Abstract
MicroRNA-130b (miR-130b) is a novel tumor-related miRNA that has been found to be involved in several biological processes. However, there is limited evidence regarding the role of miR-130b in the tumorigenesis of human gliomas. In the present study, reverse transcription-quantitative polymerase chain reaction (RT-qPCR) assays were used to quantify miR-130b expression levels in human glioma tissues and glioma cell lines (U251, U87, SNB19 and LN229). The expression level of miR-130b was found to be markedly higher in human glioma tissues than in non-neoplastic brain specimens. Specifically, higher expression levels of miR-130b were observed in the glioma cell lines, compared with those in normal human astrocytes (NHA). We also confirmed that miR-130b interacted with the 3′-untranslated region of peroxisome proliferator-activated receptor-γ (PPAR-γ), which negatively affected the protein levels of E-cadherin. Furthermore, its effects on cell proliferation and invasion were examined using CCK8, colony formation, cell cycle and Transwell assays. We found that the upregulation of miR-130b induced cell proliferation, decreased the percentage of cells in the G0/G1 phase and enhanced the invasiveness of U251 glioma cells whereas the downregulation of miR-130b exerted opposing effects. Moreover, it was demonstrated that the downregulation of miR-130b in U251 glioma cells restored the expression of PPAR-γ and E-cadherin, and inhibited the expression of β-catenin. Notably, PPAR-γ knockdown abolished the inhibitory effect of miR-130b inhibitor on the proliferation and invasivness of U251 cells. Taken together, these findings suggest that miR-130b promotes the proliferation and invasion of U251 glioma cells by inhibiting PPAR-γ.
Keywords: microRNA-130b, gliomas, proliferation, peroxisome proliferator-activated receptor-γ, invasion
Introduction
Malignant gliomas are the most common and deadly primary brain tumors of the central nervous system (1). Glioblastoma multiforme (GBM) is the most malignant and common type of glioma, which arises from astrocytes with poor differentiation and is associated with a median survival of approximately 10–14 months (2,3). Rapid proliferation and diffuse brain invasion are the histopathological hallmarks of these tumors and are likely to determine the unfavorable prognosis (4). Therefore, the recurrence of gliomas remains inevitable. Novel therapeutic targets, based on the identification of molecular events key to carcinogenesis and tumor progression, are necessary in order to improve the overall outcome of patients with GBM.
MicroRNAs (miRNAs or miRs) are a class of non-coding, regulatory RNA molecules, 21–24 nucleotides (nt) in length, that modulate the expression levels of specific proteins based on sequence complementarity with their target mRNA molecules (5). Accumulating evidence indicates that miRNAs are abnormally expressed in various types of cancer and act as both tumor suppressor genes and oncogenes by negatively regulating their mRNA targets either by degradation or translational repression (6–8). Therefore, it has been proposed that miRNAs may serve as novel targets for anticancer therapies.
It has been suggested that miR-130b is a novel tumor-related miRNA and it has been found to be significantly dysregulated in some types of cancer; this includes overexpression in melanoma (9), gastric cancer (10), bladder cancer (11), colorectal cancer (12), metastatic renal carcinoma (13) and glioma (14), as well as downregulation in papillary thyroid carcinomas (15), endometrial cancer (16), pituitary adenomas (17) and pancreatic cancer (18). Therefore, the functional significance of miR-130b in cancer development and progression appears to be specific to the type of cancer. Malzkorn et al identified the increased expression of miR-130b in four patients with WHO grade II primary gliomas that spontaneously progressed to WHO grade IV secondary glioblastomas (14). However, the precise role that miR-130b plays in the proliferation, differentiation and migration of glioma cells remains unknown. In this study, we observed that miR-130b expression was elevated in glioma tissues and cells. Moreover, we examined how miR-130b affects the proliferation and invasion of glioma cells as well as the mechanism responsible for the miRNA-mediated direct suppression of the peroxisome proliferator-activated receptor-γ (PPAR-γ) pathway in gliomas.
Materials and methods
Clinical samples and cell lines
Human glioma tumor tissue samples were obtained from patients undergoing surgical resection at the Department of Neurosurgery at Fuzhou General Hospital (Xiamen University Medical College, Fuzhou, China) in accordance with procedures approved by the Ethics Committee of our hospital and the study was performed according to the Declaration of Helsinki. Written informed consent was obtained from the family of each patient. Twelve glioma samples were thoroughly reviewed by an experienced neuropathologist according to the 2007 WHO classification, which classifies astrocytomas as i) well-differentiated low-grade diffuse astrocytoma (WHO grade II, 4 cases), ii) anaplastic astrocytoma (WHO grade III, 4 cases) and iii) glioblastoma multiforme (GBM; WHO grade IV, 4 cases). In addition, four non-neoplastic brain specimens were obtained from patients with traumatic brain injury at the site of decompression. All tissue samples were frozen in liquid nitrogen immediately after resection and were stored at −80°C. Normal human astrocytes (NHAs) were purchased from ScienCell Research Laboratories (Carlsbad, CA, USA) in 2013. The glioma cell lines U251, U87, SNB19 and LN229 that were used in this study were obtained from the Institute of Biochemistry and Cell Biology (Shanghai Institutes for Biological Sciences, Chinese Academy of Science, Shanghai, China).
