Abstract
RNA granules are dynamic cellular structures essential for proper gene expression and homeostasis. The two principle types of cytoplasmic RNA granules are stress granules (SGs), which contain stalled translation initiation complexes, and processing bodies (P-bodies, PBs), which concentrate factors involved in mRNA degradation. RNA granules are associated with gene silencing of transcripts, thus, viruses repress RNA granule functions to favor replication. This review discusses the breadth of viral interactions with cytoplasmic RNA granules, focusing on mechanisms that modulate the functions of RNA granules and that typically promote viral replication. Currently mechanisms for virus manipulation of RNA granules can be loosely grouped into three non-exclusive categories; i) cleavage of key RNA granule factors, ii) regulation of PKR activation and iii) co-opting RNA granule factors for new roles in viral replication. Viral repression of RNA granules supports productive infection by inhibiting their gene silencing functions and counteracting their role in linking stress sensing with innate immune activation.
Keywords: Stress granules, P-bodies, G3BP, PKR
INTRODUCTION
RNA granules are found in all types of eukaryotic cells and tissues and are involved in many aspects of gene regulation, homeostasis and cytopathology. As such, many disciplines of biology are now acknowledging the importance of RNA granules. RNA granules consist of condensates of mRNA and RNA-binding proteins (RBPs) found in nuclear and cytoplasmic compartments and are believed to form by dynamic reversible condensation or aggregation of key RBPs, particularly Ras-GAP-SH3-binding protein (G3BP1) and T-cell restricted intracellular antigen (TIA1). All aspects of mRNA biogenesis and expression are regulated by RBPs, including splicing, transport, subcellular localization, silencing, translation activation and RNA decay. Several of these steps involve cytoplasmic RNA granules, including mRNA transport, silencing and decay and many RBPs key to these processes are also found in RNA granules. Nuclear RNA granules include PML bodies, cajal bodies, nuclear speckles, nuclear stress bodies and others (20, 74) and have diverse functions ranging from stress responses to processing mRNA and non-coding RNAs. This review focuses on cytoplasmic RNA granules and the viruses that modify them.
The most common types of cytoplasmic RNA granules are stress granules (SGs) and processing bodies (PBs) (shown in Fig. 2A), that are defined by their function and key components. Stress granules are transient foci enriched in translation initiation factors and 40S ribosomal subunits, while P-bodies are enriched for RNA decay machinery. SGs and PBs each contain unique marker proteins, however, many proteins are found in both SGs and PBs such as eIF4E, TTP, Ago2, APOBEC3, PCBP2 and others (53, 55). Based on distributions of unique and shared marker proteins in the granules, a continuum of RNA granules has been described in yeast, C. elegans and other species that span the gulf between SG and PBs (18). Importantly, different types of RNA granules such as SGs and PBs contain many unique components associated with unique functions, but also contain some common protein constituents.
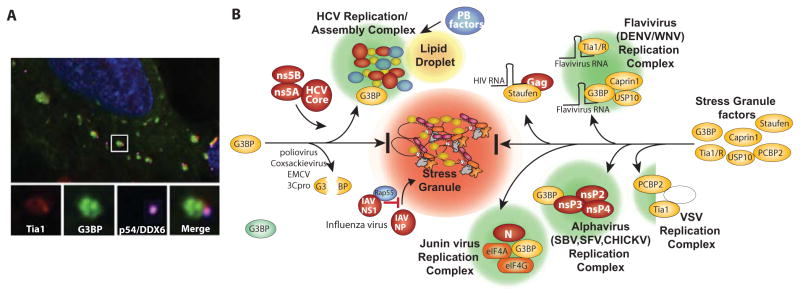
Figure 2.
Virus blockade and co-opting of stress granule responses. A. Deconvolution immunofluorescence microscopy image of SGs and PBs in uninfected oxidatively stressed U20S cells stably expressing GFP-G3BP. Boxed inset shows a typical SG (green) with a closely associated PB (magenta) and enlarged images below show individual channel antibody stains for SG markers Tia1 (red) and G3BP (green), and PB marker RCK/p54/DDX6 (magenta). Blue is DAPI stain for nuclear DNA. B. Specific points/proteins where viruses inhibit or divert the RNA granule assembly pathway are shown. Picornavirus 3Cproteinase cleaves the critical SG nucleating protein G3BP1. Several viruses co-opt G3BP and divert it into novel virus-induced foci. HCV diverts G3BP1 into replication/assembly complexes together with HCV core, ns5A and ns5B proteins that also associate with lipid droplets. HCVcomplexes also contain many P-body components detailed in Fig 3. Flaviviruses divert G3BP1 (with USP10 and caprin1) and TIA1/TIAR to replication complexes by binding the host proteins on virus RNAs. Junin virus (possibly N and G proteins) recruits G3BP1 into replication complexes with translation factors eIF4G and eIF4A. Alphaviruses recruit G3BP1 into viral replication complexes via direct interaction viral protein nsP3. Vesicular stomatitis virus diverts Tia1 and PCBP2 to virus complexes.
Translation control mechanisms dictate which mRNA transcripts gain access to ribosomes, and the process is highly regulated by the interplay of RBPs and RNA granules. Thus, stress granules and P-bodies are seen as extensions of the mRNP translation control cycle, as compartments where translationally silenced mRNPs are stored. A cytoplasmic mRNA cycle exists where mRNPs are in continual flux between active polysomes and these two silenced compartments (26, 56, 81). SGs and PBs transiently interact with each other, they rapidly exchange mRNPs with surrounding cytoplasm and can share certain protein components and specific mRNA moieties (3, 18, 22, 55, 109). Though not yet directly observed, several findings suggest that SGs and PBs exchange mRNP cargo.
SGs rapidly form when mammalian cells experience starvation, oxidative or heat stress. However, they also form under certain physiological conditions in cochlear hair cells (73, 119) and during types of virus infection (described below). Other physiological roles for RNA granules have been described in translation and development. Nondescript RNA granule assembly and disassembly temporarily controls cyclin B1 translation in zebrafish and mouse oocytes (59). Drosophila patterning is controlled by differential inclusion of mRNAs in P bodies (126). G3BP (in Drosophila called Rasputin, Rin) controls Orb translation and differentiation during Drosophila oogenesis (23).
In addition to their importance in virus systems, RNA granules are also closely linked to the pathology of neurodegenerative diseases. Mutations in key RBPs involved in RNA granules, particularly SGs, are causally associated with familial motoneuron diseases and dementias such as ALS, frontotemporal dementia and spinal muscular atrophy. These key RBPs also associate with pathological aggregates in Huntington’s chorea, Alzheimer’s disease and others. SGs share many constituents found in neuronal granules associated with disease pathology (Reviewed in (12, 120). Fused in sarcoma (FUS) and TAR DNA binding protein of 43kDa (TDP-43) are implicated as factors in amyotrophic lateral sclerosis (ALS), and FUS recruitment into SGs prevents irreversible aggregation (108). Pathological inclusions containing RBPs may arise from SGs in neurons (12). These findings have heightened interest in RNA granules and will push the field towards elucidating mechanisms that will support understanding of virus interactions with RNA granules.
RNA GRANULES PRESENT OBSTACLES FOR VIRUS REPLICATION
A basic function of cytoplasmic granules is organizing silenced mRNPs into condensed foci that serve as a type of molecular “storage closet” (Fig. 1). Retention of mRNPs in either SGs or PBs segregates those transcripts away from functioning in active polyribosomes and represses gene expression of those transcripts. However, emerging evidence indicates that RNA granules can also serve as sensing vehicles that link stress with infection and can respond by mounting antiviral innate immune responses. Depending on the strength of this response, this could be the primary reason so many viruses antagonize RNA granules.
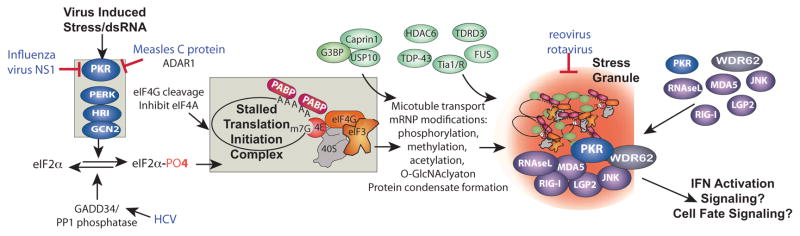
Figure 1.
PKR dependent stress granule assembly pathway, innate immune factors that enter stress granules and interference by RNA virus modulation of PKR. Virus infection causes stress at multiple levels by restricting translation through activation of eIF2 kinases, principally PKR, or by causing cleavage or inactivation of other translation initiation factors. These translation insults convert active polysome mRNPs into stalled translation initiation complex mRNPs containing 40S ribosome subunits, initiation factors and mRNAs (boxed). A complex series of events involving nucleation of multiple stress granule proteins such as G3BP1, Tia-1/TIAR, TDRD3, FUS, TDP43 and HDAC6 plus transport of mRNP complexes on microtubules leads to aggregates of translation initiation complex mRNPs in stress granules. Reovirus and rotavirus can repress SGs, but mechanisms are not known, points of interaction of other viruses with this scheme are indicated. Note that many viruses control PKR activation; only those discussed in the text are indicated.
RNA Granules Antagonize Efficient Gene Expression
Theoretically RNA granule retention of cellular mRNAs could be either deleterious or advantageous for infecting viruses, depending on the nature of the mRNA species included in granules. Inclusion of virus mRNAs themselves or key cellular gene products required to support infection would be disadvantageous, however, inclusion of mRNAs encoding innate immune factors might benefit the virus. At this point little precise information is available concerning the identity of mRNA transcripts included in various types of granules, though it is thought that certain stress-active transcripts such as heat shock protein mRNAs are excluded from SGs during heat stress. Mechanisms to distinguish these transcripts from others included in SGs are not known. In addition to silencing, inclusion of viral mRNPs into PBs carries the additional risk of RNA decay, as a significant portion of RNA turnover is carried out on transcripts within PBs. In terms of virus transcripts, inclusion into either SGs or PBs would be expected to repress the efficiency of virus replication and hinder viral spread and pathogenesis.
Finally, stress granule formation also causes sequestration of translation components into SGs, presumably reducing availability of key translation components for translation of virus transcripts. This includes small ribosome subunits, but also more critical translation initiation factors needed for both cap-dependent and cap-independent translation such as eIF3 and eIF4G.
RNA Granules Link Innate Immunity to Stress Sensing and Cell Fate Decisions
Viruses produce cell stress at multiple levels, and can trigger apoptosis, autophagy and modulate RNA granules. Thus it makes sense that sensors of cells stress may also be part of a virus sentinel network that mobilizes the innate immune system. Indeed, emerging evidence strongly supports this notion, linking innate immunity and cell stress responses at many levels, including SG and PB function.
