Abstract
The process of sister chromatid pairing, or cohesion establishment, is coupled to DNA replication and fundamental to proper chromosome segregation and cell viability. In the past year, several articles have provided important new insights into cohesion establishment, an activity predicated on the acetyltransferase Ctf7/Eco1. Here, I review new findings that the conversion of chromatid-bound cohesins into a cohesion-competent state involves Ctf7/Eco1-mediated acetylation of the cohesin subunit Smc3. These studies further explore an anti-establishment activity that involves the binding of accessory factors WAPL/Rad61 and Pds5 to the cohesin subunit Scc3/Irr1. The anti-establishment activity of WAPL/Rad61 and Pds5 is temporarily relaxed by Ctf7/Eco1 during S phase to promote sister chromatid pairing. These findings are likely to be of clinical relevance, given the role of cohesion pathways in a wide range of disease states.
Introduction
Whether an organism is uni-cellular or multi-cellular, producing viable progeny requires the execution of two fundamental processes: replication of the chromosomes, followed by segregation of the resulting replication products (termed sister chromatids) into the newly forming daughter cells. The time span between chromosome replication and segregation can be on the order of an hour to decades, depending on the cell type. Thus, it is critical for cells during S phase to identify the two sister chromatids and then maintain that identity until anaphase onset. Identification is achieved by pairing sister chromatids together (termed cohesion) and maintained by cohesin complexes that contain Smc1, Smc3, Scc3/Irr1 and Mcd1/Scc1 (Table 1). Cohesins become chromatin-associated via a Scc2–Scc4 deposition complex. At present, the structural details that explain how cohesins maintain sister chromatid pairing are not fully resolved [1]. What is clear is that the association of cohesin with chromatin is not sufficient to engender sister chromatid pairing; an additional activity termed ‘establishment’ is required to convert cohesins to a pairing-competent state. Further complicating sister identification is that a typical genome is packed with homologous chromosomes, gene duplications and a multitude of repetitive elements, creating an identity crisis of internet proportions. How do cells properly identify which chromatids to pair together? In this review, I summarize recent findings that biochemically define establishment and discuss models regarding how sister chromatid pairing may be coupled to DNA replication.
Table 1.
Cohesion-related factor nomenclature [49].
| Budding yeast |
Fission yeast |
Xenopus | Humans | Drosophila |
|---|---|---|---|---|
| Cohesins | ||||
| Smc1 | Psm1 | Smc1 | Smc1 | Smc1 |
| Smc3 | Psm3 | Smc3 | Smc3 | Smc3 |
| Mcd1/Scc1 | Rad21 | Rad21 | Rad21/Scc1 | Rad21 |
| Scc3/Irr1 | Psc3 | SA1,2 | STAG1,2 | SA |
| Anti-establishment | ||||
| Pds5 | Pds5 | Pds5A,B | Pds5A,B | Pds5 |
| Rad61 | Wap1 | WAPL | WAPL | |
| Establishment | ||||
| Ctf7/Eco1 | Eso1 | XEco1,2 | EFO1/ESCO1, EFO2/ESCO2 |
DECO |
Establishment: Sister Chromatid Pairing
The act of pairing sister chromatids together requires the establishment factor Ctf7/Eco1. The CTF7 complementation group was first identified by yeast cell mutations that exhibit elevated rates of chromosome loss and defects in heterochromatin assembly [2,3]. Subsequently, Ctf7 (also termed Eco1) was found to be highly conserved through evolution and essential in all organisms tested (Table 1). Studies in yeast first revealed that Ctf7/Eco1 is essential for cohesion but that it neither maintains sister chromatid pairing nor deposits cohesins onto chromatin. Thus, ctf7/eco1 mutant cells precociously separate their sisters despite having chromatids that are fully decorated with cohesins [4–8]. This ‘cohesin without cohesion’ phenotype defines the establishment pathway when defective. Note that ctf7/eco1 mutant cells are fully capable of cohesin deposition and subsequent DNA replication, providing one of many observations used to abate a once-dominant model that cohesion is established by passage of the DNA replication fork through huge G1-deposited cohesin rings [1].
Studies of Ctf7/Eco1 instead revealed that cohesion establishment is an active process. In part, this activity became defined by reports that Ctf7/Eco1 genetically and physically interacts with numerous DNA replication factors (Replication Factor C subunits, Chl1 DNA helicase, PCNA sliding clamp), any one of which is sufficient to produce cohesion defects when mutated [1]. Coupled with cell cycle studies that mapped the essential function of Ctf7/Eco1 specifically to S phase, an early but still robust model of establishment is that nascent chromatids emerging from the fork become tethered together by the deposition of cohesins onto each sister and subsequent Ctf7/Eco1-dependent conversion of those cohesins to a pairing state [9].
Key insight into the molecular basis of establishment was obtained by the revelation that Ctf7/Eco1 is an acetyltransferase. Subsequent studies revealed that all homologs tested similarly exhibit acetyltransferase activity [10–14]. Interestingly, the acetyltransferase domain that comprises the majority of budding yeast Ctf7/Eco1 is genetically fused to a variety of other functional amino-terminal domains, such as DNA repair polymerase (fission yeast), linker histone (human), and an uncharacterized extension (Drosophila) [11,15]. In vitro reactions using bacterially expressed yeast Ctf7/Eco1 uncovered several substrates, including multiple lysines within Ctf7/Eco1 itself, Scc3/Irr1, Pds5, and a single lysine (K210) within Mcd1/Scc1 [10]. However, Mcd1/Scc1 mutated at this key residue still provides for robust cell growth. Moreover, Ctf7/Eco1 mutant constructs that exhibit greatly diminished acetylation activity in vitro support robust cell growth and chromosome segregation when expressed in yeast cells [10,16]. These findings launched a controversy regarding whether the essential function of Ctf7/Eco1 is based on acetylation and which of the above (if any) are the relevant substrates.
Homing in on Targets
A convergence of efforts by three labs provides a clear answer — and a target. Studies published from the laboratories of Koshland, Qin, and Uhlmann [17–19] collectively demonstrate that mutations that abolish Ctf7/Eco1 acetyltransferase activity render cells inviable and produce massive cohesion defects, indicating that the essential function of Ctf7/Eco1 does in fact reside within its acetyltransferase activity. Moreover, each identified Smc3 as the Ctf7/Eco1 substrate that is essential for cohesion establishment.