Cell culture and transfection
The cells were grown in Dulbecco's modified Eagle's medium (DMEM; Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS; Invitrogen) and 1% penicillin/streptomycin (Invitrogen/Thermo Fisher Scientific Inc., Waltham, MA, USA) in 5% CO2 atmosphere at 37°C. The U251 cells (1×105) were seeded into 6-well plates and transfected with either the negative control (NC), miR-130b mimic, miR-130b inhibitor or siPPAR-γ (5′-AAUAUGACCUGAAGCUCCAAGAAUAAG-3′) which were purchased from GenePharma (Shanghai, China), using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Following a 24 h transfection, the media were removed and the cells were placed in complete medium and maintained at 37°C in an atmosphere of 5% CO2.
RNA extraction and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from the cultured cells and fresh glioma tissues using TRIzol reagent (Invitrogen) and total miRNAs were extracted using mirVana kits (Ambion, Austin, TX, USA) according to the manufacturer's instructions. Gene-specific primers were used to synthesize miR-130b cDNA from total RNA, according to the miRNA-specific TaqMan miRNA assay kit (Applied Biosystems, Foster City, CA, USA); U6 snRNA was used as an internal control. The expression levels of miR-130b and PPAR-γ were examined by performing qPCR with an SYBR-Green PCR Master Mix kit in conjunction with an ABI PRISM 7300 system (both from Applied Biosystems). The primer sequences were as follows: PPAR-γ forward, 5′-CATGCTTGTGAAGGATGCAAG-3′ and reverse, 5′-CCCATCATTAAGGAATTCATGTC-3′; GAPDH forward, 5′-TCGGAGTCAACGGATTTGG-3′ and reverse, 5′-CATGGGTGGAATCATATTGGA-3′.
Western blot analysis
Total cell lysates from different experiments were obtained by lysing the cells in RIPA buffer. The protein concentration was determined using a BCA Protein assay kit (Pierce Biotechnology, Rockford, IL, USA). Forty micrograms of protein from each sample were resolved by 10% SDS-PAGE gel and transferred to PVDF membranes (Millipore, Billerica, MA, USA). The membranes were further incubated with primary antibodies PPAR-γ (1:200 dilution), E-cadherin (1:500 dilution) and β-catenin (1:200 dilution) (all from Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) followed by incubation with an HRP-conjugated secondary antibody (1:1,000 dilution; Zymed, San Diego, CA, USA). The membranes were stripped and reprobed with a primary antibody against GAPDH. The signal intensity was determined using gel analysis software, ImageJ. GAPDH was used as an endogenous protein for normalization (1:1,000 dilution; Santa Cruz Biotechnology, Inc.).
Dual luciferase reporter assay
The 3′-untranslated region (3′-UTR) sequence of PPAR-γ predicted to interact with miR-130b was identified using TargetScan (http://www.targetscan.org) as well as a mutated sequence with the predicted target sites. They were synthesized and inserted into the XbaI and FseI sites of a pGL3 control vector (Promega Corp., Madison, WI, USA). The cells (8×103) were seeded into a 24-well plate. The cells in each well were co-transfected with either the miR-130b mimic, miR-130b inhibitor or NC. The transfections were performed using the FuGENE HD transfection reagent (Promega Corp.). A Renilla luciferase vector pRL-SV50 (Promega Corp.) was co-transfected to normalize the differences in transfection efficiency. Following a 24 h transfection, the cells were harvested and assayed using the Dual-Luciferase Reporter assay system (Promega Corp.) according to the manufacturer's instructions. Transfection was repeated in triplicate in three independent experiments.
Proliferation assays
The cells were plated at 5×103 cells/well in 96-well plates and were grown for 24, 48 and 72 h after transfection, according to the manufacturer's instructions. Cell proliferation was documented every 24 h for four days using a cell counting kit-8 (CCK8) assay (Dojindo, Tokyo, Japan). The absorbance at a wavelength of 570 nm was detected using a microplate reader (Thermo Fisher Scientific Inc.).
