Abstract
FcγRIIIa (CD16) is a low-affinity Fc receptor of IgG. As the idio-binding receptor of IgG Fc, it plays an important role in the antibody-dependent cellular cytotoxicity of natural killer cells. The aim of the present study was to investigate the distribution of Kupffer cells (KCs) and the expression of their surface receptor FcγRIIIa in hepatocellular carcinoma. Furthermore, we also aimed to observe the functional mechanism of FcγRIIIa. Immunohistochemical analysis was employed to study KCs and FcγRIIIa. In order to explore the role of FcγRIIIa in the growth of cancer cells, KCs and H22 tumor cells were co-cultured in different serum. The mRNA expression levels of tumor necrosis factor (TNF)-α and FcγRIIIa were analyzed by RT-qPCR; the TNF-α and FcγRIIIa protein expression levels were examined by enzyme-linked immunosorbent assay and western blot analysis, respectively. Our results showed that the number of Kuppfer cells in cancerous tissues (21.6±7.8) was lower than those in para-cancerous (68.8±9.1) tissues and adjacent normal hepatic tissues (62.0±1.9) (P<0.01); this decreased with the reduction in the differentiation degree of cancer (P<0.05). FcγRIIIa-positive cells were similar in morphology to KCs, and their distributive tendency was coincident (P<0.05). The increase in CD16a mRNA levels in the group treated with immune serum was 3.9-, 4.9- and 3.9-fold greater than that in the ordinary serum group at different time points, and CD16a protein expression also markedly increased (P<0.05). However, these effects were inhibited by the addition of anti-IgG Fc serum (P<0.05). The results of the present study suggested that FcγRIIIa resided in KCs, and it contributed to the inhibition of the growth of liver tumor cells.
Keywords: FcγRIIIa, Kupffer cells, hepatocellular carcinoma
Introduction
Hepatocellular carcinoma (HCC) is the fifth most common type of cancer and the third most common cause of cancer-related death globally, and it is characterized by high malignancy and poor prognosis (1). However, lack of sensitivity to therapy and multidrug resistance have resulted in poor efficacy of these treatments (2). Cancer immunotherapy has been a major research focus and has been highlighted by gene transfer, enhancing T-cell receptor-recognizing tumor antigens and the effect of antibody-dependent cellular cytotoxicity (ADCC). However, cancer immunotherapy has also failed previously, suggesting that cancer immunotherapy acts very differently from immunotherapy against infectious diseases (3). Finding more effective ways of administering immunotherapy is thus a priority.
Macrophages play an important role in immune defense. Activated macrophages suppress tumor cells through the ADCC pathway via FcγRIII (CD16) and directly or indirectly by cytokines including tumor necrosis factor (TNF)-α and interleukin (IL)-1 or certain enzymes (4). As part of the mono-nuclear phagocytic system, Kupffer cells (KCs) are the main type of hepatic macrophages, and they play an important role in the inhibition of liver tumors and their surface receptors are the mediator in this pathway (5).
CD16 is important for specific and non-specific immunity, and includes two forms, CD16a and CD16b: CD16a is the functional form of CD16 while CD16b is ancillary (6). As the specific receptor of IgG Fc, CD16a forms a bridge between cytoimmunity and humoral immunity, and has an important relationship with the maturation of dendritic cells (DCs) (7). Antibody-based protein scaffolds, immunoglobulin Fc fragments with antigen binding (Fcab) sites, have been shown to hold significant promise as next-generation protein therapeutics; in particular, Fcab (CD16a) molecules have been shown to mediate ADCC reactivity (8). Previous research has shown that the correlation between CD16a binding affinity and ADCC potency is also valid in Fcab proteins and that antigen specific Fcab molecules can be further engineered for fine tuning of immuno-effector functions (9).
CD16a functions via myeloid cells and natural killer (NK) cells, and mediates the ADCC of the mononucleus macrophage system, which is Mg2+-dependent, to kill target cells including tumor cells (10). However, the relationship between CD16a, KCs and HCC was not fully understood. The aim of the present study was to investigate whether CD16a is present in KCs in HCC. Furthermore, the function and variance of CD16a and also the ADCC pathway were investigated using cultured tumor cells for in vitro analysis.
Materials and methods
Patients and specimens
Samples of cancerous tissues, para-cancerous tissues (<10 mm distance from the tumor) and adjacent normal hepatic tissues (>10 mm distance from the tumor) (11) were obtained from 87 patients with primary HCC, in which there was no macroscopic tumor thrombi or satellite nodules and the diameter of the tumor was <10 cm. A total of 39 normal liver samples were obtained from patients who underwent hepatectomy as a result of injury in the Department of Hepatobiliary Surgery, Second Clinical College, Chongqing University of Medical Science (Yuzhong, China). The surgically resected tissues were fixed in 10% formalin, embedded in paraffin, cut into 5-mm sections and stained with hematoxylin and eosin. Histopathological diagnosis and classification were performed by the same pathologist. All specimens were handled and made anonymous according to relevant ethical and legal standards. The experimental protocols for this study were approved by the Human Research Ethical Committee and the Animal Research Ethics Committee of Chongqing Medical University, Chongqing, China. All patients provided written informed consent prior to obtaining the samples.
Experimental animals and cell line
Healthy adult male Kunming mice (20–25 g, n=48), which were purchased from the Center of Experimental Animals, Chongqing Medical University, were used in this study. The mice were kept in individual ventilated cages and were allowed access to food and water ad libitum. The mice were used accordingly for the different experiments (for the isolation of the KCs and hepatic cells, for the transplantatoin of H22 cells, and for the harvesting of ordinary serum and immune serum; 12 mice were used for each of these 4 procedures). All animals received humane care in accordance with the National Institutes of Health Guidelines and the legal requirements in China. The mouse ascitic H22 tumor cell line (HCC line) was kindly provided by Dr Ma Fang, the Institute of Ultrasonic Medical Engineering, Chongqing Medical University.
Reagents and antibodies
Collagenase IV, Percoll, special grade fetal calf serum, gadolinium chloride (GaCl3) and DMEM were all purchased from Sigma (St. Louis, MO, USA). Mouse anti-human monoclonal antibody-CD68 (sc-20060), rabbit anti-mouse CD16a polyclonal antibody (sc-20627) (both from Santa Cruz Biotechnology, Santa Cruz, CA, USA), biotinylated anti-mouse TNF-α (BS1857) and mouse anti-IgG Fc immune serum (BS9203P) (both from Bioworld, St. Louis Park, MN, USA).
Immunohistochemical analysis
The SP method was used, and the primary antibodies were mouse monoclonal anti-human CD68 (12) and CD16a (Santa Cruz Biotechnology, dilution 1:200). The operation procedure was undertaken according to the instructions of the SP kit (Santa Cruz Biotechnology). DAB was used for coloration. The brown granules in the cytoplasm were taken as CD68- and CD16a-positive signals. Negative mouse control serum and PBS were used to replace the primary antibody as negative control and blank control, respectively.