The stories in themselves are exciting. Unal and colleagues [17] focused on Ctf7/Eco1 as an acetyltransferase based on their observation that ctf7/eco1 yeast exhibit significant levels of auto-acetylation in vivo. Note that bacterial expression of this same mutated Ctf7/Eco1 protein fails to produce significant auto-acetylation, revealing that yeast contain a stabilizing factor absent in prokaryotes [10,17,19]. Analyses of deletion constructs provided evidence that acetylation indeed constitutes the essential function of Ctf7/Eco1, an observation independently confirmed in both yeast and human cells [16,18]. Unal and colleagues then isolated cohesin complexes from yeast and found that only a single subunit was acetylated — Smc3. These findings, and those described below by Ben-Shahar and colleagues [18], provide the first in vivo evidence that Ctf7/Eco1 exhibits acetyltransferase activity.
All three groups used mass spectroscopy to identify acetylated lysine residues of Smc3. Three of the lysines are conserved through evolution. Mutating Smc3 lysine 931 to arginine did not adversely affect cell growth. In contrast, mutating Smc3 lysines 112 and 113 to arginine is lethal, although acetylation of lysine 113 appears to play the more critical role. Characterization of cells that express Smc3K112R,K113R was quite informative: cell death occurred hand-in-hand with S-phase progression and correlated with severe sister chromatid cohesion defects. Despite this, chromosomes within these cells were decorated by cohesins similar to wild-type cells, identical to the cohesin without cohesion phenotype first identified in ctf7/eco1 mutant cells. In combination, these findings define establishment as occurring through Smc3 acetylation, a model supported by observations that Smc3 acetylation-mimics suppress ctf7/eco1 phenotypes [17–19].
Ben-Shahar and colleagues [18] approached cohesion establishment starting from a genetic vantage point. While temperature-sensitive ctf7/eco1 mutants are inviable when placed at the restrictive temperature, they noticed that ctf7/eco1 mutants rarely produced thermoresistant colonies and reasoned that these might constitute bypass suppressors of Ctf7/Eco1 function. Tiling array analyses of one of the thermoresistant complementation groups identified smc3 as the bypass suppressor. In a wonderful convergence of studies, Ben-Shahar et al. [18] mapped the bypass to a single lysine-to-asparagine mutation within Smc3 at position 113. Based on the notion that asparagine resembles an acetylated lysine, they isolated native Smc3 from yeast and found that lysines 112 and 113 are both acetylated. Exploiting another bypass suppressor that provides for cell viability in a ctf7/eco1-null background, Smc3 acetylation was found to indeed depend on Ctf7/Eco1.
Zhang and colleagues [19] traveled yet a third path to arrive at the mechanism of cohesion establishment. Starting from the substrate side in a human cell system, these researchers identified an electrophoretic mobility shift of hSMC3 that depends on acetylation of lysines 105 and 106 (these correspond to K112 and K113 in budding yeast). With substrate in hand, they now had a powerful assay from which to identify the acetyltransferase. Knockdown of human EFO1/ESCO1 (homolog of yeast Ctf7/Eco1; Table 1) using siRNA greatly reduced hSMC3 acetylation, an effect not exhibited by knockdown of other acetyltransferases. They next overwhelmed endogenous hSMC3 in human cells by expressing acetylation-deficient hSMC3 to very high levels. Consistent with the notion that acetylation is critical for establishment, these cells exhibited severe sister chromatid pairing defects. Notably, acetylation of endogenous hSMC3 increased upon expression of mutated hSMC3, suggesting a potential feedback mechanism in which hEFO1/ESCO1 (Ctf7/Eco1) activity is up-regulated in response to cohesion defects [19].
Ctf7/Eco1 is essential specifically during S phase, suggesting that Smc3 acetylation should be confined to this portion of the cell cycle. All three groups tested this hypothesis [17–19]. Cell cycle mapping studies in both yeast and humans indeed revealed that Smc3 is not acetylated in G1 but that acetylation rises precipitously during S phase, mirroring exactly Ctf7/Eco1 function [4,5,17–19]. Intriguingly, Smc3 acetylation persists into anaphase onset (or longer in human cells) before returning to an unacetylated G1 state. The role of Smc3 acetylation beyond S phase is puzzling, given that the establishment of sister chromaid pairing is restricted to S phase in normal cells. Other missing pieces to the puzzle include the mechanism that activates Ctf7/Eco1 during S phase and the identity of the de-acetylase that returns Smc3 to its unacetylated state during G1.
The Anti-Establishment: A Balancing Act for Ct7/Eco1 Establishment Activity
While the substrate for Ctf7/Eco1 acetylation was only recently identified, a potential mechanism for regulating establishment was uncovered quite a while ago. This story starts with PDS5 (Table 1). Yeast PDS5 originated from a collection of mutant strains that exhibit enhanced G2/M lethality and was recognized as a homolog of factors known to bind mitotic chromosomes and associate with SMC-like proteins [20,21]. Early yeast studies revealed properties of Pds5 reminiscent of cohesins: first, pds5 mutant cells exhibit cohesion defects; second, Pds5 is required to maintain cohesion during G2/M; third, Pds5 associates with cohesin complexes; and fourth, Pds5 binds the same chromosome loci as cohesins. More recent studies, however, reveal that Pds5 is not simply a cohesin subunit: Pds5 does not bind cohesin robustly and is recruited to chromatin only after cohesin deposition, a dependency conserved throughout evolution and between mitosis and meiosis. In contrast, cohesins are quite competent to bind chromatin independent of Pds5 in both budding and fission yeast, although chromatin association appears much less stable [20,22–26]. In retrospect, this latter observation signifies a role for Pds5 in regulating cohesin dynamics.