Clonogenicity assay
Transfected glioma U251 cells (1×103) were seeded into 6-well plates and cultured in cell culture medium for 2 weeks to allow colony formation. The culture medium was changed every third day. The colonies were then fixed in 100% methanol for 5 min and stained with 1.0% crystal violet solution (BioLab, Inc., Shanghai, China) for 30 sec. The number of macroscopically detectable colonies was registered.
Cell cycle assay
The cells were harvested by trypsinization 48 h after transfection. They were washed three times with ice-cold phosphate-buffered saline (PBS) and fixed with 70% ethanol overnight at 4°C. The fixed cells were rehydrated in PBS and subjected to propidium iodide/RNase staining followed by fluorescence-activated cell sorting (FACS; Becton-Dickinson, Mountain View, CA, USA). The percentage of cells in each phase of the cell cycle was estimated using PV Elite software (Intergraph Corp., Madison, AL, USA).
Transwell assay and scratch-wound assay
Cell invasion and migration was examined using Transwell and scratch-wound assays, respectively. For the Transwell assay, the appropriate oligonucleotides were transfected into the cells as described above. Following incubation for 48 h, 3×104 cells were transferred to the top of the Matrigel-coated invasion chambers (BD Biosciences, San Jose, CA, USA) in serum-free DMEM. DMEM containing 10% FBS was added to the lower chamber. After 24 h, the non-invading cells were removed, and the invading cells were fixed using 95% ethanol, stained with 0.1% crystal violet and images were captured at ×100 magnification under an inverted phase contrast microscope (Olympus CKX31/41; Olympus, Tokyo, Japan). The experiments were repeated three times independently. For the scratch-wound assay, the miR-130b mimic, miR-130b inhibitor or NC, were transfected into the cells in 6-well plates. The cell layers were then scratched using a 200 µl sterile pipette tip to form wound gaps. The wound location in the 6-well plates was marked. Images of the cells were captured to record the wound width at the marked wound locations at 0 and 24 h in order to measure the migratory ability of the cells.
Statistical analysis
All data are presented as the means ± SD. The experiments were repeated three times. All statistical analyses were performed using a two-tailed Student's t-test in SPSS 12.0 software. A P-value of <0.05 was considered to indicate a statistically significant difference.
Results
miR-130b is overexpressed in glioma tissues and cell lines
The expression of miR-310b in 12 glioma tissues and 4 non-neoplastic brain specimens was examined by RT-qPCR. The expression of miR-310b was significantly higher in the glioma tissues in comparison with that in the 4 non-neoplastic brain specimens, particularly in the grade III/IV tissues (Fig. 1A). Compared with miR-310b expression in the NHAs, it was higher in the glioma cell lines U251, U87, SNB19 and LN229 (P<0.05) (Fig. 1B). Taken together, these findings indicate that miR-310b is overexpressed in glioma tissues and glioma cell lines.
Figure 1.
miR-310b expression in glioma tissues and glioma cell lines. (A) miR-310b expression was detected in 4 non-neoplastic brain specimens and 12 glioma tissues by RT-qPCR. miR-310b expression was significantly higher in the glioma tissues compared with that in the non-neoplastic brain specimens. *P<0.05 vs. non-neoplastic brain specimens. (B) miR-310b expression was detected in several glioma cell lines (U251, U87, SNB19 and LN229) and primary normal human astrocytes (NHAs). RT-qPCR revealed that miR-310b was upregulated in all of the glioma cell lines compared with the NHA cells. *P<0.05 vs. NHAs.
miR-310b directly targets PPAR-γ in glioma cells
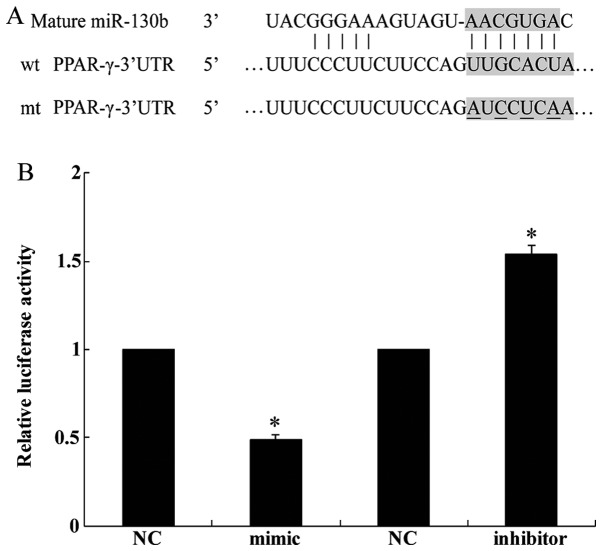
We identified the tumor suppressor gene PPAR-γ as a potential target of miR-130b by using the publicly available algorithm TargetScan (http://www.targetscan.org) (Fig. 2A). A dual luciferase reporter assay was performed in order to verify that the 3′-UTR of PPAR-γ mRNA is a direct target of miR-130b. The luciferase activity of the U251 cells transfected with miR-130b mimic was reduced when compared with the cells transfected with NC whereas miR-130b inhibitor increased the luciferase activity of the U251 cells (Fig. 2B). Collectively, this experiment demonstrated that PPAR-γ is a direct target of miR-310b.
