Abstract
Autism spectrum disorders (ASD) are common, heritable neurodevelopmental conditions. The genetic architecture of ASD is complex, requiring large samples to overcome heterogeneity. Here we broaden coverage and sample size relative to other studies of ASD by using Affymetrix 10K single nucleotide polymorphism (SNP) arrays and 1168 families with ≥ 2 affected individuals to perform the largest linkage scan to date, while also analyzing copy number variation (CNV) in these families. Linkage and CNV analyses implicate chromosome 11p12-p13 and neurexins, respectively, amongst other candidate loci. Neurexins team with previously-implicated neuroligins for glutamatergic synaptogenesis, highlighting glutamate-related genes as promising candidates for ASD.
Keywords: Chromosome Aberrations, Lod Score, Genetic Testing, Genetic Variation, Humans, Male, Risk Factors, Autistic Disorder, Chromosome Mapping, Family, Female, Genetic Linkage, Genetic Predisposition to Disease
Autism is a neurodevelopmental disorder characterized by impairments in reciprocal social interaction, communication deficits and repetitive and restricted patterns of behavior and interests. Autistic disorder is the prototypical Pervasive Developmental Disorder (PDD=ASD), which form a group that also includes Asperger’s disorder, PDD-not otherwise specified, and Rett disorder1. Population prevalence of autism is approximately 15–20 in 10,000, while all ASDs affect about 60 in 10,000 children. Occurring worldwide, males are affected fourfold more than females2. In only about 10% of individuals is autism associated with a recognized cause, most commonly fragile X syndrome, tuberous sclerosis and chromosomal abnormalities3,4.
Twin studies show concordance of 60–92% for monozygotic twins and 0–10% for dizygotic pairs, depending on phenotypic definitions5. Milder phenotypes are similarly elevated in relatives of singleton probands, consistent with a spectrum of severity6. The estimated prevalence of autism in siblings is 5–10%7,8. The ratio of sibling recurrence risk to population prevalence varies from 67 to 25, both larger than most multifactorial diseases. While familial clustering in autism could reflect shared environmental factors, twin studies5,9 and the distribution of milder phenotypes in families favors a model involving multiple interacting loci10,11. We hypothesize that liability to autism is due, in large part, to oligogenic inheritance in which combinations of susceptibility alleles contribute. Variation in phenotypic severity of sibling pairs and family members ascertained through an autistic proband are both consistent with this hypothesis. Based on numerous observations of karyotypic abnormalities in autism, we also hypothesize that submicroscopic alterations are involved.
Genome-wide linkage scans (12 and reviewed in ref 13) for autism susceptibility loci have identified chromosomal regions 2q, 7q and 17q, with 7q yielding the most positive results, including support from meta-analysis. Moreover, substantial evidence suggests that chromosomal abnormalities contribute to autism risk, but the exact prevalence is unclear because literature surveys span different diagnostic and cytogenetic approaches and sample sizes. Recent surveys3,4 show a mean rate of gross mutations and chromosomal abnormalities between 4.3% (78/1826) and 7.4% (129/1749), but many studies find rates of detected abnormalities per individual of 5–10%3. Among the most frequent findings are fra(X)(q27)(3.1%; 28/899) and anomalies involving proximal 15q (0.97%; 17/1749), specifically the Prader-Willi and Angelman region3,4. Duplications of 15q11-q13, typically of maternal origin, are observed in 1–3% of cases, either as interstitial duplications or supernumerary isodicentric marker chromosomes containing one or two extra copies of this region3. Linkage, association, and/or chromosome rearrangement studies have identified several ASD candidates including genes encoding neuroligins and their binding partners as having disease-associated mutations14–17.
In our model for autism, combinations of multiple, possibly-interacting loci and microscopic or sub-microscopic chromosomal abnormalities contribute to risk complicating the detection of individual loci. Increasing the likelihood of detecting loci requires analyzing a large sample of multiplex (≥ 2 affected individuals) families, thereby enhancing the power of linkage analysis; and controlling sources of etiologic heterogeneity (herein, the term families implies multiplex families).
We have assembled a sample of over 1400 ASD families, a resource sufficiently large to implement multiple strategies for localizing susceptibility loci (see “Power” in Supplementary Methods.) While some linkage studies have attempted to control heterogeneity attributable to chromosomal abnormalities by excluding the small number of affected families, none have attempted to merge linkage analysis with studies of fine-level chromosomal variation. We develop an approach, using comparative analysis of hybridization intensities to identify submicroscopic copy number variation (CNV) as putative risk loci, and also as a tool to stratify the sample to reduce genetic heterogeneity for linkage analyses.
RESULTS
ASD samples
The Autism Genome Project (AGP) Consortium, comprised of scientists from 50 centres in North America and Europe, collected 1,496 ASD families (7,917 family members) for this study. Diagnosis was based on the Autism Diagnostic Interview-Revised (ADI-R) and the Autism Diagnostic Observation Schedule (ADOS) or clinical evaluation (see ref 18 and Methods). Sample origins are shown in Supplementary Methods; most samples were karyotyped (≈71%) and screened for fragile X mutations (≈94%) and families were excluded if either was abnormal in at least one affected individual. Most cell lines or DNA arising from the project are available at the NIMH Center for Collaborative Genetic Studies, the European Collection of Cell Cultures, and the Autism Genetics Research Exchange. We genotyped genomic or in some cases whole genome amplified (WGA) DNA.
Linkage analysis by diagnostic group
We successfully generated genotypes from 1,491 of 1,496 nominal families (6,709 samples; Table 1) using the Affymetrix 10K v2 SNP array. WGA had no discernible impact on genotyping accuracy: for 12 duplicate samples assessed, concordance of genotypes for WGA versus blood DNA was >99.6% with no significant difference in completion rates (both ≈ 94%). From the 10,112 SNPs initially genotyped, quality control (QC) procedures resulted in marker exclusion for the following reasons: minor allele frequency < 0.05 (−749 SNPs); high rate of missing genotypes (−1,112 SNPs); selection of tag SNPs (−1,734 SNPs); and deviations from Hardy-Weinberg Equilibrium (−391 SNPs). Following QC, the discordant call rate per locus, based on 261 duplicate samples, was roughly 5/10,000.
Table 1.
Families at each stage of quality control1.
| Narrow | Broad | hASD | Total | ||
|---|---|---|---|---|---|
| Diagnostic data | ≥ 2 Affected (A) | 675 | 942 | 554 | 1496 |
| Genotypic data | ≥ 2 genotyped (G) | 675 | 942 | 549 | 1491 |
| Diagnostic and Genotypic data | ≥ 2 A&G | 597 | 829 | 488 | 1317 |
| Edit 1 | ≥ 2 A&G | 564 | 787 | 461 | 1248 |
| Edit 2 | ≥ 2 A&G | 554 | 772 | 450 | 1222 |
| Edit 3 | ≥ 2 A&G | 522 | 731 | 437 | 1168 |
Quality control of samples was based on genotypes determined by the Affymetrix10k array, which contains 10,112 SNPs. See Supplementary Methods for origin of sample by research centre. Edit 1: eliminated pedigrees/samples incompatible with linkage assumptions (monozygotic twins and inconsistent nominal/genetic relationships). Edit 2: dropped loci missing ≥ 10% of their genotype calls and then individuals missing ≥ 20% of their genotype calls (rates determined empirically based on the frequency distribution of missing genotypes). Edit 3: removed duplicate individuals/families that contributed DNA to more than one centre. Most families were nuclear; after Edit 3, the number of marriages per family was 1.26, 1.24 and 1.16 for Narrow, Broad and hASD.
QC on family data had a similar impact on reducing counts (Table 1), yielding 1,171 families for linkage analysis. Of these families, we estimate that 64% were included in smaller, published linkage studies. The families were distributed across three diagnostic categories, narrow, broad and heterogeneous ASD (or hASD), which were defined according to the distribution of diagnosis (see Methods). Linkage analyses were performed for three nested diagnostic groupings, narrow, broad and all families; for analyses focusing on one diagnostic group, a reasonable choice is broad because a substantial number of families fall in the group and the sensitivity and specificity of this diagnostic method are reliable18.
Prior to linkage analyses, we rebuilt the Affymetrix genetic map by linear interpolation from NCBI Build35 and markers of known genetic positions19 to infer genetic locations for all SNPs. We then validated the new genetic map by using the linkage data.
Linkage information, as reported by MERLIN, averaged ≈95% over the genome (minima at telomeres, ≥ 71%) because of high coverage of markers across the genome and availability of parental genotypes (3% of families have no parental genotypes while 79% have genotypes for both parents). Thus the results are also insensitive to SNPs in linkage disequilibrium (see Methods). Only for all families does statistical evidence exceed the criterion for suggestive linkage20, at 11p12-p13 (Fig. 1).
Figure 1.