The role for Pds5 in cohesion appears linked to Ctf7/Eco1. This coupling initially was predicated on two observations, the first of which is that pds5 mutant yeast cells recapitulate the cohesin without cohesion phenotype first described for ctf7/eco1 establishment mutants [20]. While mutations in either ctf7/eco1 or pds5 produce this establishment phenotype, an important difference is that Pds5 is required for cohesion maintenance during G2/M while Ctf7/Eco1 function is restricted to S phase. The second finding, originating from fission yeast, is that pds5 deletion bypasses the cell lethality associated with mutations in ESO1 (homolog of Ctf7/Eco1). These observations prompted the Okayama lab to first propose that “Pds5 p hinders the establishment of cohesion until counteracted by Eso1p” [22]. Support for this model was soon forthcoming: the synthetic rescue identified in fission yeast is conserved in budding yeast (although not all pds5 alleles bypass ctf7/eco1), and studies in both cell systems reveal that Pds5 associates with Ctf7/Eco1 [6,22,27,28]. Oddly, while PDS5 deletion bypasses Ctf7/Eco1 function in budding yeast, elevated levels of Pds5 suppress ctf7/eco1 mutant cell phenotypes. The converse is also true, suggesting that the anti-establishment activity of Pds5 is much more complicated than currently appreciated. An observation that may prove useful in unraveling this conundrum is that Pds5 is sumoylated and that this modification contributes to cohesion regulation [29].
Pds5 is not the only factor that, when mutated, bypasses the requirement for Ctf7/Eco1 function. WAPL was identified as a chromatin modifier in Drosophila [30], and two studies collectively presaged the ascension of WAPL onto the stage of cohesion establishment regulators. In human cells, siRNA knockdown of EFO2/ESCO2 produces considerable premature sister separation. Knockdown of both EFO2/ESCO2 and WAPL, however, results in normal chromosome segregation [31]. WAPL shares limited homology with budding yeast Rad61, a factor previously implicated in DNA repair and sister chromatid cohesion [32–34]. In budding yeast, mutations in RAD61 also bypass ctf7/eco1-dependent lethality [18]. Thus, the anti-establishment activity of WAPL/Rad61 is common to both vertebrate and yeast cells.
The answer to ‘How?’ is a bit more complicated. In vertebrate cell systems, both the Hirano and Peters labs report that WAPL/Rad61 depletion produces chromosomes with abnormally high cohesin levels [31,34]. Here, the net effect is to perturb chromosome condensation such that sister chromatids fail to become distinguishable as distinct (albeit paired) entities during mitosis. Consistent with the notion that WAPL/Rad61 destabilizes cohesins, elevated levels of WAPL/Rad61 reduce the level of chromatin-associated cohesins. In this case, sisters precociously separate akin to the pairing defects reported for cohesin knockdowns [31,34]. A similar role for WAPL/Rad61 in removing cohesins occurs in fission yeast [35]. More recently, the Shirahige and Nasmyth laboratories [27,28] independently assessed the effects of WAPL/Rad61, but in budding yeast. Surprisingly, their results reveal that mutations in WAPL/RAD61 reduce cohesin association to chromatin, indicating instead that WAPL/Rad61 stabilizes cohesin association with chromatin. The apparent conflict between model systems is perplexing and further studies are required to understand the molecular basis for these opposing effects. However, that the balance of cohesin dynamics is tipped in one direction or the other in various model organisms is far secondary to the features common to all: WAPL/Rad61 influences cohesin dynamics and, when mutated, bypasses the essential role of Ctf7/Eco1. WAPL, similar to Pds5, depends on cohesin for chromatin association. As mentioned previously, pds5 mutant cells also exhibit decreased cohesin association with chromatin and bypass Ctf7/Eco1 function [22,27]. WAPL/Rad61 and Pds5 also co-sediment [34]. The functional similarities between the two proteins reflect their integration into an anti-establishment complex that regulates cohesin dynamics.
Scc3/Irr1 is garnering considerable interest in its own right. Rowland and colleagues report that Pds5 and WAPL/Rad61 form a heterotrimeric complex that includes Scc3/Irr1. This observation, predicated on in vitro assembly reactions of bacterially expressed proteins, potentially repositions Scc3/Irr1 from one of cohesin to that of anti-establishment factor [28]. These studies further provided hints regarding the spatial organization of this heterotrimeric complex: WAPL/Rad61 binds both Scc3/Irr1 and Pds5, but the association of Scc3/Irr1 with Pds5 requires WAPL/Rad61. Despite these observations, the interpretation that WAPL/Rad61, Pds5 and Scc3/Irr1 form a distinct complex is premature. For instance, while WAPL/Rad61 appears to bridge Irr1/Scc3 and Pds5, WAPL/Rad61 itself is not essential. Thus, Scc3/Irr1 binding to Pds5 either does not depend on WAPL/Rad61 in vivo or is not required for cell viability. In fact, only Scc3/Irr1 is essential across species. Moreover, only Scc3/Irr1 (SA1) co-sediments with intact cohesin complexes and co-immunoprecipitates with all cohesin subunits [5,36,37]. In combination, the far more likely scenario is that WAPL/Rad61 and Pds5 are accessory factors that bind to cohesins through Scc3/Irr1.
Beyond S-Phase Establishment/Anti-Establishment
How does Ctf7/Eco1 establish cohesion and in what way do WAPL/Rad61 and Pds5 ‘buck the establishment’? The answers to both questions are complicated by the fact that the cohesin structure that maintains sister chromatid pairing is unresolved. What is clear is that Smc1 and Smc3 are elongated coiled-coil proteins that associate via ‘terminal’ hinge–hinge interactions on one end, and by binding Smc1/3 ATPase globular heads on the other end. Mcd1/Scc1 sits atop these closely apposed Smc1/3 globular heads and in turn binds Scc3/Irr1 (Figure 1). Given this structure, a popular model is that Smc1/3 coiled-coil regions bow apart to form a lumen that conceivably could capture one sister during ring opening/closing reactions (Figure 2). It is important to note, however, that multiple models regarding how cohesin associates with chromatin (cohesin dimers, oligomers, filamentous structures and laterally associated complexes) have been put forward by leaders in the field [1]. One recent electron microscopy based study of isolated DNA–cohesin complexes even suggests instead that cohesins adopt a helical rod-like structure that associates end-on with DNA [38]. Here, I defer to multiple lines of evidence that each sister is decorated with cohesins and that these subsequently become paired (Figure 2). While much has yet to be learned regarding cohesin structure, any model must account for chemical cross-linking studies that suggest that cohesin-binding to DNA survives protein denaturation, supporting some form of chromatin entrapment [39]. Readers should also be aware that more than cohesin may participate in heterochromatic pairing structures given the evidence that silencing factors and origin recognition complexes also participate in cohesion [1].