Figure 2.
Identification of peroxisome proliferator-activated receptor-γ (PPAR-γ) as the target of miR-130b. (A) Identification of target sites in the 3′-UTR of PPAR-γ was performed using TargetScan. (B) A dual luciferase reporter assay was used to determine the target relationship in U251 cells. Luciferase activity was evaluated as described in the Materials and methods. Representative of at least three independent experiments. *P<0.05. NC, negative control; mimic, miR-130b mimic; inhibitor, miR-130b inhibitor.
miR-130b regulates the expression of PPAR-γ, E-cadherin and β-catenin
We performed western blot analysis and RT-qPCR in order to elucidate the possible molecular mechanism through which miR-130b exerts effects in glioma cells. The results of western blot analysis revealed that the transfection of U251 cells with the miR-130b mimic significantly reduced the expression of PPAR-γ and E-cadherin whereas it increased the expression of β-catenin (Fig. 3A). Conversely, the protein expression levels of PPAR-γ and E-cadherin were increased whereas those of β-catenin were decreased when endogenous miR-130b was downregulated by the miR-130b inhibitor (Fig. 3A). However, the results of RT-qPCR revealed that the mRNA expression of PPAR-γ was unaffected by the miR-130b mimic and inhibitor (Fig. 3B). These results demonstrate that the overexpression of miR-130b negatively regulates the protein levels but not the mRNA levels of PPAR-γ, indicating that regulation occurs at the translational level.
Figure 3.
miR-130b regulates the expression of peroxisome proliferator-activated receptor-γ (PPAR-γ), E-cadherin and β-catenin. (A) U251 cells were transfected with the neagtive control (NC), miR-310b mimics (mimic) and miR-310b inhibitor (inhibitor). The levels of PPAR-γ, E-cadherin, and β-catenin proteins were measured by western blot analysis. (B) The levels of PPAR-γ mRNA in U251 cells were measured by RT-qPCR.
miR-130b regulates the proliferation of U251 cells
We performed the cell proliferation assay in order to explore the effect of miR-130b upregulation and downregulation on glioma cell proliferation in vitro. The cell proliferation rate was clearly increased in the miR-130b mimic-transfected group at 48 and 72 h compared with the NC groups (P<0.05) (Fig. 4A). By contrast, the cell proliferation rate was inhibited following transfection with the miR-130b inhibitor (P<0.05) (Fig. 4A). In Fig. 4B and C, stable overexpression of miR-130b markedly increased the number of surviving colonies, whereas the miR-130b inhibitor reduced the number of surviving colonies formed by U251 cells compared with the NC groups (P<0.05). This finding indicates that miR-130b significantly enhances the proliferation of glioma cells. As shown in Fig. 4D, FACS revealed that miR-130b overexpression increased the percentage of cells in the S phase and significantly decreased the percentage of cells in the G1/G0 (P<0.05). By contrast, the miR-130b inhibitor decreased the percentage of cells in the S phase and significantly increased the percentage of cells in the G1/G0 phase (P<0.05).
Figure 4.
Effect of miR-130b on the proliferation of U251 cells in vitro. (A) U251 cell proliferation was significantly induced by transfection with miR-130b mimic (mimic) and inhibited by transfection with miR-130b inhibitor (inhibitor) at 48 and 72 h. (B and C) Upregulation of miR-130b markedly increased the number of surviving colonies and downregulation of miR-130b reduced the number of surviving colonies of U251 cells compared with the negative control (NC) groups. (D) The results of the cell cycle assay revealed that the upregulation of miR-130b decreased the percentage of cells in the G0–G1 phase and downregulation of miR-130b increased the percentage of cells in the G0–G1 phase. Data are means + SD of 3 independent experiments. *P<0.05 vs. NC group; #P<0.05 vs. NC group.