Identification and counting of positive cells
Among the cells which were positive for the anti-CD68 antibody, cells in the cancerous tissue or the sinusoids of non-cancerous tissues with a stellate or spindle shape were taken as KCs, as previously discussed (12). The cells which were positive for the anti-CD16a antibody were brown, and the shape was either stellate or spindle. Image Tool 3.0 software was employed to count cells at ×200 magnification (CKX 41SF; Olympus, Tokyo, Japan) in one image randomly selected for each specimen. The expression level of CD16a was determined as the average optical density, which was measured by the Beihang CM-2000B Biological Image Analysis System (Beihang University, Beijing, China).
Preparation of KCs
KCs were isolated from mouse livers (6 mice were sacrificed for this procedure) by collagenase digestion and differential centrifugation using Percoll as previously described by Liu et al (13). Briefly, the livers were excised after perfusion via the portal vein with Ca2+ and Mg2+-free Hanks' balanced salt solution containing 0.05% collagenase IV (Sigma) at 37°C and cut into small pieces in collagenase buffer. The suspension was filtered through nylon gauze, and the filtrate was centrifuged at 500 × g for 10 min at 4°C. Cell pellets were resuspended in buffer, parenchymal cells were removed by centrifugation at 50 × g for 3 min, and the non-parenchymal cells were centrifuged on a 70:30% Percoll gradient (Sigma) at 800 × g at 4°C for 20 min. KCs concentrated at the interface of the 30 and 70% were collected and cultured at a density of 1×106 in 24-well culture plates containing DMEM supplemented with 10% FBS and antibiotics (100 U/ml of penicillin G and 100 mg/ml of streptomycin sulphate) at 37°C in the presence of 5% CO2. Nonadherent cells were removed after 1 h by replacing the buffer. All adherent cells phagocytized latex beads, indicating that they were KCs. The viability of KCs, as determined by trypan blue exclusion, was >90%.
Preparation of hepatic cells and H22 cells
Another 6 mice were sacrificed to obtain the hepatic cells. According to the above-mentioned method, using a 200 mesh stainless steel screen, the hepatic histiocyte suspension was centrifuged at 50 × g at 4°C for 2 min, the pellet was centrifuged at 50 × g at 4°C for 5 min 3 times. The cells were cultured at a density of 1×106. After 60 min, the cells were washed with PBS and centrifuged at 50 × g at 4°C, and the pellets containing the liver cells were cultured for use.
The H22 cells were then preserved in vivo in mouse ascites. The mice transplanted with H22 cells (0.2 ml, 5×106; 12 mice were used) for 2 weeks were sacrificed. The ascites were obtained and diluted 10 times with PBS and then centrifuged at 300 × g for 5 min 2–3 times. The supernatant was discarded, and the pellet was then filtrated using a 200 mesh stainless steel screen and cultured for use.
Preparation of mouse serum
Blood (2–3 ml) was obtained from mice transplanted with H22 cells for 2 weeks after the mice had been decapitated and blood collected and preserved in an incubator at 37°C for 1 h or in a refrigerator at 4°C for 3–4 h. The serum was centrifuged at 1000 × g for 15 min, and then the supernatant was obtained using a 56°C thermostatic water bath for 30 min, and 0.02% sodium azide was applied as an antiseptic, as previously described (14). The sample was the immune serum containing special IgG anti-H22 cell and was stored at 4°C for use. Ordinary serum was obtained from normal mice (not transplanted with H22 cells) by the same method mentioned above.
Co-culture of KCs and H22 cells
KCs, H22 cells and hepatic cells were mixed to culture, the cells were plated and counted and the cell count was 1×106 for each type of cell. GaCl3 (15), mouse ordinary serum, mouse immune serum, and anti-IgG Fc immune serum were employed as intervention factors. KCs and hepatic cells were set as group A; KCs and H22 cells were group B; KCs, H22 cells and 0.1 ml mouse ordinary serum were group C; KCs, H22 cells and 0.1 ml mouse immune serum were group D. GaCl3 (0.175 mg/ml) was added to the culture flask to deplete macrophages or inhibit KC functions, and thus the above groups were re-designated as group A1, B1, C1 and D1. When 0.1 ml anti-IgG Fc immune serum was added, the groups were group A2, B2, C2 and D2. At 0, 2, 4, 8, 16 and 32 h, the samples were obtained. KCs and H22 cells were separated from the feature of adherence and the KCs were eluted with 0.25% trypase and washed with PBS. The culture medium was centrifuged at 300 × g for 5 min, and 0.5 ml supernatant was collected. The pellet was H22 cells, which were then washed with PBS and diluted for the counting of living cells using the Trypan blue kit (Beyotime, Haimen, China).
Enzyme-linked immunosorbent assay (ELISA) for TNF-α
ELISA was used to determine the levels of TNF-α. All reagents, samples, and standards were prepared according to the manufacturer's instructions (ELISA kit, Santa Cruz Biotechnology). A 50 µl sample dilution was used. The reaction was performed at 37°C for 120 min, plates were incubated with biotinylated anti-mouse TNF-α (0.1 ml/well) at 37°C for 60 min. ABC solution (0.1 ml) was added into each well, and the plate was incubated at 37°C for 30 min. TMB solution (0.1 ml) was further added into each well, and the plate was incubated in the dark at 37°C for 20–25 min. TMB stop solution (0.1 ml) was added to each well. All ELISAs were read on a microplate reader at 450 nm to determine the TNF-α concentration.
RT-qPCR for TNF-α and CD16a mRNA
Total RNA was extracted from KCs using an SV Total RNA isolation kit (Promega, Madison, WI, USA). Agarose gel electrophoresis was employed to detect the three bands at 28S, 18S and 5S for total RNA. RT-qPCR was employed to amplify the CD16a mRNA and TNF-α mRNA from the mouse KCs. Specific primers synthesized by Beijing SBS Genetech Co., Ltd. (Beijing, China) were as follows: CD16a, 5′-AAG GCT GTG GTG AAA CTG GAC-3′ (sense) and 5′-GAG ATG GAG GAT GTA GTT GCT G-3′ (antisense); TNF-α, 5′-TCT ACT GAA CTT CGG GGT GA-3′ (sense) and 5′-AGT AGA CCT GCC CGG ACT C-3′ (antisense). The products were 500 and 542 bp, respectively. The primers for β-actin were 5′-CTT CTT TGC AGC TCC TTC GT-3′ (sense) and 5′-TTC TCC ATG TCG TCC CAG T-3′ (anti-sense), and the product was 300 bp. RT-qPCR was performed in a 40 µl volume consisting of 1 µM of each primer, 2 µl of 0.5 mM each dNTP, 16 µl RNase-free deionized water, 4 µl 10X RNA PCR buffer, 8 µl 5 mM MgCl2, 1 µl Tfl DNA polymerase, 2 µl 0.25 U/µl AMV reverse transcriptase, 1 µl RNA enzyme inhibitor, and 2 µl DNA template. The sample was heated to 94°C for 2 min in the first chain of cDNA, followed by amplification for 40 cycles at 94°C for 30 sec, 55°C for 45 sec, and 72°C for 60 sec, and 1 cycle at 72°C for 5 min, and 4°C for 8 min in the second chain. Subsequently, 5 µl of each product was analyzed by 1% agarose gel (containing 0.5 µg/ml EB) electrophoresis. The relative quantity of mRNA was described as the ratio of the CD16a or TNF-α mRNA to β-actin mRNA.