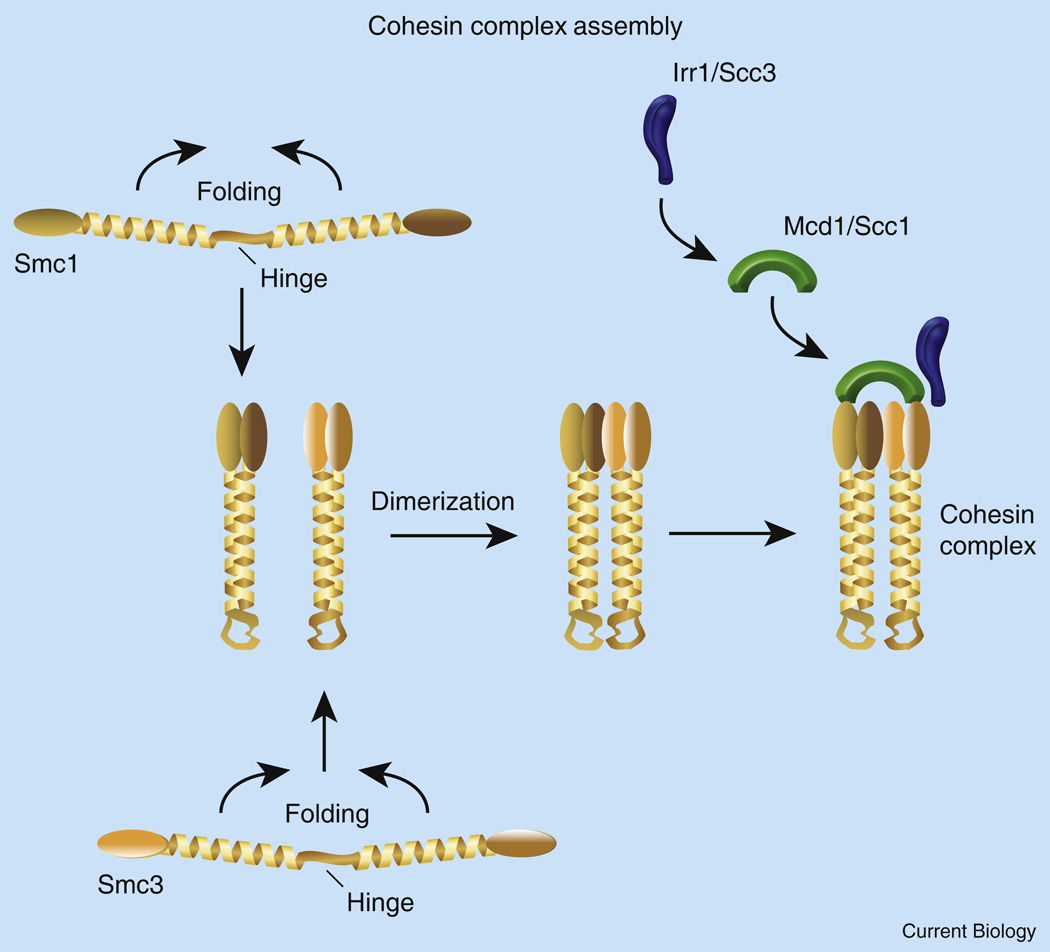
Figure 1. Cohesin complex assembly and structure.
SMCs contain long helical domains interrupted by a centrally positioned hinge. Hinge folding enables the helical domains to intertwine in an anti-parallel fashion and form a stable coiled-coil rod-like structure. Folding over also brings amino- and carboxy-terminal globular ATPase domains for each SMC into registration. Smc1/3 bind together by dimerization of hinge domains and by association of globular head domains. The Smc1/3 ATPase domains are capped by Mcd1/Scc1. Mcd1/Scc1 recruits Scc3/Irr1.
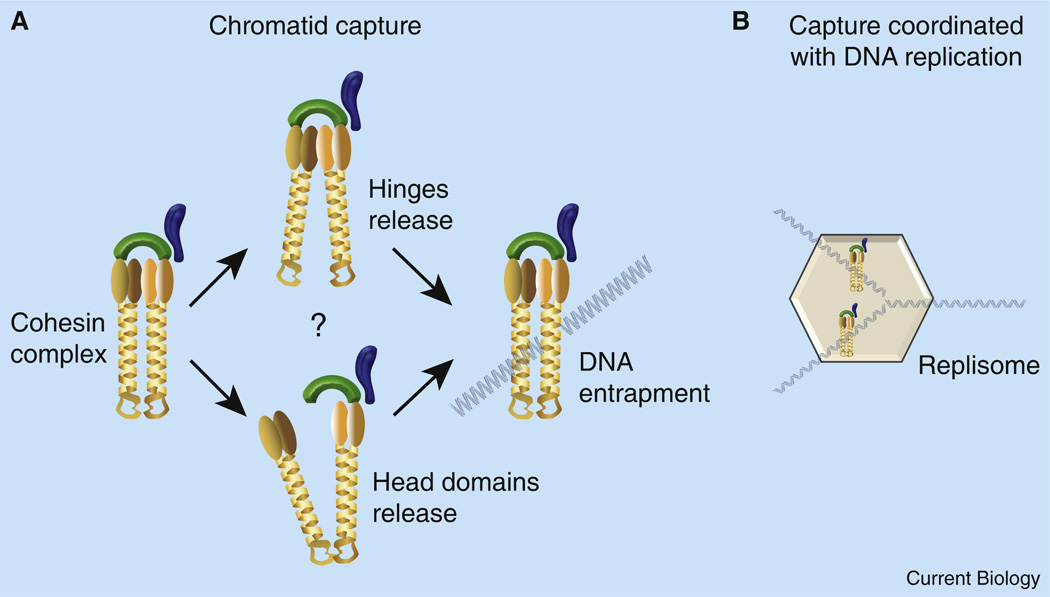
Figure 2. Chromatid capture.