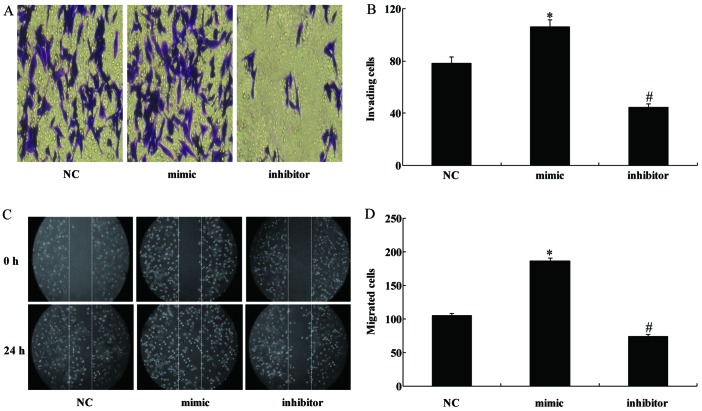
miR-130b regulates the migration and invasion of U251 cells in vitro
As invasiveness is one of the pathophysiological features of malignant human gliomas, the effects of miR-130b on the invasiveness and migration of glioma cells were examined by Transwell and scratch-wound assays, respectively. The former showed that the overexpression of miR-130b significantly enhanced the invasive ability of U251 cells whereas the downregulation of miR-130b significantly decreased the number of cells capable of invasion (Fig. 5A and B). The scratch-wound assay also demonstrated that the miR-130b mimic enhanced the invasiveness of U251 cells and the miR-130 inhibitor inhibited the invasiveness of the cells (Fig. 5C and D).
Figure 5.
miR-310b regulates the migration and invasion of U251 cells. (A and B) U251 cells were examined for cell invasion in 24-well plates with Transwell chambers. Migrated cells were stained with crystal violet. The invasiveness of U251 cells was enhanced by the upregulation of miR-310b and reduced by the downregulation of miR-310b. (C and D) The wound was photographed at 0 and 24 h, and wound gaps were analyzed by measuring the distance of migrating cells from three different areas of each wound. The number of cells migrated from the edge of the scratch was recorded. *P<0.05 vs. NC group; #P<0.05 vs. NC group.
PPAR-γ siRNA imitates the role of miR-130b in U251 glioma cells
To examine the role of PPAR-γ in the miR-130b-dependent cell proliferation and invasion, the miR-130b inhibitor was transfected into the U251 cells treated with PPAR-γ siRNA. As expected, PPAR-γ siRNA co-transfected with the miR-130b inhibitor decreased the expression levels of PPAR-γ (Fig. 6A). The effect of the miR-130b inhibitor on cell invasion (Fig. 6B and C) and proliferation (Fig. 6D) was reversed by the silencing of PPAR-γ expression. These results indicate that PPAR-γ plays an essential role in miR-130b-dependent cell proliferation and invasion.
Figure 6.
Peroxisome proliferator-activated receptor-γ (PPAR-γ) siRNA imitate the role of miR-130b in U251 glioma cells. (A) Western blot analysis revealed that the protein level of PPAR-γ was significantly increased when U251 cells were transfected with miR-310b inhibitor whereas PPAR-γ siRNA co-transfected with the miR-130b inhibitor decreased PPAR-γ expression. (B and C) Transwell assays showed that miR-130b inhibitor reduced the invasiveness of U251 cells and PPAR-γ siRNA promoted the invasion.*P<0.05; #P<0.05.(D) PPAR-γ siRNA rescued the inhibited proliferation of U251 cells induced by miR-130b inhibitor at 48 and 72 h. *P<0.05 vs. NC group; #P<0.05 vs. inhibitor group.
Discussion
Gliomas are the most common primary tumors of the central nervous system with glioblastomas as the most malignant entity (1). Despite progress in glioma therapy regimens, such as surgery, radiation, chemotherapy or combined modalities, the prognosis for malignant glioma patients remains dismal (19), and most patients with glioma die within two years of diagnosis (20). Therefore, extensive study of the biological characteristics of gliomas is necessary in order to identify an effective treatment capable of suppressing the invasiveness of glioma cells. Over the last few years, miRNAs have emerged as a new class of gene regulators which are associated with different malignancies. For example, miR-222 promoted the proliferation of epithelial ovarian cancer cells by downregulating p27Kip1 (21). miR-218 reversed the highly invasive nature of glioblastoma cells by targeting the oncogenic transcription factor LEF1 (22).