Western blot analysis
The expression of CD16a protein in the KCs was analyzed by western blot analysis. Protein extracts were obtained according to the manufacturer's instructions. Protein concentration was determined using the Bradford protein assay kit (Bio-Rad, Hercules, CA, USA). Extracted protein was separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to polyvinylidene fluoride membranes (Dupont, Wilmington, DE, USA). The membranes were washed with 0.1% Tween-20-phosphate-buffered saline and incubated with 5% dry non-fat skimmed milk powder in 0.1% Tween-20-phosphate-buffered saline, at pH 7.4, for 1 h, and subsequently with rabbit anti-mouse CD16a polyclonal antibody (dilution, 1:200 and goat anti-rabbit IgG for 1 h. Finally, the immune complexes were developed with an enhanced chemiluminescence detection kit (Pierce, Waltham, MA, USA). β-actin was used as the control. The GelGDoc2000 imaging system (Bio-Rad, Munich, Germany) was employed to analyze the bands, and the protein level was represented by the relative optical density (ROD) × areas (mm2).
Statistical analysis
Data were analyzed using SPSS 10.0 software and are expressed as the means ± SD. Statistical analysis was carried out using the t-test and bivariant analysis. A P-value <0.05 was considered to indicate a statistically significant difference.
Results
Distribution of KCs in human HCC samples
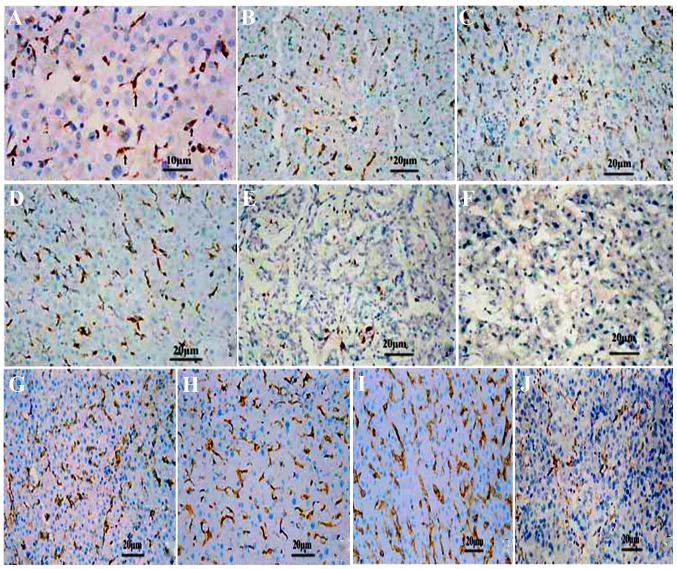
CD68 is the specific marker of the macrophage, and is a valid symbol of KCs (9). In the present study, KCs were present in all 87 samples of HCC tissues; these cells were puffy and dispersed in the liver sinusoids, and the shapes were orbicular-ovate, astro or fusiform. In the 39 normal liver tissue samples, the KCs were well distributed. In cancerous tissues, especially in the poorly differentiated tissue, the number of KCs was at a trace level (Fig. 1). The quantities of KCs were 21.6±7.8, 68.8±9.1, 62.0±1.9 and 36.2±4.1 in the HCC tissue samples, tissues around the cancer, tissues apart from the cancer and normal human hepatic tissue samples, respectively (Fig. 2). In cancerous tissues, the KCs were fewer than in tissue around the cancer, apart from the cancer and normal hepatic tissues (P<0.01). The level of KCs in tissues around and apart from the cancer was higher than in normal hepatic tissue samples (P<0.01), but there was no significant difference between these two types of tissue (P>0.05) (Fig. 2). The quantities of KC in highly, moderately and poorly differentiated tissue were 31.3±2.5, 25.3±1.5 and 14.8±2.2, respectively, and the difference between them was significant (P<0.05) (Fig. 2).
Figure 1.
The distribution and expression of Kupffer cells (KCs) and CD16a in various differentiated cancer tissues studied by immunohistochemical analysis (SP method). (A) The KCs were puffy, of orbicular-ovate and of astro or fusiform morphology in the sinusoid at ×400 magnification. (B) KCs in human hepatic tissue around the cancer, (C) normal human hepatic tissue, (D) human hepatic tissue apart from the cancer, (E) human liver cancer, and (F) poorly differentiated human liver cancer, respectively. CD16a in the human liver detected by immunohistochemical analysis in (G) the normal liver. (H) Hepatic tissue around cancer. (I) Hepatic tissue apart from cancer. (J) In liver cancer tissues, the CD16a-positive cells were puffy, and exhibited orbicular-ovate, astro or fusiform morphology in the sinusoid. Original magnification, ×200.
Figure 2.
Number of Kupffer cells in samples of liver cancer tissue, tissue around the cancer, tissue apart from the cancer (n=87) and normal human hepatic tissue (n=39). The number of Kupffer cells (KCs) in the cancerous tissue was less than in the tissue around the cancer, apart from the cancer and normal hepatic tissue (a vs. b, c, and d, P<0.01). The number of KCs was higher in the tissue around and apart from the cancer than in normal hepatic tissue (b and c vs. d, P<0.01). The number of KCs decreased as the degree of cancer differentiation decreased (* vs. each other, P<0.05).
Expression of KCs CD16a in human HCC
The CD16a-positive cells were puffy and well-distributed. In the tissues around and apart from the cancer, the number (Table I) and the distribution of CD16a-positive cells rose (Fig. 3). In the HCC tissue samples, positive cells were also observed, but the number decreased and distribution was correlated to the degree of differentiation (Table I). The CD16a-positive cells were present in the sinusoids, with a shape similar to the KCs (Fig. 1).
Table I.
