley T, Brockmann M, Engel-Riedel W, Muscarella LA, Fazio VM, Groen H, Timens W, Sietsma H, Thunnissen E, Smit E, Heideman DA, Snijders PJ, Cappuzzo F, Ligorio C, Damiani S, Field J, Solberg S, Brustugun OT, Lund-Iversen M, Sanger J, Clement JH, Soltermann A, Moch H, Weder W, Solomon B, Soria JC, Validire P, Besse B, Brambilla E, Brambilla C, Lantuejoul S, Lorimier P, Schneider PM, Hallek M, Pao W, Meyerson M, Sage J, Shendure J, Schneider R, Buttner R, Wolf J, Nurnberg P, Perner S, Heukamp LC, Brindle PK, Haas S, Thomas RK. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet. 2012;44:1104–1110. doi: 10.1038/ng.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Persson M, Andren Y, Mark J, Horlings HM, Persson F, Stenman G. Recurrent fusion of MYB and NFIB transcription factor genes in carcinomas of the breast and head and neck. Proc Natl Acad Sci U S A. 2009;106:18740–18744. doi: 10.1073/pnas.0909114106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prabowo AS, Iyer AM, Veersema TJ, Anink JJ, Schouten-van Meeteren AY, Spliet WG, van Rijen PC, Ferrier CH, Capper D, Thom M, Aronica E. BRAF V600E Mutation Is Associated with mTOR Signaling Activation in Glioneuronal Tumors. Brain Pathol. 2013 doi: 10.1111/bpa.12081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Preusser M, Hoischen A, Novak K, Czech T, Prayer D, Hainfellner JA, Baumgartner C, Woermann FG, Tuxhorn IE, Pannek HW, Bergmann M, Radlwimmer B, Villagran R, Weber RG, Hans VH. Angiocentric glioma: report of clinico-pathologic and genetic findings in 8 cases. Am J Surg Pathol. 2007;31:1709–1718. doi: 10.1097/PAS.0b013e31804a7ebb. [DOI] [PubMed] [Google Scholar]
- 45.Raabe E, Kieran MW, Cohen KJ. New strategies in pediatric gliomas: molecular advances in pediatric low-grade gliomas as a model. Clin Cancer Res. 2013;19:4553–4558. doi: 10.1158/1078-0432.CCR-13-0662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ramkissoon LA, Horowitz PM, Craig JM, Ramkissoon SH, Rich BE, Schumacher SE, McKenna A, Lawrence MS, Bergthold G, Brastianos PK, Tabak B, Ducar MD, Van Hummelen P, MacConaill LE, Pouissant-Young T, Cho YJ, Taha H, Mahmoud M, Bowers DC, Margraf L, Tabori U, Hawkins C, Packer RJ, Hill DA, Pomeroy SL, Eberhart CG, Dunn IF, Goumnerova L, Getz G, Chan JA, Santagata S, Hahn WC, Stiles CD, Ligon AH, Kieran MW, Beroukhim R, Ligon KL. Genomic analysis of diffuse pediatric low-grade gliomas identifies recurrent oncogenic truncating rearrangements in the transcription factor MYBL1. Proc Natl Acad Sci U S A. 2013;110:8188–8193. doi: 10.1073/pnas.1300252110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ramsay RG, Gonda TJ. MYB function in normal and cancer cells. Nat Rev Cancer. 2008;8:523–534. doi: 10.1038/nrc2439. [DOI] [PubMed] [Google Scholar]
- 48.Rand V, Huang J, Stockwell T, Ferriera S, Buzko O, Levy S, Busam D, Li K, Edwards JB, Eberhart C, Murphy KM, Tsiamouri A, Beeson K, Simpson AJ, Venter JC, Riggins GJ, Strausberg RL. Sequence survey of receptor tyrosine kinases reveals mutations in glioblastomas. Proc Natl Acad Sci U S A. 2005;102:14344–14349. doi: 10.1073/pnas.0507200102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reis-Filho JS, Simpson PT, Turner NC, Lambros MB, Jones C, Mackay A, Grigoriadis A, Sarrio D, Savage K, Dexter T, Iravani M, Fenwick K, Weber B, Hardisson D, Schmitt FC, Palacios J, Lakhani SR, Ashworth A. FGFR1 emerges as a potential therapeutic target for lobular breast carcinomas. Clin Cancer Res. 2006;12:6652–6662. doi: 10.1158/1078-0432.CCR-06-1164. [DOI] [PubMed] [Google Scholar]