(A) Current models suggest that Smc1/3 subunit interactions are dynamic: either hinge dimerization domains or ATPase globular head domains may transiently let go. DNA is then thought to enter the exposed lumen, becoming trapped upon reformation of Smc1/3 subunit interactions. Other models of cohesin association with chromatin (lateral binding, filamentous or spiraled structures) are not shown. DNA is depicted in a ‘naked’ state — note that a single 30 nm compacted chromatin fiber would completely fill the proposed lumen and distort the coiled-coil domains of the Smc1/3 complex (not shown). (B) Cohesin deposition and chromatid capture may be coupled to DNA replication. Deposition factors Scc2 and Scc4 are not shown; leading and lagging replisome complexes are simplified to a large hexagon.
During S phase, cohesion establishment likely proceeds along one of just a few pathways. Based on numerous studies demonstrating that Ctf7/Eco1 functions during S phase, that Ctf7/Eco1 associates with numerous DNA replication factors, and that these same DNA replication factors are required to promote efficient cohesion establishment, a reasonable model is that Ctf7/Eco1 rides the replication fork to pair together the emerging sisters [9]. It is unclear the extent to which cohesins deposited prior to or immediately after fork passage participate in cohesion. For simplicity’s sake, I limit the discussion to cohesin deposition during S phase that results in entrapment of a single sister (Figure 2). In turn, Ctf7/Eco1 drives some form of cohesin dimerization or concatenation (Figure 3). Even this establishment model has limitations. For instance, chromatin immunoprecipitation-based mapping studies fail to provide convincing evidence that Ctf7/Eco1 associates exclusively with the replication fork. Instead, Ctf7/Eco1 appears globally distributed along chromatin and at exceeding low levels [7], in support of transient Ctf7/Eco1 association with the replication fork. In addition, efforts to isolate higher-order cohesin structures for the most part have come up empty handed.
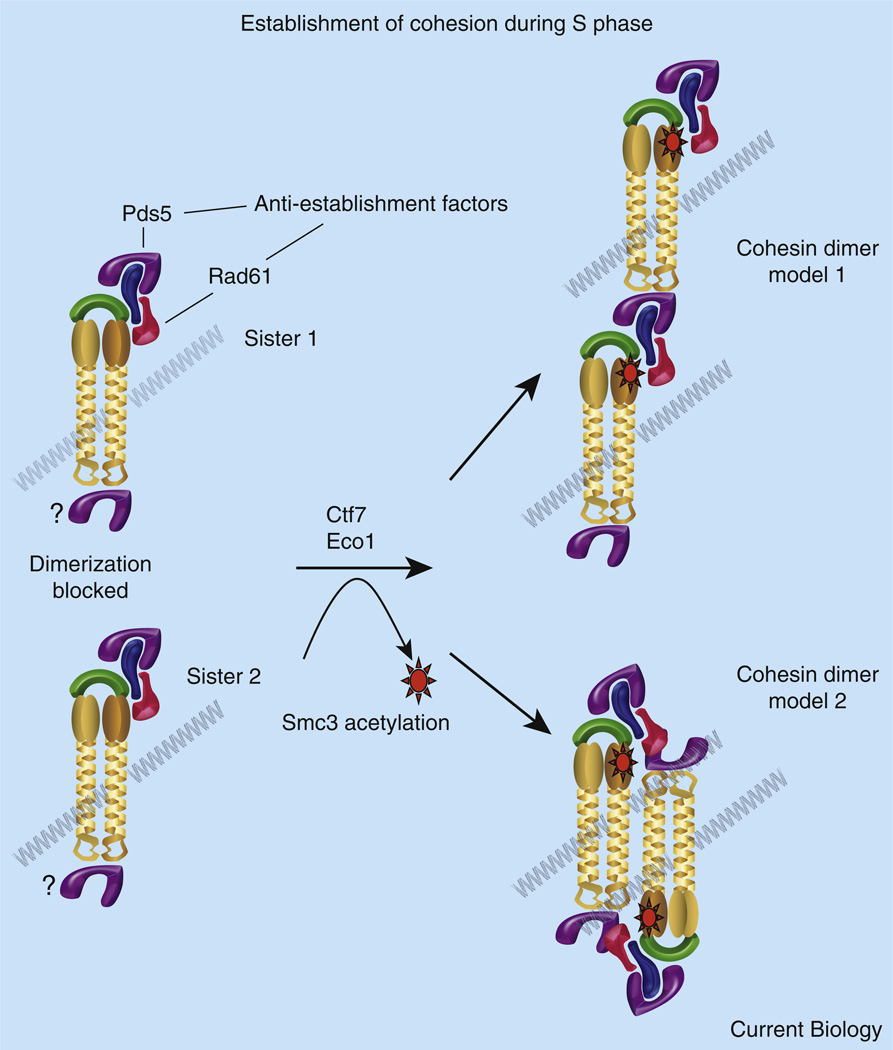
Figure 3. Cohesion establishment.
Shown is a speculative model regarding the conversion of cohesins to a cohesion-competent state during S phase. Cohesin complexes deposited onto (or entrapping) each chromatid are separate and distinct entities. Recruitment of anti-establishment factors Pds5 and WAPL/Rad61 onto Scc3/Irr1 precludes the tethering together of cohesin complexes by destabilizing cohesin–cohesin association. Lines of evidence equally support recruitment of Pds5 to both Scc3/Irr1 and to hinge dimerization domains, suggesting that either cohesin terminus may participate in pairing. Pds5 and WAPL/Rad61 anti-establishment factors may also promote chromatid capture reactions by destabilizing subunit interactions within each cohesin (not shown). During S phase, Ctf7/Eco1 acetylates Smc3 near the ATPase globular domain. Smc3 acetylation temporarily inhibits anti-establishment activity, possibly by altering the association or binding site of WAPL/Rad61 and Pds5 to the rest of the cohesin complex (note rotation of anti-establishment factors shown for instance in model 2). Dimerization of cohesins associated with each sister chromatid results in a stable structure that is resistant to antiestablishment activity and persistent Smc3 acetylation. Two of several oligomeric structures (model 1 and model 2) that account for both DNA entrapment and sister chromatid pairing are shown (see [27,28] for a single ring model). In a G2/M response to DNA damage, chromatin-bound cohesins are instead converted to a pairing state by Mcd1 phosphorylation and subsequent acetylation (not shown).