Protein kinase R (PKR), a classic interferon response protein, is a centrally located activator of multiple downstream innate immune responses. PKR helps regulate stress activated c-Jun N-terminal kinase (JNK) and coordinate pathogen sensing with cellular stress and metabolic homeostasis (84). PKR functions in insulin signaling and metabolism by phosphorylating the insulin receptor substrate IRS1 (129), but it also functions in macrophages for inflammasome activation in response to dsRNA and bacterial infections that induce release of cytokines IL-1b and HMGB1 (69). Importantly, PKR is also an activator of innate immune transcriptional responses (37, 114, 115). Thus, multiple nutrient and pathogen response systems may be integrated through PKR. Interestingly, PKR colocalizes with SGs that arise during infections with mutant influenza A virus and measles virus (89, 90). In the measles virus system PKR was found to have a functional antagonist RNA adenosine deaminase (ADAR1) that also enters SGs. ADAR1 counteracts PKR by suppressing the host innate response, PKR activation and the subsequent SG response (89).
PKR activation and stress sensing is linked to at least two RNA granule factors, RasGAP-SH3-Binding protein (G3BP1) and mRNA decapping enzyme 1a (Dcp1a). G3BP1 expression induces stress granule assembly in uninfected cells in the absence of external stress, which activates PKR and downstream eIF2-dependent translational repression. This indicates PKR may sense formation of stress granules per se through a unique mechanism of activation (102). A similar activation of PKR by Dcp1a also occurs that may involve the PKR activator PACT (28). Once activated, PKR represses translation via eIF2α phosphorylation and can signal downstream to several innate immune effectors such as transcription factor nuclear factor kappa B (NF-kB) that functions in many inflammatory responses (38). PKR also interacts with the IkB kinase complex, promoting dissociation of IkB from NF-kB and NF-kB transcriptional activation (16, 39).
A new concept is that the key SG nucleating protein G3BP1 may be antiviral. Overexpression of G3BP1 in CVB3-infected cells results in marked decreases in VP1 protein expression, and viral titers; in contrast, siRNA knockdown of G3BP1 resulted in modest increases in viral protein and RNA levels (35). The former could result simply from SG-induced activation of PKR (102), however, siRNA knockdown results support a more direct antiviral role. Similarly, EMCV disrupts SGs via cleavage of G3BP1 and expression of cleavage-resistant mutant G3BP1 restored SGs and enhanced IFN and cytokines production late in infection. Knockdown of G3BP1 also blocked SG formation, reduced IFN production and increased virus output (85). Together this supports an antiviral role of either SGs or G3BP1 but does not distinguish them.
In a new mechanistic twist on innate immune activation, cells were observed to concentrate both interferon-activating proteins and interferon-stimulated gene (ISGs) proteins within SGs. This includes PKR and viral RNA-sensing RIG-I like receptors (RIG-I, MDA-5, LGP2) that enter SGs after arsenite induction or after virus infection with mutant influenza virus (90). The well characterized ISGs RNAse L and OAS also enter SGs (90). PKR also enters P-bodies during human papilloma virus infection (44).
However innate immune activation mechanisms are likely complex and mere colocalization of factors in SGs does not necessitate activation or even protein interaction. MDA5 localizes to SGs but this localization is not required for IFN-α/β induction, as determined with a mutant mengovirus with inactive L protein that cannot suppress IFN-α/β(61). Infection of cells that are defective in SG formation yielded higher viral RNA levels, suggesting SGs are antiviral for mengovirus. SG formation induced by mengovirus was PKR-dependent, suggesting that the PKR axis of SG activation is important. As expected however, IFN activation occurs by many routes and does not absolutely require SG or PKR, as cells that were depleted of PKR and defective in SG response, still produced IFN during infection (61).
Comparing IAV and IAV with a mutant NS1 that does not block IFN induction, one group found that inhibition of IFN induction was not essential to block SG formation, which was PKR dependent (57). Another group used defective IAV ΔNS1 to show that enhanced IFN-β mRNA production was linked to appearance of SGs. Depletion of SGs via G3BP1 knockdown or PKR knockdown reduced IFN production (90). Though intriguing, these results leave open the possibility that either G3BP1 or PKR are critical for the innate immune response rather than SGs themselves. Certainly, it is well accepted that PKR is important to mount innate immune transcriptional responses.
Together, these studies suggest that stress granules (and perhaps PBs) may serve as sentinel mechanisms for interferon activation by acting as platforms that enhance IFN and cytokine signaling through condensation of IFN signaling moieties with their ligands. In this way, SGs may also be important in priming neighboring cells against infection. SGs may also serve as a platform to link innate immune activation with other stress signaling pathways. Supporting this tenet, the stress-responsive MAPK JNK is activated in a nonclassical pathway during stress granule formation involving a novel scaffold JNK-binding protein, WDR62. In this activation, both JNK and WDR62, are recruited to SGs by overexpression of SG nucleating proteins, but arsenite stress sends only WDR62 to SG while JNK localizes to PBs, indicating SGs induced by alternate pathways signal JNK differently (124).
VIRAL MECHANISMS TO CONTROL STRESS GRANULE ASSEMBLY
Viruses have evolved into highly efficient pathogens that establish successful infections by compromising host innate immune systems in a variety of ways. However, mammalian cells also have multiple strategies to cope with the myriad of virus replication schemes and viral assaults on innate immune defenses. SGs may play prominent roles in limiting virus translation and replication and perhaps buying more time for cells to activate other antiviral signaling pathways that decide cell fate. Different consequences of viral infection on SG formation have been observed: viral suppression of SG formation, induction of transient SG assembly, induction of stable SGs and oscillation between SG assembly and disassembly during the course of infection. Most viral infections will hinder SG formation at some point in the infection cycle, indicating manipulation of RNA granules is critical for viruses.
The actual mechanisms for SG formation are poorly defined but canonically start with translation suppression via phosphorylation of eIF2α, inhibition of eIF4A helicase function or cleavage of eIF4G (4, 55, 56, 76), that forces accumulation of stalled translation initiation complexes (Fig. 1). Activation or overexpression of key SG nucleating proteins such as G3BP1, TIA1, TIAR, TDP-43, and caprin1, can also drive SG assembly but specifically how they function alone or together is not understood (54). Transport of stalled mRNPs to sites of RNA granule assembly also requires mobility on microtubules and post-translational modification of factors (68, 87, 88). The actual molecular mechanisms driving mRNP aggregation into large structures while also allowing dynamic exchange of components has been elusive. Recent advances suggest transitions from dispersed to condensed phases drives formation of “liquid droplets” or condensed “gel phases” that can be considered dynamic RNA/protein droplets (13, 42, 49, 54, 125). A key molecular interaction driving the process may be multivalent weak binding interactions between key mRNP proteins that contain multiple proline-rich motifs, SRC homology domains or other regions of low complexity amino acid sequences (63). Fused in sarcoma (FUS), TDP-43, G3BP1, TIA1 and many other RNA binding proteins found in SGs have suitable weak interaction domains (9, 13, 42, 49). Post-translational modifications may figure prominently in regulation of these weak interactions and have been implicated in RNA granule assembly. For example, G3BP1 is phosphorylated and arginine methylated and TDP-43 arginine methylation and ubiquitination regulate SG assembly or coaggregation. Further, decoration of ribosomal proteins with O-linked N-acetylglucosamine is required for SG formation (15, 25, 88, 118). Virus interference in any of these steps may be sufficient to block SG formation. The discussion below is categorized into non-exclusive classes based on mechanism of virus interaction with the RNA granule machinery, namely (1) cleavage of RNA granule factors, (2) control of PKR function, and (3) co-opting of RNA granules.
Cleavage or Modification of SG Components
Plus strand RNA viruses encode viral proteinases that often cleave pivotal host proteins to favor virus replication (Fig. 2B). To date, only picornaviruses such as poliovirus (PV), coxsackievirus B3 (CVB3), and encephalomyocarditis virus (EMCV) have been found to cleave SG components, using similar mechanisms involving their conserved 3Cproteinases (3Cpro). In general, these viruses induce transient SG assembly where SGs form at the early phase of infection (~2–3 hpi), but active disassembly of SGs occurs during the mid phase of infection (~3–8 hpi) (35, 85, 127). These viruses disassemble SG through cleavage of the critical SG nucleating protein G3BP1 by the viral 3C proteinase at the glutamine residue at amino acid 326. The crucial role of G3BP1 in SG formation was demonstrated through rescue of SG formation late in poliovirus infection from expression of 3Cpro cleavage resistant G3PB1 mutant (Q326E) (127). The C-terminal fragment of G3BP1 further inhibits SG formation and promotes CVB3 viral replication, although several fragments of G3BP1 can inhibit SG formation (35, 118). A partly conflicting study reported SG stained with TIA1 persist in PV-infected cells at late times post infection (98), but these foci are compositionally distinct from bonafide SGs since they exclude key functional SG markers eIF3, eIF4G, PABP and mRNA (128). Thus, poliovirus directed cleavage of G3BP1 unlinks TIA1 aggregation, which still occurs, from assembly of stalled translation complexes in RNA granules. The residual TIA1-stained foci do not correlate with translational repression and accumulation of stalled translation initiation complex that define functional SGs. These findings imply that the composition and function of SGs can be different in virus-infected cells and SGs cannot be defined by use of only limited SG markers.
Although a viral 3C proteinase is encoded by all members of the picornavirus superfamily, not every picornavirus digests G3BP1. Theiler’s murine encephalomyelitis virus (TEMV) blocks SG formation via an undetermined function of viral leader (L) protein, while G3BP1 remains intact and enters SG during infection with a TMEV encoding mutated L (17). Cricket paralysis virus (CrPV) infection induces eIF2α phosphorylation that leads to a rapid shutoff of host translation system (58), however it does not induce SG formation, aggregation of the G3BP1 and TIA1 paralogs (Rin-8 and Rox), or cleave these markers, even in the presence of chemical SG inducers. Moreover, CrPV 3C proteinase colocalized in SG during cell stress, but not during infection, indicating other viral genes influence its cytoplasmic distribution.
PKR Regulation to Control SG Assembly
PKR is a crucial sensor of virus infection and cell stress, and is regarded as the canonical initiator of stress granule assembly. Most animal viruses provoke PKR activation, resulting in phosphorylation of eIF2α, blockage of translation initiation and accumulation of SGs. PKR-like endoplasmic reticulum kinase (PERK) is an alternative stress sensor that detects ER-unfolded protein responses caused by virus infection, potentially resulting in similar downstream SG assembly, but direct evidence has not yet been reported. Aggregation of G3BP1-mRNP complexes also results in PKR activation (102). There is a profusion of viral schemes to counteract these restrictions on translation, including mechanisms to; (i) antagonize PKR activation, (ii) avoid eIF2α phosphorylation and (iii) bypass cap dependent translation process. Some of these also apply to control of SG assembly (Fig. 1).
Influenza A virus (IAV) blocks SG formation throughout infection by expressing NS1 protein, which suppresses IFN activation and serves as a potent PKR antagonist through its dsRNA binding domain. Expression of IAV with mutated NS1 that does not bind dsRNA results in phosphorylation of eIF2α and accumulation of SGs (57). Viral replication is repressed when SG form, as determined by accumulation of viral NP protein. On the other hand, SG do not form in PKR knockout cells infected with NS1 mutant virus (57). Taken together, the repression of PKR activity via NS1 is pivotal for suppressing SG formation in this system. Other investigators have confirmed ablation of NS1 in IAV (IAV ΔNS1) promotes accumulation of SGs (90).