- 50.Ross JS, Wang K, Al-Rohil RN, Nazeer T, Sheehan CE, Otto GA, He J, Palmer G, Yelensky R, Lipson D, Ali S, Balasubramanian S, Curran JA, Garcia L, Mahoney K, Downing SR, Hawryluk M, Miller VA, Stephens PJ. Advanced urothelial carcinoma: next-generation sequencing reveals diverse genomic alterations and targets of therapy. Modern pathology: an official journal of the United States and Canadian Academy of Pathology, Inc. 2014;27:271–280. doi: 10.1038/modpathol.2013.135. [DOI] [PubMed] [Google Scholar]
- 51.Roth JJ, Santi M, Rorke-Adams LB, Harding BN, Busse TM, Tooke LS, Biegel JA. Diagnostic application of high resolution single nucleotide polymorphism array analysis for children with brain tumors. Cancer Genet. 2014;207:111–123. doi: 10.1016/j.cancergen.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sankar S, Lessnick SL. Promiscuous partnerships in Ewing’s sarcoma. Cancer Genet. 2011;204:351–365. doi: 10.1016/j.cancergen.2011.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schindler G, Capper D, Meyer J, Janzarik W, Omran H, Herold-Mende C, Schmieder K, Wesseling P, Mawrin C, Hasselblatt M, Louis DN, Korshunov A, Pfister S, Hartmann C, Paulus W, Reifenberger G, von Deimling A. Analysis of BRAF V600E mutation in 1,320 nervous system tumors reveals high mutation frequencies in pleomorphic xanthoastrocytoma, ganglioglioma and extra-cerebellar pilocytic astrocytoma. Acta Neuropathol. 2011;121:397–405. doi: 10.1007/s00401-011-0802-6. [DOI] [PubMed] [Google Scholar]
- 54.Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, Sturm D, Fontebasso AM, Quang DA, Tonjes M, Hovestadt V, Albrecht S, Kool M, Nantel A, Konermann C, Lindroth A, Jager N, Rausch T, Ryzhova M, Korbel JO, Hielscher T, Hauser P, Garami M, Klekner A, Bognar L, Ebinger M, Schuhmann MU, Scheurlen W, Pekrun A, Fruhwald MC, Roggendorf W, Kramm C, Durken M, Atkinson J, Lepage P, Montpetit A, Zakrzewska M, Zakrzewski K, Liberski PP, Dong Z, Siegel P, Kulozik AE, Zapatka M, Guha A, Malkin D, Felsberg J, Reifenberger G, von Deimling A, Ichimura K, Collins VP, Witt H, Milde T, Witt O, Zhang C, Castelo-Branco P, Lichter P, Faury D, Tabori U, Plass C, Majewski J, Pfister SM, Jabado N. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 55.Sorensen V, Zhen Y, Zakrzewska M, Haugsten EM, Walchli S, Nilsen T, Olsnes S, Wiedlocha A. Phosphorylation of fibroblast growth factor (FGF) receptor 1 at Ser777 by p38 mitogen-activated protein kinase regulates translocation of exogenous FGF1 to the cytosol and nucleus. Molecular and cellular biology. 2008;28:4129–4141. doi: 10.1128/MCB.02117-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stenman G, Andersson MK, Andren Y. New tricks from an old oncogene: gene fusion and copy number alterations of MYB in human cancer. Cell Cycle. 2010;9:2986–2995. doi: 10.4161/cc.9.15.12515. 12515 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stokland T, Liu JF, Ironside JW, Ellison DW, Taylor R, Robinson KJ, Picton SV, Walker DA. A multivariate analysis of factors determining tumor progression in childhood low-grade glioma: a population-based cohort study (CCLG CNS9702) Neuro-oncology. 2010;12:1257–1268. doi: 10.1093/neuonc/noq092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tatevossian RG, Tang B, Dalton J, Forshew T, Lawson AR, Ma J, Neale G, Shurtleff SA, Bailey S, Gajjar A, Baker SJ, Sheer D, Ellison DW. MYB upregulation and genetic aberrations in a subset of pediatric low-grade gliomas. Acta Neuropathol. 2010;120:731–743. doi: 10.1007/s00401-010-0763-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thom M, Blumcke I, Aronica E. Long-term epilepsy-associated tumors. Brain Pathol. 2012;22:350–379. doi: 10.1111/j.1750-3639.2012.00582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thon N, Eigenbrod S, Kreth S, Lutz J, Tonn JC, Kretzschmar H, Peraud A, Kreth FW. IDH1 mutations in grade II astrocytomas are associated with unfavorable progression-free survival and prolonged postrecurrence survival. Cancer. 2012;118:452–460. doi: 10.1002/cncr.26298. [DOI] [PubMed] [Google Scholar]