Number of CD16a-positive cells and the average optical density.
| Cancer tissues (n=87) |
Tissues around cancer (n=87) |
Tissues apart from cancer (n=87) |
Normal liver tissues (n=39) |
Degree of differentiation
|
|||
|---|---|---|---|---|---|---|---|
| High (n=27) |
Moderate (n=41) |
Poor (n=19) |
|||||
| Positive cell count (means ± SD) | 21.6±5.4 | 79.7±6.9 | 85.0±10.7 | 47.9±7.6 | 26.3±3.6c | 19.3±3.0d | 14.5±1.3e |
| ta | – | 21.724 | 10.693 | 6.342 | 4.427c,d | 3.307c,d | – |
| Pa | – | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | – |
| tb | – | 3.681 | 3.544 | – | – | – | – |
| Pb | – | <0.05 | <0.05 | – | – | – | – |
| Average optical | |||||||
| density | 0.78±0.1 | 0.44±0.07 | 0.35±0.02 | 0.24±0.03 | 0.59±0.05f | 0.7±0.04g | 0.86±0.03h |
| ta | – | 8.0456 | 12.38 | 15.588 | 4.282f,g | 6.025g,h | – |
| Pa | – | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | – |
| tb | – | – | 7.973 | – | – | – | – |
| Pb | – | – | <0.01 | – | – | – | – |
The number of CD16a-positive cells (means ± SD) and the average optical densities in the cancerous tissues, tissues around and apart from the cancer following human hepatocellular carcinoma (HCC) and in normal liver tissues. The amount of CD16 in cancer tissue decreased significantly compared with the tissue around and apart from the cancer, or in normal hepatic tissue samples (P<0.01). The average optical density of CD16a in the cancer tissue increased significantly compared with the tissue around the cancer, apart from the cancer and normal hepatic tissue (P<0.01). *Compared to liver cancer tissue. #Compared to normal hepatic tissue. With the decrease in degree of differentiation, the quantity of CD16a-positive cells decreased significantly (c vs. d, t=4.427, P=0.021; d vs. e, t=3.307, P=0.045); whereas, with the decrease of cancer tissue differentiation, the average optical density increased significantly (f vs. g, t=4.282, P=0.023; g vs. h, t=6.025, P=0.009). Hence, with the decrease of degree of cancer differentiation, the number of CD16a-positive cells was reduced, but the expression level of CD16a was higher (statistical analysis was carried out using t-test and bivariant analysis).
Figure 3.
The distribution of human CD68- and CD16a-positive cells as detected by immunohistochemical analysis The distributive tendency was coincident (vs. each other, coefficient of correlation=0.984, P=0.016).
The quantities of the CD16a-positive cells were 21.6±5.4, 79.7±6.9, 85.0±10.7 and 47.9±7.6 in the HCC tissue samples, tissues around the cancer, tissue apart from the cancer and normal human hepatic tissue samples, respectively. The quantities in the tissue samples around and apart from the cancer increased markedly compared with the cancerous tissue (P<0.01) and normal hepatic tissue samples (P<0.05). The average optical density of the cancerous tissue increased markedly compared with the tissue around the cancer, apart from the cancer and the normal hepatic tissue (P<0.01) (Table I). The number of CD16a-positive cells and the average optical density were significantly different in the highly, moderately and poorly differentiated cancerous tissue (P<0.05) (Table I). The distributive tendency of the CD16a-positive cells was coincident with the KCs from the associativity analysis (correlation coefficient was 0.984, P=0.016) (Fig. 3).
TNF-α expression in KCs from mice
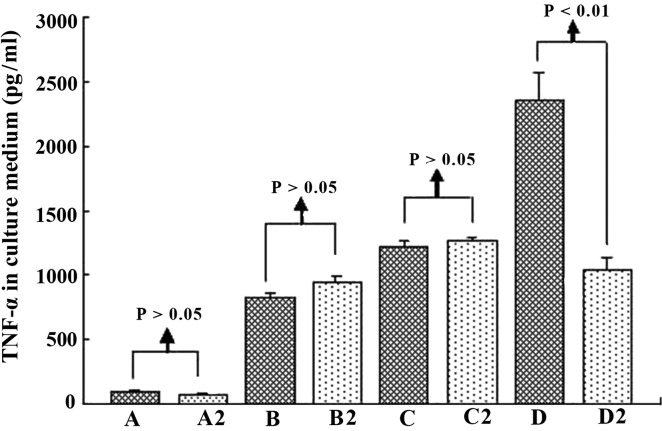
TNF-α is one of the most important indicators for the activation of KCs (16). In the present study, ELISA and RT-qPCR were employed to determine the TNF-α expression level in the culture supernatant and KCs, in order to analyze the activation of mouse KCs under different culture conditions. When the KCs were cultured with hepatic cells, the TNF-α level was stable in the supernatant (group A) and increased with the addition of H22 cells (group B), indicating that H22 cells activated KCs. When ordinary serum (group C) and immune serum (group D) were added to group B, the TNF-α level increased markedly, especially in group D (Fig. 4). Hence, we suggest that H22 cells directly activate KCs, and the ordinary and immune serum, which contained ordinary or special IgG, exerted a promotive effect. This phenomenon was noticeable especially when special IgG was added. In relation to the TNF-α level, in groups A–C the difference between cell groups was insignificant (P>0.05). Serum containing IgG activated KCs, the linkage of CD16a in KCs, and Fc in IgG was probably the key. Hence, anti-IgG Fc serum was employed to test this possibility in the present study. As expected, anti-IgG Fc effectively served as an obstacle to the functioning of the immune serum (Figs. 5 and 6).
Figure 4.
Tumor necrosis factor (TNF)-α in the culture medium of mouse Kupffer cells (KCs) under different culture conditions and at different time points. When the KCs were cultured with hepatic cells, TNF-α was stable (group A), but increased with H22 cells (group B, b vs. a, P<0.05). When the ordinary serum (group C) and immune serum (group D) were added, TNF-α increased markedly in group D (c vs. b, P<0.05; d vs. b and C, P<0.05).
Figure 5.
Tumor necrosis factor (TNF)-α in the culture medium of mouse Kupffer cells (KCs) at 32 h. When anti-IgG Fc serum was applied, TNF-α did not change markedly in groups A, B and C, but decreased significantly in D groups, indicating that anti-IgG Fc acted as an obstacle to the function of immune serum. Groups A–D and A2–D2 are as described in Materials and methods.
Figure 6.