Despite these uncertainties, it is exciting to consider mechanisms by which the anti-establishment arm of the cohesin complex might in turn regulate sister chromatid pairing. In one model, WAPL/Rad61–Pds5 stabilizes the cohesin complex, a stability transiently counteracted by Ctf7/Eco1-mediated acetylation of Smc3. Confounding this notion are observations that Smc3 acetylation persists well into anaphase onset and that Smc3 acetylation does not alter Smc3 binding to Mcd1/Scc1 or cohesin formation [17–19]. Thus, a more likely scenario may be that WAPL/Rad61 and Pds5, in association with Scc3, antagonize establishment instead of stabilizing a cohesion state (Figure 3). In this scenario, WAPL/Rad61–Pds5 inhibits the assembly of cohesins into higher-order, pairing-competent states (or promotes transient cohesin opening, consistent with changes in cohesin dynamics discussed previously). In turn, Ctf7/Eco1-dependent acetylation of Smc3 alters WAPL/Rad61–Pds5 binding to cohesins to elicit a stable cohesin-pairing state. This new and more stable cohesin assemblage is no longer sensitive to the effects of Smc3 acetylation, a modification that persists until anaphase onset.
Another twist to the story comes from the study of cells challenged by DNA damage. Thusly abused, G2/M cells up-regulate Ctf7/Eco1 to re-establish cohesion between sisters. In turn, cohesion promotes efficient DNA repair. Of particular interest is that Ctf7/Eco1-based establishment is not limited to the site of DNA damage where replication/repair factors are recruited. Instead, Ctf7/Eco1 establishes cohesion globally and on all chromosomes [40–43]. Thus, establishment during a G2/M response to DNA damage appears quite different from current models of S-phase establishment and certainly preclude models predicated on replication/repair through a cohesin ring.
While the structures that pair together sisters in S phase and G2/M are likely identical, the regulation of establishment (or anti-establishment) is quite different. As stated earlier, Smc3 acetylation is required for cohesion establishment during S phase (Figure 3). In response to DNA damage, however, Ctf7/Eco1 targets Mcd1/Scc1 for acetylation — Smc3 acetylation is completely dispensable for damage-induced cohesion establishment [44]. Critical pathways often engender multiple levels of regulation. This is borne out by the revelation that Mcd1 is not only acetylated but also phosphorylated. Mcd1/Scc1 phosphorylation at S83 results from Mec1-dependent activation of Chk1 [45], two kinases required for activation of the DNA damage checkpoint. Closing the loop on G2/M cohesion establishment, Unal and colleagues subsequently found that Mcd1 phosphorylation was critical for Ctf7/Eco1 re-activation, which in turn acetylates Mcd1/Scc1 at K84 and K210 [44]. Highlighting the differences across the cell cycle, mcd1/scc1K84R,K210R mutants are competent to establish cohesion during S phase but not during G2/M in response to DNA damage. As in S phase, Ctf7/Eco1 establishment activity in G2/M has an antagonist such that deletion of WAPL/Rad61 bypasses the requirement for Mcd1 acetylation by Ctf7/Eco1. Thus, Ctf7/Eco1 targets different substrates (Smc3 in S phase and Mcd1 in G2/M in response to DNA damage) to counteract inhibitors of cohesion establishment. Bringing the discussion full circle are findings by both the Skibbens and Kupiec laboratories [46,47] that mutations in RFC factors also suppress ctf7/eco1 mutant cell phenotypes, extending anti-establishment activity to the DNA replication fork.
Concluding Remarks
In summary, new evidence establishes a role for Ctf7/Eco1 acetylation in cohesion and that Smc3 and Mcd1/Scc1 both are Ctf7/Eco1 substrates during S phase in unchallenged cells or during G2/M in response to DNA damage, respectively. Moreover, Pds5, WAPL/Rad61, Irr1/Scc3 and at least a subset of DNA fork components appear to oppose Ctf7/Eco1 function. Future endeavors are likely to be of great clinical relevance, given the role of cohesion pathways in a wide range of disease states [1,48].
Acknowledgments
The author is indebted to the Skibbens lab group. RVS apologizes to all colleagues whose work could not be sufficiently discussed due to space limitations. RVS is supported by the National Institutes of General Medicine (1R15GM083269) and by the Susan G. Komen for the Cure Foundation (BCTR0707708). Any opinions, findings, and conclusions or recommendations expressed in this study are those of the author(s) and do not necessarily reflect the views of the National Institutes of General Medicine nor of the Susan G. Komen Foundation. Figures were generated using Powerpoint software and images obtained from http://www.biochem.wisc.edu/medialab/clipart.aspx.
References
- 1.Skibbens RV. Mechanisms of sister chromatid pairing. Int. Rev. Cell Mol. Biol. 2009;269:283–339. doi: 10.1016/S1937-6448(08)01005-8. [DOI] [PubMed] [Google Scholar]
- 2.Spencer F, Gerring SL, Connelly C, Hieter P. Mitotic chromosome transmission fidelity mutants in Saccharomyces cerevisiae. Genetics. 1990;124:237–249. doi: 10.1093/genetics/124.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doheny KF, Sorger PK, Hyman AA, Tugendreich S, Spencer F, Hieter P. Identification of essential components of the S. cerevisiae kinetochore. Cell. 1993;73:761–774. doi: 10.1016/0092-8674(93)90255-O. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Skibbens RV, Corson LB, Koshland D, Hieter P. Ctf7 p is essential for sister chromatid cohesion and links mitotic chromosome structure to the DNA replication machinery. Genes Dev. 1999;13:307–319. doi: 10.1101/gad.13.3.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Toth A, Ciosk R, Uhlmann F, Galova M, Schleffer A, Nasmyth K. Yeast cohesin complex requires a conserved protein, Eco1 p (Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev. 1999;13:320–333. doi: 10.1101/gad.13.3.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Noble D, Kenna MA, Dix M, Skibbens RV, Unal E, Guacci V. Intersection between the regulators of sister chromatid cohesion establishment and maintenance in budding yeast indicates a multi-step mechanism. Cell Cycle. 2006;21:2528–2536. doi: 10.4161/cc.5.21.3405. [DOI] [PubMed] [Google Scholar]