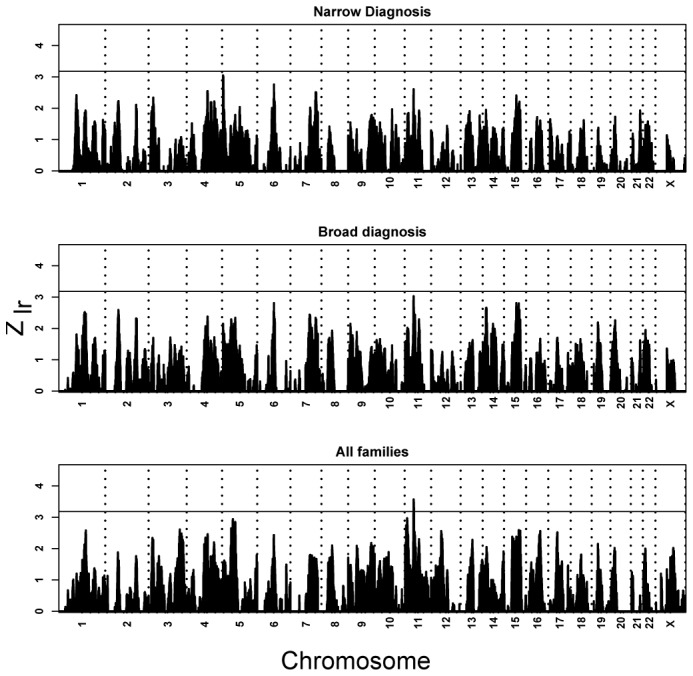
Linkage across the genome for all families and ancestries, based on levels of diagnostic certainty. Vertical reference lines separate chromosomes, which are ordered. The horizontal reference bar is given at a Zlr of 3.18, the threshold for suggestive linkage according to the Lander/Kruglyak criterion3, which is roughly accurate in this setting (4.1 is the threshold for significant linkage). The suggestive threshold would be expected to be crossed by chance once per genome scan. It is crossed once, and the peak falls within 11p12 (Zlr = 3.57 at rs2421826).
Characterization of copy number variation
We assessed our samples for CNV content using signal intensities obtained from the SNP arrays. Because the distribution of intensity is continuous, whereas copy numbers are discrete, an algorithm is required to infer copy number from signal intensity of a SNP genotype relative to intensity from other samples. Initially, to capture as many CNVs as possible, we used two approaches (termed batch and plate-by-plate) for intensity comparisons, which yielded a total of 2788 putative CNVs from 1109 samples from 715 families (Table 2). To define a more stringent set of CNV calls, we examined the raw intensity data from 42 CNV calls that were presumed real based on overlap with non-Mendelian genotype errors or by laboratory experimentation. We also assessed samples having identical CNVs within the same family, as these could also be considered to be validated calls. These analyses identified our plate-by-plate signal intensity comparison to have less background and likely contain fewer false-positive data compared with batch (see Methods and ref. 21). Therefore, we then scrutinized these intensity files to guide threshold settings to define a highly-stringent data set, called ‘filtered,’ containing 624 CNVs from 350 different families (Supplementary Table 1). Characteristics of this dataset for affected individuals are presented in Figure 2 and discussed below.
Table 2.
Characteristics of CNVs found in multiplex autism families using different stringencies of analysis.
| 1 Method | 2Batch | 3Plate-by-Plate | 4Filtered based on thresholds |
|---|---|---|---|
| Features | |||
| Total Samples | |||
| # of CNVs | 1967 | 1286 | 624 |
| # of samples | 918 | 685 | 490 |
| # of CNVs/sample | 2.14 | 1.88 | 1.28 |
| # of families | 590 | 476 | 350 |
| 5 Avg./Median size | 4.6/1.8 Mb | 3.4/1.2 Mb | 3.9/0.6 Mb |
| Gain/Loss of CNV | 1749/228 | 1064/232 | 402/222 |
| Unaffected Individuals | |||
| # of CNVs | 1186 | 802 | 370 |
| # of samples | 538 | 425 | 292 |
| # of CNVs/sample | 2.20 | 1.86 | 1.25 |
| # of families | 419 | 329 | 235 |
| 5 Avg./Median size | 4.7/1.9 | 3.6/1.3 | 4.3/0.69 |
| Gain/Loss of CNV | 1054/133 | 658/143 | 235/136 |
| Affected Individuals | |||
| # of CNVs | 781 | 495 | 254 |
| # of samples | 380 | 260 | 196 |
| # of CNVs/sample | 2.06 | 1.90 | 1.29 |
| # of families | 322 | 230 | 173 |
| 5 Avg./Median size | 4.6/1.8 | 3.2/1.2 | 3.4/0.66 |
| Gain/Loss of CNVs | 685/96 | 406/89 | 167/86 |
| 6 Inherited CNVs (#Regions) | 78 (62) | 59 (46) | 49 (39) |
| 7 Sibling CNVs (#Regions) | 68 (34) | 36 (18) | 34 (17) |
| 8 De novo CNVs (Siblings) | 33 (26) | 16 (10) | 10 (6) |
| 9 Fam. Recurrent CNVs (#Regions) | 28 (14) | 16 (8) | 14 (7) |
| 10 Recurrent CNVs (#Regions) | 209 (66) | 135 (43) | 47 (18) |
| 11 Overlapping CNVs | 422 | 251 | 79 |
| Non-overlapping CNVs | 139 | 109 | 128 |
| 12 CNVs overlapping with ACRD | 68 | 27 | 18 |
| 13 CNVs overlapping with DGV | 20 | 10 | 9 |
| 14 CNVs validated (in affected) | 326 (162) | 230 (106) | 193 (95) |
| 15# of families removed for linkage analysis modeling | 303 | 221 | 166 |
CNV analysis for the arrays was performed using three approaches. After trimming the dataset with the first pass cutoffs, arrays were re-normalized and analyzed using a ‘batch comparison’ (~1000 scans)2 or by a ‘plate-by-plate’ comparison (96 scans)3 to avoid potential plate specific batch effects. In the most stringent analysis4 thresholds were set based on validation data (see footnote 14 below) to minimize potential for false-positives. We note that while column 4 contains data from the most stringent analysis there are some CNVs found exclusively in one or both of the columns 2 and 3 that is indeed real. All data can be viewed at the Autism Chromosome Rearrangement Database (ACRD; http://projects.tcag.ca/autism/);
Range of CNV size: Batch (100bp to 240Mb), Plate-by-Plate (100bp to 134 Mb), Filtered (100bp to 134Mb); distributions are shown in Supplementary Figure 1;
CNVs in affected individuals that were inherited from either parent;
CNVs present in two or more affected siblings that are either de novo or inherited;
CNVs in affected individuals that are de novo in origin with the total number of de novo events occurring in siblings in brackets;
CNVs that are Familial and Recurrent;
CNVs gains or CNV losses with the same coordinates found in two or more unrelated families (a recurrent CNV gain and CNV loss at the same site is only counted once);
Two or more CNV gains or CNV losses with overlapping genomic coordinates (a CNV gain and CNV loss combination is not counted);
CNVs that overlap with mapped chromosome rearrangement breakpoints annotated in the Autism Chromosome Rearrangement Database (ACRD; http://projects.tcag.ca/autism/);
CNVs that overlap with other CNVs not known to be associated with disease as catalogued in the Database of Genomic Variants (DGV; http://projects.tcag.ca/variation/); We note that some CNVs found in the DGV could be predisposing or disease related;
Total number of validated CNVs. A CNV was considered validated if it overlapped with Mendelian genotype errors, or was confirmed using an independent set of experiments (eg. karyotyping, quantitative PCR, array-CGH, Affymetrix 500K Mapping arrays) (see Supplementary Table 4);
Number of families belonging to the group of 1168 families passing data cleaning that were removed from linkage analysis. Families were only removed if at least one affected individual contained a CNVs not found in the Database of Genomic Variants.
Figure 2.

Chromosome ideogram depicting 253 inferred CNVs found in 196 Autism patients These CNVs are derived from the highest stringency ‘filtered’ dataset, but many other true CNVs will also be found in the other analyses and should be examined further. Characteristics of the complete dataset are described in Table 2. Some of the larger changes could represent somatic artifacts or missed karyotypic anomalies. All data is also downloadable or viewed in Genome Browser format at the Autism Chromosome Rearrangement Database (http://projects.tcag.ca/autism/). As additional analyses and validation is performed the data will be posted at the same site.
Caveats about these data are: (i) there will be bona fide CNVs in the batch comparison data that fail to meet cutoffs in the filtered analysis; (ii) some CNVs could be somatic artifacts, such as cell culture-induced rearrangements and aneuploidy; (iii) the mapping resolution of CNV boundaries is dependent on local SNP density and is therefore non-uniform; (iv) smaller CNVs will be more likely to be missed (Supplementary Figure 1); and (v) balanced rearrangements will not be detected.