NS1 also inhibits SG formation through interaction in a complex with cellular RNA associated protein 55 (RAP55), which is a protein shared by SG and PBs that may facilitate mRNP cargo transport. Ectopic expression of RAP55 mobilized SG formation and suppressed virus replication (80). RAP55 and PKR interact with the same domain on NS1, but it is unclear if they compete. One function of NS1 prevents viral nucleoprotein (NP) colocalization with SGs, which is observed in infections where NS1 is absent (IAV ΔNS10). Interestingly, NP colocalizes with PBs during wild type virus infection (80), but the function is unknown. A contrary report indicated the levels of IAV NP produced (i.e. viral replication) did not significantly change in cells infected by IAV ΔNS1, despite significant PKR activation, production of phospho-eIF2α and SG assembly (90). These results indicate that the composition and assembly of SGs can be dependent on the context and cell type used (A549 cells versus HeLa).
Rotavirus shuts off host protein synthesis through partial induction of eIF2α phosphorylation that is mediated by viral proteins VP2, NSP2, and NSP5. Despite occurrence of this significant translation restriction, SGs are not assembled during the course of infection or after addition of external SG inducers (82). The mechanism for rotavirus inhibition of SG formation remains elusive, however, may result from fine-tuning maximal levels of phosphorylated eIF2α. Viral translation may normally be partly restricted by eIF2α phosphorylation since virus replicates more efficiently in mutant mouse embryonic fibroblasts with mutant non-phosphorylatable eIF2α (82).
Mammalian orthoreovirus (MRV) promote transient SG formation to create a competitive advantage for translating viral RNA at the early stage of infection (6 hpi), since controlled eIF2α phosphorylation is required for uncoating the virus and for translational upregulation of the transcription factor ATF4 that is proposed to facilitate virus propagation (99, 100, 112). Control of PERK and PKR may stem partly from viral manipulation of their cellular inhibitor p58IPK (112). The non-structural protein μNS strongly interacts with SG via amino acids 78 and 79, but does not induce or disrupt SG when expressed alone. However mutant μNS (78–79) no longer binds core protein (λ2) and is defective in supporting virus replication (19), so the link between μNS localization to SG and virus replication remain unclear. However, PKR and the other individual eIF2α kinases are not solely required for SG induction by MRV, indicating that SG formation requires multiple eIF2α kinase signaling or another mechanism(100). MRV restricts SG formation at late times post infection (24 hpi) at a point downstream and independent of eIF2α phosphorylation, suggesting some viral protein(s) may interfere with SG assembly.
Hepatitis C virus (HCV) is associated with slow replication kinetics and chronic infections, thus must control stress responses long term. Interestingly, HCV induces PKR-dependent SG formation but that is subsequently counteracted by DNA damage-inducible protein 34 (GADD34), the cofactor of phosphatase 1 which dephosphorylates of eIF2α. Multi-day HCV infections involve oscillating SG formation, temporarily assembling and disassembling SGs by rapidly switching eIF2α phosphorylation status(104). Disassembly of SG allows relief of translation restriction to allow viral protein synthesis and to maintain prolonged cell survival and chronic infection. In this regard, PKR and GADD34 are important gatekeepers for regulating cellular homeostasis linked to cell fate decisions. HCV also limits PKR accessibility and sensing of viral dsRNA by forming a nuclease-free membrane-wrapped vacuole compartment (93).
Coronaviruses such as Transmissible Gastroenteritis (TGEV) and Mouse Hepatitis Coronavirus (MHC) induce TIAR-containing granules throughout the infectious cycle, which correlate with an increase in eIF2α phosphorylation early in infection (MHC) or later (TGEV) (101, 113). Although it is still unclear whether these granules disassemble during the infection for either virus, SGs may restrict TGEV infection. Ablation of the SG constituent protein PTB enhanced virus replication, whereas PTB overexpression restricted virus output. MHV also replicates more efficiently in non-phosphorylatable eIF2α S51A mutant MEFs that cannot easily form SGs. The type of granules that form in both MHC and TGEV infected cells has not yet been rigorously determined since sole use of TIA1/TIAR as SG markers may not reliable (128). Hence, further research is required to demonstrate that bona fide SGs accumulate and persist in coronavirus infection or if TIA1 foci represent novel virus foci with co-opted SG components as described in the virus systems below.
Co-opting of SG Components
Plus strand RNA viruses need to recruit ribosomes to viral RNA to synthesize viral proteins and RNA-dependent RNA polymerase; however, the same RNA template must then switch to a translationally-silenced state to clear ribosomes and enable RNA replication to begin. Therefore, it is plausible that cellular RNA regulatory proteins will be utilized both for supporting translation and the shift to virus replication. Also, since SG formation involves nucleating/condensation properties of RNA binding proteins such as G3BP1 and TIA1; viruses may redirect these proteins toward new roles in building condensates of factors for virus replication factories or even aiding assembly of capsids (Fig. 2B). The alphavirus, flavivirus and coronavirus systems discussed below all coopt, but do not cleave SG proteins, unlike picornaviruses, despite the fact that each of these viruses encode proteinases.
Nonstructural proteins of several alphaviruses interact with G3BP1 in viral complexes that contain nsP3 (24, 33, 34, 40), nsP2 (8) and the viral polymerase nsP4(24) (Fig. 2B). Semliki Forest Virus (SFV) induces eIF2α phosphorylation and forms bona fide SGs at early times of infection (4 hpi). While infection progresses, SFV hijacks G3BP1 from SG and relocates it into viral replication complexes (RC) via interaction with C-terminal repeat domains of nsP3, simultaneously disassembling SGs. At this point SG formation is blocked in the presence of pateamine A, an eIF2α-independent SG inducer (77, 92). Chikungunya virus applies similar mechanism to sequester G3BP1 and redirects it into novel cytoplasmic foci that lack the SG marker eIF3 (34). SFV also contains a translational enhancer near the initiating AUG codon that permits viral RNA to escape translational repression from by eIF2α phosphorylation. However, efficient translation of viral RNAs by this enhancer also facilitates disassembly of SGs during infection (77, 92). The benefit of co-opting G3BP1 to viral RCs still needs to be explored, but maintaining SGs by mutation of the G3BP1-interacting region in nsP3 dampens viral replication (34).
The same concepts can be applied to Sindbis virus (SBV), another alphavirus, whose RNA-dependent RNA polymerase nsP4 immunoprecipitates with G3BP1 and 2 (24). Since G3BP1 is also recruited into viral complexes with nsP2 and nsP3, this may indicate overlapping interactions within a large viral RC (33, 40). Knockdown of G3BP1 only slightly altered SBV RNA levels, but significantly increased SBV polyprotein production (24). The role of G3BP1 in SGs may explain why viral translation is affected more than viral RNA replication.
Rubella virus (RUBV) is reported to induce SG-like granules containing G3BP1 late during infection (48 hpi), concomitant with peak accumulation of viral particles (75), but only in half the infected cells. These granules are compositionally distinct from normal SGs due to lack of PABP and TIA1 and resistance to dispersion by cycloheximide. The granules and G3BP1 distribution did not localize in regions with viral dsRNA but did overlap with viral ssRNA in perinuclear regions, suggesting a potential role for G3BP1 in virus encapsidation but not replication (75).
In general, flaviviruses such as West Nile virus (WNV), Dengue (DENV) and Japanese Encephalitis virus (JEV) block arsenite-induced SG formation by hijacking multiple key SG components, including, TIA1, TIAR, G3BP1 and caprin-1 (Fig. 2B). During WNV and DENV infections, TIA1/TIAR are sequestered by the 3′ stem loop structure that is a promoter for minus strand RNA synthesis, and both proteins colocalize with viral RCs (containing dsRNA and NS3) in perinuclear regions of cells (30, 31, 64). In addition, JEV virus core protein forms complexes with caprin-1 to compromise SG formation and increase viral replication (50). Proteomic analysis indicated the 3′ UTR of DENV genomic RNA interacts with G3BP1 and G3BP2, as well as G3BP1 interacting proteins USP10 and caprin1. Lacking functional studies, the exact role of these proteins in DENV replication is unknown. It is likely that recruiting SG proteins in new compartments allows virus translation or RNA replication, or prevents a strong innate immune response as a consequence of SG assembly.
Hepatitis C virus induces and controls eIF2α phosphorylation to promote SG assembly, as mentioned above, but also co-opts SG proteins and can trigger SGs through an eIF2-independent pathway (91). During infection novel foci containing HCV core protein form near cytoplasmic lipid droplets (Fig. 2B) and multiple SG (G3BP1) and PB factors (Xrn, DDX6, RCK/p54) are also repartitioned to lipid droplets (7, 91). Roles of these factors in replication and assembly are unknown and likely complex, since knockdown of G3BP1, TIA1, TIAR, PABP, USP10 and HuR affect different steps of the HCV lifecycle (7, 36, 91, 131). G3BP1 also colocalizes and interacts with NS5A and NS5B, two components of the HCV replication complex, suggesting a direct role for G3BP1 in HCV RNA replication (131). G3BP1 may also inhibit assembly and release of HCV virions (91).
The ambisense RNA arenavirus Junin virus (JUNV) encodes viral protein N and glycoprotein precursor products that repress SG assembly (67). This may occur via sequestration of G3BP1 into novel replication-transcription complexes. These complexes increase the efficiency of infection or enhance virus translation, however are not regular SGs since they contain neither PABP nor TIA1(10). Unexpectedly, some translation apparatus colocalizes in replication-transcription complex, including eIF4G, eIF4A, large ribosomal protein L10a and small ribosomal subunit protein S6, indicating the possibility that replication-transcription complexes participate in viral RNA translation. However, it is still unclear whether G3BP1 is involved in this process for replication.
The retrovirus HIV-1 actively inhibits SG formation in cells treated with arsenite (1). The mechanism of SG inhibition may involve sequestration of the stress granule protein Staufen1 to special RNPs containing the viral protein Gag and HIV1 viral RNA, called Staufen1-dependent RNPs (SHRNPs), but it is unclear because staufen is normally recruited to SGs to regulate and impair their assembly (117). Depletion of Staufen1 co-depletes Gag from infected cells and results in enhanced encapsidation of viral mRNA and residual Staufen1 (1). These results indicate that HIV1 prevents stress granule assembly to subvert Staufen1 to alternative RNPs that regulate viral encapsidation.
Tax protein of another retrovirus, HTLV, inhibits SG assembly through sequestration of a SG key component HDAC6, a histone deacetylase and important for SG formation (62) (60). Another study reports that USP10, an ubiquitin specific peptidase, is a binding partner of Tax protein in HTLV-infected T cells where it inhibits SG assembly and stimulates ROS production (116). USP10 is a binding partner of G3BP1, thus Tax may function by disrupting this interaction. Sequestration of HDAC6 may inhibit key post-translational modification of proteins required for SG formation, but this remains to be shown.