miRNAs are emerging as a novel class of regulatory molecules involved in numerous biological processes (23,24). They regulate gene expression at the transcriptional or translational level by binding to the 3′-UTR of mRNAs (25). miRNA deregulation is a common feature of human malignancies as they control the expression of oncogenes or tumor suppressors by acting as onco-miRNAs or tumor suppressor miRNAs (24). Previous studies have demonstrated the association between miR-130b with some types of solid tumors. For example, Yu et al demonstrated that miR-130b is significantly overexpressed in gastric cancer, which increased the proliferation of esophageal squamous cell carcinoma cells and enhanced their ability to migrate and invade through interactions with the 3′-UTR of phosphatase and tensin homolog (PTEN) to downregulate PTEN expression (26). On the contrary, it has been shown that miR-130b is downregulated in ovarian cancer and papillary thyroid carcinoma, and its expression inversely correlates with tumor aggressiveness and multidrug resistance in these types of cancer (15,27). In recent years, miR-130b has been found to be overexpressed in colorectal cancer and hepatocellular carcinoma (28). It has also been observed that miR-130b represses the expression of PPAR-γ protein by targeting the PPAR-γ 3′-UTR, leading to enhancement of the oncogenic capacities of cancer cells (12,28). In the present study, we used RT-qPCR to confirm that miR-130b is overexpressed in glioma tissues and cells. Furthermore, upregulated miR-130b induced cell proliferation, decreased the percentage of cells in the G0/G1 phase, and enhanced the invasiveness of U251 glioma cells whereas these abilities were weakened when miR-130b was inhibited. PPAR-γ was identified as a direct functional target of miR-130b using bioinformatics analysis, and this finding was experimentally confirmed by performing a dual luciferase reporter assay.
PPAR-γ is a ligand-activated transcription factor that belongs to the superfamily of hormone receptors (29). It is abundantly expressed in many cell types, where it regulates lipid metabolism, glucose homeostasis, tumor progression and inflammation (30). Some studies have verified that PPAR-γ is implicated in epithelial cell differentiation and anti-proliferative responses, acting as a tumor suppressor (31,32). It has also been demonstrated that PPAR-γ is frequently downregulated in human glioma cell lines including SWO-38 and U251 glioma cells (33), and PPAR-γ agonists induce growth arrest and apoptosis in glioma cells, suggesting that they may be suitable for use in the treatment of brain tumors (34). A previous study has proposed that the PPAR-signaling pathway is connected to the β-catenin pathway; β-catenin is a multifunctional, intracellular protein that binds to either E-cadherin or APC proteins; β-catenin forms a complex with the transmembrane receptor E-cadherin, becomes plasma membrane-associated and mediates intercellular adhesion (35). Wan et al found that the PPAR-γ agonist pioglitazone not only suppressed the proliferation and migration of U251 glioma cells and induced cell apoptosis, but also decreased the expression level of the β-catenin protein. The knockdown of β-catenin expression mimicked the anti-neoplastic potency of pioglitazone. These findings suggest that the PPAR-γ/β-catenin signaling pathway plays a key role in the development of glioma (36). The present study demonstrated that miR-130b binds to the 3′-UTR of PPAR-γ mRNA and downregulates the protein expression of PPAR-γ and E-cadherin and increases the expression of β-catenin. Furthermore, the silencing of PPAR-γ expression reversed the effect of the miR-130b inhibitor on cell proliferation and invasion.
Taken together, our findings reveal that miR-130b expression is markedly upregulated in glioma tissues and cells. PPAR-γ is a direct functional target of miR-130b. miR-130b repressed PPAR-γ expression which downregulated E-cadherin and increased the expression of β-catenin, thereby promoting the proliferation and invasion of glioma cells in vitro. These results suggest that miR-130b plays a critical role in promoting the development and progression of glioma, and the downregulation of miR-130b may be a useful treatment strategy for the management of glioma.