Considering solely our highest confidence data, we identified 254 CNVs in 196 ASD cases from 173 families (Table 2, Figure 2, Supplementary Tables 2–3). The average and median sizes were 3.4Mb and 0.66Mb, respectively, and the majority (66%) were CNV gains, likely owing to a greater tolerance in the genome for large gains versus deletions. The observations most relevant to ASD disease risk (Supplementary Table 1) included: (i) the identification of 10 families with apparently de novo CNVs (in 3 such families the CNV was found in both ASD sibs); (ii) 18 CNVs in unrelated cases having genomic locations coincident with published ASD chromosome rearrangements; (iii) and 126 CNVs with recurrent (47) or overlapping (79) boundaries suggesting they could be non-random events (Supplementary Tables 3–4). We also detected 7 samples from three families with known ASD-associated chromosome 15q gains all of which were maternally inherited as would be expected (including at least 2 that escaped earlier karyotypic detection).
We highlight four CNV discoveries from many other interesting ones, to demonstrate the utility and complexity of this data, and also to serve as a prototype of how this new type of genetic information can be used in mapping studies. In family AS049, two ASD female sibs were found to have apparently identical 300kb CNV losses of chromosome 2p16 not detected in either parent. Quantitative-PCR analysis confirmed the microdeletion: microsatellite analysis showed the identical maternal chromosomal segment, but no paternal DNA in the sibs, providing a likely explanation of paternal gonadal mosaicism. This hemizygous deletion eliminates coding exons from the neurexin 1 gene (NRXN1), which represents a functional candidate for ASD based on the NRXN1 role in synaptogenesis and its interaction with neuroligins. Rare NRXN1 mutations apparently generate risk for ASD and mental retardation16,22,23. Both girls presented with typical autism including characteristic developmental delays. Although too young for certainty, one appears nonverbal, while her sister had mild language regression. Neither parent had clinically important features.
The second is a recurrent 1.1 Mb CNV gain at chromosome 1q21 found in three families: AS048 with one affected male; AS039 with one affected female; and AS007 with two affected male sibs and father. It overlaps the same region implicated in mental retardation and other anomalies21,24,25.
In the third example, CNVs of ~933kb were observed at 17p12: as a de novo duplication in an affected male-female sib pair in one family (AS068); a maternally-inherited deletion in two affected male sibs (AS028); and a paternally-inherited deletion in a ASD female (AS001). This interval when duplicated causes Charcot-Marie-Tooth 1A (CMT1A) and when deleted causes hereditary neuropathy with liability to pressure palsy26. This region also overlaps with microdeletions seen in some cases of Smith-Magenis Syndrome, which itself has phenotypic overlap with ASDs27. Moreover, other microduplications of the same interval have been described in cases with mental retardation, speech and language delay, as well as autism and related phenotypes28. None of the implied disease-associated CNVs described above were observed in any known control sample database, which at the time of the study was comprised of ~500 samples from the general population21,29.
Finally, in two families with duplications of 22q11.2 further complexity is revealed. In family AS063, the male proband diagnosed with autism inherited the duplication from his father, but a brother with PDD-NOS does not have the duplication. In family AS019, the female sib diagnosed with autism carried the duplication, while an affected brother did not, and the duplication was not observed in either parent. Genotyping confirmed the biological parents. FISH analysis confirmed duplications in both families and revealed that it was de novo in the second family (data not shown.)
Exploration of linkage by subsets of the data
Linkage analysis identifies regions harboring one or more genetic variants that account for a substantial portion of risk in families. Rare de novo or familial CNVs that convey risk to ASD could be a source of noise or heterogeneity that decreases sensitivity in linkage analyses. Thus, in theory, linkage signals from major loci could be amplified if families with rare CNV risk alleles were removed. A sound strategy to evaluate subsets based on known and putative CNVs, however, is unclear. Consider our three levels of CNV discovery, namely filtered, plate and batch, which are ordered by degree of stringency of evidence required to call a CNV. Moving from filtered to batch it is reasonable to assume that the rate of false positives is increasing while the rate of false negatives potentially is decreasing. Selecting a single approach a priori favors a certain, unknown ratio of false positives to false negatives. This ratio might not be optimal, depending on how much of the attributable risk for ASD accrues to CNVs: if CNVs account for a large fraction of the risk, the false negative rate is critical; conversely, if CNVs account for a small fraction of the risk, then the false positive rate is of greater importance. For this reason, we chose to explore the effects of all three levels of CNV discovery. Within each level, we removed families in which at least one individual was diagnosed with ASD and also carried at least one putative CNV (Table 3). Using the Broad diagnostic group, we recomputed the linkage traces (see Supplementary Fig. 2).
Table 3.
Sample size for families in each level of diagnostic group and partitioned according to whether families contained ASD-diagnosed females (FC) or not (MO).
| Diagnostic Group1 | Families1 | Total | Filtered | Plate | Batch |
|---|---|---|---|---|---|
| Narrow | All | 528 | 462 | 431 | 406 |
| MO | 334 | 296 | 274 | 259 | |
| FC | 194 | 166 | 157 | 147 | |
| Broad | All | 739 | 641 | 603 | 567 |
| MO | 464 | 408 | 382 | 359 | |
| FC | 275 | 233 | 221 | 208 | |
| All | All | 1181 | 1031 | 981 | 914 |
| MO | 741 | 653 | 623 | 579 | |
| FC | 440 | 378 | 358 | 335 |
A larger fraction of families tends to be removed from the more stringently-diagnosed groups (narrow and broad) and from FC families, but neither trend approaches significance when the data are fitted to a log-linear model.
After removing ‘CNV families’ the data becomes suggestive for linkage in two regions, 11p12-p13 and 15q23-25.3 (Fig. 3), contrary to results from all broad families. The most noteworthy impact occurs for the batch method, which removes the greatest fraction of families. When families removed versus retained by the batch method are contrasted for identity-by-descent (IBD), heterogeneity is modest except for the 15q25.3 region (Fig. 3).
Figure 3.
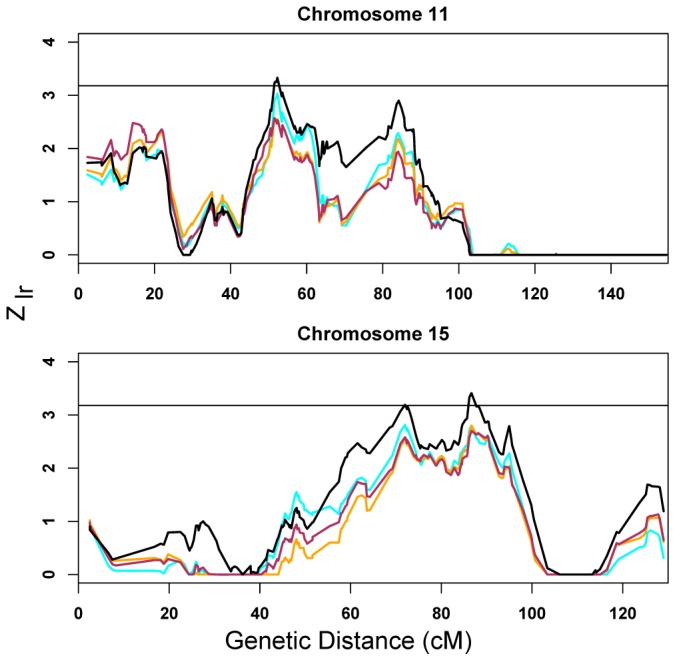
Highlighted linkage results due to removing families in which affected individuals carry putative CNV. Results from all families (ignoring CNVs) noted by the cyan line; results from the filtered set, the orange line; results from the plate set, red line; and results from the batch set, black line. Families all fall in the broad diagnostic category. Complete results are in Supplementary Figure 2. For 11p12, the maximum occurs in 11p13 (Zlr = 3.33 at rs2421826). For chromosome 15, there are two up-crossings: the smaller peak occurs at in 15q23 (Zlr = 3.19 at rs1372828) and the larger in 15q25.3 (Zlr = 3.41 near rs1433452). For families removed versus retained, heterogeneity of estimated identity-by-descent was tested in the ± 5 cM linkage region surrounding each peak and reported as regional minimum heterogeneity p-value m-p (11p12-p13, m-p = 0.074; 15q23, m-p = 0.044; 15q25.3, m-p = 0.004).