Negative strand RNA virus, measles Virus (MV) represses SG assembly though the function of viral protein C and an IFN-inducible double-stranded RNA-specific adenosine deaminase (ADAR1) during the course of infection. Cells infected with C protein mutant MV viruses (Cko) induced SGs after 24 hpi, and this process was PKR dependent. A low proportion of wild type MV infected cells could form SGs containing G3BP1 and ADAR1, a nuclear shuttling protein that is a weak PKR antagonist. In contrast, infecting ADAR1 depleted cells with wild type MV robustly stimulated PKR activation, SG formation and IFN-β induction, suggesting that ADAR1 is a SG inhibitor that antagonizes PKR activity. Further experiments also verified that the p150 isoform of ADAR1 drives this process through deaminase catalytic activity (89). The follow up study indicates (Cko) virus induces PKR efficiently via a replication defect that causes excessive accumulation of dsRNA (95). The authors reveal a new route for virus to manipulate SGs where nuclear-cytoplasmic relocation of ADAR1 inhibits PKR activation, which in turn avoids eIF2α phosphorylation, IFN-β production and eventually suppresses SG formation.
Another negative strand RNA virus, vesicular stomatitis virus, produces SG-like structures during infection containing PCBP2, TIA1, TIAR that appear simulaneously with phosphorylated-eIF2α (27). These granules were distinct from bona fide SG due to the absence of eIF3 and eIF4A. The novel granules do contain viral replicative proteins and RNAs and their formation was dependent on ongoing viral replication. siRNA depletion experiments indicated that PCBP2 and TIA1 downregulate VSV replication despite their continued inclusion into virus-induced SG-like structures (27), nonetheless, this is another example of select SG components co-opted into viral RCs, likely at the expense of formation of bona fide SGs.
In contrast to nearly all other virus systems, the negative strand virus Respiratory Syncytial Virus (RSV) is reported to induce SG-like foci that may increase viral protein production during the course of infection instead of repress it. G3BP1 knockdown in RSV-infected cells resulted in a 10-fold reduction in RSV titer, however this was mostly a delay in replication kinetics (65, 66). These data are reminiscent of results from the vaccinia virus system (see below) where, under specific conditions, SG-like aggregates correlate with virus improved replication. The RSV-induced granules were not probed for presence of multiple SG markers so their identity as bona fide SGs versus novel virus-induced G3BP1 or TIA1-containing foci are unresolved. Further, RSV was noted to deplete another SG protein HuR from G3BP1 foci, suggesting G3BP1-containing foci are defective in terms of SG composition. A contrary study using the RSV system showed that SG formation was inhibited, specifically by the trailer region of the viral genome (43). However an unspecified eIF3 subunit was used as the sole SG marker, leaving the possibility that SG-like proviral structures form, which lack that eIF3 subunit, in a trailer-dependent manner. Nevertheless, the finding was consistent with earlier findings that the related Sendai virus trailer also blocks SG formation during infection (46).
To date, herpes simplex virus (HSV), and vaccinia virus (VV) are the only DNA viruses reported to regulate SG assembly. During acute infection HSV-2 actively represses SG formation that is phospho-eIF2α-dependent but does not restrict formation of SG foci induced by eIF4A inhibitor pateamine A. In this case phosphorylated eIF2α is found to accumulate at 10 hpi without assembling SGs. The pateamine A-induced SGs were atypical, containing G3BP1 and PABP but were deficient in TIA1. TIA1 was instead partitioned into non-canonical nuclear foci containing Sam68. These data suggest that TIA1 is co-opted into novel roles that augment HSV-2 viral replication or transcription, similar to the examples for Dengue virus and WNV.
Vaccinia virus (VV) forms large viral replication factories that co-opts several key SG proteins, including G3BP1, Caprin1, eIF4E, eIF4G and poly(A) mRNAs. The functions of these SG key proteins was not to assemble bona fide SGs, but were proposed to result from coordinated transcription and translation processes in the viral replication factory (52). Indeed, VV transcription is regulated by caprin1 and G3BP1 (51). Further, translation factor and SG component eIF4G1 is recruited to single strand DNA in factories by protein I3, a ssDNA binding viral factor (134). Translation initiation factor foci that formed in and around virus factories were shown to be devoid of the translational suppressor TIA1(121). These results indicate VV subverts several components of stress granules for proviral roles in replication. Another study partly at variance with the work of Katsafanas et al. has described the colocalization of SG components (TIA1, eIF3b, G3BP1 and Usp10) in granules surrounding VV factories using cells infected with ΔE3L mutant vaccinia virus that fails to control eIF2α phosphorylation. These foci were termed “antiviral” granules, which are not canonical SG because they lack 40S ribosome subunits, but correlated with restricted virus replication similar to SGs in other systems (111).
VIRAL MECHANISMS TO CONTROL P-BODIES
Since P-bodies are constitutively present compartments for gene repression and RNA decay, it is no surprise that viruses may antagonize their functions and existence. Several viruses are known to disrupt PBs and co-opt PB components. Mechanisms of PB disruption are poorly elucidated but some details are emerging. The relationships between viruses and PBs are quite varied. The virus-PB interactions below are grouped into provisional categories that will require revision in the future after new discoveries are made. Figure 3 illustrates virus-PB interactions that are discussed.
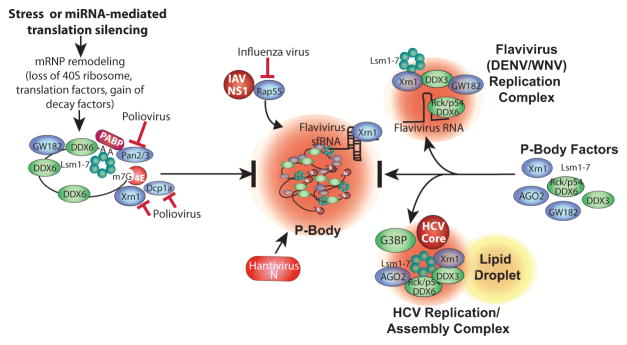
Figure 3.
Pathways of P-body disruption by viruses. P-bodies form via a complex series of events involving remodeling mRNPs by stripping of initiation factors and ribosome subunits, association with GW182, undergoing Pan2/3-mediated deadenylation, microtubule transport, and association with other RNA decay factors (e.g., Xrn1, Dcp1a, DDX6 (Rck/p54), GW182 and Lsm components of the exosome), and finally condensation in P-bodies. HCV co-opts many PB components into novel viral replication/assembly foci with viral core protein that also contain some SG components (e.g. G3BP, Fig. 2). Flaviviruses also divert PB factors into replication foci, likely bound with viral RNA through interaction with DDX6(Rck/p54). Poliovirus induces cleavage of Dcp1a and rapid degradation of Xrn1 and Pan3. Rap55 is a key PB factor that IAV protein NS1 prevents from entering PBs. Bunyavirus Junin virus viral N protein localizes to PBs to inhibit cellular Dcp1a/2 decapping function and facilitate viral cap-snatching.
Disruption of P-bodies
Enteroviruses such as poliovirus and Coxsackievirus B3 use two viral proteinases as “security” proteins that cleave select host proteins to facilitate replication. These viruses completely disrupt P-bodies during the replicative phase; however, both viral and cell proteases may contribute to their demise. Three key PB components, Dcp1a, Xrn1 and Pan3 are degraded, but only Dcp1a cleavage is linked to viral proteinase 3C cleavage, while Xrn1 and Pan3 undergo accelerated proteasomal degradation (29) (Fig. 3). Depletion of Pan3 may induce PB disruption since partial deadenylation of transcripts by the Pan2/Pan3 deadenylase complex is required for inclusion of mRNPs in PBs (135). Although Dcp1a helps regulate PB dynamics (105), more work is required to link its cleavage to PB disruption. Unlike Dcp2, Dcp1a is not a core PB protein, but rapidly exchanges with the cytoplasmic pool(2). However, Dcp1a is able to provide crosstalk with the translation regulation system by inducing PKR activation (28). Thus a potential goal of PV cleavage of Dcp1a may be to limit host defenses as well as disrupt RNA decay.
The insect dicistroviruses such as Cricket paralysis virus, which are distantly related to enteroviruses, also disrupt foci defined by PB marker proteins, but moderately and at later times in infection. In this case, PB constituents may be selectively modified since granules tagged with GFP-GW182 and GFP-Dcp1 diminished, however granules marked by inclusion of GFP-AGO1 or 2 were retained during infection, indicating formation of alternate PB-related foci of unknown function (58).
The study of influenza A virus infection indicated that cellular RAP55, found in both SG and PBs, may be required for PB formation or dynamics. Influenza viral protein NS1 can enter PBs and complexes with RAP55, coincident with slow dispersion of PBs. Overexpression of NS1 depletes PBs containing RAP55 (80). NS1-RAP55 complex formation benefits the virus by preventing viral nucleoprotein from entering PBs, which is observed early in infection and regarded as a host restriction mechanism to sequester it from translation and replication processes.
Adenovirus E4 11k protein binds DDX6, and relocates a least five PB components (DDX6, Lsm1, Ge-1, Xrn1, Ago2) to cytoplasmic aggresomes induced by the virus, sites where the proteins are inactivated. Aggresome formation correlated with PB depletion, and since PBs are sites for mRNA degradation, their modification by 11 k may explain how E4 11K stimulates late viral mRNA accumulation (41).
Co-opting of P-body components
Instead of destroying PBs, viruses may usually co-opt PB components during infection, resulting in a modest reduction in the numbers of cytoplasmic PB (Fig. 3). Flavivirus such as West Nile virus and Dengue virus that bind the SG protein TIA1 on viral RNA, also sequester many P-body constituent proteins, including DDX3, DDX6, GW182, Xrn1 and Lsm1, near or within perinuclear viral replication centers (21) while the numbers of PBs in cells decrease (30) (Fig. 3). Evidence is building that PB constituents may directly bind viral RNA to aid recruitment to viral replication factories and may functionally promote RNA replication. Depletion of GW182, Lsm1, and DDX3 by siRNA knockdown reduces virus WNV RNA output (21) and knockdown of DDX6 reduces Dengue virus output (123). Dengue virus genomic RNA has a conserved stem loop structure in the 3′ UTR that binds DDX6 (Rck/p54); this feature is adjacent to the unstructured region that binds SG proteins G3BP1 and USP10 (123). DDX6 is proposed to mask and unwind mRNAs and organize structures within PBs (32), thus it is possible that it also plays unwinding/organizational roles in viral RNA replication or packaging.
Flaviviruses also produce a novel sfRNA, a fragment of the 3′ UTR of plus strand viral RNA that results from stalling Xrn1 5′–3′ exonucleolytic decay at a highly structured pseudoknot (97, 110). sfRNA and Xrn1 colocalize in PBs, and sfRNA is important for cytopathogenicity of West Nile Virus (97). sfRNA inhibits Xrn1 activity via sequestration, thus forcing uncapped cellular mRNAs to accumulate in cells (83). sfRNA also can inhibit Dicer cleavage activity and exhibits RNAi suppressor activity (107). Thus, sfRNA has novel roles in inhibiting host nucleases involved in gene regulation and innate immunity that may indirectly affect RNA granule function.