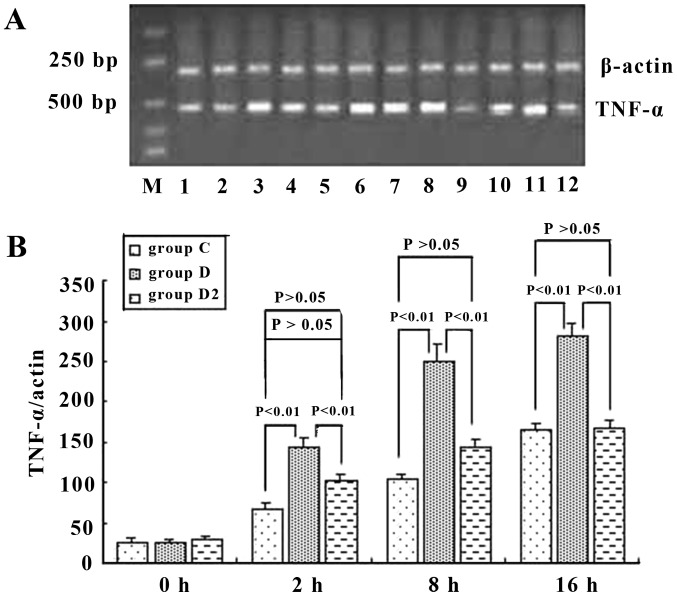
Tumor necrosis factor (TNF)-α mRNA of mouse Kupffer cells (KCs) (A) in group C, D, and D2, and semi-quantitative analysis (B) at different time points. It was shown that ordinary serum and immune serum increased the expression level of TNF-α mRNA, but the immune serum increased it more: 2.45-, 2.39- and 1.72-fold greater than that of the ordinary serum at 2, 8 and 16 h, respectively. Anti-IgG Fc blocked this effect of immune serum and reduced the mRNA expression level of TNF-α in KCs by 26.4, 42.2 and 44.1% at 2,8 and 16 h, respectively. P-values represent the difference between two indicated groups, and groups C,D and D2 are as described in the Materials and methods.
CD16a expression in KCs
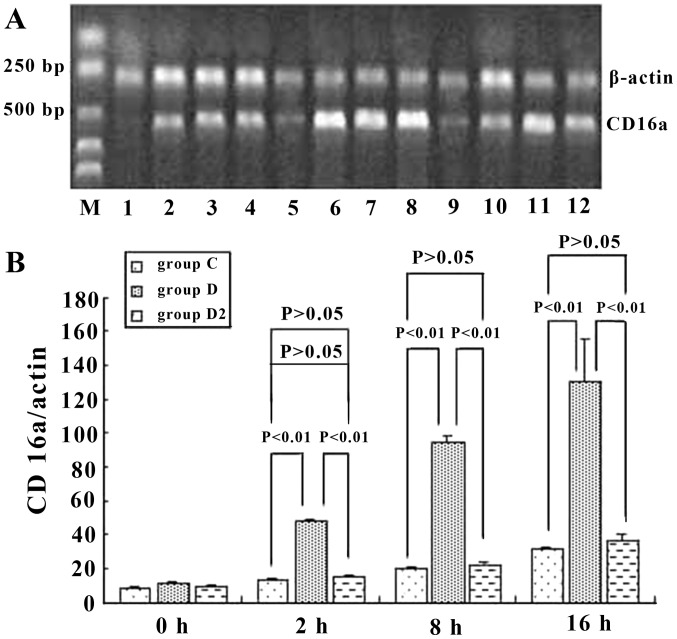
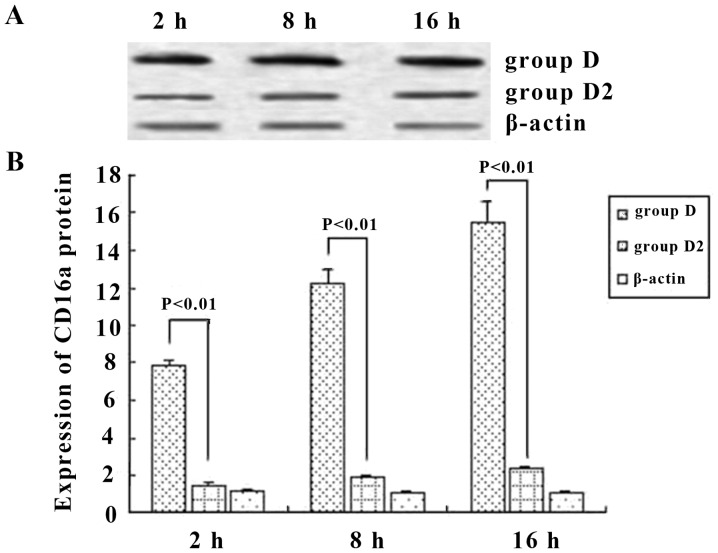
To assess the role of CD16a in KCs, we examined whether the activation of KCs was accompanied by an increase in the mRNA and protein expression of CD16a and whether this increase would be prevented by inhibition with IgG Fc. The results demonstrated that the mRNA level of CD16a in KCs was increased in ordinary serum and immune serum groups. However, the transcriptional level of CD16a in the immune serum group increased in a more marked manner and was 3.9-, 4.9- and 3.9-fold greatern than that of the ordinary serum group at 2, 8 and 16 h, respectively. Anti-IgG Fc blocked this effect of the immune serum and decreased the expression of CD16a in KCs by 59.6, 73.5 and 71.9% at 2, 8 and 16 h, respectively, which was at the same level as the ordinary serum (Fig. 7). Western blot analysis was employed to detect the production of CD16a. When immune serum was added, CD16a protein expression was increased significantly. However, the expression of CD16a protein was decreased by 77.2, 76.4 and 82.1% at 2, 8 and 16 h, respectively, when anti-IgG Fc was applied (P<0.05) (Fig. 8). Hence, at 2, 8 and 16 h, the expression of CD16a and TNF-α in KCs rose and the trend was consistent (P<0.05) when ordinary and immune serum were applied (Fig. 6 and 7), but when anti-IgG Fc was added, the expression of CD16a and TNF-α decreased (P>0.05).
Figure 7.
CD16a mRNA expression in mouse Kupffer cells (KCs) (A) in group C, D and D2 and semi-quantitative analysis (B) at different time points. Immune serum increased the expression of CD16a in KCs by 3.9-, 4.9- and 3.9-fold at 2, 8 and 16 h, respectively. Anti-IgG Fc blocked the effect of the immune serum and decreased the expression of CD16a in KCs by 59.6, 73.5 and 71.9% at 2, 8 and 16 h, respectively, and levels were similar to the ordinary serum levels. P-values represent the difference between two indicated groups, and groups are as indicated in Materials and methods.
Figure 8.
The expression of CD16a protein in Kupffer cells as shown by (A) western blot analysis and (B) semi-quantitative analysis. When immune serum was added, the protein levels increased markedly and anti-IgG Fc reduced the levels by 77.2, 76.4 and 82.1% at 2, 8 and 16 h, respectively. P-values represent the difference between two indicated groups, and groups are as indicated in Materials and methods..