The 4:1 ratio of affected males:females, higher reported recurrence risk to siblings of female versus male probands8, and the literature30,31 suggest that a useful partition of the ASD families would be whether they contained affected females (Female-Containing or FC) or only affected males (Male-Only or MO). Thus, we partitioned the families by FC/MO, and recomputed linkage traces for each of the three nested groupings of diagnosis (Fig. 4). Congruent with theory32, the FC families appear to be more informative for linkage. For instance, for the narrow diagnostic scheme, linkage traces cross the suggestive threshold three times, at 5p15.33, 9p24.1 and 11p13-12, whereas the traces do not approach this threshold for MO families. Only for the most inclusive diagnostic level do linkage traces for MO families cross the suggestive threshold (5q12.3 and 9q33.3; Fig. 4), but not the same locations as FC families. While the differences between FC and MO families in terms of linkage could be due to chance, tests of heterogeneity of IBD reveal substantial heterogeneity in 9p, 9q, and 11p, and modest heterogeneity in 5q (Fig. 4).
Figure 4.
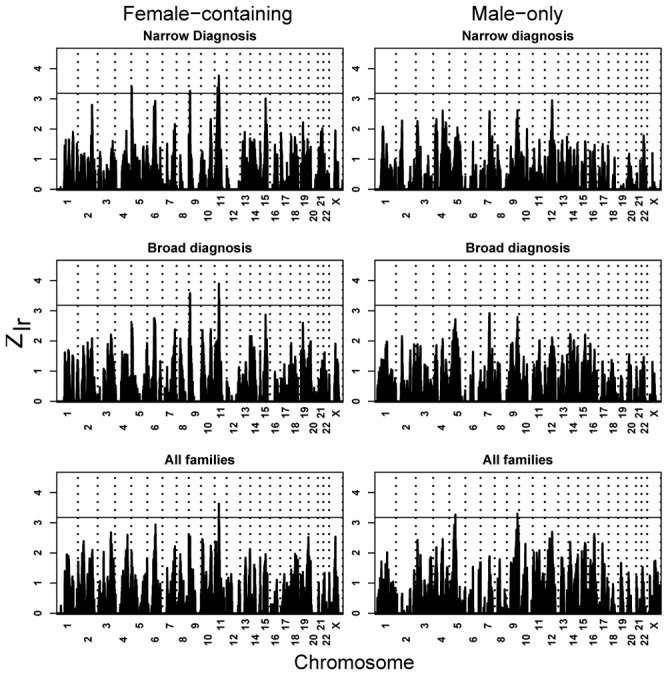
Linkage peaks by MO/FC, based on levels of diagnostic certainty. For FC families and narrow diagnosis, peaks localize to 5p14.33 (Zlr = 3.41 at rs1968011; m-p = 0.141), 9p24.1 (Zlr = 3.21 at rs1340513; m-p = 0.0007), and 11p13 (Zlr = 3.77 at rs1358054; m-p = 0.008); for FC/Broad, to 9p24.1 (Zlr = 3.59 at rs722628; m-p = 0.006) and 11p13 (Zlr = 3.90 at rs1358054; m-p = 0.015); and for FC/All, to 11p12 (Zlr = 3.63 at rs1039205; m-p = 0.078). For all MO families, peaks localize to 5q12 (Zlr = 3.26 at rs673743; m-p = 0.019), 9q33.3 (Zlr = 3.30 at rs536861; m-p = 0.0005).
Sub-setting the data according to presence of CNVs and according to sex of affected individuals appears to generate more informative linkage signals. To test whether combining both approaches to form subsets would also be useful, we used the broad diagnostic grouping. Linkage using these six subsets (Table 3), FC versus MO by three levels of CNV discovery, provides even more support for a risk locus in the vicinity of 11p12-p13 in FC families (Fig. 5). Removing families based on the batch method of CNV discovery nominates a 15q23 locus in FC families (Fig. 5), and the maximum at 11p12-p13 approaches genome-wide significance, 4.03 versus 4.1. Affected individuals in FC families have a slightly elevated rate of CNV detection (2–3%) relative to MO families, regardless of CNV-calling method, and the Broad diagnostic group of families has a similarly elevated rate of CNV detection relative to hASD; the ‘paired’ rates are not significantly different, nor is the interaction of these variables a significant predictor of the presence/absence of a detected CNV (unpublished data.)
Figure 5.
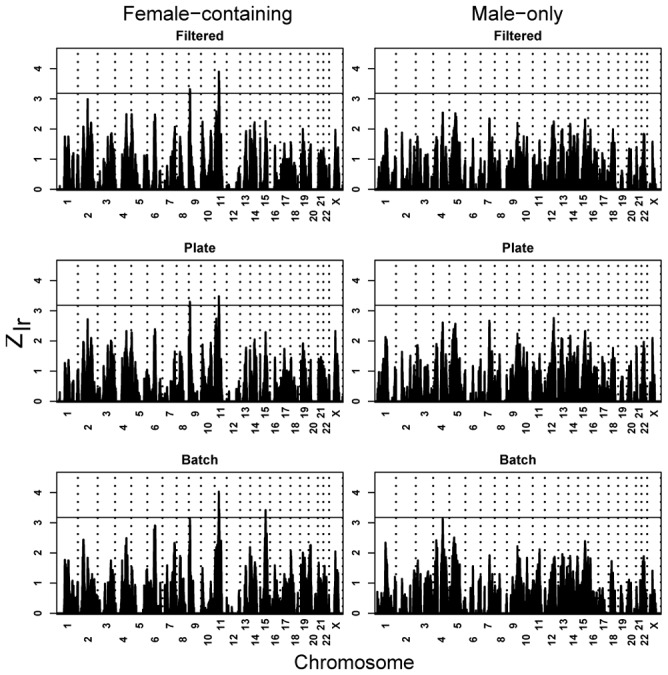
The effect on linkage of splitting families into FC and MO families while also removing families in which affected individuals putatively carry CNV. For FC families and filtered subset, peaks localize to 9p24.1 (Zlr = 3.32 at rs1575284; m-p = 0.105), and 11p12 (Zlr = 3.90 at rs1039205; m-p = 0.411); for FC/Plate, to 9p24.1 (Zlr = 3.28 at rs1821892; m-p = 0.295) and 11p12 (Zlr = 3.48 at rs1039205; m-p = 0.111); and for FC/Batch, to 11p13 (Zlr = 4.03 at rs1358054; m-p = 0.014) and 15q23 (Zlr = 3.30 at rs1433452; m-p = 0.044).
Families participating in the AGP studies vary in their ancestry. To evaluate whether linkage would be strengthened by analyzing families of relatively homogeneous ancestry, we restricted the sample to “European ancestry,” as inferred by principal components33 of SNP genotypes. All founders in 995/1168 (85.2%) families were inferred to be of European origin (Supplementary Fig. 3); inferred ancestry agreed with available self-reports (99.6%). Using this sample, we repeated all linkage analyses in Figures 1 and 3–5.
Exclusion of non-European families generally has only a modest impact on the results (see Supplementary Figure 4 for complete results). Linkage traces at chromosome 11p remain prominent, especially for FC families, although the linkage region for 11p is much broader. By using the batch results to remove “CNV families,” a new region of suggestive linkage emerges, namely 4q31.22, which also shows substantial heterogeneity of IBD (p < 0.00006) between retained and removed families. For all MO families, the linkage trace reaches Zlr=3.85 in 9q33.3. In addition, previously reported locations for linkage, especially 2q and 7q, gain more support from this “European” subset of the data. For example, linkage traces approach genome-wide suggestive linkage at 2q31.1 (FC, narrow) and 7q22.2 (MO, broad).
DISCUSSION
The results obtained from scanning the genomes of the largest cohort of ASD families yet assembled delineate a new understanding of the genetic basis for this complex disorder. That risk for ASD arises in small part from chromosomal copy number abnormalities (CNAs) was widely understood. Yet, for nuclear families containing two or more affected individuals and pre-screened for microscopic CNAs, linkage analyses have implicitly assumed other CNAs would play little if any role in the heritable component of ASD. Our CNV results lie in stark contrast with this view. Instead we find an appreciable number of families that could be assessed (68/590 or 11.5%, 36/476 or 7.6%, and 34/350 or 9.7% in the batch, plate, or filter analyses, respectively) (Table 2), in which all affected individuals share possibly-detrimental abnormalities. Due to the relatively wide and uneven spacing of SNPs and our conservative approach of calling CNVs, we have missed many other events of this kind. By contrast, we also find a number of families in which only one of the affected relatives has a detected CNA. One possible implication of this finding is that in these cases relevant CNV might be a risk factor and not the only causal event. It is also possible that closely-related individuals are etiologically heterogeneous.