Hepatitis C virus (HCV) also co-opts PB proteins and results in slow loss of PBs throughout infection (91). Key steps of HCV replication and assembly take place around lipid droplets. Although HCV core may not directly interact with DDX3 (5), HCV core protein forms complexes containing DDX3 and colocalizes in lipid droplets (72, 132). In addition, several other PB components (DDX6 (Rck/p54), Lsm1, PATL1, Ago2, and Xrn1) are co-opted to HCV core-containing assembly sites at lipid droplets (7, 91) (Fig. 3). Several of these factors are thought to play functional roles in viral assembly since knockdown of Lsm1, PatL1, DDX3, DDX6, (6, 7) or RCK/p54 and Ge-1 (91) reduced HCV replication. HCV may co-opt DDX6 helicase function to aid RNA packaging, which has been observed for the spumaretrovirus foamy virus (133). Beyond assembly, the PB components DDX6, Lsm1-7, and PatL1 also function in support of HCV translation and RNA replication (48, 106).
HCV requires microRNA-122 (miR-122) to bind genomic RNA and stimulate replication. Since miRNAs normally function through transcript silencing mechanisms involving PBs and their constituents, and since miRNAs can repress HCV experimentally, this conundrum has been investigated further. The problem is partly solved by HCV partitioning of host RNAi and viral factors into distinct cytoplasmic compartments. Viral RNA replication requires recruitment of Ago2 and miRNA-122 to lipid droplets, whereas silencing of HCV RNA by siRNA and Ago2 requires interaction with P-bodies (14). Normal RNAi biochemical roles are also separated from virus functions; DDX6 silencing decreased HCV replication and translation, but not the ability of miR-122 to stimulate HCV translation or promote HCV RNA accumulation. DDX6 does not have a role in the function of miR-122, and it appears that DDX6 and miR-122 modulate HCV through distinct pathways (45). Conversely, Lsm1 contributes to activation of HCV internal ribosome entry site (IRES)-driven translation by miR-122 in a novel interaction (103).
Despite the co-opting of numerous PB components, PB foci per se may not inhibit HCV replication. Knockdown of RAP55 diminished PBs, but did not inhibit HCV RNA or protein levels (94), which may suggest normal dynamic shuttling of PB components suffices to provide HCV with its required functions. Overall, these findings indicate that HCV co-opts some PB constituents to support replication and others for assembly; however, the existence of PB foci is not inhibitory during infection. Finally, unlike fast replicating lytic enteroviruses, HCV does not rely on cleavage and degradation of PB components, despite containing a viral proteinase (91).
The Bunyaviridae family of ambisense segmented RNA viruses acquire 5′ m7-guanosine capped oligonucleotides from cellular mRNAs by “cap-snatching” to initiate viral transcription. The nucleocapsid protein (N) of Hantavirus binds the 5′ cap of cellular mRNAs and N accumulates in PBs, resulting in inhibition of Dcp1a/Dcp2-mediated decapping (Fig. 3) and produce “snatched” 5′ caps to prime viral mRNA synthesis (78). Hantavirus transcripts readily escape PBs to reenter the cytoplasmic milieu and recruit ribosomes (79), thus subverting P-body function, rather than co-opting PB components to novel cytoplasmic locations.
Several trends described above are mirrored during replication of the plant Brome mosaic virus in a yeast system. In this case Lsm1p-7p complex, Pat1p and Dhh1p (Rck/p54, DDX6) were all required for recruitment of viral RNA into membranous replication complexes (86). Viral RNA-dependent RNA polymerase and the viral RNA can associate with Lsm1p, and also colocalize with PBs (11). Further, select PBs associate near membranes where viral replication complexes assemble (11, 122).
Human Immunodeficiency Virus-1 (HIV-1) TAT protein interacts with the PB component DDX3 (130), and is antagonized by antiviral factors APOBEC 3 and Mov10, that are PB constituents. However, depletion experiments indicate P bodies and constituent proteins DDX6 or Lsm1 do not regulate HIV-1 replication or APOBEC3 antiviral activity (47, 96).
P-bodies were also found to repress retrotransposition of mouse endogenous retroviruses known as intracisternal A-particles (IAP). Knockdown of RCK/p54 or eIF4E-T and subsequently, PBs, increased IAP retrotransposition as well as levels of IAP transcripts, Gag proteins, and reverse transcription products and promoted IAP mRNA translation. Although some IAP mRNA localized to PBs, viral Gag protein was targeted to the endoplasmic reticulum where IAP buds (70). MOV10 also inhibits IAP reverse transcription and retrotransposition, however this inhibition is independent of PBs (71).
CONCLUSIONS
The field of virus-RNA granule interactions is both in its infancy and at the same time growing rapidly. At this initial stage, a broad picture has appeared of what viruses may manipulate SGs and PBs. There is now a superficial sense how such manipulations may occur, but the molecular details of mechanisms are mostly sparse. When mammalian cells encounter stress situations the complex reprograming of gene expression that occurs is mandatory for survival and RNA granules play key roles in this process, organizing the dynamically changing pools of active versus silenced mRNPs. Viruses are potent stress inducers and have always needed to cope with cellular environments that sense and react to their presence. Thus, as we have summarized above, viruses have evolved a pleathora of schemes to block, subvert or reprogram the RNA granule responses for viral survival or more; perhaps cleverly supporting replication or assembly.
The mechanisms of virus manipulation of RNA granules need further elucidation in most virus systems. To understand how viruses control SGs, more advances in understanding how RNA granules form are also crucial. Post-translational regulation may be key to mobilize stress responses that drive the liquid droplet condensation or protein phase changes that actually form the RNA granules, but how do the major factors drive RNP condensation and what corresponding post-translational modifications on these are responsible? What proteins interact to drive the process? How many of the key SG-nucleating proteins are really required? How will viruses fit into these scenarios mechanistically? Will these mechanisms provide new antiviral drug targets?
The true deleterious (or sometimes potentially beneficial) impact of SG formation on virus replication is also unclear. Since RNA granules cannot be isolated and purified, major questions remain as to the identity and scope of mRNAs that become included in granules and the extent to which they could trap viral transcripts if virus countermeasures were absent. Do SGs and PBs serve as powerful sites for general translational repression or viral RNA decay that must be counteracted? The various types of virus-induced SGs will have unique constituents compared to SGs induced by environmental insults such as arsenite or heat shock. Do the virus-induced SGs have unique functions? For viruses that co-opt SG components into novel virus-induced foci, the roles of these proteins, if any, in virus replication and package is unclear and require investigation. When viruses relocalize contain translation factors, are these functional for translation or other purposes? Does G3BP1 perform mechanistic functions for some viruses or is it just a passenger swept into the locale of new virus-directed processes? Since the real biochemical function(s) of many of RNA granule proteins like G3BP1 is complex and enigmatic, these answers may not be easily forthcoming. However, viruses are excellent probes of molecular biology so their study may unlock secrets of RNA granule biology that might remain veiled to other approaches.
As future research progresses it is key for distinctions in compositions of RNA granules to be thoroughly characterized. Functional SGs cannot be reliably identified based on a single marker such as TIA1 and this has led to some confusion in the virus-RNA granule field as to the true nature of granules in some instances. Further, we have seen a tendency of viruses to repackage and repurpose SG components into novel foci, which require more complete characterization to discern function of components.
Lastly, it is now clear that SG formation has many consequences to cellular metabolism beyond translational arrest; that crosstalk with apopotic and innate immune sytems by recruiting select group of signaling proteins is critically important. These exciting research areas are garnering increased attention and may reveal that RNA granules serve as major platforms linking stress signals with cell fate determination and activation of innate antiviral mechanisms as part of an integrated stress response. Since stress responses and innate immunity likely crosstalk at multiple levels, it is possible that aspects of RNA granule biology could be exploited in the future for broad-spectrum antiviral strategies.
SUMMARY POINTS.
Viruses regulate RNA granule responses in diverse schemes to control gene expression
RNA granules orchestrate stress and antiviral responses that can restrict virus replication
PKR is a key factor both upstream of SG formation and downstream of SG formation linking to innate immunity
Viruses can control SG function through control of PKR
Many RNA granule components are co-opted or repurposed by viruses away from SG formation to new roles in support of virus replication
P-body assembly is disrupted by several viruses, often in conjunction with co-opting P-body factors into new roles for the virus.
Acknowledgments
We are grateful to Lucas Reineke for critical review of this work. The work in the Lloyd lab is supported by National Institutes of Health grant AI050237 and NCI Cancer Center Support Grant (P30CA125123). Additional support was provided by the Integrated Microscopy Core at Baylor College of Medicine with funding from the NIH (HD007495, DK56338, and CA125123), the Dan L. Duncan Cancer Center, and the John S. Dunn Gulf Coast Consortium for Chemical Genomics.