Living H22 cells
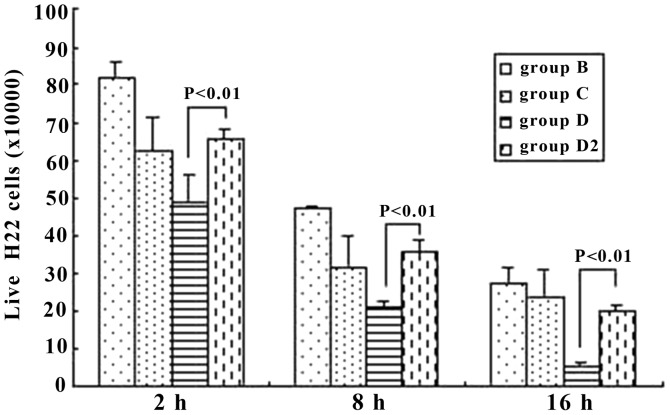
The living H22 cells were Trypan-blue resistant. From 2 h to 16 h, we demonstrated that the number of living H22 cells decreased in every group (P<0.05). However, it should be noted that at the same time points (2, 8 or 16 h), when immune serum (group D) was applied and the anti-IgG Fc was also added (group D2), the difference was significant between these two groups (P<0.01), with group D2 markedly increasing (Fig. 9).
Figure 9.
The number of living H22 cells at different time points in group B, C, D and D2. The expression of CD16a in the Kupffer cells (KCs) decreased and the inhibition of the tumor cells was attenuated when the Fc fragments of IgG were blocked. P-values represent the difference between two indicated groups, and groups are as indicated in Materials and methods.
Discussion
KCs are important defensive cells in the liver; the number and function of KCs changes markedly when the liver is diseased (17). CD68 is a specific surface marker of monocytes and macrophages, including KCs, and thus it can be used as a marker for KCs in the liver of mice. In this study, the results indicated that the number of KCs in cancerous tissue was markedly lower than that in cancer-adjacent tissue and non-cancerous hepatic tissue samples, and numbers decreased with decreasing degrees of histological grade differentiation. These results are in agreement with previous findings (18). It is likely that the irregular arrangement of liver cancer cells led to the disorganization of the hepatic lobule, and the normal sinus hepaticus on which the KCs were dependent then disappeared (19). This also explains why the quantity of KCs in poorly differentiated cancer tissue samples was less. If the cancer tissue differentiated more poorly, the heteromorphosis of the tissue was more obvious and the normal sinus hepaticus was ruined more. However, other factors likely influenced the distribution of KCs, such as immunoreactivity, the migration of KCs from outside into the liver, and splitting and activation of KCs.
Macrophages that infiltrate the tumor stroma are often referred to as tumor-associated macrophages (TAMs). Experimental models using mice have shown that TAMs facilitate tumor progression by promoting inflammation, stimulating angiogenesis and suppressing antitumor immunity (20,21). However, macrophage-activating agents suppress tumor growth and diminish tumor metastasis (22). AngII acts upstream of a potent macrophage amplification program and it has been shown that tumors remotely exploit the hormone pathway to stimulate cancer-promoting immunity (23). In the present study, the number of KCs in the para-cancerous hepatic tissue was markedly increased, indicating that the KCs were important for defending against hepatoma carcinoma cells.
ADCC has been recognized as one of several molecular mechanisms by which monoclonal antibodies of the IgG isotype class mediate anti-tumor effects (24,25). ADCC reactions are dependent on the simultaneous binding of IgG1 molecules to tumor cells and to Fc receptors such as CD16a expressed on a variety of immuno-effector cells, such as NK and monocytic cells (26,27). This indicates that the strength of antibody interaction with CD16a correlates with ADCC potencies.
As the bridge between macrophages and IgG Fc, CD16a is the key molecule of ADCC. If a promoting factor is applied between tumor cells and CD16a, further activation of macrophages and the promotion of tumor cell phagocytosis are expected to occur (7). In this study, it was shown that KCs were activated in the liver following cancer, the number of CD16a positive cells was greater than in the normal liver tissue samples, and decreased with the decrease in the degree of differentiation. CD16a-positive cells and KCs were distributed in the hepatic sinusoid and mainly had a similar shape. The expression of CD16a mRNA and protein in KCs was confirmed in this study. These results suggest the existence of CD16a in KCs and that the activation of KCs, which mainly resulted in CD16a expression, mediated ADCC reactions to secrete TNF-α.
The average optical density of CD16a in KCs in cancer tissue was more than in other hepatic tissue samples, implying that CD16a was important in the ADCC of KCs to hepatoma cells. Activated KCs kill target cells directly by swallowing and releasing lyosomal enzyme, NO and peroxydase. They also cooperate to resist tumor cells by secreting cytokines including TNF-α, IL-1, IL-6 and granulocyte-macrophage colony-stimulating factor (GM-CSF) (5). In our present study, mouse KCs were cultured with H22 tumor cells. The TNF-α level was increased in the groups treated with the ordinary and the immune serum, and we also noted that the number of H22 tumor cells decreased. After the KCs were treated with GaCl3, the TNF-α level and the number of H22 cell did not exhibit marked changes. These indicated that KCs likely killed tumor cells directly by phagocytosis, in a complementary way, and cytokines.
KCs are resident macrophages and play an important role in the defense against invading particles via phagocytosis (28). KCs can be activated by foreign stimuli, such as SiO2 nanoparticles, and release a variety of bioactive mediators, such as reactive oxygen species, TNF-α and NO (29). These findings indicate that certain other factors play an important role in the activation of KCs. In the present study, KCs and H22 cells were co-cultured. Subsequently, we noted that the number of H22 cells was reduced, demonstrating that tumor cells that contain tumor antigen were one of the activating factors of KCs.
The antibody effect is an important way of humoral immunity, while ADCC and the activation of the complement system are the main pathways (30). The ordinary mouse serum was added to the culture medium of KCs and H22 cells, and the results at 2, 8 and 16 h indicated that ordinary serum did not effectively inhibit the growth of tumor cells. On the contrary, however, Ichiyama et al found that immunoglobulin injected into the veins downregulated the immune function by inhibiting the activation of NF-κB, which was induced by TNF-α in macrophages in a dose-dependent manner, and also by blocking the surface receptor CD16a on mononuclear macrophages (31). However, we noted that the mouse serum containing tumor specific IgG boosted the clearance of H22 cells markedly, showing that the tumor-specific antibody exerted a marked effect on the immune function of the KCs. When the anti-IgG Fc immune serum was added, the expression of CD16a mRNA and protein in KCs, as well as the number of living H22 cells, decreased notably, demonstrating that the suppression of tumor cells by KCs was inhibited when the Fc fragments of IgG were blocked by the anti-IgG Fc immune serum. However, the expression of CD16a and the function of the KCs were not completely consistent, and the ordinary serum made the KCs secrete TNF-α also. These results indicated that certain other important pathways contributed to the inhibition of tumor cells, and the blockade of the Fc fragment of IgG likely caused other changes (31) and led to the functional diversity of the KCs. However, the antibodies in the anti-H22 cells immune serum were not specifically identified, which is a shortcoming of this study.