- 7.Lengronne A, McIntyre J, Katou Y, Kanoh Y, Hopfner K-P, Shirahige K, Uhlmann F. Establishment of sister chromatid cohesion at the S. cerevisiae replication fork. Mol. Cell. 2006;23:787–799. doi: 10.1016/j.molcel.2006.08.018. [DOI] [PubMed] [Google Scholar]
- 8.Milutinovich M, Unal E, Ward C, Skibbens RV, Koshland D. A multi-step pathway for the establishment of sister chromatid cohesion. PLoS Genet. 2007;3:e12–e24. doi: 10.1371/journal.pgen.0030012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Skibbens RV. Holding your own: Establishing sister chromatid cohesion. Genome Res. 2000;10:1664–1671. doi: 10.1101/gr.153600. [DOI] [PubMed] [Google Scholar]
- 10.Ivanov D, Schleiffer A, Eisenhaber F, Mechtler K, Haering CH, Nasmyth K. Eco1 is a novel acetyltransferase that can acetylate proteins involved in cohesion. Curr. Biol. 2002;12:323–328. doi: 10.1016/s0960-9822(02)00681-4. [DOI] [PubMed] [Google Scholar]
- 11.Bellows AM, Kenna MA, Cassimeris L, Skibbens RV. Human EFO1 p exhibits acetyltransferase activity and is a unique combination of linker histone and Ctf7 p/Eco1 p chromatid cohesion establishment domains. Nucleic Acids Res. 2003;31:6334–6343. doi: 10.1093/nar/gkg811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hou F, Zou H. Two human orthologues of Eco1/Ctf7 p acetyltransferases are both required for proper sister-chromatid cohesion. Mol. Biol. Cell. 2005;16:3908–3918. doi: 10.1091/mbc.E04-12-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vega H, Waisfisz Q, Gordillo M, Sakai N, Yanagirhara I, Yamada M, van Gosliga D, Kayserilii H, Xu C, Ozono K, et al. Roberts syndrome is caused by mutations in ESCO2, a human homolog of yeast ECO1 that is essential for the establishment of sister chromatid cohesion. Nat Genet. 2005;37:468–470. doi: 10.1038/ng1548. [DOI] [PubMed] [Google Scholar]
- 14.Takagi M, Bunai K, Yanagi K, Imamoto N. Cloning of Xenopus orthologs of Ctf7/Eco1 acetyltransferase and initial characterization of XEco2. FEBS J. 2008;275:6109–6122. doi: 10.1111/j.1742-4658.2008.06736.x. [DOI] [PubMed] [Google Scholar]
- 15.Tanaka K, Yonekawa T, Kawasaki Y, Kai M, Furuya K, Iwasaki M, Murakami H, Yanagida M, Okayama H. Fission yeast Eso1 p is required for establishing sister chromatid cohesion during S phase. Mol. Cell. Biol. 2000;20:3459–3469. doi: 10.1128/mcb.20.10.3459-3469.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brands A, Skibbens RV. Ctf7 p/Eco1 p exhibits acetyltransferase activity – but does it matter? Curr. Biol. 2005;15:R50–R51. doi: 10.1016/j.cub.2004.12.052. [DOI] [PubMed] [Google Scholar]
- 17.Unal E, Heidinger-Pauli JM, Kim W, Guacci V, Onn I, Gygi SP, Koshland DE. A molecular determinant for the establishment of sister chromatid cohesion. Science. 2008;321:566–569. doi: 10.1126/science.1157880. [DOI] [PubMed] [Google Scholar]
- 18.Ben-Shahar TR, Heeger S, Lehane C, East P, Flynn H, Skehel M, Uhlmann F. Eco1-dependent cohesin acetylation during establishment of sister chromatid cohesion. Science. 2008;321:563–566. doi: 10.1126/science.1157774. [DOI] [PubMed] [Google Scholar]
- 19.Zhang J, Shi X, Li Y, Kim BJ, Jia J, Huang Z, Yang T, Fu X, Jung SY, Wang Y, et al. Acetylation of Smc3 by Eco1 is required for S phase sister chromatid cohesion in both human and yeast. Mol. Cell. 2008;31:143–151. doi: 10.1016/j.molcel.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 20.Hartman T, Stead K, Koshland D, Guacci V. Pds5 p is an essential chromosomal protein required for both sister chromatid cohesion and condensation in Saccharomyces cerevisiae. J. Cell Biol. 2000;151:613–626. doi: 10.1083/jcb.151.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Panizza S, Tanaka T, Hochwagen A, Eisenhaber F, Nasmyth K. Pds5 p cooperates with cohesin in maintaining sister chromatid cohesion. Curr. Biol. 2000;10:1557–1564. doi: 10.1016/s0960-9822(00)00854-x. [DOI] [PubMed] [Google Scholar]
- 22.Tanaka K, Hao Z, Kai M, Okayama H. Establishment and maintenance of sister chromatid cohesion in fission yeast by a unique mechanism. EMBO J. 2001;20:5779–5790. doi: 10.1093/emboj/20.20.5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang SW, Read RL, Norbury CJ. Fission yeast Pds5 is required for accurate chromosome segregation and for survival after DNA damage or metaphase arrest. J. Cell Sci. 2002;115:587–598. doi: 10.1242/jcs.115.3.587. [DOI] [PubMed] [Google Scholar]
- 24.Wang F, Yoder J, Antoshechkin I, Han M. Caenorhabditis elegans EVL-14/PDS-5 and SCC-3 are essential for sister chromatid cohesion in meiosis and mitosis. Mol. Cell. Biol. 2003;23:7698–7707. doi: 10.1128/MCB.23.21.7698-7707.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Z, Ren Q, Yang H, Conrad MN, Guacci V, Kateneva A, Dresser ME. Budding yeast PDS5 plays an important role in meiosis and is required for sister chromatid cohesion. Mol. Microbiol. 2005;56:670–680. doi: 10.1111/j.1365-2958.2005.04582.x. [DOI] [PubMed] [Google Scholar]
- 26.Losada A, Yokochi T, Hirano T. Functional contribution of Pds5 to cohesin-mediated cohesion in human cells and Xenopus egg extracts. J. Cell Sci. 2005;118:2133–2141. doi: 10.1242/jcs.02355. [DOI] [PubMed] [Google Scholar]