Footnotes
DISCLOSURE STATEMENT
The author is not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Abrahamyan LG, Chatel-Chaix L, Ajamian L, Milev MP, Monette A, et al. Novel Staufen1 ribonucleoproteins prevent formation of stress granules but favour encapsidation of HIV-1 genomic RNA. J Cell Sci. 2010;123:369–83. doi: 10.1242/jcs.055897. [DOI] [PubMed] [Google Scholar]
- 2.Aizer A, Brody Y, Ler LW, Sonenberg N, Singer RH, Shav-Tal Y. The dynamics of mammalian P body transport, assembly, and disassembly in vivo. Mol Biol Cell. 2008;19:4154–66. doi: 10.1091/mbc.E08-05-0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson P, Kedersha N. Stress granules: the Tao of RNA triage. Trends Biochem Sci. 2008;33:141–50. doi: 10.1016/j.tibs.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 4.Anderson P, Kedersha N. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat Rev Mol Cell Biol. 2009;10:430–36. doi: 10.1038/nrm2694. [DOI] [PubMed] [Google Scholar]
- 5.Angus AGN, Dalrymple D, Boulant S, Mcgivern DR, Clayton RF, et al. Requirement of cellular DDX3 for hepatitis C virus replication is unrelated to its interaction with the viral core protein. J Gen Virol. 2010;91:122–32. doi: 10.1099/vir.0.015909-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ariumi Y, Kuroki M, Abe K-I, Dansako H, Ikeda M, et al. DDX3 DEAD-box RNA helicase is required for hepatitis C virus RNA replication. J Virol. 2007;81:13922–26. doi: 10.1128/JVI.01517-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ariumi Y, Kuroki M, Kushima Y, Osugi K, Hijikata M, et al. Hepatitis C Virus Hijacks P-Body and Stress Granule Components around Lipid Droplets. J Virol. 2011;85:6882–92. doi: 10.1128/JVI.02418-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Atasheva S, Gorchakov R, English R, Frolov I, Frolova E. Development of Sindbis viruses encoding nsP2/GFP chimeric proteins and their application for studying nsP2 functioning. J Virol. 2007;81:5046–57. doi: 10.1128/JVI.02746-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aulas A, Stabile S, Vande Velde C. Endogenous TDP-43, but not FUS, contributes to stress granule assembly via G3BP. Mol Neurodegener. 2012;7:54. doi: 10.1186/1750-1326-7-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baird NL, York J, Nunberg JH. Arenavirus infection induces discrete cytosolic structures for RNA replication. J Virol. 2012;86:11301–10. doi: 10.1128/JVI.01635-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beckham CJ, Light HR, Nissan TA, Ahlquist P, Parker R, Noueiry A. Interactions between brome mosaic virus RNAs and cytoplasmic processing bodies. J Virol. 2007;81:9759–68. doi: 10.1128/JVI.00844-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bentmann E, Haass C, Dormann D. Stress granules in neurodegeneration--lessons learnt from TAR DNA binding protein of 43 kDa and fused in sarcoma. FEBS Journal. 2013;280:4348–70. doi: 10.1111/febs.12287. [DOI] [PubMed] [Google Scholar]
- 13.Bentmann E, Neumann M, Tahirovic S, Rodde R, Dormann D, Haass C. Requirements for Stress Granule Recruitment of Fused in Sarcoma (FUS) and TAR DNA-binding Protein of 43 kDa (TDP-43) J Biol Chem. 2012;287:23079–94. doi: 10.1074/jbc.M111.328757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berezhna SY, Supekova L, Sever MJ, Schultz PG, Deniz AA. Dual regulation of hepatitis C viral RNA by cellular RNAi requires partitioning of Ago2 to lipid droplets and P-bodies. RNA. 2011;17:1831–45. doi: 10.1261/rna.2523911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bikkavilli RK, Malbon CC. Arginine methylation of G3BP1 in response to Wnt3a regulates {beta}-catenin mRNA. J Cell Sci. 2011;124:2310–20. doi: 10.1242/jcs.084046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bonnet MC, Weil R, Dam E, Hovanessian AG, Meurs EF. PKR stimulates NF-kappaB irrespective of its kinase function by interacting with the IkappaB kinase complex. Mol Cell Biol. 2000;20:4532–42. doi: 10.1128/mcb.20.13.4532-4542.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Borghese F, Michiels T. The leader protein of cardioviruses inhibits stress granule assembly. J Virol. 2011;85:9614–22. doi: 10.1128/JVI.00480-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buchan JR, Parker R. Eukaryotic stress granules: the ins and outs of translation. Mol Cell. 2009;36:932–41. doi: 10.1016/j.molcel.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carroll K, Hastings C, Miller CL. Amino acids 78 and 79 of Mammalian Orthoreovirus protein μNS are necessary for stress granule localization, core protein λ2 interaction, and de novo virus replication. Virology. 2014;448:133–45. doi: 10.1016/j.virol.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caudron-Herger M, Rippe K. Nuclear architecture by RNA. Curr Opin Genet Dev. 2012;22:179–87. doi: 10.1016/j.gde.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 21.Chahar HS, Chen S, Manjunath N. P-body components LSM1, GW182, DDX3, DDX6 and XRN1 are recruited to WNV replication sites and positively regulate viral replication. Virology. 2013;436:1–7. doi: 10.1016/j.virol.2012.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang T-C, Yamashita A, Chen C-YA, Yamashita Y, Zhu W, et al. UNR, a new partner of poly(A)-binding protein, plays a key role in translationally coupled mRNA turnover mediated by the c-fos major coding-region determinant. Genes Dev. 2004;18:2010–23. doi: 10.1101/gad.1219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Costa A, Pazman C, Sinsimer KS, Wong LC, McLeod I, et al. Rasputin functions as a positive regulator of orb in Drosophila oogenesis. PLoS ONE. 2013;8:e72864. doi: 10.1371/journal.pone.0072864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cristea IM, Rozjabek H, Molloy KR, Karki S, White LL, et al. Host factors associated with the Sindbis virus RNA-dependent RNA polymerase: role for G3BP1 and G3BP2 in virus replication. J Virol. 2010;84:6720–32. doi: 10.1128/JVI.01983-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dammer EB, Fallini C, Gozal YM, Duong DM, Rossoll W, et al. Coaggregation of RNA-Binding Proteins in a Model of TDP-43 Proteinopathy with Selective RGG Motif Methylation and a Role for RRM1 Ubiquitination. PLoS ONE. 2012;7:e38658. doi: 10.1371/journal.pone.0038658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dang Y, Kedersha N, Low W-K, Romo D, Gorospe M, et al. Eukaryotic initiation factor 2alpha-independent pathway of stress granule induction by the natural product pateamine A. J Biol Chem. 2006;281:32870–78. doi: 10.1074/jbc.M606149200. [DOI] [PubMed] [Google Scholar]
- 27.Dinh PX, Beura LK, Das PB, Panda D, Das A, Pattnaik AK. Induction of stress granule-like structures in vesicular stomatitis virus-infected cells. J Virol. 2013;87:372–83. doi: 10.1128/JVI.02305-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dougherty JD, Reineke LC, Lloyd RE. MRNA Decapping Enzyme 1a (Dcp1a) Induced Translational Arrest Through PKR Activation Requires the N-terminal EVH1 Domain. J Biol Chem. 2013 doi: 10.1074/jbc.M113.518191. epub Dec. 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dougherty JD, White JP, Lloyd RE. Poliovirus-mediated disruption of cytoplasmic processing bodies. J Virol. 2011;85:64–75. doi: 10.1128/JVI.01657-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Emara MM, Brinton MA. Interaction of TIA-1/TIAR with West Nile and dengue virus products in infected cells interferes with stress granule formation and processing body assembly. Proc Natl Acad Sci USA. 2007;104:9041–46. doi: 10.1073/pnas.0703348104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Emara MM, Liu H, Davis WG, Brinton MA. Mutation of mapped TIA-1/TIAR binding sites in the 3′ terminal stem-loop of West Nile virus minus-strand RNA in an infectious clone negatively affects genomic RNA amplification. J Virol. 2008;82:10657–70. doi: 10.1128/JVI.00991-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ernoult-Lange M, Baconnais S, Harper M, Minshall N, Souquere S, et al. Multiple binding of repressed mRNAs by the P-body protein Rck/p54. RNA. 2012;18:1702–15. doi: 10.1261/rna.034314.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frolova E, Gorchakov R, Garmashova N, Atasheva S, Vergara LA, Frolov I. Formation of nsP3-specific protein complexes during Sindbis virus replication. J Virol. 2006;80:4122–34. doi: 10.1128/JVI.80.8.4122-4134.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fros JJ, Domeradzka NE, Baggen J, Geertsema C, Flipse J, et al. Chikungunya Virus nsP3 Blocks Stress Granule Assembly by Recruitment of G3BP into Cytoplasmic Foci. J Virol. 2012;86:10873–79. doi: 10.1128/JVI.01506-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fung G, Ng CS, Zhang J, Shi J, Wong J, et al. Production of a Dominant-Negative Fragment Due to G3BP1 Cleavage Contributes to the Disruption of Mitochondria-Associated Protective Stress Granules during CVB3 Infection. PLoS ONE. 2013;8:e79546. doi: 10.1371/journal.pone.0079546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garaigorta U, Heim MH, Boyd B, Wieland S, Chisari FV. Hepatitis C Virus Induces the Formation of Stress Granules whose Proteins Regulate HCV RNA Replication, Virus Assembly and Egress. J Virol. 2012;86:11043–11056. doi: 10.1128/JVI.07101-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garcia MA, Gil J, Ventoso I, Guerra S, Domingo E, et al. Impact of Protein Kinase PKR in Cell Biology: from Antiviral to Antiproliferative Action. Micro Mol Bio Rev. 2006;70:1032–60. doi: 10.1128/MMBR.00027-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ghosh S, May MJ, Kopp EB. NF-kappa B and Rel proteins: evolutionarily conserved mediators of immune responses. Annu Rev Immunol. 1998;16:225–60. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- 39.Gil J, Rullas J, Garcia MA, Alcamí J, Esteban M. The catalytic activity of dsRNA-dependent protein kinase, PKR, is required for NF-kappaB activation. Oncogene. 2001;20:385–94. doi: 10.1038/sj.onc.1204109. [DOI] [PubMed] [Google Scholar]
- 40.Gorchakov R, Garmashova N, Frolova E, Frolov I. Different types of nsP3-containing protein complexes in Sindbis virus-infected cells. J Virol. 2008;82:10088–101. doi: 10.1128/JVI.01011-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Greer AE, Hearing P, Ketner G. The adenovirus E4 11k protein binds and relocalizes the cytoplasmic P-body component Ddx6 to aggresomes. Virology. 2011;417:161–68. doi: 10.1016/j.virol.2011.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Han TW, Kato M, Xie S, Wu LC, Mirzaei H, et al. Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell. 2012;149:768–79. doi: 10.1016/j.cell.2012.04.016. [DOI] [PubMed] [Google Scholar]
- 43.Hanley LL, Mcgivern DR, Teng MN, Djang R, Collins PL, Fearns R. Roles of the respiratory syncytial virus trailer region: effects of mutations on genome production and stress granule formation. Virology. 2010;406:241–52. doi: 10.1016/j.virol.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hebner CM, Wilson R, Rader J, Bidder M, Laimins LA. Human papillomaviruses target the double-stranded RNA protein kinase pathway. J Gen Virol. 2006;87:3183–93. doi: 10.1099/vir.0.82098-0. [DOI] [PubMed] [Google Scholar]