Several variants which enhanced CD16a binding affinity have been identified and have been demonstrated to enhance ADCC activity and increase phagocytosis (32). Furthermore, the gene polymorphism of CD16a is relevant to the occurrence and prognosis of many diseases, including tumor-related diseases (33,34). A previous work has shown that both glyco- and protein-engineering lead to a higher binding affinity to CD16a. From a mechanistic point of view, these data suggest that this gain in affinity is achieved by different mechanisms (35), and it has also been shown that CD16a takes effect through several mechansisms (36). In the present study, we proved that CD16a was expressed in KCs, and we also investigated a profound action mechanism of ADCC in KCs for further. As a low-affinity receptor of IgG, CD16a may be potentially used for the development of novel bioimmunotherapy for liver cancer.
Acknowledgments
This study was supported by grants from the Chongqing Health and Family Planning Commission (2015ZDXM012), the Natural Science Foundation of Hubei Provincial Department of Education (B2015468), the Natural Science Foundation of Hubei Province of China (2015CFB615) and the National Natural Scientific Foundation of China (no. 81272570).
References
- 1.Lafaro KJ, Demirjian AN, Pawlik TM. Epidemiology of hepatocellular carcinoma. Surg Oncol Clin N Am. 2015;24:1–17. doi: 10.1016/j.soc.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 2.Liu ZM, Tseng HY, Tsai HW, Su FC, Huang HS. Transforming growth factor β-interacting factor-induced malignant progression of hepatocellular carcinoma cells depends on superoxide production from Nox4. Free Radic Biol Med. 2015;84:54–64. doi: 10.1016/j.freeradbiomed.2015.03.028. [DOI] [PubMed] [Google Scholar]
- 3.Kaneda Y. Therapeutic strategies for controlling the metastasis and recurrence of cancers: contribution of drug delivery technologies. Adv Drug Deliv Rev. 2012;64:707–709. doi: 10.1016/j.addr.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 4.Leong WS, Thomas KA, Chan CH, Stevenson GT. A standardized conversion of IgG antibody to bispecific form with inversely altered affinities for Fcγ-receptors II and III. Mol Immunol. 2011;48:760–768. doi: 10.1016/j.molimm.2010.11.019. [DOI] [PubMed] [Google Scholar]
- 5.Ehling J, Tacke F. Role of chemokine pathways in hepatobiliary cancer. Cancer Lett. 2015 doi: 10.1016/j.canlet.2015.06.017. pii: S0304-3835(15)00411-5. [DOI] [PubMed] [Google Scholar]
- 6.Flaherty MM, MacLachlan TK, Troutt M, Magee T, Tuaillon N, Johnson S, Stein KE, Bonvini E, Garman R, Andrews L. Nonclinical evaluation of GMA161 - an antihuman CD16 (FcγRIII) monoclonal antibody for treatment of autoimmune disorders in CD16 transgenic mice. Toxicol Sci. 2012;125:299–309. doi: 10.1093/toxsci/kfr278. [DOI] [PubMed] [Google Scholar]
- 7.Herter S, Birk MC, Klein C, Gerdes C, Umana P, Bacac M. Glycoengineering of therapeutic antibodies enhances monocyte/macrophage-mediated phagocytosis and cytotoxicity. J Immunol. 2014;192:2252–2260. doi: 10.4049/jimmunol.1301249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kobayashi E, Motoi S, Sugiura M, Kajikawa M, Kojima S, Kohroki J, Masuho Y. Antibody-dependent cellular cytotoxicity and cytokine/chemokine secretion by KHYG-1 cells stably expressing FcγRIIIA. Immunol Lett. 2014;161:59–64. doi: 10.1016/j.imlet.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 9.Kainer M, Antes B, Wiederkum S, Wozniak-Knopp G, Bauer A, Rüker F, Woisetschläger M. Correlation between CD16a binding and immuno effector functionality of an antigen specific immunoglobulin Fc fragment (Fcab) Arch Biochem Biophys. 2012;526:154–158. doi: 10.1016/j.abb.2012.05.010. [DOI] [PubMed] [Google Scholar]
- 10.Chaigne-Delalande B, Li FY, O'Connor GM, Lukacs MJ, Jiang P, Zheng L, Shatzer A, Biancalana M, Pittaluga S, Matthews HF, et al. Mg2+ regulates cytotoxic functions of NK and CD8 T cells in chronic EBV infection through NKG2D. Science. 2013;341:186–191. doi: 10.1126/science.1240094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou XP, Yang GS, Lu JH, Zhang HB, Li QG, Han LZ, Zong M. Prospective study of liver parenchyma volume in hepatectomy of primary liver cancer. Zhonghua Wai Ke Za Zhi. 2005;43:1370–1374. In Chinese. [PubMed] [Google Scholar]
- 12.Stănculeţ N, Grigoraş A, Avădanei R, Floarea-Strat A, Amălinei C, Căruntu ID. Relationship between Kuppfer cells, inflammation, and fibrosis in chronic hepatitis B and C. Rev Med Chir Soc Med Nat Iasi. 2013;117:880–889. [PubMed] [Google Scholar]
- 13.Liu ZJ, Yan LN, Li XH, Xu FL, Chen XF, You HB, Gong JP. Up-regulation of IRAK-M is essential for endotoxin tolerance induced by a low dose of lipopolysaccharide in Kupffer cells. J Surg Res. 2008;150:34–39. doi: 10.1016/j.jss.2007.12.759. [DOI] [PubMed] [Google Scholar]
- 14.Qu YH, Li Y, Wu YF, Fang JP, Huang SL, Huang Y, Wei J. Influence of FcγRIIIa polymorphism on rituximab-dependent NK cell-mediated cytotoxicity to Raji cells. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2010;18:1269–1274. In Chinese. [PubMed] [Google Scholar]
- 15.Liu C, Yang Z, Wang L, Lu Y, Tang B, Miao H, Xu Q, Chen X. Combination of sorafenib and gadolinium chloride (GdCl3) attenuates dimethylnitrosamine (DMN)-induced liver fibrosis in rats. BMC Gastroenterol. 2015;15:159. doi: 10.1186/s12876-015-0380-5. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Yang P, Zhou W, Li C, Zhang M, Jiang Y, Jiang R, Ba H, Li C, Wang J, Yin B, et al. Kupffer-cell-expressed trans-membrane TNF-α is a major contributor to lipopolysaccharide and D-galactosamine-induced liver injury. Cell Tissue Res. 2016;363:371–383. doi: 10.1007/s00441-015-2252-2. [DOI] [PubMed] [Google Scholar]