- 61.Wang M, Tihan T, Rojiani AM, Bodhireddy SR, Prayson RA, Iacuone JJ, Alles AJ, Donahue DJ, Hessler RB, Kim JH, Haas M, Rosenblum MK, Burger PC. Monomorphous angiocentric glioma: a distinctive epileptogenic neoplasm with features of infiltrating astrocytoma and ependymoma. J Neuropathol Exp Neurol. 2005;64:875–881. doi: 10.1097/01.jnen.0000182981.02355.10. [DOI] [PubMed] [Google Scholar]
- 62.Wu G, Broniscer A, McEachron TA, Lu C, Paugh BS, Becksfort J, Qu C, Ding L, Huether R, Parker M, Zhang J, Gajjar A, Dyer MA, Mullighan CG, Gilbertson RJ, Mardis ER, Wilson RK, Downing JR, Ellison DW, Baker SJ. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nature genetics. 2012;44:251–253. doi: 10.1038/ng.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu G, Diaz AK, Paugh BS, Rankin SL, Ju B, Li Y, Zhu X, Qu C, Chen X, Zhang J, Easton J, Edmonson M, Ma X, Lu C, Nagahawatte P, Hedlund E, Rusch M, Pounds S, Lin T, Onar-Thomas A, Huether R, Kriwacki R, Parker M, Gupta P, Becksfort J, Wei L, Mulder HL, Boggs K, Vadodaria B, Yergeau D, Russell JC, Ochoa K, Fulton RS, Fulton LL, Jones C, Boop FA, Broniscer A, Wetmore C, Gajjar A, Ding L, Mardis ER, Wilson RK, Taylor MR, Downing JR, Ellison DW, Zhang J, Baker SJ St. Jude Children’s Research Hospital-Washington University Pediatric Cancer Genome P. The genomic landscape of diffuse intrinsic pontine glioma and pediatric non-brainstem high-grade glioma. Nat Genet. 2014;46:444–450. doi: 10.1038/ng.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, Friedman H, Friedman A, Reardon D, Herndon J, Kinzler KW, Velculescu VE, Vogelstein B, Bigner DD. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang J, Benavente CA, McEvoy J, Flores-Otero J, Ding L, Chen X, Ulyanov A, Wu G, Wilson M, Wang J, Brennan R, Rusch M, Manning AL, Ma J, Easton J, Shurtleff S, Mullighan C, Pounds S, Mukatira S, Gupta P, Neale G, Zhao D, Lu C, Fulton RS, Fulton LL, Hong X, Dooling DJ, Ochoa K, Naeve C, Dyson NJ, Mardis ER, Bahrami A, Ellison D, Wilson RK, Downing JR, Dyer MA. A novel retinoblastoma therapy from genomic and epigenetic analyses. Nature. 2012;481:329–334. doi: 10.1038/nature10733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D, Easton J, Chen X, Wang J, Rusch M, Lu C, Chen SC, Wei L, Collins-Underwood JR, Ma J, Roberts KG, Pounds SB, Ulyanov A, Becksfort J, Gupta P, Huether R, Kriwacki RW, Parker M, McGoldrick DJ, Zhao D, Alford D, Espy S, Bobba KC, Song G, Pei D, Cheng C, Roberts S, Barbato MI, Campana D, Coustan-Smith E, Shurtleff SA, Raimondi SC, Kleppe M, Cools J, Shimano KA, Hermiston ML, Doulatov S, Eppert K, Laurenti E, Notta F, Dick JE, Basso G, Hunger SP, Loh ML, Devidas M, Wood B, Winter S, Dunsmore KP, Fulton RS, Fulton LL, Hong X, Harris CC, Dooling DJ, Ochoa K, Johnson KJ, Obenauer JC, Evans WE, Pui CH, Naeve CW, Ley TJ, Mardis ER, Wilson RK, Downing JR, Mullighan CG. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481:157–163. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang J, Wu G, Miller CP, Tatevossian RG, Dalton JD, Tang B, Orisme W, Punchihewa C, Parker M, Qaddoumi I, Boop FA, Lu C, Kandoth C, Ding L, Lee R, Huether R, Chen X, Hedlund E, Nagahawatte P, Rusch M, Boggs K, Cheng J, Becksfort J, Ma J, Song G, Li Y, Wei L, Wang J, Shurtleff S, Easton J, Zhao D, Fulton RS, Fulton LL, Dooling DJ, Vadodaria B, Mulder HL, Tang C, Ochoa K, Mullighan CG, Gajjar A, Kriwacki R, Sheer D, Gilbertson RJ, Mardis ER, Wilson RK, Downing JR, Baker SJ, Ellison DW St. Jude Children’s Research Hospital-Washington University Pediatric Cancer Genome P. Whole-genome sequencing identifies genetic alterations in pediatric low-grade gliomas. Nat Genet. 2013;45:602–612. doi: 10.1038/ng.2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.