With the goal of minimizing heterogeneity that might confound mapping major loci conferring risk to ASD, we invested substantial effort to standardize all phases of this multi-centre project, including phenotypic assessment, sample ascertainment, genotyping, and analysis. Linkage analyses based on a three-level diagnostic scheme produce suggestive evidence for linkage in the vicinity of 11p12-p13 (Fig. 1) for one level, all families. Relative to appropriate baseline, evidence for linkage at 11p12-p13 is amplified in select subsets of the data: (i) subsets obtained by removing families containing one or more affected individuals inferred to carry CNV (Fig. 3); (ii) the set of families containing affected females (Fig. 4); and subsets of families based on both sex of affected individuals and presence of CNVs (Fig. 5). For the subset of FC families without affected individuals who carry CNVs, the maximum Zlr increases to 4.03. We believe these explorations motivate thorough fine-mapping of the 11p12-p13 region. Modest peaks for linkage have been observed previously for this region, but 11p12-p13 has not been a major focus of autism gene discovery.
Several regions have been featured prominently in previous linkage analyses, namely 2q, 7q and 17q. Of these regions, 2q and 7q garner modest linkage support from families of European ancestry (Supplementary Fig. 4), whereas the 17q linkage region does not. The largest linkage signal on 2q, Zlr = 3.1, occurs for FC families in 2q31.1; the largest signal on 7q, Zlr = 3.1, occurs for MO families in 7q22.3. Several explanations are plausible for these results. (i) The previous linkages could be false positives. (ii) For linkage studies of complex disorders, statistics for identified linkage regions tend to be biased upward relative to that expected from the linked risk loci. Because of this bias, combining samples with mixed evidence for linkage and adding new families, as done here, will often diminish previously-identified linkage signals. (iii) Studies could have collected samples that differ in heritable features tied to risk loci, exaggerating the heterogeneity already inherent in ASD. Stochastic variation combined with this heterogeneity could overwhelm the linkage signal.
None of our linkage results can be interpreted as ‘statistically significant’ because we have performed numerous analyses of the data. In fact we performed 18 linkage analyses on the full sample and 18 using families of European ancestry. Because many of these analyses were performed on overlapping subsets of the data, we effectively performed the equivalent of 4–5 independent genome scans34.
Our CNV analyses detect a hemizygous deletion of coding exons from NRXN1 for a pair of affected siblings. Absent of other information this finding might not be especially meaningful, in particular due to the prevalence of CNVs now known to exist in the genome21. However, the alteration is a de novo event and Feng et al.23 have reported rare missense variants in NRXN1 in ASD subjects not found in over 500 controls. Moreover, NRXN1 interacts with neuroligins, for which rare mutations apparently generate risk for ASD and mental retardation14,15. We, therefore, evaluated transmissions in our families for four NRXN1 SNPs(rs1363036, rs930752, rs1377238, rs2018909). Using the FBAT35 empirical statistic, we tested transmissions under additive and dominant models in all families and in the broad subset (dominant and recessive models are indistinguishable in this analysis). For all families, biased transmission was significantly noted at two loci under the dominant model, for the minor allele of rs1363036 (p = 0.0091) and the major allele of rs930752 (p = 0.025)(only rs930752 showed significantly-biased transmission under the additive model, p = 0.014). These SNPs are in modest LD (r2 = 0.048). In the broad subset, results strengthen for rs1363036 (dominant, p = 0.0041), but weaken for rs930752 (dominant, p = 0.076; additive, p = 0.072). These two SNPs, separated by 88kb, are intronic and are unlikely to convey risk directly.
Accumulating evidence thus implicate neurexins/neuroligins as having a role in risk for ASD. For communication of signal between neurons, postsynaptic receptors must oppose neurotransmitter release sites on presynaptic axons. Graf et al.36 show that neurexins induce postsynaptic differentiation in contacting dendrites, while neuroligins induce presynaptic differentiation in glutamatergic axons. The neurexin-neuroligin link is fundamentally-important for glutamatergic synaptogenesis (and, apparently, GABAergic synaptogenesis36,37). Moreover, aberrant glutamate function is often cited as an important element of risk for ASD38,39, a hypothesis compatible with its role as the major excitatory neurotransmitter and critical factor in brain development40. Autism-like behaviors and diagnoses of autism are common for individuals with either Fragile-X syndrome or tuberous sclerosis, both of which are associated with dysregulated glutamate signaling41,42.
Is oligogenic or “major gene” variation associated with other glutamate-related genes? In addition to our results, intriguing evidence for association has been found for the mitochondrial aspartate/glutamate carrier SLC25A12 (2q31) and GRIK2 13. Still the protein product of SLC25A12 is a mitochondrial aspartate-glutamate carrier not known to affect glutamatergic synaptic function. Knockout of SLC25A12 in mice impairs myelination of neuronal cells resulting from limitations to aspartate delivery, not glutamate43. The protein product of GRIK2, GluR6, is an ionotropic kainate receptor that does impact neuronal development. Based on its mapping to 6q16.3, it is not a positional candidate according to our results. For FC families, however, linkage results are modestly positive (Zlr = 1.9 for all families and families of European ancestry; Zlr ≈ 2.40 for families retained using the batch method of CNV-calling).
When the UCSC Genome Browser is searched with the keyword glutamate, it lists 168 genes. Many fall in linkage regions, including 11q13-12 (SLC1A2 and PRRG4), 2q31 (SLC25A12), 4q28.3 (SLC7A11), 7q21.3 (SLC25A13), 9p24.2 (SLC1A1), 9q34.11 (FGPS), and 15q25.2 (HOMER2). Of 10 glutamate solute carriers, half fall in the cited linkage regions (keywords = glutamate & solute & carrier), but not all are related to glutamatergic synaptic function. SLC1A1 and SLC1A2 fall close to linkage peaks, their protein products affect glutamate synapse function and brain development, and thus they are excellent targets for positional candidate gene analyses.
METHODS
Linkage screening set
A SNP-based genome scan was conducted using the Affymetrix 10K v2 SNP array. Genotyping was contracted to the Translational Genomics Research Institute (TGEN). TGEN genotyped DNA samples falling into 1496 nominal families, of which 1168 could be used for linkage.
Strategy for linkage analysis
For linkage analyses we grouped families into three diagnostic classes: narrow, broad and heterogeneous ASD (hASD). To qualify for the narrow class, two or more affected individuals had to meet criteria for autism on both the ADI-R18 and the ADOS18. For the broad category at least one individual had to meet ADI-R criteria for autism and ADOS criteria for autism or ASD. At least one other family member required criteria for impairment on the social or communication domains of the ADI-R and meet criteria for at least ASD on the ADOS. The hASD families were completely independent of the broad and narrow categories, but combined with the broad set to analyze linkage in all families. The hASD families consisted largely of (i) families meeting ADI-R criteria for ASD or autism18, but absent ADOS evaluation, (ii) multiple individuals per family meeting ASD criteria by ADOS and exhibiting impairment on the social or communication domains of the ADI-R, but not meeting full criteria for autism on the ADI-R, or (iii) both of the above. In addition to diagnostic categories, families were subset by MO/FC status.
Genetic quality control, tag SNP selection and ancestry
We evaluated three features of data quality, namely degree of missing genotypes (missingness), Mendelian errors, and Hardy Weinberg Equilibrium (HW). Individuals (20%) and loci (10%) with substantial missing data were not considered for linkage analyses because these features usually indicate poor DNA quality and problems with genotype calls, respectively. Likewise loci with minor allele frequency MAF < 0.05 were discarded.
Mendelian errors were evaluated using PEDCHECK44. Loci showing multiple Mendelian errors – for correct family structure – were discarded for linkage testing. To overcome possible problems arising due to ancestry we first selected tag SNPs and then evaluated HW with the large sample inferred to be of homogeneous European ancestry. We analyzed linkage disequilibrium (LD) using HCLUST45, selecting tag SNPs to represent clusters of others in substantial LD (r2 > 0.8). We chose those that were highly correlated with the other SNPs in the cluster; we estimated ancestry by using principal component analyses33; and evaluated HW using parental data. Loci were not used for analyses if HW was rejected at a p-value < 0.005. Finally we used MERLIN46 to infer likely genotyping errors on the basis of apparent genetic recombination. When genotypes were likely to be errors (p < 0.01) they were set to missing.
Linkage analysis
We used the BLUE method47 to estimate allele frequencies. Linkage was estimated from the entire set of SNPs using MERLIN46 and the exponential S-all statistic. Linkage was also estimated by using MERLIN46 and ALLEGRO48 from tag SNPs. We found virtually no difference in results using tag SNPs or all SNPs, with or without using the options in MERLIN to handle LD. We analyzed heterogeneity of linkage between strata by using the methods of McQueen49, which test for significant differences in shared IBD among affected siblings in families. We computed heterogeneity statistics in each region/setting in which a linkage trace crossed the threshold for suggestive linkage.