- 17.Xu FL, You HB, Li XH, Chen XF, Liu ZJ, Gong JP. Glycine attenuates endotoxin-induced liver injury by downregulating TLR4 signaling in Kupffer cells. Am J Surg. 2008;196:139–148. doi: 10.1016/j.amjsurg.2007.09.045. [DOI] [PubMed] [Google Scholar]
- 18.Liu K, He X, Lei XZ, Zhao LS, Tang H, Liu L, Lei BJ. Pathomorphological study on location and distribution of Kupffer cells in hepatocellular carcinoma. World J Gastroenterol. 2003;9:1946–1949. doi: 10.3748/wjg.v9.i9.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tanaka M, Nakashima O, Wada Y, Kage M, Kojiro M. Pathomorphological study of Kupffer cells in hepatocellular carcinoma and hyperplastic nodular lesions in the liver. Hepatology. 1996;24:807–12. doi: 10.1002/hep.510240409. [DOI] [PubMed] [Google Scholar]
- 20.Qian BZ, Pollard JW. Macrophage diversity enhances tumor progression and metastasis. Cell. 2010;141:39–51. doi: 10.1016/j.cell.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fang W, Ye L, Shen L, Cai J, Huang F, Wei Q, Fei X, Chen X, Guan H, Wang W, et al. Tumor-associated macrophages promote the metastatic potential of thyroid papillary cancer by releasing CXCL8. Carcinogenesis. 2014;35:1780–1787. doi: 10.1093/carcin/bgu060. [DOI] [PubMed] [Google Scholar]
- 22.Eljaszewicz A, Wiese M, Helmin-Basa A, Jankowski M, Gackowska L, Kubiszewska I, Kaszewski W, Michalkiewicz J, Zegarski W. Collaborating with the enemy: function of macrophages in the development of neoplastic disease. Mediators Inflamm. 2013;2013:831387. doi: 10.1155/2013/831387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cortez-Retamozo V, Etzrodt M, Newton A, Ryan R, Pucci F, Sio SW, Kuswanto W, Rauch PJ, Chudnovskiy A, Iwamoto Y, et al. Angiotensin II drives the production of tumor-promoting macrophages. Immunity. 2013;38:296–308. doi: 10.1016/j.immuni.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Westphal S, Brinkmann H, Kalupa M, Wilke A, Seitz-Merwald I, Penack O. Anti-tumor effects of anti-T-cell globulin. Exp Hematol. 2014;42:875–882. doi: 10.1016/j.exphem.2014.07.260. [DOI] [PubMed] [Google Scholar]
- 25.Liu Z, Leng EC, Gunasekaran K, Pentony M, Shen M, Howard M, Stoops J, Manchulenko K, Razinkov V, Liu H, et al. A novel antibody engineering strategy for making monovalent bispecific heterodimeric IgG antibodies by electrostatic steering mechanism. J Biol Chem. 2015;290:7535–7562. doi: 10.1074/jbc.M114.620260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poggi A, Boero S, Musso A, Zocchi MR. Selective role of mevalonate pathway in regulating perforin but not FasL and TNFalpha release in human Natural Killer cells. PLoS One. 2013;8:e62932. doi: 10.1371/journal.pone.0062932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nimmerjahn F, Ravetch JV. Fcgamma receptors as regulators of immune responses. Nat Rev Immunol. 2008;8:34–47. doi: 10.1038/nri2206. [DOI] [PubMed] [Google Scholar]
- 28.Tseng MT, Lu X, Duan X, Hardas SS, Sultana R, Wu P, Unrine JM, Graham U, Butterfield DA, Grulke E, Yokel RA. Alteration of hepatic structure and oxidative stress induced by intravenous nanoceria. Toxicol Appl Pharmacol. 2012;260:173–182. doi: 10.1016/j.taap.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 29.Chen Q, Xue Y, Sun J. Kupffer cell-mediated hepatic injury induced by silica nanoparticles in vitro and in vivo. Int J Nanomedicine. 2013;8:1129–1140. doi: 10.2147/IJN.S42242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cassard L, Cohen-Solal JF, Fournier EM, Camilleri-Broët S, Spatz A, Chouaïb S, Badoual C, Varin A, Fisson S, Duvillard P, et al. Selective expression of inhibitory Fcgamma receptor by metastatic melanoma impairs tumor susceptibility to IgG-dependent cellular response. Int J Cancer. 2008;123:2832–2839. doi: 10.1002/ijc.23870. [DOI] [PubMed] [Google Scholar]
- 31.Ichiyama T, Ueno Y, Hasegawa M, Niimi A, Matsubara T, Furukawa S. Intravenous immunoglobulin inhibits NF-kappaB activation and affects Fcgamma receptor expression in monocytes/macrophages. Naunyn Schmiedebergs Arch Pharmacol. 2004;369:428–433. doi: 10.1007/s00210-004-0877-x. [DOI] [PubMed] [Google Scholar]
- 32.Romain G, Senyukov V, Rey-Villamizar N, Merouane A, Kelton W, Liadi I, Mahendra A, Charab W, Georgiou G, Roysam B, et al. Antibody Fc engineering improves frequency and promotes kinetic boosting of serial killing mediated by NK cells. Blood. 2014;124:3241–3249. doi: 10.1182/blood-2014-04-569061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frankenberger M, Ekici AB, Angstwurm MW, Hoffmann H, Hofer TP, Heimbeck I, Meyer P, Lohse P, Wjst M, Häus-singer K, et al. A defect of CD16-positive monocytes can occur without disease. Immunobiology. 2013;218:169–174. doi: 10.1016/j.imbio.2012.02.013. [DOI] [PubMed] [Google Scholar]
- 34.Ramírez J, Fernández-Sueiro JL, López-Mejías R, Montilla C, Arias M, Moll C, Alsina M, Sanmarti R, Lozano F, Cañete JD. FCGR2A/CD32A and FCGR3A/CD16A variants and EULAR response to tumor necrosis factor-α blockers in psoriatic arthritis: a longitudinal study with 6 months of followup. J Rheumatol. 2012;39:1035–1041. doi: 10.3899/jrheum.110980. [DOI] [PubMed] [Google Scholar]
- 35.Liu Z, Gunasekaran K, Wang W, Razinkov V, Sekirov L, Leng E, Sweet H, Foltz I, Howard M, Rousseau AM, et al. Asymmetrical Fc engineering greatly enhances antibody-dependent cellular cytotoxicity (ADCC) effector function and stability of the modified antibodies. J Biol Chem. 2014;289:3571–3590. doi: 10.1074/jbc.M113.513366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Conry SJ, Meng Q, Hardy G, Yonkers NL, Sugalski JM, Hirsch A, Davitkov P, Compan A, Falck-Ytter Y, Blanton RE, et al. Genetically associated CD16(+)56(−) natural killer cell interferon (IFN)-αR expression regulates signaling and is implicated in IFN-α-induced hepatitis C virus decline. J Infect Dis. 2012;205:1131–1141. doi: 10.1093/infdis/jis027. [DOI] [PMC free article] [PubMed] [Google Scholar]