- 45.Huys A, Thibault PA, Wilson JA. Modulation of hepatitis C virus RNA accumulation and translation by DDX6 and miR-122 are mediated by separate mechanisms. PLoS ONE. 2013;8:e67437. doi: 10.1371/journal.pone.0067437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Iseni F, Garcin D, Nishio M, Kedersha N, Anderson P, Kolakofsky D. Sendai virus trailer RNA binds TIAR, a cellular protein involved in virus-induced apoptosis. EMBO J. 2002;21:5141–50. doi: 10.1093/emboj/cdf513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Izumi T, Burdick R, Shigemi M, Plisov S, Hu W-S, Pathak VK. Mov10 and APOBEC3G Localization to Processing Bodies Is Not Required for Virion Incorporation and Antiviral Activity. J Virol. 2013;87:11047–62. doi: 10.1128/JVI.02070-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jangra RK, Yi M, Lemon SM. DDX6 (Rck/p54) is required for efficient hepatitis C virus replication but not for internal ribosome entry site-directed translation. J Virol. 2010;84:6810–24. doi: 10.1128/JVI.00397-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kato M, Han TW, Xie S, Shi K, Du X, et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–67. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Katoh H, Okamoto T, Fukuhara T, Kambara H, Morita E, et al. Japanese encephalitis virus core protein inhibits stress granule formation through an interaction with Caprin-1 and facilitates viral propagation. J Virol. 2013;87:489–502. doi: 10.1128/JVI.02186-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Katsafanas GC, Moss B. Vaccinia virus intermediate stage transcription is complemented by Ras-GTPase-activating protein SH3 domain-binding protein (G3BP) and cytoplasmic activation/proliferation-associated protein (p137) individually or as a heterodimer. J Biol Chem. 2004;279:52210–17. doi: 10.1074/jbc.M411033200. [DOI] [PubMed] [Google Scholar]
- 52.Katsafanas GC, Moss B. Colocalization of transcription and translation within cytoplasmic poxvirus factories coordinates viral expression and subjugates host functions. Cell Host Microbe. 2007;2:221–28. doi: 10.1016/j.chom.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kedersha N, Anderson P. Mammalian stress granules and processing bodies. Meth Enzymol. 2007;431:61–81. doi: 10.1016/S0076-6879(07)31005-7. [DOI] [PubMed] [Google Scholar]
- 54.Kedersha N, Ivanov P, Anderson P. Stress granules and cell signaling: more than just a passing phase? Trends Biochem Sci. 2013;38:494–506. doi: 10.1016/j.tibs.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, et al. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol. 2005;169:871–84. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kedersha NL, Gupta M, Li W, Miller I, Anderson P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J Cell Biol. 1999;147:1431–42. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Khaperskyy DA, Hatchette TF, McCormick C. Influenza A virus inhibits cytoplasmic stress granule formation. The FASEB J. 2011 doi: 10.1096/fj.11-196915. [DOI] [PubMed] [Google Scholar]
- 58.Khong A, Jan E. Modulation of stress granules and P bodies during dicistrovirus infection. J Virol. 2011;85:1439–51. doi: 10.1128/JVI.02220-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kotani T, Yasuda K, Ota R, Yamashita M. Cyclin B1 mRNA translation is temporally controlled through formation and disassembly of RNA granules. J Cell Biol. 2013;202:1041–55. doi: 10.1083/jcb.201302139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kwon S, Zhang Y, Matthias P. The deacetylase HDAC6 is a novel critical component of stress granules involved in the stress response. Genes Dev. 2007;21:3381–94. doi: 10.1101/gad.461107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Langereis MA, Feng Q, van Kuppeveld FJ. MDA5 localizes to stress granules, but this localization is not required for the induction of type I interferon. J Virol. 2013;87:6314–25. doi: 10.1128/JVI.03213-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Legros S, Boxus M, Gatot JS, Van Lint C, Kruys V, et al. The HTLV-1 Tax protein inhibits formation of stress granules by interacting with histone deacetylase 6. Oncogene. 2011;30:4050–62. doi: 10.1038/onc.2011.120. [DOI] [PubMed] [Google Scholar]
- 63.Li P, Banjade S, Cheng H-C, Kim S, Chen B, et al. Phase transitions in the assembly of multivalent signalling proteins. Nature. 2012;483:336–40. doi: 10.1038/nature10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li W, Li Y, Kedersha N, Anderson P, Emara M, et al. Cell proteins TIA-1 and TIAR interact with the 3′ stem-loop of the West Nile virus complementary minus-strand RNA and facilitate virus replication. J Virol. 2002;76:11989–2000. doi: 10.1128/JVI.76.23.11989-12000.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lindquist ME, Lifland AW, Utley TJ, Santangelo PJ, Crowe JE. Respiratory syncytial virus induces host RNA stress granules to facilitate viral replication. J Virol. 2010;84:12274–84. doi: 10.1128/JVI.00260-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lindquist ME, Mainou BA, Dermody TS, Crowe JE. Activation of protein kinase R is required for induction of stress granules by respiratory syncytial virus but dispensable for viral replication. Virology. 2011;413:103–10. doi: 10.1016/j.virol.2011.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Linero FN, Thomas MG, Boccaccio GL, Scolaro LA. Junin virus infection impairs stress-granule formation in Vero cells treated with arsenite via inhibition of eIF2α phosphorylation. J Gen Virol. 2011;92:2889–99. doi: 10.1099/vir.0.033407-0. [DOI] [PubMed] [Google Scholar]
- 68.Loschi M, Leishman CC, Berardone N, Boccaccio GL. Dynein and kinesin regulate stress-granule and P-body dynamics. J Cell Sci. 2009;122:3973–82. doi: 10.1242/jcs.051383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lu B, Nakamura T, Inouye K, Li J, Tang Y, et al. Novel role of PKR in inflammasome activation and HMGB1 release. Nature. 2012;488:670–74. doi: 10.1038/nature11290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lu C, Contreras X, Peterlin BM. P bodies inhibit retrotransposition of endogenous intracisternal a particles. J Virol. 2011;85:6244–51. doi: 10.1128/JVI.02517-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lu C, Luo Z, Jäger S, Krogan NJ, Peterlin BM. Moloney leukemia virus type 10 inhibits reverse transcription and retrotransposition of intracisternal a particles. J Virol. 2012;86:10517–23. doi: 10.1128/JVI.00868-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mamiya N, Worman HJ. Hepatitis C virus core protein binds to a DEAD box RNA helicase. J Biol Chem. 1999;274:15751–56. doi: 10.1074/jbc.274.22.15751. [DOI] [PubMed] [Google Scholar]
- 73.Mangiardi DA, Mclaughlin-Williamson K, May KE, Messana EP, Mountain DC, Cotanche DA. Progression of hair cell ejection and molecular markers of apoptosis in the avian cochlea following gentamicin treatment. J Comp Neurol. 2004;475:1–18. doi: 10.1002/cne.20129. [DOI] [PubMed] [Google Scholar]
- 74.Mao YS, Zhang B, Spector DL. Biogenesis and function of nuclear bodies. Trends Genet. 2011;27:295–306. doi: 10.1016/j.tig.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Matthews JD, Frey TK. Analysis of subcellular G3BP redistribution during rubella virus infection. J Gen Virol. 2012;93:267–74. doi: 10.1099/vir.0.036780-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mazroui R, Sukarieh R, Bordeleau M-E, Kaufman RJ, Northcote P, et al. Inhibition of ribosome recruitment induces stress granule formation independently of eukaryotic initiation factor 2alpha phosphorylation. Mol Biol Cell. 2006;17:4212–19. doi: 10.1091/mbc.E06-04-0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McInerney GM, Kedersha NL, Kaufman RJ, Anderson P, Liljeström P. Importance of eIF2alpha phosphorylation and stress granule assembly in alphavirus translation regulation. Mol Biol Cell. 2005;16:3753–63. doi: 10.1091/mbc.E05-02-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mir MA, Duran WA, Hjelle BL, Ye C, Panganiban AT. Storage of cellular 5′ mRNA caps in P bodies for viral cap-snatching. Proc Natl Acad Sci USA. 2008;105:19294–99. doi: 10.1073/pnas.0807211105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mir MA, Panganiban AT. A protein that replaces the entire cellular eIF4F complex. EMBO J. 2008;27:3129–39. doi: 10.1038/emboj.2008.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mok BW-Y, Song W, Wang P, Tai H, Chen Y, et al. The NS1 Protein of Influenza A Virus Interacts with Cellular Processing Bodies and Stress Granules through RNA-Associated Protein 55 (RAP55) during Virus Infection. J Virol. 2012;86:12695–707. doi: 10.1128/JVI.00647-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mokas S, Mills JR, Garreau C, Fournier M-J, Robert F, et al. Uncoupling stress granule assembly and translation initiation inhibition. Mol Biol Cell. 2009;20:2673–83. doi: 10.1091/mbc.E08-10-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Montero H, Rojas M, Arias CF, López S. Rotavirus infection induces the phosphorylation of eIF2alpha but prevents the formation of stress granules. J Virol. 2008;82:1496–1504. doi: 10.1128/JVI.01779-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Moon SL, Anderson JR, Kumagai Y, Wilusz CJ, Akira S, et al. A noncoding RNA produced by arthropod-borne flaviviruses inhibits the cellular exoribonuclease XRN1 and alters host mRNA stability. RNA. 2012;18:2029–40. doi: 10.1261/rna.034330.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nakamura T, Furuhashi M, Li P, Cao H, Tuncman G, et al. Double-stranded RNA-dependent protein kinase links pathogen sensing with stress and metabolic homeostasis. Cell. 2010;140:338–48. doi: 10.1016/j.cell.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ng CS, Jogi M, Yoo J-S, Onomoto K, Koike S, et al. Encephalomyocarditis virus disrupts stress granules, the critical platform for triggering antiviral innate immune responses. J Virol. 2013;87:9511–22. doi: 10.1128/JVI.03248-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Noueiry AO, Díez J, Falk SP, Chen J, Ahlquist P. Yeast Lsm1p-7p/Pat1p deadenylation-dependent mRNA-decapping factors are required for brome mosaic virus genomic RNA translation. Mol Cell Biol. 2003;23:4094–4106. doi: 10.1128/MCB.23.12.4094-4106.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ohn T, Anderson P. The role of posttranslational modifications in the assembly of stress granules. WIREs RNA. 2010;1:486–93. doi: 10.1002/wrna.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ohn T, Kedersha N, Hickman T, Tisdale S, Anderson P. A functional RNAi screen links O-GlcNAc modification of ribosomal proteins to stress granule and processing body assembly. Nat Cell Biol. 2008;10:1224–31. doi: 10.1038/ncb1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Okonski KM, Samuel CE. Stress granule formation induced by measles virus is protein kinase PKR dependent and impaired by RNA adenosine deaminase ADAR1. J Virol. 2013;87:756–66. doi: 10.1128/JVI.02270-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Onomoto K, Jogi M, Yoo J-S, Narita R, Morimoto S, et al. Critical Role of an Antiviral Stress Granule Containing RIG-I and PKR in Viral Detection and Innate Immunity. PLoS ONE. 2012;7:e43031. doi: 10.1371/journal.pone.0043031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pager CT, Schütz S, Abraham TM, Luo G, Sarnow P. Modulation of hepatitis C virus RNA abundance and virus release by dispersion of processing bodies and enrichment of stress granules. Virology. 2013;435:472–84. doi: 10.1016/j.virol.2012.