CNV assessment
CNVs were inferred from Affymetrix 10K array scans using dChip 2006 software (DNA Chip Analyzer)50. We have also used other algorithms and the data will be posted at the Autism Chromosome Rearrangement Database as it is validated. Initially, 7610 scans were available (this number exceeds the 6,709 samples genotyped for linkage since the CNV experiments continued after the initial data freeze). We excluded those samples with a genotype call rate less than 92% and/or an array percentage outlier of greater than 5% leaving 5997 experiments suitable for CNV analyses. For the ‘batch’ analysis we grouped the arrays into 6 cohorts of 1000 samples each. The median probe intensities for the arrays varied greatly (<100 to >1000) indicating the need for normalization to compare signals. Arrays were normalized at the probe intensity level using invariant set normalization to a baseline array within each group of 1000 experiments50. A signal value was then calculated for each SNP using a model-based method and averaged across all samples for each SNP to obtain the mean signal of a diploid genome. The observed raw copy number was then defined, and copy number inferred for each individual/SNP using a Hidden Markov Model50. Since samples were submitted in 96-well plate formats and arrays were processed in the same manner, the ‘batch’ analysis contained plate-specific noise apparently leading to many false positive CNV calls. In an attempt to increase the signal to noise ratio, we analyzed arrays in a 96-well plate specific manner. For example, we excluded 12 plates having less than 40 samples after the initial filtering, leaving 5823 scans for the plate analysis. Arrays were normalized within each set and copy number calculated in the same manner as was for the ‘batch’ analysis. We also excluded those samples with greater than 10 CNVs per sample from all analyses to avoid high false-positive calling. Because we had family data, certain CNV could be tentatively confirmed by using the family structure and Mendelian errors (although the original CNV calls were blind to family status). Using these data as a benchmark the ‘plate’ analysis produced a cleaner dataset than the ‘batch’ analysis and was therefore parsed further using a combination of more stringent thresholds (less than 5 CNVs per sample) and manual curation of the raw data to give a ‘filtered’ dataset. The inferred CNVs for all three datasets were interpreted on several levels. We also completed similar analysis for the X-chromosome, but did not include the results since only 263 SNPs covered this segment of the genome (40 CNVs were found). Called CNVs were also examined for overlap with genomic features including mapped chromosome rearrangement breakpoints annotated in the Autism Chromosome Rearrangement Database and polymorphic CNVs in the Database of Genomic Variants. For all three analyses, affected individuals and families with CNVs that did not have complete overlap with the DGV were removed from linkage analysis.
The raw data from the Affymetrix 10k experiments are posted at the Gene Omnibus Expression Database (http://www.ncbi.nlm.nih.gov/geo/) with the Accession #GSE6754. CNV calls are released at the Autism Chromosome Rearrangement Database.
Supplementary Material
Supplementary Table 1: List of 624 CNVs in Filtered Analysis.
Supplementary Table 2: List of 254 CNVs in Affected Individuals.
Supplementary Table 3: Breakdown of CNVs in Affected Individuals.
Supplementary Table 4: List of Validated CNVs.
Supplementary Figure 1: Binned size distribution of CNVs in batch (A), plate (B) and filtered (C) analyses.
Supplementary Figure 2: Linkage results due to removing families in which affected individuals putatively carry CNV.
Supplementary Figure 3. Principal component plot used to infer ancestry.
Supplementary Figure 4: Linkage results obtained by analyzing families inferred to be of homogeneous European ancestry.
Acknowledgments
The authors are indebted to the participating families for their contribution of time and effort in support of this study. We gratefully acknowledge Autism Speaks, formerly the National Alliance for Autism Research, for financial support to pool data, SNP genotyping, and for data analysis. The individual consortia also thank the following agencies (listed alphabetically by consortia); Autism Genetics Cooperative (AGC): Assistance Publique-Hôpitaux de Paris, Canadian Institutes for Health Research (CIHR grant 11350 to P. Szatmari), Catherine and Maxwell Meighan Foundation, Fondation de France, Fondation France Télécom, Fondation pour la Recherche Médicale, Genome Canada/Ontario Genomics Institute, The Hospital for Sick Children Foundation, Howard Hughes Medical Institute (HHMI), INSERM, McLaughlin Centre for Molecular Medicine, National Institute of Child Health and Human Development, National Institute of Mental Health (MH066673 to J. Buxbaum; MH55135 to S. Folstein; MH52708 to Neil Risch; MH061009 to J. Sutcliffe), National Institute of Neurological Disorders and Stroke (NS042165 to J. Hallmayer; NS026630 and NS036738 to M. Pericak-Vance; NS049261 to J. Sutcliffe; NS043550 to T. Wassink), Swedish Science Council, Seaver Autism Research Foundation, The Centre for Applied Genomics (Toronto). S.W. Scherer is an Investigator of the CIHR and HHMI International Scholar; The Autism Genetic Resource Exchange (AGRE) Consortium gratefully acknowledges the resources provided by the participating AGRE families. The Autism Genetic Resource Exchange is a program of Cure Autism Now and is supported, in part, from the National Institute of Mental Health (MH64547 to D.H. Geschwind); The Collaborative Programs of Excellence (CPEA): National Center for Research Resources (M01-RR00064), National Institute of Child Health and Human Development (U19HD34565 G. Dawson and G. Schellenberg), NIMH (MH057881), NINDS (5 U19 HD035476 to W.M. McMahon), The Utah Autism Foundation; The International Molecular Genetic Study of Autism Consortium (IMGSAC): UK Medical Research Council, Wellcome Trust, BIOMED 2 (CT-97-2759), EC Fifth Framework (QLG2-CT-1999-0094), Telethon-Italy (GGP030227), Janus Korczak Foundation, Deutsche Forschungsgemeinschaft, Fondation France Telecom, Conseil Regional Midi-Pyrenees, Danish Medical Research Council, Sofiefonden, Beatrice Surovell Haskells Fond for Child Mental Health Research of Copenhagen, Danish Natural Science Research Council (9802210), and National Institutes of Health (U19 HD35482, MO1 RR06022, K05 MH01196, K02 MH01389). A.J. Bailey is the Cheryl and Reece Scott Professor of Psychiatry. A.P. Monaco is a Wellcome Trust Principal Research Fellow.
Autism Genome Project (AGP) Consortium: The AGP is comprised of four existing consortia of partners or countries listed alphabetically:
Autism Genetics Cooperative (AGC): Canagen Peter Szatmari 1*, Andrew D. Paterson 2*, Lonnie Zwaigenbaum3, Wendy Roberts4, Jessica Brian4, Xiao-Qing Liu2, John B. Vincent5, Jennifer L. Skaug2, Ann P. Thompson1, Lili Senman4, Lars Feuk2, Cheng Qian2, Susan E. Bryson6; Marshall B. Jones7; Christian R. Marshall2, Stephen W. Scherer 2*; Iowa Data Coordinating Center (DCC) Veronica J. Vieland 8*, Christopher Bartlett8, La Vonne Mangin8, Rhinda Goedken9, Alberto Segre9; University of Miami Margaret A. Pericak-Vance 10*, Michael L. Cuccaro10, John R. Gilbert10; University of South Carolina Harry H. Wright11, Ruth K. Abramson11; Paris Autism Research International Sibpair (PARIS) Study Catalina Betancur 12*, Thomas Bourgeron13, Christopher Gillberg14 Marion Leboyer12,15; Seaver Autism Research Center (SARC) Joseph D. Buxbaum 16*, Kenneth L. Davis16, Eric Hollander16, Jeremy M. Silverman16; Stanford University Joachim Hallmayer 17*, Linda Lotspeich17; Vanderbilt University James S. Sutcliffe 18,19*, Jonathan L. Haines 19*, Susan E. Folstein20; University of North Carolina/University of Iowa Joseph Piven 21*, Thomas H. Wassink 22*, Val Sheffield22