References
- 1.Van Meir EG, Hadjipanayis CG, Norden AD, Shu HK, Wen PY, Olson JJ. Exciting new advances in neuro-oncology: the avenue to a cure for malignant glioma. CA Cancer J Clin. 2010;60:166–193. doi: 10.3322/caac.20069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wiedemeyer R, Brennan C, Heffernan TP, Xiao Y, Mahoney J, Protopopov A, Zheng H, Bignell G, Furnari F, Cavenee WK, et al. Feedback circuit among INK4 tumor suppressors constrains human glioblastoma development. Cancer Cell. 2008;13:355–364. doi: 10.1016/j.ccr.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coras R, Hölsken A, Seufert S, Hauke J, Eyüpoglu IY, Reichel M, Tränkle C, Siebzehnrübl FA, Buslei R, Blümcke I, Hahnen E. The peroxisome proliferator-activated receptor-γ agonist troglitazone inhibits transforming growth factor-β-mediated glioma cell migration and brain invasion. Mol Cancer Ther. 2007;6:1745–1754. doi: 10.1158/1535-7163.MCT-06-0763. [DOI] [PubMed] [Google Scholar]
- 5.Wu Z, He B, He J, Mao X. Upregulation of miR-153 promotes cell proliferation via downregulation of the PTEN tumor suppressor gene in human prostate cancer. Prostate. 2013;73:596–604. doi: 10.1002/pros.22600. [DOI] [PubMed] [Google Scholar]
- 6.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 7.Kent OA, Mendell JT. A small piece in the cancer puzzle: microRNAs as tumor suppressors and oncogenes. Oncogene. 2006;25:6188–6196. doi: 10.1038/sj.onc.1209913. [DOI] [PubMed] [Google Scholar]
- 8.Wu Z, Han Y, Li Y, Li X, Sun T, Chen G, Huang Y, Zhou Y, Du Z. MiR-218-5p inhibits the stem cell properties and invasive ability of the A2B5+CD133− subgroup of human glioma stem cells. 2016;35:869–877. doi: 10.3892/or.2015.4418. [DOI] [PubMed] [Google Scholar]
- 9.Sand M, Skrygan M, Sand D, Georgas D, Gambichler T, Hahn SA, Altmeyer P, Bechara FG. Comparative microarray analysis of microRNA expression profiles in primary cutaneous malignant melanoma, cutaneous malignant melanoma metastases, and benign melanocytic nevi. Cell Tissue Res. 2013;351:85–98. doi: 10.1007/s00441-012-1514-5. [DOI] [PubMed] [Google Scholar]
- 10.Lai KW, Koh KX, Loh M, Tada K, Subramaniam MM, Lim XY, Vaithilingam A, Salto-Tellez M, Iacopetta B, Ito Y, et al. Singapore Gastric Cancer Consortium: MicroRNA-130b regulates the tumour suppressor RUNX3 in gastric cancer. Eur J Cancer. 2010;46:1456–1463. doi: 10.1016/j.ejca.2010.01.036. [DOI] [PubMed] [Google Scholar]
- 11.Scheffer AR, Holdenrieder S, Kristiansen G, von Ruecker A, Müller SC, Ellinger J. Circulating microRNAs in serum:novel biomarkers for patients with bladder cancer? World J Urol. 2014;32:353–358. doi: 10.1007/s00345-012-1010-2. [DOI] [PubMed] [Google Scholar]
- 12.Colangelo T, Fucci A, Votino C, Sabatino L, Pancione M, Laudanna C, Binaschi M, Bigioni M, Maggi CA, Parente D, et al. MicroRNA-130b promotes tumor development and is associated with poor prognosis in colorectal cancer. Neoplasia. 2013;15:1218–1231. doi: 10.1593/neo.13998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu X, Weng L, Li X, Guo C, Pal SK, Jin JM, Li Y, Nelson RA, Mu B, Onami SH, et al. Identification of a 4-microRNA signature for clear cell renal cell carcinoma metastasis and prognosis. PLoS One. 2012;7:e35661. doi: 10.1371/journal.pone.0035661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malzkorn B, Wolter M, Liesenberg F, Grzendowski M, Stühler K, Meyer HE, Reifenberger G. Identification and functional characterization of microRNAs involved in the malignant progression of gliomas. Brain Pathol. 2010;20:539–550. doi: 10.1111/j.1750-3639.2009.00328.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yip L, Kelly L, Shuai Y, Armstrong MJ, Nikiforov YE, Carty SE, Nikiforova MN. MicroRNA signature distinguishes the degree of aggressiveness of papillary thyroid carcinoma. Ann Surg Oncol. 2011;18:2035–2041. doi: 10.1245/s10434-011-1733-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dong P, Karaayvaz M, Jia N, Kaneuchi M, Hamada J, Watari H, Sudo S, Ju J, Sakuragi N. Mutant p53 gain-of-function induces epithelial-mesenchymal transition through modulation of the miR-130b-ZEB1 axis. Oncogene. 2013;32:3286–3295. doi: 10.1038/onc.2012.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leone V, Langella C, D'Angelo D, Mussnich P, Wierinckx A, Terracciano L, Raverot G, Lachuer J, Rotondi S, Jaffrain-Rea ML, et al. Mir-23b and miR-130b expression is downregulated in pituitary adenomas. Mol Cell Endocrinol. 2014;390:1–7. doi: 10.1016/j.mce.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 18.Zhao G, Zhang JG, Shi Y, Qin Q, Liu Y, Wang B, Tian K, Deng SC, Li X, Zhu S, et al. MiR-130b is a prognostic marker and inhibits cell proliferation and invasion in pancreatic cancer through targeting STAT3. PLoS One. 2013;8:e73803. doi: 10.1371/journal.pone.0073803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walbert T, Chasteen K. Palliative and supportive care for glioma patients. Cancer Treat Res. 2015;163:171–184. doi: 10.1007/978-3-319-12048-5_11. [DOI] [PubMed] [Google Scholar]