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Panas MD, Varjak M, Lulla A, Eng KE, Merits A, et al. Sequestration of G3BP coupled with efficient translation inhibits stress granules in Semliki Forest virus infection. Mol Biol Cell. 2012;23:4701–12. doi: 10.1091/mbc.E12-08-0619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Paul D, Hoppe S, Saher G, Krijnse-Locker J, Bartenschlager R. Morphological and biochemical characterization of the membranous hepatitis C virus replication compartment. J Virol. 2013;87:10612–27. doi: 10.1128/JVI.01370-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pérez-Vilaró G, Scheller N, Saludes V, Díez J. Hepatitis C virus infection alters p-body composition but is independent of p-body granules. J Virol. 2012;86:8740–49. doi: 10.1128/JVI.07167-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pfaller CK, Li Z, George CX, Samuel CE. Protein kinase PKR and RNA adenosine deaminase ADAR1: new roles for old players as modulators of the interferon response. Curr Opin Immunol. 2011;23:573–82. doi: 10.1016/j.coi.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Phalora PK, Sherer NM, Wolinsky SM, Swanson CM, Malim MH. HIV-1 replication and APOBEC3 antiviral activity are not regulated by P bodies. J Virol. 2012;86:11712–24. doi: 10.1128/JVI.00595-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pijlman GP, Funk A, Kondratieva N, Leung J, Torres S, et al. A highly structured, nuclease-resistant, noncoding RNA produced by flaviviruses is required for pathogenicity. Cell Host Microbe. 2008;4:579–91. doi: 10.1016/j.chom.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 98.Piotrowska J, Hansen SJ, Park N, Jamka K, Sarnow P, Gustin KE. Stable formation of compositionally unique stress granules in virus-infected cells. J Virol. 2010;84:3654–65. doi: 10.1128/JVI.01320-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Qin Q, Carroll K, Hastings C, Miller CL. Mammalian Orthoreovirus escape from host translational shutoff correlates with stress granule disruption and is independent of eIF2{alpha} phosphorylation and PKR. J Virol. 2011;85:8798–8810. doi: 10.1128/JVI.01831-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Qin Q, Hastings C, Miller CL. Mammalian orthoreovirus particles induce and are recruited into stress granules at early times postinfection. J Virol. 2009;83:11090–101. doi: 10.1128/JVI.01239-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Raaben M, Groot Koerkamp MJA, Rottier PJM, de Haan CAM. Mouse hepatitis coronavirus replication induces host translational shutoff and mRNA decay, with concomitant formation of stress granules and processing bodies. Cell Microbiol. 2007;9:2218–29. doi: 10.1111/j.1462-5822.2007.00951.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Reineke LC, Dougherty JD, Pierre P, Lloyd RE. Large G3BP-induced granules trigger eIF2α phosphorylation. Mol Biol Cell. 2012;23:3499–3510. doi: 10.1091/mbc.E12-05-0385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Roberts APE, Lewis AP, Jopling CL. MiR-122 activates hepatitis C virus translation by a specialized mechanism requiring particular RNA components. Nucl Acids Res. 2011;39:7716–7729. doi: 10.1093/nar/gkr426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ruggieri A, Dazert E, Metz P, Hofmann S, Bergeest J-P, et al. Dynamic oscillation of translation and stress granule formation mark the cellular response to virus infection. Cell Host Microbe. 2012;12:71–85. doi: 10.1016/j.chom.2012.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rzeczkowski K, Beuerlein K, Müller H, Dittrich-Breiholz O, Schneider H, et al. C-Jun N-terminal kinase phosphorylates DCP1a to control formation of P bodies. J Cell Biol. 2011;194:581–96. doi: 10.1083/jcb.201006089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Scheller N, Mina LB, Galão RP, Chari A, Giménez-Barcons M, et al. Translation and replication of hepatitis C virus genomic RNA depends on ancient cellular proteins that control mRNA fates. Proc Natl Acad Sci USA. 2009;106:13517–22. doi: 10.1073/pnas.0906413106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Schnettler E, Sterken MG, Leung JY, Metz SW, Geertsema C, et al. Noncoding flavivirus RNA displays RNA interference suppressor activity in insect and Mammalian cells. J Virol. 2012;86:13486–500. doi: 10.1128/JVI.01104-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Shelkovnikova TA, Robinson H, Connor-Robson N, Buchman VL. Recruitment into stress granules prevents irreversible aggregation of FUS protein mislocalized to the cytoplasm. Cell Cycle. 2013;12:3194–3202. doi: 10.4161/cc.26241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Shyu A-B, Wilkinson MF, van Hoof A. Messenger RNA regulation: to translate or to degrade. EMBO J. 2008;27:471–81. doi: 10.1038/sj.emboj.7601977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Silva PAGC, Pereira CF, Dalebout TJ, Spaan WJM, Bredenbeek PJ. An RNA pseudoknot is required for production of yellow fever virus subgenomic RNA by the host nuclease XRN1. J Virol. 2010;84:11395–406. doi: 10.1128/JVI.01047-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Simpson-Holley M, Kedersha N, Dower K, Rubins KH, Anderson P, et al. Formation of antiviral cytoplasmic granules during orthopoxvirus infection. J Virol. 2011;85:1581–93. doi: 10.1128/JVI.02247-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Smith JA, Schmechel SC, Raghavan A, Abelson M, Reilly C, et al. Reovirus induces and benefits from an integrated cellular stress response. J Virol. 2006;80:2019–33. doi: 10.1128/JVI.80.4.2019-2033.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sola I, Galán C, Mateos-Gómez PA, Palacio L, Zúñiga S, et al. The polypyrimidine tract-binding protein affects coronavirus RNA accumulation levels and relocalizes viral RNAs to novel cytoplasmic domains different from replication-transcription sites. J Virol. 2011;85:5136–49. doi: 10.1128/JVI.00195-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Steele L, Errington F, Prestwich R, Ilett E, Harrington K, et al. Pro-inflammatory cytokine/chemokine production by reovirus treated melanoma cells is PKR/NF-κB mediated and supports innate and adaptive anti-tumour immune priming. Mol Cancer. 2011;10:20–33. doi: 10.1186/1476-4598-10-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Taghavi N, Samuel CE. Protein kinase PKR catalytic activity is required for the PKR-dependent activation of mitogen-activated protein kinases and amplification of interferon beta induction following virus infection. Virology. 2012;427:208–16. doi: 10.1016/j.virol.2012.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Takahashi M, Higuchi M, Makokha GN, Matsuki H, Yoshita M, et al. HTLV-1 Tax oncoprotein stimulates ROS production and apoptosis in T cells by interacting with USP10. Blood. 2013;122:715–25. doi: 10.1182/blood-2013-03-493718. [DOI] [PubMed] [Google Scholar]
- 117.Thomas MG, Tosar LJM, Desbats MA, Leishman CC, Boccaccio GL. Mammalian Staufen 1 is recruited to stress granules and impairs their assembly. J Cell Sci. 2009;122:563–73. doi: 10.1242/jcs.038208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tourrière H, Chebli K, Zekri L, Courselaud B, Blanchard JM, et al. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J Cell Biol. 2003;160:823–31. doi: 10.1083/jcb.200212128. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 119.Towers ER, Kelly JJ, Sud R, Gale JE, Dawson SJ. Caprin-1 is a target of the deafness gene Pou4f3 and is recruited to stress granules in cochlear hair cells in response to ototoxic damage. J Cell Sci. 2011;124:1145–55. doi: 10.1242/jcs.076141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Vanderweyde T, Youmans K, Liu-Yesucevitz L, Wolozin B. Role of Stress Granules and RNA-Binding Proteins in Neurodegeneration: A Mini-Review. Gerontology. 2013;59:524–33. doi: 10.1159/000354170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Walsh D, Arias C, Perez C, Halladin D, Escandon M, et al. Eukaryotic translation initiation factor 4F architectural alterations accompany translation initiation factor redistribution in poxvirus-infected cells. Mol Cell Biol. 2008;28:2648–58. doi: 10.1128/MCB.01631-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wang X, Lee W-M, Watanabe T, Schwartz M, Janda M, Ahlquist P. Brome mosaic virus 1a nucleoside triphosphatase/helicase domain plays crucial roles in recruiting RNA replication templates. J Virol. 2005;79:13747–58. doi: 10.1128/JVI.79.21.13747-13758.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ward AM, Bidet K, Yinglin A, Ler SG, Hogue K, et al. Quantitative mass spectrometry of DENV-2 RNA-interacting proteins reveals that the DEAD-box RNA helicase DDX6 binds the DB1 and DB2 3′ UTR structures. RNA Biol. 2011;8:1173–86. doi: 10.4161/rna.8.6.17836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wasserman T, Katsenelson K, Daniliuc S, Hasin T, Choder M, Aronheim A. A novel c-Jun N-terminal kinase (JNK)-binding protein WDR62 is recruited to stress granules and mediates a nonclassical JNK activation. Mol Biol Cell. 2010;21:117–30. doi: 10.1091/mbc.E09-06-0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Weber SC, Brangwynne CP. Getting RNA and protein in phase. Cell. 2012;149:1188–91. doi: 10.1016/j.cell.2012.05.022. [DOI] [PubMed] [Google Scholar]
- 126.Weil TT, Parton RM, Herpers B, Soetaert J, Veenendaal T, et al. Drosophila patterning is established by differential association of mRNAs with P bodies. Nat Cell Biol. 2012;14:1305–15. doi: 10.1038/ncb2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.White JP, Cardenas AM, Marissen WE, Lloyd RE. Inhibition of cytoplasmic mRNA stress granule formation by a viral proteinase. Cell Host Microbe. 2007;2:295–305. doi: 10.1016/j.chom.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 128.White JP, Lloyd RE. Poliovirus unlinks TIA1 aggregation and mRNA stress granule formation. J Virol. 2011;85:12442–54. doi: 10.1128/JVI.05888-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yang X, Nath A, Opperman MJ, Chan C. The double-stranded RNA-dependent protein kinase differentially regulates insulin receptor substrates 1 and 2 in HepG2 cells. Mol Biol Cell. 2010;21:3449–58. doi: 10.1091/mbc.E10-06-0481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yasuda-Inoue M, Kuroki M, Ariumi Y. DDX3 RNA helicase is required for HIV-1 Tat function. Biochem Biophys Res Commun. 2013;44:607–11. doi: 10.1016/j.bbrc.2013.10.107. [DOI] [PubMed] [Google Scholar]
- 131.Yi Z, Pan T, Wu X, Song W, Wang S, et al. Hepatitis C Virus Co-Opts Ras-GTPase-Activating Protein-Binding Protein 1 for Its Genome Replication. J Virol. 2011;85:6996–7004. doi: 10.1128/JVI.00013-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.You LR, Chen CM, Yeh TS, Tsai TY, Mai RT, et al. Hepatitis C virus core protein interacts with cellular putative RNA helicase. J Virol. 1999;73:2841–53. doi: 10.1128/jvi.73.4.2841-2853.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yu SF, Lujan P, Jackson DL, Emerman M, Linial ML. The DEAD-box RNA helicase DDX6 is required for efficient encapsidation of a retroviral genome. PLoS Pathogens. 2011;7:e1002303. doi: 10.1371/journal.ppat.1002303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zaborowska I, Kellner K, Henry M, Meleady P, Walsh D. Recruitment of host translation initiation factor eIF4G by the Vaccinia Virus ssDNA-binding protein I3. Virology. 2012;425:11–22. doi: 10.1016/j.virol.2011.12.022. [DOI] [PubMed] [Google Scholar]
- 135.Zheng D, Ezzeddine N, Chen C-YA, Zhu W, He X, Shyu A-B. Deadenylation is prerequisite for P-body formation and mRNA decay in mammalian cells. J Cell Biol. 2008;182:89–101. doi: 10.1083/jcb.200801196. [DOI] [PMC free article] [PubMed] [Google Scholar]