The Autism Genetic Resource Exchange (AGRE) Consortium Daniel H. Geschwind 23*, Maja Bucan24, W. Ted Brown25, Rita Cantor 26*, John N. Constantino27, T. Conrad Gilliam28, Martha Herbert29, Clara LaJonchere30, David H. Ledbetter31, Christa Lese-Martin31, Janet Miller30, Stan Nelson 26* , Carol A. Samango-Sprouse32, Sarah Spence33, Matthew State34, Rudolph E. Tanzi35
The Collaborative Programs of Excellence (CPEA): Hilary Coon36, Geraldine Dawson 37,38*, Bernie Devlin 39*, Annette Estes38, Pamela Flodman40, Lambertus Klei39, William M. McMahon 36*, Nancy Minshew39, Jeff Munson37, Elena Korvatska41,42, Patricia M. Rodier43, Gerard D. Schellenberg 41,42,44*, Moyra Smith40, M. Anne Spence40, Chris Stodgell43, Ping Guo Tepper, Ellen M. Wijsman 45,46*, Chang-En Yu41,42
The International Molecular Genetic Study of Autism Consortium (IMGSAC): France Eric Fombonne47, Bernadette Rogé47; Germany Annemarie Poustka48, Bärbel Felder48, Sabine M. Klauck48,, Claudia Schuster48, Fritz Poustka49, Sven Bölte49, Sabine Feineis-Matthews49, Evelyn Herbrecht49, Gabi Schmötzer49; Greece John Tsiantis50, Katerina Papanikolaou50; Italy Elena Maestrini51, Elena Bacchelli51, Francesca Blasi51, Simona Carone51, Claudio Toma51; Netherlands Herman Van Engeland52, Maretha de Jonge52, Chantal Kemner52, Frederike Koop52, Marjolijn Langemeijer52, Channa Hijimans52, Wouter G Staal52; United Kingdom Gillian Baird53, Patrick F Bolton54, Michael L Rutter55, Emma Weisblatt56, Jonathan Green57, Catherine Aldred57, Julie-Anne Wilkinson57, Andrew Pickles58, Ann Le Couteur59, Tom Berney59, Helen McConachie59, Anthony J Bailey 60*, Kostas Francis60, Gemma Honeyman60, Aislinn Hutchinson60, Jeremy R Parr60, Simon Wallace60, Anthony P Monaco 61*, Gabrielle Barnby61, Kazuhiro Kobayashi61, Janine A Lamb61, Ines Sousa61, Nuala Sykes61; United States of America Edwin H. Cook 62*, Stephen J. Guter62, Bennett L. Leventhal62, Jeff Salt62, Catherine Lord63, Christina Corsello63, Vanessa Hus63, Daniel E. Weeks64, Fred Volkmar34
Scientific management: Andy Shih65
Footnotes
Department of Psychiatry and Behavioural Neurosciences, McMaster University, Hamilton, Canada
The Centre for Applied Genomics and Program in Genetics and Genomic Biology, The Hospital for Sick Children and University of Toronto, Toronto, Canada
Department of Pediatrics, McMaster University, Hamilton, Canada
Autism Research Unit, The Hospital for Sick Children, Toronto, Canada
Centre for Addiction and Mental Health, Clarke Institute and Department of Psychiatry, University of Toronto, Toronto, Canada
Departments of Pediatrics and Psychology, Izaak Walton Killam Health Centre – Dalhousie University, Halifax, Canada
Department of Neural and Behavioral Sciences, The Pennsylvania State University College of Medicine, Hershey, PA
Columbus Children’s Research Institute, Center for Quantitative and Computational Biology
University of Iowa, Department of Computer Science
Miami Institute for Human Genomics, University of Miami Miller School of Medicine, Miami, FL
W.S. Hall Psychiatric Institute, University of South Carolina, Columbia, SC
INSERM U513, Université Paris XII, Créteil, France
Human Genetics and Cognitive Functions, Institut Pasteur, Paris, France
Department of Child and Adolescent Psychiatry, Goteborg University, Goteborg, Sweden
Department of Psychiatry, Groupe hospitalier Henri Mondor-Albert Chenevier, AP-HP, Créteil, France
Seaver Autism Research Center and the Greater New York Autism Research Center for Excellence, Department of Psychiatry, Mount Sinai School of Medicine, New York, NY
Stanford University, Department of Psychiatry
Center for Molecular Neuroscience, Vanderbilt University, Nashville, TN.
Center for Human Genetics Research, Vanderbilt University, Nashville, TN
Department of Psychiatry, Johns Hopkins University, Baltimore, MD.
University of North Carolina, Chapel Hill, NC
University of Iowa, Iowa City, Iowa
Department of Neurology, UCLA School of Medicine, CA
University of Pennsylvania, Philadelphia, PA
N.Y.S. Institute for Basic Research in Developmental Disabilities, Staten Island, NY
Department of Human Genetics, UCLA School of Medicine
Washington University School of Medicine, St. Louis, MO
University of Chicago, Chicago, IL
Harvard Medical School, Boston, MA
Cure Autism Now, Los Angeles, CA
Emory University, Atlanta, GA
George Washington University, Washington, D.C.
UCLA, Los Angeles, CA
Yale University Child Study Center, New Haven
Massachusetts General Hospital, Boston, MA
Department of Psychiatry, University of Utah, Salt Lake City, USA 84132
Department of Psychology and the Center on Human Development and Disability, University of Washington, Seattle, WA, USA, 98195-7920
Department of Psychiatry and Behavioral Sciences, University of Washington, Seattle, WA, USA, 98195-6560
Department of Psychiatry, University of Pittsburgh, Pittsburgh, PA, USA, 15213
Department of Pediatrics, University of California, Irvine CA, USA, 92697-4036
Geriatrics Research Education and Clinical Center, Puget Sound Veterans affairs Medical Center, Seattle, WA, USA 98108
Gerontology and Geriatric Medicine, Department of Medicine, University of Washington, Seattle WA, USA, 98104-2499
Department of OB/GYN, University of Rochester Medical Center, Rochester, NY, USA, 14642
Departments of Neurology and Pharmacology, University of Washington, Seattle, WA, USA, 98195
Division of Medical Genetics, Department of Medicine, University of Washington, Seattle, WA, USA, 98195-7720
Department of Biostatistics, University of Washington, Seattle, WA, USA, 98195
Université de Toulouse Le Mirail, Centre d’Etudes et de Recherches en Psychopathologie (CERPP), Toulouse
Deutsches Krebsforschungszentrum (DKFZ), Division of Molecular Genome Analysis, Heidelberg
J W Goethe Universität Frankfurt, Klinik für Psychiatrie und Psychotherapie des Kindes- und Jugendalters, Frankfurt
National and Kapodistrian University of Athens, Department of Child Psychiatry, “Agia Sophia” Children’s Hospital, Athens
Department of Biology, University of Bologna, Bologna, Italy
University Medical Center, Utrecht, Department of Child and Adolescent Psychiatry, Utrecht
Guy’s Hospital, Newcomen Centre, London
Institute of Psychiatry, Department of Child and Adolescent Psychiatry, London
Institute of Psychiatry, Social, Genetic and Developmental Psychiatry Centre (SGDP), London
University of Cambridge Clinical School, Cambridge
University of Manchester Department of Child Psychiatry, Booth Hall Children’s Hospital, Manchester
University of Manchester School of Epidemiology and Health Science, Manchester
University of Newcastle, Child and Adolescent Mental Health, Sir James Spence Institute, Newcaste upon Tyne
University of Oxford Department of Psychiatry, Oxford
Wellcome Trust Centre for Human Genetics, Oxford
University of Illinois at Chicago, Institute for Juvenile Research, Chicago
University of Michigan Autism and Communicative Disorders Center (UMACC), Ann Arbor
University of Pittsburgh Department of Human Genetics, Pittsburgh
Autism Speaks, 2 Park Avenue, 11th Floor, New York, NY 10016
URLs. Autism Chromosome Rearrangement Database: http://projects.tcag.ca/autism/
Database of Genomic Variants: http://projects.tcag.ca/variation/
Gene Omnibus Expression Database: http://www.ncbi.nlm.nih.gov/geo/
Note: Supplementary information is available on the Nature Genetics website.
Author Contributions: Lead AGP investigators contributed equally to this project and are identified below by an asterisk.
Reprints and permissions information will be made available.
The authors declare no obvious financial interests.