- 20.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 21.Sun C, Li N, Zhou B, Yang Z, Ding D, Weng D, Meng L, Wang S, Zhou J, Ma D, Chen G. miR-222 is upregulated in epithelial ovarian cancer and promotes cell proliferation by downregulating P27(kip1) Oncol Lett. 2013;6:507–512. doi: 10.3892/ol.2013.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Y, Yan W, Zhang W, Chen L, You G, Bao Z, Wang Y, Wang H, Kang C, Jiang T. MiR-218 reverses high invasiveness of glioblastoma cells by targeting the oncogenic transcription factor LEF1. Oncol Rep. 2012;28:1013–1021. doi: 10.3892/or.2012.1902. [DOI] [PubMed] [Google Scholar]
- 23.Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol. 2008;9:219–230. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- 24.Garzon R, Calin GA, Croce CM. MicroRNAs in cancer. Annu Rev Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 25.Esquela-Kerscher A, Slack FJ. Oncomirs – microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 26.Yu T, Cao R, Li S, Fu M, Ren L, Chen W, Zhu H, Zhan Q, Shi R. MiR-130b plays an oncogenic role by repressing PTEN expression in esophageal squamous cell carcinoma cells. BMC Cancer. 2015;15:29. doi: 10.1186/s12885-015-1031-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang C, Cai J, Wang Q, Tang H, Cao J, Wu L, Wang Z. Epigenetic silencing of miR-130b in ovarian cancer promotes the development of multidrug resistance by targeting colony-stimulating factor 1. Gynecol Oncol. 2012;124:325–334. doi: 10.1016/j.ygyno.2011.10.013. [DOI] [PubMed] [Google Scholar]
- 28.Tu K, Zheng X, Dou C, Li C, Yang W, Yao Y, Liu Q. MicroRNA-130b promotes cell aggressiveness by inhibiting peroxisome proliferator-activated receptor gamma in human hepatocellular carcinoma. Int J Mol Sci. 2014;15:20486–20499. doi: 10.3390/ijms151120486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kliewer SA, Xu HE, Lambert MH, Willson TM. Peroxisome proliferator-activated receptors: from genes to physiology. Recent Prog Horm Res. 2001;56:239–263. doi: 10.1210/rp.56.1.239. [DOI] [PubMed] [Google Scholar]
- 30.Pestereva E, Kanakasabai S, Bright JJ. PPARγ agonists regulate the expression of stemness and differentiation genes in brain tumour stem cells. Br J Cancer. 2012;106:1702–1712. doi: 10.1038/bjc.2012.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Michalik L, Desvergne B, Wahli W. Peroxisome-proliferator-activated receptors and cancers: complex stories. Nat Rev Cancer. 2004;4:61–70. doi: 10.1038/nrc1254. [DOI] [PubMed] [Google Scholar]
- 32.Drori S, Girnun GD, Tou L, Szwaya JD, Mueller E, Xia K, Shivdasani RA, Spiegelman BM. Hic-5 regulates an epithelial program mediated by PPARgamma. Genes Dev. 2005;19:362–375. doi: 10.1101/gad.1240705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang MH, Zhong XY, Lin CL, Xie YK, Jia JP, Li SM, Mi C. Expression of peroxisome proliferator-activated receptor gamma in glioma. Nan Fang Yi Ke Da Xue Xue Bao. 2008;28:444–446. In Chinese. [PubMed] [Google Scholar]
- 34.Papi A, Tatenhorst L, Terwel D, Hermes M, Kummer MP, Orlandi M, Heneka MT. PPARgamma and RXRgamma ligands act synergistically as potent antineoplastic agents in vitro and in vivo glioma models. J Neurochem. 2009;109:1779–1790. doi: 10.1111/j.1471-4159.2009.06111.x. [DOI] [PubMed] [Google Scholar]
- 35.Jansson EA, Are A, Greicius G, Kuo IC, Kelly D, Arulampalam V, Pettersson S. The Wnt/β-catenin signaling pathway targets PPARgamma activity in colon cancer cells. Proc Natl Acad Sci USA. 2005;102:1460–1465. doi: 10.1073/pnas.0405928102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wan Z, Shi W, Shao B, Shi J, Shen A, Ma Y, Chen J, Lan Q. Peroxisome proliferator-activated receptor γ agonist pioglitazone inhibits β-catenin-mediated glioma cell growth and invasion. Mol Cell Biochem. 2011;349:1–10. doi: 10.1007/s11010-010-0637-9. [DOI] [PubMed] [Google Scholar]