- 27.Sutani T, Kawaguchi T, Kanno R, Itoh T, Shirahige K. Budding yeast Wpl1(Rad61)-Pds5 complex counteracts sister chromatid cohesion-establishing reaction. Curr. Biol. 2009;24:492–497. doi: 10.1016/j.cub.2009.01.062. [DOI] [PubMed] [Google Scholar]
- 28.Rowland BD, Roig MB, Nishino T, Kurze A, Uluocak P, Mishra A, Beckouët F, Underwood P, Metson J, Imre R, et al. Building sister chromatid cohesion: smc3 acetylation counteracts an antiestablishment activity. Mol. Cell. 2009;33:763–774. doi: 10.1016/j.molcel.2009.02.028. [DOI] [PubMed] [Google Scholar]
- 29.Stead K, Aguilar C, Hartman T, Drexel M, Meluh P, Guacci V. Pds5 p regulates the maintenance of sister chromatid cohesion and is sumoylated to promote the dissolution of cohesion. J. Cell Biol. 2003;163:729–741. doi: 10.1083/jcb.200305080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vernì F, Gandhi R, Goldberg ML, Gatti M. Genetic and molecular analysis of wings apart-like (WAPL), a gene controlling heterochromatin organization in Drosophila melanogaster. Genetics. 2000;154:1693–1710. doi: 10.1093/genetics/154.4.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gandhi R, Gillespie PJ, Hirano T. Human Wapl is a cohesin-binding protein that promotes sister-chromatid resolution in mitotic prophase. Curr. Biol. 2006;16:2406–2417. doi: 10.1016/j.cub.2006.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Game JC, Birrell GW, Brown JA, Shibata T, Baccari C, Chu AM, Williamson MS, Brown JM. Use of a genome-wide approach to identify new genes that control resistance of Saccharomyces cerevisiae to ionizing radiation. Radiat. Res. 2003;160:14–24. doi: 10.1667/rr3019. [DOI] [PubMed] [Google Scholar]
- 33.Warren CD, Eckley DM, Lee MS, Hanna JS, Hughes A, Peyser B, Jie C, Irizarry R, Spencer FA. S-phase checkpoint genes safeguard high-fidelity sister chromatid cohesion. Mol. Biol. Cell. 2004;15:1724–1735. doi: 10.1091/mbc.E03-09-0637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kueng S, Hegemann B, Peters BH, Lipp JJ, Schleiffer A, Mechtler K, Peters JM. Wapl controls the dynamic association of cohesin with chromatin. Cell. 2006;127:955–967. doi: 10.1016/j.cell.2006.09.040. [DOI] [PubMed] [Google Scholar]
- 35.Bernard P, Schmidt CK, Vaur S, Dheur S, Drogat J, Genier S, Ekwall K, Uhlmann F, Javerzat JP. Cell-cycle regulation of cohesin stability along fission yeast chromosomes. EMBO J. 2008;27:111–121. doi: 10.1038/sj.emboj.7601955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sumara I, Vorlaufer E, Gieffers C, Peters BH, Peters JM. Characterization of vertebrate cohesin complexes and their regulation in prophase. J. Cell Biol. 2000;151:749–762. doi: 10.1083/jcb.151.4.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Losada A, Yokochi T, Kobayashi R, Hirano T. Identification and characterization of SA/Scc3 p subunits in the Xenopus and human cohesin complexes. J. Cell Biol. 2000;150:405–416. doi: 10.1083/jcb.150.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Surcel A, Koshland D, Ma H, Simpson RT. Cohesin interaction with centromeric minichromosomes shows a multi-complex rod-shaped structure. PLoS One. 2008;3:e2453. doi: 10.1371/journal.pone.0002453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haering CH, Farcas AM, Arumugam P, Metson J, Nasmyth K. The cohesin ring concatenates sister DNA molecules. Nature. 2008;454:297–301. doi: 10.1038/nature07098. [DOI] [PubMed] [Google Scholar]
- 40.Unal E, Heidinger-Pauli JM, Koshland D. DNA double-strand breaks trigger genome-wide sister-chromatid cohesion through Eco1 (Ctf7) Science. 2007;317:245–248. doi: 10.1126/science.1140637. [DOI] [PubMed] [Google Scholar]
- 41.Ström L, Karlsson C, Lindroos HB, Wedahl S, Katou Y, Shirahige K, Sjögren C. Postreplicative formation of cohesion is required for repair and induced by a single DNA break. Science. 2007;317:242–245. doi: 10.1126/science.1140649. [DOI] [PubMed] [Google Scholar]
- 42.Unal E, Arbel-Eden A, Sattler U, Shroff R, Lichten M, Haber JE, Koshland D. DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol. Cell. 2004;16:991–1002. doi: 10.1016/j.molcel.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 43.Ström L, Lindroos HB, Shirahige K, Sjögren C. Postreplicative recruitment of cohesin to double-strand breaks is required for DNA repair. Mol. Cell. 2004;16:1003–1015. doi: 10.1016/j.molcel.2004.11.026. [DOI] [PubMed] [Google Scholar]
- 44.Heidinger-Pauli JM, Unal E, Koshland D. Distinct targets of the Eco1 acetyltransferase modulate cohesion in S phase and in response to DNA damage. Mol. Cell. 2009;34:311–321. doi: 10.1016/j.molcel.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heidinger-Pauli JM, Unal E, Guacci V, Koshland D. The kleisin subunit of cohesin dictates damage-induced cohesion. Mol. Cell. 2008;31:47–56. doi: 10.1016/j.molcel.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 46.Maradeo ME, Skibbens RV. The Elg1-RFC clamp-loading complex performs a role in sister chromatid cohesion. PLoS One. 2009;4:e4707. doi: 10.1371/journal.pone.0004707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parnas O, Zipin-Roitman A, Mazor Y, Liefshitz B, Ben-Aroya S, Kupiec M. The ELG1 clamp loader plays a role in sister chromatid cohesion. PLoS One. 2009;4:e5497. doi: 10.1371/journal.pone.0005497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dorsett D. Roles of the sister chromatid cohesion apparatus in gene expression, development, and human syndromes. Chromosoma. 2007;116:1–13. doi: 10.1007/s00412-006-0072-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Onn I, Heidinger-Pauli JM, Guacci V, Unal E, Koshland DE. Sister chromatid cohesion: A simple concept with a complex reality. Annu. Rev. Cell Dev. Biol. 2008;24:105–129. doi: 10.1146/annurev.cellbio.24.110707.175350. [DOI] [PubMed] [Google Scholar]