References
- 1.Association, A.P. Diagnostic and statistical manual of mental disorders. Washington, D.C: 1994. [Google Scholar]
- 2.Chakrabarti S, Fombonne E. Pervasive developmental disorders in preschool children: confirmation of high prevalence. Am J Psychiatry. 2005;162:1133–1141. doi: 10.1176/appi.ajp.162.6.1133. [DOI] [PubMed] [Google Scholar]
- 3.Veenstra-Vanderweele J, Christian SL, Cook EH., Jr Autism as a paradigmatic complex genetic disorder. Annu Rev Genomics Hum Genet. 2004;5:379–405. doi: 10.1146/annurev.genom.5.061903.180050. [DOI] [PubMed] [Google Scholar]
- 4.Xu J, Zwaigenbaum L, Szatmari P, Scherer SW. Molecular Cytogenetics of Autism. Current Genomics. 2004;5:347–364. [Google Scholar]
- 5.Bailey A, et al. Autism as a strongly genetic disorder: evidence from a British twin study. Psychol Med. 1995;25:63–77. doi: 10.1017/s0033291700028099. [DOI] [PubMed] [Google Scholar]
- 6.Piven J. The broad autism phenotype: a complementary strategy for molecular genetic studies of autism. Am J Med Genet. 2001;105:34–35. [PubMed] [Google Scholar]
- 7.Jones MB, Szatmari P. Stoppage rules and genetic studies of autism. J Autism Dev Disord. 1988;18:31–40. doi: 10.1007/BF02211816. [DOI] [PubMed] [Google Scholar]
- 8.Ritvo ER, et al. The UCLA-University of Utah epidemiologic survey of autism: prevalence. Am J Psychiatry. 1989;146:194–199. doi: 10.1176/ajp.146.2.194. [DOI] [PubMed] [Google Scholar]
- 9.Hallmayer J, et al. On the twin risk in autism. Am J Hum Genet. 2002;71:941–946. doi: 10.1086/342990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pickles A, et al. Latent-class analysis of recurrence risks for complex phenotypes with selection and measurement error: a twin and family history study of autism. Am J Hum Genet. 1995;57:717–726. [PMC free article] [PubMed] [Google Scholar]
- 11.Risch N, et al. A genomic screen of autism: Evidence for a multilocus etiology. Am J Hum Genet. 1999;65:493–507. doi: 10.1086/302497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schellenberg GD, et al. Evidence for genetic linkage of autism to chromosomes 7 and 4. Mol Psychiatry. 2006;11:979. [Google Scholar]
- 13.Freitag CM. The genetics of autistic disorders and its clinical relevance: a review of the literature. Mol Psychiatry. 2007;12:2–22. doi: 10.1038/sj.mp.4001896. [DOI] [PubMed] [Google Scholar]
- 14.Jamain S, et al. Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat Genet. 2003;34:27–29. doi: 10.1038/ng1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laumonnier F, et al. X-linked mental retardation and autism are associated with a mutation in the NLGN4 gene, a member of the neuroligin family. Am J Hum Genet. 2004;74:552–557. doi: 10.1086/382137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chubykin AA, et al. Dissection of synapse induction by neuroligins: effect of a neuroligin mutation associated with autism. J Biol Chem. 2005;280:22365–22374. doi: 10.1074/jbc.M410723200. [DOI] [PubMed] [Google Scholar]
- 17.Durand CM, et al. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat Genet. 2006 doi: 10.1038/ng1933. advanced online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Risi S, et al. Combining information from multiple sources in the diagnosis of autism spectrum disorders. J Am Acad Child Adolesc Psychiatry. 2006;45:1094–1103. doi: 10.1097/01.chi.0000227880.42780.0e. [DOI] [PubMed] [Google Scholar]
- 19.Kong X, et al. A combined linkage-physical map of the human genome. Am J Hum Genet. 2004;75:1143–1148. doi: 10.1086/426405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- 21.Redon R, et al. Global variation in copy number in the human genome. Nature. 2006;444:445–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Persico AM, Bourgeron T. Searching for ways out of the autism maze: genetic, epigenetic and environmental clues. Trends Neurosci. 2006;29:349–358. doi: 10.1016/j.tins.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 23.Feng J, et al. High Frequency of Neurexin 1β Signal Peptide Structural Variants in Patients with Autism. Neurosci Lett. 2006;409:10–13. doi: 10.1016/j.neulet.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 24.de Vries BB, et al. Diagnostic genome profiling in mental retardation. Am J Hum Genet. 2005;77:606–616. doi: 10.1086/491719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sharp AJ, et al. Discovery of previously unidentified genomic disorders from the duplication architecture of the human genome. Nat Genet. 2006;38:1038–1042. doi: 10.1038/ng1862. [DOI] [PubMed] [Google Scholar]
- 26.Houlden H, Reilly MM. Molecular genetics of autosomal-dominant demyelinating Charcot-Marie-Tooth disease. NeuroMolecular Med. 2006;8:43–62. doi: 10.1385/nmm:8:1-2:43. [DOI] [PubMed] [Google Scholar]
- 27.Potocki L, et al. Molecular mechanism for duplication 17p11.2- the homologous recombination reciprocal of the Smith-Magenis microdeletion. Nat Genet. 2000;24:84–87. doi: 10.1038/71743. [DOI] [PubMed] [Google Scholar]
- 28.Moog U, et al. Hereditary motor and sensory neuropathy (HMSN) IA, developmental delay and autism related disorder in a boy with duplication (17)(p11.2p12) Genet Couns. 2004;15:73–80. [PubMed] [Google Scholar]
- 29.Iafrate AJ, et al. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 30.Lamb JA, et al. Analysis of IMGSAC autism susceptibility loci: evidence for sex limited and parent of origin specific effects. J Med Genet. 2005;42:132–137. doi: 10.1136/jmg.2004.025668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stone JL, et al. Evidence for sex-specific risk alleles in autism spectrum disorder. Am J Hum Genet. 2004;75:1117–1123. doi: 10.1086/426034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Falconer DS. Introduction to quantitative genetics. Longman; London; New York: 1981. [Google Scholar]
- 33.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 34.Camp NJ, Farnham JM. Correcting for multiple analyses in genomewide linkage studies. Ann Hum Genet. 2001;65:577–582. doi: 10.1017/S0003480001008922. [DOI] [PubMed] [Google Scholar]
- 35.Lange C, Laird NM. On a general class of conditional tests for family-based association studies in genetics: the asymptotic distribution, the conditional power, and optimality considerations. Genet Epidemiol. 2002;23:165–180. doi: 10.1002/gepi.209. [DOI] [PubMed] [Google Scholar]
- 36.Graf ER, Zhang X, Jin SX, Linhoff MW, Craig AM. Neurexins induce differentiation of GABA and glutamate postsynaptic specializations via neuroligins. Cell. 2004;119:1013–1026. doi: 10.1016/j.cell.2004.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Varoqueaux F, et al. Neuroligins determine synapse maturation and function. Neuron. 2006;51:741–754. doi: 10.1016/j.neuron.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 38.Purcell AE, Jeon OH, Zimmerman AW, Blue ME, Pevsner J. Postmortem brain abnormalities of the glutamate neurotransmitter system in autism. Neurology. 2001;57:1618–1628. doi: 10.1212/wnl.57.9.1618. [DOI] [PubMed] [Google Scholar]
- 39.Shinohe A, et al. Increased serum levels of glutamate in adult patients with autism. Prog Neuropsychopharmacol Biol Psychiatry. 2006 doi: 10.1016/j.pnpbp.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 40.Kugler P, Schleyer V. Developmental expression of glutamate transporters and glutamate dehydrogenase in astrocytes of the postnatal rat hippocampus. Hippocampus. 2004;14:975–985. doi: 10.1002/hipo.20015. [DOI] [PubMed] [Google Scholar]
- 41.Belmonte MK, Bourgeron T. Fragile X syndrome and autism at the intersection of genetic and neural networks. Nat Neurosci. 2006;9:1221–1225. doi: 10.1038/nn1765. [DOI] [PubMed] [Google Scholar]
- 42.Tavazoie SF, Alvarez VA, Ridenour DA, Kwiatkowski DJ, Sabatini BL. Regulation of neuronal morphology and function by the tumor suppressors Tsc1 and Tsc2. Nat Neurosci. 2005;8:1727–1734. doi: 10.1038/nn1566. [DOI] [PubMed] [Google Scholar]
- 43.Jalil MA, et al. Reduced N-acetylaspartate levels in mice lacking aralar, a brain- and muscle-type mitochondrial aspartate-glutamate carrier. J Biol Chem. 2005;280:31333–31339. doi: 10.1074/jbc.M505286200. [DOI] [PubMed] [Google Scholar]
- 44.O’Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rinaldo A, et al. Characterization of multilocus linkage disequilibrium. Genet Epidemiol. 2005;28:193–206. doi: 10.1002/gepi.20056. [DOI] [PubMed] [Google Scholar]
- 46.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 47.McPeek MS, Wu X, Ober C. Best linear unbiased allele-frequency estimation in complex pedigrees. Biometrics. 2004;60:359–367. doi: 10.1111/j.0006-341X.2004.00180.x. [DOI] [PubMed] [Google Scholar]
- 48.Gudbjartsson DF, Jonasson K, Frigge ML, Kong A. Allegro, a new computer program for multipoint linkage analysis. Nat Genet. 2000;25:12–13. doi: 10.1038/75514. [DOI] [PubMed] [Google Scholar]
- 49.McQueen MB, Blacker D, Laird NM. Variance Calculations for Identity-by-Descent Estimation. Am J Hum Genet. 2006;78:914–921. doi: 10.1086/503920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li C, Wong WH. DNA-Chip Analyzer (dChip) In: Parmigiani G, Garrett ES, Irizarry R, Zeger SL, editors. The analysis of gene expression data: methods and software. Springer; New York: 2001. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1: List of 624 CNVs in Filtered Analysis.
Supplementary Table 2: List of 254 CNVs in Affected Individuals.
Supplementary Table 3: Breakdown of CNVs in Affected Individuals.
Supplementary Table 4: List of Validated CNVs.
Supplementary Figure 1: Binned size distribution of CNVs in batch (A), plate (B) and filtered (C) analyses.
Supplementary Figure 2: Linkage results due to removing families in which affected individuals putatively carry CNV.
Supplementary Figure 3. Principal component plot used to infer ancestry.
Supplementary Figure 4: Linkage results obtained by analyzing families inferred to be of homogeneous European ancestry.


